Summary
Microbial research in space is being conducted for almost 50 years now. The closed system of the International Space Station (ISS) has acted as a microbial observatory for the past 10 years, conducting research on adaptation and survivability of microorganisms exposed to space conditions. This adaptation can be either beneficial or detrimental to crew members and spacecraft. Therefore, it becomes crucial to identify the impact of two primary stress conditions, namely, radiation and microgravity, on microbial life aboard the ISS. Elucidating the mechanistic basis of microbial adaptation to space conditions aids in the development of countermeasures against their potentially detrimental effects and allows us to harness their biotechnologically important properties. Several microbial processes have been studied, either in spaceflight or using devices that can simulate space conditions. However, at present, research is limited to only a few microorganisms, and extensive research on biotechnologically important microorganisms is required to make long-term space missions self-sustainable.
Subject areas: Microbiology, Space Sciences
Graphical abstract
Microbiology ; Space Sciences
Introduction
The International Space Station (ISS) harbors a variety of microorganisms, originating as contaminants from Earth, components of experiments, or the normal microbiota of crew members (Castro et al., 2004; Checinska Sielaff et al., 2019; Pierson, 2001; Singh et al., 2018a; Venkateswaran et al., 2014; Voorhies et al., 2019). Research on microbial survivability upon exposure to space conditions began more than 50 years ago (Horneck et al., 2010; Taylor, 1974). Initial experiments involving exposure of microorganisms to spaceflight conditions started as early as 1935 with balloon flights and rocket payloads (Dickson, 1991; Nickerson et al., 2004). It was in 1966 that the National Aeronautics and Space Administration (NASA), during the Gemini IX and XII missions, began experiments in which bacteriophage T1 and spores of the fungi Penicillium roqueforti were directly exposed to space conditions for 16.8 and 6.5 h, respectively. However, they found that microbial survivability was almost negligible. This finding was later learned to be due to non-penetrating radiation in space, including solar UV radiation or soft X-rays, because covering the samples with a thin layer (0.4 mm) of aluminum resulted in 3,000-fold higher survival of bacteriophage T1 and nearly 100% survival of fungal spores (Hotchin et al., 1968). This study was one of the first to assess the survival limits of microorganisms upon exposure to space conditions.
Since the advent of manned space missions, several procedures, such as using freeze-dried food and preflight crew quarantine, have been implemented to monitor microbial population in the ISS (Castro et al., 2004, 2013; Rogers, 1986). However, exposure to spaceflight conditions and the immunocompromised state of the individuals might still result in otherwise commensal microorganisms becoming pathogenic (Klaus and Howard, 2006). It is well known that microorganisms undergo certain genetic and physiological changes to adapt to stress conditions, and the ISS, likewise, poses a stressful environment to biological life (Horneck et al., 2010). Several studies have been conducted to understand how microorganisms adapt to these space conditions, but these studies are limited to a few species and also require researchers to further understand changes at the genetic level (Nickerson et al., 2004). The microbial species that have been isolated from the ISS include both potentially pathogenic and industrially important microorganisms (Checinska Sielaff et al., 2019). Therefore, understanding how these microorganisms adapt to space conditions will aid in developing strategies to mitigate the risk posed by pathogenic microorganisms to the health of crew members, who might be in an immunocompromised state in the ISS (Lesnyak et al., 1996; Taylor et al., 1997). In addition, understanding how space conditions alter microbial processes in industrially important microorganisms will provide us information on how to engineer these microbes for efficient production of important compounds (Blachowicz et al., 2019b; Romsdahl et al., 2018, 2019). The knowledge gained from these studies will be of utmost importance in implementing safety measures for long-duration spaceflights and interplanetary explorations involving humans. This undertaking will also inform how physical forces influence microorganisms at the cellular, molecular, and evolutionary level, which can be translated to studies on Earth.
In this review, we first discuss the different extreme conditions that are experienced by microorganisms in space and how these conditions influence microbial processes. We also introduce some devices that have been developed to simulate and study the individual effect of microgravity or radiation on microbial processes. Finally, we discuss the applied aspects of space microbiology and how the advent of omics technology has contributed to the generation of data. In addition, we present some perspectives that warrant future investigation.
Influence of extreme space conditions on microorganisms
The ISS has a stable orbit in the thermosphere of the Earth's upper atmosphere and circles the Earth at an average altitude of 400 km (Castro et al., 2013). The microbial species in outer space, that is, in the low earth orbit (LEO), thus, experience a plethora of stress conditions. These include altered gravity (10−3 to 10−6 g), temperature (153–393 K), space vacuum (pressure 10−7 to 10−4 Pa), and elevated CO2 levels (partial pressure of 0.2–0.5 kPa) (Checinska Sielaff et al., 2019; Horneck et al., 2010; Huang et al., 2018; Senatore et al., 2018). In addition, LEO features galactic cosmic radiation (GCR), solar cosmic radiation (SCR), and a radiation belt trapped by the Earth's magnetosphere that can cause elements to ionize (Horneck et al., 2010). These radiations are composed of high-energy particles; for instance, GCR comprises high-energy protons (90%), α-particles (9%), and heavy particles (1%); SCR is primarily composed of protons and electrons, α-particles, and heavy particles (Ohnishi and Ohnishi, 2004; Senatore et al., 2018). In addition, the radiations trapped by Earth's magnetosphere consists of protons and electrons (Ohnishi and Ohnishi, 2004; Senatore et al., 2018). Most of the experiments on the ISS are conducted in a controlled environment within a space capsule, which is a pressurized module with life support systems where microorganisms are protected from most of the extreme conditions in space except for microgravity and cosmic radiations. Therefore, the differences observed in microbial physiology at the cellular or genetic levels are mostly attributed to the effect of microgravity and cosmic radiation (Senatore et al., 2018), unless the studies are conducted outside the space station (Horneck and Zell, 2012). Microgravity refers to the condition of weightlessness that occurs because of reduced physical force exerted by gravity, where gravity is not equal to zero but ranges from 10−3 to 10−6 g (Herranz et al., 2013; Huang et al., 2018; Nickerson et al., 2004). Convection currents are almost absent in microgravity, thereby resulting in low-shear and low-turbulence conditions; it is the alteration of these physical forces that affect microbial cell physiology (Hammond et al., 2000; Klaus et al., 1998). On the basis of results obtained from several experiments conducted under spaceflight conditions, it was hypothesized that the effect of microgravity on microbial cells is dependent on their motility (Benoit and Klaus, 2007). In the case of non-motile cells, a reduction in lag phase and an increase in cell density under spaceflight microgravity conditions were observed compared with ground controls (Klaus et al., 2004; Zea et al., 2017). This finding was attributed to the fact that the fluid around the non-motile cells remains quiescent, resulting in reduced mass transfer (nutrient uptake and release of metabolic byproducts) between the cell and the surrounding microenvironment. A result might be an altered chemical makeup surrounding the cell, eliciting a biological response. On the contrary, in motile cells, flagellar movement results in continuous mixing of the cell and surrounding environment, allowing improved mass transfer and counteracting the effect of microgravity or a low-shear environment (Thevenet et al., 1996). Thus, microgravity alters microbial responses indirectly because of the physical forces surrounding the cells that modify the extracellular environment of the cells, which in turn affects mass transfer between cells and their surrounding environment.
The influence of microgravity on the cellular properties of microorganisms appears to be multifaceted. For instance, simulated microgravity resulted in an increase in membrane fluidity in Escherichia coli, which could be responsible for increased drug resistance (Baker et al., 2004). However, results obtained for Pseudomonas aeruginosa showed no change in membrane fluidity under simulated microgravity (England et al., 2003). Some recent reports have focused on the effect of microgravity at the genetic level; for instance, expression of around 100 genes was altered in Salmonella under simulated microgravity (Wilson et al., 2002a). These gene products included transcription regulators, virulence factors, lipopolysaccharide synthesis enzymes, and iron utilization enzymes. Another study reported an increase in the rate of plasmid exchange under spaceflight conditions compared with ground controls in Gram-positive bacteria (DeBoever et al., 2007), implying that genetic exchange might play a role in adaptation to spaceflight stress conditions. Further research is warranted to understand and correlate the specific genetic factors responsible for adaptation to microgravity conditions. The results from these studies can also be translated to infection by microbial pathogens in certain areas of the human body where they experience similar low-shear conditions, like microvilli of epithelial cells in the gastrointestinal, urogenital, and respiratory tracts of humans (Nickerson et al., 2004; Senatore et al., 2018).
Cosmic radiation is another important parameter that influences microbial processes in the ISS. Radiation results in biological effects via generating mutations either through direct energy absorption by biomolecules (proteins and nucleic acids) or indirectly through the interaction of biomolecules with radiation-induced radicals that are generated by different surrounding processes (Horneck et al., 2010; Huang et al., 2018). A Biostack experiment was performed with the spores of Bacillus subtilis, where a sandwich of monolayers of bacterial spores was mounted on cellulose nitrate foils and exposed to the outer space radiations in a spaceflight experiment (Bucker and Horneck, 1975; Horneck, 1993). The results obtained from this experiment led to the conclusion that the spores (1 μm diameter) on the outer areas were not affected by the high-energy particles of the galactic cosmic radiations; instead, a “bystander effect” was observed in which the effect of charged particles traversed from the outer layer of spores to the surrounding spores, where a biological effect was observed. Additionally, DNA is more prone to be damaged by radiation through the formation of double-stranded breaks that ultimately result in mutations, as observed in E. coli, B. subtilis, and Deinococcus radiodurans (Micke et al., 1994; Schafer et al., 1994; Zimmermann et al., 1994). Microorganisms possess several mechanisms to repair DNA, either by homologous recombination or non-homologous end joining (NHEJ). However, the ability to tolerate radiation depends on the extent to which the organism can repair DNA using DNA repair pathways; for example, D. radiodurans is about 5 times more resistant to ionizing radiation than B. subtilis spores (Baumstark-Khan and Facius, 2002; Senatore et al., 2018). The extent of this ability to repair DNA, mostly via NHEJ, results in mutations and is thus responsible for the scope of survivability of microorganisms in the presence of space radiations.
In addition to cosmic radiation, direct exposure to solar UV-A radiation and visible light in outer space can result in the production of reactive oxygen species and cause damage to nucleic acids, proteins, and lipids (Senatore et al., 2018). It can also result in the synthesis of photolyase, that is, a specific light-dependent repair enzyme (Senatore et al., 2018). The absorption of photons from solar UV radiation and its excitation results in the formation of bipyrimidine lesions in DNA. These lesions lead to the generation of cyclobutane pyrimidine dimers and pyrimidine (6-4) pyrimidone photoproducts between adjacent pyrimidine residues on the same DNA strand in vegetative cells (Cadet et al., 1992). Another type of bipyrimidine lesion, 5,6-dihydro-5(α-thyminyl) thymine, known as spore photoproduct (SP), was observed to be generated in bacterial spores. Formation of SP was likely due to the effects of spore desiccation, the presence of dipicolinic acid, and the binding of small, acid-soluble spore proteins to the DNA, in addition to the UV radiation (Nicholson et al., 2000). For instance, endospore monolayers of different Bacillus sp. exposed to simulated Mars-UV radiation exhibited differential inactivation kinetics, with Bacillus pumilus being most resistant, thereby indicating differences in their ability to activate repair pathways (Newcombe et al., 2005; Schuergera et al., 2006).
Similar to bacteria, many fungal species and their spores are highly resistant to radiation. In most fungi, the major pathway responsible for the repair of double-stranded breaks in DNA is NHEJ. Most double-stranded breaks are thus repaired without using a homologous DNA sequence, eventually resulting in mutations (Arentshorst et al., 2012). Interestingly, conidial monolayers of some fungi, when exposed to simulated Martian UV-C radiation, displayed enhanced UV resistance compared with Bacillus endospores (Blachowicz et al., 2019a). In another study, Aspergillus sp. and Penicillium sp. were found to dominate among different fungal species sampled from the ISS over a period of 6 years (Novikova et al., 2006). In addition to this component, some fungi, for example, Saccharomyces cerevisiae, exhibit large-scale genomic rearrangements under stress conditions that assist its survival (Chan et al., 2007). Thus, exposure to space radiation can result in an increase in mutation frequency throughout the microbial genome, thereby affecting several microbial processes.
Ground simulation of space microgravity conditions
Spaceflight experiments are particularly challenging with respect to spacecraft and astronaut health. There are several limitations associated with conducting experiments on spacecraft, including concerns regarding crew health, limited flight opportunities, significant costs, the requirement of specialized equipment to perform experiments, and limitations in power, work area mass, and crew time (Castro et al., 2013; National Academies of Sciences and Medicine, 2018; Nickerson et al., 2004). All these limitations restrict the range of experiments that can be conducted on spaceflight. Therefore, devices that can simulate space conditions on Earth are necessary to expand our understanding of the effects of space conditions on microorganisms. It is important to note that the ground-based simulators are not an exact representation of spaceflight conditions. However, they can still provide us with key information on how microorganisms adapt to these conditions (Baker and Leff, 2005; Baker et al., 2004; Benoit and Klaus, 2005; Huitema et al., 2002). Importantly, such devices can be used to conduct preliminary experiments before conducting the spaceflight experiments, which will be discussed later in this review. In addition, these simulator devices help to delineate the effect of individual conditions on the adaptation and evolution of microorganisms, rather than a combined effect of different conditions experienced during spaceflight.
