Summary
Individuals with coronavirus disease 2019 (COVID-19) frequently develop neurological symptoms, but the biological underpinnings of these phenomena are unknown. Through single-cell RNA sequencing (scRNA-seq) and cytokine analyses of cerebrospinal fluid (CSF) and blood from individuals with COVID-19 with neurological symptoms, we find compartmentalized, CNS-specific T cell activation and B cell responses. All affected individuals had CSF anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies whose target epitopes diverged from serum antibodies. In an animal model, we find that intrathecal SARS-CoV-2 antibodies are present only during brain infection and not elicited by pulmonary infection. We produced CSF-derived monoclonal antibodies from an individual with COVID-19 and found that these monoclonal antibodies (mAbs) target antiviral and antineural antigens, including one mAb that reacted to spike protein and neural tissue. CSF immunoglobulin G (IgG) from 5 of 7 patients showed antineural reactivity. This immune survey reveals evidence of a compartmentalized immune response in the CNS of individuals with COVID-19 and suggests a role of autoimmunity in neurologic sequelae of COVID-19.
Keywords: COVID-19, neurological infection, autoimmunity, cerebrospinal fluid, SARS-CoV-2
Graphical abstract

Highlights
Immune cell scRNA-seq showed divergent T cell activation in the CNS during COVID-19
Individuals with COVID-19 had a compartmentalized cytokine response in the CNS
All individuals with COVID-19 had anti-SARS-CoV-2 antibodies in their CSF
Five of seven individuals with COVID-19 had antineural autoantibodies in their CSF
Neurological symptoms are frequent in hospitalized individuals with acute COVID-19. Song et al. find that, compared with control individuals, those with COVID-19 with neurologic symptoms have divergent immune responses between the CNS and periphery, including high rates of antineural autoantibodies in their CSF.
Introduction
The causative pathogen of pandemic coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), primarily causes respiratory illness. However, in some people, SARS-CoV-2 infection is associated with severe and debilitating neurological symptoms.1 About a third of individuals with moderate to severe COVID-19 experience neurological sequelae, including anosmia, dysgeusia, headache, impaired consciousness, and seizures, only some of which are explained by systemic complications, including hypercoagulability.2 Rarely, SARS-CoV-2 RNA is detected in the cerebrospinal fluid (CSF) of individuals with COVID-19, and some studies have found evidence of SARS-CoV-2 protein in brain parenchyma. However, there is little evidence that SARS-CoV-2 directly damages neural tissue.3, 4, 5, 6, 7 These observations suggest that mechanisms other than direct cytopathic effects of SARS-CoV-2 contribute to neurological symptoms. Therefore, a broad characterization of CNS immunity may provide further insight into the causes of neurologic impairment in COVID-19. In this exploratory study, we profiled intrathecal and peripheral immune responses in individuals with COVID-19 complicated by diverse neurological symptoms.
Results
Overview
Hospitalized individuals with COVID-19 with various neurological symptoms who underwent clinically indicated lumbar puncture consented to collection of surplus CSF to be used for research. Six participants with acute COVID-19, based on positive SARS-CoV-2 qRT-PCR of nasopharyngeal swabs, were enrolled (Table S1). Neurological symptoms included encephalopathy, intractable headaches, and seizures. All participants donated paired blood and CSF, except for one individual who did not donate blood. Lumbar punctures were performed on median hospital day 12.5 (range, 2–43 days). Pre-pandemic CSF from age- and gender-matched healthy control individuals (n = 3) was obtained from a neuroinfectious disease biorepository at Yale. Because fresh CSF is required for single-cell transcriptomics, we recruited additional uninfected control participants during the COVID-19 pandemic (n = 3); 2 were healthy community-dwelling adults, and 1 was hospitalized for work-up of frequent falls. Additional blood and CSF single-cell sequencing data were included from publicly available data derived from healthy control individuals (n = 8).8 Recruited control individuals tested negative for SARS-CoV-2 by RT-PCR of nasopharyngeal swabs. Fresh CSF and blood samples were processed into CSF-resident cells, CSF supernatant, peripheral blood mononuclear cells (PBMCs), and plasma (Figure 1A). CSF (n = 5) and plasma (n = 6) samples from individuals with COVID-19 were negative for SARS-CoV-2 RNA by qRT-PCR using Centers of Disease Control and Prevention (CDC) primer-probe sets.9
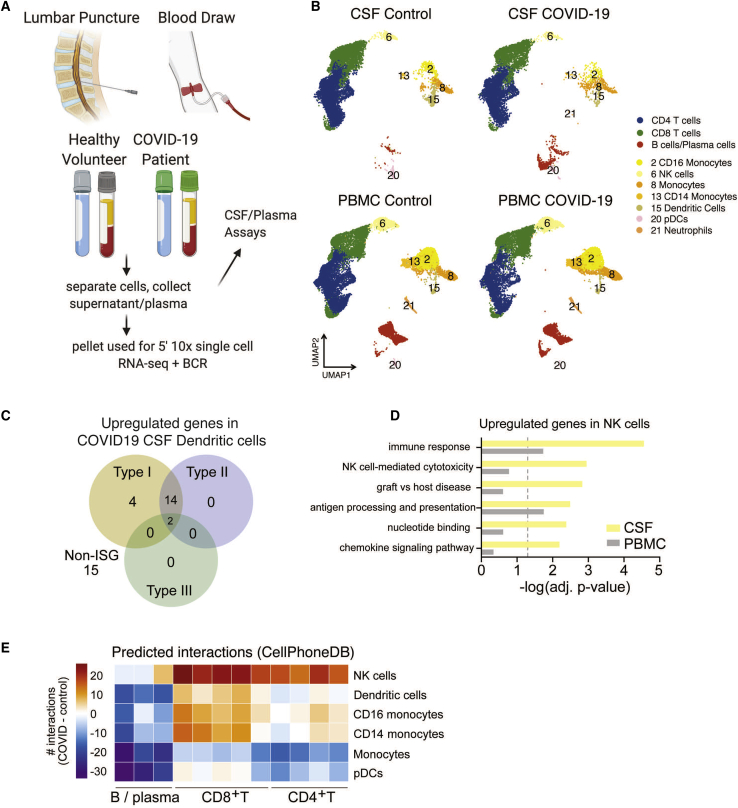
Figure 1.
Distinct immunological landscape of CSF and PBMCs in individuals with COVID-19 with neurological symptoms
(A) Schematic of the study design. CSF and blood were collected from individuals with COVID-19 and healthy control individuals. PBMCs and CSF cells were isolated, along with the CSF supernatant and plasma, for downstream analysis.
(B) UMAP (Uniform Manifold Approximation and Projection) projections of 10x single-cell RNA sequencing of CSF and PBMCs of individuals with COVID-19 and healthy control individuals.
(C) Venn diagram depicting upregulated interferon-stimulated genes (ISGs) and non-ISGs in dendritic cells in COVID-19 CSF compared with healthy control CSF based on the Interferome database.10
(D) Gene Ontology enrichment of genes upregulated in NK cells of individuals with COVID-19 in the CSF and peripheral blood.
(E) Heatmap depicting cell-cell interactions between innate immune cells and adaptive immune cells by clustering shown in (B). The difference in interaction strength (COVID-19 interaction minus control interaction) is color coded and derived from log-scaled interaction counts using the CellphoneDB repository of ligands, receptors, and their interactions.11 Single-cell RNA-seq is derived from a total of 16 libraries plus 8 additional controls from Gate et al.8 (n = 3 for control CSF and PBMCs, n = 5 for COVID-19 CSF and PBMCs, and n = 8 from Gate et al.8).
Transcriptional analysis reveals a coordinated innate immune cell response to COVID-19 in the CNS
To investigate the effect of SARS-CoV-2 infection on host immune cell gene expression, we performed single-cell RNA sequencing with the 10x Genomics platform on 76,473 immune cells from the CSF and blood of individuals hospitalized with acute COVID-19 and uninfected control individuals. To test for the presence of intracellular virus, open reading frames of SARS-CoV-2 (spike, ORF3a, envelope, membrane glycoprotein, ORF6, ORF7a, ORF8, nucleocapsid, and ORF10) were added to the reference genome before alignment of RNA sequencing data with CellRanger. Viral transcripts were not detected in any CSF immune cells or PBMCs. We performed unsupervised cluster analysis of CSF cells and PBMCs and identified distinct T cell, B cell, and myeloid cell populations (Figure 1B), characterized by expression patterns of canonical immune cell marker genes (Figure S1A). Next we identified genes that were differentially expressed in CSF—but not peripheral blood—immune cells of individuals with COVID-19 compared with control individuals (Data S2).
In the CSF of individuals with COVID-19, dendritic cells had an activated transcriptional profile; 57% and 47% of their upregulated genes were classified as type 1 and type 2 interferon-stimulated genes, respectively (Figure 1C). Genes associated with natural killer (NK) cell activation were also upregulated in the CSF of individuals with COVID-19 (Figure 1D). Although NK cells in the CSF and peripheral blood demonstrated comparable changes in the number of differentially expressed genes in individuals with COVID-19 compared with control individuals, the affected genes were mostly unique to each compartment (Figure S1C). Using CellphoneDB signaling network analysis,11,12 in individuals with COVID-19, CSF dendritic and NK cells were predicted to have significantly increased interactions with CSF CD8 and CD4 T cells relative to healthy controls, whereas interactions between CD4 T cells and monocytes were diminished, suggesting a dysregulated innate-to-adaptive immune interface (Figure 1E).
T cells in the CSF display increased cellular activation during COVID-19
Because signaling network analysis predicted that T cells were the main recipients of altered innate-adaptive cross-talk, we isolated and re-clustered CSF and peripheral T cells for targeted transcriptional analysis of T cell subsets (Figures 2A and S2A–S2C). Among peripheral T cells, there was a decrease in the frequency of naive CD4 T cells (mean: COVID-19, 9.65%; healthy, 18.219%; p = 0.001) and an increase in effector CD8 T cells (mean: COVID-19, 30.9%; healthy, 16.245%; p = 0.02) (Figure 2B). In contrast, in CSF, the relative proportions of T cell populations were conserved in individuals with COVID-19 compared with control individuals, but we found significant COVID-19-associated transcriptional changes in CSF T cells (Figure S2D). After excluding any genes that were also differentially expressed between T cells in the CSF and periphery of healthy control individuals, we identified genes that were upregulated in Th1 and Th2 CD4 T cells from COVID-19 CSF (Figure 2C). These genes were enriched for interleukin-1 (IL-1)- and IL-12-mediated signaling pathways and several genetic pathways important for T cell activation (Figure 2D). Effector CD8 T cells in the CSF were similarly enriched for genes involved in canonical immune response pathways, including (1) increased motility and cell adhesion, (2) differentiation/proliferation, and (3) effector programming (responses to IL-12, IL-1, interferon-g, and T cell co-stimulation), indicating the presence of a coordinated T cell-based immune response in the CNS (Figures 2E and 2F). Transcriptional changes in individuals with COVID-19 were observed in CD4 and CD8 T cells in the CSF and predicted cell-cell interactions that were unique to the CSF in individuals with COVID-19, including T cell co-stimulation factors and trafficking interactions (Figures S3A and S3B). Analysis of T cell receptor (TCR) sequences in CSF and blood revealed clonal expansion of unique but not shared CD4 T cell clones in the CSF of individuals with COVID-19 (Figure S4), further suggesting a compartmentalized T cell response to CNS antigen.
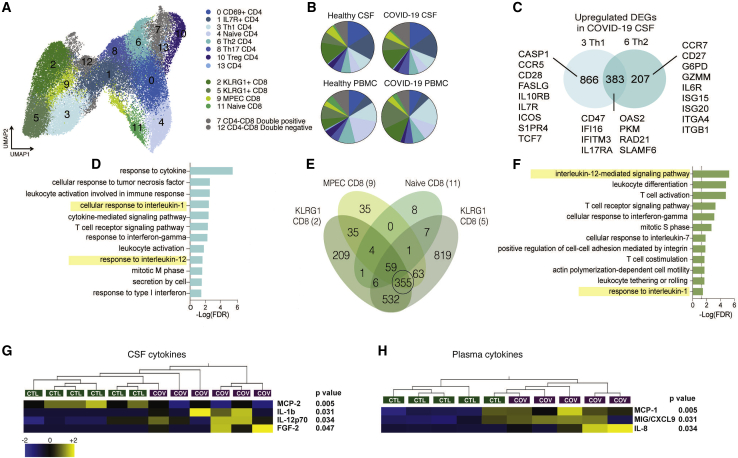
Figure 2.
