Many leaf-colonizing bacteria produce surfactants and are able to degrade aliphatic compounds; however, whether surfactant production provides a competitive advantage during leaf colonization is unclear. Furthermore, it is unclear if leaf colonizers take advantage of the aliphatic compounds that constitute the leaf cuticle and cuticular waxes.
KEYWORDS: plant-microbe interactions, phyllosphere-inhabiting microbes, leaf wax, leaf cuticle, aliphatics, viscosin, massetolide A
ABSTRACT
Biosurfactant production is a common trait in leaf surface-colonizing bacteria that has been associated with increased survival and movement on leaves. At the same time, the ability to degrade aliphatics is common in biosurfactant-producing leaf colonizers. Pseudomonads are common leaf colonizers and have been recognized for their ability to produce biosurfactants and degrade aliphatic compounds. In this study, we investigated the role of biosurfactants in four non-plant-pathogenic Pseudomonas strains by performing a series of experiments to characterize their surfactant properties and their role during leaf colonization and diesel degradation. The biosurfactants produced were identified using mass spectrometry. Two strains produced viscosin-like biosurfactants, and the other two produced massetolide A-like biosurfactants, which aligned with the phylogenetic relatedness between the strains. To further investigate the role of surfactant production, random Tn5 transposon mutagenesis was performed to generate knockout mutants. The knockout mutants were compared to their respective wild types with regard to their ability to colonize gnotobiotic Arabidopsis thaliana and to degrade diesel or dodecane. It was not possible to detect negative effects during plant colonization in direct competition or individual colonization experiments. When grown on diesel, knockout mutants grew significantly slower than their respective wild types. When grown on dodecane, knockout mutants were less impacted than during growth on diesel. By adding isolated wild-type biosurfactants, it was possible to complement the growth of the knockout mutants.
IMPORTANCE Many leaf-colonizing bacteria produce surfactants and are able to degrade aliphatic compounds; however, whether surfactant production provides a competitive advantage during leaf colonization is unclear. Furthermore, it is unclear if leaf colonizers take advantage of the aliphatic compounds that constitute the leaf cuticle and cuticular waxes. Here, we tested the effect of surfactant production on leaf colonization, and we demonstrate that the lack of surfactant production decreases the ability to degrade aliphatic compounds. This indicates that leaf surface-dwelling, surfactant-producing bacteria contribute to degradation of environmental hydrocarbons and may be able to utilize leaf surface waxes. This has implications for plant-microbe interactions and future studies.
INTRODUCTION
The leaf cuticle is a hydrophobic barrier which consists of cutin, a polymer of very-long-chain aliphatics, interspersed and overlaid by very-long-chain monomeric aliphatics, cuticular waxes (1, 2). The cuticle reduces water loss, provides protection against UV radiation, and is the primary interface for plant microorganism and insect interactions (3–5). The cutin is a biopolymer which consists mainly of ω and midchain hydroxy and epoxy fatty acids (C16 to C18) as well as glycerol (6–8). The cutin forms the structural backbone of the cuticle, as it is known to prevent mechanical damage. The cuticular waxes are the second major component of the leaf cuticle, mostly consisting of alkanes, alcohols, acids, and aldehydes with chain lengths between C16 and C32. Cuticular waxes may also include secondary metabolites, such as flavonoids, triterpenoids, and phenylpropanoids (9). Cuticular waxes can be separated into two distinct waxes. The intracuticular wax within the cutin polymer is clearly distinct from the epicuticular wax, which is on the outer surface of the cutin polymer (10, 11). These differences thus affect the physical properties of the plant surfaces. The composition of the cuticular waxes is dependent on plant species and environmental conditions (12, 13). Wax monomers are very energy rich and a potential source of energy and carbon if they are bioavailable. However, it is still unclear whether bacteria are able to utilize these aliphatic compounds constituting the cuticle of living leaves as a source of carbon and whether surfactants facilitate their utilization.
Leaves are home to a wide variety of bacteria and can be covered by up to 5% bacterial biomass (14, 15). Many leaf surface-colonizing genera were previously shown to degrade hydrocarbons, e.g., Rhodococcus spp., Sphingomonas spp., Pantoea spp., Methylobacterium spp., and pseudomonads (16–19). Pseudomonads are common leaf colonizers and have many different ecological roles; e.g., many Pseudomonas syringae strains can be bona fide and host-specific pathogens (20), while others may act as antagonists against agents of plant disease (21, 22) or have unknown, tritagonistic (23) functions in the microbiota (24, 25). Pseudomonads have the ability to produce so-called biosurfactants in common (26). Biosurfactants are biologically produced amphiphilic molecules consisting of a hydrophilic head group and a hydrophobic moiety.
Leaf-colonizing pseudomonads produce cyclic peptide biosurfactants (26). Their ecophysiological role is not always clear, but it has been shown that pseudomonads may gain different fitness advantages by producing surfactants, including increasing survival during fluctuating humidity conditions on leaves (27) and increasing local water availability due to the hygroscopic nature of their surfactants (28). On agar plates, it has been shown that biosurfactants increase surface mobility by swarming, and it has been assumed that they may have similar functions on leaves (29).
In this study, we characterized the physiological effect of biosurfactants in four different pseudomonads that were isolated from leaves of spinach (Pseudomonas sp. strain FF1) or romaine lettuce (Pseudomonas sp. strains FF2, FF3, and FF4). Their biosurfactants were characterized using mass spectrometry, and their physical properties were analyzed. Furthermore, we investigated the ecophysiological functions of the biosurfactants for the bacteria. To that end, random insertion libraries were produced, and biosurfactant knockout mutants were identified. The knockout mutants were characterized in a series of experiments that investigated fitness changes in vitro and in planta.
RESULTS
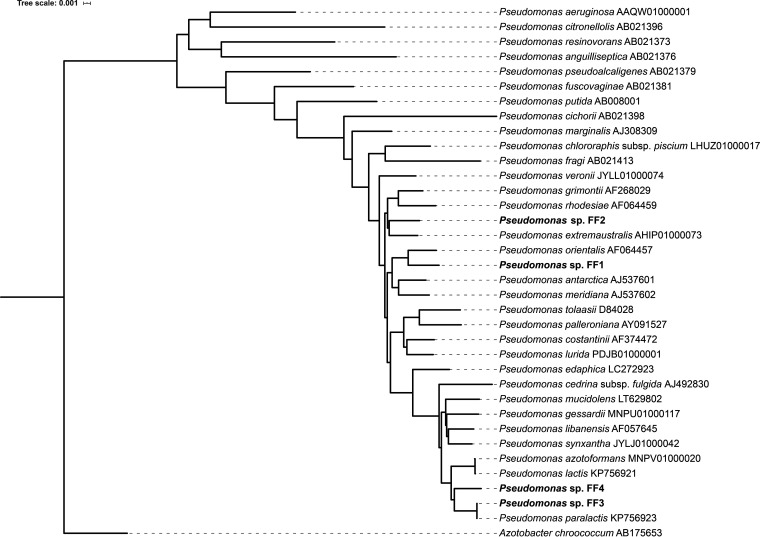
Phylogenetic placement of Pseudomonas sp. strains FF1, FF2, FF3, and FF4.
