Enterotoxigenic Escherichia coli (ETEC) contributes significantly to the substantial burden of infectious diarrhea among children living in low- and middle-income countries. In the absence of a vaccine for ETEC, children succumb to acute dehydration as well as nondiarrheal sequelae related to these infections, including malnutrition.
KEYWORDS: diarrhea, enteric pathogens, pathogenesis, surface antigens, vaccines
ABSTRACT
Enterotoxigenic Escherichia coli (ETEC) contributes significantly to the substantial burden of infectious diarrhea among children living in low- and middle-income countries. In the absence of a vaccine for ETEC, children succumb to acute dehydration as well as nondiarrheal sequelae related to these infections, including malnutrition. The considerable diversity of ETEC genomes has complicated canonical vaccine development approaches defined by a subset of ETEC pathovar-specific antigens known as colonization factors (CFs). To identify additional conserved immunogens unique to this pathovar, we employed an “open-aperture” approach to capture all potential conserved ETEC surface antigens, in which we mined the genomic sequences of 89 ETEC isolates, bioinformatically selected potential surface-exposed pathovar-specific antigens conserved in more than 40% of the genomes (n = 118), and assembled the representative proteins onto microarrays, complemented with known or putative colonization factor subunit molecules (n = 52) and toxin subunits. These arrays were then used to interrogate samples from individuals with acute symptomatic ETEC infections. Surprisingly, in this approach, we found that immune responses were largely constrained to a small number of antigens, including individual colonization factor antigens and EtpA, an extracellular adhesin. In a Bangladeshi cohort of naturally infected children <2 years of age, both EtpA and a second antigen, EatA, elicited significant serologic responses that were associated with protection from symptomatic illness. In addition, children infected with ETEC isolates bearing either etpA or eatA genes were significantly more likely to develop symptomatic disease. These studies support a role for antigens not presently targeted by vaccines (noncanonical) in virulence and the development of adaptive immune responses during ETEC infections. These findings may inform vaccine design efforts to complement existing approaches.
INTRODUCTION
Enterotoxigenic Escherichia coli (ETEC) is one of the most common causes of childhood diarrhea, accounting for hundreds of millions of cases annually (1). This high burden of disease is associated with a substantial risk of increased childhood morbidity and mortality (2–4). Repeated diarrheal infections, including those caused by ETEC, lead to the development of growth stunting and environmental enteropathy, which are lifelong consequences of these enteric infections (5). Therefore, preventative efforts, including vaccination, could have a tremendous impact on global health (6). Despite the lack of a licensed ETEC vaccine, two important lines of evidence suggest that ETEC vaccine development is feasible. First, controlled human infection models (CHIMs) demonstrate that protective immunity develops following ETEC challenge (7, 8). In addition, the frequency of symptomatic infections in young children living in regions of endemicity wanes substantially with age (9, 10), suggesting that natural infections afford subsequent protection.
ETEC biology, and the extraordinary genetic plasticity of E. coli, has complicated the development of a broadly protective vaccine. Canonical approaches have focused primarily on surface features known as colonization factors (CFs) or Coli Surface (CS) antigens. However, the structural and antigenic diversity of these targets has proved challenging (11). Although toxoids that can elicit neutralizing antibodies against the heat-labile toxin (LT) (12) and heat-stable toxin (ST) (13) that define the ETEC pathovar are currently under development (14, 15), it is not yet clear whether these alone will afford sufficient, long-lasting protection.
While the ETEC pathovar exhibits high genetic diversity, the recent availability of multiple genomic sequences from globally diverse ETEC strains affords the ability to apply reverse-vaccinology approaches to the identification of conserved, surface-expressed antigens (16, 17). In addition, microarray-based profiling of immune responses in human volunteers to ETEC challenge has recently highlighted noncanonical antigens (those not targeted in ETEC vaccines to date) that are recognized during controlled experimental infection (7, 18).