Transcriptional characterization of T cells in CSF and PBMCs of individuals with COVID-19
(A) Reclustered UMAP projection of combined CSF and peripheral blood T cells, demonstrating CD4 and CD8 T cell subsets (two KLRG1+ clusters are distinguished by GZMB and IFNG expression; Figure S3).
(B) Pie charts depicting the relative population frequency of different T cell subtypes found in CSF and PBMCs of control individuals and those with COVID-19.
(C) Venn diagram depicting genes upregulated (adjusted p < 0.05) in CSF of individuals with COVID-19 compared with PBMCs of individuals with COVID-19 in Th1 and Th2 CD 4 T cells.
(D) Gene Ontology analysis of genes that are upregulated in Th1 and Th2 cells, as depicted in (C).
(E) Quad-Venn diagram of genes upregulated in CSF of individuals with COVID-19 compared with CSF of control individuals in CD8 T cells. Genes shared by the three effector CD8 T cell subtypes are circled.
(F) Gene Ontology analysis of genes shared between the three effector CD8 T cell subtypes in (F).
(G and H) Heatmap of Luminex-based cytokine profiling of CSF (G) and plasma (H) from individuals with COVID-19 and control individuals showing cytokines that were increased significantly increased in individuals with COVID-19 compared with control individuals (n = 6 CSF, n = 5 plasma). For each cytokine, two-tailed p values were calculated using Student’s t test. Data for each row were mean centered; each column shows data from one sample.
Unique cytokine profiles exist in CSF of individuals with COVID-19 compared with serum
To validate the transcriptional enrichment in IL-12 and IL-1 signaling in the CSF of individuals with COVID-19 with neurologic symptoms, we measured inflammatory cytokine levels in the CSF and plasma using a Luminex cytokine panel (Figures 2G and 2H). Consistent with the single-cell RNA sequencing results, IL-1b and IL-12 were elevated in the CSF of individuals with COVID-19 compared with healthy control individuals but were not elevated in the plasma of individuals with COVID-19. Conversely, CCL2, CXCL9, and IL-8 were increased significantly in the plasma of individuals with COVID-19 compared with control individuals but not in their CSF. Because IL-12 is thought to be produced by activated antigen-presenting cells to orchestrate Th1 responses through T and NK cell activation, we examined the cellular source of IL-12 in individuals with COVID-19. The innate immune cells with the highest IL12A expression were CSF NK and dendritic cells (Figure S1E). These data support the single-cell RNA sequencing analyses that identified IL-12 as differentially expressed in CSF but not blood innate immune cells of individuals with COVID-19. Moreover, they suggest a distinct effect of COVID-19 in the CNS on cytokines important for innate immunity and induction of cell-mediated immunity, including IL-1 and IL-12.
CNS B cell responses to SARS-CoV-2 differ from those in the periphery
We found significant enrichment of B cells in the CSF of individuals with COVID-19 cases compared with CSF of healthy control individuals (Figure 3A). Single-cell RNA sequencing identified several subtypes among peripheral and CSF B cells (Figures 3B and S5A–S5C), including distinct CSF plasma cell clusters. We therefore wanted to find out whether antibody-secreting B cells in the CSF exhibit a different anti-SARS-CoV-2 antibody profile than the those in the periphery. To do so, we utilized a recently developed SARS-CoV-2 epitope Luminex panel13 to screen for anti-SARS-CoV-2 antibodies in the CSF and plasma of individuals with COVID-19 and control individuals. As expected, anti-SARS-CoV-2 antibodies were not detected in any control individuals. In contrast, all individuals with COVID-19 had anti-SARS-CoV-2 antibodies in the CSF and plasma. However, although all individuals with COVID-19 developed antibodies to SARS-CoV-2 spike and nucleocapsid in the plasma and CSF, anti-receptor binding domain (RBD) antibodies were rare in CSF but uniformly present in the plasma (Figure 3C). In addition, we found that, in all individuals with COVID-19, the relative prevalence (rank score: 12, most frequent; 1, least frequent; Figure 3D) and levels of antibody (Figure S5D) diverged between the CSF and plasma, indicating a different anti-SARS-CoV-2 antibody profile between the CSF and plasma of the same individual.
Figure 3.
Localized central nervous system B cell responses in individuals with COVID-19
(A) Frequency of B cells as a percentage of all CSF cells in control individuals and those with COVID-19. Colors represent different individuals.
(B) Re-clustered UMAP projection of B cells from CSF and blood.
(C) Heatmap showing antibody binding in plasma (left) and CSF (right) to nine peptides from immunogenic regions of S, N, and ORF3a as well as whole S and N protein along with the RBD of the S protein. All data are represented as fold change of the fluorescent anti-IgG antibody signal over intra-assay negative controls. HC, healthy control.
(D) Epitope frequency was ranked in each sample individually, and a difference in rank number for each cluster was graphed to determine CSF-enriched (positive) or plasma-enriched (negative) antibody epitopes. Two-tailed unpaired t test, ∗∗p < 0.01.
A mouse model of SARS-CoV-2 brain infection demonstrates compartmentalized CNS antibody secretion in response to CNS infection
Direct detection of SARS-CoV-2 in the CSF is extremely rare in reported cases of neurological complications of COVID-19,14 and SARS-CoV-2 RNA was not detected in the CSF our cohort. However, we detected intrathecal antiviral antibodies in all cases. With some other encephalitis-causing viruses, including West Nile virus, Japanese encephalitis virus, and measles virus,15, 16, 17 the presence of antiviral antibodies is consistent with viral neuroinvasion even in the absence of viral nucleic acid. To determine whether CNS infection is sufficient to stimulate a CNS humoral response during COVID-19, we used a recently developed mouse model that reliably dissociates pulmonary and neurological infection of SARS-CoV-2.18
We used an adeno-associated virus (AAV) to express the human ACE2 (hACE2) receptor in the lungs or brain or the lungs and brain, allowing us to target SARS-CoV-2 infection to specific tissue. First, we used mice that express hACE2 in the lung and brain and administered SARS-CoV-2 intranasally (Figure 4A). This permits SARS-CoV-2 to infect the lungs and brain. In these mice, we detected increased titers of SARS-CoV-2 RNA in lung and brain tissue following inoculation. However, despite robust brain infection, we did not detect SARS-CoV-2 RNA in the CSF of these mice (Figure 4B). This suggests that direct detection of SARS-CoV-2 RNA in CSF at a single time point may be insensitive to parenchymal or short-lived SARS-CoV-2 neuroinvasion.
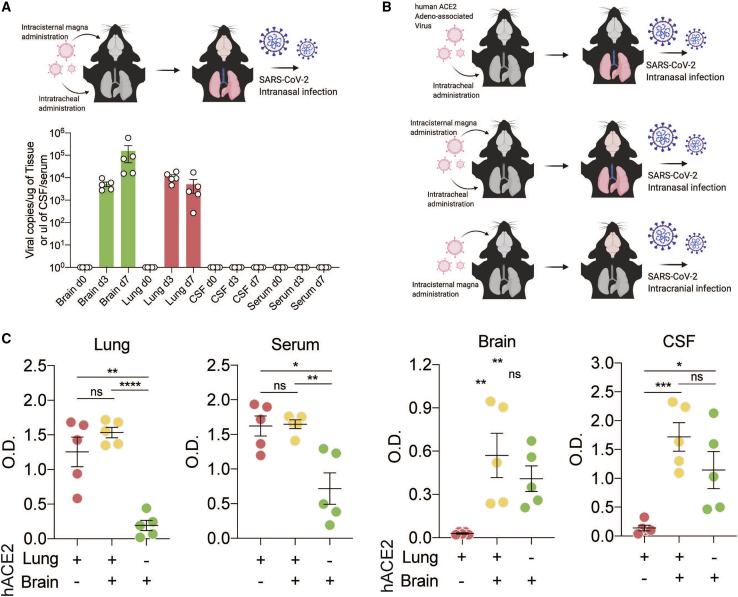
Figure 4.
CSF antibodies reflect localized CNS infection
(A) Mice were transduced with AAV-hACE2 intrathecally and intratracheally for expression in the brain and lungs, and SARS-CoV-2 was introduced intranasally to establish brain and lung infection. Mouse brains, lungs, CSF, and serum were collected on day 0 (before infection) and on days 3 and 7 after infection, and qPCR was performed to detect SARS-CoV-2 RNA.
(B) Schematic of the experimental procedure for (C). Mice were given localized AAV-hACE2 to overexpress human ACE2 in the lungs (top), brain and lungs (center), or brain only (bottom). After 2 weeks, mice were infected with SARS-CoV-2.
(C) ELISA against SARS-CoV-2 spike protein was performed with lung homogenates, serum, brain homogenates, and CSF. n = 5 for all three conditions. Two-tailed unpaired t test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.0001) and one-way ANOVA were performed (lungs, p < 0.0001; serum, p = 0.002; brain, p = 0.0082; CSF, p = 0.0016).
We next used the mouse model to evaluate whether detection of intrathecal anti-SARS-CoV-2 antibodies in our individuals with COVID-19 was more likely triggered by a local antigen (i.e., as a consequence of SARS-CoV-2 neuroinvasion) or reflected passive transfer of antibody from the systemic circulation.14 When SARS-CoV-2 was administered intranasally to mice expressing hACE2 only in the lungs (generating mice with pulmonary but not brain infection), we detected significantly elevated anti-spike SARS-CoV-2 immunoglobulin G (IgG) in the lungs and serum but not in the brain or CSF (Figure 4B, red). When SARS-CoV-2 was administered intranasally to mice expressing hACE2 in the brain and lungs (generating mice with pulmonary and brain infection), we detected increased anti-spike antibodies in all four compartments: lungs, serum, brain, and CSF (Figure 4B, orange). Finally, when hACE2 was expressed in the brain only and SARS-CoV-2 was administered intracranially (causing infection in the brain but not in the lungs), we detected increased anti-spike antibodies in the brain and CSF but not in the lungs or serum (Figure 4B, green). These data support the hypothesis that CSF antibodies do not solely reflect passive transfer of antibodies from the systemic circulation. Indeed, in these mice, anti-spike antibodies in the CSF and brain were only observed in the setting of brain infection, independent of whether there was an accompanying systemic SARS-CoV-2 infection.
Monoclonal antibodies from CSF B cells from individuals with COVID-19 are self reactive
Although our individuals with COVID-19 did not have detectable SARS-CoV-2 RNA in their CSF, CSF serology suggested that CSF-expanded B cell populations might be reactive to SARS-CoV-2 antigen(s). To address this, we cloned individual monoclonal antibodies from the affected individual with the largest number of clonally expanded B cell receptor (BCR) sequences in the CSF (n = 5) and blood (n = 4) (case 1; Data S1 and S5). In this individual, the most prevalent BCR sequences comprised ∼25% and ∼10% of total B cells in the blood and CSF, respectively (Figure 5A). Notably, the most prevalent BCR sequences detected in the CSF of case 1 did not overlap with the most prevalent blood BCR sequences (Figure 5B), supporting the hypothesis that a subset of CSF antibodies targets antigens within the CNS.
Figure 5.
Antigenic specificity of CSF- and PBMC-derived monoclonal antibodies
(A) Bar graph depicting the frequency of the top five most expanded clones in PBMCs and CSF of affected individual 1.
(B) Graph depicting overlap of clones found in CSF and PBMCs of an affected individual. Green indicates clones only found in CSF, orange clones shared between the CSF and PBMC, and red clones unique to PBMCs. The yellow box indicates clones that would fall under the top 10 most frequent clones in each compartment.
(C) Heatmap showing CSF-derived (mAbs C1–C5) and PBMC-derived (mAbs P1–P3 and P5) mAb binding to nine peptides from immunogenic regions of S, N, and ORF3a as well as whole S and N protein along with the RBD of the S protein. mAb numbers correspond to the clone numbers from (A) and (B); PBMC clone 4 (mAb P4) did not express well as a mAb and was not used for subsequent studies. mAbs were screened in technical replicates. Heatmap values are mean fold change of the fluorescent anti-IgG antibody signal over intra-assay negative controls.
