Preterm labor precedes premature birth, the leading cause of neonatal morbidity and mortality worldwide. Preterm labor can either occur in the context of microbe-associated intra-amniotic inflammation (i.e., intra-amniotic infection) or intra-amniotic inflammation in the absence of detectable microorganisms (i.e., sterile intra-amniotic inflammation).
KEYWORDS: acute histologic chorioamnionitis, alarmin, fetal inflammatory response, inflammasome, intra-amniotic infection, transcriptome
ABSTRACT
Preterm labor precedes premature birth, the leading cause of neonatal morbidity and mortality worldwide. Preterm labor can occur in the context of either microbe-associated intra-amniotic inflammation (i.e., intra-amniotic infection) or intra-amniotic inflammation in the absence of detectable microorganisms (i.e., sterile intra-amniotic inflammation). Both intra-amniotic infection and sterile intra-amniotic inflammation trigger local immune responses that have deleterious effects on fetal life. Yet, the extent of such immune responses in the fetal tissues surrounding the amniotic cavity (i.e., the chorioamniotic membranes) is poorly understood. By using RNA sequencing (RNA seq) as a discovery approach, we found that there were significant transcriptomic differences involving host response to pathogens in the chorioamniotic membranes of women with intra-amniotic infection compared to those from women without inflammation. In addition, the sterile or microbial nature of intra-amniotic inflammation was associated with distinct transcriptomic profiles in the chorioamniotic membranes. Moreover, the immune response in the chorioamniotic membranes of women with sterile intra-amniotic inflammation was milder in nature than that induced by microbes and involved the upregulation of alarmins and inflammasome-related molecules. Lastly, the presence of maternal and fetal inflammatory responses in the placenta was associated with the upregulation of immune processes in the chorioamniotic membranes. Collectively, these findings provide insight into the immune responses against microbes or alarmins that take place in the fetal tissues surrounding the amniotic cavity, shedding light on the immunobiology of preterm labor and birth.
INTRODUCTION
Preterm birth is the leading cause of neonatal morbidity and mortality worldwide (1–4) and is commonly preceded by the syndrome of spontaneous preterm labor (5, 6). Among the etiologies proposed for spontaneous preterm labor (6), only intra-amniotic inflammation is an established causal link (7–18). The inflammatory milieu taking place in the amniotic cavity was traditionally associated with microbes, a clinical condition known as intra-amniotic infection (19–25). Yet, consistent evidence has now shown that endogenous danger signals derived from cellular stress or necrosis, known as damage-associated molecular patterns (DAMPs) or alarmins (26–29), can also elicit intra-amniotic inflammation and lead to preterm birth in mice (11, 14, 18, 30). The latter is termed sterile intra-amniotic inflammation (23, 31–35) and is observed in women with different pregnancy complications (23–25, 36, 37). Importantly, sterile intra-amniotic inflammation is more frequently diagnosed than intra-amniotic infection in women with spontaneous preterm labor and intact membranes who ultimately delivered preterm (23). Hence, both intra-amniotic infection and sterile intra-amniotic inflammation are associated with adverse perinatal outcomes (23, 38). Nevertheless, cytokine network analysis has suggested that these two clinical conditions are unique in nature since specific immune mediators are enriched in the amniotic cavity of each subset of women (31, 35). Indeed, the severity of the cellular immune response in the amniotic cavity largely depends on the detection of culturable microorganisms (39–43). However, the extent of such immune responses in the fetal tissues surrounding the amniotic cavity (i.e., the chorioamniotic membranes) (44), and whether these differ between intra-amniotic infection and sterile intra-amniotic inflammation, is poorly understood.
The chorioamniotic membranes, which are composed of the amnion and chorion and attached to the decidua parietalis, serve to protect the fetus through anatomical (44–46), biochemical (47, 48), and antibacterial (49–52) functions. Herein, we hypothesize that the intra-amniotic inflammatory status (microbe-associated or sterile) can differentially modulate immune responses in the chorioamniotic membranes. Consistent with this hypothesis, descriptive studies have shown that inflammatory lesions of the chorioamniotic membranes (i.e., acute histologic chorioamnionitis) are associated with intra-amniotic inflammation as well as preterm labor and birth (53–57). Yet the transcriptomic changes underlying the immune responses to intra-amniotic infection and sterile intra-amniotic inflammation in the chorioamniotic membranes have not been investigated.
Furthermore, by using systems biology approaches, we recently provided evidence that the maternal and fetal acute inflammatory responses in the chorioamniotic membranes and umbilical cord influence the transcriptome of the immune cells in the amniotic cavity (58). Therefore, herein, we also investigated whether the severity of the immune responses in the amniotic cavity (intra-amniotic infection and sterile intra-amniotic inflammation) is influenced by the presence of maternal or fetal acute inflammatory responses in the placenta.
The aim of this study was to utilize systems immunobiology approaches to investigate the transcriptome of the chorioamniotic membranes in the context of intra-amniotic infection or sterile intra-amniotic inflammation. Specifically, by using RNA sequencing (RNA seq) and confirmatory reverse transcription-quantitative PCR (qRT-PCR), determinations of the inflammatory and microbiological status of amniotic fluid, and placental histopathological examination, we revealed specific biological processes enriched in the chorioamniotic membranes from women with microbe-associated or sterile intra-amniotic inflammation and how such processes differed based on the presence of acute maternal and fetal inflammatory responses in the placenta.
RESULTS
RNA seq demonstrates upregulation of immunological processes in the chorioamniotic membranes from women with preterm labor and intra-amniotic infection.
First, to determine how the intra-amniotic inflammatory responses elicited by microbes affect the transcriptome of the chorioamniotic membranes, we performed RNA seq on such tissues from women who underwent preterm labor with intra-amniotic infection compared to those without intra-amniotic inflammation (Fig. 1A). The demographic and clinical characteristics of women whose chorioamniotic membrane samples were analyzed by RNA seq are shown in Table 1. The presence of microbes in the amniotic cavity induced a variety of local inflammatory responses that differed in severity according to amniotic fluid interleukin-6 (IL-6) concentrations (Fig. 1B). Such variation was also observed within individual bacterial species, as Ureaplasma species, the most commonly detected microbes in amniotic fluid (19, 38, 59–61), were associated with a range of inflammatory responses (Fig. 1B). Despite the variable intra-amniotic inflammatory responses related to different microbes, RNA seq data revealed 6,608 differentially expressed genes (DEGs) in the chorioamniotic membranes from cases of intra-amniotic infection compared to those without, of which 3,476 DEGs were upregulated (red dots) and 3,132 DEGs were downregulated (blue dots) as displayed in the volcano plot (Fig. 1C). Such transcriptomic differences are clearly emphasized in the heatmap representation (Fig. 1D). Next, the DEGs were analyzed to determine KEGG database pathways impacted based on differential expression (see Materials and Methods for details). Fifty-six significantly impacted pathways were identified, of which the top 10 are displayed in Fig. 1E. Of interest, pathways such as “chemokine signaling pathway,” “complement and coagulation cascades,” “cell adhesion molecules,” “phagosome,” “leukocyte transendothelial migration,” and “natural killer cell mediated cytotoxicity” were identified (Fig. 1E). Therefore, transcriptomic changes in the chorioamniotic membranes associated with intra-amniotic infection involve host immune responses. We then utilized upstream regulator analysis to predict the activation or inhibition of specific upstream molecules based on the identified DEGs. Toll-like receptor 3 (TLR3) and IL-3 were predicted as activated, while G protein signaling modulators (GPSM) 1, 2, and 3 as well as Purkinje cell protein 2 (PCP 2), which are implicated in chemokine signaling (62–64), were predicted as inhibited in the chorioamniotic membranes of women with intra-amniotic infection compared to those without (Fig. 1F). Next, we evaluated the gene ontology (GO) biological processes that were enriched among the upregulated or downregulated DEGs. Among the genes upregulated in the chorioamniotic membranes with intra-amniotic infection compared to those without, enriched biological processes included immunological GO terms such as “adaptive immune response,” “leukocyte activation involved in immune response,” “neutrophil activation,” “regulation of T cell proliferation,” “defense response,” “interferon gamma production,” and “macrophage activation” (Fig. 1G). Downregulated DEGs were associated with enrichment of biological processes such as “intermediate filament cytoskeleton organization,” “cell motility,” “cell-matrix adhesion,” and “cell-substrate junction assembly” (Fig. 1H).
FIG 1.
RNA seq demonstrates upregulation of immunological processes in the chorioamniotic membranes from women with preterm labor and intra-amniotic infection. (A) Experimental design showing the transcriptomic comparison between the chorioamniotic membranes from women with intra-amniotic infection (n = 10) and those without intra-amniotic inflammation (n = 11). (B) Heatmap showing the different species of bacteria detected in amniotic fluid from women diagnosed with intra-amniotic infection. Each column represents one patient, stratified according to the log10-transformed amniotic fluid concentrations of IL-6. (C) Volcano plot showing the differentially expressed genes (DEGs) between the chorioamniotic membranes from women with intra-amniotic infection and those without intra-amniotic inflammation. (D) Heatmap showing the chorioamniotic membrane expression (log2 thereof after eventual adjusting for covariates and subtracting the mean in the reference group) of the top 100 DEGs ranked by magnitude of change between groups. (E) KEGG pathway impact analysis evidence plot showing gene overrepresentation evidence (pORA) and total pathway accumulation evidence (pAcc) based on differential expression between the chorioamniotic membranes from women with intra-amniotic infection and those without intra-amniotic inflammation. Significantly impacted pathways are shown as red dots. (F) Predicted activated and inhibited upstream regulators of DEGs in the chorioamniotic membranes from women with intra-amniotic infection compared to those without intra-amniotic inflammation. (G and H) Gene ontology (GO) analysis using genes significantly upregulated (G) or downregulated (H) in the chorioamniotic membranes from women with intra-amniotic infection compared to those without intra-amniotic inflammation. The top 10 most enriched biological processes are shown. N.S., not significant.
TABLE 1.
Clinical and demographic characteristics of patients included in the first cohort for RNA seq
| Characteristic | Preterm labor and delivery without intra-amniotic inflammation (n = 11) | Preterm labor and delivery with sterile intra-amniotic inflammation (n = 15) | Preterm labor and delivery with intra-amniotic infection (n = 10) | P value |
|---|---|---|---|---|
| Maternal age (median [IQR] [yrs])a,f | 23 (19.5–23) | 26 (23.5–28.5) | 21 (20–24.5) | 0.09 |
| Body mass index (median [IQR] [kg/m2])a | 21.8 (19.4–25.1) | 30 (26–33.9) | 29.7 (22.6–36.3) | 0.02 |
| Primiparityb (% [no. of patients/total no. of patients]) | 36.4 (4/11) | 26.7 (4/15) | 10 (1/10) | 0.4 |
| Race/ethnicityb (% [no. of patients/total no. of patients]) | 0.7 | |||
| African-American | 90.9 (10/11) | 86.7 (13/15) | 90 (9/10) | |
| White | 9.1 (1/11) | 13.3 (2/15) | 0 (0/10) | |
| Other | 0 (0/11) | 0 (0/15) | 10 (1/10) | |
| Gestational age at amniocentesis (median [IQR] [wks])a | 32.4 (30–33.8) | 26.4 (23.9–29.7) | 29.7 (25.7–31.2) | 0.03 |
| IL-6 (median [IQR] [ng/mL])a | 0.3 (0.2–0.4) | 10.9 (5.6–17.1) | 149.1 (31.2–195.4) | <0.001 |
| Amniotic fluid glucose (median [IQR] [mg/dL])a | 23 (18–29.5)c | 24 (21–27)d | 5 (1–13.5)d | 0.001 |
| Amniotic fluid WBC (median [IQR] [cells/mm3])a | 0 (0–0)c | 2 (1–5)d | 310 (114.5–330)e | <0.001 |
| Gestational age at delivery (median [IQR] [wks])a | 34.3 (33.9–34.7) | 26.7 (24.5–30.5) | 30 (25.7–31.3) | 0.007 |
| Cesarean sectionb (% [no. of patients/total no. of patients]) | 27.3 (3/11) | 33.3 (5/15) | 30 (3/10) | 1.0 |
| Birth weight (median [IQR] [g])a | 2,155 (1,940–2,542.5) | 917 (625–1,545) | 1,220 (815.5–1,486.3) | 0.01 |
| Acute maternal inflammatory responseb (% [no. of patients/total no. of patients]) | ||||
| Stage 1 (early acute subchorionitis or chorionitis) | 0 (0/11) | 20 (3/15) | 10 (1/10) | 0.3 |
| Stage 2 (acute chorioamnionitis) | 18.2 (2/11) | 13.3 (2/15) | 30 (3/10) | 0.6 |
| Stage 3 (necrotizing chorioamnionitis) | 0 (0/11) | 20 (3/15) | 60 (6/10) | 0.006 |
| Acute fetal inflammatory responseb (% [no. of patients/total no. of patients]) | ||||
| Stage 1 (chorionic vasculitis or umbilical phlebitis) | 0 (0/11) | 20 (3/15) | 30 (3/10) | 0.2 |
| Stage 2 (umbilical arteritis) | 9.1 (1/11) | 6.7 (1/15) | 60 (6/10) | 0.004 |
| Stage 3 (necrotizing funisitis) | 0 (0/11) | 0 (0/15) | 0 (0/10) | 1.0 |
Kruskal-Wallis/ANOVA test with correction for multiple comparisons.
Fisher’s exact test.
One missing datum.
Two missing data.
Three missing data.
IQR, interquartile range; yrs, years; wks, weeks; WBC, white blood cells.
Given the large number of differentially expressed transcripts revealed by RNA seq, we chose specific genes for further validation by qRT-PCR using only those samples that were also included in RNA seq studies (Table 1). These results are summarized in Table S1. A subset of these genes are related to the inflammasome pathway and are known to be expressed in the chorioamniotic membranes of women with intra-amniotic infection (65) (“Expected and RNA seq-discovered” genes; Table S1). The other genes chosen for further validation were selected based on their adjusted P values as revealed in the current RNA seq data together with their known association with immunological, cell biological, or pregnancy-related processes (“RNA seq-discovered” genes; Table S1). qRT-PCR-derived fold changes for the comparison of preterm labor with intra-amniotic infection group versus the preterm labor without inflammation group were significantly correlated with those obtained by RNA seq using the same samples (internal validation) (P < 0.0001; r = 0.85; direction of change agreement for 95.6% [43/45] of genes tested) (Fig. S1A). The validation rate (consistent DEGs between RNA seq and qRT-PCR) was 56.3%.
