The Staphylococcus aureus Tet38 membrane protein has distinct functions, including drug efflux and host cell attachment and internalization mediated by interaction with host cell CD36. Using structural modeling and site-directed mutagenesis, we identified key amino acids involved in different functions.
KEYWORDS: CD36, resistance to tetracycline, Staphylococcus aureus, Tet38, efflux pumps, internalization
ABSTRACT
The Staphylococcus aureus Tet38 membrane protein has distinct functions, including drug efflux and host cell attachment and internalization mediated by interaction with host cell CD36. Using structural modeling and site-directed mutagenesis, we identified key amino acids involved in different functions. Tet38, a member of the major facilitator superfamily, is predicted to have 14 transmembrane segments (TMS), 6 cytoplasmic loops, and 7 external loops. Cysteine substitutions of arginine 106 situated at the junction of TMS 4 and external loop L2, and glycine 151 of motif C on TMS 5, resulted in complete or near-complete (8- to 16-fold) reductions in Tet38-mediated resistance to tetracycline, with minimal to no effect on A549 host cell internalization. In contrast, a three-amino-acid deletion, F411P412G413, in external loop L7 situated between TMS 13 and 14 led to a decrease of 4-fold in S. aureus internalization by A549 cells and a partial effect on tetracycline resistance (4-fold reduction). A three-amino-acid deletion, D38D39L40, in external loop L1 situated between TMS-1 and TMS-2, had a similar partial effect on tetracycline resistance but did not affect cell internalization. Using an Ni column retention assay, we showed further that the L7, but not the L1, deletion impaired binding to CD36. Thus, the L7 domain of Tet38 is key for interaction with CD36 and host cell internalization, and amino acids R106 and G151 (TMSs 4 and 5) are particularly important for tetracycline resistance without affecting internalization.
INTRODUCTION
Staphylococcus aureus Tet38 is a major facilitator superfamily (MFS) efflux membrane protein that has at least two important functions, extrusion of organic compounds and antibiotics (1–5), and invasion of epithelial cells via binding to the host receptor CD36 (2, 6). The structural determinants of these two functions have not been previously studied.
MFS transporters comprise six well-characterized families of proteins which include the families of multidrug efflux pumps with 12 or 14 transmembrane segments (TMS). The 12-TMS proteins include TetA/B of Escherichia coli and NorA of S. aureus, and the 14-TMS proteins include TetK of S. aureus and TetL of Bacillus subtilis efflux pumps. Multiple-sequence alignment and analysis among these efflux pumps reveal several conserved motifs that play essential roles in the structure and function of the MFS transporters (7). Motif A, which is located in the cytoplasmic loop between TMS-2 and TMS-3 and is essential in controlling the passage of substrates across the cytoplasm, is present in all six families of MFS transporters. Motif B, located in TMS-4 and predicted to play a role in energy coupling, is present in the 12- and 14-TMS and family 3 of MFS transporters. Motif C, located in TMS-5, is only carried by the 12-TMS and the 14-TMS efflux pumps. It plays an essential role in the direction of the transport of substrates. Other motifs with unknown function are also found in 12-TMS proteins (motif G) and 14-TMS proteins (motifs E and F) (7).
Two 14-TMS tetracycline resistance proteins, TetK of S. aureus and TetL of B. subtilis, showed the highest similarity in amino acid sequences to that of Tet38. TetL and TetK encode two antiporters of 50.4 kDa and 50.7 kDa, respectively, that extrude metal-tetracycline complexes coupled to proton exchange (8–10). TetL and TetK function in alkaline pH homeostasis and sodium Na+ resistance by serving as antiporters for the transport of sodium/potassium/hydrogen protons (Na+/K+/H+) (11). Tet38 also showed limited homology to the MFS 12-TMS tetracycline resistance proteins TetA and TetB of E. coli (12, 13).
TetL and TetK shared a series of conserved and family-specific motifs located in different transmembrane segments such as motifs D1 (TMS-1), H (TMS-6), E (TMS-7), and F (TMS-13). They also carried family-specific motifs of 14-TMS and 12-TMS members, such as motif C, located within the transmembrane segment TMS-5 of TetL and TetK. Motif C harbors a consensus sequence with a conserved glycine-proline (G155P156) dipeptide situated at the permeability barrier on TMS-5, a region undergoing conformational switching during transport of substrates. These tetracycline resistance proteins also carried other MFS common motifs such as motif A (between TMS-2 and TMS-3) and motif B (TMS-4). The conserved residues E152G155P156 of the motif C (TMS-5) and the residue R110 of motif B (TMS-4) were important for efflux of tetracycline by TetL and TetK. The 12-TMS tetracycline resistance protein TetA/B of E. coli also carries motif B with the conserved residue R103 (arginine) and motif C with the conserved residue G147 (glycine) (11, 13). Mutations at these four amino acids led to reduction or loss of tetracycline efflux and reduced resistance to tetracycline (7, 11). Early studies on MFS transporters suggested that their N-terminal regions played a role in the energization of the transport event, the C-terminal regions are involved in substrate specificity, and the conserved motifs participated in the structural and functional roles of the efflux pumps (7).
Tet38 is a 14-TMS MFS transporter based on homology with TetK/L (1) and topology modeling. It was first identified to confer resistance to tetracycline similar to TetK/L but was unexpectedly found also to confer resistance to other compounds, including fosfomycin, palmitoleic acid, tunicamycin, and Congo red (3–5), and to play a role in S. aureus invasion and survival in epithelial cells mediated in part by binding to host cell CD36 (2, 5, 6). In this study, we investigated the structural determinants of resistance to tetracycline and other substrates of this efflux pump at the protein level, as well as structural components important for the role of Tet38 in host cell internalization and binding to CD36. We have focused this work on the roles of motif B, motif C, and external loops L1 and L7 in Tet38 functions, finding that there are differential roles for these domains.
RESULTS
Putative membrane topology of S. aureus Tet38.