Analysis of the 16S rRNA genes of all four isolates revealed that they are all members of the genus Pseudomonas and members of the Pseudomonas fluorescens lineage and subgroup (30). Pseudomonas sp. strain FF1 (PFF1) clusters closely with Pseudomonas orientalis, and Pseudomonas sp. strain FF2 (PFF2) clusters closely with Pseudomonas extremaustralis, while Pseudomonas sp. strain FF3 (PFF3) and Pseudomonas sp. strain FF4 (PFF4) cluster closely with Pseudomonas paralactis (Fig. 1). PFF1 and PFF2 are more closely related to each other than to PFF3 and PFF4. PFF3 and PFF4 are closely related.
FIG 1.
Phylogenetic placement of the four isolated pseudomonads. The newly sequenced isolates are highlighted in bold. NCBI accession numbers of the respective sequences are given after the species names. Azotobacter chroococcum was used as an outgroup.
Surfactant production of tested pseudomonads.
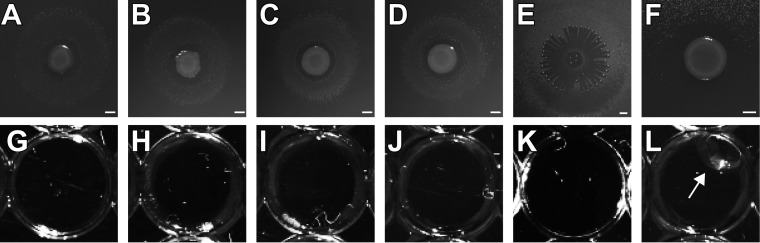
All four wild-type pseudomonads were tested for their production of surfactants on agar plates using the atomized-oil assay (16, 31). All four strains produced clear halos where the reflection of the light by the oil, indicating production of surfactants (Fig. 2A to D). Similarly, the positive control, Tween 20, showed a halo (Fig. 2E), while the negative control, Escherichia coli DH5α, lacked a halo (Fig. 2F). The drop collapse assay was used as a secondary test for surfactant production. All tested wild-type-culture supernatants collapsed into the engine oil (Fig. 2G to J). The collapse is due to a change in surface tension of the supernatant.
FIG 2.
(A to F) Atomized-oil assays to demonstrate the production of surfactants. (A to D) Wild-type colonies of PFF1, PFF2, PFF3, and PFF4, respectively, exhibiting a halo indicative of surfactant production. (E) Tween 20. (F) E. coli DH5α. (G to L) Drop collapse assays to demonstrate the production of surfactants. (G to J) Culture supernatants of wild-type PFF1, PFF2, PFF3, and PFF4, respectively, collapsed into oil, indicative of surfactant production. (K) Collapsed drop containing Tween 20. (J) Noncollapsed drop of E. coli culture supernatant (arrow).
Mass-spectrometric analysis of surfactants.
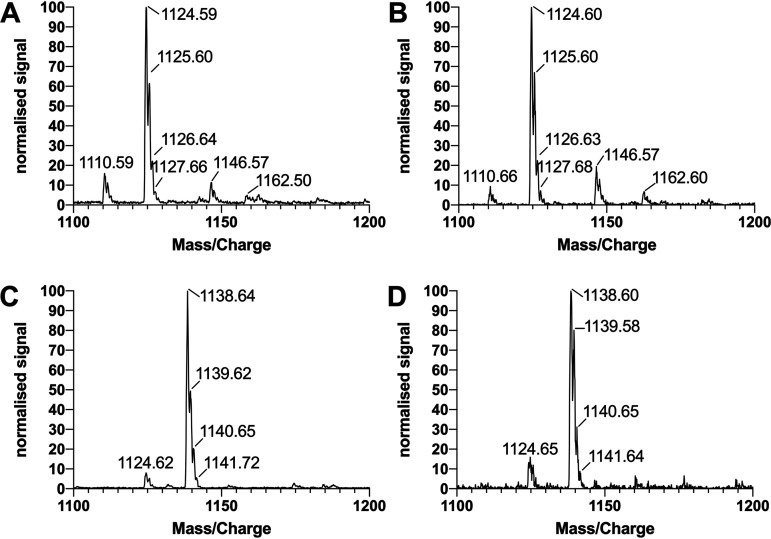
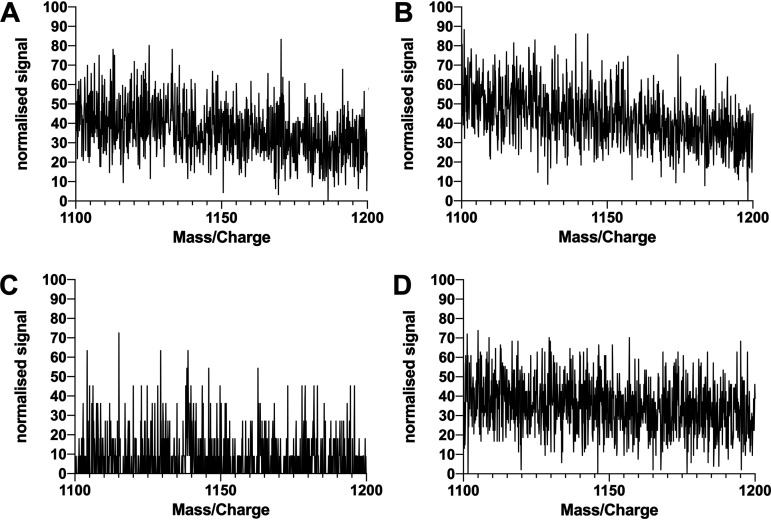
The analysis of surfactants harvested from the pseudomonads using liquid chromatography-mass spectrometry (LC-MS) with electrospray ionization (ESI) in negative mode revealed that PFF1 and PFF2 produced the same compounds with a characteristic main peak at an m/z of 1,124.59 (Fig. 3A and B), which can be attributed to the deprotonated molecular ion [M-H]−. The analogous pattern for the protonated molecular ion [M+H]+ has been previously described for the cyclic lipopeptide viscosin with ESI in positive mode for detection (32, 33). Similarly, PFF3 and PFF4 share the same characteristic main peak at an m/z of 1,138.60 (Fig. 3C and D), and the analogous pattern has previously been described for the cyclic lipopeptide massetolide A (32).
FIG 3.
MS/MS spectra of extracted surfactants of PFF1, PFF2, PFF3, and PFF4 (A to D, respectively). PFF1 and PFF2 produce viscosin-like surfactants; PFF3 and PFF4 produce massetolide A-like surfactants. Spectra were normalized against the maximal intensity.
Random Tn5 mutagenesis and mutant characterization.
