Klebsiella pneumoniae is an opportunistic pathogen that mostly affects patients with weakened immune systems, but a few serotypes (especially K1 and K2) are highly invasive and result in systemic infection in healthy persons. The ability to evade and survive the components of the innate immune system is critical in infection. To investigate the role and mechanism of transcription regulator KP1_RS12260 (KbvR) in virulence and defense against the innate immune response, kbvR deletion mutant and complement strains were constructed. The in vivo animal infection assay and in vitro antiphagocytosis assay demonstrate K. pneumoniae KbvR is an important regulator that contributes to virulence and the defense against phagocytosis of macrophages.
KEYWORDS: Klebsiella pneumoniae, KbvR, virulence, antiphagocytosis, capsule polysaccharide
ABSTRACT
Klebsiella pneumoniae is an opportunistic pathogen that mostly affects patients with weakened immune systems, but a few serotypes (especially K1 and K2) are highly invasive and result in systemic infection in healthy persons. The ability to evade and survive the components of the innate immune system is critical in infection. To investigate the role and mechanism of transcription regulator KP1_RS12260 (KbvR) in virulence and defense against the innate immune response, kbvR deletion mutant and complement strains were constructed. The in vivo animal infection assay and in vitro antiphagocytosis assay demonstrate K. pneumoniae KbvR is an important regulator that contributes to virulence and the defense against phagocytosis of macrophages. The transcriptome analysis and phenotype experiments demonstrated that deletion of kbvR decreased production of capsular polysaccharide (CPS) and biosynthesis of partly outer membrane proteins (OMPs). The findings suggest that KbvR is a global regulator that confers pathoadaptive phenotypes, which provide several implications for improving our understanding of the pathogenesis of K. pneumoniae.
INTRODUCTION
Klebsiella pneumoniae is an opportunistic pathogen that causes nosocomial infections in the urinary tract, respiratory tract, and blood (1, 2). This pathogen has more than 134 distinct capsular K loci (3). Its hypervirulent variants (hvKp), especially K1 and K2 serotypes, can cause serious community-acquired invasive infections, including pyogenic liver abscess (PLA) and meningitis, even among healthy persons (4, 5). In nature, K. pneumoniae isolates are mostly observed in the mouth, skin, and intestine of mammals and in environmental sources, such as soils and water surfaces (6). However, the sources of endogenous and exogenous PLA infections are being debated. Epidemiological studies suggest that most PLA infections are preceded by colonization of the gastrointestinal tract and can metastasize to distant sites such as the eyes and central nervous system (5, 7, 8). The ability to evade host immune defense contributes to metastatic infection of hypervirulent K. pneumoniae (9).
The innate immune response is the first line of host defense against pathogens, depending on phagocytic cells, molecules such as complement proteins, cytokines, and antimicrobial peptides (AMPs). K. pneumoniae employs various surface structures (e.g., capsular polysaccharide [CPS], lipopolysaccharide [LPS], outer membrane protein A [OmpA], OmpK36/OmpK35, AcrAB, etc.) to evade host immune defenses, enabling the bacteria to resist complement-mediated killing, the action of host-derived antimicrobial peptides, and the phagocytosis of epithelial cells, macrophages, and neutrophils (10–16). The expression of these surface structures is regulated in response to environmental cues to permit K. pneumoniae negate the host immune defense. Despite having many regulators, regulators such as CpxAR, KvrAB, RamR, RmpA, and cAMP receptor protein (CRP) have been proven to regulate the pathogenicity of K. pneumoniae (17–19). The regulatory mechanism of K. pneumoniae for defense against the innate immune system needs to be studied further.
A bioinformatics analysis reveals that the gene KP1_RS12260 contains a LuxR-type helix-turn-helix DNA-binding domain, suggesting it would encode a regulator. Through signature-tagged mutagenesis, KP1_RS12260 is associated with biofilm formation and intranasal infection in K. pneumoniae K2 serotype strain ATCC 43816 (20). We named this regulator KbvR (Klebsiella biofilm and virulence regulator), and KbvR was found in 90 different sequenced K. pneumoniae classical and hypervirulent strains. However, the specific role of this regulator in pathogenicity remains unclear. In this study, the K. pneumoniae K1 serotype strain NTUH-K2044, its kbvR in-frame deletion mutant (ΔkbvR), and the cis- and trans-complemented strain (C-kbvR) were used to examine the role of KbvR in K. pneumoniae. Our results demonstrate that KbvR plays an important role in pathogenicity by regulating bacterial evasion of phagocytes, antimicrobial peptides, and complement-mediated killing abilities. The transcriptome analysis and phenotype experiments with the wild-type (WT) and ΔkbvR strains revealed that KbvR regulates an expanded list of surface structures, including capsular polysaccharide (CPS) gene clusters, outer membrane protein A, OmpK36/OmpK35, AcrAB, etc. Taken together, this study demonstrates that KbvR is an essential regulator that contributes to capsule production, outer membrane protein (OMP) biosynthesis, antiphagocytosis, and virulence in K. pneumoniae.
RESULTS
KbvR does not affect bacterial growth in LB broth.
The kbvR gene deletion mutant and complementation strains were successfully constructed and were confirmed by reverse transcription-PCR (RT-PCR) (see Fig. S1 in the supplemental material). The growth curves for the WT, ΔkbvR, and C-kbvR sttrains cultured in the LB broth were compared: although the ΔkbvR and C-kbvR strains appeared to have slower growth than the WT strain in the early stage, the difference was not statistically significant (P = 0.3768), thereby suggesting that the kbvR gene does not affect bacterial growth in LB broth (Fig. 1).
FIG 1.
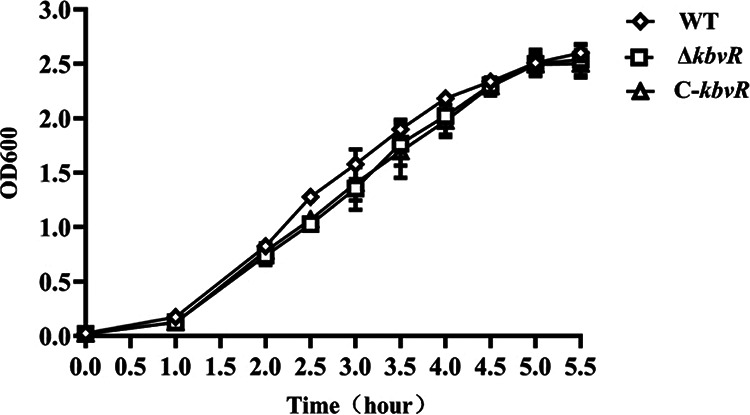
Bacterial growth curves. Bacteria were precultivated overnight in LB broth shaken at 200 rpm and 37°C, and the resulting cell cultures were diluted (1:100) into fresh LB broth and grown to an OD600 of 1.6. Then the bacteria were diluted (1:100) into fresh LB broth and cultured under the aforementioned conditions. The OD600 values were monitored at intervals of every 0.5 or 1 h.