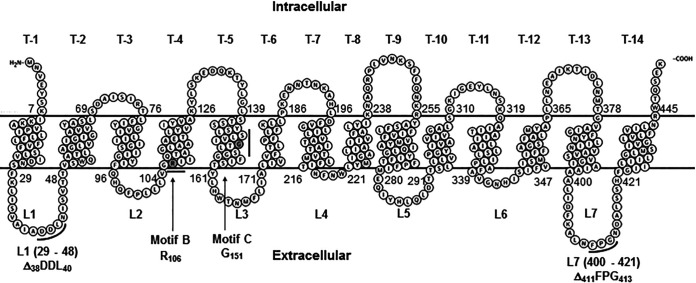
The amino acid sequence of Tet38 (GenPept accession no. BAB41352.1) with the predicted location of each residue based on analysis using the ConSurf server (https://consurf.tau.ac.il/) (14) was evaluated with Protter interactive feature visualization (https://wlab.ethz.ch/protter/start/) (15) to generate a putative membrane topology. Tet38 belongs to the MFS family of transporters with predicted 14 transmembrane segments (TMS), 6 internal loops, and 7 external loops, L1 through L7. Among the external loops, loop L4 is the shortest loop with 5 amino acids (216 to 221), and the longest loop, L7, has 21 amino acids (400 to 421) (Fig. 1).
FIG 1.
Topological model of Tet38 was generated with the Protter interactive feature visualization (https://wlab.ethz.ch/protter/start/). The 14 transmembrane segments (T-1 to T-14) are indicated with their respective residues and positions. The 7 external loops are indicated with their respective positions (L1 to L7). The motifs B and C and the conserved residues R106 and G151 (shaded gray) are indicated based on the alignment of Tet38, TetK/L, and TetA/B using the ClustaL Omega and ConSurf servers. The motifs are as reported in Paulsen et al. (7).
Amino acid sequence alignment using the Clustal Omega web server program from EMBL-EBI (www.ebi.ac.uk/Tools/msa/clustalo/) and the published amino acid sequences of Tet38 (GenPept accession no. BAB41352.1), TetK of S. aureus (GenPept accession no. KII19949.1), TetL of B. subtilis (GenPept accession no. ADN51953.1), TetA of E. coli (GenPept accession no. QBQ69130.1), and TetB of E. coli (GenPept accession no. WP_001089068.1), taken from the protein database of NCBI (www.ncbi.nlm.nih.gov), showed that S. aureus Tet38 shared 29% identity and 50% similarity with TetK of S. aureus, and it shared 28% identity and 51% similarity with the TetL of B. subtilis. The homologous region of Tet38 in the alignment with TetK and TetL covered 415 residues out of a 450-amino-acid sequence in length. Tet38 showed 23% identity and 45% similarity within a short region spanning residues A62 (Ala) to Y161 (Tyr) with plasmid-encoded E. coli TetA, a 23% identity and 42% similarity in the region covering residues A66 (Ala) to D197 (Asp), and a 23% identity and 58% similarity in the region that includes L387 (Leu) to V434 (Val) with the TetB of E. coli.
Compared with the amino acid sequences of TetK of S. aureus, TetL of B. subtilis, TetA of E. coli, and TetB of E. coli, Tet38 displayed the identical residues R106 (Arg) and G151 (Gly) that were essential to the tetracycline efflux function of the Tet transporters (10, 13, 16, 17) (Tables 1 and 2). The consensus sequence GX8GX3GPX2GG of the motif C of TetK and TetL shows 5 matching residues compared with the putative motif C of Tet38 (G138, S142, G151, G155, and G156). Among these amino acids, the residue Gly at position 151 (G151, equivalent with G155 of TetK/L and G147 of TetA/B) was essential in the extrusion of the metal-tetracycline complex by tetracycline resistance proteins TetL/TetK (11).
TABLE 1.
Homology in amino acid sequence between Tet38, TetL, TetK, TetA, and TetB
| Protein | Sequence ID (GenPept accession no.) | Length (aa)a | Covered region | Identity/similarity (%) | Function |
|---|---|---|---|---|---|
| Tet38 | BAB41352.1 | 450 | 100 | Tetracycline pump | |
| TetK | KII19949.1 | 460 | 1–415 | 29/50 (Tet38/TetK) | Tetracycline exporter |
| TetL | ADN51953.1 | 460 | 1–415 | 28/51 (Tet38/TetL) | Tetracycline exporter |
| TetA | QBQ69130.1 | 399 | 62–161 | 23/42 (Tet38/TetA) | Tetracycline exporter |
| TetB | WP_001089068.1 | 401 | 66–197; 387–434 | 23/58 (Tet38/TetB) | Tetracycline exporter |
aa, amino acids.
TABLE 2.
Essential amino acids for the tetracycline efflux function of the exporter
| Protein | Residues | Locations | Identity (%) | Function |
|---|---|---|---|---|
| Tet38 | Arg, Gly | R106, G151 | 100 | Tetracycline efflux |
| TetK/L | Arg, Gly | R110, G155 | 100 | Tetracycline efflux |
| TetA/B | Arg, Gly | R103, G147 | 100 | Tetracycline efflux |
Similarly, the conserved residue Arg at position 106 (R106, equivalent with R110 of TetK/L and R103 of TetA/B), located on motif B of Tet38, was also an essential amino acid in tetracycline efflux (11). Previous studies showed that replacement of residues R110 and G155 of TetK/L by cysteine (R to C; G to C) leads to a loss of bacterial resistance to tetracycline (11, 16, 18) (Tables 1 and 2). We modified Tet38 cloned on plasmid pLI50 with R106C and G151C mutations by site-directed mutagenesis.
In the absence of information on TetK or TetL interaction with external proteins, we tested two external loops for their role in interaction with CD36 and epithelial cell invasion, creating 3-amino-acid deletions on external loops L1 and L7 [pLI50-tet38-ΔL1(38DDL40) and pLI50-tet38-ΔL7(411FPG413), respectively] for further investigation (Fig. 1).