(D) Sagittal mouse brain sections were immunostained with mAbs 1–9, and a representative whole-brain sagittal image is shown for PBMC-derived mAbs (mAb 7) and CSF-derived mAbs (mAb 4). An anti-hemagglutinin (anti-HA) antibody in the same IgG1 backbone was used as a negative control. Scale bars, 500 μm.
(E) Select regions of immunostaining from mAbs 1–4. (i) mAb 1 immunostaining of cerebellar Purkinje cells (arrow) and the overlying molecular layer. (ii) mAb C2 immunostaining of cortical neuropil and occasional staining of neuron-like somata (arrow). (iii) mAb C3 immunostaining of large cells within the hilus of the hippocampus. (iv) mAb C4 immunostaining of mitral-like cells of the olfactory bulb (arrow). (v) mAb C4 immunostaining of pyramidal neurons (arrow) in CA3 of the hippocampus. (vi) mAb C4 immunostaining of neuronal cell bodies in layer II of the cortex (arrow). Scale bars, 10 μm.
We found that one of five CSF-derived (monoclonal antibody [mAb] C2) and two of four peripherally derived mAbs (mAbs P1 and P2) targeted the SARS-CoV-2 spike protein (Figure 5C). None of the other mAbs recognized other SARS-CoV-2 antigens in the Luminex panel. Using biolayer interferometry, we determined that all three anti-spike mAbs derived from individuals with COVID-19 bound 2P-stabilized spike protomers with high affinity (KD[mAb C2] = 2 nM, KD[mAb P1] = 0.2 nM, KD[mAb P2] = 2 nM) (Data S3). Because these binding affinities are similar to those of SARS-CoV-2-neutralizing antibodies, we tested each anti-spike mAb for neutralizing activity against wild-type SARS-CoV-2.19 None of the mAbs exhibited neutralizing activity at concentrations ranging from 2.5–25μg /mL (Figure S6).
Given reports of new-onset humoral autoimmunity in COVID-19, we wondered whether any of the mAbs from CSF-expanded B cells were autoreactive to neural tissue. Therefore, we tested all mAbs using a standard and validated screening method for antineural autoreactivity: anatomic mouse brain tissue staining.20 mAbs were used as a primary antibodies to immunostain mouse brain tissue and labeled with an anti-human IgG secondary antibody. An anti-influenza antibody targeting the hemagglutinin antigen (anti-HA) was used as a negative control. Similar to anti-HA, none of the PBMC-derived mAbs recognized mouse brain tissue. In contrast, four of five CSF-derived mAbs exhibited some degree of antineural immunoreactivity, including the anti-spike mAb (mAb C2) (Figures 5D and 5E). Notably, mAb C2 produced a neuropil-predominant immunostaining pattern, suggesting that the antigen may be enriched in neuronal process or harbor an extracellular epitope.
Intrathecal humoral autoimmunity in individuals with COVID-19 with neurological symptoms
The emergence of inflammatory and humoral autoimmune disorders of the nervous system during the para- or post-infectious period in COVID-19 is increasingly recognized and includes acute disseminated encephalomyelitis (ADEM), autoimmune encephalitis associated with known autoantibodies, transverse myelitis, and Guillain-Barré syndrome and one of its variants, Miller Fisher syndrome.21, 22, 23, 24, 25 Given this literature and the autoreactivity of CSF-derived monoclonal antibodies from case 1, we hypothesized that our other individuals with COVID-19 might harbor intrathecal autoantibodies. To test this, we screened our cohort of individuals with COVID-19 for intrathecal antineural antibodies using a suite of complementary autoantigen detection platforms: anatomic mouse brain tissue immunostaining, immunoprecipitation-mass spectrometry (IP-MS), and pan-human proteome phage display IP sequencing (PhIP-seq).26, 27, 28 In these screens, we included one additional individual with post-COVID-19 seizures and cognitive impairment who had been recruited after completion of the transcriptomics and cytokine analyses were completed (case 7; Table S1; Data S1). Like the other six individuals with acute COVID-19, this subject had SARS-CoV-2 antibodies in his CSF (Figure S7).
Anatomic mouse brain immunostaining demonstrates the presence of intrathecal antineural autoantibodies in most individuals with COVID-19
More COVID-19 CSF samples (5 of 7) were immunoreactive to mouse brain tissue at 1:10 dilution than control CSF (2 of 6) (Figures 5A and S8A). Control CSF staining was not specific to any anatomic region, weakly pan-nuclear, or primarily subpial (Figure S8B). None of the control CSF samples were immunoreactive beyond 1:10 dilution, indicating the absence of high-titer or high-affinity antineural autoantibodies. In contrast, at 1:10 dilution, COVID-19 CSF produced immunoreactive staining of specific anatomic regions, including cortical neurons (n = 4), the olfactory bulb (n = 3), the thalamus (n = 3), the CA3 field of the hippocampus (n = 3), the cerebellum (n = 3), the brain stem (n = 4), and cerebral vasculature (n = 2) (Figures 5B, 5C, and S8A). Four and three COVID-19 CSF samples showed continuing immunoreactivity at 1:25 and 1:50 dilution, respectively. The data indicate that an unexpectedly high proportion of CSF samples from individuals with COVID-19 with neurological impairment harbor high titers of antineural autoantibodies of unknown pathogenic significance.
IP-MS identifies intrathecal candidate autoantigens in a subset of individuals with COVID-19
To screen for the neural protein targets of intrathecal autoantibodies, we immunoprecipitated mouse whole-brain lysate using CSF and plasma, trypsinized precipitated proteins, and analyzed the resulting peptides by MS. IPs was performed in technical replicates by different individuals using different mice as input. First we searched the resulting spectra against the SARS-CoV-2 proteome (Uniprot SARS-CoV-2 reference proteome, June 2020). SARS-CoV-2 proteins were not detected. We then searched for human proteins. Consistent with circulating protein expression patterns29,30 and circulating prothrombotic autoantibodies31 in COVID-19, complement, coagulation, and platelet degranulation pathway proteins of human origin were significantly overrepresented in the IgG-bound protein fraction of plasma from our individuals (Figure S9).
To specifically identify candidate antineural autoantibodies in the CSF, we searched for mouse proteins that were observed in both technical replicates and significantly enriched by spectral counting and/or 1.5× enriched by MS1 peak area in COVID-19 CSF IP relative to controls.32 Between 5 and 56 (median = 20) proteins were enriched by the CSF, but not by the plasma, of the same case or control CSF, indicating that they were unique to the CSF compartment of individuals with COVID-19. Gene Ontology pathway analysis indicated that COVID-19 CSF and plasma IP-MS fractions were enriched for brain-enriched and synaptic proteins. In some cases, neural antigens were statistically enriched by CSF but not plasma (e.g., NEFM and NEFH by cases 3 and 4 and APP by case 4).
PhIP-seq identifies intrathecal candidate autoantigens in a subset of individuals with COVID-19
COVID-19 CSF was also screened for autoantibodies using a previously described PhIP-seq platform (T7 bacteriophage display) displaying ∼730,000 overlapping 49-amino-acid peptides spanning all human proteins,26 including all known and predicted isoforms.27,28 To identify peptides that were significantly enriched by COVID-19 CSF compared with controls, we first established an empirical enrichment threshold using a validated commercial antibody targeting the protein GFAP (Data S4). For COVID-19 CSF samples, peptides with suprathreshold enrichment in both technical replicates were considered candidate autoantigens. COVID-19 CSF enriched between 2 and 40 CSF-specific proteins (median = 18; Data S4). By Gene Ontology, synaptic proteins were also enriched by COVID-19 CSF (e.g., NRG3, SYNJ2, and DPYSL2; Bonferroni-corrected p = 1.6 × 10−1). COVID-19 plasma PhIP-seq candidates were enriched for transcriptional activators of catabolism (Bonferroni-corrected p = 2.6 × 10−2) but not synaptic or brain-enriched proteins. COVID-19 CSF that was immunoreactive to mouse brain tissue at a 1:50 dilution was associated with greater enrichment of candidate autoantigens by PhIP-seq (19–40, median = 37) than CSF that immunostained at a lower dilution or was not immunoreactive (1–16, median = 6), suggesting a correlation between immunostaining status and the burden of CSF autoantibodies. In some instances, the same candidate autoantigen was detected in individuals with COVID-19 by IP-MS and PhIP-seq: UHRF1BP1 (case 2), NUAK1 (case 3), and DBN1 (case 7).
Validation of two candidate autoantigens in the CSF of individuals with COVID-19
To validate autoantibodies identified in CSF, we selected two COVID-19 candidate autoantigens that were enriched more by CSF than plasma on PhIP-seq: intraflagellar transport protein 88 homolog (IFT88) and THAP domain-containing 3 (THAP3). To validate IFT88, HEK293T cells were transfected with a plasmid encoding RFP-IFT88, methanol fixed, and immunostained with CSF from case 1. Case 1 CSF IgG, but not control CSF IgG, colocalized with an anti-RFP and an anti-IFT88 commercial antibody (Figures 6E and S10). To validate THAP3, lysate from HEK293T cells overexpressing FLAG-tagged THAP3 was separated by SDS-PAGE, transferred to a polyvinylidene fluoride (PVDF) membrane, and sequentially probed with case 3 CSF (1:60 dilution) and a commercial anti-FLAG antibody. CSF IgG and anti-FLAG recognized the same band of ∼25 kDa in the THAP3 overexpression lane but not in the untransfected lane (Figure 6F). Therefore, anti-IFT88 and anti-THAP3 are bona fide autoantibodies in the CSF of COVID-19 cases 1 and 3, respectively (Figures 6E, 6F, and S10).
Figure 6.
Autoantibodies in the CSF of individuals with COVID-19
(A) Sagittal mouse brain sections were immunostained with CSF at 1:10 dilution (green) and the nuclear stain DAPI (blue). Anti IgG secondary only antibody negative control (left) and 4/6 control CSF (example CTRL 6, center) were not immunoreactive. In contrast, 5 of 7 COVID-19 CSF samples were immunoreactive (example CASE 3, right). Scale bars, 100 μm. CTRL, control.
(B) Binary matrix indicating anatomic immunoreactivity of COVID-19 CSF at 1:10 dilution.
(C) Select examples of COVID-19 CSF anatomic immunostaining of the hippocampus (n = 3; arrows, CA3; left column; scale bar, 100 μm), cerebrovasculature (top panel, second column arrow indicates endothelial staining [scale bar, 50 μm]; bottom panel arrow indicates a perivascular cell [scale bar, 10 μm]), olfactory bulb (n = 3, two shown; third column, top panel shows neuron-like cells; bottom panel, mitral cells; scale bars, 10 μM), and cortical neuron-like cells (n = 4, two cases shown; fourth column; scale bars, 10 μM).
(D) Heatmaps of sequence-sharing peptides mapping to IFT88 (case 1, top) and THAP3 (case 3, bottom) that were enriched by CSF, shown with their corresponding enrichment by plasma. Rows, individual peptides; left two columns, technical replicates for case 1 (top) and case 3 (bottom). For COVID-19 and control columns, cell values represent the mean of log10(fold change enrichment) of technical replicates. For case 1 and case 3, candidate IFT88 and THAP3 peptides, respectively, were enriched more by CSF than plasma.
(E) The HEK293 overexpression cell-based assay was performed in technical replicates. A representative example demonstrates that case 1 CSF is immunoreactive to overexpressed RFP-IFT88 (CSF, green; anti-RFP, red; anti-IFT88 antibody, magenta). Scale bars, 10 μM.
(F) Western blot validation of anti-THAP3 autoantibodies in CSF of case 3. CSF IgG (green) and anti-FLAG (red) recognize the same ∼25-kDa band in THAP3-overexpressing (OE) lysate but not untransfected (UN) lysate (arrow).
Discussion
Neurotropic viruses, such as herpes simplex virus, West Nile virus, and HIV, are common pathogens of the CNS, but it is increasingly recognized that respiratory viruses, including influenza virus and human respiratory syncytial virus, also lead to neurological complications in a minority of individuals.33, 34, 35, 36, 37, 38, 39 The biological underpinnings of neurological complications of respiratory viruses are diverse and include neuronal damage because of direct viral neuroinvasion as well as parainfectious processes, including elevation of pro-inflammatory cytokines, and post-viral autoimmune reactions In this exploratory study, we performed an extensive set of immunologic investigations to assess CNS-specific immune responses in a series of individuals with COVID-19 with neurological symptoms. Although systemic multi-organ dysfunction is almost certainly a driver of neurological complications in a proportion of individuals with COVID-19, here we identified innate and adaptive antiviral immune responses as well as humoral autoimmunity, which appears to be unique to the CNS and may therefore contribute to COVID-19 neuropathology.