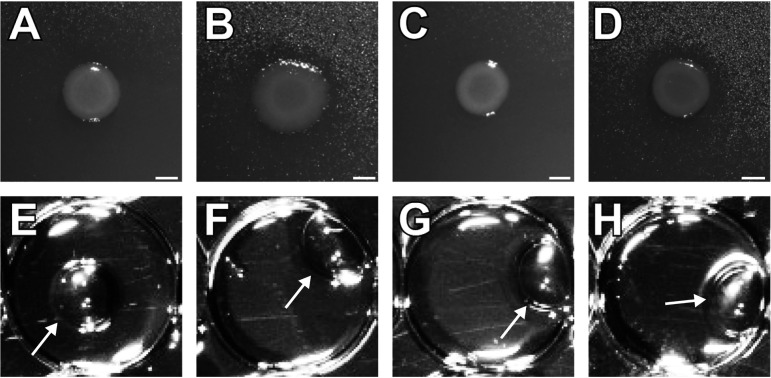
The surfactant-producing wild types were subjected to random insertion mutagenesis using the EZ-Tn5 transposon system. The screen resulted in a transposon mutant library with several hundred transposon mutants for each of the four isolates. We obtained 3 in 168, 4 in 1,100, 26 in 1,725, and 1 in ∼200 surfactant negative mutants for PFF1, PFF2, PFF3, and PFF4, respectively. Pseudomonas genomes are between 6 Mbp and 6.5 Mbp; based on the previously published sizes of viscosin and massetolide A gene clusters (each ∼30 kbp) (32, 34), we were expecting approximately 1 surfactant-negative mutant every ∼200 to 220 clones, not considering essential genes. Each of the mutant libraries was screened with the atomized-oil assay for the lack of surfactant production mutants. For each strain, we selected one of the obtained surfactant mutants for further characterization studies (Fig. 4A to D). The drop collapse assay was conducted and confirmed the results of the atomized-oil assay (Fig. 4E to H). The insertion site of each mutant was determined by digesting the genomic DNA of the mutants and cloning it into pUC19 before selecting for kanamycin resistance encoded in the transposon (see Table S1 in the supplemental material).
FIG 4.
(A to D) Atomized-oil assay to demonstrate the production of surfactants. Tn5 insertion mutant colonies of PFF1::ezTn5-visB, PFF2::ezTn5-visB, PFF3::ezTn5-massB, and PFF4::ezTn5-massB, respectively, lacking a halo indicative of surfactant production. (E to H) Drop collapse assays to demonstrate the production of surfactants. Culture supernatants of Tn5 insertion mutants PFF1::ezTn5-visB, PFF2::ezTn5-visB, PFF3::ezTn5-massB, and PFF4::ezTn5-massB, respectively, showing a beaded bubble swimming on top of the oil, indicative of the lack of surfactants. The noncollapsed droplets are indicated by arrows.
The PFF1 mutant we investigated carried an insertion in a gene with 97% similarity to a nonribosomal peptide synthetase in P. orientalis F9 (GenPept accession number AUZ46831 [locus tag BOP93_14875] (21), which has 80% peptide similarity to the viscB gene of P. fluorescens SBW25 (UniProtKB ID C3K9G2) (34, 35). The PFF2 mutant carried an insertion in a gene with 86% similarity to the viscB gene (GenPept no. CAY48788.1) of P. fluorescens SBW25. Therefore, these mutants are designated PFF1::ezTn5-viscB and PFF2::ezTn5-viscB, respectively. The viscB gene encodes a nonribosomal peptide synthetase that, in conjunction with viscA and viscC, produces the cyclic lipopeptide biosurfactant viscosin (34). The PFF3 Tn5 transposon mutant carried an insertion in a gene with 99% similarity to the massB gene in Pseudomonas fluorescens SS101 (GenPept no. ABH06368.2). The PFF4 Tn5 transposon mutant carried an insertion in a gene with 95% similarity to the massB gene in Pseudomonas fluorescens SS101 (32). The massB gene is part of the massetolide A synthesis gene cluster. Therefore, the mutants were designated PFF3::ezTn5-massB and PFF4::ezTn5-massB.
After the surfactant extraction from mutant-inoculated agar plates, no surfactants could be detected (Fig. 5A to D). The effect of the transposon insertions and the lack of surfactant production was tested in shaking liquid cultures in two different conditions, either KB complex medium (see Fig. S1A) or M9 minimal medium supplemented with glucose as the sole source of carbon (see Fig. S1B). None of the tested insertion mutants exhibited significantly changed doubling times under the two sets of conditions tested.
FIG 5.
Knockout mutants show no sign of surfactant production. MS/MS spectra of extracts of PFF1::ezTn5-viscB, PFF2::ezTn5-viscB, PFF3::ezTn5-massB, and PFF4::ezTn5-massB (A to D, respectively), are shown. None of the random knockout mutants produced detectable surfactant peaks at the corresponding wild-type m/z values. Spectra were normalized against the maximal intensity.
Growth on diesel oil or dodecane as the sole carbon source.
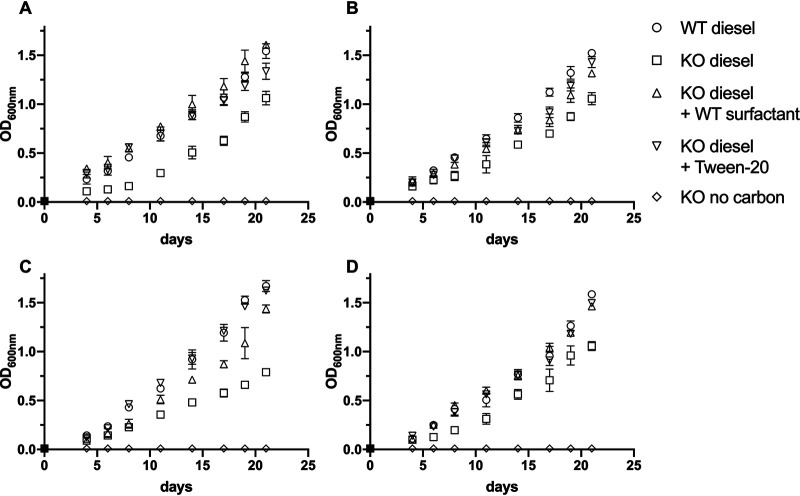
To investigate if the lack of surfactant production could impact the ability of the pseudomonad strains to degrade alkanes, the different wild types and transposon mutants were grown on Bushnell-Haas broth with diesel as the sole carbon source. This experiment revealed that all surfactant mutants, even though they were still able to grow on diesel, had a reduced growth rate and a reduced final optical density after up to 21 days of growth (Fig. 6). No growth could be observed on Bushnell-Haas broth without a carbon source for either the wild types or the surfactant mutants. In general, the growth on diesel oil was slower than growth on complex medium or minimal medium supplemented with glucose as the sole carbon source and was better described by a linear function than an exponential growth function. By supplementing knockout mutants with biosurfactants harvested from respective wild-type strains or with the synthetic surfactant Tween 20, growth on diesel could be complemented, in part or completely, compared to the wild type. The knockout mutants were not able to grow on surfactants alone to a degree that explains the increased growth on diesel (Fig. S2).
FIG 6.