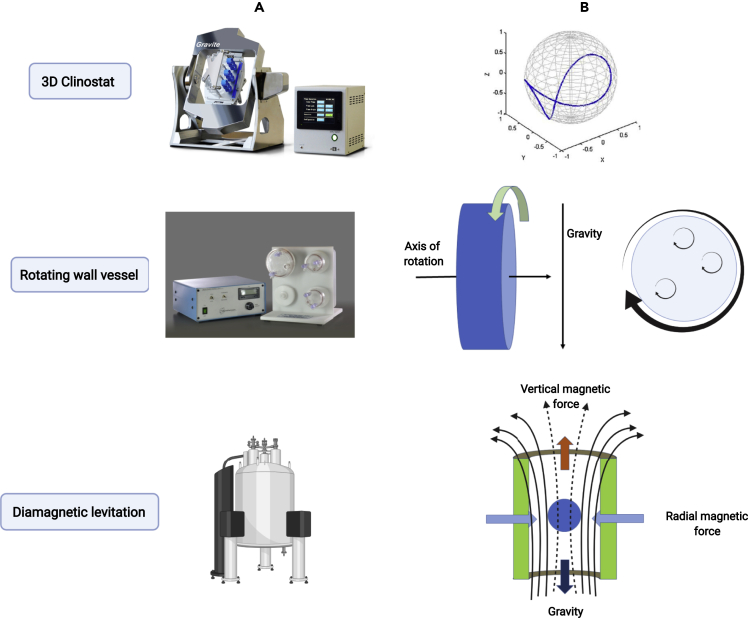
Several ground-based systems are available to simulate space microgravity that can be selected based on the exposure required. In cases where short-duration exposure to microgravity, ranging from a few seconds to a few minutes, is required, space stations can be replaced with sounding rockets (3–13 min; 10−4–10−3 g), parabolic flights (20–30 s; 10−3–10−2 g), or drop towers (5 s; 10−5–10−2 g) that provide free fall (Herranz et al., 2013; Huang et al., 2018; Senatore et al., 2018; Thomas et al., 2000). Long-duration exposure, on the other hand, requires special devices that can simulate microgravity conditions ranging from a few days to a few months (Castro et al., 2013; Herranz et al., 2013). Microgravity Simulation Support Facility (MSSF) at Kennedy Space Center (KSC) harbors such simulators that can be accommodated within controlled environmental chambers to conduct experiments, mimicking the controlled environment in the ISS (NASA document SP-2017-10-1096-KSC). Such ground-based simulators cannot be used to achieve real weightlessness because the magnitude of Earth's gravity cannot be changed. Nevertheless, they are still able to achieve “functional near-weightlessness” by changing the gravity vector continuously and constantly, with the gravity averaged to near zero (Herranz et al., 2013; Huang et al., 2018; Briegleb, 1992). These ground-based microgravity simulator devices (Figure 1) are described in the following sections.
Figure 1.
Ground-based microgravity simulators
(A and B) Different (A) ground-based simulators that are commonly used and (B) their mode of action to mimic space microgravity conditions. The figure has been generated using BioRender (https://biorender.com/). The source of images for 3D clinostat and rotating wall vessel is As One International, Inc. and Synthecon Inc., respectively. The image for the mode of action of 3D clinostat is adapted from elsewhere (Borst and van Loon, 2009).
Clinostats
A clinostat is a device that enables the rotation of a sample along one or two axes such that the cells remain in a constantly suspended state while continuously changing direction, therefore preventing the sample from perceiving the gravity vector (Huang et al., 2018; Klaus, 2001; Briegleb, 1992). A classical 2D clinostat was developed in 1879 by Julius Sachs, rotating along one axis, perpendicular to the gravity vector (Senatore et al., 2018). A 2D clinostat is now rarely used, and its derivative, a 3D clinostat, was later developed to improve the quality of simulations. A 3D clinostat uses two independent frames rotating along two different axes (Herranz et al., 2013), changing its orientation at a constant speed and direction relative to the gravity vector such that the sample remains in a suspended state by eliminating the effect of gravity (Huang et al., 2018). The 3D clinostats are appropriately called random positioning machines (RPMs), because the two frames can be rotated with different speeds and different directions, thus resulting in randomly changing speed and direction (Borst and van Loon, 2009; van Loon, 2007). A clinostat can thus be used to achieve a state of relative motionless of a cell with respect to its extracellular environment, as the cells remain in a suspended state. Therefore, 3D clinostats can be used to mimic the quiescent, unstirred fluid conditions characteristic of samples in orbit (Horneck et al., 2010). The use of RPMs has been found to be particularly suitable for studying the effect of simulated microgravity on higher organisms like plants, because they provide a suitable environment for 3D tissue morphogenesis (Hoson et al., 1997).
Rotating Wall Vessels (RWVs)
Rotating wall vessels (RWVs) consists of a hollow disk or cylinder that is completely filled with a liquid medium, without any bubbles, that rotates on one axis, perpendicular to the gravity vector. As opposed to clinostats, upon solid-body rotation, RWVs maintain the cells in a continuous fall state without being allowed to sediment at the bottom, thus achieving a low-shear, low-turbulence environment (Barrila et al., 2010; Horneck et al., 2010; Nickerson et al., 2004). This free-fall state allows the mass transfer and localized mixing with the extracellular environment, and air exchange via a gas-permeable membrane on one side (Nickerson et al., 2004). Various analogs of RWVs that are available include rotating wall bioreactors, rotating cell culture systems, and high-aspect rotating vessels. These RWV analogs function on the same principle of creating conditions of low-shear modeled microgravity (LSMMG), where cells remain in a free-fall state upon rotation, similar to space microgravity (Huang et al., 2018).
RWV was first developed by the NASA Johnson Space Center (Houston, Texas, USA) to study the effect of simulated microgravity on cell cultures, as it was shown that the low-shear environment supports the generation of 3D differentiated tissue-like assemblies (Freed et al., 1997; Freed and Vunjak-Novakovic, 1997; Unsworth and Lelkes, 1998; Vunjak-Novakovic et al., 2002). Thereafter, they have been used to study the impact of microgravity on different cell types, ranging from microorganisms to higher organisms (Crabbe et al., 2010; Fang et al., 2000; Fang et al., 1997a, b; Hammond and Hammond, 2001; Nickerson et al., 2000; Schwarz et al., 1992).
An important parameter to be considered while using clinostats or RWVs to maintain the cells in a continuous free-fall state is the rotation rate. A comparatively higher rotation speed will result in moving the cells outward near the wall of the container, and a lower rotation speed will result in sedimentation of the cells in a rolling motion on the bottom of the container (Klaus et al., 1998). A rotation speed higher or lower than the required speed will thus result in impairing the free-fall state or simulated microgravity effect. The changes in cell density and mass transfer between the cells and the extracellular environment should also be considered while defining the rotation speed (Begley and Kleis, 2002; Klaus et al., 1998).
Diamagnetic levitation
Diamagnetic levitation technology uses a strong, spatially varied magnetic field produced by a Bitter solenoid, or a superconducting solenoid magnet, to simulate near weightlessness. The diamagnetic force opposes the gravitational force on a levitating object and thereby reduces internal stresses induced by the gravitational force to nearly zero (Herranz et al., 2013). This technology has been previously used for studying the effect of microgravity on many types of organisms, including protozoans, microorganisms, plants, and animals (Coleman et al., 2007; Guevorkian and Valles, 2006; Kuznetsov and Hasenstein, 1996; Liu et al., 2010). However, the major limitation to its use is that the magnetic field can itself influence the organism's physiology, in addition to microgravity (Liu et al., 2011). However, if the experiments are performed using appropriate controls for the influence of the magnetic field, it can still be practical.
Ground simulation of space radiation conditions
Another important parameter that influences microbial life in outer space is the high level of radiation. Although it is not feasible to simulate space radiation conditions on Earth because of the presence of multiple radiations with different energies in space, some devices with controlled radiation exposures have been developed to study their effect on microorganisms. Heavy-ion accelerators and polychromatic UV sources have been used to simulate cosmic rays and solar UV radiation, respectively (Horneck et al., 2010). For a long time, single ion beam accelerators with fixed energies were used to simulate space radiation conditions. However, its use was later limited because space radiation comprises a variety of ions with different energies (Kim et al., 2015; Norbury et al., 2016). In more recent times, this technology has developed substantially, such that it is now feasible to accelerate multiple beams with different energies, with beam switching times of less than 2 minutes. This technology has been implemented at the NASA Space Radiation Laboratory (NSRL) at the Brookhaven National Laboratory (BNL) (Kim et al., 2015; Norbury et al., 2016). Heavy-ion accelerators to simulate space radiation condition are also available at Deutsches Zentrum für Luft-und Raumfahrt Planetary and Space Simulation facilities (PSI) in Germany (Rabbow et al., 2012) and the National Institute of Radiological Sciences in Japan (Cortesao et al., 2020). Despite recent advancements, single-beam experiments are still important in understanding the effect of each radiation component and in deciphering whether the radiation components act additively or synergistically to produce an effect on a biological system (Norbury et al., 2016). These technologies allow one to simulate GCR and to study its effect on microorganisms and other biological life forms using ground-based facilities. Nevertheless, there are still some challenges associated with the use of these accelerators, such as determining the appropriate radiation dose, creating multiple experiments with multiple beams, and costs associated with the experiments (Norbury et al., 2016). Moreover, the use of a heavy-ion accelerator is not an exact representation of space radiation, but it can still be used to generate preliminary data before conducting spaceflight experiments.
Application of microorganisms in space
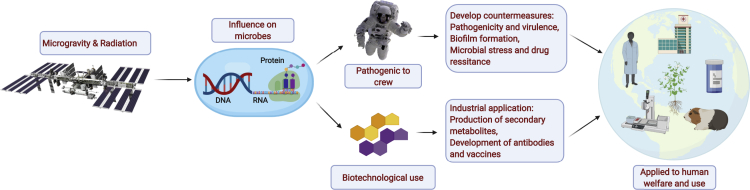
Up to now, results from experiments conducted on spaceflight or using ground-based simulators described above have shown that microgravity conditions can alter microbial processes, including growth, morphology, gene expression, virulence, drug resistance, biofilm formation, and secondary metabolism (Baker et al., 2004; Crabbe et al., 2010; Demain and Fang, 2001; Kacena et al., 1999; Kim et al., 2013; Lawal et al., 2013; Van Mulders et al., 2011). A plausible explanation for these alterations is the loss of gravitational force, resulting in reduced mass transfer and thereby changing various cellular processes (Zea et al., 2016). It is important to study how these conditions alter the cellular processes for two main reasons. First, microorganisms that acquire or display increased virulence or drug resistance properties can pose a threat to the health of crew members, especially in the closed habitat of the ISS (Crabbe et al., 2010, 2013; Rosenzweig et al., 2010; Wilson et al., 2007). Second, microorganisms with altered secondary metabolism under spaceflight conditions can produce novel compounds or display increased production of known compounds that are pharmaceutically or industrially important (Benoit et al., 2006; Lam et al., 1998, 2002). Some of the aspects of microbial species that are altered under spaceflight or simulated microgravity conditions are detailed in the following sections (Figure 2).
Figure 2.
A brief overview of the need to study microbes on the ISS
The figure has been generated using BioRender (https://biorender.com/).
Production of secondary metabolites
For decades, it has been known that microgravity can affect cellular processes and functions in microorganisms (Taylor, 1974). Alterations in the production of secondary metabolites under space conditions have proved particularly intriguing because of their biotechnological implications. In fact, several spaceflight and ground-simulated experiments have demonstrated enhanced production of pharmaceutically relevant secondary metabolites from microorganisms (Blachowicz et al., 2019a; Knox et al., 2016; Romsdahl et al., 2019). Some of these gene clusters synthesizing secondary metabolites are either silent or expressed at very low concentrations under normal, terrestrial conditions. Moreover, in space, cells can produce certain secondary metabolites in suspension and in the absence of shear forces (Klaus, 2004). Hence, spaceflight could offer unique advantages for bioprocessing applications.
Several studies indicated that secondary metabolism is likewise generally enriched in response to heightened extracellular environmental signals and stresses. For instance, Humicola fuscoatra samples aboard the US Space Shuttle mission STS-77 exhibited considerably higher production of the antibiotic monorden compared with ground controls (Lam et al., 1998). Specifically, the space samples displayed a 30% increase and a 190% increase in monorden production in T8 and PG media, respectively. Follow-up paired spaceflight and ground control experiments with Streptomyces plicatus samples showed an increased productivity of the antibiotic actinomycin D (Lam et al., 2002). After 17 days, comparisons of specific productivity in terms of the amount of antibiotic produced per viable cell showed a 296% increase and a 577% increase in defined and complex media, respectively. Similarly, several Aspergillus species exposed to spaceflight conditions or isolated from the ISS showed enhanced production of known or novel metabolites of economic importance. For instance, Aspergillus fumigatus isolates from the ISS exhibited increased production of the antibacterial compound fumigaclavine A compared with the reference strain (Knox et al., 2016). Subsequent genomic analyses revealed a distinct frameshift mutation in fgaPT1 that likely accounts for the increased fumigaclavine A production. In another study, Aspergillus nidulans nkuA mutant exposed to spaceflight conditions resulted in significant production of asperthicin, an anthraquinone pigment (Romsdahl et al., 2019). Aspergillus niger strain isolated from the ISS exhibited a 6,000% increase in the production of an antioxidant pyranonigrin, compared with the reference strain ATCC 1015 (Romsdahl et al., 2020). Genomic analysis indicated the presence of INDELs within the promoter of flbA, a regulator of pyranonigrin production that might contribute to its enhanced production (Aerts et al., 2018). The induction of asperthicin synthesis in A. nidulans, and increased production of pyranonigrin in A. niger, can be explained as a response to provide protection to the fungi from space radiations. Collectively, these studies highlight the potential of space conditions to enhance the production of useful secondary metabolites.