Contribution of KbvR to K. pneumoniae virulence and live abscess formation in vivo.
To determine whether KbvR is involved in K. pneumoniae virulence in vivo, BALB/c mice were infected intraperitoneally with the WT, ΔkbvR, and C-kbvR strains, and the number of dead mice was counted for 14 days to calculate the 50% lethal dose (LD50). The LD50s were equal to 500, more than 5 × 105 CFU, and 1,000 CFU for the WT, ΔkbvR, and C-kbvR strains, respectively. Then the murine survival curves for intraperitoneal (i.p.) infection at a dose of 104 CFU were measured for 10 days. Eighty percent of mice were dead within 3 days after being infected with the WT strain. However, all mice survived the injection of the ΔkbvR strain at 10 day after infection. The lethality of the ΔkbvR strain in mice was much lower than that of the WT (Fig. 2a [P < 0.01]). For both LD50 and murine survival curve assays, the C-kbvR strain exhibited a restored virulence phenotype in mice (P = 0.384). The lung tissues of mice infected with 103 CFU of the WT were filled with blood, and inflammatory lesions of different sizes were observed in their livers (Fig. 2b). The mice treated with the ΔkbvR strain and phosphate-buffered saline (PBS [negative control]) showed no obvious pathological changes in their livers and lungs. The results illustrate the contribution of the KbvR regulator in bacterial virulence and dissemination.
FIG 2.
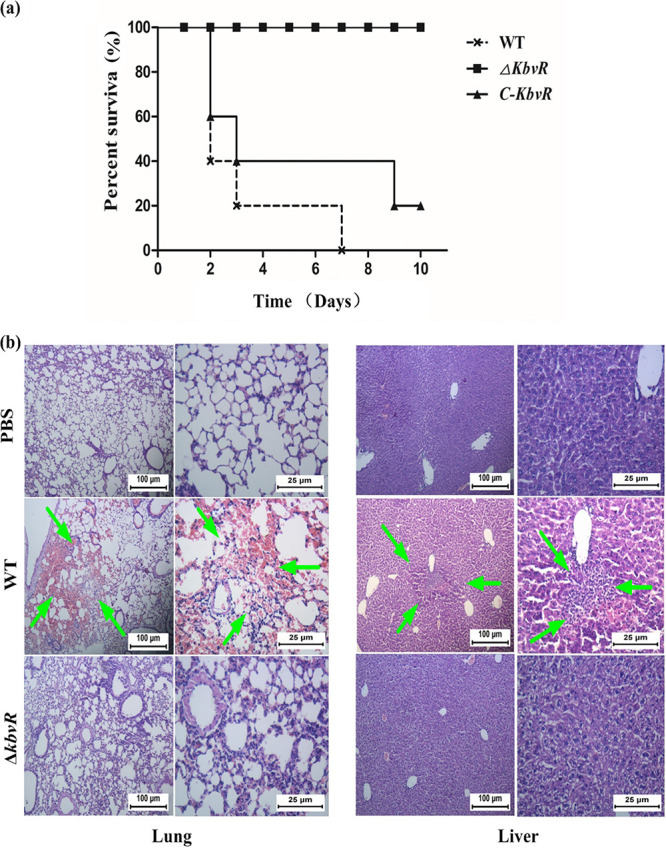
Virulence of three K. pneumoniae strains in mice. (a) Murine survival curves in the intraperitoneal infection model. Each 10 mice of 3 groups were injected intraperitoneally with 104 CFU of the WT, ΔkbvR, and C-kbvR strains, respectively. The murine survival rate for 10 days was measured. (b) Histopathology of mouse lung and liver after infection with PBS (negative control), the WT, and the ΔkbvR strain. Sixty hours after the infection of 103 CFU of the WT, ΔkbvR strain, or PBS, the lungs and livers of different groups of mice were dissected and stained. The lung tissues of mice infected by the WT strain were damaged and filled with blood. Inflammatory lesions of different sizes were observed on their livers. The mice treated with the ΔkbvR strain and PBS did not show any obvious pathological changes. The arrows indicate the blood cell or lymphocyte infiltration in lung and liver tissues, respectively. The figures are representative of at least three sections in three mice that showed similar pathology in the tissues.
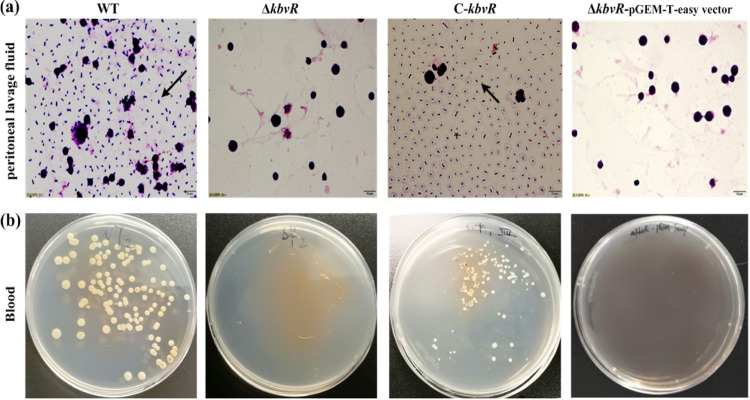
Enhanced clearance of the KbvR mutant by the peritoneal immune system.
To deeply understand the role of KbvR in the defense against the immune responses of the abdominal cavity, the abilities of the WT, ΔkbvR, and C-kbvR strains to survive in the mouse peritoneal cavity were examined. After 12 h of infection via intraperitoneal injection, the peritoneal washes were smeared, stained, and observed by microscope. No K. pneumoniae cells were observed in the peritoneal lavage fluids from mice infected with the ΔkbvR and ΔkbvR–pGEM-T Easy vector strains. However, plenty of K. pneumoniae cells were found in the abdominal cavities of the mice infected with the WT and C-kbvR strains (Fig. 3). The WT and C-kbvR but not ΔkbvR cells were cultured from the blood of mice infected with the corresponding strains. Therefore, the ability of the ΔkbvR strain to defend against the peritoneal immune system and its ability to invade the blood obviously decreased, thereby partly explaining the decreased virulence of the ΔkbvR strain.
FIG 3.
Survival in mouse abdominal cavity and invasion into blood of K. pneumoniae strains. Five-week-old BALB/c female mice were intraperitoneally injected with 105 CFU of the WT, ΔkbvR, C-kbvR, or ΔkbvR-pGEM-T Easy vector strain, respectively. (a) At 12 h postinfection, the mice were sacrificed, and their peritoneal lavage fluid was collected, stained, and observed with a microscope. The arrows indicate the bacteria. (b) The blood was plated onto an LB agar medium to calculate the bacteria. No bacteria were found in the blood of ΔkbvR strain- and ΔkbvR-pGEM-T Easy vector strain-infected mice.
KbvR contributes to bacterial defense against phagocytosis by macrophages.