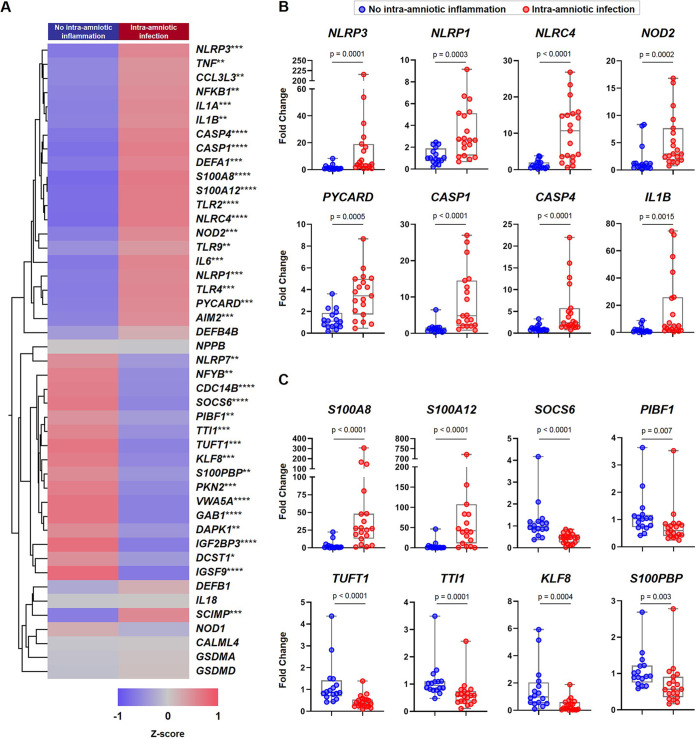
Next, to increase statistical power, we performed qRT-PCR analysis of the same 45 genes described above in an extended set of samples, which included those used for RNA seq profiling as well as new samples. The demographic and clinical characteristics of women whose chorioamniotic membrane samples were utilized for qRT-PCR validation, including all the patients shown in Table 1 together with new patients, are shown in Table 2. The qRT-PCR results are illustrated in the representative heatmap shown in Fig. 2A. Specifically, qRT-PCR validation in the extended sample set confirmed the significant upregulation of inflammasome-related genes such as NLRP3, NLRP1, NLRC4, NOD2, PYCARD, CASP1, CASP4, and IL1B in the chorioamniotic membranes of women with intra-amniotic infection compared to those without (Fig. 2B). Among the RNA seq-discovered genes, the expression of the alarmins S100A8 and S100A12 was increased in cases of intra-amniotic infection, whereas the expression of several transcripts was downregulated, including SOCS6, PIBF1, TUFT1, TTI1, KLF8, and S100PBP (Fig. 2C). There was also a significant correlation between the expression of these target genes in samples used for RNA seq and the qRT-PCR data from the newly added samples (external validation) (P < 0.0001; r = 0.70; direction of change agreement for 82.2% [37/45] of genes tested) (Fig. S1B). These findings suggest that the presence of microbes in the amniotic cavity (i.e., intra-amniotic infection) correlates with the upregulation of immunological processes related to antimicrobial host defense in the chorioamniotic membranes of women with preterm labor and birth.
TABLE 2.
Clinical and demographic characteristics of patients included in the second cohort for qRT-PCR validation
| Characteristic | Preterm labor and delivery without intra-amniotic inflammation (n = 16) | Preterm labor and delivery with sterile intra-amniotic inflammation (n = 19) | Preterm labor and delivery with intra-amniotic infection (n = 19) | P value |
|---|---|---|---|---|
| Maternal age (median [IQR] [yrs])a | 23 (19.8–23.8) | 26 (22.5–28.5) | 23 (20–26) | 0.07 |
| Body mass index (median [IQR] [kg/m2])a | 21.8 (20.1–27)c | 29.8 (25.4–33.8)d | 26.9 (22.5–34)d | 0.03 |
| Primiparityb (% [no. of patients/total no. of patients]) | 31.3 (5/16) | 21.1 (4/19) | 21.1 (4/19) | 0.8 |
| Race/ethnicityb (% [no. of patients/total no. of patients]) | 0.4 | |||
| African-American | 87.5 (14/16) | 89.5 (17/19) | 94.7 (18/19) | |
| White | 12.5 (2/16) | 10.5 (2/19) | 0 (0/19) | |
| Other | 0 (0/16) | 0 (0/19) | 5.3 (1/19) | |
| Gestational age at amniocentesis (median [IQR] [wks])a | 31.8 (29.9–33.6) | 26.4 (23.9–29.7) | 26.7 (23.7–31.2) | 0.02 |
| IL-6 (median [IQR] [ng/mL])a | 0.2 (0.2–0.5) | 11.2 (5.7–33.2) | 121.8 (59.3–153.9) | <0.001 |
| Amniotic fluid glucose (median [IQR] [mg/dL])a | 24 (18–30)c | 21 (18–26)d | 5 (1–17)e | 0.002 |
| Amniotic fluid WBC (median [IQR] [cells/mm3])a | 0 (0–0)c | 3 (1–14)d | 292 (43.3–467.5)e | <0.001 |
| Gestational age at delivery (median [IQR] [wks])a | 34.3 (33.3–34.9) | 26.7 (24.5–30.5) | 26.7 (23.8–31.2) | <0.001 |
| Cesarean sectionb (% [no. of patients/total no. of patients]) | 18.8 (3/16) | 26.3 (5/19) | 26.3 (5/19) | 0.9 |
| Birth weight (median [IQR] [g])a | 2,230 (1,807–2,575) | 917 (635–1,545) | 1,040 (587.5–1,447.5) | <0.001 |
| Acute maternal inflammatory responseb (% [no. of patients/total no. of patients]) | ||||
| Stage 1 (early acute subchorionitis or chorionitis) | 12.5 (2/16) | 21.1 (4/19) | 10.5 (2/19) | 0.7 |
| Stage 2 (acute chorioamnionitis) | 18.8 (3/16) | 10.5 (2/19) | 21.1 (4/19) | 0.7 |
| Stage 3 (necrotizing chorioamnionitis) | 0 (0/16) | 26.3 (5/19) | 63.2 (12/19) | <0.001 |
| Acute fetal inflammatory responseb (% [no. of patients/total no. of patients]) | ||||
| Stage 1 (chorionic vasculitis or umbilical phlebitis) | 6.3 (1/16) | 31.6 (6/19) | 26.3 (5/19) | 0.2 |
| Stage 2 (umbilical arteritis) | 6.3 (1/16) | 5.3 (1/19) | 57.9 (11/19) | <0.001 |
| Stage 3 (necrotizing funisitis) | 0 (0/16) | 0 (0/19) | 0 (0/19) | 1.0 |
Kruskal-Wallis/ANOVA test with correction for multiple comparisons.
Fisher’s exact test.
One missing datum.
Two missing data.
Three missing data.
IQR, interquartile range; yrs, years; wks, weeks; WBC, white blood cells.
FIG 2.
qRT-PCR validation of expected and RNA seq-discovered genes in the chorioamniotic membranes from women with preterm labor and intra-amniotic infection. (A) Heatmap representation showing the mean gene expression in the chorioamniotic membranes from women with intra-amniotic infection (n = 19) and those without intra-amniotic inflammation (n = 16). Red indicates upregulation; blue indicates downregulation. Asterisks indicate significant differences between study groups: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (B) Fold changes in the expression of NLRP3, NLRP1, NLRC4, NOD2, PYCARD, CASP1, CASP4, and IL1B in the chorioamniotic membranes from women with intra-amniotic infection and those without intra-amniotic inflammation. (C) Fold changes in the expression of S100A8, S100A12, SOCS6, PIBF1, TUFT1, TTI1, KLF8, and S100PBP in the chorioamniotic membranes from women with intra-amniotic infection and those without intra-amniotic inflammation. Data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum/maximum ranges. P values were obtained by using Mann-Whitney U-tests.
RNA seq reveals distinct immune responses in the chorioamniotic membranes of women with preterm labor and sterile intra-amniotic inflammation.
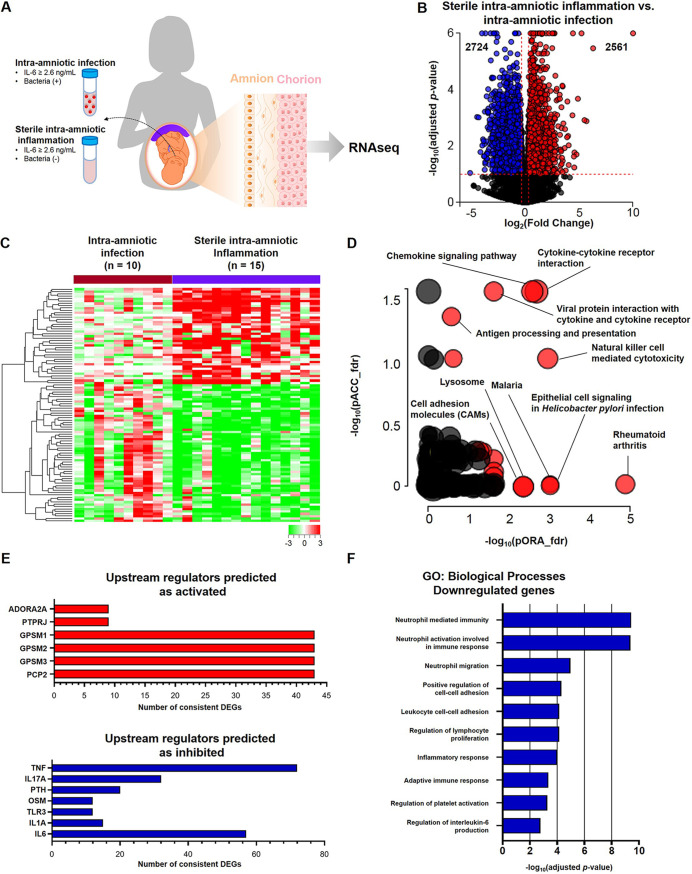
Sterile intra-amniotic inflammation is driven by endogenous danger signals or alarmins (11, 14, 18, 23, 30), and has been proposed to be immunologically distinct from microbe-associated inflammatory responses (31, 35). Both sterile intra-amniotic inflammation and intra-amniotic infection have been shown to affect the chorioamniotic membranes (23–25, 57, 66–72). Therefore, we next evaluated the transcriptome of the chorioamniotic membranes from women with sterile intra-amniotic inflammation compared to those with intra-amniotic infection (Fig. 3A). The chorioamniotic membranes from women with sterile intra-amniotic inflammation displayed differential expression of 5,285 genes compared to those with intra-amniotic infection, of which 2,561 DEGs were upregulated (red dots) and 2,724 DEGs (blue dots) were downregulated (Fig. 3B). Heatmap visualization illustrated the distinct transcriptomic profiles of the chorioamniotic membranes with sterile intra-amniotic inflammation or intra-amniotic infection, further indicating the divergence of these intra-amniotic inflammatory responses (Fig. 3C). KEGG pathway analysis revealed significant enrichment of 21 pathways, of which “cytokine-cytokine receptor interaction,” “chemokine signaling pathway,” “natural killer cell mediated cytotoxicity,” “lysosome,” “antigen processing and presentation,” and “cell adhesion molecules” were of interest (Fig. 3D). The upstream regulators predicted to be activated in sterile intra-amniotic inflammation included adenosine A2A receptor (ADORA2A); protein tyrosine phosphatase receptor type J (PTPRJ); GPSM 1, 2, and 3; and PCP 2, whereas the upstream regulators predicted as inhibited included tumor necrosis factor (TNF), IL17A, TLR3, IL1A, and IL-6 (Fig. 3E), suggesting less severe inflammatory responses in sterile intra-amniotic inflammation. Among the genes upregulated in the chorioamniotic membranes of women with sterile intra-amniotic inflammation, no GO biological processes were determined to be enriched. Among the downregulated genes, enriched GO processes included immunological terms such as “neutrophil mediated immunity,” “leukocyte cell-cell adhesion,” “regulation of lymphocyte proliferation,” “inflammatory response,” “adaptive immune response,” and “regulation of interleukin-6 production” (Fig. 3F), further supporting that cases of sterile intra-amniotic inflammation exhibit milder acute immune responses. The opposite comparison (using the chorioamniotic membranes from women with sterile intra-amniotic inflammation as the reference group) was also performed, and consistent results were obtained (Fig. S2).
FIG 3.
RNA seq reveals distinct immune responses in the chorioamniotic membranes of women with preterm labor and sterile intra-amniotic inflammation. (A) Experimental design showing the transcriptomic comparison between the chorioamniotic membranes from women with sterile intra-amniotic inflammation (n = 15) and those with intra-amniotic infection (n = 10). (B) Volcano plot showing the differentially expressed genes (DEGs) between the chorioamniotic membranes from women with sterile intra-amniotic inflammation and those with intra-amniotic infection. (C) Heatmap showing the chorioamniotic membrane expression (log2 thereof after eventual adjusting for covariates and subtracting the mean in the reference group) of the top 100 DEGs ranked by magnitude of change between groups. (D) KEGG pathway impact analysis evidence plot showing gene overrepresentation evidence (pORA) and total pathway accumulation evidence (pAcc) based on differential expression between the chorioamniotic membranes from women with sterile intra-amniotic inflammation and those with intra-amniotic infection. Significantly impacted pathways are shown as red dots. (E) Predicted activated and inhibited upstream regulators of DEGs in the chorioamniotic membranes from women with sterile intra-amniotic inflammation compared to those with intra-amniotic infection. (F) Gene ontology (GO) analysis using genes significantly downregulated in the chorioamniotic membranes from women with sterile intra-amniotic inflammation compared to those with intra-amniotic infection. The top 10 most significantly enriched biological processes are shown.
Similar to the previous comparison (Fig. 1 and 2), we chose specific genes for further validation by qRT-PCR, using only those samples that were also included in RNA seq studies (Table 1). These results are summarized in Table S2. qRT-PCR-derived fold changes for the comparison of the preterm labor with sterile intra-amniotic inflammation group versus the preterm labor with intra-amniotic infection group were significantly correlated with those obtained by RNA seq using the same samples (internal validation) (P < 0.0001; r = 0.84; direction of change agreement for 88.9% [40/45] of genes tested) (Fig. S3A). The validation rate (consistent DEGs between RNA seq and qRT-PCR) was 73.1%.