Despite these promising results, other experiments suggest that microorganisms may respond to spaceflight and simulated microgravity conditions in diverse ways. For instance, the production of the immunosuppressive agent rapamycin by Streptomyces hygroscopicus was inhibited by simulated microgravity conditions (Fang et al., 2000). In fact, rapamycin production decreased by about 90% compared with normal gravity controls. Furthermore, longer-term studies have suggested that increased productivity by microorganisms in space conditions may be time-sensitive. Interestingly, Benoit et al. (2006) found that whereas increased productivity of actinomycin D by S. plicatus was observed on days 8 and 12, as reported previously (Lam et al., 2002), ground control productivity of actinomycin D surpassed that of spaceflight samples for the remainder of the mission (Benoit et al., 2006). Similarly, simulated microgravity experiments using RWVs showed increased production of the polyester polymer poly-b-hydroxybutyrate by Cupriavidus metallidurans after 24 h, but not after 48 h, compared with controls (De Gelder et al., 2009). These studies suggest that the effects of microgravity on secondary metabolism may be strain-, growth condition-, or pathway-specific and may be time-dependent. Therefore, additional studies in simulated microgravity conditions are needed to better characterize the microbial response to space conditions and, thus, maximize bioprocessing efficiency in this application.
Pathogenicity and virulence
The effect of space conditions on microorganisms spans beyond altered secondary metabolite production. Studies conducted with pathogens like P. aeruginosa (Crabbe et al., 2010), A. fumigatus (Knox et al., 2016), and Fusarium oxysporum (Urbaniak et al., 2019) reported that space conditions affect regulatory signals resulting in changes in gene expression profile and physiology, and as an extension pathogenicity. Enhanced pathogenicity and virulence of microorganisms in space could pose a significant risk to immunocompromised astronauts during long-term missions. Therefore the identification of microbial strains that can become pathogenic in spaceflight and gaining insight into the underlying mechanisms is crucial.
Owing to the increased duration and frequency of manned space missions, studies aimed at defining the pathogenicity and virulence of microorganisms in space have surged. In one of the studies conducted, A. fumigatus isolates from the ISS were shown to be significantly more lethal than clinical controls in a larval zebrafish model (Knox et al., 2016). Subsequent proteomic analyses revealed an increased abundance of proteins involved in the biosynthesis of trypacidin, a potent mycotoxin, in the ISS strains that could account for the increased virulence (Blachowicz et al., 2019b). Trypacidin has been previously reported to be cytotoxic toward alveolar lung cells (Gauthier et al., 2012). Therefore it becomes crucial to characterize the virulence of common microorganisms in space and understand its impact on crew health. Similarly, it was shown that fruit flies infected with the bacterial pathogen Serratia marcescens aboard the ISS displayed significantly shorter survival times than controls (Gilbert et al., 2020). The fact that this trend was upheld for strains grown on the ground in simulated microgravity conditions suggests that microgravity is the contributing virulence factor. In another study, Salmonella enterica serovar Typhimurium grown in LSMMG was found to be more virulent in a murine model of infection compared with its control culture, exhibiting an LD50 value 5.2 times lower than the strain grown under normal gravity (Nickerson et al., 2000). A follow-up study surprisingly showed a decrease in the expression levels of pathogenicity islands (SPI-1 and SPI-2) in the LSMMG-grown S. enterica culture, indicating a role of a previously unknown virulence mechanism or regulatory pathways (Wilson et al., 2002b). In the case of yeasts like Candida albicans, pathogenicity and virulence are known to be associated with its morphological transition from yeast to hyphal form. This transition has been reported in studies conducted with C. albicans in spaceflight or simulated microgravity (Altenburg et al., 2008). However, exposure of C. albicans to spaceflight conditions did now show an altered virulence compared with ground controls, as determined using an intra-peritoneal mouse model of infection. This result led to the conclusion that either short-term exposure to spaceflight did not significantly affect its virulence or the mode of infection was ineffective because intra-peritoneal is not the standard method for infecting C. albicans (Crabbe et al., 2013). Collectively, these studies exemplify the importance of defining microbial pathogenicity and virulence in space.
Biofilm formation
Several studies with bacterial and fungal species reported the formation of biofilms on spaceflight surfaces and its water systems, potentially corroding surfaces or becoming detrimental to crew health (Novikova, 2004; Ott et al., 2004; Schultz et al., 1989; Song and Leff, 2005). Microbial growth has been observed previously on different locations, including piping, water recycling system, electrical connectors, air-conditioning, and thermal control, aboard Mir space station (Klintworth et al., 1999). Among the 234 microbial species isolated from the Mir space station, most of the fungal species exhibited potential polymer biodegradation activity, thereby posing a threat to the structural integrity of the space station (Novikova, 2004). Several bacterial pathogens, mostly staphylococci and enterococci, have been shown to form biofilms on the surfaces of the ISS (Schiwon et al., 2013; Sobisch et al., 2019). Some bacterial species like Bacillus and coliforms that are present in wastewater or soil were isolated from water and waste lines in the ISS (Koenig and Pierson, 1997; La Duc et al., 2004; Ott et al., 2004). Sphingomonas sp. and Methylobacterium sp. were found to be dominant in potable water on the ISS (Novikova et al., 2006). Therefore, in the future, more emphasis should be given to materials or surface coatings that are resistant to biodegradation and biofilm formation (Zea et al., 2018).
The formation of biofilm under microgravity conditions was first reported by McLean et al. (2001); they showed that P. aeruginosa, when exposed to spaceflight conditions on polycarbonate filters, remained viable and formed a biofilm (McLean et al., 2001). Another report showed that P. aeruginosa formed a biofilm that was typical in structure, consisting of columns overlaid with a canopy under spaceflight conditions. Flagella-driven motility was found to be essential for this typical biofilm structure (Kim et al., 2013). Interestingly, it had been previously shown that a global regulator, Hfq, plays a role in P. aeruginosa to respond to microgravity conditions, resulting in differential regulation of 167 genes and 28 proteins (Crabbe et al., 2010). In another study, S. enterica serovar Typhimurium exhibited increased cell clumping, aggregation, and production of an extracellular matrix, indicating increased biofilm formation under spaceflight conditions. The transcriptome and proteome analysis of S. enterica revealed the role of Hfq under spaceflight conditions, similar to that identified in P. aeruginosa (Wilson et al., 2007). Hfq is an RNA-binding protein that is conserved in both prokaryotes and eukaryotes (Valentin-Hansen et al., 2004). It is known to be important for regulating the expression of various genes involved in virulence and stress resistance in several opportunistic pathogens, by stabilizing small regulatory RNAs and interfering with their interaction with mRNA molecules (Ding et al., 2004; Sittka et al., 2007; Sonnleitner et al., 2003; Valentin-Hansen et al., 2004). Another study involving a different species, Micrococcus luteus, reported an increase in production of extracellular polysaccharides and enhanced biofilm formation under spaceflight conditions (Mauclaire and Egli, 2010). Further studies are warranted to understand the mechanistic basis of typical biofilm structures that are formed by other microbial species under microgravity conditions.
The ability of microbial species to form biofilms has also been demonstrated in LSMMG, similar to spaceflight. For instance, some bacterial species like Klebsiella pneumoniae, P. aeruginosa, E. coli, Staphylococcus aureus, and Streptocococcus mutans, also showed increased aggregation and biofilm formation when grown under simulated microgravity or LSMMG conditions in RWV (Castro et al., 2011; Crabbe et al., 2010; Lynch et al., 2006; Orsini et al., 2017; Tucker et al., 2007; Wang et al., 2016, 2017). Thus far, studies on biofilm formation with lower eukaryotes have focused on only a few species. The experiments with the model yeast S. cerevisiae under simulated microgravity conditions showed that yeasts undergo random budding instead of bipolar budding, resulting in an increase in cell clumping (Purevdorj-Gage et al., 2006). A subsequent study conducted on S. cerevisiae in LSMMG showed that shear stress resulted in differential expression of genes involved in various cellular processes, including chromosome organization (Johanson et al., 2006). Further analysis of these findings will aid in understanding their role in the adaptation and survival of yeasts both under microgravity conditions and similar low-shear environments encountered in the human body. Another yeast, C. albicans, also showed a similar response of increased filamentation, cells undergoing random budding clustered with the filamentous forms, and changes in expression levels of two genes associated with the yeast-hyphae transition under LSMMG conditions (Altenburg et al., 2008; Searles et al., 2011). A subsequent report on C. albicans grown under spaceflight conditions further confirmed this finding (Crabbe et al., 2013). Microarray analysis indicated that random budding can be a consequence of altered expression of genes involved in the synthesis of the actin cytoskeleton and its polymerization because the actin cytoskeleton is essential for bud site selection (Crabbe et al., 2013; Ni and Snyder, 2001). These features might confer an advantage to virulent properties of C. albicans in long-term spaceflights in a species that is otherwise a human commensal.
Microbial stress and drug resistance
Several studies have indicated differential expression of stress response genes under spaceflight conditions, some of which result in altered drug susceptibility/resistance profiles in microorganisms. These microbial characteristics are of importance, particularly because of their impact on the health of crew members and the integrity of spacecraft. Some of the early studies conducted on E. coli and S. aureus exhibited increased resistance to all the antibiotics tested under spaceflight conditions when compared with ground controls (Lapchine et al., 1986; Tixador et al., 1985). Another experiment showed that this difference was observed only when cells were grown in liquid media and not when grown on agar, indicating a physical effect rather than a biological effect (Kacena and Todd, 1999). S. typhimurium and E. coli cells grown under simulated microgravity conditions showed enhanced resistance to acid stress, thermal stress, osmotic stress, and an increased ability to survive in macrophages, independent of the expression levels of global regulator RpoS. This finding indicates that the response is likely due to another stress response pathway (Lynch et al., 2004; Wilson et al., 2002a). In stationary-phase cells of E. coli, however, rpoS mRNA was found to be more stable under simulated microgravity, resulting in increased RpoS protein levels that might be responsible for enhanced stress resistance in the stationary phase (Lynch et al., 2004). In another study, E. coli were grown in the presence of increasing concentrations of the antibiotic gentamicin during spaceflight, which revealed that the cells adapted to the antibiotic on spaceflight earlier compared with ground controls. Transcriptome analysis showed that 50 stress response genes were upregulated in microgravity, including antibiotic stress-responsive genes (Aunins et al., 2018). Microbial cells present in a biofilm are often more resistant to antibiotics and other stresses, and this trend holds for spaceflight samples. For instance, E. coli biofilm exhibited increased resistance to salt stress, ethanol stress, and the antibiotics penicillin and chloramphenicol under LSMMG conditions. Interestingly, the E. coli rpoS mutant showed impaired resistance to salt and ethanol stress but not antibiotic resistance, indicating the role of separate pathways for different stress conditions (Lynch et al., 2006).
In addition to mutations in the chromosome, some studies indicated the role of horizontal gene transfer in imparting drug resistance. A report showed that all Gram-positive bacteria were resistant to at least one antibiotic, the most abundant resistant element being the plasmid-borne erythromycin and tetracycline resistance genes that likely spread via horizontal gene transfer from staphylococci (Sobisch et al., 2019; Vaishampayan and Grohmann, 2019). Bacillus thuringiensis also exhibited an increased probability of transfer of genetic material (plasmids or chromosome fragments) in spaceflight conditions, which might explain its increased antibiotic resistance and virulence (Beuls et al., 2009; Senatore et al., 2018). In contrast to all these reports, some studies showed no difference in the antibiotic resistance profile postflight compared with the ground control, probably because of transient changes under spaceflight conditions (Juergensmeyer et al., 1999; Mora et al., 2019).
In the case of lower eukaryotic microorganisms, studies on responses to different stress conditions and antifungal drugs under spaceflight conditions are limited to only a few species. In a study on S. cerevisiae exposed to LSMMG, microarray analysis of the yeast revealed that among the genes differentially expressed, 26% of genes were involved in general environmental stress responses, whereas the remaining were unique to microgravity conditions (Sheehan et al., 2007). In another study on C. albicans exposed to spaceflight conditions, cells in the biofilm exhibited increased expression of ABC transporters and multidrug efflux proteins and downregulation of ergosterol-encoding genes, which are all involved in imparting drug resistance to the cells. In addition, several genes involved in oxidative stress resistance were also differentially regulated under spaceflight conditions (Crabbe et al., 2013). Among the Lsm family of proteins that are orthologs of Hfq in eukaryotes, expression of Lsm2 was significantly affected under spaceflight conditions. However, its role in the spaceflight response is yet to be determined (Crabbe et al., 2013). Similarly, C. albicans cells exposed to the LSMMG condition formed a complex biofilm structure and exhibited significantly more resistance to amphotericin B compared with ground control (Altenburg et al., 2008). Further research is needed to understand the physiological or genetic basis of drug resistance in human pathogens that might vary among different species.