Macrophages are the primary mechanism of the peritoneal immune system to defend against and eliminate the invading bacteria. To assess whether the decreased survival ability of the ΔkbvR strain in the peritoneal cavity was partly due to increased susceptibility to macrophage phagocytosis, the antiphagocytosis abilities of the WT, ΔkbvR and C-kbvR strains were investigated. As shown in Fig. 4a, the uptake of the kbvR mutant by the macrophages was significantly higher than those of the WT and C-kbvR strains. The WT strain was rarely present inside macrophages, but more than 200 ΔkbvR strain cells were internalized within 300 macrophages (Fig. 4b). Thus, in K. pneumoniae, KbvR contributes to bacterial defense against phagocytosis by macrophages.
FIG 4.
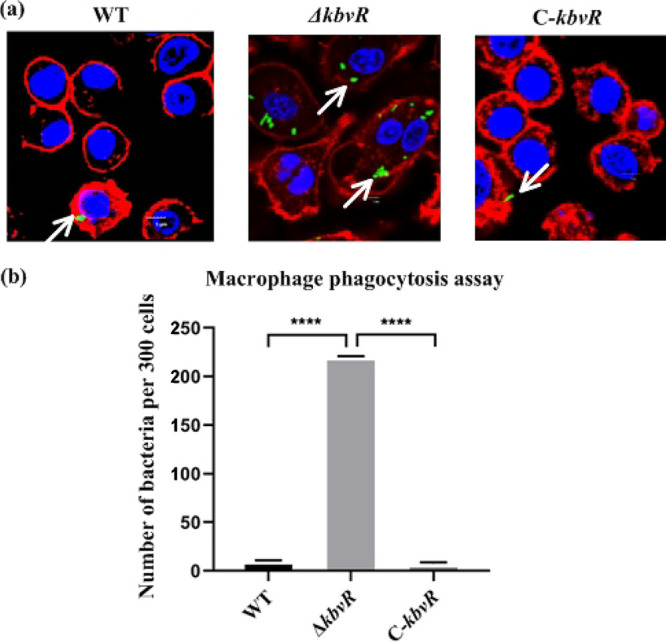
Determination of phagocytosis resistance of the wild type, the kbvR gene mutant, and complement strains by macrophages. (a) Confocal micrograph showing invasion into RAW264.7 macrophages by K. pneumoniae. Cells were infected with the WT, ΔkbvR mutant, and a C-kbvR strain carrying pLac-EGFP with a gene encoding green fluorescence protein for 120 min, and then extracellular bacteria were washed away, and macrophages were incubated for 90 min in the presence of gentamicin to eliminate the remaining extracellular bacteria. The cells were fixed, stained with biotin-XX phalloidin (red) for actin cytoskeleton and DAPI (blue) for host cell nuclei, and observed by confocal microscopy. Arrows denote intracellular bacteria. Scale bar, 5 μm. (b) Bacteria phagocytosed by macrophages were counted under a confocal microscope. Data are presented as the intracellular bacterial numbers in 300 cells from 3 independent trials.
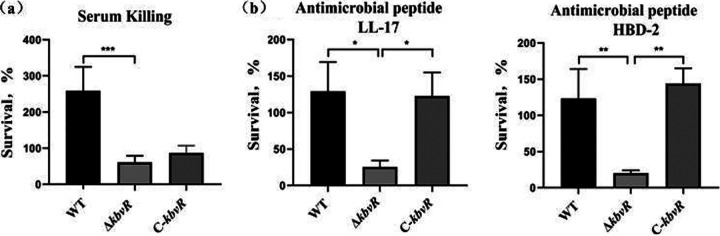
Influence of KbvR on serum killing and antimicrobial peptide resistance.
Complement proteins and antimicrobial peptides (AMPs) are important components of host innate immunity. We characterized the functions of the K. pneumoniae KbvR in complement and AMP interactions. Susceptibilities to nonimmune human serum and the AMPs cathelicidin LL-37 and human β-defensin 2 (HBD-2) were tested by the survival assay. After 3 h of incubation, the survival of the kbvR deletion mutant was significantly decreased compared with those of the WT and C-kbvR strains when exposed to serum and the AMPs LL-37 and HBD-2. These results indicated that KbvR was needed for protection against serum and antimicrobial peptide killing in K. pneumoniae (Fig. 5).
FIG 5.
Resistance of three K. pneumoniae strains against the killing by complement and antimicrobial peptides. (a) Serum sensitivity assays of the K. pneumoniae WT, ΔkbvR, and C-kbvR strains. The data represent the means from 3 independent trials; the error bars represent the standard deviations. A mean survival ratio of ≥1 corresponds to serum resistance. (b) Survival of the WT, ΔkbvR, and C-kbvR strains against the killing by antimicrobial peptide human LL37 (left panel) and defensin β2 (right panel). Strains were incubated with 2-μg/ml concentrations of LL37 or 0.5 μg/ml defensin β2 at 37°C for 3 h. The numbers of viable cells were determined at the baseline and 3 h after incubation. The survival rate of bacteria is shown as follows: (CFU exposed to AMPs for 3 h/not exposed to AMPs) × 100%.
Transcriptome analysis of the kbvR deletion mutant.
To identify the genes regulated by KbvR, the transcriptomes of the ΔkbvR and WT strains were comparatively analyzed via transcriptome sequencing (RNA-seq). The results showed that a substantial proportion of reads of each library were matched to K. pneumoniae genes. Of them, the majority of the mapped reads were those with a 1-bp mismatch or indel. After mapping to the gene models of the K. pneumoniae reference genome, more than 5,000 putative genes were identified from the reads of the WT and KbvR libraries, respectively, of which approximately 90% of K. pneumoniae genes predicted in the reference genome were expressed.
The differentially expressed genes (DEGs) between the WT and ΔkbvR strains were studied by filtering with the IDEG6 program. Compared with the ΔkbvR strain, the numbers of up- and downregulated genes in the library pair in the WT were 758 and 562, respectively (log2 fold change value of >2) (see Table S3 in the supplemental material). In order to reveal the global functional annotation of DEGs, Gene Ontology (GO) assignments were used to classify the functions. Based on sequence homology, the sequences were categorized into several groups (see Fig. S2 in the supplemental material). In each of the three main categories (biological process, cellular component, and molecular function) of the GO classification, the major subcategories were as follows. The five subcategories for the pair WT and KbvR for biological process included “macromolecule biosynthetic process,” “cellular macromolecule biosynthetic process,” “growth,” “translation,” and “macromolecule localization”. The four subcategories for the WT and KbvR for cellular component were “external encapsulating structure,” “cytoplasmic part,” “cytosol,” and “cell wall”. The three categories for the WT and KbvR for molecular function were “structural molecule activity,” “structural constituent of ribosome,” and “RNA binding.” Only a few genes were clustered in terms of “antioxidant activity,” “nickel cation binding,” “small ribosome subunit,” “cytosolic small ribosome subunit,” and “ribonucleoprotein complex binding.” KEGG pathway enrichment analysis was performed to categorize the biological functions of DEGs. We mapped all of the genes to terms in the KEGG database. In the comparison of the pair WT and KvR, obvious enrichment of genes was observed for 39 pathways, which included the capsular polysaccharide synthesis, quorum sensing, and ribosome and bacterial secretion system pathways as the top 4 affected by differentially expressed genes, respectively. The first nine enriched pathways for the library pair are shown in Fig. S3 in the supplemental material. In this figure, the secretion system and quorum sensing pathways enriched the most genes, which represent the first and second enrichment pathways.