Utilization of diesel by biosurfactant knockout mutants and wild types. (A) PFF1; (B) PFF2; (C) PFF3; (D) PFF4. Each wild type and knockout mutant was grown in Bushnell-Haas broth supplemented with diesel as the sole source of carbon (circles and squares, respectively). Knockout mutants were complemented with either wild-type surfactant (triangles) or Tween 20 (inverted triangles) or were incubated with no additional carbon source (diamonds). The statistical analysis can be found in Table S3. Error bars depict the standard deviations of the means. Experiments were performed in triplicate.
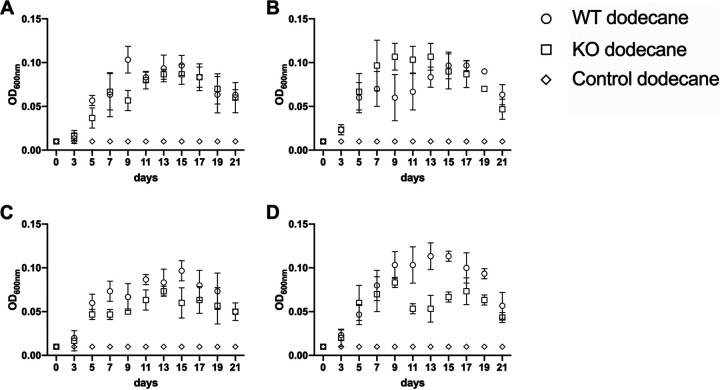
While growing on dodecane, surfactant production had a less dramatic effect than that during growth on diesel (Fig. 7 and Table S3). The overall growth was lower and the maximal optical density at 600 nm (OD600) of all tested strains was more than an order of magnitude lower than during growth on diesel, and the maximal OD600 was reached after 9 to 13 days. After 21 days, the optical densities were already markedly reduced, and the cultures were in their death phase. For all wild-type strains and combinations with all mutants but PFF2, we were able to detect periods where the wild type achieved significantly higher optical densities than the knockout mutants. The PFF2 surfactant knockout mutant reached its maximal optical density earlier than the PFF2 wild type.
FIG 7.
Growth of Pseudomonas knockout strains on dodecane as the sole carbon source. Bushnell-Haas broth (BHB) supplemented with dodecane was inoculated with either wild-type strains (circles) or surfactant knockout mutant (squares) or was left noninoculated (diamonds). (A) Growth of PFF1 and the respective surfactant mutant. The growth of the surfactant mutant was significantly lower than that of the wild type on days 5 and 9. (B) Growth of PFF2 and the respective surfactant mutant. From day 7 to day 13, the growth of the surfactant mutant was significantly lower than that of the wild type. (C) Growth of PFF3 and the respective surfactant mutant. The growth of the surfactant mutant was significantly lower than that of the wild type on days 7, 11, and 15. (D) Growth of PFF4 and the respective surfactant mutant. From day 9 to day 19, the growth of the surfactant mutant was significantly lower than that of the wild type. The statistical analysis can be found in Table S3. Error bars depict the standard deviations of the means from three replicates.
Fitness in planta.
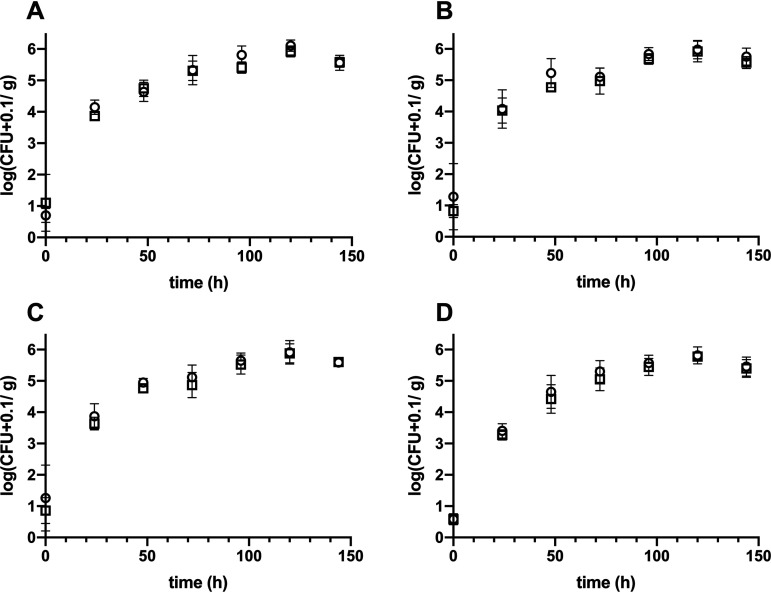
To investigate changes in the ability of the transposon mutants to colonize leaf surfaces, the mutants were coinoculated with the respective wild types by airbrushing. Whole above-ground plant material was sampled daily for 6 days, and numbers of CFU of the wild type and transposon mutants were determined (Fig. 8). The initial bacterial densities were similar between wild types and knockout mutants. Wild types (PFF1, PFF2, PFF3, and PFF4) and corresponding mutants (PFF1::ezTn5-viscB, PFF2::ezTn5-viscB, PFF3::ezTn5-massB, and PFF4::ezTn5-massB) colonized Arabidopsis at similar rates. PFF1, PFF2, and their mutants reached approximately 107 CFU per g of plant weight, whereas PFF3, PFF4, and their mutants reached approximately 106 CFU per g of plant weight. Thus, no differences between the plant colonization of wild types and mutants were found. Furthermore, growth in planta of all strains was tested individually, and no significant differences in plant colonization could be determined (Fig. S3).
FIG 8.
In planta competition of wild types (circles) and mutants (squares). (A) PFF1 versus PFF1::ezTn5-visB; (B) PFF2 versus PFF2::ezTn5-visB; (C) PFF3 versus PFF3::ezTn5-massB; (D) PFF4 versus PFF2::ezTn5-massB. Symbols represent the mean number of CFU on five plants per measurement. The statistical analysis can be found in Table S3. Error bars depict the standard deviations of the means. Experiments were performed in quintuplicate.
DISCUSSION
All four pseudomonads isolated from either spinach or romaine lettuce leaf material (36) belong to the fluorescent pseudomonads (37). PFF1 and PFF2 are phylogenetically more closely related to each other than to PFF3 and PFF4. PFF3 and PFF4 are very closely related. All four strains produced surfactants on agar plates and in liquid culture, as shown by the atomized-oil assay and the drop collapse assay. As the ability to produce surfactants is widely distributed in the genus Pseudomonas, this result was not surprising (38, 39). The relatedness of the strains is also reflected in the surfactants that each of the strains produces: PFF1 and PFF2 produce the viscosin-like surfactants, while PFF3 and PFF4 produced massetolide A-like surfactants. The production of viscosin and massetolide A by pseudomonads has been demonstrated previously (32). Both viscosin and massetolide A are the product of nonribosomal peptide synthetase genes. Viscosin production depends on a gene cluster which encompasses the genes viscA, viscB, and viscC and which spans approximately 32 kb (34). Massetolide A production depends on a gene cluster which encompasses the genes massA, massB, and massC and spans approximately 30 kb (32).