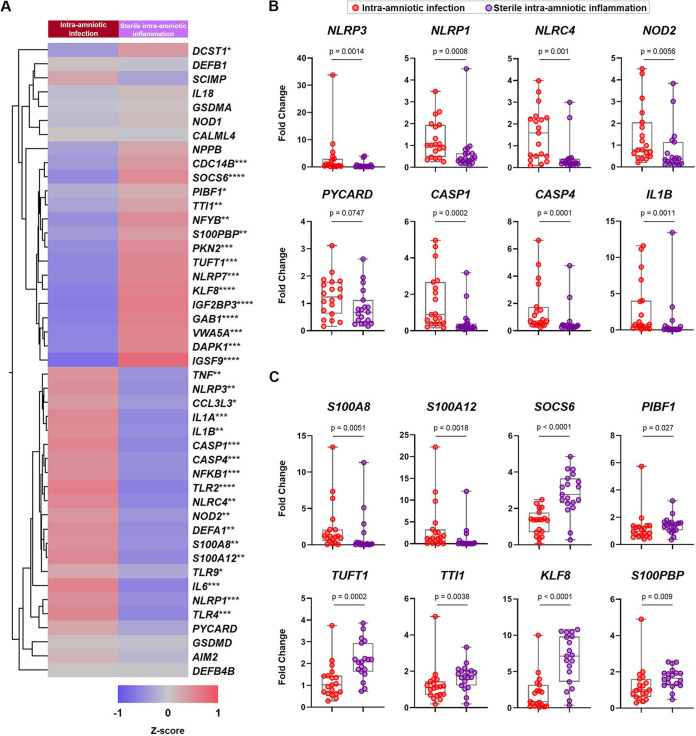
Next, to increase statistical power, we performed qRT-PCR analysis of the same 45 genes described above in an increased sample set, which included those used for RNA seq profiling as well as new samples (Table 2). The qRT-PCR results are illustrated in the representative heatmap in Fig. 4A. Specifically, inflammasome-related genes such as NLRP3, NLRP1, NLRC4, NOD2, CASP1, CASP4, and IL1B showed greater upregulation in the chorioamniotic membranes from women with intra-amniotic infection than those from women with sterile intra-amniotic inflammation, and expression of PYCARD tended to be greater as well (Fig. 4B). Among the RNA seq-discovered genes, the expression of the alarmins S100A8 and S100A12 was elevated in the chorioamniotic membranes from women with intra-amniotic infection compared to those from women with sterile intra-amniotic inflammation (Fig. 4C). Conversely, the expression of SOCS6, TUFT1, TTI1, KLF8, and S100PBP was greater in the chorioamniotic membranes from women with sterile intra-amniotic inflammation than those from women with intra-amniotic infection, and PIBF1 expression followed the same tendency (Fig. 4C). There was also a significant correlation between the expression of these target genes in samples used for RNA seq and the qRT-PCR data from the newly added samples (external validation) (P < 0.0001; r = 0.77; direction of change agreement for 84.4% [38/45] of genes tested) (Fig. S3B). Together, these findings show that the chorioamniotic membranes of women with preterm labor and sterile intra-amniotic inflammation display a transcriptomic profile that is distinct from that observed in women with intra-amniotic infection.
FIG 4.
qRT-PCR validation of expected and RNA seq-discovered genes in the chorioamniotic membranes from women with preterm labor and sterile intra-amniotic inflammation. (A) Heatmap representation showing the mean gene expression in the chorioamniotic membranes from women with intra-amniotic infection (n = 19) and those with sterile intra-amniotic inflammation (n = 19). Red indicates upregulation; blue indicates downregulation. Asterisks indicate significant differences between study groups: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (B) Fold changes in the expression of NLRP3, NLRP1, NLRC4, NOD2, PYCARD, CASP1, CASP4, and IL1B. (C) Fold changes in the expression of S100A8, S100A12, SOCS6, PIBF1, TUFT1, TTI1, KLF8, and S100PBP. Data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum/maximum ranges. P values were obtained by using Mann-Whitney U-tests.
The transcriptomic profile of the chorioamniotic membranes of women with preterm labor and sterile intra-amniotic inflammation involves the upregulation of inflammasome-related molecules.
Until now, we have shown that the transcriptome of the chorioamniotic membranes from women with intra-amniotic infection is distinct from those with sterile intra-amniotic inflammation. Therefore, we next investigated whether the chorioamniotic membrane transcriptomes from women with intra-amniotic infection, with sterile intra-amniotic inflammation, or without inflammation could be distinguished (Fig. 5A). Principal component analysis (PCA) revealed that the chorioamniotic membrane transcriptomes from cases of intra-amniotic infection formed the most distinct cluster, whereas those from chorioamniotic membranes with sterile intra-amniotic inflammation and without inflammation were less easily distinguished (Fig. 5B). The comparison between chorioamniotic membranes with sterile intra-amniotic inflammation and without inflammation demonstrated differential expression of the following 13 transcripts: IZUMO1R, NOS2, MT1X, ODAPH, CNKSR3, MT1F, and INA (downregulated, blue dots) as well as FAM236B, LYPLAL1, LIAS, LCNL1, GSTT2, and PPP1R27 (upregulated, red dots) (Fig. 5C). Next, we performed qRT-PCR validation of most of the RNA seq-discovered differentially expressed genes (11/13). Yet, the validation did not strongly confirm the RNA seq results: only two genes (LIAS and GSTT2) tended to be upregulated, and two genes (IZUMO1R and ODAPH) tended to be downregulated in the chorioamniotic membranes of women with sterile intra-amniotic inflammation (Fig. 5D). Therefore, we performed qRT-PCR analysis of expected genes. Interestingly, this analysis revealed the upregulation of the alarmin S100A8, as well as the inflammasome-related molecules PYCARD, AIM2, and NLRC4, which is in line with prior studies suggesting a role for the inflammasome in sterile intra-amniotic inflammation (14, 18, 30, 32) (Fig. 5E). The gene SCIMP, associated with antigen presentation (73), was also upregulated in sterile intra-amniotic inflammation (data not shown). These results suggest that the immune response in the chorioamniotic membranes of women with preterm labor and sterile intra-amniotic inflammation is milder in nature than that induced by microbes, yet this may also involve the upregulation of alarmins and inflammasome-related molecules. These findings confirm our in vivo studies showing that alarmins can trigger the activation of the inflammasome in the amniotic cavity (14, 18).
FIG 5.
The transcriptomic profile of the chorioamniotic membranes of women with preterm labor and sterile intra-amniotic inflammation involves the upregulation of alarmins and inflammasome-related molecules. (A) Experimental design showing the transcriptomic comparison among the chorioamniotic membranes from women with intra-amniotic infection (n = 10), those with sterile intra-amniotic inflammation (n = 15), and those without intra-amniotic inflammation (n = 11). (B) Principal-component analysis (PCA) based on the transcriptomes of the chorioamniotic membrane samples without intra-amniotic inflammation, with sterile intra-amniotic inflammation, and with intra-amniotic infection. The PCA plot shows the samples on the PC1-PC2-PC3 coordinates. (C) Volcano plot showing the differentially expressed genes (DEGs) between the chorioamniotic membranes from women with sterile intra-amniotic inflammation and those without intra-amniotic inflammation. (D) Fold changes in the expression of LIAS, GSTT2, IZUMO1R, and ODAPH in the chorioamniotic membranes from women with sterile intra-amniotic inflammation (n = 19) and those without intra-amniotic inflammation (n = 16). (E) Fold changes in the expression of S100A8, PYCARD, AIM2, and NLRC4 in the chorioamniotic membranes from women with sterile intra-amniotic inflammation and those without intra-amniotic inflammation. Data are shown as box-and-whisker plots, where midlines indicate medians, boxes indicate interquartile ranges, and whiskers indicate minimum/maximum ranges. P values were obtained by using Mann-Whitney U-tests. N.S., not significant.
The transcriptomic profile of the chorioamniotic membranes is largely influenced by the presence of a maternal acute inflammatory response in the placenta.
Intra-amniotic inflammation, induced by either microbes or alarmins, is strongly associated with maternal inflammatory responses in the placenta, defined as acute histologic chorioamnionitis (57). Thus, we determined the frequency of maternal inflammatory lesions of the placenta in our study groups (Table 1). Lesions of acute histologic chorioamnionitis are diagnosed by the presence of infiltrating maternal neutrophils in the placental tissues and are classified into stages based on severity (see Materials and Methods for details) (57). Among the study groups, two patients (2/11 [18.2%]) without intra-amniotic inflammation displayed a maternal inflammatory response in the placenta (Table 1). A greater proportion of those with intra-amniotic infection presented a severe maternal inflammatory response (stage 2/3, 9/10 [90.0%]) than those with sterile intra-amniotic inflammation (stage 2/3, 5/15 [33.3%]) (Table 1), which is consistent with prior studies (23).
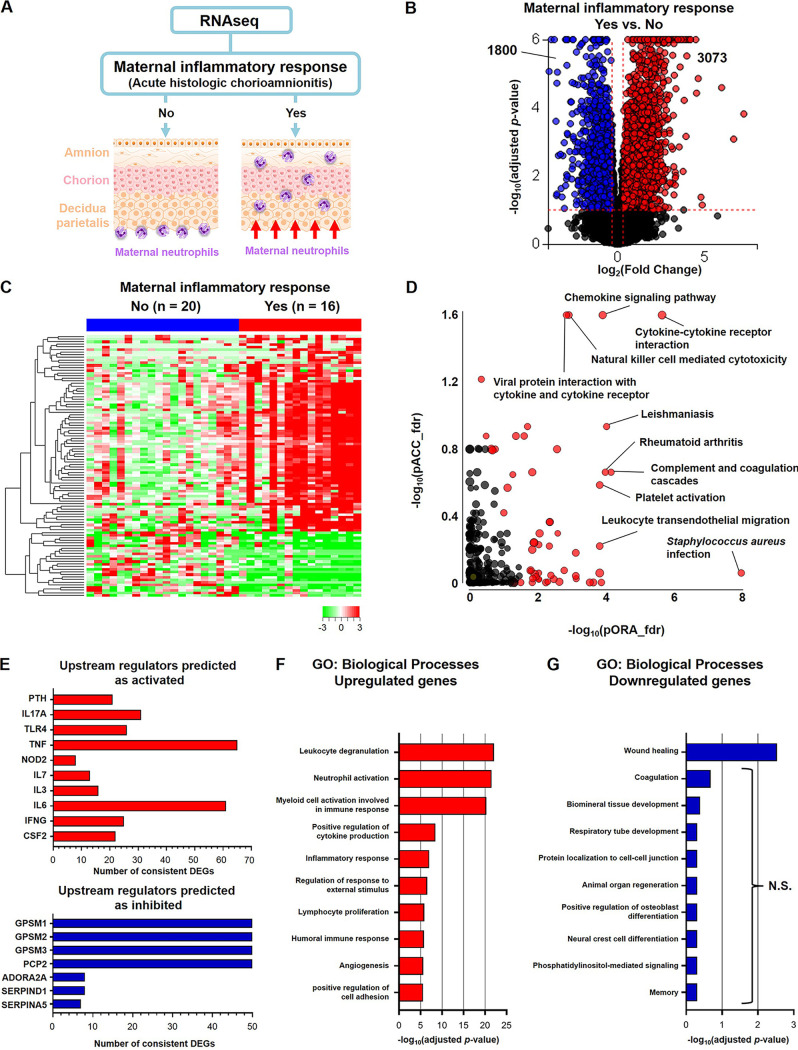
Next, we divided our total study population into those patients who presented a maternal inflammatory response in the placenta and those without such lesions to investigate whether the presence of acute histologic chorioamnionitis is associated with transcriptomic changes in the chorioamniotic membranes (Fig. 6A). The number of patients from each study group that showed histologic signs of a maternal inflammatory response are shown in Fig. S4A. Differential expression of 4,873 genes was determined, of which 3,073 DEGs were upregulated (red dots) and 1,800 DEGs were downregulated (blue dots) in the chorioamniotic membranes with acute histologic chorioamnionitis compared to those without this lesion (Fig. 6B). Heatmap visualization revealed the transcriptomic changes occurring in the chorioamniotic membranes with the presence of acute histologic chorioamnionitis (Fig. 6C). Furthermore, impact analysis of KEGG pathways revealed significant enrichment of 61 pathways based on DEGs, which included relevant processes such as “cytokine-cytokine receptor interaction,” “chemokine signaling pathway,” “complement and coagulation cascades,” “natural killer cell mediated cytotoxicity,” and “leukocyte transendothelial migration” (Fig. 6D), suggesting that the DEGs affected by acute histologic chorioamnionitis are largely associated with inflammatory processes. Upstream regulators predicted as activated in the chorioamniotic membranes with acute histologic chorioamnionitis included IL17A, TLR4, TNF, IL-6, and IFNG, all of which are implicated in intra-amniotic inflammation (31, 74–78) (Fig. 6E). Upstream regulators predicted as inhibited were GPSM 1, 2, and 3; PCP 2; ADORA2A; serine proteinase inhibitor, clade D, member 1 (SERPIND1); and SERPINA5 (Fig. 6E). Among the genes upregulated in the chorioamniotic membranes with acute histologic chorioamnionitis, enriched GO biological processes included immunological terms such as “leukocyte degranulation,” “neutrophil activation,” “myeloid cell activation involved in immune response,” “positive regulation of cytokine production,” “inflammatory response,” and “lymphocyte proliferation” (Fig. 6F). Among the downregulated genes, the only significantly enriched term was “wound healing” (Fig. 6G). Taken together, these findings demonstrate that the presence of maternal inflammatory responses in the placenta is associated with the upregulation of immune processes in the chorioamniotic membranes.
FIG 6.
The transcriptomic profile of the chorioamniotic membranes is largely influenced by the presence of a maternal acute inflammatory response in the placenta. (A) Experimental design showing the transcriptomic comparison between the chorioamniotic membranes from women with a maternal inflammatory response (acute histologic chorioamnionitis) (n = 16) and those without a maternal inflammatory response (no acute histologic chorioamnionitis) (n = 20). (B) Volcano plot showing the differentially expressed genes (DEGs) between the chorioamniotic membranes from women with acute histologic chorioamnionitis and those without acute histologic chorioamnionitis. (C) Heatmap showing the chorioamniotic membrane expression (log2 thereof after eventual adjusting for covariates and subtracting the mean in the reference group) of the top 100 DEGs ranked by magnitude of change between groups. (D) KEGG pathway impact analysis evidence plot showing gene overrepresentation evidence (pORA) and total pathway accumulation evidence (pAcc) based on differential expression between the chorioamniotic membranes from women with acute histologic chorioamnionitis and those without acute histologic chorioamnionitis. Significantly impacted pathways are shown as red dots. (E) Predicted activated and inhibited upstream regulators of DEGs in the chorioamniotic membranes from women with acute histologic chorioamnionitis compared to those without acute histologic chorioamnionitis. (F and G) Gene ontology (GO) analysis using genes significantly upregulated (F) or downregulated (G) in the chorioamniotic membranes from women with acute histologic chorioamnionitis compared to those without acute histologic chorioamnionitis. The top 10 most enriched biological processes are shown. N.S., not significant.