In this study, those surface structures associated with antiphagocytosis and resistance to antimicrobial peptides were of interest. The genes with transcription linked to CPS and outer membrane proteins were obviously affected by KbvR (Table 1). The expression levels of all 21 genes associated with CPS synthesis and outer membrane proteins OmpK35/Ompk36, YfgL, Lpp, Pal, AcrAB, and OmpA, the factors associated with the antiphagocytosis or the resistance to antimicrobial peptides and complement-mediated killing, were decreased in the ΔkbvR strain, thereby partly accounting for the reduced virulence and resistance to the immune system in the ΔkbvR strain. Apart from the structure genes of CPS, some regulators, including CRP, RcsB, OxyR, and Fur, which have been reported to positively regulate CPS synthesis, were also downregulated in the ΔkbvR strain (Table 1).
TABLE 1.
Partly selected differentially expressed genes associated with antiphagocytosis between the WT and ΔkbvR strains
| Category | Gene ID | Gene name | Log2 fold change for ΔkbvR vs WT | Gene function |
|---|---|---|---|---|
| cps gene clusters | KP1_RS17255 | uge | −1.60986029 | Uridine diphosphate galacturonate 4-epimerase |
| KP1_RS17270 | ugd | −2.527969041 | UDP-glucose 6-dehydrogenase | |
| KP1_RS17275 | manB | −1.405835586 | Phosphomannomutase | |
| KP1_RS17280 | manC | −1.42636699 | Mannose-1-phosphate guanylyltransferase | |
| KP1_RS17285 | gnd | −2.6593155 | 6-Phosphogluconate dehydrogenase | |
| KP1_RS17295 | wcaI | −2.289617278 | Putative colanic biosynthesis glycosyltransferase | |
| KP1_RS17300 | wcaH | −2.530934376 | GDP-mannose mannosyl hydrolase | |
| KP1_RS17305 | wcaG | −2.615464837 | GDP-fucose synthetase | |
| KP1_RS17310 | gmd | −2.597771967 | GDP-d-mannose dehydratase | |
| KP1_RS17315 | ORF10 | −2.556989904 | Putative glycosyltransferase | |
| KP1_RS17320 | magA | −2.57577744 | Mucoviscosity-associated protein | |
| KP1_RS17325 | ORF8 | −2.313703585 | Polysaccharide pyruvyl transferase | |
| KP1_RS28055 | wzx | −2.299838097 | Repeat unit exporter | |
| KP1_RS17330 | wzc | −2.439647037 | Inner membrane tyrosine autokinase | |
| KP1_RS17335 | wzb | −2.694320682 | Putative protein tyrosine phosphatase | |
| KP1_RS17340 | wza | −2.613250868 | Putative capsule polysaccharide export protein precursor | |
| KP1_RS17355 | ORF2 | −2.712270704 | Putative acid phosphatase | |
| KP1_RS17360 | galF | −3.238383282 | UTP-glucose-1-phosphate uridylyltransferase | |
| KP1_RS17345 | wzi | −2.076971824 | Outer membrane protein | |
| KP1_RS17290 | wcaJ | −1.908522304 | Probable CPS biosynthesis glycosyltransferase | |
| Regulators associated with CPS synthesis | KP1_RS23625 | crp | −3.657727207 | Catabolite activator protein |
| KP1_RS18010 | rcsB | −2.58690418 | DNA-binding response regulator in two-component regulatory system | |
| KP1_RS00535 | oxyR | −2.525981601 | Activator of hydrogen peroxide-inducible genes | |
| KP1_RS07790 | fur | −3.122422839 | Ferric uptake regulation protein | |
| KP1_RS16895 | rmpA | −1.470982131 | Regulator of mucoid phenotype | |
| Outer membrane proteins | KP1_RS09230 | ompA | −4.349287 | Outer membrane protein A |
| KP1_RS15075 | lpp | −2.963897995 | Murein lipoprotein | |
| KP1_RS08010 | pal | −3.085186099 | Peptidoglycan-associated lipoprotein | |
| KP1_RS10700 | ompW | −3.028696848 | Outer membrane protein W | |
| KP1_RS08505 | ompX | −3.387507606 | Outer membrane protein X | |
| KP1_RS18000 | ompC | −4.388425403 | Outer membrane porin protein OmpK36 | |
| KP1_RS09095 | ompF | −1.069932592 | Outer membrane protein 1A/OmpK35 porin | |
| KP1_RS06240 | acrB | −2.103374805 | Acriflavine resistance protein B | |
| KP1_RS06245 | acrA | −2.06558227 | Acriflavine resistance protein A | |
| KP1_RS19070 | yfgL | −2.510301493 | Outer membrane lipoprotein | |
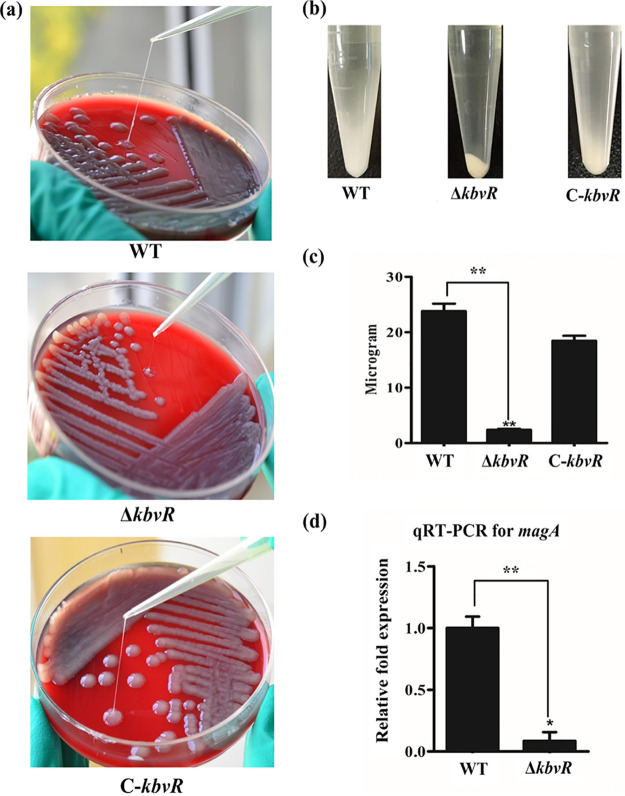
KbvR positively regulates CPS.
