The symbiotic N2 fixation carried out between prokaryotic rhizobia and legume plants performs a substantial contribution to the N cycle in the biosphere. This symbiotic association is initiated when rhizobia infect and penetrate the root hairs, which is followed by the growth and development of root nodules, within which the infective rhizobia are established and protected.
KEYWORDS: stringent response, type 3 secretion system, plant defense, regulation of nodulation
ABSTRACT
When subjected to nutritional stress, bacteria modify their amino acid metabolism and cell division activities by means of the stringent response, which is controlled by the Rsh protein in alphaproteobacteria. An important group of alphaproteobacteria are the rhizobia, which fix atmospheric N2 in symbiosis with legume plants. Although nutritional stress is common for rhizobia while infecting legume roots, the stringent response has scarcely been studied in this group of soil bacteria. In this report, we obtained a mutant with a kanamycin resistance insertion in the rsh gene of Bradyrhizobium diazoefficiens, the N2-fixing symbiont of soybean. This mutant was defective for type 3 secretion system induction, plant defense suppression at early root infection, and nodulation competition. Furthermore, the mutant produced smaller nodules, although with normal morphology, which led to lower plant biomass production. Soybean (Glycine max) genes GmRIC1 and GmRIC2, involved in autoregulation of nodulation, were upregulated in plants inoculated with the mutant under the N-free condition. In addition, when plants were inoculated in the presence of 10 mM NH4NO3, the mutant produced nodules containing bacteroids, and GmRIC1 and GmRIC2 were downregulated. The rsh mutant released more auxin to the culture supernatant than the wild type, which might in part explain its symbiotic behavior in the presence of combined N. These results indicate that the B. diazoefficiens stringent response integrates into the plant defense suppression and regulation of nodulation circuits in soybean, perhaps mediated by the type 3 secretion system.
IMPORTANCE The symbiotic N2 fixation carried out between prokaryotic rhizobia and legume plants performs a substantial contribution to the N cycle in the biosphere. This symbiotic association is initiated when rhizobia infect and penetrate the root hairs, which is followed by the growth and development of root nodules, within which the infective rhizobia are established and protected. Thus, the nodule environment allows the expression and function of the enzyme complex that catalyzes N2 fixation. However, during early infection, the rhizobia find a harsh environment while penetrating the root hairs. To cope with this nuisance, the rhizobia mount a stress response known as the stringent response. In turn, the plant regulates nodulation in response to the presence of alternative sources of combined N in the surrounding medium. Control of these processes is crucial for a successful symbiosis, and here we show how the rhizobial stringent response may modulate plant defense suppression and the networks of regulation of nodulation.
INTRODUCTION
Symbiotic N2 fixation accounts for most of N inputs to the N cycle in terrestrial environments. This process is of agroecological importance because of its sustainability as a biofertilization method in diverse legume crops, and thanks to its low cost and ease of application, it is extensively used worldwide (1). The symbiotic N2 fixation is carried out by the association of legume plants and a group of bacteria collectively known as rhizobia, which form and occupy nodules in the legume roots (2). An important member of this group is Bradyrhizobium diazoefficiens, an alphaproteobacterium that fixes N2 in symbiosis with soybean (Glycine max), a pulse harvested in more than 128 million hectares in 2019, mainly in the Americas (3).
Legume root nodulation is a multistep process that starts with the rhizobium-induced deformation of growing root hairs (4). This early manifestation is followed by an invagination of the root hair cell wall to produce a tubular structure toward root cortical cell layers. The rhizobia advance and proliferate within this tubular structure, known as the infection thread (IT), and finally they are released to the plant cell cytoplasm entrapped inside vesicles that bud into the root cortical cell layers where root nodules are growing (4, 5). In this way, the nodule-infected cells are filled with these rhizobium-containing vesicles known as symbiosomes, within which the rhizobia differentiate to bacteroids, express the nitrogenase complex, and fix N2. Because ITs are formed by invagination of the root hair cell wall (6), the IT interior is similar to the external root surface in terms of the polarity of its transport systems (7). Hence, during early infection the rhizobium-legume association might be harmful for both partners. On the one hand, the rhizobia behave like pathogens that proliferate at the expense of plant energy while they grow in the IT and during nodule cell invasion before N2 fixation starts. On the other hand, ITs are N-limited environments (7) into which the plant releases antifungal and antibacterial substances and provokes osmotic and oxidative stresses (8). In agreement with these data, a B. diazoefficiens extracytoplasmic function sigma factor (σEcfG) required for general stress response is upregulated in ITs, and a mutant defective in σEcfG cannot progress in ITs toward the nodules’ primordia (9). In this scenario, unless both partners evolved mechanisms to cope with this mutual unkindness, the rhizobium-legume symbiosis would be rapidly doomed. However, the rhizobia are able to suppress the plant defense reaction, and the plant, in turn, may recognize the rhizobia as beneficial partners even before the symbiotic N2 fixation is activated (10). As part of this mutual adaptation, rhizobia adjust their metabolism in response to the stress conditions prevailing in the IT to preserve viability while gaining access to the inside of the developing root nodule.
Bacteria respond to these kinds of stresses by using various transcriptionally regulated systems, the stringent response being among the most important (11–16). The stringent response is a pleiotropic adaptation of bacteria to stressful and starvation conditions, involving changes in translation and transcription rates, cell cycle control, and DNA repair (for review, see reference 17). This response is triggered by accumulation of the second messenger alarmones guanosine 5′-diphosphate 3′-diphosphate (ppGpp) and guanosine 5′-triphosphate 3′-diphosphate (pppGpp), collectively known as (p)ppGpp, and has been thoroughly studied in Escherichia coli. In response to stressing or starvation stimuli, (p)ppGpp is synthesized from GTP by the RelA GTP pyrophosphokinase and, after the normal conditions are restored, the alarmone is degraded by the bifunctional SpoT protein, which has both pyrophosphokinase and phosphodiesterase activities. Hence, the timing and magnitude of the response depend on the different steady-state levels of (p)ppGpp inside the cell under the particular conditions encountered (18). In alphaproteobacteria, like B. diazoefficiens, these roles are fulfilled by a bifunctional enzyme called Rsh (from “RelA-SpoT homologous”).
Accumulation of (p)ppGpp in response to C and N starvation was reported decades ago in rhizobia (19, 20). However, the possible involvement of the stringent response in legume root infection and nodulation was recently studied only in Ensifer meliloti and Rhizobium etli. Nodulation and N2 fixation of Medicago sativa plants inoculated with E. meliloti relA or dksA mutants unable to trigger the stringent response are severely affected (21–23). However, the relA E. meliloti mutant nodulates Medicago truncatula, although N2 fixation remains altered (23). Likewise, R. etli relA mutants produce small nodules with bacteroids devoid of polyhydroxybutyrate in Phaseolus vulgaris, and accordingly, the N2 fixation activity is substantially diminished in this plant (24, 25). In agreement with these results, nifA and rpoN, which are the main regulators of symbiotic genes, were reported as part of the relA regulon in these rhizobial species (25, 26).
B. diazoefficiens is a slow-growing rhizobial species distantly related to the above-mentioned fast-growing rhizobia. Therefore, to better understand the role of the stringent response in the rhizobium-legume interaction, in this study we mutated the rsh homolog of the B. diazoefficiens type strain USDA 110 and investigated its effects on the regulation of plant defense and nodulation in soybean.
RESULTS
The B. diazoefficiens rsh mutant is impaired for survival under stress conditions.