The transcriptomic profile of the chorioamniotic membranes is influenced by the presence of a fetal inflammatory response in the placenta.
Intra-amniotic inflammation, either induced by microbes or alarmins, is also associated with a fetal inflammatory response in the placenta (57). Thus, we determined the frequency of a fetal inflammatory response in our study groups (Table 1). Lesions associated with a fetal inflammatory response are diagnosed by the presence of infiltrating fetal neutrophils in the umbilical cord and chorionic plate and are classified into stages based on severity (see Materials and Methods for details) (57). Among the study groups, one patient (1/11 [9.1%]) without intra-amniotic inflammation displayed a fetal inflammatory response (Table 1). As expected, a greater proportion of patients with intra-amniotic infection presented a fetal inflammatory response than those with sterile intra-amniotic inflammation (stage 1/2, 9/10 [90.0%] compared to 4/15 [26.7%]) (Table 1).
Lastly, we divided our total study population into those patients who presented a fetal inflammatory response and those without such lesions to determine whether the presence of fetal placental inflammation was associated with transcriptomic changes in the chorioamniotic membranes (Fig. 7A). The number of patients from each study group that showed histologic signs of a fetal inflammatory response are shown in Fig. S4B. Differential expression of 4,073 genes was observed in the chorioamniotic membranes from cases with a fetal inflammatory response compared to those without this placental lesion, of which 2,245 DEGs were upregulated (red dots) and 1,828 DEGs were downregulated (blue dots) (Fig. 7B). The chorioamniotic membrane transcriptomes from cases with a fetal inflammatory response were clearly distinguished from those without this lesion, as revealed by heatmap visualization (Fig. 7C). Impact analysis of KEGG pathways of the DEGs showed enrichment of 54 pathways based on DEGs, of which the most relevant were “chemokine signaling pathway,” “cytokine-cytokine receptor interaction,” “B cell receptor signaling pathway,” and “NF-kappa B signaling pathway” (Fig. 7D). Upstream regulators predicted to be activated in the chorioamniotic membranes with a fetal inflammatory response included TLR4, TNF, IL17A, IL1B, and IL-6 (Fig. 7E), while upstream regulators predicted as inhibited included GPSM 1, 2, and 3; PCP 2; and ADORA2A (Fig. 7E). Among the genes upregulated in the chorioamniotic membranes with a fetal inflammatory response, the enriched GO biological processes included immunological terms such as “leukocyte degranulation,” “neutrophil activation involved in immune response,” “neutrophil migration,” “inflammatory response,” “cellular response to cytokine stimulus,” and “positive regulation of cytokine production” (Fig. 7F). Among the downregulated genes, no significantly enriched GO processes were identified. These findings indicate that the presence of a fetal inflammatory response in the placenta correlates with upregulation of immune processes in the chorioamniotic membranes.
FIG 7.
The transcriptomic profile of the chorioamniotic membranes is influenced by the presence of a fetal inflammatory response in the placenta. (A) Experimental design showing the transcriptomic comparison between chorioamniotic membranes from women with a fetal inflammatory response (n = 14) and those without a fetal inflammatory response (n = 22). (B) Volcano plot showing the differentially expressed genes (DEGs) between the chorioamniotic membranes from women with a fetal inflammatory response and those without a fetal inflammatory response. (C) Heatmap showing the chorioamniotic membrane expression (log2 thereof after eventual adjusting for covariates and subtracting the mean in the reference group) of the top 100 DEGs ranked by magnitude of change between groups. (D) KEGG pathway impact analysis evidence plot showing gene overrepresentation evidence (pORA) and total pathway accumulation evidence (pAcc) based on differential expression between the chorioamniotic membranes from women with a fetal inflammatory response and those without a fetal inflammatory response. Significantly impacted pathways are shown as red dots. (E) Predicted activated and inhibited upstream regulators of DEGs in the chorioamniotic membranes from women with a fetal inflammatory response compared to those without a fetal inflammatory response. (F) Gene ontology (GO) analysis using genes significantly upregulated in the chorioamniotic membranes from women with a fetal inflammatory response compared to those without a fetal inflammatory response. The top 10 most significantly enriched biological processes are shown.
DISCUSSION
The traditional view is that the amniotic cavity, and therefore the fetus and placenta, are maintained under sterile conditions throughout gestation (79–88). Hence, microbial invasion into the amniotic cavity from the lower genital tract causes disease and intra-amniotic infection (89–93), thereby triggering soluble and cellular local immune responses (31, 35, 39, 41–43). The most common microorganisms identified in the amniotic cavity of women with intra-amniotic infection are Ureaplasma species, Mycoplasma hominis, Gardnerella vaginalis, and Streptococcus agalactiae, among others (19, 38, 59–61, 94, 95), all of which were identified in our amniotic fluid samples. Accordingly, we recently demonstrated that the intra-amniotic inoculation of Ureaplasma species in mice induces inflammatory responses in the fetus, placenta, and at the maternal-fetal interface, leading to preterm birth and neonatal mortality (17). Consistent with the findings herein, the intra-amniotic inoculation of Ureaplasma species induced the upregulation of multiple inflammatory mediators in the fetal membranes (17). Moreover, our data demonstrated that Ureaplasma species elicit a range of inflammatory responses among patients, as previously reported (17, 96, 97). Thus, microbes elicit immune responses in the membranes surrounding the amniotic cavity in humans and mice.
Our transcriptomic analyses showed that enriched biological processes in the chorioamniotic membranes from women with intra-amniotic infection were largely related to the host immune response against pathogens. This finding is relevant since microbes invading the amniotic cavity are not typically found in the chorioamniotic membranes (92), yet whether the chorioamniotic membranes can harbor a microbiome that is distinct from that detected in amniotic fluid in the context of intra-amniotic infection requires further investigation. Regardless, our findings indicate that these fetal tissues contribute to the host response mechanisms against intra-amniotic infection. These mechanisms are governed by maternal neutrophils infiltrating the chorioamniotic membranes (98, 99), a placental lesion referred to as “acute histologic chorioamnionitis” (57, 100, 101). Consistently, we found that the biological processes enriched in the chorioamniotic membranes of women with acute histologic chorioamnionitis include neutrophil functions. Previous studies have shown that neutrophils infiltrating the chorioamniotic membranes from women with intra-amniotic infection can form neutrophil extracellular traps (NETs) (102), perform phagocytosis (103), and express proinflammatory mediators (104, 105). Elegant in vivo studies have also shown that Gram-positive microbes such as Streptococcus agalactiae, also detected in amniotic fluid in the current study, induce neutrophil infiltration and NET formation in the chorioamniotic membranes (106). Such a process of NET formation seems to be partially mediated by factors released by the chorioamniotic membranes (107). In line with this concept, the chorioamniotic membranes also orchestrate the migration of maternal neutrophils from the decidual vessels toward the amnion and chorion (108–112), a process that is enhanced in cases with intra-amniotic infection (57). This process is mediated by chemokines such as CXCL8, which is released by the chorioamniotic membranes (108–113). Interestingly, our results also showed that neutrophil degranulation, phagocytosis, and cytokine release, as well as adaptive immune responses, are biological processes enriched in the chorioamniotic membranes of women with intra-amniotic infection and/or acute histologic chorioamnionitis. However, mechanistic studies are required to confirm the participation of such pathways in the chorioamniotic membrane host immune response against microbes invading the amniotic cavity.
Consistent with our previous studies (32, 33, 65, 114–116), women with preterm labor and microbial-associated intra-amniotic inflammation displayed upregulation of inflammasome-related molecules in the chorioamniotic membranes. These findings are in tandem with in vivo studies showing that the intra-amniotic injection of a microbial product (endotoxin) induces preterm birth by activating the NLRP3 inflammasome in the fetal membranes, which was mitigated by the administration of a specific inhibitor of inflammasome assembly (15). Furthermore, the intra-amniotic inoculation of Ureaplasma species also induces preterm birth, a process that includes activation of the NLRP3 inflammasome in the fetal membranes (17). In such a model, preterm birth can be prevented by treatment with clarithromycin (17), an effective treatment for intra-amniotic infection in women (117–120), suggesting that this antibiotic interferes with the inflammasome pathway.
Interestingly, our RNA seq analysis revealed that the expression of the known alarmins S100A8 and S100A12 (121, 122) was increased in the chorioamniotic membranes of women with intra-amniotic infection. These findings are consistent with previous reports showing increased concentrations of these alarmins in amniotic fluid of women with intra-amniotic infection (31, 123). Therefore, it is tempting to suggest that the alarmins found in the amniotic cavity upon microbial invasion are derived from the chorioamniotic membranes. Intriguingly, we also found that the chorioamniotic membranes of women with intra-amniotic infection displayed downregulation of immunosuppressive mediators such as SOCS6 (124, 125) and PIBF1 (126–129), indicating that microbial invasion of the amniotic cavity disrupts regulatory immune mechanisms in the chorioamniotic membranes. Moreover, the senescence-associated genes TUFT1 (130) and TTI1 (131) were also downregulated in cases with intra-amniotic infection, supporting the hypothesis that mammalian target of rapamycin (mTOR)-related cellular senescence mechanisms are implicated in the pathogenesis of preterm labor (132–134); yet, such a process can also occur in the absence of acute histologic chorioamnionitis (135). Lastly, the chorioamniotic membranes of women with intra-amniotic infection downregulated the expression of KLF8 (an inducer of epithelial-to-mesenchymal transition) (136) and S100PBP (a protein that seems to be implicated in tumorigenesis) (137); however, the role of such molecules in these fetal tissues requires further investigation.
Intra-amniotic infection can also initiate a range of fetal inflammatory responses in the placenta, which are known as chorionic vasculitis, umbilical phlebitis, umbilical arteritis, and necrotizing funisitis (57, 71). These inflammatory lesions are characterized by the infiltration of fetal neutrophils into the surrounding tissues (57, 71). Consistently, we found herein that neutrophil degranulation and chemotaxis were among the top biological processes enriched in the chorioamniotic membranes of women whose placentas displayed fetal inflammatory responses. This finding supports the concept that fetal inflammation in the umbilical cord and chorionic plate occurs in tandem with acute histologic chorioamnionitis upon microbial invasion of the amniotic cavity (71, 72, 138, 139). Indeed, it has been proposed that chemokines released into the amniotic cavity (i.e., fetal compartment) participate in the active recruitment of maternal neutrophils from the decidua throughout the chorioamniotic membranes (57). Hence, microbes invading the amniotic cavity trigger maternal and fetal inflammatory responses in the intra-amniotic space, a process that likely initiates the premature activation of the parturition cascade leading to preterm birth (89–91).
Notably, our current study provides the first demonstration in humans that alarmin- and inflammasome-related inflammatory processes in the chorioamniotic membranes are associated with the pathogenesis of preterm labor with sterile intra-amniotic inflammation, which is in tandem with our previous study (32). The novelty of this study arises from the difficulty of investigating host immune processes in both amniotic fluid and the placenta from the same woman: the former is collected during the episode of preterm labor via the invasive procedure of transabdominal amniocentesis, and the latter is obtained at the time of delivery, which can occur several days later. Yet, the concept that alarmins initiate sterile intra-amniotic inflammatory processes mediated by the inflammasome in the fetal membranes is an ongoing line of research for our group, and thus our transcriptomic findings are consistent with previous in vitro experiments using human chorioamniotic membranes (30) and in vivo studies using a translational combination of animal models and high-dimensional ultrasound (11, 14, 18). In such studies, we have also revealed that the inhibition of the inflammasome can serve as a novel strategy to prevent preterm birth induced by alarmins (14, 18), a pregnancy complication that currently lacks treatment.
It is worth mentioning that both acute histologic chorioamnionitis and fetal inflammation of the umbilical cord can occur in the context of sterile intra-amniotic inflammation (23–25, 32, 37). However, these placental lesions are less common in women with sterile intra-amniotic inflammation than in those with intra-amniotic infection (23–25, 32, 37), which is consistent with the concept that the inflammatory responses triggered by alarmins are less severe than those driven by microbes (140). Such reduced incidence of placental lesions in our study population prevented the adequate transcriptomic comparison among women with sterile intra-amniotic inflammation.
In conclusion, in the current study, by utilizing RNA seq as a discovery approach, we investigated the transcriptomic profiles of the chorioamniotic membranes from women who underwent preterm labor in the context of microbial or sterile intra-amniotic inflammation as well as maternal and fetal inflammatory responses in the placenta. There were significant transcriptomic differences involving host response to pathogens in the chorioamniotic membranes of women with intra-amniotic infection compared to those from women without inflammation. In addition, the sterile or microbial nature of intra-amniotic inflammation was associated with distinct transcriptomic profiles in the chorioamniotic membranes. Moreover, the immune response in the chorioamniotic membranes of women with sterile intra-amniotic inflammation appeared milder in nature than that induced by microbes and may involve the upregulation of alarmins and inflammasome-related molecules. Furthermore, the presence of maternal or fetal inflammatory responses in the placenta was associated with an upregulation of immune processes in the chorioamniotic membranes. Collectively, these findings provide insight into the immune responses against microbes or alarmins that take place in the fetal tissues surrounding the amniotic cavity.
MATERIALS AND METHODS
Human subjects and clinical specimens.