Effects of mutations on Tet38 conferring resistance to tetracycline and other substrates.
We used S. aureus RN6390, QT7, and QT7 transformed with plasmids carrying the wild-type or mutated tet38 gene to determine the MICs of tetracycline, minocycline, fosfomycin, tunicamycin, and palmitoleic acid, agents to which Tet38 confers resistance. The effect of the efflux inhibitor reserpine on resistance was also tested using the same S. aureus strains.
The mutant QT7, which contains tet38 disrupted by a cat gene insertion, was created from the wild-type reference strain RN6390 (6). We used QT7 as a background S. aureus recipient strain lacking a functional Tet38 to study the effect of various mutated and plasmid-expressed Tet38 on the susceptibility of S. aureus to antibiotics and palmitoleic acid.
QT7 showed a 2-fold decrease in the MICs to tetracycline and fosfomycin compared to that of RN6390 (Table 3). These data were similar to those in our previous study (3, 6) and suggest low levels of expression of tet38 in RN6390 under these conditions. QT7 was similarly 2-fold more susceptible to tunicamycin and palmitoleic acid than RN6390, but no difference was seen with minocycline susceptibilities. The addition of reserpine reduced MICs of all drugs 2-fold in RN6390 but had no effect in QT7.
TABLE 3.
Susceptibility of S. aureus to antibiotics and palmitoleic acid in the presence or absence of reserpine at 50 μg/ml
| S. aureus strain or mutantb,c | MIC (μg/ml) of:a |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TET | MINO | FOS | TUNI | PAL | TET | MINO | FOS | TUNI | PAL | |
| Without reserpine | With reserpine at 50 μg/ml | |||||||||
| RN6390 | 0.5 | 0.5 | 8 | 8 | 125 | 0.25 | 0.25 | 4 | 4 | 62.5 |
| QT7 (tet38::cat) | 0.25 | 0.5 | 4 | 4 | 62.5 | 0.25 | 0.5 | 4 | 4 | 62.5 |
| QT7 (pLI50) | 0.25 | 0.5 | 4 | 4 | 62.5 | 0.25 | 0.5 | 4 | 4 | 62.5 |
| QT7 (pLI50-tet38) | 4 | 2 | 16 | 16 | 500 | 1 | 0.5 | 4 | 4 | 125 |
| QT7 (pLI50-tet38-R106C) | 0.25 | 0.5 | 4 | 16 | 500 | 0.25 | 0.5 | 4 | 4 | 250 |
| QT7 (pLI50-tet38-G151C) | 0.5 | 0.5 | 4 | 4 | 250 | 0.25 | 0.5 | 4 | 4 | 125 |
| QT7 (pLI50-tet38-ΔL1) | 0.5 | 1 | 4 | 8 | 250 | 0.25 | 0.5 | 4 | 4 | 125 |
| QT7 (pLI50-tet38-ΔL7) | 1 | 1 | 8 | 16 | 500 | 0.5 | 0.5 | 4 | 8 | 250 |
TET, tetracycline; MINO, minocycline; FOS, fosfomycin; TUNI, tunicamycin; PAL, palmitoleic acid.
All strains harboring plasmid pLI50 were grown in the presence of 20 μg/ml chloramphenicol at 37°C.
QT7, tet38 mutant.
QT7 was transformed with plasmid constructs carrying the tet38 gene with or without mutations R106C and G151C. Introduction of pLI50 alone had no effect on drug susceptibilities. Introduction of pLI50-tet38 produced 4- to 16-fold increases in MICs across the five drugs. These increases were reduced by reserpine either completely or 4-fold (tetracycline) or 2-fold (palmitoleic acid) higher than that of QT7 pLI50.
Mutants of Tet38 were similarly cloned into and expressed from pLI50. In contrast to pLI50-tet38, pLI50-tet38-R106C conferred no increase in MICs of tetracycline, minocycline, or fosfomycin in QT7 but conferred increases in MICs of tunicamycin (4-fold) and palmitoleic acid (8-fold). These increases were eliminated by reserpine with the exception of an incomplete reduction of the MIC of palmitoleic acid.
pLI50-tet38-G151C produced only a partial increase in MICs of palmitoleic acid (4-fold) and tetracycline (2-fold) in QT7, without increases for minocycline, fosfomycin, and tunicamycin. These remaining resistance phenotypes were reduced by the addition of reserpine. Thus, arginine 106 and glycine 151 are both important for the ability of Tet38 to confer resistance to some but not all compounds, suggesting different structural determinants of efflux for different substrates of Tet38.
In comparison with the increases in the MICs of QT7 complemented with the pLI50-tet38 plasmid (16-fold for tetracycline; 4-fold for minocycline, fosfomycin, and tunicamycin; and 8-fold for palmitoleic acid) (Table 3), small deletions Asp-Asp-Leu (DDL) of predicted external loop L1 and Phe-Pro-Gly (FPG) of predicted external loop L7 (Fig. 1) also retained the ability to confer residual resistance phenotypes to the tested antibiotics and palmitoleic acid with a small difference in the MICs of fosfomycin. QT7(pLI50-tet38-ΔL1) conferred an increase of 2-fold in the MICs of tetracycline, minocycline, and tunicamycin; an increase of 4-fold in the MIC of palmitoleic acid; and no change in the MIC of fosfomycin. QT7(pLI50-tet38-ΔL7) conferred an increase of 4-fold in the MICs of tetracycline and tunicamycin, a 2-fold increase in the MICs of minocycline and fosfomycin, and an increase of 8-fold in the MIC of palmitoleic acid. The loop L7 mutant retained the ability to confer resistance to tunicamycin and palmitoleic acid equivalent to that of wild-type Tet38 and the mutant QT7(pLI50-tet38-R106C) (Table 3). Thus, these external loops appear to have less important roles in the resistance properties of Tet38 than do arginine 106 and glycine 151 in motifs B and C (Table 3).