To further investigate the ecological function of the surfactants in the leaf-colonizing pseudomonads, random Tn5 transposon insertion mutants were produced and further characterized. The screen yielded complete loss of surfactant production mutants for every strain, indicating that each strain encodes only one surfactant that is active during the selection conditions. The insertion sites were mapped to genes which matched previously characterized nonribosomal peptide synthase clusters responsible for surfactant production and which matched the surfactants that were identified using mass spectrometry. PFF1 and PFF2 knockout mutants were mapped to viscB gene homologues, and PFF3 and PFF4 knockout mutants were mapped to massB gene homologues (32, 34).
The assumption that only one surfactant is produced by each strain was corroborated by a sequence of experiments during which the surfactant mutants consistently failed to produce signs of surfactant production independent of their growth conditions. The surfactant mutants failed to produce halos in the atomized-oil assay, and the culture supernatant did not collapse into motor oil in the drop collapse assay. Mass-spectrometric analysis of the knockout mutants showed that the production of surfactants was completely abolished, and no detectable peak pattern was found after the surfactant extraction procedure (Fig. 5).
Despite the loss of surfactant production and the additional burden of expressing the kanamycin resistance gene from the Tn5 transposon, the insertions had no detectable fitness effects in either complex KB medium or minimal M9 medium supplemented with glucose. In shaking liquid cultures, surfactants did not provide critical functions for growth (Fig. S1). We hypothesize that surfactants may enable bacteria to utilize parts of the plant cuticle as a source for carbon. To that end, we tested the strains’ abilities to utilize diesel as a proxy for aliphatic compounds also found in cuticular waxes. A clear difference in the abilities of wild types and mutants to utilize diesel for growth was demonstrated (Fig. 6). Even though growth was not completely abolished, it was significantly reduced (Table S2). Due to the size of the nonribosomal peptide synthetase genes, it was not possible to construct rescue mutants. However, we attempted to complement the reduced ability of the knockout mutants to degrade diesel oil by adding harvested wild-type surfactant or Tween 20 to growing cultures. Indeed, both surfactants were able to complement the growth phenotype either in part or completely (Fig. 6 and Table S2), indicating that the lack of surfactants was the causal reason for reduced growth. Despite the chain length differences between the diesel (40) and the alkane monomers in waxes of leaf cuticles (2), both aliphatic mixtures contain similar monomers. It is thus not unthinkable that, under nutrient-limiting conditions, the Pseudomonas strains tested here are able to utilize aliphatic components of leaf cuticles in a surfactant-dependent manner. However, we failed to provide a final proof of this relationship.
To investigate the role of the surfactants during plant colonization, we inoculated axenically grown Arabidopsis with mixtures of wild types and knockout mutants or with individual strains. During coinoculation with their respective wild types onto axenic Arabidopsis, no fitness disadvantages for the knockout mutants were detected. This might be a consequence of the surfactant acting as a public good that increases the fitness of wild types and coinoculated mutants alike (41). However, single-strain inoculations also did not result in a diminished ability of the knockout mutants to colonize Arabidopsis. This is in contrast to previous experiments that demonstrated that surfactants do indeed have a positive effect on plant colonization (27). It is noteworthy that the experimental setup used in our study was markedly different, including a different plant host as well as incubation conditions under constant relative humidities, whereas previously, it was shown that fluctuating humidities are a prerequisite to result in a fitness advantage. Therefore, it might still be possible that the surfactants in the strains tested here impact plant colonization, for example, under fluctuating relative humidities, by increasing mobility of the strains on the phylloplane (42, 43) or increasing permeability of leaf cuticles (44). Furthermore, the antimicrobial activity of many surfactants may provide fitness advantages in microbial communities (43).
Conclusion.
The experiments reported here demonstrated that the biosurfactants produced by four different leaf-colonizing pseudomonads impacted their ability to degrade aliphatic compounds. However, the ability to produce biosurfactants had no measurable impact on the ability of the strains to colonize axenic Arabidopsis leaves in competition or after individual strain inoculations. We gathered additional evidence that the bacteria may utilize aliphatic compounds originating from leaf cuticles but failed to conclusively demonstrate a relationship between surfactant production and leaf colonization ability. Future studies will have to be performed to address this hypothesis.
MATERIALS AND METHODS
Bacterial strains used in this study.
Bacteria used in this study were Pseudomonas sp. strain FF1 (PFF1), PFF2, PFF3, and PFF4 (45); all pseudomonads were kind gifts from Adrien Burch and Steven Lindow (University of California, Berkeley). E. coli Stellar (Lucigen) was used for cloning. PFF1 was isolated from spinach, and PFF2, PFF3, and PFF4 were isolated from romaine lettuce. Pseudomonads were routinely grown on liquid King’s B (KB) (20 g proteose peptone, 1.15 g K2HPO4, 1.5 g MgSO4·7H2O, 10 g glycerol per liter [pH 7]; for agar medium [KBA], 15 g agar was added per liter) or lysogeny broth (LB) (5 g yeast extract, 10 g tryptone, 10 g NaCl per liter [pH 7]; for agar medium, 15 g agar was added per liter). E. coli was routinely grown on LB and LBA. For in planta competition experiments, spontaneous streptomycin-resistant mutants of the wild-type pseudomonads were selected (46). Where appropriate, the media were supplemented with kanamycin (50 μg ml−1) or streptomycin (50 μg ml−1).
16S rRNA gene sequencing.
To determine the phylogeny of the strains, their 16S rRNA genes were amplified from genomic DNA that was extracted using the NucleoSpin microbial DNA kit (Macherey-Nagel) following the manufacturer's recommendations. A PCR using KAPA2G Fast 2× ready mix with dye (Kapa) was performed using the manufacturer’s recommendation, with 1 μl of the genomic and 16S rRNA gene-targeting primers SLK8-F 5′-AGAGTTTGATCATGGCTCAGAT-3′ and SRK1506-R 5′-TACCTTGTTACGACTTCACCCC-3′. The resulting ∼1.5-kbp fragments were sequenced (Eurofins Genomic) and then curated and assembled using Geneious prime (Geneious). The assembled fragments were uploaded to ezbiocloud (47), and the 30 best matches of organisms that were validly named were recovered for each of the four strains. Additional Pseudomonas 16S sequences and outgroup sequences were recovered from the silva database (48). All sequences were compiled into a FASTA file and aligned and visualized using the FastME/OneClick option of ngphylogeny.fr (49). The resulting tree was imported into iTOL, edited for publication, and then exported (50).
Preparation of electrocompetent pseudomonads.
Electrocompetent pseudomonads were produced as explained elsewhere (51). Briefly, bacteria were grown overnight in 6 ml KB in a shaking incubator at 25°C. Three milliliters of the overnight culture was then used to inoculate 100 ml KB, which was incubated at 25°C in a shaking incubator until the culture reached a mid-exponential-growth-phase OD600 of approximately 0.6. The culture was then split into 50-ml aliquots and cooled on ice for 30 min. Bacteria were then harvested by centrifugation at 6,000 × g and 4°C for 10 min. The supernatant was discarded, and the aliquots were washed twice with 50 ml ice-cold sterile water. Then the aliquots were washed in 25 ml ice-cold water and combined again. After a final centrifugation, the cell pellet was resuspended in 250 μl sterile 10% glycerol and distributed in 50-μl aliquots that were stored at −80°C.