CPS is one of the prominent virulence factors of K. pneumoniae that can protect pathogens from opsonization and phagocytosis. The RNA-seq results show that the mRNA levels of genes associated with CPS synthesis have decreased in the ΔkbvR strain. To confirm this result, CPS production was first examined via a mucoviscosity assay. Given that bacteria with thick, highly mucoid capsules were pelleted less readily, a precipitation test was performed on mid-log cell cultures to determine the effect of kbvR deletion on CPS production. After centrifugation at 5,000 × g for 5 min, the ΔkbvR strain formed denser pellets than the WT and C-kbvR strains (Fig. 6a). The string test results showed that the ΔkbvR strain was significantly shorter than the WT and C-kbvR strains (Fig. 6b), which suggests that KbvR positively controls capsule biosynthesis. Moreover, the amount of capsule, as a function of uronic acid content, was quantified. There was statistical difference in the amounts of uronic acid detected in overnight cultures of the wild-type and kbvR mutant strains, indicating that mutation of KbvR decreased capsule production. The magA (wzyKpK1) gene, which encodes a capsular K1-specific Wzy polymerase has been associated with virulent hypermucoviscous K. pneumoniae strains (9, 10, 21). The quantitative RT-PCR (qRT-PCR) results show the expression of magA decreased in the ΔkbvR strain, which confirmed the RNA-seq results.
FIG 6.
Detection of CPS. (a) String test. Bacteria were cultivated in a blood plate medium, and the strings of three strains were longer than 5 mm. Compared with the lengths of the WT and C-kbvR strains, the ΔkbvR strain was significantly shorter. (b) Centrifugal precipitation test. The 1-ml mid-log cell cultures were subjected to a centrifugal precipitation test to measure the effect of kbvR deletion on CPS production. (c) Quantification of CPS. CPS was isolated from the bacterial cells and then determined as μg of glucuronic acid per 109 CFU of bacterial cells. (d) qRT-PCR of magA. The expression levels of magA of K. pneumoniae strains were monitored by qRT-PCR. The magA transcript level decreased in the ΔkbvR strain.
DISCUSSION
K. pneumoniae is a member of the Enterobacteriaceae, bacteria that cause community and nosocomial infections, especially in immunocompromised hosts. Serotypes K1 and K2 have been identified as the virulent bacteria that can cause PLA among healthy adults without underlying diseases (5). In an animal model, the LD50 of the K1 serotype produced approximately 100 CFU through abdominal infection (22). The pathogenicity of K. pneumoniae is associated with some virulence factors that are essential in different interaction processes with the host (9). Governing this infection process is a regulatory network that allows for a concomitant expression of virulence factors that are coordinated at the genetic level. The virulence of the mutant strains of the regulators CRP, RamA, and KvrAB was obviously lower than that of the WT strain (2, 18). In this study, we demonstrated the gene KP1_RS12260, which encoded the KbvR regulator, was also essential for the virulence of K. pneumoniae. In a mouse intraperitoneal infection model, the survival rate of mice infected with the ΔkbvR strain increased greatly compared with that of mice infected with the WT strain, suggesting that kbvR deletion attenuates the bacterial virulence in vivo. The ability to avoid phagocytosis was obviously impaired in the kbvR mutant strain in both an in vivo infection animal model and in vitro antiphagocytosis experimentation, suggesting KbvRs regulate the virulence and the ability of K. pneumoniae to defend against macrophage uptake. Meanwhile, the resistance to killing by complement and antimicrobial peptides was reduced. These results confirm that KbvR is important in the virulence of K. pneumoniae through regulation of the ability to evade the innate immune defenses of the host.
The innate immune response, including phagocytosis by phagocytic cells, complement-mediated lysis, and host-derived antimicrobial peptides, is the first line of host defense to discriminate and eliminate the invading bacteria. The ability to attach to the host epithelial cell, resist phagocytosis, and impede bacterial killing by the complement serum and antimicrobial peptides contributes to the pathogenicity of the hypervirulent K. pneumonia (2, 18, 23). In this process, the polysaccharide capsules (CPS), lipopolysaccharide (LPS), and outer membrane proteins (OMPs) play important roles (16). A wealth of evidence demonstrates that the CPS plays a crucial role in resistance to phagocytosis, complement-mediated killing, and antimicrobial peptides (10, 24–26). Acapsular K. pneumoniae strains are dramatically decreased in terms of the ability of the bacteria to spread systemically (27, 28). Capsule protects K. pneumoniae by reducing adherence to phagocytic cells and reducing uptake by phagocytic cells. The uptake of the kbvR mutant was significantly higher than that of the WT strain by the macrophages. The data from transcriptome profiling revealed that the levels of expression of all 21 genes in the CPS gene clusters were decreased. The phenotype experiments and quantification of capsule confirmed the expression of CPS was decreased in the ΔkbvR strain. The qRT-PCR result found the expression level of mucoviscosity-associated gene A (magA; wzyKpK1), the gene mostly found in invasive strains, was obviously lower than that of the WT. The magA gene deletion mutant also lost the ability to resist killing by nonimmune healthy human serum (29). These results partly explain the decrease in the antiphagocytosis and virulence of the ΔkbvR mutant. The capsule regulatory network of K. pneumoniae NTUH-K2044 has been defined by density-TraDISort, and a lot of regulators have been identified to influence capsule production (30, 31). However, the role of KbvR regulator in capsule biosynthesis had not been described. Apart from its effect on the genes in the CPS gene clusters, the levels of expression of regulators Crp, RcsB, OxyR, Fur, and RmpA, which have positive regulatory roles in CPS synthesis, were decreased in the ΔkbvR mutant. Therefore, KbvR may directly regulate the expression of CPS structure genes or indirectly regulate the regulators of CPS synthesis to reduce CPS production. The accurate regulatory mechanism of KbvR in CPS synthesis warrants further study.
In K. pneumoniae, the outer membrane (OM) serves as the barrier, and the major components of it, outer membrane proteins, including outer membrane protein A (OmpA), murein lipoprotein (Lpp), peptidoglycan-associated lipoprotein (Pal), outer membrane porins, and efflux pumps, play important roles in resistance to phagocytosis and antimicrobial agent (14, 32–34). OmpA is one of the major outer membrane proteins of K. pneumoniae and protects bacteria against the bactericidal action of antimicrobial peptides (35). Loss of OmpA makes K. pneumoniae more susceptible to antimicrobial peptides and serum components. Except for OmpA, K. pneumoniae produces two major outer membrane porins—OmpK35 and OmpK36—which may contribute to virulence by preventing phagocytosis (32). AcrAB is an efflux pump in the outer membrane, and the acrB deletion mutant was more sensitive to exposure to antimicrobial peptides (15). OmpA, Pal, Lpp, and AcrAB are needed for protection against antimicrobial humoral components such as antimicrobial peptides and serum complement. In this study, the levels of expression of OMPs, such as OmpA, Lpp, Pal, YfgL, AcrAB, and OmpK35/OmpK36, were significantly decreased in the kbvR mutant. Therefore, the K. pneumoniae KbvR protein is important for the regulation of outer membrane protein assembly, which influences the ability of bacterium to defend against the innate immune system.