Effects of Tet38 mutations on S. aureus internalization by A549 host cells.
In prior work (2, 5, 6), we showed that Tet38, in addition to its resistance properties, contributed to the ability of S. aureus to internalize in host epithelial cells by its direct interaction with host cell CD36. To evaluate the structural determinants of these properties of Tet38, we tested the four tet38 mutants for their effects on S. aureus internalization in A549 cells. Consistent with our previous findings, QT7 had a 6-fold reduction in internalization relative to RN6390 (Table 4). QT7(pLI50) also had a similar reduced internalization as QT7, and pLI50-tet38 in QT7 restored internalization to wild-type levels. pLI50-tet38-R106C, pLI50-tet38-G151C, and pLI50-tet38-ΔL1 each also restored internalization of QT7 to near-wild-type levels. In contrast, pLI50-tet38-ΔL7 produced a minimal increase in internalization in QT7. Thus, loop L7 is a domain important for Tet38’s ability to invade epithelial cells (Table 4).
TABLE 4.
Internalization of S. aureus by A549 cellsa
| S. aureus strain or mutantb,c | CFU/monolayer | CFU/monolayer (S. aureus plus Res)d |
|---|---|---|
| RN6390 | (2.4 ± 0.005) × 106 | (2.2 ± 0.008) × 106 |
| QT7 (tet38::cat) | (0.4 ± 0.001) × 106 | (0.3 ± 0.015) × 106 |
| QT7 (pLI50) | (0.3 ± 0.015) × 106 | (0.3 ± 0.010) × 106 |
| QT7 (pLI50-tet38) | (2.5 ± 0.001) × 106 | (1.2 ± 0.015) × 106 |
| QT7 (pLI50-tet38-R106C) | (2.0 ± 0.002) × 106 | (1.5 ± 0.020) × 106 |
| QT7 (pLI50-tet38-G151C) | (2.3 ± 0.008) × 106 | (1.3 ± 0.012) × 106 |
| QT7 (pLI50-tet38-ΔL1) | (2.1 ± 0.001) × 106 | (2.0 ± 0.011) × 106 |
| QT7 (pLI50-tet38-ΔL7) | (0.5 ± 0.002) × 106 | (0.4 ± 0.020) × 106 |
Experiments were done in triplicate and with three separate biological samples. The differences between the tet38 overexpressor and tet38-ΔL7 overexpressor in QT7 were statistically significant as determined by a Student's t test (P < 0.05). The differences between the tet38 overexpressor in QT7 in the absence or presence of reserpine were statistically significant as determined by a Student's t test (P < 0.05).
All strains harboring plasmid pLI50 were grown in the presence of chloramphenicol 20 μg/ml at 37°C.
QT7, tet38 mutant. QT7 transformed with plasmids carrying the wild-type or mutated tet38 gene are given in parentheses.
Res, reserpine at 50 μg/ml final concentration.
Effects of Tet38 loop deletions on binding to CD36.
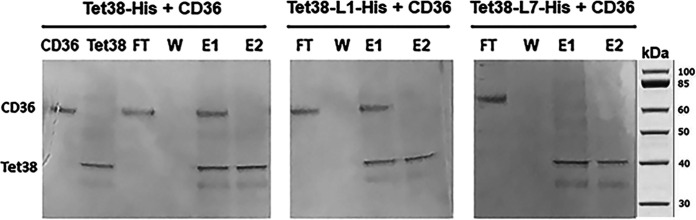
To further assess the role of Tet38 external loops L1 and L7 in its binding to host cell CD36, we performed column retention assays previously developed in our study of the Tet38-CD36 binding complex (5). In this assay, histidine-tagged purified Tet38 protein (Tet38-His) is loaded on an Ni affinity column followed by CD36 protein from which the histidine tag has been removed with enterokinase. CD36 is also loaded on a separate control Ni affinity column without Tet38. CD36 was not bound to the control column and could be found in the flowthrough initial wash. In contrast, CD36 was retained on the Tet38-anchored column and eluted together with Tet38 in the imidazole elution buffer (Fig. 2), as previously reported.
FIG 2.
SDS-PAGE of the binding assays between Tet38-His, Tet38-ΔL1(38DDL40)-His, Tet38-ΔL7(411FPG413)-His, and CD36. Enterokinase-treated CD36 was applied to an Ni column that had been previously loaded with Tet38-His, Tet38-ΔL1(38DDL40)-His, or Tet38-ΔL7(411FPG413)-His. Flowthrough (FT), wash (W), and elution fractions (E1 and E2) were collected and submitted to SDS-PAGE. Reference proteins included Tet38 and CD36. Tet38-L1-His is equal to Tet38-ΔL1(38DDL40)-His, and Tet38-L7-His is equal to Tet38-ΔL7(411FPG413)-His. Molecular weights of proteins in the assays are ∼45 kDa for Tet38 and ∼66 kDa for CD36.
We purified histidine-tagged Tet38-ΔL1 and histidine-tagged Tet38-ΔL7 proteins and loaded them on separate Ni columns followed by CD36. Flowthrough, wash, and imidazole elution fractions were evaluated by SDS-PAGE. For Tet38-ΔL1, CD36 was identified in the initial flowthrough, was absent in the first wash, and was identified again in the imidazole elution fraction together with Tet38, indicating column retention and physical binding of CD36 to Tet38-ΔL1 (Fig. 2). In contrast, for Tet38-ΔL7, CD36 was only identified in the flowthrough fraction, and Tet38 alone was identified in the elution fraction, indicating that Tet38-ΔL7 did not retain CD36 on the column, a finding consistent with its loss of the ability to facilitate S. aureus internalization into epithelial cells.