Random transposon mutagenesis.
Random knockout mutants were produced using the EZ::Tn5Tm <KAN-2> Tnp transposome kit (Epicentre) following the manufacturer’s recommendations. In brief, 50 μl electrocompetent pseudomonads were thawed on ice and 1 μl Tn5 transposome and 1 μl endonuclease inhibitor were mixed with the cells. The mixture was incubated for 5 min on ice before the cells were pipetted into a prechilled 0.1-cm-gap electroporation cuvette. A Gene Pulser (Bio-Rad) was used to pulse the cells (2.5 kV, 200 Ω, 25 μF). Immediately after that, 1 ml SOC (SOB is 20 g tryptone, 5 g yeast extract, 0.5 g NaCl, 10 ml 250 mM KCl per liter [pH 7]; SOC is SOB supplemented with 5 ml 2 M MgCl2 and 20 ml 1 M glucose) was added, and the cells were incubated for 1 h at 30°C and 150 rpm. Transposon insertion mutants were selected on minimal medium agar plates (15 ml glycerol, 5 g l-glutamine, 1.5 g K2HPO4, 1.15 g MgSO4 · 7H2O, 15 g agar per liter [pH 7]) supplemented with kanamycin. Minimal medium was used to prevent the growth of auxotrophic mutants. Transposon mutants could be detected after 2 days.
To determine the site of transposon integration, genomic DNA of knockout mutants was isolated using the Isolate II kit (Bioline). Genomic DNA was cut using KpnI (New England Biolabs) or EcoRI and ligated into similarly digested and dephosphorylated vector pUC19 (New England Biolabs) using T4 ligase (New England Biolabs) following the recommendations of the manufacturer. A 5-μl portion of each ligation mixture was transformed into chemically competent E. coli Stellar using the manufacturer’s recommendations. Clones harboring plasmids containing the transposon were selected on LB supplemented with kanamycin. Inserts of the plasmids were sequenced using the transposon-specific primer kan2_RP-1 (5′-GCAATGTAACATCAGAGATTTTGAG-3′). Sequencing results were compared to the NCBI database using NCBI BLAST restricted to the genus Pseudomonas (52).
Screens for surfactant production.
To screen for surfactant production, the atomized-oil assay was performed (31). To that end, agar plates containing transposon mutants were sprayed with hydrophobic dodecane using an airbrush. Bacterial colonies that produced surfactants resulted in a halo around the colony where the surfactant in the agar changed the surface angle of oil droplets on the surface. Colonies that lacked this characteristic halo were further characterized. Presumptive surfactant mutants were tested in the drop collapse assay as described previously (16). Briefly, 2 μl of Magnatec 10W-40 oil (Castrol) was pipetted into each well of a 96-well plate lid (Corning Incorporated) and allowed to equilibrate for 2 h to ensure that each well was evenly coated. Bacterial overnight cultures were centrifuged at 2,600 × g for 10 min. Five microliters of the culture supernatant was pipetted into the center of an oil-filled well. Drops that collapsed into the oil, i.e., decreased their contact angle, were positive for surfactant production, while drops that remained intact and stayed on top of the oil were negative for surfactant production. All experiments were performed in at least 8 biological replicates.
Extraction of surfactants.
Bacterial strains were grown as crude streaks on five separate KBA plates for 48 h at 25°C. Afterwards, bacterial biomass was harvested using 5 ml of sterile water per plate, and the cell suspensions of all 5 plates were combined in a 50-ml centrifugation tube. Ethyl acetate (25 ml) was added to the suspension, and the tube was vortexed for 3 min. The mixture was then centrifuged for 10 min at 1,000 × g to facilitate separation of the aqueous and organic phase. The organic phase was recovered using a glass pipette and transferred to a glass vessel before the ethyl acetate was evaporated off under constant nitrogen flow. The result was resolved in ethanol and sterile filtered through a 0.22-μm filter. The filtered solution was then dried under constant nitrogen flow and weighed before it was resuspended to 5 μg ml−1 in ethyl acetate.
Mass-spectrometric analysis.
Mass-spectrometric analysis of the biosurfactants was performed using a QTRAP 4500 (Applied Biosystems, AB Sciex) triple-quadrupole mass spectrometer, operated in negative electrospray ionization (ESI)-Q1 scan mode. The surfactant solution with a concentration of 5 μg ml−1 was injected via a syringe pump set to a flow rate of 10 μl min−1 directly into the MS. The analytes were detected in negative mode within a mass-over-charge (m/z) range of 1,000 to 1,200.
Plant growth and in planta experiments.
Arabidopsis thaliana was grown axenically as described previously (53). Briefly, Arabidopsis seeds were sterilized in a 1.5-ml Eppendorf tube by adding 1 ml 70% ethanol and 0.1% Triton X-100. The seeds were vortexed and then incubated for 1 min. The supernatant was removed by pipetting, followed by the addition of 1 ml of 10% bleach and 10 μl of 0.1% Triton X-100 for 12 min. After the bleach was removed, the seeds were rinsed three times with 1 ml of sterile distilled water and stratified for 48 h at 4°C. Stratified seeds were pipetted onto Murashige and Skoog agar (MS agar; 2.2 g of Murashige and Skoog medium including vitamins [Duchefa] and 10 g plant agar [Duchefa] per liter of Milli-Q water; pH 5.8) with filled 200-μl pipette tips that were shortened by 1 cm to allow the plant’s roots to easily pass the tip. The tips were placed pointed end first into an MS agar plate. The seeds were germinated for 7 days under short-day conditions (11 h day/13 h night). After the germination period, the seedling-filled tips were transferred to autoclaved Magenta GA-7 (bioWORLD) plant culture boxes filled with 90 g finely ground zeolite clay (Purrfit Clay Litter; Vitapet) and 60 ml MS medium. Four seedlings were transferred into each Magenta box, and the plants were grown for an additional 3 weeks under short-day conditions (11 h day/13 h night; chamber set to 85% relative humidity). To prepare bacterial inocula, bacteria were cultured on LB broth overnight. Bacteria were then harvested by 10 min centrifugation at 2,600 × g and washed with 1× phosphate-buffered saline (PBS; 0.2 g liter−1 NaCl, 1.44 g liter−1 Na2HPO4, and 0.24 g liter−1 KH2PO4). Bacteria were resuspended to an OD600 of 0.5 and then serially diluted to and OD600 of 0.00005. For competition experiments, wild-type and surfactant knockout strains were mixed at a ratio of 1:1. A 100-μl portion of the mixtures or the monocultures were inoculated onto 3-week-old Arabidopsis plants using a T-180 airbrush (KKmoon).
Bacteria were recovered by harvesting the leaf material of individual plants and placing the samples in a 1.5 ml Eppendorf vial. The plants were weighed, and 1 ml of 1× PBS was added. The vial was vortexed for 2 min and then sonicated for 5 min in a sonication bath (Elmasonic) before being vortexed for another 2 min. The supernatant was serially diluted, and CFU of wild types and surfactant mutants were enumerated by growing the strains on LB agar containing appropriate antibiotics to select for either the spontaneous streptomycin-resistant wild-type strain or the kanamycin-resistant mutants.