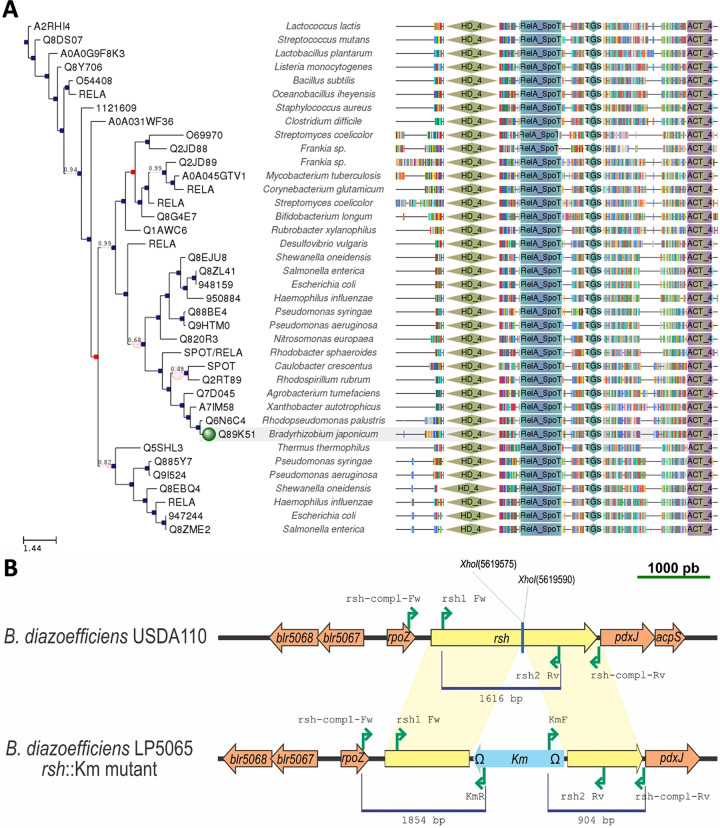
The locus bll5065 of B. diazoefficiens USDA 110 (2,289 bp) is annotated as relA (27). Furthermore, no other paralogs were found in this genome, whereby we considered that this open reading frame (ORF) is actually an rsh gene. Its gene product is predicted with high score as a bifunctional (p)ppGpp synthetase II/guanosine 3′,5′-bis pyrophosphatase and possesses a domain structure similar to relA. Specifically, the domains HD (metal-dependent phosphohydrolase), RelA/SpoT, and TGS (ThrRS, GTPase, and SpoT), which are characteristic of RelA/SpoT or Rsh proteins in other bacteria, are present in the predicted order in bll5065 (Fig. 1A). A mutant was obtained by inserting a kanamycin resistance (Kmr) cassette in reverse orientation between amino acid positions 343 and 348 (Fig. 1B), which lie in the middle of the RelA/SpoT domain of bll5065 (amino acid positions 270 to 373). This insertional mutant was named LP 5065, and hereafter it will be referred as the rsh mutant.
FIG 1.
Scheme of rsh and its mutant derivative. (A) Domain structure of various rsh, relA or spoT analogs from different species, obtained from Phylome DB-Tree Explorer on 16 August 2019. Blue squares indicate speciation events; red squares indicate duplication events. The green dot indicates the target sequence. Since the species name was not updated in this database, it still appears as “Bradyrhizobium japonicum USDA 110,” but it should be referred to as “Bradyrhizobium diazoefficiens USDA 110.” The following domains (according to Pfam) are shown: the metal-dependent phosphohydrolase (HD_4 [green diamonds]); the characteristic RelA/SpoT protein domain (RelA_SpoT [blue-green rectangles]); ThrRS, GTPase, and SpoT, which is a possible nucleotide-binding region (TGS [blue diamonds]); and the ACT domain, which binds to amino acids and regulates associated enzyme domains (ACT_4 [violet and yellow rectangles]). (B) Comparison of the wild type (upper row) and rsh insertional mutant (lower row) from B. diazoefficiens. The positions and directions of the primers used to obtain and validate the mutation are indicated by green arrows. The sizes of the fragments used for the mutation are indicated below the blue lines. The insertion occurred between the XhoI sites at bases 5619575 and 5619590 in reverse orientation.
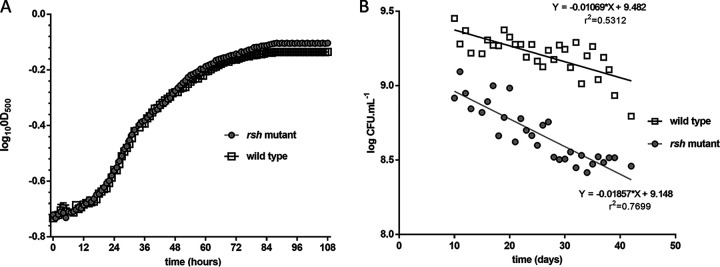
To compare the effect of the mutation, late-exponential PSY (peptone-salts-yeast extract) cultures of the wild type and the rsh mutant were centrifuged and suspended in either PSY rich medium or MOPS [3-(N-morpholino)propanesulfonic acid] buffer (hereafter referred to as starvation conditions). Then, (p)ppGpp was extracted from these cultures and measured by anion-exchange chromatography. The signal obtained with the wild type under starvation conditions was about 2 nM in the culture extract, indicating that B. diazoefficiens produced far smaller quantities of (p)ppGpp than other well-characterized species in which the alarmone was measured with the same strategy (26). Nevertheless, (p)ppGpp could be detected neither in the wild type cultured in rich medium nor in the rsh mutant incubated in either starvation or rich medium. Growth kinetics and final culture turbidity were similar between the wild type and the mutant in rich medium, indicating that the rsh mutation did not alter growth under these conditions (Fig. 2A). However, bacterial survival under starvation conditions was compromised in the rsh mutant (Fig. 2B). In an ongoing study performed as previously described (28) to compare the proteomes of the rsh mutant and the wild type, it was observed that neither PdxJ nor AcpS—the proteins encoded in the genes downstream of rsh (Fig. 1B)—were produced differentially between these strains whatever the condition, indicating that Kmr insertion in rsh did not alter their expression (J. Pérez-Giménez, unpublished data). Taken together, the above results confirmed that the locus bll5065 encodes an Rsh protein, which is involved in the stringent response of B. diazoefficiens to starvation.
FIG 2.
Growth and survival of B. diazoefficiens USDA 110 (wild type) and its rsh mutant derivative. (A) Growth as evaluated by optical density at 500 nm (OD500) in PSY. (B) Remnant viable bacteria (as CFU) from a suspension in MOPS shaken at 28°C after the indicated times.
The rsh mutation precludes induction of the type 3 secretion system.
Induction of the type 3 secretion system (T3SS) is an important cellular process affected by the stringent response (29). Previously, it was shown that in B. diazoefficiens, the T3SS is inducible by genistein, a flavonoid released by soybean roots (30). To determine if there is a link between the stringent response and T3SS in this bacterium, the levels of expression of the T3SS structural gene rhcJ and T3SS regulator ttsI were measured by reverse transcription-quantitative PCR (RT-qPCR) in the wild type or the rsh mutant incubated under starvation conditions. As shown in Table 1, both genes were strongly induced by genistein in the wild type, but there was no response in the rsh mutant. Thus, in B. diazoefficiens unable to trigger the stringent response, the T3SS cannot be induced by genistein.
TABLE 1.
Fold change in expression of selected genes from the B. diazoefficiens T3SS in starvation cultures under genistein-induced relative to noninduced conditionsa
| Gene | Fold change in expression vs noninducedb |
|
|---|---|---|
| Wild type | rsh mutant | |
| ttsI | 29.66 ± 3.72 A | 2.83 ± 1.26 B |
| rhcJ | 42.60 ± 22.00 A | −1.45 ± 0.02 B |
Induction was performed with 2 μM genistein in starvation medium. The housekeeping gene sigA was used as an internal control.
Values are the average ± SE from three independent experiments. Statistical analysis was performed by ANOVA. Values followed by different letters in the same row are significantly different (P < 0.0001).
The rsh mutant elicits a stronger early plant defense response.
Previously, Jiménez-Guerrero et al. (31) observed that a T3SS-defective Ensifer fredii mutant is unable to suppress the soybean plant defense reaction. To see if a similar situation exists for the B. diazoefficiens rsh mutant, the expression of the Glycine max “pathogenesis-related 1” (GmPR1) marker was measured by RT-qPCR in the soybean roots. As shown in Fig. 3 (black bars), the induction of GmPR1 in plants at 12 h after inoculation with the rsh mutant was significantly higher than in plants inoculated with the wild type. Later, at 48 h, there was no statistically significant difference in levels of GmPR1 transcript accumulation between both strains, indicating that rsh was required to inhibit GmPR1 transcription during the first 12 h of rhizobium-plant interaction. A related plant defense marker is the production of cytoplasmic reactive oxygen species (cytROS). In agreement with the changes observed in GmPR1 expression levels, the cytROS levels in the tip of root hair cells, where polar growth is highly active, were significantly higher at 12 h in plants inoculated with the rsh mutant than in plants inoculated with the wild type, without difference at 48 h (Fig. 3, gray bars). Therefore, these results suggest that early suppression of plant defense responses (12 h after inoculation) was compromised in the rsh mutant.
FIG 3.