DISCUSSION
Chromosomally encoded Tet38 is an important membrane protein in S. aureus that contributes to S. aureus’s resistance to antibacterial drugs and natural compounds such as antibacterial fatty acids. Tet38 also contributes to S. aureus’s ability to survive in skin abscesses, and to invade epithelial cells by interaction with host cell CD36 (3–5). We thus sought to evaluate some of the structural determinants of Tet38’s diverse functions, with a focus on efflux-mediated resistance, invasion of epithelial cells, and interaction with CD36.
S. aureus Tet38 is related to the tetracycline proteins TetL of B. subtilis and plasmid-encoded TetK of S. aureus. It shares with these proteins motif C, which plays an important role in substrate efflux of the TetL/TetK antiporters due to its location near the permeability barrier on TMS-5. The consensus sequence GX8GX3GPX2GG of motif C contains highly conserved residues, including E152 (glutamate), G155 (glycine), P156 (proline), and A157 (alanine) (TetL) or S157 (serine) (TetK), that are crucial for the efflux function and tetracycline resistance phenotype of TetL and TetK (11, 16).
Alignment between Tet38, TetL, TetK, TetA, and TetB showed that glycine residue at position 151 in Tet38 is equivalent to the residue G155 (glycine) of TetL and TetK and residue G147 of TetA and TetB. G155 of TetL/K has been hypothesized to influence the conformation of TMS-5 and, in conjunction with other glycine residues in its vicinity, forms a pocket on one face of TMS-5 that influences the transport of TetL/TetK substrates. A G155C mutant exhibits significant alteration in the transport of metal-tetracycline by the TetL/TetK proteins (9, 11, 18).
Arginine R110 of TetL and TetK, located at TMS-4 (motif B) and interacting with motif C, also played an important role in influencing the extrusion of metal-tetracycline by the antiporters. Substitution of arginine by a cysteine (R110C) resulted in a complete loss of efflux function of the protein and a loss of tetracycline resistance phenotype in bacteria (11). In Tet38, R106 is equivalent to R110 of TetL and TetK and R103 of TetA and TetB.
We found that the tetracycline and, to a lesser extent, minocycline resistance phenotypes of Tet38 were similarly affected by the R106C and G151C mutations, homologous to those in TetL/K. Notably, the fosfomycin resistance phenotype was also eliminated in both mutants, but the tunicamycin resistance phenotype was only eliminated in the G151C mutant, and there was little or no effect of either mutant on palmitoleic acid resistance. Thus, the R106C and G151C mutations have selective effects on different Tet38 substrates, suggesting that tunicamycin and palmitoleic acid may have different points of critical interaction for their efflux to occur. Although it is possible that resistance to these compounds could be due to mechanisms other than efflux, it is notable that reserpine, a competitive efflux inhibitor, does reduce Tet38-mediated resistance to these compounds, thereby supporting a probable role for efflux as the mechanism of resistance, as it is for the tetracyclines.
We had previously demonstrated that Tet38 bound directly to the host scavenger receptor CD36 using a column retention assay (5). The mechanism behind this binding event is still not fully understood. Other S. aureus proteins that mediate binding to host cells include adhesins SraP and Eap (19, 20). SraP promotes S. aureus adhesion and invasion to A549 cells via its conserved residue, Y3670 (tyrosine), that bound to sialylated receptors (19). Eap is a secreted extracellular adhesin that remains attached to the bacterial cell wall via a repeat region. Domain EapH1 has three conserved amino acids, R89, E94, and K95, that play crucial roles in human neutrophil adhesion (20). Eap and SraP have not been shown to bind directly to CD36 of A549 cells, but their essential amino acids displayed external positions similar to the amino acids 411FPG413 of the external loop L7 of Tet38 (data not shown) (19, 20).
We selected two external loops, L1 and L7, of Tet38 to investigate further. The external loop L1 of Tet38 spans from residues 29 to 48 and is located between TMS-1 and TMS-2. Tet38 external loop L7 spans from residues 400 to 421 and is located between TMS-13 and TMS-14. Based on a putative membrane topology of Tet38 (Fig. 1), the three central amino acids, D38D39L40 of loop L1 and F411P412G413 of loop L7, that we deleted were positioned at an angle comparable with that of the three residues, R89, E94, and K95 of EapH1, based on the crystal structure of the binding domain EapH1 and its conserved amino acids (20).
To delineate the contribution of Tet38 and its mutants to internalization in A549 cells, we focused on the effects of plasmid expression of tet38 seen in QT7, which lacks a functional chromosomally encoded Tet38. In QT7, Tet38 with a deletion in loop L7 (F411P412G413) expressed from pLI50 produced only a slight increase in S. aureus internalization above that of pLI50 alone. In contrast, pLI50-tet38-ΔL1(38DDL40) produced an increase which was similar to that of pLI50-tet38. In keeping with the role of Tet38 interaction with CD36 in S. aureus internalization (5), purified Tet38-ΔL1 bound to CD36 in the column retention assay, whereas Tet38-ΔL7 did not. Thus, loop L7 is both important for internalization and binding to CD36. Notably the R106C and G151C mutants enabled internalization with a similar magnitude to that of wild-type Tet38.
The plasmid-expressed L1 and L7 deletion mutants in QT7, however, conferred less resistance to tetracyclines and fosfomycin than wild-type Tet38, with a somewhat greater reduction in the L1 mutant than the L7 mutant. These data suggest that these deletions may generate changes in the conformation of Tet38, which, in turn, affected the transport efficiency of these substrates without affecting transport efficiency for tunicamycin and palmitoleic acid. The specific mechanism of these differential effects on substrate efficiency, however, will require additional structural studies of Tet38 with and without drug substrates. Notably, as was seen for the R106C mutant, the L7 deletion (FPG) had little effect on the ability of Tet38 to confer resistance to tunicamycin and palmitoleic acid. Mutants thus exhibited specific functional defects, distinguishing the importance of different domains in Tet38 for resistance, epithelial cell internalization, and binding to CD36. Our findings thus provide initial insight into how Tet38 effects its several functions and highlight the importance of its ability to link resistance and reduced response to antibiotics (21), with the ability of S. aureus to survive within epithelial cells, colonize skin (4), and survive within abscesses (2, 6, 22, 23).