Space conditions influence astronaut microbiome
So far, this review discusses the effect of space or simulated-space conditions on microorganisms and their impact on spacecraft and human health. However, in addition to being present in the space environment, microbes are an essential constituent of the human body that is also affected by space conditions. Studies on the effect of space conditions on human microflora have been ongoing since the Skylab mission. During the Skylab mission, although the microbial counts in the gastrointestinal (GI) samples of astronauts increased after the spaceflight, there was a reduction in microbial diversity (Taylor et al., 1971). The study also concluded an increase in pathogenic species, including S. aureus and S. marcescens, with S. aureus capable of being transferred from astronaut to astronaut. Later on, several culture-based studies showed that the space conditions result in alteration of the astronauts' oral, nasal, and GI microbial composition (Brown et al., 1976; Decelle and Taylor, 1976; Lencner et al., 1984). One of the studies utilized culture-independent methods to analyze the changes in microbial communities of GI tract, skin, nose, and tongue of astronauts during long-term space missions (Voorhies et al., 2019). The results showed that during 6 months flight, there was an increase in GI microbial diversity in four of five astronauts under observation; however, intestinal microbiota's composition became similar across astronauts with time in space. A slight increase in the inflammatory immune response during spaceflight was correlated to the altered GI microbiome in astronauts. In addition to GI, although the skin microbiome varied between different astronauts, common microbial shifts were observed among them during spaceflight. Crew members have reported allergies because they are continuously exposed to dust in a closed system of the ISS. Several human-associated bacterial commensals and a few opportunistic fungal pathogens were isolated from the ISS debris, which might be responsible for causing allergies (Venkateswaran et al., 2014). A different study also reported a higher abundance of Malassezia, lipophilic skin fungi, compared with other fungal species from astronauts' skin surfaces, during their 6 months stay on the ISS (Sugita et al., 2016). Another report showed differences in the gut microbiota of astronauts and humans on the earth, with astronauts exhibiting a significant decrease in microbial composition, mostly attributed to differences in their lifestyle and diet (Hao et al., 2018). Significant differences between an astronaut's gut microbiome during 1-year mission aboard the ISS, compared with his twin on Earth, were also observed (Garrett-Bakelman et al., 2019). The human gut microbiome can have an effect on general physiological responses, immunity, and health of the body (Heyde and Ruder, 2015; Siddiqui et al., 2021). Therefore, it becomes crucial to maintain a healthy microbiome, especially in astronauts who are immunocompromised in space.
The advent of omics technology in space
Our understanding of microbial diversity in the upper atmosphere and aboard spacecraft has relied mostly on the culturable microorganisms. In recent times, the advent of genomics has allowed for the identification of new species (Bijlani et al., 2021; Singh et al., 2019; Venkateswaran et al., 2017) and several other microorganisms found in the ISS that are otherwise unculturable. It is well known that only 0.1%–1% of microorganisms from any environment are culturable (Lloyd et al., 2018). In a study conducted by Singh et al., metagenomic data generated from the ISS samples were used to retrieve nearly complete whole-genome sequences (WGSs) of bacterial species that led to the identification of a novel genus. The metagenome-assembled genome sequences of the novel species were shown to be identical to the WGS obtained from previously isolated ISS cultures. Thus, metagenomic data can provide nearly complete WGS that can be used to accurately identify novel species. This study was the first wherein the approach known as “metagenome to phenome” was used on the ISS samples and resulted in the identification of a novel genus (Singh et al., 2019). A different study developed Growth Rate InDex (GRiD) to determine the growth rates of ultra-low coverage or de novo-assembled metagenomes (Emiola and Oh, 2018). This represents an important parameter that can be applied to the metagenomics data generated from the ISS samples to determine the growth rate of different bacterial species, especially those that are lower in abundance. In the future, this approach can be extended to identify unculturable species as well.
The use of multi-omics, including genomics, transcriptomics, proteomics, and metabolomics, has particularly aided in studying the variations in microbial diversity aboard spaceflight. Several studies have utilized amplicon and shot-gun sequencing to study changes in microbial diversity at different time-points in different areas aboard the ISS (Checinska et al., 2015; Checinska Sielaff et al., 2019; Ichijo et al., 2016; Lang et al., 2017; Mora et al., 2016, 2019). In all the studies conducted, microbial diversity was found to vary at different time points aboard the spaceflight. The results showed that the ISS is chiefly composed of human-associated microbiomes, including Biosafety Level-2 (BSL-2) opportunistic pathogens. These opportunistic pathogens isolated from the ISS exhibited antibiotic resistance, but it was not found significant to result in an increase in pathogenicity under space conditions; however, the presence of most of the microorganisms indicated a risk toward damage to the integrity of spaceflight material (Mora et al., 2016, 2019). Further experiments are warranted to understand their antibiotic resistance profile and pathogenicity while aboard the ISS. A more recent study utilized metagenomics, in combination with a technique to analyze only viable cells on the ISS, to study the spacecraft's active microbial diversity (Singh et al., 2018b). The study led to the similar conclusion that microbial diversity varied at different time points at different locations aboard the ISS. The data indicated that BSL-2 microorganisms, for instance, Klebsiella pneumonia, persisted in the ISS over different time points. The metagenome-based analysis provides an advantage of higher sequence coverage and improved resolution compared with amplicon sequencing. Metagenomic sequences also helped in predicting that there was an increase in the virulence factors and antimicrobial resistance genes associated with the microorganisms over a time period in the ISS, which needs to be further validated when compared with appropriate ground controls (Singh et al., 2018b).
Recently, nanopore sequencing technology MinION, along with streamlined sample preparation protocol, was first tested during a parabolic flight, which was also followed by data analysis (McIntyre et al., 2016). This allowed researchers to understand the effect of microgravity on sequencing protocol and tools, and to further modify the procedure to implement on the ISS. Thereafter, MinION DNA sequencer was successfully established in-flight aboard the ISS that has further paved the way for conducting molecular biology experiments on spaceflight (Castro-Wallace et al., 2017). The use of this technology offers a substantial potential for monitoring the microbial population aboard the spaceflight in real time. Several other compact third-generation sequencing technologies can also be adapted to microgravity conditions and deployed in the ISS (Goodwin et al., 2016; McGinn and Gut, 2013). Furthermore, a nucleic acid extraction technology has been recently tested and adapted to operate under microgravity conditions (Urbaniak et al., 2020b). In addition, the ISS harbors several hardware components to conduct microbiology experiments, including growth chambers and instruments to control the exposure of microbes to space radiation (Castro et al., 2013). Therefore, the inclusion of omics technologies in addition to the existing facilities available on the ISS will allow researchers to conduct experiments followed by sequencing and data analysis in the spaceflight, rather than waiting for sample return to Earth.
A previous report suggested the use of omics, including genomics, proteomics, transcriptomics, metabolomics, epigenomics, exposomics, and metallomics, to establish markers specific to simulated microgravity conditions (Schmidt et al., 2016). To date, the use of genomics, transcriptomics, metabolomics, and proteomics has been of great importance in studying gene regulation at the cellular level, thereby providing insight into altered cellular processes upon exposure to spaceflight or microgravity conditions. For instance, genomic comparison of ISS Enterobacter bugandensis isolates with their Earth counterparts revealed the presence of antimicrobial drug resistance genes, including upregulation of efflux pumps (Singh et al., 2018a). These results indicate potential drug resistance properties of these ISS isolates that need to be further confirmed by phenotypic characterization. Another study reported a multi-omics analysis of a model fungus, A. nidulans, exposed to space conditions when compared with ground controls. Correlation of the genomics, proteomics, and metabolomics data of spaceflight-exposed A. nidulans showed that the introduction of a stop codon in a regulatory gene, laeA, resulted in reduced LaeA protein levels, thereby affecting secondary metabolite production (Romsdahl et al., 2019). In another study, proteomic analysis of A. fumigatus ISS isolates and its comparison to clinical isolates identified the proteins that are responsible for its adaptation to the space environment. Most of the proteins that were upregulated were involved in stress responses, carbohydrate metabolism, and secondary metabolism (Blachowicz et al., 2019b). However, all these studies relied on the ISS sample return to Earth for analysis. Therefore, several different alternatives that can be adapted to the ISS, for instance, those discussed previously to analyze transcriptome and proteome (Grimm et al., 2014; reviewed in Karouia et al., 2017), should be emphasized for future space research. Furthermore, in addition to genomics, nanopore sequencing has enabled the identification of epigenetic changes in bacteria via detection of DNA base modification at N6-methyladenine (McIntyre et al., 2019). The availability of nanopore sequencer on the ISS can help address epigenetic questions in addition to genetic analysis. Therefore, the inclusion of other omics technologies to study microbial physiology under spaceflight or simulated microgravity conditions can help develop microbial markers under space microgravity conditions. Overall, the correlation of omics with microbial phenotypic responses can help us gain insight into the global changes responsible for the observed phenotype in space conditions. This will further aid in developing precision medicines against pathogenic microbes, similar to that hypothesized for humans in space (Schmidt and Goodwin, 2013), both of which are critical for the safety of future long-term manned space missions.
Future directions
The major goal of the microbiology element in the NASA Science plan involves understanding the effect of spaceflight on microbial life, processes, and dynamics (NASA, 2016). Some of the guiding questions include how the spaceflight influences microbial pathways, microbe-microbe interactions, biofilm formation, virulence, and microbial interactions with other organisms (NASA, 2016). Based on the NASA decadal survey, several microbial tracking experiments have been initiated to understand the microbial persistence on the spacecraft (Checinska Sielaff et al., 2019; Singh et al., 2018b; Venkateswaran et al., 2014). In addition, several experiments have been conducted to understand microbial processes, including biofilm formation (Kim et al., 2013), pathogenicity, and virulence (Crabbe et al., 2013; Ding et al., 2004), as discussed previously. An overview of the key space microbiology experiments is shown in Figure 3. However, the research in spaceflight or simulated microgravity has been limited to only a few microorganisms. This is partly due to the extensive time, resources, and efforts required to conduct experiments on the spaceflight followed by transporting the biological materials back to Earth for further analysis. Therefore, in the future, more efforts should be put toward adapting different technologies, including omics (Bayram et al., 2018; Karouia et al., 2017; Schmidt et al., 2016), to the ISS. Incorporating “omics” in space would provide an answer to many different questions and help understand different aspects of microbial life on the ISS. Further on, to simplify the experiments onboard, emphasis should be given to conducting preliminary experiments on Earth using devices that can simulate spaceflight conditions, which will allow researchers to plan experiments meticulously on the ISS. Another aspect that needs attention is the scarcity of information available on the persistence of viral load and their interaction with other organisms on the spacecraft. Some of the previous studies have reported reactivation of viruses in astronauts, including herpesvirus and Epstein-Barr virus, while onboard the ISS (Mehta et al., 2017; Rooney et al., 2019). Omics can also help obtain information about the persistence of viruses on the ISS so that effective techniques to detect those viruses and countermeasures for the safety of crew health can be developed.
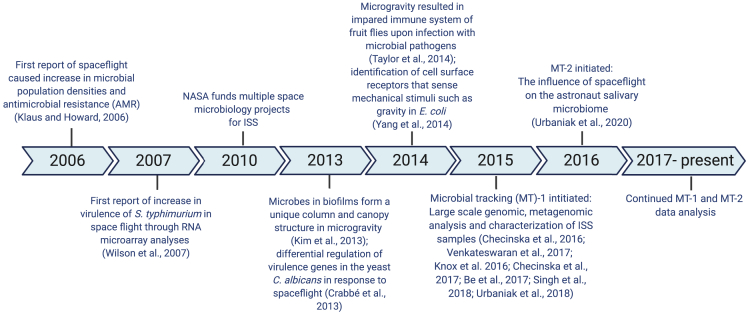
Figure 3.
TImeline of key space microbiology experiments
An overview of the space microbiology experiments leading to Microbial Tracking (MT) experiments (MT-1 and MT-2) (Be et al., 2017; Checinska Sielaff et al., 2017; Checinska Sielaff et al., 2016; Crabbe et al., 2013; Kim et al., 2013; Klaus and Howard, 2006; Knox et al., 2016; Singh et al., 2018b; Taylor et al., 2014; Urbaniak et al., 2018, 2020a; Venkateswaran et al., 2017; Wilson et al., 2007; Yang et al., 2013b). The figure has been generated using BioRender (https://biorender.com/).
Despite promising applications of microorganisms in space, information regarding how bacteria and fungi adapt to microgravity and radiation conditions, genetically or physiologically, remains limited. Although increased pathogenicity and virulence are a concern for crew health, microbial activity aboard spacecraft can be beneficial. Therefore, studies moving forward should be aimed at characterizing the advantages of microbial life in space. Life support functions, such as waste degradation, water recovery, and oxygen production, can be carried out by microorganisms, thus, sustaining life in space. In fact, several studies have shown that fungi, bacteria, and cyanobacteria enhance vitamin production, water recycling, air decontamination, and waste management under space conditions (Acevedo-Rocha et al., 2019; Aydogan-Cremaschi et al., 2009; Carillo et al., 2020; Lindeboom et al., 2020; Roberts et al., 2004). In particular, fungal biotechnology can be harnessed such that space stations are self-sustaining with respect to food, pharmaceuticals, waste recycling, and plastic degradation for long-term missions (Cortesão et al., 2020). Challenges ahead may involve developing a balanced microflora that maximizes life support functions and minimizes the threat to crew health in closed space systems.
Another important property of fungi that can be harnessed is their ability to synthesize pigments in the presence of radiations that helps them survive. Fungal species belonging to genera Cladosporium, Paecilomyces, and Penicillium were observed to grow toward the source of radiation at the Chernobyl nuclear accident site, and this property was termed “radiotropism” (Zhdanova et al., 2004). Cryptococcus neoformans and Cladosporium sphaerospermum exhibited increased growth in the presence of ionizing radiations due to changes in melanin synthesis (Dadachova and Casadevall, 2008). Microorganisms exposed to space conditions encounter similar extreme radiation conditions and therefore develop protective mechanisms to adapt to these conditions. Proteomic characterization of A. fumigatus and A. niger isolated from the ISS surface showed an increased abundance of Arp1 and AlbA proteins, respectively, that are involved in the synthesis of DHN-melanin (1.8-dihydroxynaphthalene melanin) (Blachowicz et al., 2019b; Romsdahl et al., 2018). Another study as a part of CASIS-NPμG project exposed eight radiation-resistant fungal strains isolated from Chernobyl nuclear accident site to spaceflight condition followed by their genome sequencing and assembly (Singh et al., 2017). Further omics analysis is ongoing to understand the mechanistic basis of the production of bioactive compounds by these fungal species exposed to space conditions, compared with ground controls (unpublished data). This knowledge can help us harness their potential to protect life forms, including protecting humans from harmful radiations on space as well as on Earth.