CSF, although not identical to brain, is produced by the choroid plexus and bathes the CNS. It is the only CNS tissue surrogate that can be sampled readily in living humans. Analysis of CSF immune cells has shed light on immune mechanisms of neuronal injury during other infections, including HIV, neurosyphilis, and neuroborreliosis.40, 41, 42 By assessing CSF and blood in individuals with acute COVID-19 and neurological symptoms, we find evidence of a compartmentalized CNS immune response to SARS-CoV-2. Through transcriptional and cytokine analyses, we find an increase in CSF but not plasma IL-12 and IL-1b, factors that are central for coordinating innate and adaptive immune responses to invading pathogens. Notably, neuroinvasion of mouse hepatitis virus, a coronavirus of laboratory mice, also leads to IL-12 production by astrocytes and microglia.43
Our data identified increased and divergent humoral responses in the CNS. This humoral response included indirect evidence of neuroinvasion of SARS-CoV-2 through the presence of antiviral antibodies in the CNS during acute SARS-CoV-2 infection. Using an animal model, we show that SARS-CoV-2 infection in the CNS stimulates production of intrathecal antibodies and that an isolated systemic infection is not sufficient to do so. These data suggest that the presence of anti-SARS-CoV-2 antibodies in the CSF of individuals with COVID-19 may similarly reflect viral infection of the CNS. Further supporting this contention, we generated a mAb from the CSF of an individual with COVID-19 (case 1) with marked B cell clonal expansion in the CSF that was specific for the SARS-CoV-2 spike protein. This B cell clone was not detected in the peripheral blood of the same individual. Notably, the other CSF-derived mAbs were variably immunoreactive to mouse brain tissue, which motivated and expanded our search for autoantibodies in the CSF of our other individuals with COVID-19.
Our extended studies of the CNS humoral response suggest that a subset of individuals with COVID-19 with neurological symptoms has an elevated burden of autoreactive antibodies in the CSF. As seen by anatomic immunostaining, an unexpectedly high proportion of our individuals with COVID-19 harbored intrathecal autoantibodies, including antineural autoantibodies. Hippocampal immunostaining from three individuals was primarily restricted to the CA3 region, similar to a recent report of SARS-CoV-2-associated encephalitis.44 Other immunoreactive anatomic regions included the olfactory bulb in three individuals and cerebrovasculature in two—anatomic regions with prima facie relevance to common neurologic sequelae of COVID-19 (i.e., anosmia and stroke). Subsequent unbiased protein and peptide screens of CSF identified a diversity of candidate autoantibodies, and two of these autoantigens, THAP3 and IFT88, were subsequently validated. IFT88 is a ciliary protein whose mutation causes a ciliopathy in humans and anosmia in mice.45 THAP3 is expressed in the brain, among other organs, and may be implicated in genetic causes of dystonia.46
However, the mere presence of an intrathecal autoantibody does not mean that a person has autoimmune encephalitis. Indeed, the individuals in our exploratory cohort lacked evidence of active inflammation upon neuroimaging and/or did not have elevated conventional CSF markers of neuroinflammation (i.e., white blood cell count, IgG index, and CSF-restricted oligoclonal bands) that are typically, but not always, found in people with autoimmune encephalitis. Notably, individuals with COVID-19 with neurological symptoms appear to have immune responses to multiple autoantigens, implying that the increased compartmental humoral immune response may reflect a broader immune activation syndrome. This is particularly true given that humoral autoimmunity has been observed to target other organ systems in COVID-19 and may also contribute to neuropathology during COVID-19.47, 48, 49, 50
Our exploratory data suggest that, even in individuals with COVID-19 with neurologic symptoms who lack overt evidence of neuroinflammation on MRI or conventional CSF testing, there is a compartmentalized immune response involving the innate and adaptive arms of the immune system. Future research involving careful clinical phenotyping and timely investigations of the CSF will help place these findings into a broader clinical context and inform whether antiviral and/or immunomodulatory therapies might be indicated for carefully selected, neurologically impaired individuals with COVID-19.
Limitations of study
Our study is complicated by the diverse range of neurological symptoms in our study participants. A comparison group with a different systemic viral infection would help determine which aspects of the findings are specific to COVID-19. Moreover, our study did not include control CSF from individuals with COVID-19 without neurological symptoms because these people do not undergo clinical LP and did not consent to research-only LP. Thus we do not know whether auto-reactive antibodies in the CNS are present in individuals with COVID-19 who do not experience neurological symptoms.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-IFT88 | Proteintech | Cat# 13967-1-AP, RRID:AB_2121979 |
| Anti-FLAG | Cell Signaling Technology | Cat# 14793, RRID:AB_2572291 |
| Anti-FLAG | Cell Signaling Technology | Cat# 2368, RRID:AB_2217020 |
| Anti-RFP | Abcam | Cat# ab65856, RRID:AB_1141717 |
| Anti-GFAP | Agilent | Cat# # Z033429-2 |
| Anti-SARS-CoV nucleocapsid protein | Sino Biological | Cat #40143-R001, RRID:AB_2827974 |
| Goat-anti-mouse 488 | Invitrogen | Cat# A-11001, RRID:AB_2534069 |
| Goat-anti-rabbit 488 | Invitrogen | Cat# A-11008, RRID:AB_143165 |
| Donkey-anti-human 488 | Jackson ImmunoResearch | Cat# 709-545-149, RRID:AB_2340566 |
| Donkey-anti-mouse 594 | Jackson ImmunoResearch | Cat# 715-585-151, RRID:AB_2340855 |
| Donkey-anti-rabbit 594 | Jackson ImmunoResearch | Cat# 711-585-152, RRID:AB_2340621 |
| Donkey-anti-human 594 | Jackson ImmunoResearch | Cat# 709-585-149, RRID:AB_2340572 |
| Donkey-anti-mouse Cy5 | Jackson ImmunoResearch | Cat# 715-175-151, RRID:AB_2340820 |
| Donkey-anti-rabbit Cy5 | Jackson ImmunoResearch | Cat# 711-175-152, RRID:AB_2340607 |
| Goat-anti-human IRDye 800CW | Li-Cor | Cat# 926-32232, RRID:AB_10806644 |
| Goat-anti-rabbit IRDye 680RD | Li-Cor | Cat# 926-68071, RRID:AB_10956166 |
| Mouse IgG-specific secondary antibody | BioLegend | Cat# 405306 |
| Bacterial and virus strains | ||
| SARS-CoV-2 isolate USA-WA1/2020 | N/A | RRID: N/A |
| SARS-CoV-2/human/USA/CA-UCSF-0001H/2020 | N/A | RRID: N/A |
| hACE2-AAV9 | Vector biolabs | RRID: N/A |
| Biological samples | ||
| 96-well MaxiSorp plates | Thermo Scientific | #442404 |
| Chemicals, peptides, and recombinant proteins | ||
| ProLong Gold | ThermoFisher Scientific | Cat# P10144 RRID: N/A |
| HEK293 THAP3 overexpression lysate | Novus Biologicals | Cat# NBP2-05659 RRID: N/A |
| Histopaque | Sigma-Aldrich | Cat#10771-500ML |
| SARS Cov-2 S1 protein | ACROBiosystems | #S1N-C52H3-100ug |
| Critical commercial assays | ||
| CDC RT-qPCR assay for N1 N2 human RNase P | CDC | N/A |
| NEB Luna Universal Probe One-Step RT-qPCR kit | NEB | N/A |
| 10x Chromium single-cell 5¢ Chip (V3 technology) | 10x Genomics | N/A |
| HD71 Human Cytokine Array/Chemokine Array | Eve Technologies, Calgary | N/A |
| Deposited data | ||
| Raw mass spectrometry files and data analysis output | MassIVE and Proteome Exchange | MassIVE identifier: MSV000087115 Proteome Exchange identifier: PXD025035 RRIDs: N/A |
| Experimental models: Cell lines | ||
| HEK293T/17 cells | UCSF Cell and Genome Engineering Core | Cat# CCLZR076 RRID: N/A |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | The Jackson Laboratory | Cat# 000664 RRID:IMSR_JAX:000664 |
| FVB/NJ mice | The Jackson Laboratory | Cat# 001800 RRID:IMSR_JAX:001800 |
| Recombinant DNA | ||
| pEF5-FRT-TagRFP-T-IFT88 | Addgene | Cat# 61684, RRID:Addgene_61684 |
| AbVec2.0-IGHG1 | Addgene | Cat# 80795, RRID:Addgene_80795 |
| AbVec1.1-IGKC | Addgene | Cat# 80796, RRID:Addgene_80796 |
| AbVec1.1-IGLC2-XhoI | Addgene | Cat# 99575, RRID:Addgene_99575 |
| Software and algorithms | ||
| ImageJ | https://imagej.net/ | Version 2.1.0/1.53c, Build:5f23140693 RRID:SCR_003070 |
| R | https://cran.r-project.org/ | Version 3.6.3 RRID:SCR_001905 |
| Zen 3.2 (blue edition) | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html; RRID:SCR_013672 |
| Image Studio Lite | Li-Cor | https://www.licor.com/bio/image-studio-lite/download; RRID: RRID:SCR_013715 |
| Quandenser | Open Source | https://github.com/statisticalbiotechnology/quandenser-pipeline; RRID: N/A |
| RAPSearch | Open Source | https://github.com/zhaoyanswill/RAPSearch2; RRID: N/A |
| Seurat v3.0 | Open Source | https://satijalab.org/seurat/; RRID: N/A |
| CellPhoneDB v2 | Open Source | https://www.cellphonedb.org/; RRID: N/A |
| Interferome | Open Source | http://www.interferome.org/; RRID: N/A |
| UpSetR | Open Source |
http://gehlenborglab.org/research/projects/upsetr/#:∼:text=UpSetR%20is%20an%20R%20package,based%20on%20groupings%20and%20queries; RRID: N/A |
| Change-O v0.3.4 | Open Source | N/A |
| Qlucore Omics Software, version 3.6 | Open Source | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shelli F. Farhadian (Shelli.Farhadian@yale.edu).
Materials availability
Immunoglobulin sequencing data for patient-derived monoclonal antibodies are available in Data S5.
Data and code availability
Gene expression and repertoire data in the study are available in the NCBI repository SRA PRJNA717310. Raw mass spectrometry files and data analysis output are available from MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) and Proteome Exchange (http://www.proteomexchange.org/) under MassIVE dataset identifier # MSV000087099 and Proteome Exchange # PXD025016.
Experimental model and subject details
Human Subjects
COVID-19 patients admitted to Yale New Haven Hospital in March-May 2020 were recruited to the IRB approved Yale IMPACT study (Implementing Medical and Public Health Action Against Coronavirus CT; HIC#200027690). COVID-19 patients who were undergoing clinical lumbar puncture for evaluation of neurological symptoms were included. Negative controls were recruited from the surrounding community via IMPACT and the Yale University IRB approved HARC study (HIC# 1502015318). Healthy control participants recruited after March 2020 were confirmed to be negative for SARS-CoV-2 infection by nasopharyngeal swab PCR. All participants or their designated surrogate consented to donate 25cc of CSF for research studies. Blood was collected within one hour of lumbar puncture. For autoantibody characterization assays, additional pre-pandemic healthy volunteers (CTRLS 7 - 12; Table S2) were recruited from the general population via word of mouth, newspaper and internet advertisements, and poster flyers. Based on a questionnaire, they were excluded if they had an Axis I psychiatric diagnosis, a first-degree relative with a known or suspected Axis I psychiatric disorder, or any neurologic disorder. CSF samples were collected at The Zucker Hillside Hospital, Glen Oaks, NY.
Animals
Mice for SARS-CoV-2 model
Six to twelve-week-old mixed sex C57BL/6 (B6J) purchased from Jackson laboratories were subsequently bred and housed at Yale University. All procedures used in this study (sex-matched, age-matched) complied with federal guidelines and the institutional policies of the Yale School of Medicine Animal Care and Use Committee.
Mice for anatomic immunostaining
Postnatal day 40 – 60 mice from the F1 cross of FVB (Jackson Laboratory, Cat. No. #001800) x C57BL/6J (The Jackson Laboratory, Cat. No. #000664) mice were used for anatomic immunostaining. All procedures used in this study complied with federal guidelines and the institutional policies of the University of California San Francisco Institutional Animal Care and Use Committee.