Chorioamniotic membrane and amniotic fluid samples were retrospectively obtained from the Biological Bank of the Perinatology Research Branch, an intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services, Wayne State University (Detroit, MI, USA), and the Detroit Medical Center (Detroit, MI, USA). The collection and use of human materials for research purposes were approved by the Institutional Review Boards of Wayne State University and the National Institute of Child Health and Human Development. All participating women provided written informed consent prior to sample collection. The study included two patient cohorts. The first cohort comprised women who underwent spontaneous preterm labor with intact membranes and delivered preterm, whose chorioamniotic membranes were used for discovery RNA seq; this cohort was divided into the following groups (see “Clinical definitions and sample collection” below): (i) women without intra-amniotic inflammation (n = 11), (ii) women with sterile intra-amniotic inflammation (n = 15), and (iii) women with proven intra-amniotic infection (n = 10). The demographic and clinical characteristics of the first cohort are shown in Table 1. The second cohort comprised women with spontaneous preterm labor and birth, whose chorioamniotic membranes were used for qRT-PCR validation assays, some of which had also been utilized for RNA seq (n = 16 to 19 each group). The demographic and clinical characteristics of the second cohort are shown in Table 2. Women with preterm prelabor rupture of membranes (pPROM), multiple gestations, or those who had a fetus with chromosomal and/or sonographic abnormalities were excluded. Maternal and neonatal data were obtained by retrospective clinical chart review.
Clinical definitions and sample collection.
Gestational age was established based on the last menstrual period and confirmed by ultrasound examination. Spontaneous preterm labor was defined as the presence of regular uterine contractions with a frequency of at least 2 every 10 min and cervical ripening between 20 and 36 (6/7) weeks of gestation. Preterm delivery was defined as birth at <37 weeks of gestation.
Standard clinical laboratory determinations in amniotic fluid included the evaluation of the white blood cell count (141), glucose concentration (142), Gram stain (143), and microbiological culture of aerobic/anaerobic bacteria and genital mycoplasmas (19, 22, 144). A previous study from our group sought to apply molecular microbiological techniques for the broad detection of microorganisms in amniotic fluid and showed that the use of combined broad-range real-time PCR with electrospray ionization-time of flight mass spectrometry (PCR/ESI-MS) could detect microbial signals in samples that did not yield a positive microbial culture (22). Accordingly, in this study, we have used the following process to assign patients to the study groups. If a sample has a positive culture/Gram stain and/or PCR/ESI-MS test, this is considered indicative of the presence of microbes in the amniotic cavity. When both the culture and PCR/ESI-MS results are negative, this confirms the absence of microbes in the amniotic cavity. The study group classifications used in this study were determined by combining the presence or absence of microbes together with the evaluation of amniotic fluid IL-6 concentrations as an indicator of intra-amniotic inflammation, as previously established (20). Thus, a positive microbial signal (either by culture/Gram stain or PCR/ESI-MS), together with elevated IL-6 (≥2.6 ng/mL), indicates intra-amniotic infection (22–25); a negative microbial signal indicated by both culture/Gram stain and PCR/ESI-MS, together with elevated IL-6, indicates sterile intra-amniotic inflammation (22–25); and the absence of microbial signals (indicated by both culture/Gram stain and PCR/ESI-MS), together with low IL-6 levels, indicates no intra-amniotic inflammation/infection. The detection of microbes in amniotic fluid is considered the most reliable means of evaluating infection of the amniotic cavity since prior studies have established that the most common route of infection is by ascension from the lower genital tract (92, 93).
For each patient from which amniotic fluid was obtained, chorioamniotic membrane samples were collected from the placenta within 30 min after delivery. A section of chorioamniotic membrane that extended the entire length of the membrane was cut, rolled, and snap-frozen using liquid nitrogen. Collected chorioamniotic membrane samples were stored at −80°C until use for RNA isolation. Due to the inherent difficulty of obtaining matched chorioamniotic membrane and amniotic fluid from well-characterized patients, the collection of samples included in this study spanned a period of several years.
Detection of microorganisms in amniotic fluid using PCR/ESI-MS.
The PCR/ESI-MS (Ibis Technology-Athogen, Carlsbad, CA, USA) platform includes a broad bacteria and Candida (BAC) detection assay that can identify 3,400 bacteria and 40 Candida species, as described in the platform’s signature database (145). In addition, the platform includes a viral detection assay that identifies the following viruses: herpes simplex virus 1 (HHV-1), herpes simplex virus 2 (HHV-2), varicella-zoster virus (HHV-3), Epstein-Barr virus (HHV-4), cytomegalovirus (HHV-5), roseolovirus (HHV-6), Kaposi’s sarcoma-associated herpesvirus (HHV-8), human adenoviruses, human enteroviruses, BK polyomavirus, JC polyomavirus, and parvovirus B19.
Determination of interleukin-6 in amniotic fluid.
Interleukin-6 concentrations in the amniotic fluid samples were measured with a sensitive and specific enzyme immunoassay from R&D Systems (Minneapolis, MN, USA) as previously established (20). The IL-6 concentrations were determined by interpolation from the standard curves. The inter- and intra-assay coefficients of variation for IL-6 were 8.7% and 4.6%, respectively. The detection limit of the IL-6 assay was 0.09 pg/mL.
Placental histopathological examination.
Histopathological examination of the placenta was performed by perinatal pathologists blinded to clinical diagnoses and obstetrical outcomes according to standardized Perinatology Research Branch protocols (57). Acute inflammatory lesions of the placenta (maternal inflammatory response), defined as the invasion of maternal neutrophils, were diagnosed according to established criteria, including staging and grading (57). Severity is reported as stages, where stage 1 indicates early acute subchorionitis or chorionitis, stage 2 indicates acute chorioamnionitis, and stage 3 indicates necrotizing chorioamnionitis (57). Early acute subchorionitis or chorionitis (stage 1) may not be representative of infection but rather the inflammatory processes associated with labor (146–148). Stages 2 and 3 were considered as acute histologic chorioamnionitis.
Fetal inflammatory responses were histologically evaluated by the presence and severity of acute lesions (i.e., neutrophil invasion) in the umbilical cord (funisitis) or chorionic plate (chorionic vasculitis) (57, 149). Severity is reported as stages, where stage 1 indicates umbilical phlebitis and/or chorionic vasculitis (mild inflammation), stage 2 indicates umbilical arteritis (severe inflammation), and stage 3 indicates necrotizing funisitis (total inflammation and necrosis in the umbilical cord) (57).
RNA seq and qRT-PCR.
RNA was isolated from snap-frozen chorioamniotic membrane samples using the Qiagen RNeasy mini kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. The purity, concentration, and integrity of the RNA samples were assessed using the NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and the Bioanalyzer 2100 (Agilent Technologies, Wilmington, DE, USA). For RNA seq, the RNA seq library was prepared by the Beijing Genomics Institute (BGI, Shenzhen, China) using the TruSeq RNA library prep kits (Illumina, San Diego, CA, USA). Paired-end sequence reads (100 million total reads) of 150 bp length were generated using the NovaSeq sequencer (Illumina), and raw data were provided by BGI. For qRT-PCR validation, cDNA was synthesized using SuperScript IV VILO master mix (Invitrogen, Thermo Fisher Scientific Baltics UAB, Vilnius, Lithuania). Gene expression profiling was performed on the BioMark system for high-throughput qRT-PCR (Fluidigm, San Francisco, CA, USA) with the TaqMan gene expression assays (Applied Biosystems, Life Technologies Corporation, Pleasanton, CA, USA) listed in Table S3. The schema of the high-throughput qRT-PCR is shown in Fig. S5.
RNA seq data analysis.
Transcript abundance from RNA seq reads was quantified with Salmon (150) and used to test for differential expression with a negative binomial distribution model implemented in the DESeq2 (151) package from Bioconductor (152). Maternal body mass index (BMI) and the gestational age at time of amniocentesis were included as covariates in between-group comparisons when these covariates were significantly different between the groups. Gestational age at amniocentesis was chosen because our focus is based on the inflammatory/microbiological status of the amniotic cavity. Genes with a minimum fold change of 1.25-fold and an adjusted P value of <0.1 were considered differentially expressed. The differentially expressed genes (DEGs) for each between-group comparison were used as input in iPathwayGuide (Advaita Bioinformatics, Ann Arbor, MI, USA) (153–155) to determine the significantly impacted pathways. Volcano plots were used to display the evidence of differential expression for each comparison. iPathwayGuide pathway annotations were derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, release 90.0+/05-29 (156, 157). The top 10 most significantly enriched GO biological processes in strictly upregulated or downregulated genes are also shown. Heatmaps were generated on log2 read counts for the top 100 genes selected by magnitude of change. For data visualization using principal-component analysis (PCA) and heatmaps, read count data were first adjusted to remove the effect of BMI and gestational age at amniocentesis based on the effects of these covariates in the control group. Upstream regulator analysis ranks genes using a P value that combines with Fisher’s method with two other P values, one P value corresponding to the z-score computed on the number of incoming edges of the differentially expressed target genes downstream of the regulator, and another P value based on the number of differentially expressed targets consistent with the type of incoming signal and the selected hypothesis type (regulator activation or inhibition). The interactions in the regulatory network are taken from BioGRID (158, 159) and iPathwayGuide's proprietary knowledge base.
qRT-PCR data analysis.
Delta threshold cycle (ΔCt) values were determined using multiple reference genes (RPLP0, GAPDH, and ACTB) averaged within each sample to determine gene expression levels. The −ΔCt values were normalized by calculating the z-score of each gene, and heatmaps were created using the Subio platform (Subio, Inc, Kagoshima, Japan; https://www.subioplatform.com/). Relative fold changes for mRNA expression were calculated the no intra-amniotic inflammation group in the comparison between intra-amniotic inflammation and no intra-amniotic inflammation (Fig. 2), the intra-amniotic infection group in the comparison between sterile intra-amniotic inflammation and intra-amniotic infection (Fig. 4), and the no intra-amniotic inflammation group in the comparison between sterile intra-amniotic inflammation and no intra-amniotic inflammation (Fig. 5) as the references using the 2−ΔΔCt method (160). The Mann-Whitney U-test was used to compare gene expression fold changes obtained by utilizing qRT-PCR. A P value <0.05 was considered statistically significant for all tests. The validation rate was calculated as the percentage of genes that were shown to be significantly upregulated or downregulated, utilizing both RNA seq and qRT-PCR, among the total number of significant DEGs chosen for validation.
Statistical analysis of the patient demographics.
Statistical analyses were conducted using GraphPad Prism version 8.0.1 for Windows (GraphPad Software, San Diego, CA, USA). The Kruskal-Wallis tests or one-way analysis of variance (ANOVA) with post hoc multiple comparisons were used to compare continuous variables based on the normality of the distribution of the groups. Fisher’s exact test was used for nominal variables. A P value of <0.05 was considered statistically significant for all tests.
Data availability.
The majority of the data underlying this article are available in the article. The RNA seq data generated herein are available from the Gene Expression Omnibus (GEO accession number GSE155526).
Supplementary Material
ACKNOWLEDGMENTS
We thank the physicians and nurses from the Center for Advanced Obstetrical Care and Research and the Intrapartum Unit for their help in collecting human samples. We also thank the staff members of the PRB Clinical Laboratory and the PRB Histology/Pathology Unit for the processing and examination of the pathological sections. Lastly, we thank Derek Miller for his critical readings of the manuscript.