Although enhanced virulence under space conditions has the potential to negatively impact crew health, it can also be used as a tool to guide the development of novel therapeutics. Drug discovery, modeling of drug pathways, and vaccine development are important areas that necessitate attention for the benefit of human life on space and Earth. Specifically, the discovery of new factors involved in pathogenicity via microgravity experiments can inform antibiotic drug and vaccine development. For example, the identification of novel virulence genes influenced by microgravity could allow us to more precisely attenuate bacteria using live-attenuated vaccines (Higginson et al., 2016). Traditionally, live-attenuated vaccines reduce virulence by introducing strains with metabolic mutations that slow or inhibit bacterial growth. This approach will help us identify novel therapeutic targets that allow the strain to remain more metabolically fit, thereby resulting in a heightened host immune response. Currently, research is in progress aboard the ISS by National Lab Pathfinder (NLP) missions to develop a vaccine against diarrhea-causing strains of Salmonella (Horneck et al., 2010). In addition to virulence genes, the identification of microgravity-sensitive global regulators could guide the development of novel antibiotics. In particular, compounds capable of disrupting these global regulators could inhibit bacteria from sensing their environment and activating virulence factors (Higginson et al., 2016). This approach to disarm bacterial virulence has been successfully utilized against Citrobacter rodentium in mice using Regicin, a novel inhibitor of the global regulator RegA (Yang et al., 2013a). Hence, employing such an approach with microgravity-sensitive global regulators could prove beneficial for the design of novel antibiotics. Independent studies have shown that hepatocytes grown in RWVs form three-dimensional colonies characterized by increased and prolonged functionality (Brown et al., 2003; Chang and Hughes-Fulford, 2014). Not only the fact that such 3D culture systems better mimic the microenvironment of the intact liver but also the fact that they can sustain themselves longer than traditional cultures makes them optimal candidates for pre-clinical drug testing (Hammond et al., 2016). These 3D culture systems that mimic human tissues can also be used to study host-pathogen interactions to identify drug targets and, additionally, drug testing. These novel vaccines or drug targets can be used for treatment in spaceflight conditions, and further can be translated for use on Earth to treat an infected individual where the pathogenic organism encounters a similar low-shear environment.
In summary, several research projects are being carried out to utilize the ISS as a test bed for microbiological studies and established MSSF to support microgravity-related scientific experiments. Furthermore, with the advancement of multi-omics, a better understanding of the microbial process is being obtained that will help devise strategies for sustaining healthy human life in future long-term manned missions to Moon, Mars, and Beyond.
Acknowledgments
The authors acknowledge funds from NASA’s 2018 Space Biology (ROSBio) NNH18ZTT001N-FG App B: Flight and Ground Space Biology Research Grant 80NSSC19K1501 awarded to C.C.C.W and K.V. All figures including graphical abstract were prepared using BioRender (www.biorender.com).
Author contributions
S.B., E.S., and N.K.S. wrote and revised the manuscript. K.V. and C.C.C.W. revised and provided feedback on the manuscript. S.B. and C.C.C.W. prepared an outline for the manuscript.
Declaration of interests
All authors declare no competing interests.
References
- Acevedo-Rocha C.G., Gronenberg L.S., Mack M., Commichau F.M., Genee H.J. Microbial cell factories for the sustainable manufacturing of B vitamins. Curr. Opin. Biotechnol. 2019;56:18–29. doi: 10.1016/j.copbio.2018.07.006. [DOI] [PubMed] [Google Scholar]
- Aerts D., Hauer E.E., Ohm R.A., Arentshorst M., Teertstra W.R., Phippen C., Ram A.F.J., Frisvad J.C., Wosten H.A.B. The FlbA-regulated predicted transcription factor Fum21 of Aspergillus niger is involved in fumonisin production. Antonie Van Leeuwenhoek. 2018;111:311–322. doi: 10.1007/s10482-017-0952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenburg S.D., Nielsen-Preiss S.M., Hyman L.E. Increased filamentous growth of Candida albicans in simulated. Microgravity. Genomics Proteomics Bioinformatics. 2008;6:42–50. doi: 10.1016/S1672-0229(08)60019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arentshorst M., Ram A.F.J., Meyer V. Using non-homologous end-joining-deficient strains for functional gene analyses in filamentous fungi. In: Bolton M., Thomma B., editors. Plant Fungal Pathogens. Humana Press; 2012. pp. 133–150. [DOI] [PubMed] [Google Scholar]
- Aunins T.R., Erickson K.E., Prasad N., Levy S.E., Jones A., Shrestha S., Mastracchio R., Stodieck L., Klaus D., Zea L. Spaceflight modifies Escherichia coli gene expression in response to antibiotic exposure and reveals role of oxidative stress response. Front. Microbiol. 2018;9:310. doi: 10.3389/fmicb.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydogan-Cremaschi S., Orcun S., Blau G., Pekny J.F., Reklaitis G.V. A novel approach for life-support-system design for manned space missions. Acta Astronautica. 2009;65:330–346. [Google Scholar]
- Baker P.W., Leff L.G. Intraspecific differences in bacterial responses to modelled reduced gravity. J. Appl. Microbiol. 2005;98:1239–1246. doi: 10.1111/j.1365-2672.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- Baker P.W., Meyer M.L., Leff L.G. Escherichia coli growth under modeled reduced gravity. Microgravity Sci. Technol. 2004;15:39–44. doi: 10.1007/BF02870967. [DOI] [PubMed] [Google Scholar]
- Barrila J., Radtke A.L., Crabbe A., Sarker S.F., Herbst-Kralovetz M.M., Ott C.M., Nickerson C.A. Organotypic 3D cell culture models: using the rotating wall vessel to study host-pathogen interactions. Nat. Rev. Microbiol. 2010;8:791–801. doi: 10.1038/nrmicro2423. [DOI] [PubMed] [Google Scholar]
- Baumstark-Khan C., Facius R. Life under conditions of ionizing radiation. In: Horneck G., Baumstark-Khan C., editors. Astrobiology, the Quest for the Conditions of Life. Springer; Berlin, Germany: 2002. pp. 261–284. [Google Scholar]
- Bayram M., Asar R., Ozdemir V. Is space the new frontier for omics? Mars-omics, planetary science, and the next-generation technology futurists. OMICS. 2018;22:696–699. doi: 10.1089/omi.2018.0167. [DOI] [PubMed] [Google Scholar]
- Be N.A., Avila-Herrera A., Allen J.E., Singh N., Checinska Sielaff A., Jaing C., Venkateswaran K. Whole metagenome profiles of particulates collected from the International Space Station. Microbiome. 2017;5:81. doi: 10.1186/s40168-017-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley C.M., Kleis S.J. RWPV bioreactor mass transport: earth-based and in microgravity. Biotechnol. Bioeng. 2002;80:465–476. doi: 10.1002/bit.10395. [DOI] [PubMed] [Google Scholar]
- Benoit M., Klaus D. Can genetically modified Escherichia coli with neutral buoyancy induced by gas vesicles be used as an alternative method to clinorotation for microgravity studies? Microbiology. 2005;151:69–74. doi: 10.1099/mic.0.27062-0. [DOI] [PubMed] [Google Scholar]
- Benoit M., Klaus D.M. Microgravity, bacteria, and the influence of motility. Adv. Space Res. 2007;39:1225–1232. [Google Scholar]
- Benoit M.R., Li W., Stodieck L.S., Lam K.S., Winther C.L., Roane T.M., Klaus D.M. Microbial antibiotic production aboard the international space station. Appl. Microbiol. Biotechnol. 2006;70:403–411. doi: 10.1007/s00253-005-0098-3. [DOI] [PubMed] [Google Scholar]
- Beuls E., Van Houdt R., Leys N., Dijkstra C., Larkin O., Mahillon J. Bacillus thuringiensis conjugation in simulated microgravity. Astrobiology. 2009;9:797–805. doi: 10.1089/ast.2009.0383. [DOI] [PubMed] [Google Scholar]
- Bijlani S., Singh N.K., Eedara V.V.R., Podile A.R., Mason C.E., Wang C.C.C., Venkateswaran K. Methylobacterium ajmalii sp. nov., isolated from the international space station. Front. Microbiol. 2021;12:639396. doi: 10.3389/fmicb.2021.639396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachowicz A., Chiang A.J., Elsaesser A., Kalkum M., Ehrenfreund P., Stajich J.E., Torok T., Wang C.C.C., Venkateswaran K. Proteomic and metabolomic characteristics of Extremophilic fungi under simulated Mars conditions. Front. Microbiol. 2019;10:1013. doi: 10.3389/fmicb.2019.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachowicz A., Chiang A.J., Romsdahl J., Kalkum M., Wang C.C.C., Venkateswaran K. Proteomic characterization of Aspergillus fumigatus isolated from air and surfaces of the International Space Station. Fungal Genet. Biol. 2019;124:39–46. doi: 10.1016/j.fgb.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A.G., van Loon J.J.W.A. Technology and developments for the random positioning machine. RPM. Microgravity Sci. Technology. 2009;21:287–292. [Google Scholar]
- Briegleb W. Some qualitative and quantitative aspects of the fast-rotating clinostat as a research tool. ASGSB Bull. 1992;5:23–30. [PubMed] [Google Scholar]
- Brown L.R., Fromme W.J., Handler S.F., Wheatcroft M.G., Johnston D.A. Effect of Skylab missions on clinical and microbiologic aspects of oral health. J. Am. Dent Assoc. 1976;93:357–363. doi: 10.14219/jada.archive.1976.0502. [DOI] [PubMed] [Google Scholar]
- Brown L.A., Arterburn L.M., Miller A.P., Cowger N.L., Hartley S.M., Andrews A., Silber P.M., Li A.P. Maintenance of liver functions in rat hepatocytes cultured as spheroids in a rotating wall vessel. Vitro Cell Dev Biol Anim. 2003;39:13–20. doi: 10.1290/1543-706X(2003)039<0013:MOLFIR>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bucker H., Horneck G. Studies on the effects of cosmic HZEparticles in different biological systems in the Biostack experiments I and II, flown on board of Apollo 16 and 17. In: Nygaard O.F., Adler H.I., Sinclair W.K., editors. Radiation Research. Academic Press; 1975. pp. 1138–1151. [Google Scholar]
- Cadet J., Anselmino C., Douki T., Voituriez L. Photochemistry of nucleic acids in cells. J. Photochem. Photobiol. B. 1992;15:277–298. doi: 10.1016/1011-1344(92)85135-h. [DOI] [PubMed] [Google Scholar]
- Carillo P., Morrone B., Fusco G.M., De Pascale S., Rouphael Y. Challenges for a sustainable food production system on board of the international space station: a Technical review. Agronomy. 2020;10:687. [Google Scholar]
- Castro V.A., Thrasher A.N., Healy M., Ott C.M., Pierson D.L. Microbial characterization during the early habitation of the international space station. Microb. Ecol. 2004;47:119–126. doi: 10.1007/s00248-003-1030-y. [DOI] [PubMed] [Google Scholar]
- Castro S.L., Nelman-Gonzalez M., Nickerson C.A., Ott C.M. Induction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl. Environ. Microbiol. 2011;77:6368–6378. doi: 10.1128/AEM.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro S.L., Smith D.J., Ott C.M. NASA ISS Program Science Office; 2013. A Researcher's Guide to International Space Station: Microbial Research. [Google Scholar]
- Castro-Wallace S.L., Chiu C.Y., John K.K., Stahl S.E., Rubins K.H., McIntyre A.B.R., Dworkin J.P., Lupisella M.L., Smith D.J., Botkin D.J. Nanopore DNA sequencing and genome assembly on the international space station. Sci. Rep. 2017;7:18022. doi: 10.1038/s41598-017-18364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.Y., Kiechle M., Manivasakam P., Schiestl R.H. Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:5051–5059. doi: 10.1093/nar/gkm442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.T., Hughes-Fulford M. Molecular mechanisms underlying the enhanced functions of three-dimensional hepatocyte aggregates. Biomaterials. 2014;35:2162–2171. doi: 10.1016/j.biomaterials.2013.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checinska A., Probst A.J., Vaishampayan P., White J.R., Kumar D., Stepanov V.G., Fox G.E., Nilsson H.R., Pierson D.L., Perry J. Microbiomes of the dust particles collected from the international space station and spacecraft assembly facilities. Microbiome. 2015;3:50. doi: 10.1186/s40168-015-0116-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checinska Sielaff A., Singh N.K., Allen J.E., Thissen J., Jaing C., Venkateswaran K. Draft genome sequences of biosafety level 2 opportunistic pathogens isolated from the environmental surfaces of the international space station. Genome Announc. 2016;4:e01263-16. doi: 10.1128/genomeA.01263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checinska Sielaff A., Kumar R.M., Pal D., Mayilraj S., Venkateswaran K. Solibacillus kalamii sp. nov., isolated from a high-efficiency particulate arrestance filter system used in the International Space Station. Int. J. Syst. Evol. Microbiol. 2017;67:896–901. doi: 10.1099/ijsem.0.001706. [DOI] [PubMed] [Google Scholar]