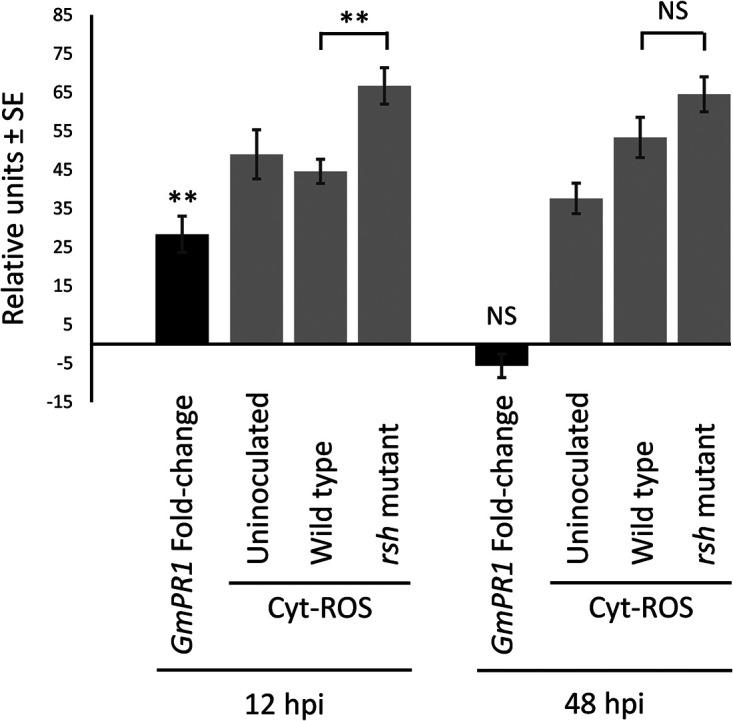
Plant defense response in soybean. Plants inoculated with B. diazoefficiens USDA 110 (wild type) or its rsh mutant derivative were cultivated for 12 or 48 h postinoculation (hpi). As controls, a group of plants were kept uninoculated. The significance of the differences between wild-type and rsh mutant strains was evaluated by ANOVA and is indicated as follows: NS, nonsignificant (P > 0.05); **, significant (P < 0.01). Data were collected from three independent experiments, each with at least four different plants for each condition. Black bars show relative expression of the plant defense marker GmPR1 in the rsh mutant with respect to the wild type (fold change). GmEF1 was used as an internal constitutive control. Gray bars show cytoplasmic reactive oxygen species (Cyt-ROS) measured in the root hair tips of soybean plants with a H2DCF-DA probe.
The rsh mutant has diminished symbiotic capacity.
The defect in suppressing the plant defense reaction might lead to impaired nodulation; therefore, we studied the nodulation capacity of our B. diazoefficiens strains.
Soybean plants inoculated with the rsh mutant produced 30 nodules per plant at 21 days postinoculation (dpi), similarly to the wild type (Table 2). However, the nodules produced by the rsh mutant were smaller than those of the wild type, albeit they were histologically normal and contained symbiosomes occupied by bacteroids (Fig. 4A to D). Accordingly, the shoot dry weight of plants nodulated by the rsh mutant was intermediate between those of plants nodulated by the wild type and uninoculated controls, but the ureide contents in leaves were similar (Table 2). These results suggest that nodulation and N2 fixation were somehow compromised in the mutant, although not substantially inhibited.
TABLE 2.
Symbiotic parameters from soybean plants inoculated with the B. diazoefficiens USDA 110 wild type or its rsh mutant derivative
| Parameter | Result fora: |
||
|---|---|---|---|
| Wild type | rsh mutant | Uninoculated control | |
| Nodules per plant | 31 ± 12 A | 30 ± 14 A | 0 B |
| Nodule dry wt (mg) | 1.9 ± 0.2 A | 1.2 ± 0.2 B | 0 C |
| Shoot dry wt (g) | 0.46 ± 0.04 A | 0.31 ± 0.04 B | 0.20 ± 0.04 C |
| Ureides (μmol g−1 leaf dry wt) | 23.7 ± 3.7 A | 17.8 ± 2.1 A | 13.1 ± 1.0 B |
| Nodule occupation in competition (%)b | 74 ± 15 A | 26 ± 12 B | 0 C |
Values are the average ± SE from three independent experiments. Statistical analysis was performed by ANOVA followed by Tukey’s test. Values followed by different letters in the same row are significantly different (P < 0.05).
Plants were coinoculated with equal numbers of cells of the wild type and the rsh mutant. Nodules were then recovered, and the percentage of occupation was recorded by antibiotic resistance. Nodules containing Kmr bacteria harbored the rsh mutant, but some of them might also contain the wild type (double occupation).
FIG 4.
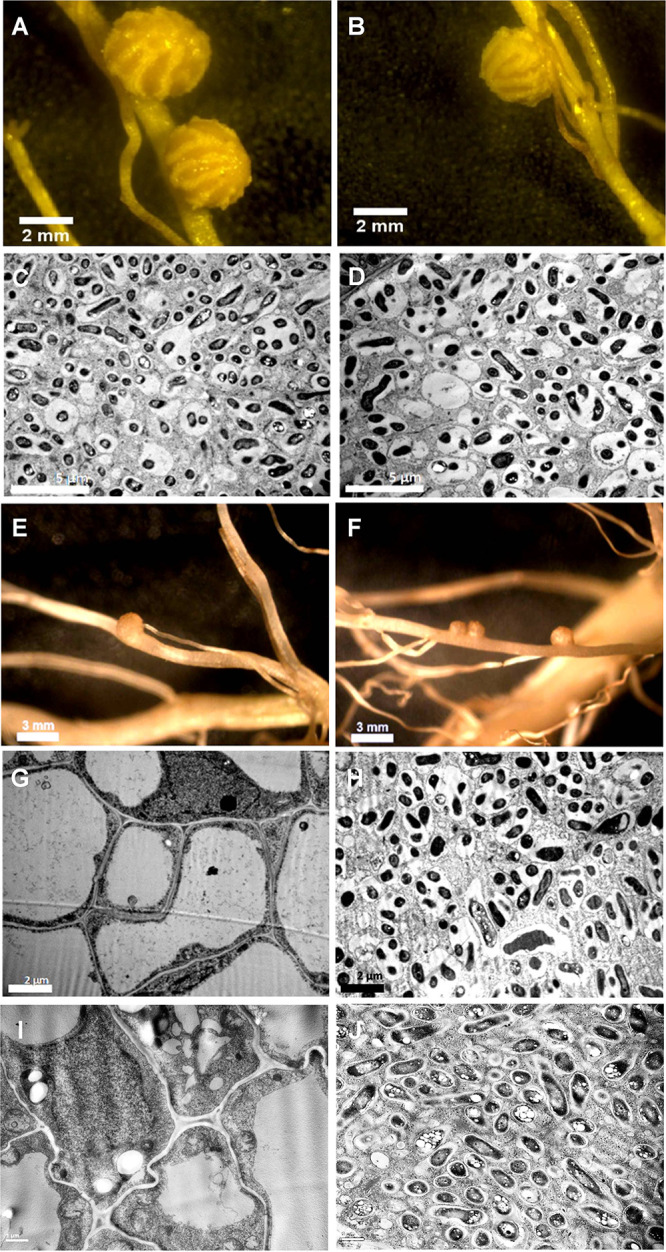
Nodules from soybean inoculated with B. diazoefficiens USDA 110 in the absence (A to D) or the presence (E to J) of 10 mM NH4NO3. The wild type (A, C, E, and G) and its rsh mutant derivative (B, D, F, and H to J) are compared. Entire nodules were viewed under a stereomicroscope (A, B, E, and F). Ultrathin cuts from these nodules were observed with a transmission electron microscope (C, D, and G to J). To confirm the role of rsh in the phenotypes observed, the rsh mutant was complemented with the wild-type rsh complete gene carried in the replicative vector pFAJ1708 (I) or the empty vector (J). The micrograph set is representative of results from three independent experiments.
When two different rhizobial genotypes are coinoculated on the same plant, both genotypes have to compete for root infection and nodule occupation. Thus, the proportion of nodules occupied by each genotype is a good indicator of its intrinsic infectivity and competitiveness during these early symbiotic processes (32). When the rsh mutant was coinoculated with the wild type in a 1:1 proportion, only 26% of the nodules were occupied by the mutant (Table 2), indicating that intrinsic infectivity and competitiveness for nodulation were affected by the loss of the stringent response.
The rsh mutant nodulates soybean in the presence of a high concentration of combined N.