This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS) and, in part, with federal funds from NICHD/NIH/DHHS under contract no. HHSN275201300006C. R.R. contributed to this work as part of his official duties as an employee of the United States Federal Government. This research was also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal and Child Health. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare no potential conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Blencowe H, Lee AC, Cousens S, Bahalim A, Narwal R, Zhong N, Chou D, Say L, Modi N, Katz J, Vos T, Marlow N, Lawn JE. 2013. Preterm birth-associated neurodevelopmental impairment estimates at regional and global levels for 2010. Pediatr Res 74:17–34. 10.1038/pr.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, Black RE. 2015. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 385:430–440. 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 3.Chawanpaiboon S, Vogel JP, Moller AB, Lumbiganon P, Petzold M, Hogan D, Landoulsi S, Jampathong N, Kongwattanakul K, Laopaiboon M, Lewis C, Rattanakanokchai S, Teng DN, Thinkhamrop J, Watananirun K, Zhang J, Zhou W, Gulmezoglu AM. 2019. Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob Health 7:e37–e46. 10.1016/S2214-109X(18)30451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AC, Blencowe H, Lawn JE. 2019. Small babies, big numbers: global estimates of preterm birth. Lancet Glob Health 7:e2–e3. 10.1016/S2214-109X(18)30484-4. [DOI] [PubMed] [Google Scholar]
- 5.Goldenberg RL, Culhane JF, Iams JD, Romero R. 2008. Epidemiology and causes of preterm birth. Lancet 371:75–84. 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero R, Dey SK, Fisher SJ. 2014. Preterm labor: one syndrome, many causes. Science 345:760–765. 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gravett MG, Witkin SS, Haluska GJ, Edwards JL, Cook MJ, Novy MJ. 1994. An experimental model for intraamniotic infection and preterm labor in rhesus monkeys. Am J Obstet Gynecol 171:1660–1667. 10.1016/0002-9378(94)90418-9. [DOI] [PubMed] [Google Scholar]
- 8.Gravett MG, Adams KM, Sadowsky DW, Grosvenor AR, Witkin SS, Axthelm MK, Novy MJ. 2007. Immunomodulators plus antibiotics delay preterm delivery after experimental intraamniotic infection in a nonhuman primate model. Am J Obstet Gynecol 197:518.e1–518.e8. 10.1016/j.ajog.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novy MJ, Duffy L, Axthelm MK, Sadowsky DW, Witkin SS, Gravett MG, Cassell GH, Waites KB. 2009. Ureaplasma parvum or Mycoplasma hominis as sole pathogens cause chorioamnionitis, preterm delivery, and fetal pneumonia in rhesus macaques. Reprod Sci 16:56–70. 10.1177/1933719108325508. [DOI] [PubMed] [Google Scholar]
- 10.Grigsby PL, Novy MJ, Sadowsky DW, Morgan TK, Long M, Acosta E, Duffy LB, Waites KB. 2012. Maternal azithromycin therapy for Ureaplasma intraamniotic infection delays preterm delivery and reduces fetal lung injury in a primate model. Am J Obstet Gynecol 207:475.e1–475.e14. 10.1016/j.ajog.2012.10.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez-Lopez N, Romero R, Plazyo O, Panaitescu B, Furcron AE, Miller D, Roumayah T, Flom E, Hassan SS. 2016. Intra-amniotic administration of HMGB1 induces spontaneous preterm labor and birth. Am J Reprod Immunol 75:3–7. 10.1111/aji.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Lopez N, Romero R, Arenas-Hernandez M, Panaitescu B, Garcia-Flores V, Mial TN, Sahi A, Hassan SS. 2018. Intra-amniotic administration of lipopolysaccharide induces spontaneous preterm labor and birth in the absence of a body temperature change. J Matern Fetal Neonatal Med 31:439–446. 10.1080/14767058.2017.1287894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Flores V, Romero R, Miller D, Xu Y, Done B, Veerapaneni C, Leng Y, Arenas-Hernandez M, Khan N, Panaitescu B, Hassan SS, Alvarez-Salas LM, Gomez-Lopez N. 2018. Inflammation-induced adverse pregnancy and neonatal outcomes can be improved by the immunomodulatory peptide exendin-4. Front Immunol 9:1291. 10.3389/fimmu.2018.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Lopez N, Romero R, Garcia-Flores V, Leng Y, Miller D, Hassan SS, Hsu CD, Panaitescu B. 2019. Inhibition of the NLRP3 inflammasome can prevent sterile intra-amniotic inflammation, preterm labor/birth, and adverse neonatal outcomes. Biol Reprod 100:1306–1318. 10.1093/biolre/ioy264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faro J, Romero R, Schwenkel G, Garcia-Flores V, Arenas-Hernandez M, Leng Y, Xu Y, Miller D, Hassan SS, Gomez-Lopez N. 2019. Intra-amniotic inflammation induces preterm birth by activating the NLRP3 inflammasome. Biol Reprod 100:1290–1305. 10.1093/biolre/ioy261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman M, Orvis A, Wu TY, Dacanay M, Merillat S, Ogle J, Baldessari A, Kretzer NM, Munson J, Boros-Rausch AJ, Shynlova O, Lye S, Rajagopal L, Adams WK. 2020. A broad spectrum chemokine inhibitor prevents preterm labor but not microbial invasion of the amniotic cavity or neonatal morbidity in a non-human primate model. Front Immunol 11:770. 10.3389/fimmu.2020.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motomura K, Romero R, Xu Y, Theis KR, Galaz J, Winters AD, Slutsky R, Garcia-Flores V, Zou C, Levenson D, Para R, Ahmad MM, Miller D, Hsu CD, Gomez-Lopez N. 2020. Intra-amniotic infection with Ureaplasma parvum causes preterm birth and neonatal mortality that are prevented by treatment with clarithromycin. mBio 11:e00797-20. 10.1128/mBio.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motomura K, Romero R, Garcia-Flores V, Leng Y, Xu Y, Galaz J, Slutsky R, Levenson D, Gomez-Lopez N. 2020. The alarmin interleukin-1alpha causes preterm birth through the NLRP3 inflammasome. Mol Hum Reprod 26:712–726. 10.1093/molehr/gaaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero R, Sirtori M, Oyarzun E, Avila C, Mazor M, Callahan R, Sabo V, Athanassiadis AP, Hobbins JC. 1989. Infection and labor. V. Prevalence, microbiology, and clinical significance of intraamniotic infection in women with preterm labor and intact membranes. Am J Obstet Gynecol 161:817–824. 10.1016/0002-9378(89)90409-2. [DOI] [PubMed] [Google Scholar]
- 20.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, Jun JK. 2001. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol 185:1130–1136. 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 21.Gravett MG, Novy MJ, Rosenfeld RG, Reddy AP, Jacob T, Turner M, McCormack A, Lapidus JA, Hitti J, Eschenbach DA, Roberts CT, Jr., Nagalla SR. 2004. Diagnosis of intra-amniotic infection by proteomic profiling and identification of novel biomarkers. JAMA 292:462–469. 10.1001/jama.292.4.462. [DOI] [PubMed] [Google Scholar]
- 22.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L. 2014. A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes. Am J Reprod Immunol 71:330–358. 10.1111/aji.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero R, Miranda J, Chaiworapongsa T, Korzeniewski SJ, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Yeo L. 2014. Prevalence and clinical significance of sterile intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Reprod Immunol 72:458–474. 10.1111/aji.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM. 2015. Sterile intra-amniotic inflammation in asymptomatic patients with a sonographic short cervix: prevalence and clinical significance. J Matern Fetal Neonatal Med 28:1343–1359. 10.3109/14767058.2014.954243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero R, Miranda J, Chaemsaithong P, Chaiworapongsa T, Kusanovic JP, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM. 2015. Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med 28:1394–1409. 10.3109/14767058.2014.958463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matzinger P. 1998. An innate sense of danger. Semin Immunol 10:399–415. 10.1006/smim.1998.0143. [DOI] [PubMed] [Google Scholar]
- 27.Oppenheim JJ, Yang D. 2005. Alarmins: chemotactic activators of immune responses. Curr Opin Immunol 17:359–365. 10.1016/j.coi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 28.Bianchi ME. 2007. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol 81:1–5. 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 29.Lotze MT, Deisseroth A, Rubartelli A. 2007. Damage associated molecular pattern molecules. Clin Immunol 124:1–4. 10.1016/j.clim.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plazyo O, Romero R, Unkel R, Balancio A, Mial TN, Xu Y, Dong Z, Hassan SS, Gomez-Lopez N. 2016. HMGB1 induces an inflammatory response in the chorioamniotic membranes that is partially mediated by the inflammasome. Biol Reprod 95:130. 10.1095/biolreprod.116.144139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero R, Grivel JC, Tarca AL, Chaemsaithong P, Xu Z, Fitzgerald W, Hassan SS, Chaiworapongsa T, Margolis L. 2015. Evidence of perturbations of the cytokine network in preterm labor. Am J Obstet Gynecol 213:836.e1–836.e18. 10.1016/j.ajog.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-Lopez N, Romero R, Panaitescu B, Leng Y, Xu Y, Tarca AL, Faro J, Pacora P, Hassan SS, Hsu CD. 2018. Inflammasome activation during spontaneous preterm labor with intra-amniotic infection or sterile intra-amniotic inflammation. Am J Reprod Immunol 80:e13049. 10.1111/aji.13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomez-Lopez N, Romero R, Tarca AL, Miller D, Panaitescu B, Schwenkel G, Gudicha DW, Hassan SS, Pacora P, Jung E, Hsu CD. 2019. Gasdermin D: evidence of pyroptosis in spontaneous preterm labor with sterile intra-amniotic inflammation or intra-amniotic infection. Am J Reprod Immunol 82:e13184. 10.1111/aji.13184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peiris HN, Romero R, Vaswani K, Reed S, Gomez-Lopez N, Tarca AL, Gudicha DW, Erez O, Maymon E, Mitchell MD. 2019. Preterm labor is characterized by a high abundance of amniotic fluid prostaglandins in patients with intra-amniotic infection or sterile intra-amniotic inflammation. J Matern Fetal Neonatal Med :1–16. 10.1080/14767058.2019.1702953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatti G, Romero R, Rice GE, Fitzgerald W, Pacora P, Gomez-Lopez N, Kavdia M, Tarca AL, Margolis L. 2020. Compartmentalized profiling of amniotic fluid cytokines in women with preterm labor. PLoS One 15:e0227881. 10.1371/journal.pone.0227881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musilova I, Kolackova M, Andrys C, Drahosova M, Baranova I, Chmelarova M, Stranik J, Jacobsson B, Kacerovsky M. 2019. Nicotinamide phosphoribosyltransferase and intra-amniotic inflammation in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med 34:736–746. 10.1080/14767058.2019.1615049. [DOI] [PubMed] [Google Scholar]
- 37.Gomez-Lopez N, Romero R, Maymon E, Kusanovic JP, Panaitescu B, Miller D, Pacora P, Tarca AL, Motomura K, Erez O, Jung E, Hassan SS, Hsu CD. 2019. Clinical chorioamnionitis at term IX: in vivo evidence of intra-amniotic inflammasome activation. J Perinat Med 47:276–287. 10.1515/jpm-2018-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Combs CA, Gravett M, Garite TJ, Hickok DE, Lapidus J, Porreco R, Rael J, Grove T, Morgan TK, Clewell W, Miller H, Luthy D, Pereira L, Nageotte M, Robilio PA, Fortunato S, Simhan H, Baxter JK, Amon E, Franco A, Trofatter K, Heyborne K, ProteoGenix/Obstetrix Collaborative Research Network. 2014. Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am J Obstet Gynecol 210:125.e1–125.e15. 10.1016/j.ajog.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Varea A, Romero R, Xu Y, Miller D, Ahmed AI, Chaemsaithong P, Chaiyasit N, Yeo L, Shaman M, Lannaman K, Cher B, Hassan SS, Gomez-Lopez N. 2017. Clinical chorioamnionitis at term VII: the amniotic fluid cellular immune response. J Perinat Med 45:523–538. 10.1515/jpm-2016-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez-Lopez N, Romero R, Xu Y, Miller D, Arenas-Hernandez M, Garcia-Flores V, Panaitescu B, Galaz J, Hsu CD, Para R, Berry SM. 2019. Fetal T cell activation in the amniotic cavity during preterm labor: a potential mechanism for a subset of idiopathic preterm birth. J Immunol 203:1793–1807. 10.4049/jimmunol.1900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomez-Lopez N, Romero R, Galaz J, Xu Y, Panaitescu B, Slutsky R, Motomura K, Gill N, Para R, Pacora P, Jung E, Hsu CD. 2019. Cellular immune responses in amniotic fluid of women with preterm labor and intra-amniotic infection or intra-amniotic inflammation. Am J Reprod Immunol 82:e13171. 10.1111/aji.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galaz J, Romero R, Xu Y, Miller D, Slutsky R, Levenson D, Hsu CD, Gomez-Lopez N. 2020. Cellular immune responses in amniotic fluid of women with preterm clinical chorioamnionitis. Inflamm Res 69:203–216. 10.1007/s00011-019-01308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galaz J, Romero R, Slutsky R, Xu Y, Motomura K, Para R, Pacora P, Panaitescu B, Hsu CD, Kacerovsky M, Gomez-Lopez N. 2020. Cellular immune responses in amniotic fluid of women with preterm prelabor rupture of membranes. J Perinat Med 48:222–233. 10.1515/jpm-2019-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bourne G. 1962. The foetal membranes. A review of the anatomy of normal amnion and chorion and some aspects of their function. Postgrad Med J 38:193–201. 10.1136/pgmj.38.438.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLaren J, Malak TM, Bell SC. 1999. Structural characteristics of term human fetal membranes prior to labour: identification of an area of altered morphology overlying the cervix. Hum Reprod 14:237–241. 10.1093/humrep/14.1.237. [DOI] [PubMed] [Google Scholar]
- 46.Lowe DE, Robbins JR, Bakardjiev AI. 2018. Animal and human tissue models of vertical Listeria monocytogenes transmission and implications for other pregnancy-associated infections. Infect Immun 86:e00801-17. 10.1128/IAI.00801-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawdy RJ, Dennes WJ, Allport V, Slater DM, Elder MG, Sullivan MH, Bennett PR. 1999. Region and labour-dependent synthesis of prostaglandin E2 by human fetal membranes. Placenta 20:181–184. 10.1053/plac.1998.0372. [DOI] [PubMed] [Google Scholar]
- 48.Lappas M, Odumetse TL, Riley C, Reti NG, Holdsworth-Carson SJ, Rice GE, Permezel M. 2008. Pre-labour fetal membranes overlying the cervix display alterations in inflammation and NF-kappaB signalling pathways. Placenta 29:995–1002. 10.1016/j.placenta.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Talmi YP, Sigler L, Inge E, Finkelstein Y, Zohar Y. 1991. Antibacterial properties of human amniotic membranes. Placenta 12:285–288. 10.1016/0143-4004(91)90010-d. [DOI] [PubMed] [Google Scholar]
- 50.Kjaergaard N, Hein M, Hyttel L, Helmig RB, Schonheyder HC, Uldbjerg N, Madsen H. 2001. Antibacterial properties of human amnion and chorion in vitro. Eur J Obstet Gynecol Reprod Biol 94:224–229. 10.1016/s0301-2115(00)00345-6. [DOI] [PubMed] [Google Scholar]