- Checinska Sielaff A., Urbaniak C., Mohan G.B.M., Stepanov V.G., Tran Q., Wood J.M., Minich J., McDonald D., Mayer T., Knight R. Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces. Microbiome. 2019;7:50. doi: 10.1186/s40168-019-0666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman C.B., Gonzalez-Villalobos R.A., Allen P.L., Johanson K., Guevorkian K., Valles J.M., Hammond T.G. Diamagnetic levitation changes growth, cell cycle, and gene expression of Saccharomyces cerevisiae. Biotechnol. Bioeng. 2007;98:854–863. doi: 10.1002/bit.21526. [DOI] [PubMed] [Google Scholar]
- Cortesao M., de Haas A., Unterbusch R., Fujimori A., Schutze T., Meyer V., Moeller R. Aspergillus niger spores are highly resistant to space radiation. Front. Microbiol. 2020;11:560. doi: 10.3389/fmicb.2020.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortesão M., Schütze T., Marx R., Moeller R., Meyer V. Fungal biotechnology in space: Why and how? In: Nevalainen H., editor. Grand Challenges in Fungal Biotechnology. Springer; 2020. pp. 501–535. [Google Scholar]
- Crabbe A., Pycke B., Van Houdt R., Monsieurs P., Nickerson C., Leys N., Cornelis P. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 2010;12:1545–1564. doi: 10.1111/j.1462-2920.2010.02184.x. [DOI] [PubMed] [Google Scholar]
- Crabbe A., Nielsen-Preiss S.M., Woolley C.M., Barrila J., Buchanan K., McCracken J., Inglis D.O., Searles S.C., Nelman-Gonzalez M.A., Ott C.M. Spaceflight enhances cell aggregation and random budding in Candida albicans. PLoS One. 2013;8:e80677. doi: 10.1371/journal.pone.0080677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadachova E., Casadevall A. Ionizing radiation: how fungi cope, adapt, and exploit with the help of melanin. Curr. Opin. Microbiol. 2008;11:525–531. doi: 10.1016/j.mib.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever P., Mergeay M., Ilyin V., Forget-Hanus D., Van der Auwera G., Mahillon J. Conjugation-mediated plasmid exchange between bacteria grown under space flight conditions. Microgravity Sci. Technol. 2007;19:138–144. [Google Scholar]
- Decelle J.G., Taylor G.R. Autoflora in the upper respiratory tract of Apollo astronauts. Appl. Environ. Microbiol. 1976;32:659–665. doi: 10.1128/aem.32.5.659-665.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain A.L., Fang A. Secondary metabolism in simulated microgravity. Chem. Rec. 2001;1:333–346. doi: 10.1002/tcr.1018. [DOI] [PubMed] [Google Scholar]
- Dickson K.J. Summary of biological spaceflight experiments with cells. ASGSB Bull. 1991;4:151–260. [PubMed] [Google Scholar]
- Ding Y., Davis B.M., Waldor M.K. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- La Duc M.T., Sumner R., Pierson D., Venkat P., Venkateswaran K. Evidence of pathogenic microbes in the International Space Station drinking water: reason for concern? Habitation (Elmsford) 2004;10:39–48. doi: 10.3727/154296604774808883. [DOI] [PubMed] [Google Scholar]
- Emiola A., Oh J. High throughput in situ metagenomic measurement of bacterial replication at ultra-low sequencing coverage. Nat. Commun. 2018;9:4956. doi: 10.1038/s41467-018-07240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England L.S., Gorzelak M., Trevors J.T. Growth and membrane polarization in Pseudomonas aeruginosa UG2 grown in randomized microgravity in a high aspect ratio vessel. Biochim. Biophys. Acta. 2003;1624:76–80. doi: 10.1016/j.bbagen.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Fang A., Pierson D.L., Mishra S.K., Koenig D.W., Demain A.L. Gramicidin S production by Bacillus brevis in simulated microgravity. Curr. Microbiol. 1997;34:199–204. doi: 10.1007/s002849900168. [DOI] [PubMed] [Google Scholar]
- Fang A., Pierson D.L., Mishra S.K., Koenig D.W., Demain A.L. Secondary metabolism in simulated microgravity: beta-lactam production by Streptomyces clavuligerus. J. Ind. Microbiol. Biotechnol. 1997;18:22–25. doi: 10.1038/sj.jim.2900345. [DOI] [PubMed] [Google Scholar]
- Fang A., Pierson D.L., Mishra S.K., Demain A.L. Growth of Steptomyces hygroscopicus in rotating-wall bioreactor under simulated microgravity inhibits rapamycin production. Appl. Microbiol. Biotechnol. 2000;54:33–36. doi: 10.1007/s002539900303. [DOI] [PubMed] [Google Scholar]
- Freed L.E., Vunjak-Novakovic G. Microgravity tissue engineering. Vitro Cell Dev Biol Anim. 1997;33:381–385. doi: 10.1007/s11626-997-0009-2. [DOI] [PubMed] [Google Scholar]
- Freed L.E., Langer R., Martin I., Pellis N.R., Vunjak-Novakovic G. Tissue engineering of cartilage in space. Proc. Natl. Acad. Sci. U S A. 1997;94:13885–13890. doi: 10.1073/pnas.94.25.13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Bakelman F.E., Darshi M., Green S.J., Gur R.C., Lin L., Macias B.R., McKenna M.J., Meydan C., Mishra T., Nasrini J. The NASA Twins Study: a multidimensional analysis of a year-long human spaceflight. Science. 2019;364:eaau8650. doi: 10.1126/science.aau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier T., Wang X., Sifuentes Dos Santos J., Fysikopoulos A., Tadrist S., Canlet C., Artigot M.P., Loiseau N., Oswald I.P., Puel O. Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS One. 2012;7:e29906. doi: 10.1371/journal.pone.0029906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gelder J., Vandenabeele P., De Boever P., Mergeay M., Moens L., De Vos P. Raman Spectroscopic analysis of Cupriavidus metallidurans LMG 1195 (CH34) cultured in low-shear microgravity conditions. Microgravity Sci. Technol. 2009;21:217–223. [Google Scholar]
- Gilbert R., Torres M., Clemens R., Hateley S., Hosamani R., Wade W., Bhattacharya S. Spaceflight and simulated microgravity conditions increase virulence of Serratia marcescens in the Drosophila melanogaster infection model. NPJ Microgravity. 2020;6:4. doi: 10.1038/s41526-019-0091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Pietsch J., Wehland M., Richter P., Strauch S.M., Lebert M., Magnusson N.E., Wise P., Bauer J. The impact of microgravity-based proteomics research. Expert Rev. Proteomics. 2014;11:465–476. doi: 10.1586/14789450.2014.926221. [DOI] [PubMed] [Google Scholar]
- Guevorkian K., Valles J.M., Jr. Swimming Paramecium in magnetically simulated enhanced, reduced, and inverted gravity environments. Proc. Natl. Acad. Sci. U S A. 2006;103:13051–13056. doi: 10.1073/pnas.0601839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.G., Hammond J.M. Optimized suspension culture: the rotating-wall vessel. Am. J. Physiol. Ren. Physiol. 2001;281:F12–F25. doi: 10.1152/ajprenal.2001.281.1.F12. [DOI] [PubMed] [Google Scholar]
- Hammond T.G., Benes E., O'Reilly K.C., Wolf D.A., Linnehan R.M., Taher A., Kaysen J.H., Allen P.L., Goodwin T.J. Mechanical culture conditions effect gene expression: gravity-induced changes on the space shuttle. Physiol. Genomics. 2000;3:163–173. doi: 10.1152/physiolgenomics.2000.3.3.163. [DOI] [PubMed] [Google Scholar]
- Hammond T., Allen P., Birdsall H. Is there a space-based technology Solution to problems with preclinical drug Toxicity testing? Pharm. Res. 2016;33:1545–1551. doi: 10.1007/s11095-016-1942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z., Li L., Fu Y., Liu H. The influence of bioregenerative life-support system dietary structure and lifestyle on the gut microbiota: a 105-day ground-based space simulation in Lunar Palace 1. Environ. Microbiol. 2018;20:3643–3656. doi: 10.1111/1462-2920.14358. [DOI] [PubMed] [Google Scholar]
- Herranz R., Anken R., Boonstra J., Braun M., Christianen P.C., de Geest M., Hauslage J., Hilbig R., Hill R.J., Lebert M. Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiology. 2013;13:1–17. doi: 10.1089/ast.2012.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyde K.C., Ruder W.C. Exploring host-microbiome interactions using an in silico model of biomimetic robots and engineered living cells. Sci. Rep. 2015;5:11988. doi: 10.1038/srep11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginson E.E., Galen J.E., Levine M.M., Tennant S.M. Microgravity as a biological tool to examine host-pathogen interactions and to guide development of therapeutics and preventatives that target pathogenic bacteria. Pathog. Dis. 2016;74:ftw095. doi: 10.1093/femspd/ftw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horneck G. The biostack concept and its application in space and at accelerators: studies on Bacillus subtilis spores. In: Swenberg C.E., Horneck G., Stassinopoulos E.G., editors. Biological Effects and Physics of Solar and Galactic Cosmic Radiation. Springer; 1993. pp. 99–115. [Google Scholar]
- Horneck G., Zell M. Introduction to the EXPOSE-E mission. Astrobiology. 2012;12:373. doi: 10.1089/ast.2012.0831. [DOI] [PubMed] [Google Scholar]
- Horneck G., Klaus D.M., Mancinelli R.L. Space microbiology. Microbiol. Mol. Biol. Rev. 2010;74:121–156. doi: 10.1128/MMBR.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoson T., Kamisaka S., Masuda Y., Yamashita M., Buchen B. Evaluation of the three-dimensional clinostat as a simulator of weightlessness. Planta. 1997;203(Suppl):S187–S197. doi: 10.1007/pl00008108. [DOI] [PubMed] [Google Scholar]
- Hotchin J., Lorenz P., Hemenway C. The survival of terrestrial microorganisms in space at orbital altitudes during Gemini satellite experiments. Life Sci. Space Res. 1968;6:108–114. [PubMed] [Google Scholar]
- Huang B., Li D.G., Huang Y., Liu C.T. Effects of spaceflight and simulated microgravity on microbial growth and secondary metabolism. Mil. Med. Res. 2018;5:18. doi: 10.1186/s40779-018-0162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema C., Beaudette L.A., Trevors J.T. Simulated microgravity (SMG) and bacteria. Riv Biol. 2002;95:497–503. [PubMed] [Google Scholar]
- Ichijo T., Yamaguchi N., Tanigaki F., Shirakawa M., Nasu M. Four-year bacterial monitoring in the international space station-Japanese experiment module "Kibo" with culture-independent approach. NPJ Microgravity. 2016;2:16007. doi: 10.1038/npjmgrav.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson K., Allen P.L., Gonzalez-Villalobos R.A., Baker C.B., D'Elia R., Hammond T.G. Gene expression and survival changes in Saccharomyces cerevisiae during suspension culture. Biotechnol. Bioeng. 2006;93:1050–1059. doi: 10.1002/bit.20810. [DOI] [PubMed] [Google Scholar]
- Juergensmeyer M.A., Juergensmeyer E.A., Guikema J.A. Long-term exposure to spaceflight conditions affects bacterial response to antibiotics. Microgravity Sci. Technol. 1999;12:41–47. [PubMed] [Google Scholar]
- Kacena M.A., Todd P. Gentamicin: effect on E. coli in space. Microgravity Sci. Technol. 1999;12:135–137. [PubMed] [Google Scholar]
- Kacena M.A., Merrell G.A., Manfredi B., Smith E.E., Klaus D.M., Todd P. Bacterial growth in space flight: logistic growth curve parameters for Escherichia coli and Bacillus subtilis. Appl. Microbiol. Biotechnol. 1999;51:229–234. doi: 10.1007/s002530051386. [DOI] [PubMed] [Google Scholar]
- Karouia F., Peyvan K., Pohorille A. Toward biotechnology in space: high-throughput instruments for in situ biological research beyond Earth. Biotechnol. Adv. 2017;35:905–932. doi: 10.1016/j.biotechadv.2017.04.003. [DOI] [PubMed] [Google Scholar]
- Kim W., Tengra F.K., Young Z., Shong J., Marchand N., Chan H.K., Pangule R.C., Parra M., Dordick J.S., Plawsky J.L. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS One. 2013;8:e62437. doi: 10.1371/journal.pone.0062437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Rusek A., Cucinotta F.A. Issues for simulation of galactic cosmic ray exposures for Radiobiological research at ground-based accelerators. Front. Oncol. 2015;5:122. doi: 10.3389/fonc.2015.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus D.M. Clinostats and bioreactors. Gravit. Space Biol. Bull. 2001;14:55–64. [PubMed] [Google Scholar]
- Klaus D.M. Gravitational influence on biomolecular engineering processes. Gravit. Space Biol. 2004;17:51–66. [Google Scholar]
- Klaus D.M., Howard H.N. Antibiotic efficacy and microbial virulence during space flight. Trends Biotechnol. 2006;24:131–136. doi: 10.1016/j.tibtech.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Klaus D.M., Todd P., Schatz A. Functional weightlessness during clinorotation of cell suspensions. Adv. Space Res. 1998;21:1315–1318. doi: 10.1016/s0273-1177(97)00404-3. [DOI] [PubMed] [Google Scholar]
- Klaus D.M., Benoit M.R., Nelson E.S., Hammond T.G. Extracellular mass transport considerations for space flight research concerning suspended and adherent in vitro cell cultures. J. Gravit. Physiol. 2004;11:17–27. [PubMed] [Google Scholar]