Soybean plants were inoculated with the wild type or the rsh mutant and then cultured for 21 days with or without 10 mM NH4NO3, a combined N concentration in the range that is inhibitory to nodulation (33–39). As expected, the plants inoculated with the wild type produced normal nodules in the absence of the combined N source but produced only a few and small pseudonodules when cultured with 10 mM NH4NO3 (Fig. 4A and E). Accordingly, when these pseudonodules were observed with a transmission electron microscope, their ultrastructure showed cells with large vacuoles and absence of rhizobia (Fig. 4G). In contrast, in the plants inoculated with the rsh mutant, normal and red but small nodules were produced in the presence of 10 mM NH4NO3 (Fig. 4F), which possessed symbiosomes with normal bacteroids (Fig. 4H). When plants cultured with 10 mM NH4NO3 were inoculated with the rsh mutant complemented with the wild-type rsh allele carried in a replicative plasmid, empty pseudonodules similar to those of plants inoculated with the wild type were produced at 21 dpi (Fig. 4I). Meanwhile, the rsh mutant carrying the empty vector produced nodules that contained symbiosomes occupied by bacteroids (Fig. 4J). These results indicated that the rsh mutation was responsible for the anomalous nodulation in the presence of NH4NO3.
The rsh mutation alters the regulation of nodulation.
The above observations indicated that regulation of nodulation might be altered in plants inoculated with the rsh mutant. Two ways of such regulation have evolved in plants. One involves the regulation of root nodules in response to the nitrate concentration. This strategy allows the plant to avoid excessive photosynthate expenditures in nodulation when alternative N sources are available (nitrogen-dependent regulation of nodulation). The second one, autoregulation of nodulation (AON), occurs according to the development of a number of nodules that is enough to satisfy plant N requirements (40). These regulatory responses are mediated by signals in which a class of peptides, named “clavata/endosperm-surrounding region” (CLE) peptides, play the main role. In soybean (G. max), monitoring of combined N and nodule quantity is exerted by related CLE peptides, named “nitrogen-induced CLE” (GmNIC) for the combined N response and “rhizobium-induced CLE” (GmRIC) for the nodule quantity response (41). Thus, we measured the levels of these markers.
Soybean plants inoculated with the wild type or the rsh mutant were cultivated for 72 h in the presence or absence of 10 mM NH4NO3. Then, the levels of GmNIC1, GmRIC1, and GmRIC2 transcripts were measured in plant roots by RT-qPCR. As previously reported, in plants cultivated in the presence of NH4NO3, the GmNIC1 transcript accumulated, while GmRIC1 and GmRIC2 transcripts diminished with both strains (41, 42). Moreover, the GmNIC1 transcript was more abundant in plants inoculated with the rsh mutant than with the wild type, irrespective of the presence or absence of NH4NO3 (Table 3). On the contrary, the behavior of GmRIC1 and GmRIC2 was dependent on the presence of NH4NO3: both transcripts were upregulated in plants inoculated with the rsh mutant in the absence of combined N, but were downregulated in plants inoculated with the rsh mutant in the presence of 10 mM NH4NO3 (Table 3).
TABLE 3.
Relative expression of genes encoding CLE peptides in soybeans inoculated with the B. diazoefficiens USDA 110 wild type or its rsh mutant derivative
| Gene | Relative expression of genea |
|||
|---|---|---|---|---|
| 10 mM NH4NO3/N-free |
rsh mutant/wild type |
|||
| Wild type | rsh mutant | N-free | 10 mM NH4NO3 | |
| GmNIC1 | 251.14 ± 36.65 A | 338.41 ± 116.60 A | 2.90 ± 1.08 A | 3.27 ± 0.55 A |
| GmRIC1 | −7.38 ± 3.19 A | −13.10 ± 4.29 A | 3.41 ± 2.95 A | −3.34 ± 2.93 B |
| GmRIC2 | −6.33 ± 4.79 A | −3.03 ± 2.63 A | 3.21 ± 0.93 A | −1.79 ± 0.90 B |
The plants were cultivated in either N-free plant nutrient solution or plant nutrient solution supplemented with 10 mM NH4NO3. The housekeeping gene GmEF1 was used as an internal control. The values shown are averages ± SE from three independent experiments. Statistical analysis was performed by ANOVA followed by Tukey’s test. Values followed by different letters in the same row for each parameter (10 mM NH4NO3/N-free and rsh mutant/wild type) are significantly different (P < 0.05).
Higher production of extracellular auxins by the rsh mutant.
Nodulation requires a certain balance of the auxin/cytokinin ratio, as well as reduction of auxin transport to the root tips (43–45). To see whether the rsh mutant might exogenously produce auxin imbalances in the plants, we measured extracellular auxins produced by the wild type and the rsh mutant grown in culture medium. We observed that the rsh mutant released a significantly larger quantity of auxins to the culture supernatant than the wild type (Fig. 5).
FIG 5.
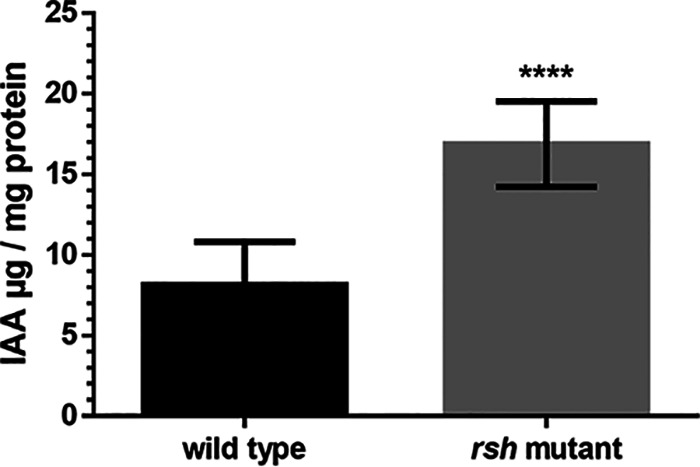
Auxins released to the culture medium by B. diazoefficiens USDA 110 (wild type) or its rsh mutant derivative. Auxins were measured by a colorimetric method using indoleacetic acid (IAA) as a standard. Error bars indicate standard deviation (SD). Statistical analysis was carried out by ANOVA (****, significant difference at P < 0.0001). Representative results from three independent experiments are shown.
DISCUSSION
The IT seems a harsh environment for the rhizobia (7–9). Therefore, we speculated that the stringent response should be active at this infection step and that such activity may act to regulate host responses to infection. Thus, in this report we obtained and characterized a B. diazoefficiens rsh mutant and compared the soybean responses to infection by either this mutant or the wild type.
We observed that, under starvation conditions, the rsh mutant did not induce the expression of the T3SS regulator ttsI and the rhcJ structural gene in the presence of genistein. These results are in agreement with previous studies indicating that the (p)ppGpp alarmone is required for activation of the T3SS in Erwinia amylovora (29). Insertion and deletion mutations in ttsI and rhcJ abolished T3SS activity in B. diazoefficiens (46, 47). Hence, if the stringent response is triggered in the IT, it might promote the injection of Nop effectors (48) into the root hair cell, which were reported as suppressors of early plant defense response (31, 49). Here we observed that the rsh mutant strongly induced GmPR1 in our soybean roots, in parallel with a transient increase in cytROS. A similar relationship was observed by Fernandez-Göbel et al. (50), who described a transient increase in cytROS in soybean leaves soon after inoculation of roots with wild-type Bradyrhizobium japonicum. Taken together, these results suggest that the stringent response is required to induce T3SS-mediated injection of Nops into the root hair cells and suppress the transient plant defense reaction a few hours after inoculation.
During N2 fixation, the plant incurs an energetic cost to maintain the nodules’ functioning. To avoid excessive proliferation of nodules, the plant exerts autoregulatory control through the CLE peptides GmRIC1 and GmRIC2 (40, 51), whose expression was increased by the rsh mutant in the absence of combined N. It is tempting to speculate that Nops participate in inhibiting the induction of these CLE peptides, but up to now, it has been unclear whether such inhibition is carried out directly by these effectors in parallel with plant defense suppression. Alternatively, a possible timeline of events triggered by the rsh mutant might be an early accumulation of ROS and GmPR1 followed by plant defense, which in turn induces GmRIC1 and GmRIC2. In this scenario, plant defense suppression by T3SS-delivered Nop effectors from the wild type might preclude GmRIC1 and GmRIC2 induction, thus explaining the different kinetics of these processes.