MATERIALS AND METHODS
Cell lines, bacterial strains, and culture media.
The bacterial strains, plasmids, and cell lines used in this study are listed in Table 5. The human lung adenocarcinoma cell line, A549, was purchased from ATCC (CCL-185) and cultivated in assay medium which contained modified Eagle’s medium (DMEM; Life Technologies, Grand Island, NY), supplemented with 10% fetal bovine serum (FBS) and 4 mM l-glutamine (Fisher Scientific, Waltham, MA) at 37°C in 5% CO2, as previously reported (6).
TABLE 5.
Bacterial strains, plasmids, cell line, and plasmids used in this study
| Strain, plasmid, cell line, or primer | Genotype, relevant characteristic(s), or sequence | Reference no. or source |
|---|---|---|
| S. aureus and E. coli strains, plasmids, and cell line | ||
| S. aureus RN6390 | Wild type | 6 |
| S. aureus QT7 | RN6390 (tet38::cat) mutant | 6 |
| S. aureus RN6390 or QT7 (pLI50-tet38) | tet38 overexpressor; Cmr | 6 |
| S. aureus RN6390 or QT7 (pLI50-tet38-ΔL1) | tet38(L1-ΔDDL) overexpressor; Cmr | This study |
| S. aureus RN6390 or QT7 (pLI50-tet38-ΔL7) | tet38(L7-ΔFPG) overexpressor; Cmr | This study |
| S. aureus RN6390 or QT7 (pLI50-tet38-R106C) | tet38(R106C) overexpressor; Cmr | This study |
| S. aureus RN6390 or QT7 (pLI50-tet38-G151C) | tet38(G151C) overexpressor; Cmr | This study |
| E. coli BL21 | F− ompT gal dcm lon hsdSB(rB−mB−) [malB+]K-12(λS) | Novagen |
| pLI50 | Shuttle plasmid E. coli-S. aureus; Cmr | 6 |
| pTrcHisA | Cloning plasmid for protein expression | Invitrogen |
| pTrcHisA-tet38 | Tet38 protein expression | 4 |
| pTrcHisA-tet38-ΔL1 | Tet38-ΔL1 protein expression | This study |
| pTrcHisA-tet38-ΔL7 | Tet38-ΔL7 protein expression | This study |
| A549 | Human lung adenocarcinoma cells | ATCC |
| Primers for site-directed mutagenesis | ||
| Tet38-1F | Loop L1; 5′-AACATATCTGTAACGACAGTAAGT-3′ | |
| Tet38-1R | Loop L1; 3′-AAATTGATTTCTGTAGCCATTGCT-5′ | |
| Tet38-7F | Loop L7; 5′-AATGATGCATTAAGTTCACATTTC-3′ | |
| Tet38-7R | Loop L7; 3′-CTAATCGATTTTAAAGCATTAAAT-5′ | |
| Tet38-R106CF | R106C; 5′-CTTTTAGTTGGACGTATTATTCAAACT-3′ | |
| Tet38-R106CR | R106C; 3′-ATTTTCCAACATCAATTCCCATTA-5′ | |
| Tet38-R151CF | G151C; 5′-TCATTAGTTATCGGTACATTATCAGGT-3′ | |
| Tet38-R151CR | G151C; 3′-TTAAGTACGAGCAGTTATTCCTTG-5′ |
Gentamicin, ampicillin, chloramphenicol, Triton X-100, and lysostaphin were purchased from Sigma-Aldrich (St. Louis, MO).
S. aureus containing plasmid pLI50 and its various constructs were grown at 37°C in Luria-Bertani media (LB) supplemented with chloramphenicol at 20 μg/ml. Expression plasmids pTrcHisA-tet38 and the loop deletion variants pTrcHisA-tet38-ΔL1 and pTrcHisA-tet38-ΔL7 were introduced into E. coli BL21 for protein expression.
E. coli transformants were grown at 37°C in the presence of ampicillin at 100 μg/ml.
Drug susceptibility determinations.
Tetracycline, minocycline, fosfomycin, tunicamycin, palmitoleic acid, and reserpine were purchased from Sigma Chemical Co. (St. Louis, MO). The MICs were determined by broth microdilution at 37°C for 24 h as previously described (1, 6). A log-phase culture of S. aureus (optical density at 600 nm [OD600], 0.5) grown in LB media was diluted 100-fold and inoculated into microtiter plates (Fisher Scientific, Pittsburgh, PA) containing serial 2-fold dilutions of drugs.
Reserpine was added when specified at a concentration of 50 μg/ml, which was chosen after preliminary testing at a range of concentrations of 10 to 100 μg/ml.
Putative membrane topology of Tet38 TMSs and external loops.
We used the web server ConSurf to analyze and predict the Tet38 TMS regions and their external loops (https://consurf.tau.ac.il/) (14). The server generated an alignment between the Tet38 amino acid sequence and the sequences of homologous proteins of the same MFS family. Based on the evolution rate and the conservation score of each amino acid, the generated ConSurf sequence data indicated the position and the degree of conservation of each amino acid of the Tet38 protein.
We then created a putative membrane topology of Tet38 using the ConSurf data and the web server Protter interactive feature visualization (https://wlab.ethz.ch/protter/start/) (15).
The putative Tet38 membrane structure showed 14 TMS, 6 internal loops, and 7 external loops, with loop L4 as the shortest with 5 amino acids and L7 as the longest with 21 amino acids. The first external loop, L1, and the last external loop, L7, were chosen for further analyses.
Construction of tet38 mutants.
We designed specific forward and reverse primers for the external loops L1 and L7 following the recommendation of the Phusion site-directed mutagenesis kit (Thermo Scientific, Waltham, MA). Primers used in this study to generate mutants with deletion in the external loops L1 and L7 are listed in Table 5. All primers were 5′ phosphorylated and synthesized by Eton Bioscience Inc. (Boston, MA).