Hydrocarbon utilization assay.
To measure the ability of wild types and surfactant knockout mutants to grow on diesel or dodecane as the sole source of carbon, Bushnell-Haas broth (0.2 g liter−1 MgSO4, 0.02 g liter−1 CaCl2, 1.0 g liter−1 KH2PO4, 1.0 g liter−1 K2HPO4, 1.0 g liter−1 NH4NO3, and 0.05 g liter−1 FeCl3; pH 7.2) was supplemented with 1% diesel (commercial diesel, locally sourced) or 1% dodecane (for synthesis; Merck) (16). Bushnell-Haas broth without an additional carbon source was used as a negative control. In control experiments, to complement surfactant knockout mutants, between 0.23 and 0.265 mg ml−1 of isolated wild-type (WT) surfactants or 0.1 mg ml−1 Tween 20 was used as a supplement. Bacteria were grown overnight in LB, diluted 100 × using Bushnell-Haas broth without a carbon source. The diluted bacterial suspensions were inoculated into 50-ml broth cultures in 250-ml Erlenmeyer flasks. Cultures were incubated at 30°C and 200 rpm for up to 17 days. Cell density was regularly measured by determining the optical density at 600 nm using a spectrophotometer (Biochrom WPA CO8000; Biowave). All experiments were performed in three biological replicates.
Statistical analysis.
Statistical analysis was performed using Prism 9 (GraphPad). To analyze growth data in liquid culture, two-way analyses of variance (ANOVA) with Tukey’s multiple-comparison test were performed. To analyze wild-type and corresponding knockout mutant growth in planta, two-way ANOVA with Šídák’s multiple-comparison test was performed.
Data availability.
Newly determined 16S rRNA gene sequence data have been deposited in the NCBI database under accession numbers MW603784 to MW603787.
Supplementary Material
ACKNOWLEDGMENTS
We thank Adrien Burch and Steven Lindow, UC Berkeley, for the kind gift of the Pseudomonas strains. We thank Paula Jameson for critically reading the manuscript and Thomas Evans for technical support.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kolattukudy PE. 1980. Biopolyester membranes of plants: cutin and suberin. Science 208:990–1000. doi: 10.1126/science.208.4447.990. [DOI] [PubMed] [Google Scholar]
- 2.Zeisler-Diehl VV, Barthlott W, Schreiber L. 2018. Plant cuticular waxes: composition, function, and interactions with microorganisms, p 1–16. In Wilkes H (ed), Hydrocarbons, oils and lipids: diversity, origin, chemistry and fate. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 3.Yeats TH, Buda GJ, Wang Z, Chehanovsky N, Moyle LC, Jetter R, Schaffer AA, Rose JKC. 2012. The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J 69:655–666. doi: 10.1111/j.1365-313X.2011.04820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riederer M, Schreiber L. 2001. Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52:2023–2032. doi: 10.1093/jexbot/52.363.2023. [DOI] [PubMed] [Google Scholar]
- 5.Serrano M, Coluccia F, Torres M, L’Haridon F, Métraux J-P. 2014. The cuticle and plant defense to pathogens. Front Plant Sci 5:274. doi: 10.3389/fpls.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wattendorff J, Holloway PJ. 1980. Studies on the ultrastructure and histochemistry of plant cuticles: the cuticular membrane of Agave americana L. in situ. Ann Bot 46:13–28. doi: 10.1093/oxfordjournals.aob.a085891. [DOI] [Google Scholar]
- 7.Pollard M, Beisson F, Li Y, Ohlrogge JB. 2008. Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Graça J, Schreiber L, Rodrigues J, Pereira H. 2002. Glycerol and glyceryl esters of ω-hydroxyacids in cutins. Phytochemistry 61:205–215. doi: 10.1016/s0031-9422(02)00212-1. [DOI] [PubMed] [Google Scholar]
- 9.Jeffree CE. 2006. The fine structure of the plant cuticle, p 11–125. In Riederer M, Müller C (ed), Biology of the plant cuticle. Blackwell Publishing Ltd., Oxford, United Kingdom. [Google Scholar]
- 10.Buschhaus C, Jetter R. 2011. Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J Exp Bot 62:841–853. doi: 10.1093/jxb/erq366. [DOI] [PubMed] [Google Scholar]
- 11.Samuels L, Kunst L, Jetter R. 2008. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu Rev Plant Biol 59:683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- 12.Jetter R, Kunst L, Samuels AL. 2006. Composition of plant cuticular waxes, p 145–181. In Riederer M, Müller C (ed), Biology of the plant cuticle. Blackwell Publishing Ltd., Oxford, United Kingdom. [Google Scholar]
- 13.Shepherd T, Wynne Griffiths D. 2006. The effects of stress on plant cuticular waxes. New Phytol 171:469–499. doi: 10.1111/j.1469-8137.2006.01826.x. [DOI] [PubMed] [Google Scholar]
- 14.Remus-Emsermann MNP, Lücker S, Müller DB, Potthoff E, Daims H, Vorholt JA. 2014. Spatial distribution analyses of natural phyllosphere-colonizing bacteria on Arabidopsis thaliana revealed by fluorescence in situ hybridization. Environ Microbiol 16:2329–2340. doi: 10.1111/1462-2920.12482. [DOI] [PubMed] [Google Scholar]
- 15.Schlechter RO, Miebach M, Remus-Emsermann MNP. 2019. Driving factors of epiphytic bacterial communities: a review. J Adv Res 19:57–65. doi: 10.1016/j.jare.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oso S, Walters M, Schlechter RO, Remus-Emsermann MNP. 2019. Utilisation of hydrocarbons and production of surfactants by bacteria isolated from plant leaf surfaces. FEMS Microbiol Lett 366:fnz061. doi: 10.1093/femsle/fnz061. [DOI] [PubMed] [Google Scholar]
- 17.Kertesz MA, Kawasaki A. 2010. Hydrocarbon-degrading sphingomonads: Sphingomonas, Sphingobium, Novosphingobium, and Sphingopyxis, p 1693–1705. In Timmis KN (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 18.Salam LB, Obayori OS, Raji SA. 2015. Biodegradation of used engine oil by a methylotrophic bacterium, Methylobacterium mesophilicum isolated from tropical hydrocarbon-contaminated soil. Pet Sci Technol 33:186–195. doi: 10.1080/10916466.2014.961610. [DOI] [Google Scholar]
- 19.Pizzolante G, Durante M, Rizzo D, Di Salvo M, Tredici SM, Tufariello M, De Paolis A, Talà A, Mita G, Alifano P, De Benedetto GE. 2018. Characterization of two Pantoea strains isolated from extra-virgin olive oil. AMB Express 8:113. doi: 10.1186/s13568-018-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin X-F, Kvitko B, He SY. 2018. Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16:316–328. doi: 10.1038/nrmicro.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zengerer V, Schmid M, Bieri M, Müller DC, Remus-Emsermann MNP, Ahrens CH, Pelludat C. 2018. Pseudomonas orientalis F9: a potent antagonist against phytopathogens with phytotoxic effect in the apple flower. Front Microbiol 9:145. doi: 10.3389/fmicb.2018.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabrefiga J, Bonaterra A, Montesinos E. 2007. Mechanisms of antagonism of Pseudomonas fluorescens EPS62e against Erwinia amylovora, the causal agent of fire blight. Int Microbiol 10:123–132. [PubMed] [Google Scholar]