In addition, it was reported that GmRIC1 and GmRIC2 levels are modulated by Nod factors (NFs), with participation of the transcription factor “nodule inception” (GmNINa), a plant microRNA called MiR1/2c, and the transcriptional repressor “nodule number control 1” (GmNNC1) (52). Nevertheless, in E. meliloti and R. etli cultured in the presence of flavonoid inducers, there were no differences in expression of nod genes or NF production between wild-type strains and rsh mutants (23, 24), and the same was observed for nodC expression in E. meliloti between the wild type and a dksA mutant (22). Moreover, in this report the numbers of nodules obtained with the rsh mutant and the wild type were similar, in agreement with the notion that increases in GmRIC1 and GmRIC2 expression might not be due to a direct effect of rsh mutation on NF production.
GmRIC1 and GmRIC2 are translocated and perceived in the shoot by the Glycine max nodule autoregulation receptor kinase (GmNARK) (53). Similarly, another CLE peptide, GmNIC1, is induced by nitrogenous compounds and perceived locally in the roots by GmNARK. The NARK orthologue in Lotus japonicus is “hypernodulation aberrant root formation 1” (LjHAR1). After perception of such CLE peptides, LjHAR1 induces the synthesis of cytokinin, which is translocated as a shoot-derived signal to the roots, where it inhibits nodulation (54). Therefore, the circuit of CLE peptides, GmNARK, and cytokinin might integrate the signals of excess infections or availability of combined N to negatively control the number of active nodules. In the present report, we observed that, in the absence of combined N, the expression of the three CLE peptides was higher in plants inoculated with the rsh mutant, while in the presence of combined N, GmNIC1 was upregulated and GmRIC1/2 were downregulated by the rsh mutant. As GmRIC1/2 are involved in a negative control of nodulation (51), this result correlates with the ability of the rsh mutant to nodulate in the presence of 10 mM NH4NO3. Although these nodules were scarce and small, they were red and did possess infected cells with bacteroids inside normal symbiosomes. This might suggest that, although initial infection was precluded by combined N as for the wild type, the rsh mutant bacteria that managed to travel all along ITs were successful in budding from ITs once the cortical cell layers were reached. In addition, these results are in agreement with the proposal of a cross-regulation between nitrogen control of nodulation and AON (42).
Although upregulation of GmRIC1/2 by the rsh mutant in the absence of combined N did not lead to diminished nodulation, nodule size was reduced in plants inoculated with the rsh mutant. The lack of nodulation inhibition might be due to the weak GmRIC1/2 upregulation (40). In addition, N2 fixation was somewhat affected, but not at the level previously observed in common beans inoculated with an R. etli rsh mutant (24, 25). These results indicate that the effects of rsh mutation in B. diazoefficiens on soybean nodule morphology and function are not as severe as those in R. etli on common bean nodules, despite both plants producing determinate nodules. This difference might be related to the low level of (p)ppGpp production in B. diazoefficiens. For the case of E. meliloti, nodulation was compromised in M. sativa but not in M. truncatula, although in this host, nodule cell infection and N2 fixation were lower with the rsh mutant than with the wild type (23). In addition, our B. diazoefficiens rsh mutant was intrinsically less competitive than the wild type to occupy soybean nodules, indicating that, as observed in E. meliloti and M. sativa (23), root infection might have been detracted.
We observed that the rsh mutant secreted a higher quantity of auxin to the surrounding medium. It is known that auxin synthesis and transport regulate root hair elongation (55), and local auxin accumulation is essential for nodule development (56). Therefore, these processes might be altered by the increased auxin production by the rsh mutant. This hypothesis might be confirmed with an rsh mutant unable to secrete auxins, together with in situ measures of auxin levels in the root infection zone.
Taken together, our results indicate that the stringent response of B. diazoefficiens is integrated into the circuits of regulation of plant defense and nodulation, with an impact on plant biomass production and response to the presence of combined N in the rhizosphere.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. diazoefficiens was grown at 28°C in a rotary shaker at 180 rpm in liquid peptone-salts-yeast extract medium (PSY [57]). Growth was measured by optical density at 500 nm (OD500), and the number of viable bacteria was determined by counting the number of CFU on yeast extract-mannitol agar (YMA) plates (58). Escherichia coli was grown in LB medium at 37°C (59). Media were supplemented with antibiotics at the following concentrations: for E. coli, gentamicin (Gm) at 10 μg ml−1, kanamycin (Km) at 25 μg ml−1, and tetracycline (Tc) at 20 μg ml−1; for B. diazoefficiens, chloramphenicol (Cm) at 20 μg ml−1, Km at 150 μg ml−1, and Tc at 100 μg ml−1.
DNA manipulation.
Standard protocols were used for DNA manipulations (59). To obtain the rsh::Km insertional mutant, a central region of 1,616 bp was amplified by PCR with Pfx polymerase (Thermo Fisher Scientific, Buenos Aires, Argentina) using the primers rsh1 Fw and rsh2 Rv (Table 4). The PCR cycling was started at 94°C for 3 min, followed by 35 cycles at 94°C for 20 s, 56°C for 20 s, and 68°C for 2 min, with a final elongation step at 68°C for 4 min. The fragment was cloned into a previously SmaI-digested plasmid, pG18mob2 (60). The resulting plasmid was named pG18mob2::rsh. The construction was confirmed by sequencing. Subsequently, the Kmr cassette was obtained as a SalI fragment from the pUC4K (61) and cloned into XhoI-digested pG18mob2::rsh. The latter plasmid was called pG18mob2::rsh::Km and was introduced by conjugation into B. diazoefficiens USDA 110 to generate the rsh::Km insertional mutant. To assess the correct integration by double crossing over into rsh, PCRs were carried out with primers rsh-compl-Fw and KmR, and KmF and rsh-compl-Rv (Table 4). The PCR products evaluated are shown in Fig. 1B. Once the correctness of the integration was confirmed, the rsh::Km strain was named LP 5065.
TABLE 4.
Primers used in this study
| Name | Sequence | Source or reference |
|---|---|---|
| rsh mutant construction and complementation | ||
| rsh1 Fw | TACAATCCCAACACCAACGA | This study |
| rsh2 Rv | ACGCGCTCCTCCTTGTAGT | This study |
| rsh-compl-Fw | AAAAATCTAGAATACCGAAGTTGCCGTTGAG | This study |
| rsh-compl-Rv | AAAAACTGCAGCACAAATTCGAAGGA | This study |
| Km F | CATCGGGCTTCCCATACA | 73 |
| Km R | TGCCATTCTCACCGGATT | 73 |
| Plant defense | ||
| PR1_Fw | ATGTTGCCTACGCTCAAGATTCAGCA | This study |
| PR1_Rv | AGCAACCGTATCATCCCAAGCCAAACT | This study |
| T3SS | ||
| blr1813_Fw | GACGGCTTCGCGTGTTTC | This study |
| blr1813_Rv | GGACAGCATCAACGCCCT | This study |
| bll1843_Fw | ACCTCACGCCGAGCAATC | This study |
| bll1843_Rv | CGGGCCATTTTGCGAAGG | This study |
| Regulation of nodulation | ||
| RIC1_Fw | GTCGGCAGTAGTAGTTGTATGG | This study |
| RIC1_Rv | AGCCTCCTCACTGAACTTGT | This study |
| NIC1_Fw | CTAAGCCCTGGAGGACCTGA | This study |
| NIC1_Rv | CAGCATACGTGGTAATTGCCTG | This study |
| RIC2_Fw | TGTGGGTGATGTAGGGTGGAT | This study |
| RIC2_Rv | CCTGGTTCGTCTTCATTGATCTCC | This study |
| Soybean reference gene | ||
| Ef1-α_Fw | GGTCATTGGTCATGTCGACTCTGG | 74 |
| Ef1-α_Rv | GCACCCAGGCATACTTGAATGACC | 74 |
| B. diazoefficiens reference gene | ||
| sigA-1533F | CTGATCCAGGAAGGCAACATC | 75 |
| sigA-1617R | TGGCGTAGGTCGAGAACTTGT | 75 |
To complement the mutation, we amplified a 2,653-bp fragment containing the wild-type rsh and its putative promoter with Pfx DNA polymerase (Thermo Fisher Scientific, Buenos Aires, Argentina) using the primers rsh-compl-Fw and rsh-compl-Rv (Table 1). This amplicon was digested with XbaI and PstI and cloned in the pFAJ1708 vector (62) digested with the same enzymes, yielding pFAJ1708::rsh. In this way, the rsh gene was under the promoter nptII to ensure the expression of the gene as described previously (63).