In summary, this work describes the roles of KbvR in the pathogenesis of highly virulent K. pneumoniae strains. The results show that the deletion of KbvR mediates the decreased expression of CPS and partly outer membrane protein biosynthesis, thereby reducing antiphagocytosis, resistance to killing by complement and antimicrobial peptides, and finally modulating bacterial virulence. KbvR positively regulates virulence by affecting both capsule production and outer membrane protein biosynthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. K. pneumoniae NTUH-K2044, a capsular serotype K1 strain isolated from a patient with community-acquired PLA, was used as the WT strain in this study (36). Strains were grown in Luria-Bertani (LB) medium at 37°C on an orbital shaker (200 rpm). For biochemical and phenotypic assays, strains were streaked from a frozen stock (–80°C), precultivated overnight in LB broth at 37°C, and diluted 100-fold into fresh medium. When appropriate, 100 μg/ml kanamycin (Km) or ampicillin (Amp) was added to the LB medium.
Construction of kbvR deletion mutant and complementation strain of K. pneumoniae NTUH-K2044.
The whole kbvR gene deletion mutant (ΔkbvR) was constructed via in-frame deletion by using a double homologous recombinant as previously described (37). Sequences of regions flanking the kbvR gene were cloned into the temperature-sensitive suicide pKO3-Km vector containing a sacB counterselectable marker. For the complementation of the ΔkbvR strain (C-kbvR), a PCR-generated 1,876-bp DNA fragment containing the kbvR coding region was cloned together with its promoter-proximal region and putative transcriptional terminator into the km-pGEM-T Easy vector and was transformed into the mutant strain (38). The empty km-pGEM-T Easy plasmid was transformed into the ΔkbvR mutant strain as a vector-only control (ΔkbvR–pGEM-T Easy vector). All strains were grown in Luria-Bertani (LB) broth containing appropriate antibiotics unless otherwise noted. Reverse transcription-PCR (RT-PCR) experiments were performed to detect transcription of the kbvR gene in the WT, ΔkbvR, and C-kbvR strains to validate the mutation and complementation. Information on the primers used in this study can be found in Table S2 in the supplemental material.
Bacterial growth rate.
To examine the bacterial growth rate in vitro, overnight cultures of the WT, ΔkbvR, and C-kbvR strains were diluted 100 times into fresh LB broth and were grown into an optical density at 600 nm (OD600) of 1.6. Afterward, the cells were diluted 100 times into another 30 ml of fresh LB broth and cultured at 37°C and 200 rpm. The OD600 was measured using a UV spectrophotometer every 0.5 or 1 h to quantify the growth curve.
Mouse infection assay.
Virulence was evaluated by mortality in a murine model by intraperitoneal (i.p.) injection according to the methods described previously (21). Briefly, the WT, ΔkbvR, and C-kbvR strains were grown to an OD600 of 1.6 and subjected to serial 10-fold dilutions with phosphate-buffered saline (PBS), and appropriate dilutions were plated onto LB agar plates to calculate the number of CFU. A total of 5 mice from each group of 5- to 6-week-old female BALB/c mice were injected intraperitoneally with 100 μl of bacterial suspension containing 102, 103, 104, 105, and 106 CFU. The mice were monitored daily for 14 days, and the 50% lethal dose (LD50) was calculated as described previously (39). Then each group of 10 mice was injected intraperitoneally with 104 CFU of the WT, ΔkbvR, and C-kbvR strains to calculate the corresponding survival curve for 10 days. Groups of 3 mice were inoculated intraperitoneally with PBS or 103 CFU of the WT or ΔkbvR strain as described above. At 60 h after injection, the livers and lungs of mice from each group were removed, embedded, and stained with hematoxylin and eosin for histopathological examination to understand the metastatic infection and pathological change. All animal experiments were established by the Animal Care and Use Committee at Hubei University of Medicine and complied with all ethical and husbandry regulations (no. 2019-076).
Survival assay against the mouse peritoneal immunity system.
To investigate the role of KbvR in bacterial defense against the immunity system of the abdominal cavity and the ability to invade blood, 3 mice from each group of 5- to 6-week-old female BALB/c mice were intraperitoneally injected with 105 CFU of the WT, ΔkbvR, C-kbvR, or ΔkbvR–pGEM-T Easy vector strain. At 12 h postinfection, the mice were sacrificed, and blood samples were obtained and plated onto an LB agar plate to observe the number of bacteria. Afterward, 3 ml LB medium was injected into the abdominal cavity of the mice, and slides were coated with 10 μl peritoneal lavage fluid and stained via the Gram stain method. Photographs were taken via optical microscopy.
In vitro assay for bacterial phagocytosis by macrophages.
Murine RAW264.7 macrophage cells were cultured in RPMI 1640 medium containing 2 mM glutamine and 10% (vol/vol) fetal calf serum in 24-well glass bottom culture plates. For the phagocytosis assay, plasmid pLac-EGFP, which contains a gene encoding enhanced green fluorescence protein (EGFP), was electroporated into the wild-type and kbvR mutant strains. The bacteria were cultured in LB at 37°C to an OD600 of 1.6, washed with PBS, and adjusted to approximately 107 CFU/ml in RPMI 1640 medium. Phagocytosis assays were performed as previously described (29). Briefly, macrophages were incubated with bacteria (50 bacteria/macrophage) for 120 min, then extracellular bacteria were washed away, and macrophages were incubated for 90 min in the presence of gentamicin to eliminate the remaining extracellular bacteria. Then the cells were washed, fixed, and stained with biotin-XX phalloidin (red) for actin cytoskeleton and DAPI (4′,6-diamidino-2-phenylindole [blue]) for host cell nuclei. After preparation, the cells were observed by confocal microscopy, and the numbers of intracellular bacteria in at least 9 fields were counted (10 to 40 cells in each field). The sums of the intracellular bacteria in the total number of cells in these fields were calculated and quantified within 300 cells. Experiments were carried out in three independent occasions.
Serum killing assay.
The survival of K. pneumoniae WT, ΔkbvR, and C-kbvR strains in nonimmune human serum was measured as previously described (40). Overnight cultures of strains were diluted 100-fold in LB broth and incubated at 37°C to an OD600 of 1.6. A total of 106 bacterial cells and 75% (vol/vol) nonimmune human serum donated by healthy volunteers were incubated at 37°C for 3 h. The colony count was determined by the serial dilution method, and the mean survival ratio was plotted. A mean survival ratio of ≥1 corresponds to serum resistance.
Antimicrobial peptide susceptibility assays.
The bacteria were cultured by a method like that used for the serum killing assay. A volume of 100 μl diluted bacteria with 106 CFU was mixed with human AMP LL-37 (2 μg/ml) or human β-defensin 2 (HBD-2; 0.5 μg/ml), respectively. At 0 h and 3 h after incubation at 37°C, 20-μl mixtures were taken out for serial dilution. The inoculum CFU at the baseline and recovered CFU at 3 h were determined by plating of serial dilutions on LB agar, and the survival ratio of bacteria exposed to AMPs was expressed as (recovered CFU/inoculum CFU) × 100% (41).