SARS-CoV-2
SARS-CoV-2 for mouse model
SARS-CoV-2 isolate USA-WA1/2020
SARS-CoV-2 for neutralization assays
SARS-CoV-2/human/USA/CA-UCSF-0001H/2020
Cell Lines
HEK293T/17 cells (293 cells) were obtained from the UCSF Cell and Genome Engineering Core (Cat. No. #CCLZR076). 293 cells were cultured in DMEM (DME H-21, UCSF Cell Culture Facility, Cat. No #CCFAA005) + 10% FBS (Fisher Scientific, Cat. No. MT35010CV) without antibiotics. Cells were maintained at 37°C in 5% CO2 and split 1:5 every 3 – 5 days based on having reached 80 – 100% confluence by visual inspection. Cells were not passaged more than 50 times. For overexpression cell-based assays, 0.5mL of 293 cells at 200,000 cells / mL were added to each well of a 24-well plate the day prior to transfection.
Method details
SARS-CoV-2 RT-qPCR
RNA was extracted from nasopharyngeal swabs, CSF, and plasma using the MagMax Viral/Pathogen Nucleic Acid Isolation kit. A modified CDC RT-qPCR assay was used to detect SARS-CoV-2 with the N1, N2, and human RNase P (RP) primer-probe sets and the NEB Luna Universal Probe One-Step RT-qPCR kit on the Bio-Rad CFX96 Touch Real-Time PCR Detection System.9
PMBC and CSF cell preparation
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized whole blood after 1:1 PBS dilution. The blood was layered over a Histopaque (Sigma-Aldrich, #10771-500ML) gradient in a SepMate tube (StemCell Technologies, Inc.; #85460) and isolated according to manufacturer’s instructions. The PBMCs were then aliquoted and stored at −80 for subsequent analysis. CSF was centrifuged at 400G for 8 minutes, with cell-free supernatant removed for cytokine and antibody assays, and cell pellet processed for single cell RNA sequencing, as below.
Single cell RNA sequencing
Approximately 8,000 single cells from CSF and from PBMC from each participants were loaded into each channel of a Chromium single-cell 5′ Chip (V3 technology). 5′ 10X libraries were sequenced on Illumina Novaseq at approximately 50,000 reads per cell. Raw sequencing reads were aligned to the human GRCh38 genome and gene counts were quantified as UMIs using Cell Ranger count v3.0 (10x Genomics). We removed cells with > 10% mitochondrial RNA content, and included cells with > 500 and < 2000 genes expressed per cell. Dimensionality reduction, clustering, and visualization was performed using Seurat. Clusters were identified based on expression of canonical immune cell markers (heatmap Figure S1a). Interferon regulated genes were identified using Interferome.10 Differential expression analysis was performed in Seurat v351 using the two-tailed Wilcoxon test, comparing cells from COVID-19 patients versus healthy controls. Significant differentially expressed genes (DEGs) were defined as: adjusted p < 0.05 and |log fold change| ³ 0.25. Gene ontology and pathway enrichment analysis was performed using DAVID.52 DEGs in PBMC and CSF samples were compared using the UpSetR package.
To identify potential intercellular interactions between different cell types in the scRNA-seq data, we utilized CellPhoneDB v2.11 Normalized count matrices and associated cell type labels were provided to CellPhoneDB and analyzed under both the statistical mode and the thresholding mode. Of note, since the statistical mode of CellPhoneDB seeks to assess the specificity of a given interaction, a lack of statistical significance does not necessarily mean a given interaction is not present. Therefore, when comparing the number of potential intercellular interactions in COVID-19 patients versus healthy controls, the simpler threshold-based analysis mode was used. In contrast, for pinpointing the top candidate cell-cell interactions in each dataset, the statistical analysis mode was used, with a significance threshold of p < 0.05.
T and B cell clustering
We initially combined all 76,473 CSF and blood cells and generated clusters using Seurat. For each cluster we assigned a cell-type label using statistical enrichment for sets of marker genes, and manual evaluation of gene expression for small sets of known marker genes. We then created a separate Seurat object consisting only of T cells clusters from the original analysis, and a separate Seurat object consisting only of plasma and B cells. We then re-clustered these T and B cells and annotated sub-clusters using previously annotated marker genes.
BCR analysis
Single cell V(D)J sequences were generated using CellRanger vdj function. Assignments of V(D)J sequences were performed using IgBLAST v.1.6.1 with the September 12, 2018 version of the IMGT gene database (as described previously).53,54 Non-functional V(D)J sequences were removed. Cells with multiple IGH V(D)J sequences were assigned to the most abundant V(D)J sequence by unique molecular identifier count (and based on numbers of sequenced reads in instances with ties). B cell clones in the CSF and circulation were identified using an approach described previously using hierarchical clustering implemented using the DefineClones.py function of Change-O v0.3.4 and a junctional sequence hamming dissimilarity threshold of 0.17.54 To account for the presence of light chains, heavy chain-based clones were corrected for using an approach described previously.55
Cytokine assays
Soluble chemokines and cytokines were assessed in CSF supernatant and paired plasma using the HD71 Human Cytokine Array/Chemokine Array (Eve Technologies, Calgary, AB). Statistical analysis was carried out using Qlucore Omics Software, version 3.6 (Lund, Sweden). Cytokines that were absent from CSF or plasma under both COVID and controls conditions were excluded from the respective analyses. Heatmaps were plotted using Z-scores, with the color scale set to range from −2 to +2. Hierarchical clustering was applied to samples.
AAV infection (Intratracheal and Intracisternal magna injection)
Adeno-associated virus 9 encoding hACE2 were purchased from Vector biolabs (AAV-CMV-hACE2).
Intratracheal injection
Animals were anaesthetized using a mixture of ketamine (50 mg kg−1) and xylazine (5 mg kg−1), injected intraperitoneally. The rostral neck was shaved and disinfected. A 5mm incision was made and the salivary glands were retracted, and trachea was visualized. Using a 500 μL insulin syringe a 50 μL bolus injection of 1011GC of AAV-CMV-hACE2 was injected into the trachea. The incision was closed with VetBond skin glue. Following intramuscular administration of analgesic (Meloxicam and buprenorphine, 1 mg kg−1), animals were placed in a heated cage until full recovery.
Intracisternal magna injection
Mice were anesthetized using ketamine and xylazine, and the dorsal neck was shaved and sterilized. A 2 cm incision was made at the base of the skull, and the dorsal neck muscles were separated using forceps. After visualization of the cisterna magna, a Hamilton syringe with a 15 degree 33 gauge needle was used to puncture the dura. 3 μL of AAV9 (3.1013 viral particles/mouse) or mRNA (4-5 μg) was administered per mouse at a rate of 1 μL min-1. Upon completion of the injection, needle was left in to prevent backflow for an additional 3 minutes. The skin was stapled, disinfected and same post-operative procedures as intratracheal injections were performed.
Generation of SARS-CoV-2 virus
To generate SARS-CoV-2 viral stocks, Huh7.5 cells were inoculated with SARS-CoV-2 isolate USA-WA1/2020 (BEI Resources #NR-52281) to generate a P1 stock. To generate a working stock, VeroE6 cells were infected at a MOI 0.01 for four days. Supernatant was clarified by centrifugation (450 g x 5min) and filtered through a 0.45 micron filter. To concentrate virus, one volume of cold (4°C) 4x PEG-it Virus Precipitation Solution (40% (w/v) PEG-8000 and 1.2M NaCl) was added to three volumes of virus-containing supernatant. The solution was mixed by inverting the tubes several times and then incubated at 4°C overnight. The precipitated virus was harvested by centrifugation at 1,500 x g for 60 minutes at 4°C. The pelleted virus was then resuspended in PBS then aliquoted for storage at −80°C. Virus titer was determined by plaque assay using Vero E6 cells.
SARS-CoV-2 infection (intranasal)
Mice were anesthetized using 30% v/v Isoflurane diluted in propylene glycol. Using a pipette, 50 μL of SARS-CoV-2 (3x107 PFU/ml) was delivered intranasally.
SARS-CoV-2 infection (intraventricular)
Animals were anaesthetized using a mixture of ketamine (50 mg kg−1) and xylazine (5 mg kg−1), injected intraperitoneally. After sterilization of the scalp with alcohol and betadine, a midline scalp incision was made to expose the coronal and sagittal sutures, and a burr holes were drilled 1 mm lateral to the sagittal suture and 0.5 mm posterior to the bregma. A 10 μl Hamilton syringe loaded with virus, and was inserted into the burr hole at a depth of 2 mm from the surface of the brain and left to equilibrate for 1 min before infusion. Once the infusion was finished, the syringe was left in place for another minute before removal of the syringe. Bone wax was used to fill the burr hole and skin was stapled and cleaned. Following intramuscular administration of analgesic (Meloxicam and buprenorphine, 1 mg kg−1), animals were placed in a heated cage until full recovery. For high condition, 5 μL of SARS-CoV-2 (3x107 PFU/ml) and for low condition 5 μL of SARS-CoV-2 (3x106 PFU/ml) was used.
Enzyme-linked immunosorbent assay
ELISAs were performed as previously reported.56 In short, Triton X-100 and RNase A were added to serum samples at final concentrations of 0.5% and 0.5mg/ml respectively and incubated at room temperature (RT) for 3 hours before use to reduce risk from any potential virus in serum. 96-well MaxiSorp plates (Thermo Scientific #442404) were coated with 50 μl/well of recombinant SARS Cov-2 S1 protein (ACROBiosystems #S1N-C52H3-100ug) at a concentration of 2 μg/ml in PBS and were incubated overnight at 4°C. The coating buffer was removed, and plates were incubated for 1h at RT with 200 μL of blocking solution (PBS with 0.1% Tween-20, 3% milk powder). Serum was diluted 1:50 in dilution solution (PBS with 0.1% Tween-20, 1% milk powder) and 100 μL of diluted serum was added for two hours at RT. Plates were washed three times with PBS-T (PBS with 0.1% Tween-20) and 50 μL of mouse IgG-specific secondary antibody (BioLegend #405306, 1:10,000) diluted in dilution solution added to each well. After 1h of incubation at RT, plates were washed three times with PBS-T. Samples were developed with 100 μL of TMB Substrate Reagent Set (BD Biosciences #555214) and the reaction was stopped after 15 min by the addition of 2 N sulfuric acid.
Statistical methods
Statistical analyses were performed using commercially available software (Prism or Excel) except as noted. Differences in means between two groups were analyzed using unpaired two-sided t tests, unless otherwise noted. For scRNA-seq analyses, we corrected for multiple comparisons and report adjusted P values using Benjamini–Hochberg correction. For pathway analyses, Fisher’s exact test was used with Bonferroni correction for multiple testing.
SARS-CoV-2 Serological Assay
Highly immunogenic linear regions of the SARS-CoV-2 proteome were isolated by ReScan and conjugated to Luminex beads as previously described.57 Briefly, high concentration T7 phage stocks displaying immunodominant epitopes of the S, N and ORF3a proteins were propagated and grown to high (> 1011 PFU/mL) titer then were each conjugated to unique bead IDs according to manufacturer’s Antibody Coupling Kit instructions (Luminex). Whole N protein (RayBiotech) beads were conjugated similarly using manufacturer instructions with 5μg of protein per 1 million beads. For other whole protein Luminex-based beads, MagPlex-Avidin Microspheres (Luminex) were coated with either the S protein RBD (residues 328-533) or the trimeric S protein ectodomain (residues 1-1213).
All beads were blocked overnight before use and pooled on day of use. 2000-2500 beads per ID were pooled per incubation with patient plasma at a final dilution of 1:500, patient CSF at a final dilution of 1:20, or 1 μg of human-derived monoclonal antibodies for 1 hour, washed, then stained with an anti-IgG pre-conjugated to phycoerythrin (Thermo Scientific, #12-4998-82) for 30 minutes at 1:2000. Primary incubations were done in PBST supplemented with 2% nonfat milk and secondary incubations were done in PBST. Beads were processed in 96 well format and analyzed on a Luminex LX 200 cytometer.
Median Fluorescence Intensity from each set of beads within each bead ID were retrieved directly from the LX200 and log transformed after normalizing to the mean signal across two intra-assay negative controls (glial fibrillary acidic protein (GFAP) and tubulin phage peptide conjugated beads).