Plasmid transfer to B. diazoefficiens was carried out by conjugation using E. coli S17-1 as the donor (64). Matings were made in PSY.
(p)ppGpp extraction and determination.
The procedure described by Krol and Becker (26) was adapted to B. diazoefficiens. Briefly, 300-ml late-exponential PSY cultures were centrifuged at 4,200 × g for 10 min at 4°C. Each sample was separated and resuspended in the same volume with 3-(N-morpholino)propanesulfonic acid (MOPS; 48 mM at pH 7.2) or PSY to generate starvation or control conditions, respectively. The incubation continued under the same conditions for an additional hour, after which the nucleotide was extracted with 4 M formic acid and five freeze-thaw cycles. Then, samples were incubated 30 min on ice and centrifuged at 15,000 × g for 10 min at 4°C, and the supernatants were filtered through 0.2-μm-pore cellulose acetate filters. Finally, the amount of (p)ppGpp was determined by anion-exchange chromatography on Mono Q 5/50 GL column as described previously (65).
Plant assays.
Glycine max cv. Williams soybean seeds were surface sterilized and germinated as previously described (66). Then, each germinated seedling was planted in 650-ml pots containing sterile perlite-sand (2:1) and watered with 250 ml sterile modified Fåhraeus solution (MFS [67]). To inoculate the plants, bacterial colonies from YMA were diluted in 1 ml sterile MFS and deposited onto each seedling. Control plants were inoculated with 1 ml of sterile MFS without bacteria.
Plants were grown and analyzed for nodulation, biomass production, and ureide contents as described previously (68). To observe the effects of combined N, MFS was supplemented with 10 mM NH4NO3. These results were subjected to analysis of variance (ANOVA) followed by Tukey’s test with a significance level of P < 0.05.
To evaluate competition for nodule occupancy, soybean plants were coinoculated with a mixture of B. diazoefficiens USDA 110 and LP 5065 in a 1:1 proportion. For this purpose, pots were inoculated by watering with MFS containing 106 CFU ml−1 of each strain. Twenty-one days after inoculation, all nodules were collected, surface sterilized, and analyzed as described previously (69). For statistical analysis, these values were transformed to the arcsine root square, and analysis of variance was done with the transformed values, followed by Tukey’s test employing a significance level of P < 0.05.
RNA extractions and quantification of transcript levels by RT-qPCR.
For RNA extraction from roots, germinated seeds were grown in liquid MFS with the same light and temperature conditions described above. Roots were cut and then frozen in liquid N2, and RNA was extracted with TRIzol (Thermo Fisher Scientific, Buenos Aires, Argentina) following the manufacturer’s instructions. For bacterial RNA extraction, B. diazoefficiens late-exponential PSY liquid cultures were induced with genistein dissolved in methanol at a final concentration of 2 μM or the same volume of methanol as a control. After 48 h of growth under the same conditions, cells were collected and RNA extractions were performed as described previously (70).
For RT-qPCR from both plant and bacterial extracts, two-step cDNA was obtained using a Moloney murine leukemia virus (MMLV) reverse transcriptase (Thermo Fisher Scientific, Buenos Aires, Argentina) and quantified by qPCR using iQ SYBR green Supermix (Bio-Rad, Hercules, CA, USA), according to the manufacturer’s instructions. Gene-specific primers used in RT-qPCRs are listed in Table 4. qPCRs were performed in a qTower 2.0 (Analytik Jena, Germany). Normalized expression values were calculated as the ratio between the relative quantities of the gene of interest (GOI) and the relative quantities of the housekeeping genes indicated in the figure captions.
cytROS determinations.
Two-day-old soybean seedlings were inoculated with 106 CFU ml−1 B. diazoefficiens wild type or rsh mutant in MFS or in MFS alone as control. After 12 or 48 h, roots were incubated with 500 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA) for 30 min in darkness at room temperature. Images were obtained by using a Zeiss Imager A2 epifluorescence microscope with excitation at 488 nm and emission at 517 to 527 nm. The objective used was 10×, NA = 0.3, and the exposure time was 80 to 500 ms. The images were analyzed using the ImageJ program. To measure cytROS levels, a circular region of interest (ROI; r = 2.5 μm) was selected at the root tip, and the average of the intensities of the pixels included in the ROI was calculated. The reported values are the mean ± standard error (SE), and statistical analysis was performed by analysis of variance followed by Tukey’s test.
IAA determinations.
Indoleacetic acid (IAA) was measured as described by Gravel et al. (71). B. diazoefficiens was grown in PSY liquid medium for 7 days at 28°C and then centrifuged at 13,000 × g for 3 min. The Salkowski reagent was added to the supernatant (72), and the culture was left in the dark for 20 min. After this incubation time, auxin concentrations were estimated by optical density at 535 nm against a calibration curve made with known IAA concentrations.
Microscopy.
Microscopy analyses were performed at the Microscopy Service of Facultad de Ciencias Veterinarias (UNLP, La Plata, Argentina) as previously described (73). Briefly, nodules were transversally cut into halves and fixed in 2% (vol/vol) glutaraldehyde. Samples were dehydrated, infiltrated with epoxy resin, and sectioned. For optical microscopy, glutaraldehyde-fixed 2-μm-thick nodule sections were dried onto glass slides, stained with a saturated solution of toluidine blue, and analyzed using a Nikon Eclipse E200 microscope (Melville, NY). For transmission electron microscopy, ultrathin nodule sections were stained with 0.5% to 1% (wt/vol) uranyl acetate for 10 min and 1% (wt/vol) lead citrate for 5 min, washed in distilled water, and air dried. Then the cuts were analyzed with a JEM 1200 EX (JEOL, Japan Electron Optics Laboratory Co., Ltd.). Micrographs were obtained with an ES500W Erlangshen charge-coupled device camera (Gatan, Inc., Pleasanton, CA).
ACKNOWLEDGMENTS
J.P.-G., G.T.T., J.I.Q., J.M.E., and A.R.L. are members of the Scientific Research Career of CONICET. E.T.I. is a postdoctoral fellow of CONICET.
The authors are grateful to Paula Giménez, Silvana Tongiani, Claudio Mazo, and Abel Bortolameotti for technical assistance and to Susana Jurado from the UNLP Microscopy Service for help with transmission electron microscopy.
This work was supported by ANPCyT (PICT2017-2456, PICT2016-0604, and PICT2017-0066), CONICET PIP 11220150100700CO, ICGEB CRP/ARG16-03, Instituto Milenio iBio—Iniciativa Científica Milenio, MINECON, and SPP 1879—BE 2121/8-2 (DFG).
We declare that there are no conflicts of interest.
This article is dedicated to Federico Sánchez, our friend and teacher, who was an inexhaustible source of inspiration for this work.
REFERENCES
- 1.Terpolilli JJ, Hood GA, Poole PS. 2012. What determines the efficiency of N2-fixing Rhizobium-legume symbioses? Adv Microb Physiol 60:325–389. 10.1016/B978-0-12-398264-3.00005-X. [DOI] [PubMed] [Google Scholar]
- 2.Martínez-Romero E. 2009. Coevolution in Rhizobium-legume symbiosis? DNA Cell Biol 28:361–370. 10.1089/dna.2009.0863. [DOI] [PubMed] [Google Scholar]
- 3.Food and Agriculture Organization of the United Nations. 2020. http://www.fao.org/faostat/en/#data/QC. Accessed 15 January 2021.