- 23.Freimoser FM, Pelludat C, Remus-Emsermann MNP. 2016. Tritagonist as a new term for uncharacterised microorganisms in environmental systems. ISME J 10:1–3. doi: 10.1038/ismej.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remus-Emsermann MNP, Schmid M, Gekenidis M-T, Pelludat C, Frey JE, Ahrens CH, Drissner D. 2016. Complete genome sequence of Pseudomonas citronellolis P3B5, a candidate for microbial phyllo-remediation of hydrocarbon-contaminated sites. Stand Genomic Sci 11:75. doi: 10.1186/s40793-016-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid M, Frei D, Patrignani A, Schlapbach R, Frey JE, Remus-Emsermann MNP, Ahrens CH. 2018. Pushing the limits of de novo genome assembly for complex prokaryotic genomes harboring very long, near identical repeats. Nucleic Acids Res 46:8953–8965. doi: 10.1093/nar/gky726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'aes J, De Maeyer K, Pauwelyn E, Höfte M. 2010. Biosurfactants in plant-Pseudomonas interactions and their importance to biocontrol. Environ Microbiol Rep 2:359–372. doi: 10.1111/j.1758-2229.2009.00104.x. [DOI] [PubMed] [Google Scholar]
- 27.Burch AY, Zeisler V, Yokota K, Schreiber L, Lindow SE. 2014. The hygroscopic biosurfactant syringafactin produced by Pseudomonas syringae enhances fitness on leaf surfaces during fluctuating humidity. Environ Microbiol 16:2086–2098. doi: 10.1111/1462-2920.12437. [DOI] [PubMed] [Google Scholar]
- 28.Hernandez MN, Lindow SE. 2019. Pseudomonas syringae increases water availability in leaf microenvironments via production of hygroscopic syringafactin. Appl Environ Microbiol 85:e01014-19. doi: 10.1128/AEM.01014-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883. doi: 10.1128/aem.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peix A, Ramírez-Bahena M-H, Velázquez E. 2018. The current status on the taxonomy of Pseudomonas revisited: an update. Infect Genet Evol 57:106–116. doi: 10.1016/j.meegid.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 31.Burch AY, Shimada BK, Browne PJ, Lindow SE. 2010. Novel high-throughput detection method to assess bacterial surfactant production. Appl Environ Microbiol 76:5363–5372. doi: 10.1128/AEM.00592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bruijn I, de Kock MJD, de Waard P, van Beek TA, Raaijmakers JM. 2008. Massetolide A biosynthesis in Pseudomonas fluorescens. J Bacteriol 190:2777–2789. doi: 10.1128/JB.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laycock MV, Hildebrand PD, Thibault P, Walter JA, Wright JLC. 1991. Viscosin, a potent peptidolipid biosurfactant and phytopathogenic mediator produced by a pectolytic strain of Pseudomonas fluorescens. J Agric Food Chem 39:483–489. doi: 10.1021/jf00003a011. [DOI] [Google Scholar]
- 34.de Bruijn I, de Kock MJD, Yang M, de Waard P, van Beek TA, Raaijmakers JM. 2007. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol Microbiol 63:417–428. doi: 10.1111/j.1365-2958.2006.05525.x. [DOI] [PubMed] [Google Scholar]
- 35.Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang X-X, Moon CD, Gehrig SM, Godfrey SAC, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR. 2009. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burch AY, Do PT, Sbodio A, Suslow TV, Lindow SE. 2016. High-level culturability of epiphytic bacteria and frequency of biosurfactant producers on leaves. Appl Environ Microbiol 82:5997–6009. doi: 10.1128/AEM.01751-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E. 2015. Phylogenomics and systematics in Pseudomonas. Front Microbiol 6:214. doi: 10.3389/fmicb.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nybroe O, Sørensen J. 2004. Production of cyclic lipopeptides by fluorescent Pseudomonads, p 147–172. In Ramos J-L (ed), Pseudomonas, vol 3. Biosynthesis of macromolecules and molecular metabolism. Springer US, Boston, MA. [Google Scholar]
- 39.Geudens N, Martins JC. 2018. Cyclic lipodepsipeptides from Pseudomonas spp.—biological Swiss-army knives. Front Microbiol 9:1867. doi: 10.3389/fmicb.2018.01867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wante SP, Leung DWM. 2018. Phytotoxicity testing of diesel-contaminated water using Petunia grandiflora Juss. Mix F1 and Marigold-Nemo Mix (Tagetes patula L.). Environ Monit Assess 190:408. doi: 10.1007/s10661-018-6790-4. [DOI] [PubMed] [Google Scholar]
- 41.Lyons NA, Kolter R. 2017. Bacillus subtilis protects public goods by extending kin discrimination to closely related species. mBio 8:e00723-17. doi: 10.1128/mBio.00723-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burch AY, Shimada BK, Mullin SWA, Dunlap CA, Bowman MJ, Lindow SE. 2012. Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. J Bacteriol 194:1287–1298. doi: 10.1128/JB.06058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. 2010. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber L, Krimm U, Knoll D, Sayed M, Auling G, Kroppenstedt RM. 2005. Plant-microbe interactions: identification of epiphytic bacteria and their ability to alter leaf surface permeability. New Phytol 166:589–594. doi: 10.1111/j.1469-8137.2005.01343.x. [DOI] [PubMed] [Google Scholar]
- 45.Burch AY, Browne PJ, Dunlap CA, Price NP, Lindow SE. 2011. Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Environ Microbiol 13:2681–2691. doi: 10.1111/j.1462-2920.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- 46.Newcombe HB, Hawirko R. 1949. Spontaneous mutation to streptomycin resistance and dependence in Escherichia coli. J Bacteriol 57:565–572. doi: 10.1128/JB.57.5.565-572.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, Seo H, Chun J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W. 2017. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol 261:169–176. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 49.Lemoine F, Correia D, Lefort V, Doppelt-Azeroual O, Mareuil F, Cohen-Boulakia S, Gascuel O. 2019. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res 47:W260–W265. doi: 10.1093/nar/gkz303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Artiguenave F, Vilaginès R, Danglot C. 1997. High-efficiency transposon mutagenesis by electroporation of a Pseudomonas fluorescens strain. FEMS Microbiol Lett 153:363–369. doi: 10.1111/j.1574-6968.1997.tb12597.x. [DOI] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Miebach M, Schlechter RO, Clemens J, Jameson PE, Remus-Emsermann MNP. 2020. Litterbox—a gnotobiotic zeolite-clay system to investigate Arabidopsis-microbe interactions. Microorganisms 8:464. doi: 10.3390/microorganisms8040464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Newly determined 16S rRNA gene sequence data have been deposited in the NCBI database under accession numbers MW603784 to MW603787.