Generation of Human Monoclonal Antibodies
Expression Vector Cloning
Select heavy and light chain variable region fragments, or framework 1 through 4 as defined by the IMGT human V gene database, were synthesized by Integrated DNA Technologies, Inc. Heavy, kappa, and lambda fragments were cloned into AbVec2.0-IGHG1, AbVec1.1-IGKC, and AbVec1.1-IGLC2-XhoI linearized expression vectors, respectively. All expression vectors were a gift from Hedda Wardeman.58 Cloning was performed in a total of 20 μl with 60ng of linearized vectors, 18 ng heavy or light chain fragment and 10 μL of Gibson assembly master mix (New England Biolabs, Cat# E2611). 5-alpha competent E. coli (New England Biolabs, Cat# c2987) were transfected at 42°C with 2 μL unpurified, assembled plasmid. Colonies were sequenced to confirm correct assembly, and then purified from 3mL of E. coli in LB Broth with 100 μg/mL ampicillin using QIAprep Spin columns (QIAGEN).
Monoclonal Antibody Production
Monoclonal antibodies (mAbs) were produced by Celltheon Co. by transfecting Chinese hamster ovarian cells with equal amounts of heavy chain and light chain plasmids. Culture supernatant was harvested and mAbs isolated by affinity purification.
Determination of Monoclonal Antibody Binding Affinity to Purified Spike Protein
The binding affinities of purified antibodies to recombinant SARS-CoV-2 spike protein59 were measured on an Octet QK system using Anti-Human Fc-Capture (AHC) biosensor tips (Sartorius). Purified monoclonal antibodies were diluted to 5 μg/mL in phosphate buffered saline containing 0.1% bovine serum albumin and 0.02% Tween-20 at pH 7.4 (PBSTB). Antibodies were loaded and analyzed according to the following protocol: (0) Tips pre-equilibrated in PBSTB for 10 min; (1) Equilibration in PBSTB for 60 s; (2) Antibody loaded on tips for 300 s; (3) Tips washed in PBSTB to reach baseline for 300 s; (4) Tips dipped in spike-containing wells to allow for spike association for 300 s; (5) Tips dipped in PBSTB to allow for dissociation for 1800 s. All steps were carried out at 30°C with shaking at 1000 rpm. Binding curves for each antibody were recorded at spike concentrations ranging from 0-316 nM. A single affinity value for each antibody was calculated using a 1:1 global fit binding model (Octet Data Analysis HT software) with all R2 values > 0.95.
SARS-CoV-2 Neutralization Assays
SARS-CoV-2/human/USA/CA-UCSF-0001H/2020 from a UCSF clinical specimen was isolated and titered by standard plaque assay as described previously.60 To assess neutralization capacity, monoclonal antibodies were incubated with SARS-CoV-2 for one hour at 37°C and virus/antibody dilutions were used to infect Vero E6 cells and Huh7.5.1 cells overexpressing ACE2-TMPRSS2 at an MOI of 1.61 19 hours post infection, cells were fixed in 4% paraformaldehyde and permeabilized in 0.2% Triton-X. Fixed cells were blocked in 5% BSA, stained with a primary anti-SARS-CoV nucleocapsid protein antibody and subsequently stained with a secondary goat anti-rabbit IgG conjugated to Alexa Fluor 488. Slides were mounted with Fluoromount (SouthernBiotech, Cat# 0100-01).
Anatomic Mouse Brain Tissue Staining
Postnatal day 40 – 60 mice (F1 generation of FVB x C57BL/6J cross) were transcardially perfused with 4% paraformaldehyde (PFA) and brains post-fixed in PFA overnight. After sucrose equilibration, brains were blocked in OCT and sectioned at 12 μm on a standard cryostat. For screening and determination of anti-neural autoreactivity, sections were permeabilized and blocked in PBS containing 10% lamb serum and 0.1% triton x-100. Sections were then incubated with patient-derived monoclonal antibodies (18 μg/mL) or CSF at 1:10, 1:25, and 1:50 overnight at 4C. In some cases, CSF that was immunoreactive at 1:10 was repeated at 1:4 for additional confocal imaging. Sections were rinsed at least 5x with PBS and counterstained with anti-human IgG (Alexafluor 488). Nuclei were stained with DAPI at 1:2000 and stained sections were coversliped with ProLong Gold. Studies were approved the UCSF IACUC committee.
Imaging
Panoramic images of immunostained sagittal mouse brain sections were captured at a 20x on a Zeiss Axio Scan.Z1. Confocal images of sagittal mouse brain sections and HEK293T cell-based assays were captured at 60X at the UCSF Nikon Imaging Center using a Nikon CSU-W1 spinning disk confocal microscope, equipped with an Andor Zyla sCMOS camera. Images of the SARS-CoV-2 neutralizing assay were collected on a Nikon Ti inverted fluorescence microscope.
Immunoprecipitation Mass Spectrometry
Sample preparation
For IP-MS, all sample handling through protein digestion was performed in a BSL2 biosafety hood. Technical replicates were performed on the same day, by different individuals, using different mice. Plasma samples were unbuffered. CSF samples were stored 1:1 in 40% glycerol storage buffer. Samples were thawed on ice. Immunoprecipitations were performed in unblocked, thin-walled 96-well Hard-Shell® PCR plates (Bio-Rad Cat. No. HSP9641). For immunoprecipitations using patient plasma as the source of IgG, 2 μL of plasma was diluted in 200 μL of 1x PBS (GIBCO Cat. No. 10010-023). For CSF immunoprecipitations, 75 – 200 μL of CSF (depending on the amount of remaining biospecimen) was added to individual wells. For IgG conjugation, magnetic protein A/G beads were washed once in 1x PBS and suspended in an equal volume of PBS. To each well, 10 μL of washed protein A/G bead slurry was added and plates were placed on a rocker at 4C for one hour followed by incubation on a shaker at room temperature for one hour.
Postnatal day 40 – 60 mice (F1 generation of FVB x C57BL/6J cross) were used as the source of antigen. Mice anesthetized in isofluorane and sacrificed by cervical dislocation. For each set, 3 brains (two males and one female for one replicate and two females and one male for the other replicate) were rapidly dissected in ice cold PBS. For each replicate, 3 brains were homogenized in ice cold tissue lysis buffer (7 mL) using a dounce homogenizer (approximately 20 strokes). Homogenized brain lysate was transferred to 1.5 mL microcentrifuge tubes and centrifuged at 4°C for 10 minutes at 10,000 rcf. The supernatant from each set of brains was pooled yielding two separately prepared stocks of brain lysate. After BCA protein concentration determination, brain lysate stocks were diluted to 5 ug / uL in lysis buffer.
After IgG conjugation, the IP plate was placed on a magnetic plate, and the supernatant aspirated and discarded into 10% bleach. To each well, 200 μL of brain lysate (5 μg / μL) was added. Plates were sealed with adhesive aluminumized plate covers (Bio-Rad, Microseal® ‘F’ PCR Plate Seal, foil, pierceable Cat. No. #MSF1001). Antibody-bead-lysate complexes were incubated for 1 hour at room temperature under constant gentle agitation.
After 1 hour, IP plates were placed on magnetic plates and the lysate was aspirated and discarded into 10% bleach. Beads and their respective immune complexes were washed twice with 180 μL of detergent wash buffer, then once in high salt wash buffer, then once in nondetergent wash buffer, and finally once with ammonium bicarbonate buffer. For each well, washed beads were then resuspended in 35 μL of ammonium bicarbonate buffer to which 1 μL sequencing grade porcine trypsin was added (Promega, Cat. No. V5111). Immune complexes were digested on-bead for 1 hr at 37°C. After digestion, IP plates were placed on magnetic plates and the digestion reaction containing trypsinized peptides was transferred to a protein LoBind Eppendorf tube (Eppendorf, Cat. No. 022431081) and stored at −80°C until liquid chromatography (LC) was performed.
Mass Spectrometry
LC separation was done on a Dionex nano Ultimate 3000 (Thermo Scientific) with a Thermo Easy-Spray source . The digested peptides were reconstituted in 2% acetonitrile /0.1% trifluoroacetic acid and 1 μg in 5 μl of each sample was loaded onto a PepMap 100Å 3U 75 um x 20 mm reverse phase trap where they were desalted online before being separated on a 100 Å 2U 50 micron x 150 mm PepMap EasySpray reverse phase column. Peptides were eluted using a 60 minute gradient of 0.1% formic acid and 80% acetonitrile with a flow rate of 200nL/min. The separation gradient was ran with 2% to 5% acetonitrile over 1 minutes, 5% to 10% acetonitrile over 7 minutes, 10% to 55% acetonitrile over for 43 minutes, 55% acetonitrile to 99% acetonitrile over 1 minutes, a 4 minute hold at 99% acetonitrile, and finally 99% acetonitrile to 2% acetonitrile held at 2% acetonitrile for 10 minutes. Mass spectra were collected on a Fusion Lumos mass spectrometer (Thermo Fisher Scientific) in a data-dependent top speed mode with one MS precursor scan followed by MS/MS spectra for 3 s. A dynamic exclusion of 60 s was used. MS spectra were acquired with an isolation window of 1.2 Da, a resolution of 60,000 and a target of 4 × 105 ions or a maximum injection time of 50ms. MS/MS spectra were acquired with a resolution of 15K and a target of 1 × 104 ions or a maximum injection time of 35ms with maximum parallelizable time turned on. Peptide fragmentation was performed using collisionally induced dissociation with a normalized collision energy value of 30. Unassigned charge states as well as +1 and ions > +5 were excluded from MS/MS fragmentation.
Phage Display Immunoprecipitation Sequencing (PhIP-Seq)
The design of our human proteome phage display library26 containing 731,724 peptides of 49 amino acids has been previously described.28 Each peptide overlaps with its N-terminal by 25 amino acids. Preparation and titering phage libraries from stocks was as previously described.62 For our studies, phage libraries were incubated with 1 μL of patient biofluid (CSF or serum, both diluted 1:1 in 40% glycerol storage buffer) or 0.06μg of anti-GFAP antibody overnight at 4°C. IgG and bound phage were then isolated with protein A/G beads and phage were expanded by inoculating E. coli BLT5403 with bead-bound phage as previously described.62 E. coli lysate from this inoculation was used as the input phage library for a second round of immunoprecipitation using patient biofluids or anti-GFAP. Enrichment of phage DNA, and barcoding of individual IP reactions was performed using a single nested PCR reaction using panOME and multiplexing primers as previously described.62 PCR products were pooled and bead cleaned (SPRISelect, Beckman Coulter). Resultant libraries were sequenced on the NovaSeq 6000 or the iSeq (Illumina) using 150nt paired-end reads with a 20% PhiX spike in.
HEK293T/17 Cell-Based Assay Autoantigen Screening
HEK293T cells were plated onto 10mm poly-d-lysine coated (50μg/mL) coverslips in 24-well plates. 293 cells were transfected overnight with pEF5-FRT-TagRFP-T-IFT88 using Lipofectamine 3000 (ThermoFisher, Cat# L3000001). The following day, after two rinses with ice cold 1X PBS, RFP-IFT88 transfected cells were fixed with ice-cold methanol for 10 minutes. The fixed cells were rinsed with PBS, blocked with 5% lamb serum in PBS (blocking buffer), and permeabilized for 30 minutes using with blocking buffer containing 0.5% Triton. RFP-IFT88 HEK293T overexpressing cells were stained overnight using mouse anti-RFP at 1:100, rabbit anti-IFT88 at 1:100, and undiluted CSF. The cells were rinsed with PBS four times, and stained with anti-human 488, anti-mouse 594, and anti-rabbit Cy5 each at a 1:1,000 dilution in 5% blocking buffer. Nuclei were stained with DAPI at 1:2,000 in PBS for 5 minutes. Stained slides were then mounted onto microscope slides with Prolong Gold antifade.