- Klintworth R., Reher H.J., Viktorov A.N., Bohle D. Biological induced corrosion of materials II: new test methods and experiences from MIR station. Acta Astronaut. 1999;44:569–578. doi: 10.1016/s0094-5765(99)00069-7. [DOI] [PubMed] [Google Scholar]
- Knox B.P., Blachowicz A., Palmer J.M., Romsdahl J., Huttenlocher A., Wang C.C., Keller N.P., Venkateswaran K. Characterization of Aspergillus fumigatus isolates from air and surfaces of the international space station. mSphere. 2016;1 doi: 10.1128/mSphere.00227-16. e00227–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig D.W., Pierson D.L. Microbiology of the space shuttle water system. Water Sci. Technol. 1997;35:59–64. [PubMed] [Google Scholar]
- Kuznetsov O.A., Hasenstein K.H. Intracellular magnetophoresis of amyloplasts and induction of root curvature. Planta. 1996;198:87–94. doi: 10.1007/BF00197590. [DOI] [PubMed] [Google Scholar]
- Lam K.S., Mamber S.W., Pack E.J., Forenza S., Fernandes P.B., Klaus D.M. The effects of space flight on the production of monorden by Humicola fuscoatra WC5157 in solid-state fermentation. Appl. Microbiol. Biotechnol. 1998;49:579–583. doi: 10.1007/s002530051216. [DOI] [PubMed] [Google Scholar]
- Lam K.S., Gustavson D.R., Pirnik D.L., Pack E., Bulanhagui C., Mamber S.W., Forenza S., Stodieck L.S., Klaus D.M. The effect of space flight on the production of actinomycin D by Streptomyces plicatus. J. Ind. Microbiol. Biotechnol. 2002;29:299–302. doi: 10.1038/sj.jim.7000312. [DOI] [PubMed] [Google Scholar]
- Lang J.M., Coil D.A., Neches R.Y., Brown W.E., Cavalier D., Severance M., Hampton-Marcell J.T., Gilbert J.A., Eisen J.A. A microbial survey of the international space station (ISS) PeerJ. 2017;5:e4029. doi: 10.7717/peerj.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapchine L., Moatti N., Gasset G., Richoilley G., Templier J., Tixador R. Antibiotic activity in space. Drugs Exp. Clin. Res. 1986;12:933–938. [PubMed] [Google Scholar]
- Lawal A., Kirtley M.L., van Lier C.J., Erova T.E., Kozlova E.V., Sha J., Chopra A.K., Rosenzweig J.A. The effects of modeled microgravity on growth kinetics, antibiotic susceptibility, cold growth, and the virulence potential of a Yersinia pestis ymoA-deficient mutant and its isogenic parental strain. Astrobiology. 2013;13:821–832. doi: 10.1089/ast.2013.0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencner A.A., Lencner C.P., Mikelsaar M.E., Tjuri M.E., Toom M.A., Väljaots M.E., Silov V.M., Liz'ko N.N., Legenkov V.I., Reznikov I.M. The quantitative composition of the intestinal lactoflora before and after space flights of different lengths. Nahrung. 1984;28:607–613. doi: 10.1002/food.19840280608. [DOI] [PubMed] [Google Scholar]
- Lesnyak A., Sonnenfeld G., Avery L., Konstantinova I., Rykova M., Meshkov D., Orlova T. Effect of SLS-2 spaceflight on immunologic parameters of rats. J. Appl. Physiol. 1996;81:178–182. doi: 10.1152/jappl.1996.81.1.178. [DOI] [PubMed] [Google Scholar]
- Lindeboom R.E.F., Paepe J.D., Vanoppen M., Alonso-Fariñas B., Coessens W., Alloul A., Christiaens M.E.R., Dotremont C., Beckers H., Lamaze B. A five-stage treatment train for water recovery from urine and shower water for long-term human Space missions. Desalination. 2020;495:114634. [Google Scholar]
- Liu Y.M., Zhu D.M., Strayer D.M., Israelsson U.E. Magnetic levitation of large water droplets and mice. Adv. Space Res. 2010;45:208–213. [Google Scholar]
- Liu M., Gao H., Shang P., Zhou X., Ashforth E., Zhuo Y., Chen D., Ren B., Liu Z., Zhang L. Magnetic field is the dominant factor to induce the response of Streptomyces avermitilis in altered gravity simulated by diamagnetic levitation. PLoS One. 2011;6:e24697. doi: 10.1371/journal.pone.0024697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd K.G., Steen A.D., Ladau J., Yin J., Crosby L. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems. 2018;3 doi: 10.1128/mSystems.00055-18. e00055–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon J.J.W.A. Some history and use of the random positioning machine, RPM, in gravity related research. Adv. Space Res. 2007;39:1161–1165. [Google Scholar]
- Lynch S.V., Brodie E.L., Matin A. Role and regulation of sigma S in general resistance conferred by low-shear simulated microgravity in Escherichia coli. J. Bacteriol. 2004;186:8207–8212. doi: 10.1128/JB.186.24.8207-8212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch S.V., Mukundakrishnan K., Benoit M.R., Ayyaswamy P.S., Matin A. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl. Environ. Microbiol. 2006;72:7701–7710. doi: 10.1128/AEM.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauclaire L., Egli M. Effect of simulated microgravity on growth and production of exopolymeric substances of Micrococcus luteus space and earth isolates. FEMS Immunol. Med. Microbiol. 2010;59:350–356. doi: 10.1111/j.1574-695X.2010.00683.x. [DOI] [PubMed] [Google Scholar]
- McGinn S., Gut I.G. DNA sequencing - spanning the generations. N. Biotechnol. 2013;30:366–372. doi: 10.1016/j.nbt.2012.11.012. [DOI] [PubMed] [Google Scholar]
- McIntyre A.B.R., Rizzardi L., Yu A.M., Alexander N., Rosen G.L., Botkin D.J., Stahl S.E., John K.K., Castro-Wallace S.L., McGrath K. Nanopore sequencing in microgravity. NPJ Microgravity. 2016;2:16035. doi: 10.1038/npjmgrav.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre A.B.R., Alexander N., Grigorev K., Bezdan D., Sichtig H., Chiu C.Y., Mason C.E. Single-molecule sequencing detection of N6-methyladenine in microbial reference materials. Nat. Commun. 2019;10:579. doi: 10.1038/s41467-019-08289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean R.J., Cassanto J.M., Barnes M.B., Koo J.H. Bacterial biofilm formation under microgravity conditions. FEMS Microbiol. Lett. 2001;195:115–119. doi: 10.1111/j.1574-6968.2001.tb10507.x. [DOI] [PubMed] [Google Scholar]
- Mehta S.K., Laudenslager M.L., Stowe R.P., Crucian B.E., Feiveson A.H., Sams C.F., Pierson D.L. Latent virus reactivation in astronauts on the international space station. NPJ Microgravity. 2017;3:11. doi: 10.1038/s41526-017-0015-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micke U., Horneck G., Kozubek S. Double strand breaks in the DNA of Bacillus subtilis cells irradiated by heavy ions. Adv. Space Res. 1994;14:207–211. doi: 10.1016/0273-1177(94)90469-3. [DOI] [PubMed] [Google Scholar]
- Mora M., Perras A., Alekhova T.A., Wink L., Krause R., Aleksandrova A., Novozhilova T., Moissl-Eichinger C. Resilient microorganisms in dust samples of the International Space Station-survival of the adaptation specialists. Microbiome. 2016;4:65. doi: 10.1186/s40168-016-0217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M., Wink L., Kogler I., Mahnert A., Rettberg P., Schwendner P., Demets R., Cockell C., Alekhova T., Klingl A. Space Station conditions are selective but do not alter microbial characteristics relevant to human health. Nat. Commun. 2019;10:3990. doi: 10.1038/s41467-019-11682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mulders S.E., Stassen C., Daenen L., Devreese B., Siewers V., van Eijsden R.G., Nielsen J., Delvaux F.R., Willaert R. The influence of microgravity on invasive growth in Saccharomyces cerevisiae. Astrobiology. 2011;11:45–55. doi: 10.1089/ast.2010.0518. [DOI] [PubMed] [Google Scholar]
- NASA . 2016. Space Biology Science Plan 2016-2025. [Google Scholar]
- National Academies of Sciences, E., and Medicine . The National Academies Press; 2018. A Midterm Assessment of Implementation of the Decadal Survey on Life and Physical Sciences Research at NASA. [PubMed] [Google Scholar]
- Newcombe D.A., Schuerger A.C., Benardini J.N., Dickinson D., Tanner R., Venkateswaran K. Survival of spacecraft-associated microorganisms under simulated martian UV irradiation. Appl. Environ. Microbiol. 2005;71:8147–8156. doi: 10.1128/AEM.71.12.8147-8156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W.L., Munakata N., Horneck G., Melosh H.J., Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson C.A., Ott C.M., Mister S.J., Morrow B.J., Burns-Keliher L., Pierson D.L. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect Immun. 2000;68:3147–3152. doi: 10.1128/iai.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson C.A., Ott C.M., Wilson J.W., Ramamurthy R., Pierson D.L. Microbial responses to microgravity and other low-shear environments. Microbiol. Mol. Biol. Rev. 2004;68:345–361. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury J.W., Schimmerling W., Slaba T.C., Azzam E.I., Badavi F.F., Baiocco G., Benton E., Bindi V., Blakely E.A., Blattnig S.R. Galactic cosmic ray simulation at the NASA space radiation laboratory. Life Sci. Space Res. (Amst) 2016;8:38–51. doi: 10.1016/j.lssr.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova N.D. Review of the knowledge of microbial contamination of the Russian manned spacecraft. Microb. Ecol. 2004;47:127–132. doi: 10.1007/s00248-003-1055-2. [DOI] [PubMed] [Google Scholar]
- Novikova N., De Boever P., Poddubko S., Deshevaya E., Polikarpov N., Rakova N., Coninx I., Mergeay M. Survey of environmental biocontamination on board the international space station. Res. Microbiol. 2006;157:5–12. doi: 10.1016/j.resmic.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Ohnishi T. The biological effects of space radiation during long stays in space. Biol. Sci. Space. 2004;18:201–205. doi: 10.2187/bss.18.201. [DOI] [PubMed] [Google Scholar]
- Orsini S.S., Lewis A.M., Rice K.C. Investigation of simulated microgravity effects on Streptococcus mutans physiology and global gene expression. NPJ Microgravity. 2017;3:4. doi: 10.1038/s41526-016-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C.M., Bruce R.J., Pierson D.L. Microbial characterization of free floating condensate aboard the Mir space station. Microb. Ecol. 2004;47:133–136. doi: 10.1007/s00248-003-1038-3. [DOI] [PubMed] [Google Scholar]
- Pierson D.L. Microbial contamination of spacecraft. Gravit. Space Biol. Bull. 2001;14:1–6. [PubMed] [Google Scholar]
- Purevdorj-Gage B., Sheehan K.B., Hyman L.E. Effects of low-shear modeled microgravity on cell function, gene expression, and phenotype in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006;72:4569–4575. doi: 10.1128/AEM.03050-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbow E., Rettberg P., Barczyk S., Bohmeier M., Parpart A., Panitz C., Horneck G., von Heise-Rotenburg R., Hoppenbrouwers T., Willnecker R. EXPOSE-E: an ESA astrobiology mission 1.5 years in space. Astrobiology. 2012;12:374–386. doi: 10.1089/ast.2011.0760. [DOI] [PubMed] [Google Scholar]
- Roberts M.S., Garland J.L., Mills A.L. Microbial astronauts: assembling microbial communities for advanced life support systems. Microb. Ecol. 2004;47:137–149. doi: 10.1007/s00248-003-1060-5. [DOI] [PubMed] [Google Scholar]
- Rogers E. NASA Technical Memorandum; 1986. The Ecology of Microorganisms in a Small Closed System: Potential Benefits and Problems for Space Station. [Google Scholar]
- Romsdahl J., Blachowicz A., Chiang A.J., Singh N., Stajich J.E., Kalkum M., Venkateswaran K., Wang C.C.C. Characterization of Aspergillus Niger isolated from the International Space Station. mSystems. 2018;3 doi: 10.1128/mSystems.00112-18. e00112–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romsdahl J., Blachowicz A., Chiang A.J., Chiang Y.M., Masonjones S., Yaegashi J., Countryman S., Karouia F., Kalkum M., Stajich J.E. International Space Station conditions alter genomics, proteomics, and metabolomics in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2019;103:1363–1377. doi: 10.1007/s00253-018-9525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romsdahl J., Blachowicz A., Chiang Y.M., Venkateswaran K., Wang C.C.C. Metabolomic analysis of Aspergillus niger isolated from the international space station reveals enhanced production levels of the antioxidant pyranonigrin A. Front. Microbiol. 2020;11:931. doi: 10.3389/fmicb.2020.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney B.V., Crucian B.E., Pierson D.L., Laudenslager M.L., Mehta S.K. Herpes virus reactivation in astronauts during spaceflight and its application on earth. Front. Microbiol. 2019;10:16. doi: 10.3389/fmicb.2019.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig J.A., Abogunde O., Thomas K., Lawal A., Nguyen Y.U., Sodipe A., Jejelowo O. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl. Microbiol. Biotechnol. 2010;85:885–891. doi: 10.1007/s00253-009-2237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer M., Schmitz C., Bucker H. Heavy ion induced DNA double strand breaks in cells of E. coli. Adv. Space Res. 1994;14:203–206. doi: 10.1016/0273-1177(94)90468-5. [DOI] [PubMed] [Google Scholar]