Mapping of RNA-seq reads and quantitative analysis of gene expression.
The RNA-seq technology was employed to determine the transcription profiles of the WT and ΔkbvR strains. The strains were cultivated to log phase, and the cultures were collected. The total RNAs of the WT or ΔkbvR strain were extracted by using the EZNA Fungal RNA minikit (Omega Bio-tek, Frederick, CO, USA). The quality of the RNAs was preliminarily examined by 1.0% (wt/vol) agarose gel electrophoresis and confirmed by an Agilent 2100 Bioanalyzer. The RNA concentrations and purity were determined by NanoDrop 2000 spectrophotometry. Samples for the transcriptome analysis were prepared by using Illumina’s kit. Specifically, mRNA was purified from total RNA and was fragmented into small pieces. The cleaved RNA fragments were used for the first-strand cDNA synthesis, the second-strand cDNA was synthesized, and the products were purified and enriched to produce the final cDNA libraries, respectively. These libraries were sequenced via the Illumina HiSeq X platform at the Wuhan Bioacme Biological Technologies Corporation (Wuhan, China), and 150-bp paired-end reads were obtained. The qualified raw sequence data in the quality control were filtrated via deletion of the reads containing adaptors, more than 10% “N,” and low-quality reads (i.e., where a Q score of ≤10 of the bases accounts for more than 50%). The remaining reads by filtration called “clean” reads, were used for informational analysis. Then the clean data were mapped to the genome of K. pneumoniae (36) using SOAPaligner/SOAP2 (Short Oligonucleotide Alignment Program), allowing up to 2 bases of mismatches (42). For quantification, gene expression levels were calculated using the reads per kilobase of transcriptome per million mapped reads (RPKM) method (43), thereby limiting the effects of different gene lengths and sequencing levels. Rigorous algorithms were applied to identify differentially expressed genes (DEGs) between every two treated samples referring to previously described methods (44). DEGs were identified using a false-discovery rate (FDR) of ≤0.001 and an absolute value of the log2 ratio of ≥1 as the threshold (45).
For further identification of the function of differentially expressed genes, GO and KEGG enrichment analyses were carried out. Based on the Gene Ontology database (http://www.geneontology.org/), GO enrichment analysis was calculated by referring to the software GO::TermFinder (http://smd.stanford.edu/help/GO-TermFinder/GOTermFinderhelp.shtml/) (46). All of the P values were subjected to Bonferroni correction for GO enrichment analysis. A corrected P value of <0.05 was selected as the threshold for determining significant enrichment of the gene sets. The KEGG enrichment was analyzed via the KEGG database similar to the GO enrichment (47), and a pathway with an FDR of ≤0.05 was identified as showing significant enrichment of the pathway.
Quantitative real-time RT-PCR analysis.
To validate the results of RNA-seq, total RNA from appropriate cultures of K. pneumoniae WT and ΔkbvR strains were isolated using an RNeasy Midi column (Qiagen) according to the manufacturer’s instructions. Purified RNA was DNase treated with RNase-free DNase I (Qiagen) to eliminate DNA contamination. RNA was reverse transcribed with a SuperScript III first-strand synthesis system (Invitrogen) using random primers in a reaction mixture of 20 μl. The differences in the expressions of magA between the WT and ΔkbvR strains were quantified via qRT-PCR. Relative expression levels were quantified using the comparative threshold cycle (2−△△CT) method with 16S rRNA as the endogenous reference.
Mucoviscosity assay.
The mucoviscosity levels of the WT, ΔkbvR, and C-kbvR strains were determined by performing centrifugation precipitation and the string test. For the centrifugal precipitation test of mucoviscosity, aliquots of 1 ml of mid-log cultured bacteria were centrifuged at 5,000 × g for 5 min to measure the degree of mucoidy. For the string test, the WT, ΔkbvR, and C-kbvR strains were inoculated onto 5% sheep blood plates for 16 h. A tip was used to stretch a mucoviscous string from the colony. The formation of viscous strings with lengths of >5 mm was viewed as a positive result.
Extraction and quantification of capsule.
To quantify CPS, the K. pneumoniae CPS was isolated by the hot phenol-water method and quantified following the procedures described by Ou et al. (37). The relative CPS concentration was represented as the amount (μg) of glucuronic acid per 109 CFU of bacterial cells.
Experimental replicates and statistical methods.
All experiments were performed with three independent bacterial cultures, and the values were expressed as mean ± standard deviation. Paired Student's t test, one-way analysis of variance (ANOVA) and mixed repeated-measures ANOVA with Bonferroni correction, and Kaplan-Meier estimator and log-rank test were performed to determine statistically significant differences, and P < 0.05 and P < 0.01 were treated as the levels of statistical significance.
Data availability.
The transcriptomic read data from the WT strain and ΔkbvR strain samples recovered in this study have been deposited in the NCBI GenBank Short Read Archive (SRA) under accession no. SAMN17192750 to SAMN17192755.
Supplementary Material
ACKNOWLEDGMENTS
Financial support was provided by the Natural Science Foundation of Hubei Province for Distinguished Young Scholars (2018CFA046), Hubei Province’s Outstanding Medical Academic Leader Program, Cultivating Project for Young Scholars at Hubei University of Medicine (2017QDJZR16), and Natural Science Foundation of Hubei Province for Young Scholars (2019CFB143).