The template for site-directed mutagenesis was the plasmid-borne tet38 gene that was previously created by cloning the chromosomal tet38 gene of S. aureus RN6390 into the shuttle E. coli-S. aureus plasmid pLI50 to generate the construct pLI50-tet38. This construct was first introduced into E. coli Top10 by electroporation, then subsequently introduced into S. aureus RN4220, and finally introduced into S. aureus RN6390 for our tet38 overexpression studies (5).
PCR amplification of the tet38 gene using site-directed mutagenesis primers (Tables 1 and 2) and the plasmid template pLI50-tet38 was based on the protocol provided with the Phusion site-directed mutagenesis kit. The PCR products were ligated following the manufacturer’s instructions and were introduced into E. coli Top10 by electroporation. E. coli transformants were selected on ampicillin at 100 μg/ml, and DNA sequencing was performed to verify the correct deletion constructs.
The new constructs pLI50-tet38-ΔL1 and pLI50-tet38-ΔL7 were extracted from E. coli and introduced into S. aureus RN4220 and then reintroduced into RN6390 and QT7 by electroporation for further evaluations and assays. Reference strains included RN6390 and QT7 carrying plasmids pLI50 and pLI50-tet38. The construction of Tet38 with mutations R106C and G151C followed the same protocol as above with primers listed in Table 5.
Internalization of S. aureus in A549 epithelial cells.
The internalization assays were performed as previously described (6). A549 lung epithelial cells were cultured in 5 ml of assay medium until 85% confluence in a 25-ml tissue culture flask and then seeded into 24-well plates (Costar) in assay medium and grown again to 85% confluence. The A549 cell concentration was adjusted to 104cells/ml. S. aureus RN6390 and QT7 with appropriate plasmid constructs were prepared from overnight cultures and grown to an OD600 of 0.5. The bacteria were washed twice with 1× PBS, and the bacterial pellet was resuspended in 10 ml of fresh assay medium to a concentration of 106 CFU/ml. A549 cells were infected with S. aureus at a multiplicity of infection (MOI) of 100 bacteria per epithelial cell (MOI, 100:1 or 106 washed bacteria/104 cells). The 24-well plates were centrifuged quickly for 30 s at 500 × g to allow bacterial adhesion to the cell monolayer. The bacterium-cell mixtures were incubated at 37°C in 5% CO2 for 120 min, and then the infected monolayers were washed three times with 1× PBS to remove residual nonadherent bacteria. The washed monolayers were incubated for 120 min at 37°C in 5% CO2 in assay medium with 200 μg/ml gentamicin and 20 μg/ml lysostaphin. Monolayers were again washed three times with 1× PBS, and the A549 epithelial cells were lysed with 200 μl of Triton X-100 (0.1%). The bacteria were diluted in PBS and plated on LB agar plates, and colony counts were performed to determine the number of viable intracellular bacteria.
Protein purification of Tet38, Tet38-ΔL1(38DDL40), and Tet38-ΔL7(411FGP413).
Histidine-tagged Tet38 and its variants, Tet38-ΔL1(38DDL40) and Tet38-ΔL7(411FGP413), were expressed from E. coli BL21 and purified by Ni affinity chromatography, as previously described (5). tet38 of S. aureus RN6390 with and without modifications were cloned into the expression vector pTrcHis2A and introduced into E. coli BL21 by electroporation. The transformants were cultured in 500 ml of LB medium supplemented with ampicillin (100 μg/ml) under shaking at 37°C until an OD600 of 0.6 was reached. Isopropyl-β-D-thiogalactopyranoside (IPTG) at 1 mM was then added, and the incubation was continued at 30°C for 20 h under shaking. The bacteria were harvested by centrifugation at 6,000 × g for 20 min at 4°C, then the pellet was resuspended in 10 ml of lysis buffer and processed as previously reported (5). Protein concentration was measured in a NanoDrop spectrophotometer (ND-1000 336 spectrophotometer V3.6.0) by direct absorbance at 280 nm, and the homogeneity of the protein was assessed by SDS-PAGE.
SDS-PAGE electrophoresis was carried out using NuPAGE 12% Bis-Tris precast SDS-PAGE protein gels and morpholinepropanesulfonic acid (MOPS) SDS running buffer (1×) as suggested by the manufacturer (Thermo Scientific, Waltham, MA). Running conditions were 150 V for 60 min following by staining in Coomassie blue G250 and destaining in water.
Affinity column retention assay.
The histidine tag was cleaved from commercial His-CD36 (Sino Biological, Inc., Wayne, PA) using 1 U of enterokinase in a reaction mixture of 10 μg of CD36 protein in 25 μl incubated at 25°C for 16 h. Enterokinase was purchased from New England BioLabs (Beverly, MA).
We used 5 μg each of CD36 and Tet38 proteins wild type and mutants Tet38-ΔL1(38DDL40) or Tet38-ΔL7(411FGP413) for the column retention assay, as described previously (5). Tet38-His was first applied to an Ni column, and then enterokinase-treated CD36 was loaded onto the same column. The column was washed with 5 column volumes of buffer A (10 mM Tris-HCl [pH 7.6], 500 mM NaCl, and 10 mM imidazole); then the proteins were eluted with 100 mM imidazole in buffer A. In parallel, the enterokinase-treated CD36 protein was loaded onto a separate Ni column, washed with buffer A, and then eluted with 100 mM imidazole to verify the absence of nonspecific binding of CD36 to the column. This step was used as as control for the specificity of CD36 binding to the affinity column-anchored histidine-tagged protein Tet38.
ACKNOWLEDGMENT
This work was supported by U.S. Public Health Service grants P01-AI083214 (M. Gilmore, principal investigator; subproject to D.C.H.) and R37-AI23988 (to D.C.H.) from the National Institutes of Health.