- 51.King AE, Paltoo A, Kelly RW, Sallenave JM, Bocking AD, Challis JR. 2007. Expression of natural antimicrobials by human placenta and fetal membranes. Placenta 28:161–169. 10.1016/j.placenta.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 52.Boldenow E, Jones S, Lieberman RW, Chames MC, Aronoff DM, Xi C, Loch-Caruso R. 2013. Antimicrobial peptide response to group B Streptococcus in human extraplacental membranes in culture. Placenta 34:480–485. 10.1016/j.placenta.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russell P. 1979. Inflammatory lesions of the human placenta: clinical significance of acute chorioamnionitis. Am J Diagn Gynecol Obstet 2:127–137. [Google Scholar]
- 54.Guzick DS, Winn K. 1985. The association of chorioamnionitis with preterm delivery. Obstet Gynecol 65:11–16. [PubMed] [Google Scholar]
- 55.Srinivas SK, Ma Y, Sammel MD, Chou D, McGrath C, Parry S, Elovitz MA. 2006. Placental inflammation and viral infection are implicated in second trimester pregnancy loss. Am J Obstet Gynecol 195:797–802. 10.1016/j.ajog.2006.05.049. [DOI] [PubMed] [Google Scholar]
- 56.Srinivas SK, Ernst LM, Edlow AG, Elovitz MA. 2008. Can placental pathology explain second-trimester pregnancy loss and subsequent pregnancy outcomes? Am J Obstet Gynecol 199:402.e1-5. 10.1016/j.ajog.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM. 2015. Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance. Am J Obstet Gynecol 213:S29–S52. 10.1016/j.ajog.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomez-Lopez N, Romero R, Varrey A, Leng Y, Miller D, Done B, Xu Y, Bhatti G, Motomura K, Gershater M, Pique-Regi R, Tarca AL. 2020. RNA sequencing reveals diverse functions of amniotic fluid neutrophils and monocytes/macrophages in intra-amniotic infection. J Innate Immun :1–20. 10.1159/000509718:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoon BH, Chang JW, Romero R. 1998. Isolation of Ureaplasma urealyticum from the amniotic cavity and adverse outcome in preterm labor. Obstet Gynecol 92:77–82. 10.1016/S0029-7844(98)00122-7. [DOI] [PubMed] [Google Scholar]
- 60.DiGiulio DB, Romero R, Kusanovic JP, Gomez R, Kim CJ, Seok KS, Gotsch F, Mazaki-Tovi S, Vaisbuch E, Sanders K, Bik EM, Chaiworapongsa T, Oyarzun E, Relman DA. 2010. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol 64:38–57. 10.1111/j.1600-0897.2010.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romero R, Miranda J, Kusanovic JP, Chaiworapongsa T, Chaemsaithong P, Martinez A, Gotsch F, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM. 2015. Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques. J Perinat Med 43:19–36. 10.1515/jpm-2014-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giguere PM, Billard MJ, Laroche G, Buckley BK, Timoshchenko RG, McGinnis MW, Esserman D, Foreman O, Liu P, Siderovski DP, Tarrant TK. 2013. G-protein signaling modulator-3, a gene linked to autoimmune diseases, regulates monocyte function and its deficiency protects from inflammatory arthritis. Mol Immunol 54:193–198. 10.1016/j.molimm.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Branham-O'Connor M, Robichaux WG, III, Zhang XK, Cho H, Kehrl JH, Lanier SM, Blumer JB. 2014. Defective chemokine signal integration in leukocytes lacking activator of G protein signaling 3 (AGS3). J Biol Chem 289:10738–10747. 10.1074/jbc.M113.515031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Billard MJ, Gall BJ, Richards KL, Siderovski DP, Tarrant TK. 2014. G protein signaling modulator-3: a leukocyte regulator of inflammation in health and disease. Am J Clin Exp Immunol 3:97–106. [PMC free article] [PubMed] [Google Scholar]
- 65.Gomez-Lopez N, Romero R, Xu Y, Plazyo O, Unkel R, Leng Y, Than NG, Chaiworapongsa T, Panaitescu B, Dong Z, Tarca AL, Abrahams VM, Yeo L, Hassan SS. 2017. A role for the inflammasome in spontaneous preterm labor with acute histologic chorioamnionitis. Reprod Sci 24:1382–1401. 10.1177/1933719116687656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blanc WA. 1959. Amniotic infection syndrome; pathogenesis, morphology, and significance in circumnatal mortality. Clin Obstet Gynecol 2:705–734. 10.1097/00003081-195902030-00010. [DOI] [PubMed] [Google Scholar]
- 67.Blanc WA. 1979. Pathology of the placenta and cord in ascending and in haematogenous infection. Ciba Found Symp 17–38. 10.1002/9780470720608.ch3:17-38. [DOI] [PubMed] [Google Scholar]
- 68.Hillier SL, Martius J, Krohn M, Kiviat N, Holmes KK, Eschenbach DA. 1988. A case-control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. N Engl J Med 319:972–978. 10.1056/NEJM198810133191503. [DOI] [PubMed] [Google Scholar]
- 69.Hillier SL, Krohn MA, Kiviat NB, Watts DH, Eschenbach DA. 1991. Microbiologic causes and neonatal outcomes associated with chorioamnion infection. Am J Obstet Gynecol 165:955–961. 10.1016/0002-9378(91)90447-y. [DOI] [PubMed] [Google Scholar]
- 70.Romero R, Salafia CM, Athanassiadis AP, Hanaoka S, Mazor M, Sepulveda W, Bracken MB. 1992. The relationship between acute inflammatory lesions of the preterm placenta and amniotic fluid microbiology. Am J Obstet Gynecol 166:1382–1388. 10.1016/0002-9378(92)91609-e. [DOI] [PubMed] [Google Scholar]
- 71.Redline RW, Faye-Petersen O, Heller D, Qureshi F, Savell V, Vogler C, The Society for Pediatric Pathology, Perinatal Section, Amniotic Fluid Infection Nosology Committee. 2003. Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol 6:435–448. 10.1007/s10024-003-7070-y. [DOI] [PubMed] [Google Scholar]
- 72.Romero R, Chaemsaithong P, Docheva N, Korzeniewski SJ, Kusanovic JP, Yoon BH, Kim JS, Chaiyasit N, Ahmed AI, Qureshi F, Jacques SM, Kim CJ, Hassan SS, Chaiworapongsa T, Yeo L, Kim YM. 2016. Clinical chorioamnionitis at term VI: acute chorioamnionitis and funisitis according to the presence or absence of microorganisms and inflammation in the amniotic cavity. J Perinat Med 44:33–51. 10.1515/jpm-2015-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Draber P, Vonkova I, Stepanek O, Hrdinka M, Kucova M, Skopcova T, Otahal P, Angelisova P, Horejsi V, Yeung M, Weiss A, Brdicka T. 2011. SCIMP, a transmembrane adaptor protein involved in major histocompatibility complex class II signaling. Mol Cell Biol 31:4550–4562. 10.1128/MCB.05817-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ito M, Nakashima A, Hidaka T, Okabe M, Bac ND, Ina S, Yoneda S, Shiozaki A, Sumi S, Tsuneyama K, Nikaido T, Saito S. 2010. A role for IL-17 in induction of an inflammation at the fetomaternal interface in preterm labour. J Reprod Immunol 84:75–85. 10.1016/j.jri.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 75.Gillaux C, Mehats C, Vaiman D, Cabrol D, Breuiller-Fouche M. 2011. Functional screening of TLRs in human amniotic epithelial cells. J Immunol 187:2766–2774. 10.4049/jimmunol.1100217. [DOI] [PubMed] [Google Scholar]
- 76.Kacerovsky M, Andrys C, Hornychova H, Pliskova L, Lancz K, Musilova I, Drahosova M, Bolehovska R, Tambor V, Jacobsson B. 2012. Amniotic fluid soluble Toll-like receptor 4 in pregnancies complicated by preterm prelabor rupture of the membranes. J Matern Fetal Neonatal Med 25:1148–1155. 10.3109/14767058.2011.626821. [DOI] [PubMed] [Google Scholar]
- 77.Abrahams VM, Potter JA, Bhat G, Peltier MR, Saade G, Menon R. 2013. Bacterial modulation of human fetal membrane Toll-like receptor expression. Am J Reprod Immunol 69:33–40. 10.1111/aji.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Romero R, Chaemsaithong P, Korzeniewski SJ, Tarca AL, Bhatti G, Xu Z, Kusanovic JP, Dong Z, Docheva N, Martinez-Varea A, Yoon BH, Hassan SS, Chaiworapongsa T, Yeo L. 2016. Clinical chorioamnionitis at term II: the intra-amniotic inflammatory response. J Perinat Med 44:5–22. 10.1515/jpm-2015-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, Leite R, Elovitz MA, Parry S, Bushman FD. 2016. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4:29. 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rowlands S, Danielewski JA, Tabrizi SN, Walker SP, Garland SM. 2017. Microbial invasion of the amniotic cavity in midtrimester pregnancies using molecular microbiology. Am J Obstet Gynecol 217:71.e1–71.e5. 10.1016/j.ajog.2017.02.051. [DOI] [PubMed] [Google Scholar]
- 81.Lim ES, Rodriguez C, Holtz LR. 2018. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 6:87. 10.1186/s40168-018-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leiby JS, McCormick K, Sherrill-Mix S, Clarke EL, Kessler LR, Taylor LJ, Hofstaedter CE, Roche AM, Mattei LM, Bittinger K, Elovitz MA, Leite R, Parry S, Bushman FD. 2018. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome 6:196. 10.1186/s40168-018-0575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Theis KR, Romero R, Winters AD, Greenberg JM, Gomez-Lopez N, Alhousseini A, Bieda J, Maymon E, Pacora P, Fettweis JM, Buck GA, Jefferson KK, Strauss JF, III, Erez O, Hassan SS. 2019. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am J Obstet Gynecol 220:267.e1–267.e39. 10.1016/j.ajog.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, Parkhill J, Charnock-Jones DS, Smith GCS. 2019. Human placenta has no microbiome but can contain potential pathogens. Nature 572:329–334. 10.1038/s41586-019-1451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Theis KR, Romero R, Greenberg JM, Winters AD, Garcia-Flores V, Motomura K, Ahmad MM, Galaz J, Arenas-Hernandez M, Gomez-Lopez N. 2020. No consistent evidence for microbiota in murine placental and fetal tissues. mSphere 5:e00933-19. 10.1128/mSphere.00933-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Theis KR, Romero R, Winters AD, Jobe AH, Gomez-Lopez N. 2020. Lack of evidence for microbiota in the placental and fetal tissues of rhesus macaques. mSphere 5:e00210-20. 10.1128/mSphere.00210-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuperman AA, Zimmerman A, Hamadia S, Ziv O, Gurevich V, Fichtman B, Gavert N, Straussman R, Rechnitzer H, Barzilay M, Shvalb S, Bornstein J, Ben-Shachar I, Yagel S, Haviv I, Koren O. 2020. Deep microbial analysis of multiple placentas shows no evidence for a placental microbiome. BJOG 127:159–169. 10.1111/1471-0528.15896. [DOI] [PubMed] [Google Scholar]
- 88.Burnham P, Gomez-Lopez N, Heyang M, Cheng AP, Lenz JS, Dadhania DM, Lee JR, Suthanthiran M, Romero R, De Vlaminck I. 2020. Separating the signal from the noise in metagenomic cell-free DNA sequencing. Microbiome 8:18. 10.1186/s40168-020-0793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Romero R, Mazor M. 1988. Infection and preterm labor. Clin Obstet Gynecol 31:553–584. 10.1097/00003081-198809000-00006. [DOI] [PubMed] [Google Scholar]
- 90.Romero R, Gomez R, Chaiworapongsa T, Conoscenti G, Kim JC, Kim YM. 2001. The role of infection in preterm labour and delivery. Paediatr Perinat Epidemiol 15(Suppl 2):41–56. 10.1046/j.1365-3016.2001.00007.x. [DOI] [PubMed] [Google Scholar]
- 91.Romero R, Espinoza J, Kusanovic JP, Gotsch F, Hassan S, Erez O, Chaiworapongsa T, Mazor M. 2006. The preterm parturition syndrome. BJOG 113(Suppl 3):17–42. 10.1111/j.1471-0528.2006.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim MJ, Romero R, Gervasi MT, Kim JS, Yoo W, Lee DC, Mittal P, Erez O, Kusanovic JP, Hassan SS, Kim CJ. 2009. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab Invest 89:924–936. 10.1038/labinvest.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Romero R, Gomez-Lopez N, Winters AD, Jung E, Shaman M, Bieda J, Panaitescu B, Pacora P, Erez O, Greenberg JM, Ahmad MM, Hsu CD, Theis KR. 2019. Evidence that intra-amniotic infections are often the result of an ascending invasion - a molecular microbiological study. J Perinat Med 47:915–931. 10.1515/jpm-2019-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Vornhagen J, Quach P, Boldenow E, Merillat S, Whidbey C, Ngo LY, Adams Waldorf KM, Rajagopal L. 2016. Bacterial hyaluronidase promotes ascending GBS infection and preterm Birth. mBio 7:e00781-16. 10.1128/mBio.00781-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Flaherty RA, Magel M, Aronoff DM, Gaddy JA, Petroff MG, Manning SD. 2019. Modulation of death and inflammatory signaling in decidual stromal cells following exposure to group B streptococcus. Infect Immun 87:e00729-19. 10.1128/IAI.00729-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oh KJ, Lee KA, Sohn YK, Park CW, Hong JS, Romero R, Yoon BH. 2010. Intraamniotic infection with genital mycoplasmas exhibits a more intense inflammatory response than intraamniotic infection with other microorganisms in patients with preterm premature rupture of membranes. Am J Obstet Gynecol 203:211.e1-8. 10.1016/j.ajog.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Oh KJ, Romero R, Park JY, Hong JS, Yoon BH. 2019. The earlier the gestational age, the greater the intensity of the intra-amniotic inflammatory response in women with preterm premature rupture of membranes and amniotic fluid infection by Ureaplasma species. J Perinat Med 47:516–527. 10.1515/jpm-2019-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McNamara MF, Wallis T, Qureshi F, Jacques SM, Gonik B. 1997. Determining the maternal and fetal cellular immunologic contributions in preterm deliveries with clinical or subclinical chorioamnionitis. Infect Dis Obstet Gynecol 5:273–279. 10.1155/S1064744997000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Steel JH, O'Donoghue K, Kennea NL, Sullivan MH, Edwards AD. 2005. Maternal origin of inflammatory leukocytes in preterm fetal membranes, shown by fluorescence in situ hybridisation. Placenta 26:672–677. 10.1016/j.placenta.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 100.Redline RW. 2004. Placental inflammation. Semin Neonatol 9:265–274. 10.1016/j.siny.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 101.Redline RW. 2015. Classification of placental lesions. Am J Obstet Gynecol 213:S21–8. 10.1016/j.ajog.2015.05.056. [DOI] [PubMed] [Google Scholar]
- 102.Gomez-Lopez N, Romero R, Leng Y, Garcia-Flores V, Xu Y, Miller D, Hassan SS. 2017. Neutrophil extracellular traps in acute chorioamnionitis: a mechanism of host defense. Am J Reprod Immunol 77:e12617. 10.1111/aji.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matsubara S, Yamada T, Minakami H, Watanabe T, Takizawa T, Sato I. 1999. Polymorphonuclear leukocytes in the fetal membranes are activated in patients with preterm delivery: ultrastructural and enzyme-histochemical evidence. Placenta 20:185–188. 10.1053/plac.1998.0366. [DOI] [PubMed] [Google Scholar]