- 4.Goodchild DJ, Bergersen FJ. 1966. Electron microscopy of the infection and subsequent development of soybean nodule cells. J Bacteriol 92:204–213. 10.1128/JB.92.1.204-213.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turgeon BG, Bauer WD. 1985. Ultrastructure of infection-thread development during the infection of soybean by Rhizobium japonicum. Planta 163:328–349. 10.1007/BF00395142. [DOI] [PubMed] [Google Scholar]
- 6.Fournier J, Timmers AC, Sieberer BJ, Jauneau A, Chabaud M, Barker DG. 2008. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol 148:1985–1995. 10.1104/pp.108.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patriarca EJ, Tatè R, Iaccarino M. 2002. Key role of bacterial NH4+ metabolism in Rhizobium-plant symbiosis. Microbiol Mol Biol Rev 66:203–222. 10.1128/mmbr.66.2.203-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brechenmacher L, Lei Z, Libault M, Findley S, Sugawara M, Sadowsky MJ, Sumner LW, Stacey G. 2010. Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol 153:1808–1822. 10.1104/pp.110.157800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ledermann R, Bartsch I, Müller B, Wülser J, Fischer HM. 2018. A functional general stress response of Bradyrhizobium diazoefficiens is required for early stages of host plant infection. Mol Plant Microbe Interact 31:537–547. 10.1094/MPMI-11-17-0284-R. [DOI] [PubMed] [Google Scholar]
- 10.Gourion B, Berrabah F, Ratet P, Stacey G. 2015. Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci 20:186–194. 10.1016/j.tplants.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Meury J. 1988. Glycine betaine reverses the effects of osmotic stress on DNA replication and cellular division in Escherichia coli. Arch Microbiol 149:232–239. 10.1007/BF00422010. [DOI] [PubMed] [Google Scholar]
- 12.Okada Y, Makino S, Tobe T, Okada N, Yamazaki S. 2002. Cloning of rel from Listeria monocytogenes as an osmotolerance involvement gene. Appl Environ Microbiol 68:1541–1547. 10.1128/aem.68.4.1541-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pezzoni M, Pizarro RA, Costa CS. 2012. Protective effect of low UVA irradiation against the action of lethal UVA on Pseudomonas aeruginosa: role of the relA gene. J Photochem Photobiol B 116:95–104. 10.1016/j.jphotobiol.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 14.Potamitou A, Neubauer P, Holmgren A, Vlamis-Gardikas A. 2002. Expression of Escherichia coli glutaredoxin 2 is mainly regulated by ppGpp and sigmaS. J Biol Chem 277:17775–17780. 10.1074/jbc.M201306200. [DOI] [PubMed] [Google Scholar]
- 15.Rallu F, Gruss A, Maguin E. 1996. Lactococcus lactis and stress. Antonie Van Leeuwenhoek 70:243–251. 10.1007/BF00395935. [DOI] [PubMed] [Google Scholar]
- 16.Wells DH, Gaynor EC. 2006. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J Bacteriol 188:3726–3729. 10.1128/JB.188.10.3726-3729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronneau S, Hallez R. 2019. Make and break the alarmone: regulation of (p)ppGpp synthetase/hydrolase enzymes in bacteria. FEMS Microbiol Rev 43:389–400. 10.1093/femsre/fuz009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinchen W, Zegarra V, Bange G. 2020. (p)ppGpp: magic modulators of bacterial physiology and metabolism. Front Microbiol 11:2072. 10.3389/fmicb.2020.02072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belitsky B, Kari C. 1982. Absence of accumulation of ppGpp and RNA during amino acid starvation in Rhizobium meliloti. J Biol Chem 257:4677–4679. 10.1016/S0021-9258(18)34574-5. [DOI] [PubMed] [Google Scholar]
- 20.Howorth SM, England RR. 1999. Accumulation of ppGpp in symbiotic and free-living nitrogen-fixing bacteria following amino acid starvation. Arch Microbiol 171:131–134. 10.1007/s002030050689. [DOI] [PubMed] [Google Scholar]
- 21.Wells DH, Long SR. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol Microbiol 43:1115–1127. 10.1046/j.1365-2958.2002.02826.x. [DOI] [PubMed] [Google Scholar]
- 22.Wippel K, Long SR. 2016. Contributions of Sinorhizobium meliloti transcriptional regulator DksA to bacterial growth and efficient symbiosis with Medicago sativa. J Bacteriol 198:1374–1383. 10.1128/JB.00013-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wippel K, Long SR. 2019. Symbiotic performance of Sinorhizobium meliloti lacking ppGpp depends on the Medicago host species. Mol Plant Microbe Interact 32:717–728. 10.1094/MPMI-11-18-0306-R. [DOI] [PubMed] [Google Scholar]
- 24.Calderón-Flores A, Du Pont G, Huerta-Saquero A, Merchant-Larios H, Servín-González L, Durán S. 2005. The stringent response is required for amino acid and nitrate utilization, Nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J Bacteriol 187:5075–5083. 10.1128/JB.187.15.5075-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moris M, Braeken K, Schoeters E, Verreth C, Beullens S, Vanderleyden J, Michiels J. 2005. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J Bacteriol 187:5460–5469. 10.1128/JB.187.15.5460-5469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krol E, Becker A. 2011. ppGpp in Sinorhizobium meliloti: biosynthesis in response to sudden nutritional downshifts and modulation of the transcriptome. Mol Microbiol 81:1233–1254. 10.1111/j.1365-2958.2011.07752.x. [DOI] [PubMed] [Google Scholar]
- 27.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, Kohara M, Matsumoto M, Shimpo S, Tsuruoka H, Wada T, Yamada M, Tabata S. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 9:189–197. 10.1093/dnares/9.6.189. [DOI] [PubMed] [Google Scholar]
- 28.Cogo C, Pérez-Giménez J, Rajeswari CB, Luna MF, Lodeiro AR. 2018. Induction by Bradyrhizobium diazoefficiens of different pathways for growth in d-mannitol or l-arabinose leading to pronounced differences in CO2 fixation, O2 consumption, and lateral-flagellum production. Front Microbiol 9:1189. 10.3389/fmicb.2018.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ancona V, Lee JH, Chatnaparat T, Oh J, Hong JI, Zhao Y. 2015. The bacterial alarmone (p)ppGpp activates the type III secretion system in Erwinia amylovora. J Bacteriol 197:1433–1443. 10.1128/JB.02551-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause A, Doerfel A, Göttfert M. 2002. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol Plant Microbe Interact 15:1228–1235. 10.1094/MPMI.2002.15.12.1228. [DOI] [PubMed] [Google Scholar]
- 31.Jiménez-Guerrero I, Pérez-Montano F, Monreal JA, Preston GM, Fones H, Vioque B, Ollero FJ, López-Baena FJ. 2015. The Sinorhizobium (Ensifer) fredii HH103 type 3 secretion system suppresses early defense responses to effectively nodulate soybean. Mol Plant Microbe Interact 28:790–799. 10.1094/MPMI-01-15-0020-R. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Giménez J, Quelas JI, Lodeiro AR. 2011. Competition for nodulation, p 139–166. In El-Shemy HA (ed), Soybean physiology and biochemistry. IntechOpen, Rijeka, Croatia. [Google Scholar]
- 33.Carroll BJ, McNeil DL, Gresshoff PM. 1985. A supernodulation and nitrate-tolerant symbiotic (nts) soybean mutant. Plant Physiol 78:34–40. 10.1104/pp.78.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streeter J, Wong PP. 1988. Inhibition of legume nodule formation and N2 fixation by nitrate. Crit Rev Plant Sci 7:1–23. 10.1080/07352688809382257. [DOI] [Google Scholar]
- 35.Ligero F, Poveda JL, Gresshoff PM, Caba JM. 1999. Nitrate- and inoculation-enhanced ethylene biosynthesis in soybean roots as a possible mediator of nodulation control. J Plant Physiol 154:482–488. 10.1016/S0176-1617(99)80287-9. [DOI] [Google Scholar]
- 36.Zahran HH. 1999. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63:968–989. 10.1128/MMBR.63.4.968-989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbulova A, Rogato A, D'Apuzzo E, Omrane S, Chiurazzi M. 2007. Differential effects of combined N sources on early steps of the Nod factor-dependent transduction pathway in Lotus japonicus. Mol Plant Microbe Interact 20:994–1003. 10.1094/MPMI-20-8-0994. [DOI] [PubMed] [Google Scholar]