Li Xu and Meng Wang performed all major experiments. Jie Yuan finished the animal experiments.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Podschun R, Ullmann U. 1998. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11:589–603. 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102. 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pomakova DK, Hsiao CB, Beanan JM, Olson R, MacDonald U, Keynan Y, Russo TA. 2012. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis 31:981–989. 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 5.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagley ST. 1985. Habitat association of Klebsiella species. Infect Control 6:52–58. 10.1017/s0195941700062603. [DOI] [PubMed] [Google Scholar]
- 7.Fung CP, Lin YT, Lin JC, Chen TL, Yeh KM, Chang FY, Chuang HC, Wu HS, Tseng CP, Siu LK. 2012. Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg Infect Dis 18:1322–1325. 10.3201/eid1808.111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada S, Tateda K, Mitsui H, Hattori Y, Okubo M, Kimura S, Sekigawa K, Kobayashi K, Hashimoto N, Itoyama S, Nakai T, Suzuki T, Ishii Y, Yamaguchi K. 2011. Familial spread of a virulent clone of Klebsiella pneumoniae causing primary liver abscess. J Clin Microbiol 49:2354–2356. 10.1128/JCM.00034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B, Zhao Y, Liu C, Chen Z, Zhou D. 2014. Molecular pathogenesis of Klebsiella pneumoniae. Future Microbiol 9:1071–1081. 10.2217/fmb.14.48. [DOI] [PubMed] [Google Scholar]
- 10.Cortés G, Borrell N, de Astorza B, Gómez C, Sauleda J, Albertí S. 2002. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect Immun 70:2583–2590. 10.1128/iai.70.5.2583-2590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llobet E, Tomás JM, Bengoechea JA. 2008. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology (Reading) 154:3877–3886. 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 12.Alvarez D, Merino S, Tomás JM, Benedí VJ, Albertí S. 2000. Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect Immun 68:953–955. 10.1128/iai.68.2.953-955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llobet E, Campos MA, Giménez P, Moranta D, Bengoechea JA. 2011. Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect Immun 79:3718–3732. 10.1128/IAI.05226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, Siu LK. 2011. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother 55:1485–1493. 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bengoechea JA, Sa Pessoa J. 2019. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev 43:123–144. 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan VB, Vaidyanathan V, Mondal A, Rajamohan G. 2012. Role of the two component signal transduction system CpxAR in conferring cefepime and chloramphenicol resistance in Klebsiella pneumoniae NTUH-K2044. PLoS One 7:e33777. 10.1371/journal.pone.0033777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palacios M, Miner TA, Frederick DR, Sepulveda VE, Quinn JD, Walker KA, Miller VL. 2018. Identification of two regulators of virulence that are conserved in Klebsiella pneumoniae classical and hypervirulent strains. mBio 9:e01443-18. 10.1128/mBio.01443-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Finn S, Spence S, Monahan A, Monaghan A, Kissenpfennig A, Ingram RJ, Bengoechea J, Gally DL, Fanning S, Elborn JS, Schneiders T. 2015. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 11:e1004627. 10.1371/journal.ppat.1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boddicker JD, Anderson RA, Jagnow J, Clegg S. 2006. Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect Immun 74:4590–4597. 10.1128/IAI.00129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fung CP, Chang FY, Lin JC, Ho DM, Chen CT, Chen JH, Yeh KM, Chen TL, Lin YT, Siu LK. 2011. Immune response and pathophysiological features of Klebsiella pneumoniae liver abscesses in an animal model. Lab Invest 91:1029–1039. 10.1038/labinvest.2011.52. [DOI] [PubMed] [Google Scholar]
- 22.Tsai FC, Huang YT, Chang LY, Wang JT. 2008. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis 14:1592–1600. 10.3201/eid1410.071254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. 2017. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence 8:1111–1123. 10.1080/21505594.2017.1317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu JH, Wu AM, Tsai CG, Chang XY, Tsai SF, Wu TS. 2008. Contribution of fucose-containing capsules in Klebsiella pneumoniae to bacterial virulence in mice. Exp Biol Med (Maywood) 233:64–70. 10.3181/0706-RM-170. [DOI] [PubMed] [Google Scholar]
- 25.Campos MA, Vargas MA, Regueiro V, Llompart CM, Albertí S, Bengoechea JA. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect Immun 72:7107–7114. 10.1128/IAI.72.12.7107-7114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merino S, Camprubí S, Albertí S, Benedí VJ, Tomás JM. 1992. Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect Immun 60:2529–2535. 10.1128/IAI.60.6.2529-2535.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshida K, Matsumoto T, Tateda K, Uchida K, Tsujimoto S, Yamaguchi K. 2000. Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J Med Microbiol 49:1003–1010. 10.1099/0022-1317-49-11-1003. [DOI] [PubMed] [Google Scholar]
- 28.Lawlor MS, Handley SA, Miller VL. 2006. Comparison of the host responses to wild-type and cpsB mutant Klebsiella pneumoniae infections. Infect Immun 74:5402–5407. 10.1128/IAI.00244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh PF, Hsu CR, Chen CT, Lin TL, Wang JT. 2016. The Klebsiella pneumoniae YfgL (BamB) lipoprotein contributes to outer membrane protein biogenesis, type-1 fimbriae expression, anti-phagocytosis, and in vivo virulence. Virulence 7:587–601. 10.1080/21505594.2016.1171435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorman MJ, Feltwell T, Goulding DA, Parkhill J, Short FL. 2018. The capsule regulatory network of Klebsiella pneumoniae defined by density-TraDISort. mBio 9:e01863-18. 10.1128/mBio.01863-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker KA, Miller VL. 2020. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr Opin Microbiol 54:95–102. 10.1016/j.mib.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JH, Siu LK, Fung CP, Lin JC, Yeh KM, Chen TL, Tsai YK, Chang FY. 2010. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother 65:986–990. 10.1093/jac/dkq056. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh PF, Liu JY, Pan YJ, Wu MC, Lin TL, Huang YT, Wang JT. 2013. Klebsiella pneumoniae peptidoglycan-associated lipoprotein and murein lipoprotein contribute to serum resistance, antiphagocytosis, and proinflammatory cytokine stimulation. J Infect Dis 208:1580–1589. 10.1093/infdis/jit384. [DOI] [PubMed] [Google Scholar]
- 34.Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llobet E, March C, Giménez P, Bengoechea JA. 2009. Klebsiella pneumoniae OmpA confers resistance to antimicrobial peptides. Antimicrob Agents Chemother 53:298–302. 10.1128/AAC.00657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, Tsai HC, Fung CP, Chen HJ, Liu YM, Wang JT, Fang CT, Chang SC, Shu HY, Liu TT, Chen YT, Shiau YR, Lauderdale TL, Su IJ, Kirby R, Tsai SF. 2009. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol 191:4492–4501. 10.1128/JB.00315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou Q, Fan J, Duan D, Xu L, Wang J, Zhou D, Yang H, Li B. 2017. Involvement of cAMP receptor protein in biofilm formation, fimbria production, capsular polysaccharide biosynthesis and lethality in mouse of Klebsiella pneumoniae serotype K1 causing pyogenic liver abscess. J Med Microbiol 66:1–7. 10.1099/jmm.0.000391. [DOI] [PubMed] [Google Scholar]
- 38.Xu L, Lin DS, Yang J, Li J, Li B. 2016. Effect of Klebsiella pneumoniae KbvR regulator on bacterial biofilm formation and capsular synthesis. Nan Fang Yi Ke Da Xue Xue Bao 36:1435–1439. (In Chinese.) [PubMed] [Google Scholar]
- 39.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 40.Hsieh PF, Lin TL, Yang FL, Wu MC, Pan YJ, Wu SH, Wang JT. 2012. Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS One 7:e33155. 10.1371/journal.pone.0033155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu CR, Chang IW, Hsieh PF, Lin TL, Liu PY, Huang CH, Li KT, Wang JT. 2019. A novel role for the Klebsiella pneumoniae Sap (sensitivity to antimicrobial peptides) transporter in intestinal cell interactions, innate immune responses, liver abscess, and virulence. J Infect Dis 219:1294–1306. 10.1093/infdis/jiy615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967. 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 43.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 44.Audic S, Claverie JM. 1997. The significance of digital gene expression profiles. Genome Res 7:986–995. 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 45.Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Statist 29:1165–1188. 10.1214/aos/1013699998. [DOI] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomic read data from the WT strain and ΔkbvR strain samples recovered in this study have been deposited in the NCBI GenBank Short Read Archive (SRA) under accession no. SAMN17192750 to SAMN17192755.