- Schiwon K., Arends K., Rogowski K.M., Furch S., Prescha K., Sakinc T., Van Houdt R., Werner G., Grohmann E. Comparison of antibiotic resistance, biofilm formation and conjugative transfer of Staphylococcus and Enterococcus isolates from international space station and antarctic research station Concordia. Microb. Ecol. 2013;65:638–651. doi: 10.1007/s00248-013-0193-4. [DOI] [PubMed] [Google Scholar]
- Schmidt M.A., Goodwin T.J. Personalized medicine in human space flight: using Omics based analyses to develop individualized countermeasures that enhance astronaut safety and performance. Metabolomics. 2013;9:1134–1156. doi: 10.1007/s11306-013-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.A., Goodwin T.J., Pelligra R. Incorporation of omics analyses into artificial gravity research for space exploration countermeasure development. Metabolomics. 2016;12:36. doi: 10.1007/s11306-015-0942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuergera A.C., Richards J.T., Newcombe D.A., Venkateswaran K. Rapid inactivation of seven Bacillus spp. under simulated Mars UV irradiation. Icarus. 2006;181:52–62. [Google Scholar]
- Schultz J., Taylor R., Flanagan D., Gibbons R., Brown H., Sauer R., Pierson D. biofilm formation and control in a simulated spacecraft water system: Interim results. SAE Trans. 1989;98:964–973. [Google Scholar]
- Schwarz R.P., Goodwin T.J., Wolf D.A. Cell culture for three-dimensional modeling in rotating-wall vessels: an application of simulated microgravity. J. Tissue Cult. Methods. 1992;14:51–57. doi: 10.1007/BF01404744. [DOI] [PubMed] [Google Scholar]
- Searles S.C., Woolley C.M., Petersen R.A., Hyman L.E., Nielsen-Preiss S.M. Modeled microgravity increases filamentation, biofilm formation, phenotypic switching, and antimicrobial resistance in Candida albicans. Astrobiology. 2011;11:825–836. doi: 10.1089/ast.2011.0664. [DOI] [PubMed] [Google Scholar]
- Senatore G., Mastroleo F., Leys N., Mauriello G. Effect of microgravity & space radiation on microbes. Future Microbiol. 2018;13:831–847. doi: 10.2217/fmb-2017-0251. [DOI] [PubMed] [Google Scholar]
- Sheehan K.B., McInnerney K., Purevdorj-Gage B., Altenburg S.D., Hyman L.E. Yeast genomic expression patterns in response to low-shear modeled microgravity. BMC Genomics. 2007;8:3. doi: 10.1186/1471-2164-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui R., Akbar N., Khan N.A. Gut microbiome and human health under the space environment. J. Appl. Microbiol. 2021;130:14–24. doi: 10.1111/jam.14789. [DOI] [PubMed] [Google Scholar]
- Singh N.K., Blachowicz A., Romsdahl J., Wang C., Torok T., Venkateswaran K. Draft genome sequences of several fungal strains selected for exposure to microgravity at the international space station. Genome Announc. 2017;5 doi: 10.1128/genomeA.01602-16. e01602–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.K., Bezdan D., Checinska Sielaff A., Wheeler K., Mason C.E., Venkateswaran K. Multi-drug resistant Enterobacter bugandensis species isolated from the International Space Station and comparative genomic analyses with human pathogenic strains. BMC Microbiol. 2018;18:175. doi: 10.1186/s12866-018-1325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.K., Wood J.M., Karouia F., Venkateswaran K. Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome. 2018;6:204. doi: 10.1186/s40168-018-0585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.K., Wood J.M., Mhatre S.S., Venkateswaran K. Metagenome to phenome approach enables isolation and genomics characterization of Kalamiella piersonii gen. nov., sp. nov. from the International Space Station. Appl. Microbiol. Biotechnol. 2019;103:4483–4497. doi: 10.1007/s00253-019-09813-z. [DOI] [PubMed] [Google Scholar]
- Sittka A., Pfeiffer V., Tedin K., Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobisch L.Y., Rogowski K.M., Fuchs J., Schmieder W., Vaishampayan A., Oles P., Novikova N., Grohmann E. Biofilm forming antibiotic resistant gram-positive pathogens isolated from surfaces on the international space station. Front. Microbiol. 2019;10:543. doi: 10.3389/fmicb.2019.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Leff L.G. Identification and characterization of bacterial isolates from the Mir space station. Microbiol. Res. 2005;160:111–117. doi: 10.1016/j.micres.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Sonnleitner E., Hagens S., Rosenau F., Wilhelm S., Habel A., Jager K.E., Blasi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Sugita T., Yamazaki T., Makimura K., Cho O., Yamada S., Ohshima H., Mukai C. Comprehensive analysis of the skin fungal microbiota of astronauts during a half-year stay at the International Space Station. Med. Mycol. 2016;54:232–239. doi: 10.1093/mmy/myv121. [DOI] [PubMed] [Google Scholar]
- Taylor G.R. Space microbiology. Annu. Rev. Microbiol. 1974;28:121–137. doi: 10.1146/annurev.mi.28.100174.001005. [DOI] [PubMed] [Google Scholar]
- Taylor G.R., Graves R.C., Brockett R.M., Ferguson J.K., Mieszkuc B.J. NASA Johnson Space Center; 1971. Skylab Environmental and Crew Microbiology Studies. [Google Scholar]
- Taylor G.R., Konstantinova I., Sonnenfeld G., Jennings R. Changes in the immune system during and after spaceflight. Adv. Space Biol. Med. 1997;6:1–32. doi: 10.1016/s1569-2574(08)60076-3. [DOI] [PubMed] [Google Scholar]
- Taylor K., Kleinhesselink K., George M.D., Morgan R., Smallwood T., Hammonds A.S., Fuller P.M., Saelao P., Alley J., Gibbs A.G. Toll mediated infection response is altered by gravity and spaceflight in Drosophila. PLoS One. 2014;9:e86485. doi: 10.1371/journal.pone.0086485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenet D., D'Ari R., Bouloc P. The SIGNAL experiment in BIORACK: Escherichia coli in microgravity. J. Biotechnol. 1996;47:89–97. doi: 10.1016/0168-1656(96)01384-3. [DOI] [PubMed] [Google Scholar]
- Thomas V., Prasad N., Reddy C. Microgravity research platforms – a study. Curr. Sci. 2000;79:336–340. [Google Scholar]
- Tixador R., Richoilley G., Gasset G., Templier J., Bes J.C., Moatti N., Lapchine L. Study of minimal inhibitory concentration of antibiotics on bacteria cultivated in vitro in space (Cytos 2 experiment) Aviat Space Environ. Med. 1985;56:748–751. [PubMed] [Google Scholar]
- Tucker D.L., Ott C.M., Huff S., Fofanov Y., Pierson D.L., Willson R.C., Fox G.E. Characterization of Escherichia coli MG1655 grown in a low-shear modeled microgravity environment. BMC Microbiol. 2007;7:15. doi: 10.1186/1471-2180-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth B.R., Lelkes P.I. Growing tissues in microgravity. Nat. Med. 1998;4:901–907. doi: 10.1038/nm0898-901. [DOI] [PubMed] [Google Scholar]
- Urbaniak C., Sielaff A.C., Frey K.G., Allen J.E., Singh N., Jaing C., Wheeler K., Venkateswaran K. Detection of antimicrobial resistance genes associated with the International Space Station environmental surfaces. Sci. Rep. 2018;8:814. doi: 10.1038/s41598-017-18506-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., van Dam P., Zaborin A., Zaborina O., Gilbert J.A., Torok T., Wang C.C.C., Venkateswaran K. Genomic characterization and virulence potential of two fusarium oxysporum isolates cultured from the international space station. mSystems. 2019;4 doi: 10.1128/mSystems.00345-18. e00021–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Lorenzi H., Thissen J., Jaing C., Crucian B., Sams C., Pierson D., Venkateswaran K., Mehta S. The influence of spaceflight on the astronaut salivary microbiome and the search for a microbiome biomarker for viral reactivation. Microbiome. 2020;8:56. doi: 10.1186/s40168-020-00830-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Wong S., Tighe S., Arumugam A., Liu B., Parker C.W., Wood J.M., Singh N.K., Skorupa D.J., Peyton B.M. Validating an automated nucleic acid extraction device for omics in space using whole cell microbial reference standards. Front. Microbiol. 2020;11:1909. doi: 10.3389/fmicb.2020.01909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishampayan A., Grohmann E. Multi-resistant biofilm-forming pathogens on the international space station. J. Biosci. 2019;44:125. [PubMed] [Google Scholar]
- Valentin-Hansen P., Eriksen M., Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Venkateswaran K., Vaishampayan P., Cisneros J., Pierson D.L., Rogers S.O., Perry J. International Space Station environmental microbiome - microbial inventories of ISS filter debris. Appl. Microbiol. Biotechnol. 2014;98:6453–6466. doi: 10.1007/s00253-014-5650-6. [DOI] [PubMed] [Google Scholar]
- Venkateswaran K., Singh N.K., Checinska Sielaff A., Pope R.K., Bergman N.H., van Tongeren S.P., Patel N.B., Lawson P.A., Satomi M., Williamson C.H.D. Non-Toxin-producing Bacillus cereus strains belonging to the B. Anthracis clade isolated from the international space station. mSystems. 2017;2 doi: 10.1128/mSystems.00021-17. e00021–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhies A.A., Mark Ott C., Mehta S., Pierson D.L., Crucian B.E., Feiveson A., Oubre C.M., Torralba M., Moncera K., Zhang Y. Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci. Rep. 2019;9:9911. doi: 10.1038/s41598-019-46303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vunjak-Novakovic G., Searby N., De Luis J., Freed L.E. Microgravity studies of cells and tissues. Ann. N. Y Acad. Sci. 2002;974:504–517. doi: 10.1111/j.1749-6632.2002.tb05927.x. [DOI] [PubMed] [Google Scholar]
- Wang H., Yan Y., Rong D., Wang J., Wang H., Liu Z., Wang J., Yang R., Han Y. Increased biofilm formation ability in Klebsiella pneumoniae after short-term exposure to a simulated microgravity environment. Microbiologyopen. 2016;5:793–801. doi: 10.1002/mbo3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li W., Gu L., Gao X., Ni B., Deng H., Yang R., Han Y. Emergence of two distinct subpopulations from Klebsiella pneumoniae grown in the stimulated microgravity environment. Future Microbiol. 2017;12:939–951. doi: 10.2217/fmb-2017-0032. [DOI] [PubMed] [Google Scholar]
- Wilson J.W., Ott C.M., Ramamurthy R., Porwollik S., McClelland M., Pierson D.L., Nickerson C.A. Low-Shear modeled microgravity alters the Salmonella enterica serovar typhimurium stress response in an RpoS-independent manner. Appl. Environ. Microbiol. 2002;68:5408–5416. doi: 10.1128/AEM.68.11.5408-5416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.W., Ramamurthy R., Porwollik S., McClelland M., Hammond T., Allen P., Ott C.M., Pierson D.L., Nickerson C.A. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc. Natl. Acad. Sci. U S A. 2002;99:13807–13812. doi: 10.1073/pnas.212387899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.W., Ott C.M., Honer zu Bentrup K., Ramamurthy R., Quick L., Porwollik S., Cheng P., McClelland M., Tsaprailis G., Radabaugh T. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl. Acad. Sci. U S A. 2007;104:16299–16304. doi: 10.1073/pnas.0707155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Hocking D.M., Cheng C., Dogovski C., Perugini M.A., Holien J.K., Parker M.W., Hartland E.L., Tauschek M., Robins-Browne R.M. Disarming bacterial virulence through chemical inhibition of the DNA binding domain of an AraC-like transcriptional activator protein. J. Biol. Chem. 2013;288:31115–31126. doi: 10.1074/jbc.M113.503912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.-M., Zhong D., Blount P. Chimeras reveal a single lipid-interface residue that controls MscL channel kinetics as well as mechanosensitivity. Cell Rep. 2013;3:520–527. doi: 10.1016/j.celrep.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zea L., Prasad N., Levy S.E., Stodieck L., Jones A., Shrestha S., Klaus D. A molecular genetic basis explaining altered bacterial behavior in space. PLoS One. 2016;11:e0164359. doi: 10.1371/journal.pone.0164359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zea L., Larsen M., Estante F., Qvortrup K., Moeller R., Dias de Oliveira S., Stodieck L., Klaus D. Phenotypic changes exhibited by E. coli cultured in space. Front. Microbiol. 2017;8:1598. doi: 10.3389/fmicb.2017.01598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zea L., Nisar Z., Rubin P., Cortesao M., Luo J., McBride S.A., Moeller R., Klaus D., Muller D., Varanasi K.K. Design of a spaceflight biofilm experiment. Acta Astronaut. 2018;148:294–300. doi: 10.1016/j.actaastro.2018.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova N.N., Tugay T., Dighton J., Zheltonozhsky V., McDermott P. Ionizing radiation attracts soil fungi. Mycol. Res. 2004;108:1089–1096. doi: 10.1017/s0953756204000966. [DOI] [PubMed] [Google Scholar]
- Zimmermann H., Schafer M., Schmitz C., Bucker H. Effects of heavy ions on inactivation and DNA double strand breaks in Deinococcus radiodurans R1. Adv. Space Res. 1994;14:213–216. doi: 10.1016/0273-1177(94)90470-7. [DOI] [PubMed] [Google Scholar]