- 104.Hamilton SA, Tower CL, Jones RL. 2013. Identification of chemokines associated with the recruitment of decidual leukocytes in human labour: potential novel targets for preterm labour. PLoS One 8:e56946. 10.1371/journal.pone.0056946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Presicce P, Park CW, Senthamaraikannan P, Bhattacharyya S, Jackson C, Kong F, Rueda CM, DeFranco E, Miller LA, Hildeman DA, Salomonis N, Chougnet CA, Jobe AH, Kallapur SG. 2018. IL-1 signaling mediates intrauterine inflammation and chorio-decidua neutrophil recruitment and activation. JCI Insight 3:e98306. 10.1172/jci.insight.98306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Boldenow E, Gendrin C, Ngo L, Bierle C, Vornhagen J, Coleman M, Merillat S, Armistead B, Whidbey C, Alishetti V, Santana-Ufret V, Ogle J, Gough M, Srinouanprachanh S, MacDonald JW, Bammler TK, Bansal A, Liggitt HD, Rajagopal L, Waldorf KM. 2016. Group B Streptococcus circumvents neutrophils and neutrophil extracellular traps during amniotic cavity invasion and preterm labor. Sci Immunol 1:eaah4576. 10.1126/sciimmunol.aah4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tong M, Potter JA, Mor G, Abrahams VM. 2019. Lipopolysaccharide-stimulated human fetal membranes induce neutrophil activation and release of vital neutrophil extracellular traps. J Immunol 203:500–510. 10.4049/jimmunol.1900262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gomez-Lopez N, Estrada-Gutierrez G, Jimenez-Zamudio L, Vega-Sanchez R, Vadillo-Ortega F. 2009. Fetal membranes exhibit selective leukocyte chemotaxic activity during human labor. J Reprod Immunol 80:122–131. 10.1016/j.jri.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 109.Gomez-Lopez N, Vadillo-Perez L, Nessim S, Olson DM, Vadillo-Ortega F. 2011. Choriodecidua and amnion exhibit selective leukocyte chemotaxis during term human labor. Am J Obstet Gynecol 204:364.e9-16. 10.1016/j.ajog.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 110.Gomez-Lopez N, Hernandez-Santiago S, Lobb AP, Olson DM, Vadillo-Ortega F. 2013. Normal and premature rupture of fetal membranes at term delivery differ in regional chemotactic activity and related chemokine/cytokine production. Reprod Sci 20:276–284. 10.1177/1933719112452473. [DOI] [PubMed] [Google Scholar]
- 111.Gomez-Lopez N, Tanaka S, Zaeem Z, Metz GA, Olson DM. 2013. Maternal circulating leukocytes display early chemotactic responsiveness during late gestation. BMC Pregnancy Childbirth 13(Suppl 1):S8. 10.1186/1471-2393-13-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gomez-Lopez N, Tong WC, Arenas-Hernandez M, Tanaka S, Hajar O, Olson DM, Taggart MJ, Mitchell BF. 2015. Chemotactic activity of gestational tissues through late pregnancy, term labor, and RU486-induced preterm labor in guinea pigs. Am J Reprod Immunol 73:341–352. 10.1111/aji.12333. [DOI] [PubMed] [Google Scholar]
- 113.Gomez-Lopez N, Vega-Sanchez R, Castillo-Castrejon M, Romero R, Cubeiro-Arreola K, Vadillo-Ortega F. 2013. Evidence for a role for the adaptive immune response in human term parturition. Am J Reprod Immunol 69:212–230. 10.1111/aji.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gotsch F, Romero R, Chaiworapongsa T, Erez O, Vaisbuch E, Espinoza J, Kusanovic JP, Mittal P, Mazaki-Tovi S, Kim CJ, Kim JS, Edwin S, Nhan-Chang CL, Hamill N, Friel L, Than NG, Mazor M, Yoon BH, Hassan SS. 2008. Evidence of the involvement of caspase-1 under physiologic and pathologic cellular stress during human pregnancy: a link between the inflammasome and parturition. J Matern Fetal Neonatal Med 21:605–616. 10.1080/14767050802212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gomez-Lopez N, Motomura K, Miller D, Garcia-Flores V, Galaz J, Romero R. 2019. Inflammasomes: their role in normal and complicated pregnancies. J Immunol 203:2757–2769. 10.4049/jimmunol.1900901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Theis KR, Romero R, Motomura K, Galaz J, Winters AD, Pacora P, Miller D, Slutsky R, Florova V, Levenson D, Para R, Varrey A, Kacerovsky M, Hsu CD, Gomez-Lopez N. 2020. Microbial burden and inflammasome activation in amniotic fluid of patients with preterm prelabor rupture of membranes. J Perinat Med 48:115–131. 10.1515/jpm-2019-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee J, Romero R, Kim SM, Chaemsaithong P, Yoon BH. 2016. A new antibiotic regimen treats and prevents intra-amniotic inflammation/infection in patients with preterm PROM. J Matern Fetal Neonatal Med 29:2727–2737. 10.3109/14767058.2015.1103729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oh KJ, Romero R, Park JY, Lee J, Conde-Agudelo A, Hong JS, Yoon BH. 2019. Evidence that antibiotic administration is effective in the treatment of a subset of patients with intra-amniotic infection/inflammation presenting with cervical insufficiency. Am J Obstet Gynecol 221:140.e1–140.e18. 10.1016/j.ajog.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yoon BH, Romero R, Park JY, Oh KJ, Lee J, Conde-Agudelo A, Hong JS. 2019. Antibiotic administration can eradicate intra-amniotic infection or intra-amniotic inflammation in a subset of patients with preterm labor and intact membranes. Am J Obstet Gynecol 221:142.e1–142.e22. 10.1016/j.ajog.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kacerovsky M, Romero R, Stepan M, Stranik J, Maly J, Pliskova L, Bolehovska R, Palicka V, Zemlickova H, Hornychova H, Spacek J, Jacobsson B, Pacora P, Musilova I. 2020. Antibiotic administration reduces the rate of intraamniotic inflammation in preterm prelabor rupture of the membranes. Am J Obstet Gynecol 223:114.e1–114.e20. 10.1016/j.ajog.2020.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bertheloot D, Latz E. 2017. HMGB1, IL-1alpha, IL-33 and S100 proteins: dual-function alarmins. Cell Mol Immunol 14:43–64. 10.1038/cmi.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shank JM, Kelley BR, Jackson JW, Tweedie JL, Franklin D, Damo SM, Gaddy JA, Murphy CN, Johnson JG. 2018. The host antimicrobial protein calgranulin C participates in the control of Campylobacter jejuni growth via zinc sequestration. Infect Immun 86:e00234-18. 10.1128/IAI.00234-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park SJ, Yoon WG, Song JS, Jung HS, Kim CJ, Oh SY, Yoon BH, Jung G, Kim HJ, Nirasawa T. 2006. Proteome analysis of human amnion and amniotic fluid by two-dimensional electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proteomics 6:349–363. 10.1002/pmic.200500084. [DOI] [PubMed] [Google Scholar]
- 124.Choi YB, Son M, Park M, Shin J, Yun Y. 2010. SOCS-6 negatively regulates T cell activation through targeting p56lck to proteasomal degradation. J Biol Chem 285:7271–7280. 10.1074/jbc.M109.073726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kazi JU, Sun J, Phung B, Zadjali F, Flores-Morales A, Ronnstrand L. 2012. Suppressor of cytokine signaling 6 (SOCS6) negatively regulates Flt3 signal transduction through direct binding to phosphorylated tyrosines 591 and 919 of Flt3. J Biol Chem 287:36509–36517. 10.1074/jbc.M112.376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Szekeres-Bartho J, Csernus V, Hadnagy J, Pacsa AS. 1983. Immunosuppressive effect of serum progesterone during pregnancy depends on the progesterone binding capacity of the lymphocytes. J Reprod Immunol 5:81–88. 10.1016/0165-0378(83)90003-7. [DOI] [PubMed] [Google Scholar]
- 127.Szekeres-Bartho J, Kilaŕ F, Falkay G, Csernus V, Török A, Pacsa AS. 1985. The mechanism of the inhibitory effect of progesterone on lymphocyte cytotoxicity: I. Progesterone-treated lymphocytes release a substance inhibiting cytotoxicity and prostaglandin synthesis. Am J Reprod Immunol Microbiol 9:15–18. 10.1111/j.1600-0897.1985.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 128.Szekeres-Bartho J, Wegmann TG. 1996. A progesterone-dependent immunomodulatory protein alters the Th1/Th2 balance. J Reprod Immunol 31:81–95. 10.1016/0165-0378(96)00964-3. [DOI] [PubMed] [Google Scholar]
- 129.Kozma N, Halasz M, Polgar B, Poehlmann TG, Markert UR, Palkovics T, Keszei M, Par G, Kiss K, Szeberenyi J, Grama L, Szekeres-Bartho J. 2006. Progesterone-induced blocking factor activates STAT6 via binding to a novel IL-4 receptor. J Immunol 176:819–826. 10.4049/jimmunol.176.2.819. [DOI] [PubMed] [Google Scholar]
- 130.Kawasaki N, Isogaya K, Dan S, Yamori T, Takano H, Yao R, Morishita Y, Taguchi L, Morikawa M, Heldin CH, Noda T, Ehata S, Miyazono K, Koinuma D. 2018. TUFT1 interacts with RABGAP1 and regulates mTORC1 signaling. Cell Discov 4:1. 10.1038/s41421-017-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura S, Natsume T, Mizushima N. 2010. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem 285:20109–20116. 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. 2010. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest 120:803–815. 10.1172/JCI40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hirota Y, Cha J, Yoshie M, Daikoku T, Dey SK. 2011. Heightened uterine mammalian target of rapamycin complex 1 (mTORC1) signaling provokes preterm birth in mice. Proc Natl Acad Sci U S A 108:18073–18078. 10.1073/pnas.1108180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cha J, Hirota Y, Dey SK. 2012. Sensing senescence in preterm birth. Cell Cycle 11:205–206. 10.4161/cc.11.2.18781. [DOI] [PubMed] [Google Scholar]
- 135.Gomez-Lopez N, Romero R, Plazyo O, Schwenkel G, Garcia-Flores V, Unkel R, Xu Y, Leng Y, Hassan SS, Panaitescu B, Cha J, Dey SK. 2017. Preterm labor in the absence of acute histologic chorioamnionitis is characterized by cellular senescence of the chorioamniotic membranes. Am J Obstet Gynecol 217:592.e1–592.e17. 10.1016/j.ajog.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang X, Zheng M, Liu G, Xia W, McKeown-Longo PJ, Hung MC, Zhao J. 2007. Kruppel-like factor 8 induces epithelial to mesenchymal transition and epithelial cell invasion. Cancer Res 67:7184–7193. 10.1158/0008-5472.CAN-06-4729. [DOI] [PubMed] [Google Scholar]
- 137.Lines KE, Chelala C, Dmitrovic B, Wijesuriya N, Kocher HM, Marshall JF, Crnogorac-Jurcevic T. 2012. S100P-binding protein, S100PBP, mediates adhesion through regulation of cathepsin Z in pancreatic cancer cells. Am J Pathol 180:1485–1494. 10.1016/j.ajpath.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 138.Park CW, Moon KC, Park JS, Jun JK, Romero R, Yoon BH. 2009. The involvement of human amnion in histologic chorioamnionitis is an indicator that a fetal and an intra-amniotic inflammatory response is more likely and severe: clinical implications. Placenta 30:56–61. 10.1016/j.placenta.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Redline RW. 2012. Inflammatory response in acute chorioamnionitis. Semin Fetal Neonatal Med 17:20–25. 10.1016/j.siny.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 140.Bezbradica JS, Coll RC, Schroder K. 2017. Sterile signals generate weaker and delayed macrophage NLRP3 inflammasome responses relative to microbial signals. Cell Mol Immunol 14:118–126. 10.1038/cmi.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Romero R, Quintero R, Nores J, Avila C, Mazor M, Hanaoka S, Hagay Z, Merchant L, Hobbins JC. 1991. Amniotic fluid white blood cell count: a rapid and simple test to diagnose microbial invasion of the amniotic cavity and predict preterm delivery. Am J Obstet Gynecol 165:821–830. 10.1016/0002-9378(91)90423-o. [DOI] [PubMed] [Google Scholar]
- 142.Romero R, Jimenez C, Lohda AK, Nores J, Hanaoka S, Avila C, Callahan R, Mazor M, Hobbins JC, Diamond MP. 1990. Amniotic fluid glucose concentration: a rapid and simple method for the detection of intraamniotic infection in preterm labor. Am J Obstet Gynecol 163:968–974. 10.1016/0002-9378(90)91106-m. [DOI] [PubMed] [Google Scholar]
- 143.Romero R, Emamian M, Quintero R, Wan M, Hobbins JC, Mazor M, Edberg S. 1988. The value and limitations of the Gram stain examination in the diagnosis of intraamniotic infection. Am J Obstet Gynecol 159:114–119. 10.1016/0002-9378(88)90503-0. [DOI] [PubMed] [Google Scholar]
- 144.Oh KJ, Kim SM, Hong JS, Maymon E, Erez O, Panaitescu B, Gomez-Lopez N, Romero R, Yoon BH. 2017. Twenty-four percent of patients with clinical chorioamnionitis in preterm gestations have no evidence of either culture-proven intraamniotic infection or intraamniotic inflammation. Am J Obstet Gynecol 216:604.e1–604.e11. 10.1016/j.ajog.2017.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ecker DJ, Sampath R, Li H, Massire C, Matthews HE, Toleno D, Hall TA, Blyn LB, Eshoo MW, Ranken R, Hofstadler SA, Tang YW. 2010. New technology for rapid molecular diagnosis of bloodstream infections. Expert Rev Mol Diagn 10:399–415. 10.1586/erm.10.24. [DOI] [PubMed] [Google Scholar]
- 146.Seong HS, Lee SE, Kang JH, Romero R, Yoon BH. 2008. The frequency of microbial invasion of the amniotic cavity and histologic chorioamnionitis in women at term with intact membranes in the presence or absence of labor. Am J Obstet Gynecol 199:375.e1-5. 10.1016/j.ajog.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Park HS, Romero R, Lee SM, Park CW, Jun JK, Yoon BH. 2010. Histologic chorioamnionitis is more common after spontaneous labor than after induced labor at term. Placenta 31:792–795. 10.1016/j.placenta.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Romero R, Kim YM, Pacora P, Kim CJ, Benshalom-Tirosh N, Jaiman S, Bhatti G, Kim JS, Qureshi F, Jacques SM, Jung EJ, Yeo L, Panaitescu B, Maymon E, Hassan SS, Hsu CD, Erez O. 2018. The frequency and type of placental histologic lesions in term pregnancies with normal outcome. J Perinat Med 46:613–630. 10.1515/jpm-2018-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pacora P, Chaiworapongsa T, Maymon E, Kim YM, Gomez R, Yoon BH, Ghezzi F, Berry SM, Qureshi F, Jacques SM, Kim JC, Kadar N, Romero R. 2002. Funisitis and chorionic vasculitis: the histological counterpart of the fetal inflammatory response syndrome. J Matern Fetal Neonatal Med 11:18–25. 10.1080/jmf.11.1.18.25. [DOI] [PubMed] [Google Scholar]
- 150.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. 2007. A systems biology approach for pathway level analysis. Genome Res 17:1537–1545. 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Khatri P, Draghici S, Tarca AL, Hassan SS, Romero R. 2007. A system biology approach for the steady-state analysis of gene signaling networks. In Rueda L, Mery D, Kittler J (ed), Lecture notes in computer science, vol 4756. Springer, Berlin, Germany. [Google Scholar]
- 155.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, Kim CJ, Kusanovic JP, Romero R. 2009. A novel signaling pathway impact analysis. Bioinformatics 25:75–82. 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114. 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34:D535–D539. 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Kolas N, O'Donnell L, Leung G, McAdam R, Zhang F, Dolma S, Willems A, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M. 2019. The BioGRID interaction database: 2019 update. Nucleic Acids Res 47:D529–D541. 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The majority of the data underlying this article are available in the article. The RNA seq data generated herein are available from the Gene Expression Omnibus (GEO accession number GSE155526).