The application of these approaches to antigen discovery has reinforced the importance of several surface-expressed molecules common to the ETEC pathovar that are not currently targeted in classical vaccine approaches (19). These include two novel secreted molecules, the EtpA adhesin (20) and the EatA autotransporter (21), both originally identified in H10407, an ETEC strain isolated from a case of severe cholera-like diarrhea in Bangladesh. Recent work demonstrates that both antigens are globally distributed in the ETEC pathovar and are more highly conserved than the most common CFs (19, 22). Moreover, they are protective in murine models of infection (23–26) and immunogenic in human challenge trials (7, 18), suggesting that these molecules could provide additional antigenic targets for vaccine development. While much is known about EatA and EtpA under experimental conditions, less is known about their respective roles in natural infections. The present studies were designed to explore the roles of these and other potential noncanonical antigens in shaping the adaptive immune response to ETEC infection and to examine their contribution to virulence.
RESULTS
Antibodies following natural infection recognize a finite repertoire of ETEC proteins.
Both human experimental models (7) as well as natural infections (10) demonstrate that prior infection with ETEC affords substantial protection against symptomatic disease. Elucidation of the nature of protective adaptive immune responses to these mucosal pathogens can therefore inform vaccine development. While the majority of previous ETEC vaccinology efforts have centered on a canonical approach focused on colonization factor (CF) antigens, the present studies were designed to broadly profile antigenic responses to CFs as well as noncanonical antigens that have not been specifically targeted in vaccines to date. To assess the breadth of immune responses to ETEC during acute natural infection, we designed protein microarrays containing all proteins bioinformatically predicted (see Fig. S1 in the supplemental material) to be both surface-expressed proteins and conserved in more than 40% of the ETEC pathovar strains. These included EtpA and EatA, secreted antigens expressed by a majority of ETEC strains in a global collection of isolates (19). Although no single CF antigen was conserved in at least 40% of ETEC strains, each of the known CF protein subunits was also included on the arrays.
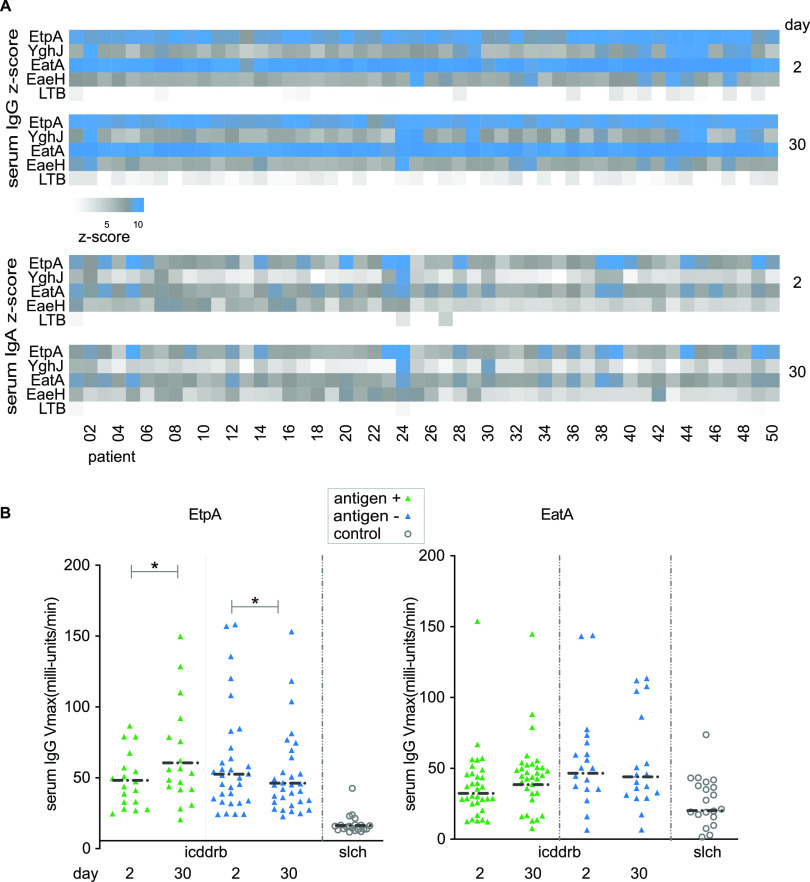
Despite the inclusion of multiple candidate surface molecules on the array predicted to be conserved among strains in Bangladesh from our in silico analysis, immune responses following infection were largely constrained to a small group of antigens, including EtpA and EatA (Fig. 1A), LT (Fig. S2), and select colonization factor subunits (Fig. S3). The latter included CssB, one of two components of the CS6 polymer (27), a predominant immunogenic antigen among strains circulating in Bangladesh (28). Compared to control specimens obtained outside the area where ETEC is endemic, both EatA and EtpA exhibited high levels of reactivity. Notably, for patients infected with EtpA-expressing strains, EtpA responses were significantly higher at day 30 following infection than those observed immediately following admission, whereas the converse was true in patients admitted with EtpA-negative strains (Fig. 1B).