- 38.Saito A, Tanabata S, Tanabata T, Tajima S, Ueno M, Ishikawa S, Ohtake N, Sueyoshi K, Ohyama T. 2014. Effect of nitrate on nodule and root growth of soybean (Glycine max (L.) Merr.). Int J Mol Sci 15:4464–4480. 10.3390/ijms15034464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iturralde ET, Stocco MC, Faura A, Mónaco CI, Cordo C, Pérez-Giménez J, Lodeiro AR. 2020. Coinoculation of soybean plants with Bradyrhizobium japonicum and Trichoderma harzianum: coexistence of both microbes and relief of nitrate inhibition of nodulation. Biotechnol Rep (Amst) 26:e00461. 10.1016/j.btre.2020.e00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishida H, Suzaki T. 2018. Two negative regulatory systems of root nodule symbiosis: how are symbiotic benefits and costs balanced? Plant Cell Physiol 59:1733–1738. 10.1093/pcp/pcy102. [DOI] [PubMed] [Google Scholar]
- 41.Reid DE, Ferguson BJ, Gresshoff PM. 2011. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact 24:606–618. 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- 42.Lim CW, Lee YW, Lee SC, Hwang CH. 2014. Nitrate inhibits soybean nodulation by regulating expression of CLE genes. Plant Sci 229:1–9. 10.1016/j.plantsci.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 43.Caba JM, Centeno ML, Fernández B, Gresshoff PM, Ligero F. 2000. Inoculation and nitrate alter phytohormone levels in soybean roots: differences between a supernodulating mutant and the wild type. Planta 211:98–104. 10.1007/s004250000265. [DOI] [PubMed] [Google Scholar]
- 44.Jin J, Watt M, Mathesius U. 2012. The autoregulation gene SUNN mediates changes in root organ formation in response to nitrogen through alteration of shoot-to-root auxin transport. Plant Physiol 159:489–500. 10.1104/pp.112.194993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng JL, Hassan S, Truong TT, Hocart CH, Laffont C, Frugier F, Mathesius U. 2015. Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell 27:2210–2226. 10.1105/tpc.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suss C, Hempel J, Zehner S, Krause A, Patschkowski T, Gottfert M. 2006. Identification of genistein-inducible and type III-secreted proteins of Bradyrhizobium japonicum. J Biotechnol 126:69–77. 10.1016/j.jbiotec.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 47.Tsukui T, Eda S, Kaneko T, Sato S, Okazaki S, Kakizaki-Chiba K, Itakura M, Mitsui H, Yamashita A, Terasawa K, Minamisawa K. 2013. The type III secretion system of Bradyrhizobium japonicum USDA122 mediates symbiotic incompatibility with Rj2 soybean plants. Appl Environ Microbiol 79:1048–1051. 10.1128/AEM.03297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staehelin C, Krishnan HB. 2015. Nodulation outer proteins: double-edged swords of symbiotic rhizobia. Biochem J 470:263–274. 10.1042/BJ20150518. [DOI] [PubMed] [Google Scholar]
- 49.López-Baena FJ, Monreal JA, Pérez-Montaño F, Guasch-Vidal B, Bellogín RA, Vinardell JM, Ollero FJ. 2009. The absence of Nops secretion in Sinorhizobium fredii HH103 increases GmPR1 expression in Williams soybean. Mol Plant Microbe Interact 22:1445–1454. 10.1094/MPMI-22-11-1445. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Göbel TF, Deanna R, Muñoz NB, Robert G, Asurmendi S, Lascano R. 2019. Redox systemic signaling and induced tolerance responses during soybean-Bradyrhizobium japonicum interaction: involvement of Nod Factor receptor and autoregulation of nodulation. Front Plant Sci 10:141. 10.3389/fpls.2019.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferguson BJ, Mens C, Hastwell AH, Zhang M, Su H, Jones CH, Chu X, Gresshoff PM. 2019. Legume nodulation: the host controls the party. Plant Cell Environ 42:41–51. 10.1111/pce.13348. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Sun Z, Su C, Wang Y, Yan Q, Chen J, Ott T, Li X. 2019. A GmNINa-miR172c-NNC1 regulatory network coordinates the nodulation and autoregulation of nodulation pathways in soybean. Mol Plant 12:1211–1226. 10.1016/j.molp.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299:109–112. 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- 54.Sasaki T, Suzaki T, Soyano T, Kojima M, Sakakibara H, Kawaguchi M. 2014. Shoot-derived cytokinins systemically regulate root nodulation. Nat Commun 5:4983. 10.1038/ncomms5983. [DOI] [PubMed] [Google Scholar]
- 55.Velásquez SM, Barbez E, Kleine-Vehn J, Estévez JM. 2016. Auxin and cellular elongation. Plant Physiol 170:1206–1215. 10.1104/pp.15.01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Zhang C, Yang J, Yu N, Wang E. 2018. Hormone modulation of legume-rhizobial symbiosis. J Integr Plant Biol 60:632–648. 10.1111/jipb.12653. [DOI] [PubMed] [Google Scholar]
- 57.Regensburger B, Hennecke H. 1983. RNA polymerase from Rhizobium japonicum. Arch Microbiol 135:103–109. 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 58.Vincent JM. 1970. A manual for the practical study of the root-nodule bacteria. IBP Handbook No. 15. Blackwell Scientific, Oxford, United Kingdom. [Google Scholar]
- 59.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 60.Kirchner O, Tauch A. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J Biotechnol 104:287–299. 10.1016/s0168-1656(03)00148-2. [DOI] [PubMed] [Google Scholar]
- 61.Vieira J, Messing J. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268. 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 62.Dombrecht B, Vanderleyden J, Michiels J. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in Gram-negative bacteria. Mol Plant Microbe Interact 14:426–430. 10.1094/MPMI.2001.14.3.426. [DOI] [PubMed] [Google Scholar]
- 63.Quelas JI, Mongiardini EJ, Pérez Giménez J, Parisi G, Lodeiro AR. 2013. Analysis of two polyhydroxyalkanoate synthases in Bradyrhizobium japonicum USDA 110. J Bacteriol 195:3145–3155. 10.1128/JB.02203-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 65.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.López-García SL, Vázquez TE, Favelukes G, Lodeiro AR. 2001. Improved soybean root association of N-starved Bradyrhizobium japonicum. J Bacteriol 183:7241–7252. 10.1128/JB.183.24.7241-7252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lodeiro AR, González P, Hernández A, Balagué LJ, Favelukes G. 2000. Comparison of drought tolerance in nitrogen-fixing and inorganic nitrogen-grown common beans. Plant Sci 154:31–41. 10.1016/s0168-9452(99)00246-0. [DOI] [PubMed] [Google Scholar]
- 68.Iturralde ET, Covelli JM, Alvarez F, Pérez-Giménez J, Arrese-Igor C, Lodeiro AR. 2019. Soybean-nodulating strains with low intrinsic competitiveness for nodulation, good symbiotic performance, and stress-tolerance isolated from soybean-cropped soils in Argentina. Front Microbiol 10:1061. 10.3389/fmicb.2019.01061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Althabegoiti MJ, Covelli JM, Pérez-Giménez J, Quelas JI, Mongiardini EJ, López MF, López-García SL, Lodeiro AR. 2011. Analysis of the role of the two flagella of Bradyrhizobium japonicum in competition for nodulation of soybean. FEMS Microbiol Lett 319:133–139. 10.1111/j.1574-6968.2011.02280.x. [DOI] [PubMed] [Google Scholar]
- 70.Mongiardini EJ, Quelas JI, Dardis C, Althabegoiti MJ, Lodeiro AR. 2017. Transcriptional control of the lateral-flagellar genes of Bradyrhizobium diazoefficiens. J Bacteriol 199:e00253-17. 10.1128/JB.00253-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gravel V, Antoun H, Tweddell RJ. 2007. Growth stimulation and fruit yield improvement of greenhouse tomato plants by inoculation with Pseudomonas putida or Trichoderma atroviride: possible role of indole acetic acid (IAA). Soil Biol Biochem 39:1968–1977. 10.1016/j.soilbio.2007.02.015. [DOI] [Google Scholar]
- 72.Gordon SA, Weber RP. 1951. Colorimetric estimation of indoleacetic acid. Plant Physiol 26:192–195. 10.1104/pp.26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quelas JI, Mongiardini EJ, Casabuono A, López-García SL, Althabegoiti MJ, Covelli JM, Pérez-Giménez J, Couto A, Lodeiro AR. 2010. Lack of galactose or galacturonic acid in Bradyrhizobium japonicum USDA 110 exopolysaccharide leads to different symbiotic responses in soybean. Mol Plant Microbe Interact 23:1592–1604. 10.1094/MPMI-05-10-0122. [DOI] [PubMed] [Google Scholar]
- 74.Díaz-Camino C, Annamalai P, Sánchez F, Kachroo A, Ghabrial SA. 2011. An effective virus-based gene silencing method for functional genomics studies in common bean. Plant Methods 7:16. 10.1186/1746-4811-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hauser F, Lindemann A, Vuilleumier S, Patrignani A, Schlapbach R, Fischer HM, Hennecke H. 2006. Design and validation of a partial-genome microarray for transcriptional profiling of the Bradyrhizobium japonicum symbiotic gene region. Mol Genet Genomics 275:55–67. 10.1007/s00438-005-0059-7. [DOI] [PubMed] [Google Scholar]