Western Blotting
Untransfected control HEK293 cell lysate (30 μg) and HEK293 cell lysate overexpressing recombinant human C-MYC/FLAG-tagged THAP3 (30 μg)(Novus, #NBP2-05659) were separated by 4 – 12% BIS-Tris PAGE (Bio-Rad, #3450123). Chameleon Duo (7.5 μL) was used as the protein ladder (LI-COR, #928-60000). Proteins were separated for 1 hour at 180 V at room temperature. The protein transfer chamber was placed in an ice bucket, filled with 4°C transfer buffer (15% methanol and 85% 1x Tris/Glycine transfer buffer, Bio-Rad, #1610734), and proteins were transferred to Immun-Blot Low Fluorescence PVDF membrane at 100 V for 1 hour (Bio-Rad, #1620264). Membranes were blocked in undiluted blocking buffer (LI-COR, #927-70001) for 30 minutes at room temperature. The PVDF membrane was then incubated in CSF from Case 3 (1:60 in 1:1 LI-COR blocking buffer:TBST) overnight at room temperature. Membranes were rinsed five times in TBST and probed with goat-anti-human IRDye 800CW at 1:5,000 at room temperature for 1 hour. Membrane was rinsed five times in TBST and imaged on a LI-COR Odyssey. The same membrane was then probed with rabbit anti-FLAG antibody at 1:1,000 (Cell Signal, #2368) for 1 hour at room temperature, rinsed five times in TBST, probed with goat-anti-rabbit IgG at for 1 hour at room temperature, rinsed five times in TBST and reimaged on a LI-COR Odyssey.
Quantification and statistical analysis
Analysis of Mass Spectral Data
Spectral Counting
Tandem mass spectra were extracted by Proteome Explorer v1.4 (Thermo Scientific). Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version X! Tandem Alanine (2017.2.1.4)). X! Tandem was set up to search the Uniprot Mouse reference proteome (version 01/2020, 258832 entries) and Uniprot SARS-CoV-2 Database (version 06/2020, 262 entries) assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 20 PPM. Glu- > pyro-Glu of the n-terminus, ammonia-loss of the n-terminus, gln- > pyro-Glu of the n-terminus, deamidated of asparagine and glutamine, oxidation of methionine and tryptophan and dioxidation of methionine and tryptophan were specified in X! Tandem as variable modifications. Scaffold (version Scaffold_4.9.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Scaffold Local FDR algorithm. Peptide identifications were also required to exceed specific database search engine thresholds. X! Protein identifications were accepted if they could be established at greater than 5.0% probability to achieve an FDR less than 5.0% and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.63 Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. For identification of significant peptides, Scaffold settings were as such: protein threshold = FDR < 5%, minimum peptides = 1, peptide threshold = MRS_otIt. Mouse proteins that were significantly enriched after Benjamini-Hochberg corrected t test (alpha = 0.05) and that were observed in both replicates were considered candidate autoantigens.
Quandenser (MS1 peak area)
The Quandenser pipeline was used with its default settings on the raw mass spectrometry files. The Quandenser pipeline consists of Quandenser, Crux, and Triqler.32
PhIP-Seq Bioinformatic Analysis
Raw FASTQ reads generated from the PhIP-Seq peptidome assay were aligned to our reference PhIP-Seq database using RAPSearch (v2.2). Peptide counts outputted from this workflow were normalized to reads per 100 thousand (RPK) for every sample by dividing each peptide count by the sum and multiplying by 100,000. The resulting peptide RPK count matrices were analyzed in R as described below.
For the analysis, these data were divided into disease and reference groups for both CSF and plasma samples. The disease group contained COVID-19 patient samples. The reference group contained healthy control (HC) and A/G bead samples. Peptide fold change (FC) was calculated for each sample. Peptide counts for COVID-19 patient samples were divided by the mean RPK of the reference group, and healthy control samples were divided by the mean RPK of the combined set of COVID-19 samples and A/G beads. In addition, the FC for GFAP samples were calculated in the same way using the mean of all A/G bead samples.
To identify enriched peptides, results from each sample were filtered using a set of thresholds that, when using a commercial antibody to GFAP, consistently identified GFAP peptides while minimizing nonspecific off-target peptide identification. Each peptide was required to have a minimum of 1 RPK as well as a FC > 10. In addition, thresholds were applied at the gene level. Genes were kept if at least one peptide had a FC > 100 and a total (summed) RPK > 20 across all peptides in the gene. A Kmer analysis was applied to amino acid sequences of all peptides that passed the previous filters. Using a sliding window algorithm, with a window size of 7 amino acids and a step size of 1, all 7-mers were compared across COVID-19 and HC samples. Proteins for which peptides containing at least one 7-mer overlap with another peptide whose total rpK was ³ 20 were carried forward in the analysis. Additionally, proteins with nonoverlapping peptides with an individual rpK ³ 20 an FC ³ 100 were also carried forward. Proteins that passed these thresholds in both technical replicates but were not enriched by reference samples were considered candidate autoantibodies. This workflow was repeated on a per sample level, and the results for each sample were stored separately.
Gene Ontology
ToppGene was used for gene ontology analyses of IP-MS and PhIP-Seq Data.
Acknowledgments
We thank James A. Wells, James R. Byrnes, and Jayant V. Rajan for contribution of reagents to assist with the serological assays. We are grateful to the participants who volunteered to be part of this study, Santos Bermejo and Allison Nelson for study assistance, Patrick Wong and Orr-el Weizman for helpful discussions, the Yale environmental health and safety team, and the Yale Center for Genome Analysis. This work was supported by NIH K23MH118999 (to S.F.F.), F30CA239444 (to E.S.), F30CA250249 (to R.D.C.), R01AI157488 (to S.F.F. and A.I.), R01AI104739 (to S.H.K.), U19AI089992 (to S.H.K. and A.I.), T32GM136651 (to E.S., R.D.C., and R.J.), K08NS096117 (to M.R.W.), R01MH122471 (to J.L.D., S.J.P., and M.R.W.), and R21MH118109 (to S.S.); George Mason University (to A.I.); the Chan Zuckerberg Biohub (to J.L.D.); the Brain Research Foundation (to S.J.P.); the Program for Breakthroughs in Biomedical Research (to S.J.P. and M.R.W.); a Hanna H. Gray Fellowship, Howard Hughes Medical Institute (to C.M.B.); the President’s Postdoctoral Fellowship Program, University of California (to C.M.B.); the John A. Watson Scholar Program, University of California, San Francisco (to C.M.B.); the Beatrice Kleinberg Neuwirth Fund (to A.I.K.); and Fast Grant funding support from Emergent Ventures at the Mercatus Center. LC-MS was supported by NIH shared instrumentation grant S10OD021801.
Author contributions
E.S. devised and executed single-cell RNA sequencing and mouse SARS-CoV-2 experiments, performed analysis and interpretation of data resulting from the assays, and drafted the manuscript. C.M.B. designed and assisted with anatomic immunostaining, IP-MS, PhIP-seq, and analysis and interpretation of data resulting from the assays, and wrote and edited the manuscript. R.D.C., R.J., and S.H.K. assisted with analysis of single-cell RNA sequencing. C.R.Z. designed and performed experiments with the SARS-CoV-2 Luminex assay. A.C.-M., J.C., C.L., J.K., H.W., T.M., B.G.-I., and J.O. assisted with human subject recruitment and sample preparation. C.B.F.V. and N.D.G. contributed to SARS-CoV-2 RT-PCR and edited the manuscript. L.M. interpreted clinical data and edited the manuscript. S.S. and A.I.K. provided human samples, edited the manuscript, and contributed useful discussions. F.L., Y.D., and A.R. assisted with SARS-CoV-2 ELISA and edited the manuscript. N.-T.R. performed bioinformatics analyses and edited the manuscript. T.T.N. assisted with performing anatomic immunostaining, cloning of overexpression plasmids, overexpression cell-based assays, microscopy, and interpretation of the resulting data. R.P.L. assisted with IP-MS, the SARS-CoV-2 Luminex assay, cloning of mAbs, and anatomic immunostaining. R.D. assisted with development of the PhIP-seq bioinformatics pipeline and PhIP-seq data presentation. I.A.H. and B.D.A. assisted with PhIP-seq. B.S.P. and M.S. assisted with IP-MS design, data acquisition, and analyses. J.A.G. and T.L. recruited pre-pandemic human subjects. J.L.D. assisted with development of the PhIP-seq platform, edited the manuscript, and contributed useful discussions. A.I. assisted with data analysis, edited the manuscript, and contributed useful discussions. S.F.F., S.J.P., and M.R.W. conceived and supervised the project and wrote and edited the manuscript.
Declaration of interests
The authors declare to competing interests.
Published: April 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100288.
Contributor Information
Eric Song, Email: eric.song@yale.edu.
Samuel J. Pleasure, Email: samuel.pleasure@ucsf.edu.
Michael R. Wilson, Email: michael.wilson@ucsf.edu.
Shelli F. Farhadian, Email: shelli.farhadian@yale.edu.
Supplemental information
References
- 1.Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D. Neurologic Manifestations of Hospitalized Patients With Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020;77:683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann B., Schmidbauer M.L., Dimitriadis K., Otto S., Knier B., Niesen W.D., Hosp J.A., Günther A., Lindemann S., Nagy G., PANDEMIC and the IGNITE study groups Cerebrospinal fluid findings in COVID-19 patients with neurological symptoms. J. Neurol. Sci. 2020;418:117090. doi: 10.1016/j.jns.2020.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellon M., Schweblin C., Lambeng N., Cherpillod P., Vazquez J., Lalive P.H., Schibler M., Deffert C. Cerebrospinal fluid features in SARS-CoV-2 RT-PCR positive patients. Clin. Infect. Dis. 2020:ciaa1165. doi: 10.1093/cid/ciaa1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espíndola O.M., Siqueira M., Soares C.N., Lima M.A.S.D., Leite A.C.C.B., Araujo A.Q.C., Brandão C.O., Silva M.T.T. Patients with COVID-19 and neurological manifestations show undetectable SARS-CoV-2 RNA levels in the cerebrospinal fluid. Int. J. Infect. Dis. 2020;96:567–569. doi: 10.1016/j.ijid.2020.05.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Destras G., Bal A., Escuret V., Morfin F., Lina B., Josset L., COVID-Diagnosis HCL Study Group Systematic SARS-CoV-2 screening in cerebrospinal fluid during the COVID-19 pandemic. Lancet Microbe. 2020;1:e149. doi: 10.1016/S2666-5247(20)30066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matschke J., Lütgehetmann M., Hagel C., Sperhake J.P., Schröder A.S., Edler C., Mushumba H., Fitzek A., Allweiss L., Dandri M. Neuropathology of patients with COVID-19 in Germany: a post-mortem case series. Lancet Neurol. 2020;19:919–929. doi: 10.1016/S1474-4422(20)30308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gate D., Saligrama N., Leventhal O., Yang A.C., Unger M.S., Middeldorp J., Chen K., Lehallier B., Channappa D., De Los Santos M.B. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature. 2020;577:399–404. doi: 10.1038/s41586-019-1895-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., Hertzog P.J. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040-6. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 12.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 13.Zamecnik C.R., Rajan J.V., Yamauchi K.A., Mann S.A., Sowa G.M., Zorn K.C., Alvarenga B.D., Stone M., Norris P.J., Gu W. ReScan, a Multiplex Diagnostic Pipeline, Pans Human Sera for SARS-CoV-2 Antigens. Cell Rep. Med. 2020;1:100123. doi: 10.1016/j.xcrm.2020.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexopoulos H., Magira E., Bitzogli K., Kafasi N., Vlachoyiannopoulos P., Tzioufas A., Kotanidou A., Dalakas M.C. Anti-SARS-CoV-2 antibodies in the CSF, blood-brain barrier dysfunction, and neurological outcome: Studies in 8 stuporous and comatose patients. Neurol. Neuroimmunol. Neuroinflamm. 2020;7:e893. doi: 10.1212/NXI.0000000000000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marfin A.A., Gubler D.J. West Nile encephalitis: an emerging disease in the United States. Clin. Infect. Dis. 2001;33:1713–1719. doi: 10.1086/322700. [DOI] [PubMed] [Google Scholar]