REFERENCES
- 1.Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. 2005. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol 187:2395–2405. doi: 10.1128/JB.187.7.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Truong-Bolduc QC, Khan N, Vyas JM, Hooper DC. 2017. Tet38 efflux pump affects Staphylococcus aureus internalization by epithelial cells through interaction with CD36 and contributes to bacterial escape from acidic and non-acidic phagolysosomes. Infect Immun 85:e00862-16. doi: 10.1128/IAI.00862-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Truong-Bolduc QC, Wang Y, Hooper DC. 2018. Tet38 efflux pump contributes to fosfomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 62:e00927-18. doi: 10.1128/AAC.00927-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Truong-Bolduc QC, Villet RA, Estabrooks ZA, Hooper DC. 2014. Native efflux pumps contribute resistance to antimicrobials of skin and the ability of Staphylococcus aureus to colonize skin. J Infect Dis 209:1485–1493. doi: 10.1093/infdis/jit660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truong-Bolduc QC, Wang Y, Hooper DC. 2019. Tet38 of Staphylococcus aureus binds to host cell receptor complex CD36-toll-like receptor 2 and protects from teichoic acid synthesis inhibitors tunicamycin and Congo red. Infect Immun 87:e00194-19. doi: 10.1128/IAI.00194-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truong-Bolduc QC, Bolduc GR, Medeiros H, Vyas JM, Wang Y, Hooper DC. 2015. Role of the Tet38 efflux pump in Staphylococcus aureus internalization and survival in epithelial cells. Infect Immun 83:4362–4372. doi: 10.1128/IAI.00723-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulsen IT, Brown MH, Skurray RA. 1996. Proton-dependent multidrug efflux systems. Microbiol Rev 60:575–608. doi: 10.1128/MR.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginn SL, Brown MH, Skurray RA. 1997. Membrane topology of the metal-tetracycline/H+ antiporter TetA(K) from Staphylococcus aureus. J Bacteriol 179:3786–3789. doi: 10.1128/jb.179.11.3786-3789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirata T, Fujihira E, Kimura-Someya T, Yamaguchi A. 1998. Membrane topology of the Staphylococcal tetracycline efflux protein tet(K) determined by antibacterial resistance gene fusion. J Biochem 124:1206–1211. doi: 10.1093/oxfordjournals.jbchem.a022239. [DOI] [PubMed] [Google Scholar]
- 10.Guay GG, Khan SA, Rothstein DM. 1993. The tet(K) gene of plasmid pT181 of Staphylococcus aureus encodes an efflux protein that contains 14 transmembrane helices. Plasmid 30:163–166. doi: 10.1006/plas.1993.1045. [DOI] [PubMed] [Google Scholar]
- 11.De Jesus M, Jin J, Guffanti AA, Krulwich TA. 2005. Importance of the GP dipeptide of the antiporter motif and other membrane-embedded proline and glycine residues in tetracycline efflux protein Tet(L). Biochemistry 44:12896–12904. doi: 10.1021/bi050762c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujihira E, Tamura N, Yamaguchi A. 2002. Membrane topology of a multidrug efflux transporter, AcrB, in Escherichia coli. J Biochem 131:145–151. doi: 10.1093/oxfordjournals.jbchem.a003069. [DOI] [PubMed] [Google Scholar]
- 13.Tamura N, Konishi S, Iwaki S, Kimura-Someya T, Nada S, Yamaguchi A. 2001. Complete cysteine-scanning mutagenesis and site-directed chemical modification of the Tn10-encoded metal-tetracycline/H+ antiporter. J Biol Chem 276:20330–20339. doi: 10.1074/jbc.M007993200. [DOI] [PubMed] [Google Scholar]
- 14.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44:W344–50. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omasits U, Ahrens CH, Muller S, Wollscheid B. 2014. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 16.Jin J, Guffanti AA, Bechhofer DH, Krulwich TA. 2002. Tet(L) and tet(K) tetracycline-divalent metal/H+ antiporters: characterization of multiple catalytic modes and a mutagenesis approach to differences in their efflux substrate and coupling ion preferences. J Bacteriol 184:4722–4732. doi: 10.1128/jb.184.17.4722-4732.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guay GG, Rothstein DM. 1993. Expression of the tetK gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and TetK efflux proteins. Antimicrob Agents Chemother 37:191–198. doi: 10.1128/aac.37.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginn SL, Brown MH, Skurray RA. 2000. The TetA(K) tetracycline/H+ antiporter from Staphylococcus aureus: mutagenesis and functional analysis of motif C. J Bacteriol 182:1492–1498. doi: 10.1128/jb.182.6.1492-1498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YH, Jiang YL, Zhang J, Wang L, Bai XH, Zhang SJ, Ren YM, Li N, Zhang YH, Zhang Z, Gong Q, Mei Y, Xue T, Zhang JR, Chen Y, Zhou CZ. 2014. Structural insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS Pathog 10:e1004169. doi: 10.1371/journal.ppat.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herdendorf TJ, Geisbrecht BV. 2018. Investigation of human neutrophil elastase inhibition by Staphylococcus aureus EapH1: the key role played by arginine 89. Biochemistry 57:6888–6896. doi: 10.1021/acs.biochem.8b01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Hooper DC. 2018. Effect of Staphylococcus aureus Tet38 native efflux pump on in vivo response to tetracycline in a murine subcutaneous abscess model. J Antimicrob Chemother 73:720–723. doi: 10.1093/jac/dkx432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y, Onodera Y, Lee JC, Hooper DC. 2008. NorB, an efflux pump in Staphylococcus aureus MW2, contributes to bacterial fitness in abscesses. J Bacteriol 190:7123–7129. doi: 10.1128/JB.00655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, Fu Y, Lee JC, Hooper DC. 2012. Staphylococcus aureus NorD, a putative efflux pump coregulated with the Opp1 oligopeptide permease, contributes selectively to fitness in vivo. J Bacteriol 194:6586–6593. doi: 10.1128/JB.01414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]