- 16.Djukic M., Schmidt-Samoa C., Lange P., Spreer A., Neubieser K., Eiffert H., Nau R., Schmidt H. Cerebrospinal fluid findings in adults with acute Lyme neuroborreliosis. J. Neurol. 2012;259:630–636. doi: 10.1007/s00415-011-6221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagel M.A., Forghani B., Mahalingam R., Wellish M.C., Cohrs R.J., Russman A.N., Katzan I., Lin R., Gardner C.J., Gilden D.H. The value of detecting anti-VZV IgG antibody in CSF to diagnose VZV vasculopathy. Neurology. 2007;68:1069–1073. doi: 10.1212/01.wnl.0000258549.13334.16. [DOI] [PubMed] [Google Scholar]
- 18.Song E., Zhang C., Israelow B., Lu P., Weizman O.-E., Liu F., Dai Y., Szigeti-Buck K., Yasumoto Y., Wang G. Neuroinvasive potential of SARS-CoV-2 revealed in a human brain organoid model. bioRxiv. 2020 doi: 10.1101/2020.06.25.169946. [DOI] [Google Scholar]
- 19.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricken G., Schwaiger C., De Simoni D., Pichler V., Lang J., Glatter S., Macher S., Rommer P.S., Scholze P., Kubista H. Detection Methods for Autoantibodies in Suspected Autoimmune Encephalitis. Front. Neurol. 2018;9:841. doi: 10.3389/fneur.2018.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delamarre L., Gollion C., Grouteau G., Rousset D., Jimena G., Roustan J., Gaussiat F., Aldigé E., Gaffard C., Duplantier J., NeuroICU Research Group COVID-19-associated acute necrotising encephalopathy successfully treated with steroids and polyvalent immunoglobulin with unusual IgG targeting the cerebral fibre network. J. Neurol. Neurosurg. Psychiatry. 2020;91:1004–1006. doi: 10.1136/jnnp-2020-323678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutiérrez-Ortiz C., Méndez-Guerrero A., Rodrigo-Rey S., San Pedro-Murillo E., Bermejo-Guerrero L., Gordo-Mañas R., de Aragón-Gómez F., Benito-León J. Miller Fisher syndrome and polyneuritis cranialis in COVID-19. Neurology. 2020;95:e601–e605. doi: 10.1212/WNL.0000000000009619. [DOI] [PubMed] [Google Scholar]
- 23.Toscano G., Palmerini F., Ravaglia S., Ruiz L., Invernizzi P., Cuzzoni M.G., Franciotta D., Baldanti F., Daturi R., Postorino P. Guillain-Barré Syndrome Associated with SARS-CoV-2. N. Engl. J. Med. 2020;382:2574–2576. doi: 10.1056/NEJMc2009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valiuddin H., Skwirsk B., Paz-Arabo P. Acute transverse myelitis associated with SARS-CoV-2: A Case-Report. Brain Behav Immun Health. 2020;5:100091. doi: 10.1016/j.bbih.2020.100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varatharaj A., Thomas N., Ellul M.A., Davies N.W.S., Pollak T.A., Tenorio E.L., Sultan M., Easton A., Breen G., Zandi M., CoroNerve Study Group Neurological and neuropsychiatric complications of COVID-19 in 153 patients: a UK-wide surveillance study. Lancet Psychiatry. 2020;7:875–882. doi: 10.1016/S2215-0366(20)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larman H.B., Zhao Z., Laserson U., Li M.Z., Ciccia A., Gakidis M.A., Church G.M., Kesari S., Leproust E.M., Solimini N.L., Elledge S.J. Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 2011;29:535–541. doi: 10.1038/nbt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandel-Brehm C., Dubey D., Kryzer T.J., O’Donovan B.D., Tran B., Vazquez S.E., Sample H.A., Zorn K.C., Khan L.M., Bledsoe I.O. Kelch-like Protein 11 Antibodies in Seminoma-Associated Paraneoplastic Encephalitis. N. Engl. J. Med. 2019;381:47–54. doi: 10.1056/NEJMoa1816721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donovan B., Mandel-Brehm C., Vazquez S.E., Liu J., Parent A.V., Anderson M.S., Kassimatis T., Zekeridou A., Hauser S.L., Pittock S.J. High-resolution epitope mapping of anti-Hu and anti-Yo autoimmunity by programmable phage display. Brain Commun. 2020;2:fcaa059. doi: 10.1093/braincomms/fcaa059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrill J.T., Erkan D., Winakur J., James J.A. Emerging evidence of a COVID-19 thrombotic syndrome has treatment implications. Nat. Rev. Rheumatol. 2020;16:581–589. doi: 10.1038/s41584-020-0474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen B., Yi X., Sun Y., Bi X., Du J., Zhang C., Quan S., Zhang F., Sun R., Qian L. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020;182:59–72.e15. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuo Y., Estes S.K., Ali R.A., Gandhi A.A., Yalavarthi S., Shi H., Sule G., Gockman K., Madison J.A., Zuo M. Prothrombotic autoantibodies in serum from patients hospitalized with COVID-19. Sci. Transl. Med. 2020;12:eabd3876. doi: 10.1126/scitranslmed.abd3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The M., Käll L. Focus on the spectra that matter by clustering of quantification data in shotgun proteomics. Nat. Commun. 2020;11:3234. doi: 10.1038/s41467-020-17037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonucci R., Fanos V. Acute encephalopathy associated with respiratory syncytial virus infections in childhood. A literature review. Minerva Pediatr. 2005;57:137–142. [PubMed] [Google Scholar]
- 34.Kawasaki Y., Suyama K., Go H., Hosoya M. Clinical manifestations of respiratory syncytial virus-associated encephalopathy in Fukushima, Japan. Pediatr. Int. (Roma) 2019;61:802–806. doi: 10.1111/ped.13928. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura K., Kato M., Sasaki A., Shiihara T., Hayasaka K. Respiratory syncytial virus-associated encephalopathy complicated by congenital myopathy. Pediatr. Int. 2012;54:709–711. doi: 10.1111/j.1442-200X.2012.03594.x. [DOI] [PubMed] [Google Scholar]
- 36.Newland J.G., Romero J.R., Varman M., Drake C., Holst A., Safranek T., Subbarao K. Encephalitis associated with influenza B virus infection in 2 children and a review of the literature. Clin. Infect. Dis. 2003;36:e87–e95. doi: 10.1086/368184. [DOI] [PubMed] [Google Scholar]
- 37.Simon M., Hernu R., Cour M., Casalegno J.S., Lina B., Argaud L. Fatal influenza A(H1N1)pdm09 encephalopathy in immunocompetent man. Emerg. Infect. Dis. 2013;19:1005–1007. doi: 10.3201/eid1906.130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sivadon-Tardy V., Orlikowski D., Porcher R., Sharshar T., Durand M.C., Enouf V., Rozenberg F., Caudie C., Annane D., van der Werf S. Guillain-Barre syndrome and influenza virus infection. Clin. Infect. Dis. 2009;48:48–56. doi: 10.1086/594124. [DOI] [PubMed] [Google Scholar]
- 39.Sugimoto M., Morichi S., Kashiwagi Y., Suzuki S., Nishimata S., Yamanaka G., Sawada A., Kawashima H. A case of respiratory syncytial virus-associated encephalopathy in which the virus was detected in cerebrospinal fluid and intratracheal aspiration despite negative rapid test results. J. Infect. Chemother. 2020;26:393–396. doi: 10.1016/j.jiac.2019.11.008. [DOI] [PubMed] [Google Scholar]
- 40.Kessing C.F., Spudich S., Valcour V., Cartwright P., Chalermchai T., Fletcher J.L.K., Nichols C., Josey B.J., Slike B., Krebs S.J. High Number of Activated CD8+ T Cells Targeting HIV Antigens are Present in Cerebrospinal Fluid in Acute HIV Infection. J. Acquir. Immune Defic. Syndr. 2017;75:108–117. doi: 10.1097/QAI.0000000000001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marra C.M., Maxwell C.L., Dunaway S.B., Sahi S.K., Tantalo L.C. Cerebrospinal Fluid Treponema pallidum Particle Agglutination Assay for Neurosyphilis Diagnosis. J. Clin. Microbiol. 2017;55:1865–1870. doi: 10.1128/JCM.00310-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pachner A.R., Steere A.C., Sigal L.H., Johnson C.J. Antigen-specific proliferation of CSF lymphocytes in Lyme disease. Neurology. 1985;35:1642–1644. doi: 10.1212/wnl.35.11.1642. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Fu L., Gonzales D.M., Lavi E. Coronavirus neurovirulence correlates with the ability of the virus to induce proinflammatory cytokine signals from astrocytes and microglia. J. Virol. 2004;78:3398–3406. doi: 10.1128/JVI.78.7.3398-3406.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mulder J., Feresiadou A., Fallmar D., Frithiof R., Virhammar J., Rasmusson A., Rostami E., Kumlien E., Cunningham J.L. Autoimmune Encephalitis Presenting with Acute Excited Catatonia in a 40-Year-Old Male Patient with Covid-19. medRxiv. 2020 doi: 10.1101/2020.07.23.20160770. [DOI] [PubMed] [Google Scholar]
- 45.Chekuri A., Guru A.A., Biswas P., Branham K., Borooah S., Soto-Hermida A., Hicks M., Khan N.W., Matsui H., Alapati A. IFT88 mutations identified in individuals with non-syndromic recessive retinal degeneration result in abnormal ciliogenesis. Hum. Genet. 2018;137:447–458. doi: 10.1007/s00439-018-1897-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LeDoux M.S., Xiao J., Rudzińska M., Bastian R.W., Wszolek Z.K., Van Gerpen J.A., Puschmann A., Momčilović D., Vemula S.R., Zhao Y. Genotype-phenotype correlations in THAP1 dystonia: molecular foundations and description of new cases. Parkinsonism Relat. Disord. 2012;18:414–425. doi: 10.1016/j.parkreldis.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowles L., Platton S., Yartey N., Dave M., Lee K., Hart D.P., MacDonald V., Green L., Sivapalaratnam S., Pasi K.J., MacCallum P. Lupus Anticoagulant and Abnormal Coagulation Tests in Patients with Covid-19. N. Engl. J. Med. 2020;383:288–290. doi: 10.1056/NEJMc2013656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., Tan Z., Zicari S., Ruggiero A., Pascucci G.R., CACTUS Study Team The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell. 2020;183:968–981.e7. doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodruff M., Ramonell R.P., Lee F.E.-H., Sanz I. Clinically identifiable autoreactivity is common in severe SARS-CoV-2 Infection. medRxiv. 2020 doi: 10.1101/2020.10.21.20216192. [DOI] [Google Scholar]
- 51.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lefranc M.P., Giudicelli V., Ginestoux C., Bodmer J., Muller W., Bontrop R., Lemaitre M., Malik A., Barbie V., Chaume D. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 1999;27:209–212. doi: 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang R., Fichtner M.L., Hoehn K.B., Pham M.C., Stathopoulos P., Nowak R.J., Kleinstein S.H., O’Connor K.C. Single-cell repertoire tracing identifies rituximab-resistant B cells during myasthenia gravis relapses. JCI Insight. 2020;5:e136471. doi: 10.1172/jci.insight.136471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oh J.E., Iijima N., Song E., Lu P., Klein J., Jiang R., Kleinstein S.H., Iwasaki A. Migrant memory B cells secrete luminal antibody in the vagina. Nature. 2019;571:122–126. doi: 10.1038/s41586-019-1285-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Madel Alfajaro M., Wei J., Dong H., Homer R.J. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. bioRxiv. 2020 doi: 10.1101/2020.05.27.118893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zamecnik C.R., Rajan J.V., Yamauchi K.A., Mann S.A., Loudermilk R.P., Sowa G.M., Zorn K.C., Alvarenga B.D., Gaebler C., Caskey M. ReScan, a Multiplex Diagnostic Pipeline, Pans Human Sera for SARS-CoV-2 Antigens. Cell Rep Med. 2020;1:100123. doi: 10.1016/j.xcrm.2020.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pilarowski G., Lebel P., Sunshine S., Liu J., Crawford E., Marquez C., Rubio L., Chamie G., Martinez J., Peng J. Performance characteristics of a rapid SARS-CoV-2 antigen detection assay at a public plaza testing site in San Francisco. J. Infect. Dis. 2021;223:1139–1144. doi: 10.1093/infdis/jiaa802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schubert R.D., Hawes I.A., Ramachandran P.S., Ramesh A., Crawford E.D., Pak J.E., Wu W., Cheung C.K., O’Donovan B.D., Tato C.M. Pan-viral serology implicates enteroviruses in acute flaccid myelitis. Nat. Med. 2019;25:1748–1752. doi: 10.1038/s41591-019-0613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression and repertoire data in the study are available in the NCBI repository SRA PRJNA717310. Raw mass spectrometry files and data analysis output are available from MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) and Proteome Exchange (http://www.proteomexchange.org/) under MassIVE dataset identifier # MSV000087099 and Proteome Exchange # PXD025016.