FIG 1.
Serologic response to noncanonical antigens following natural infection. (A) Heat map of log2-transformed Z-score data indicating ETEC protein microarray responses from days 2 and 30 following presentation to the icddr,b to four noncanonical antigens, EtpA, YghJ, the passenger domain of EatA, and EaeH, and the B subunit of ETEC heat-labile toxin (LT-B). (B) Kinetic ELISA responses to EtpA and EatA following infection. Data are segregated by the presence or absence of the respective antigen in the strain recovered at presentation. Negative-control samples from St. Louis Children’s Hospital (slch) are shown as open circles. *, P < 0.05 by a Wilcoxon matched-pairs signed-rank test.
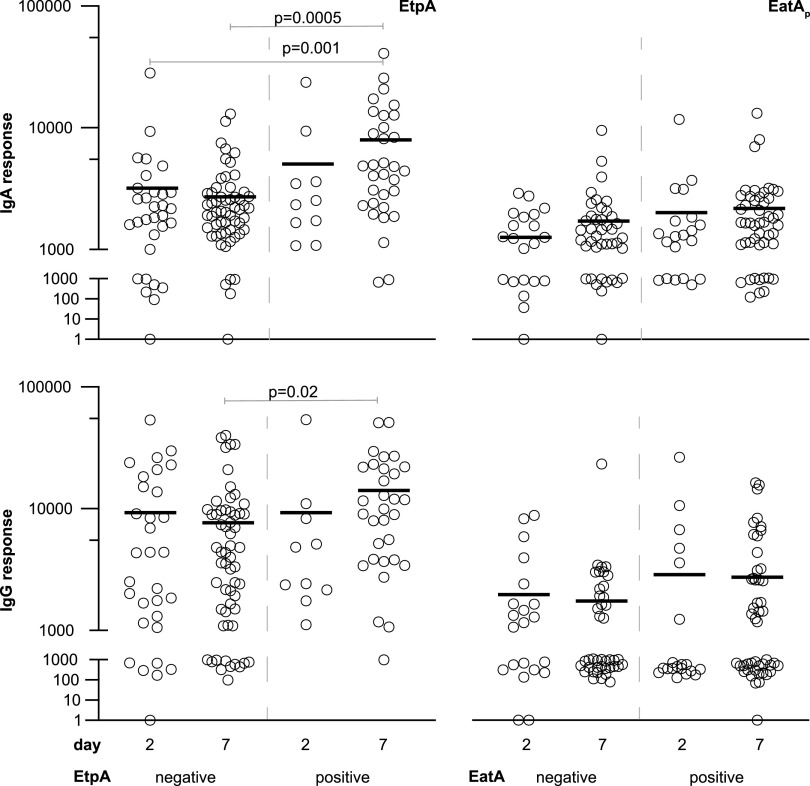
In an open-aperture assessment of antibody lymphocyte supernatant (ALS) specimens (29, 30) obtained from adults hospitalized at the International Centre for Diarrheal Disease Research, Bangladesh (icddr,b), Hospital in Dhaka, Bangladesh, or from patients recruited at the Mirpur field site with acute symptomatic diarrheal illness, we again noted that immune responses following infection were largely constrained to a relatively small group of antigens, including CS6, EtpA, and EatA (Data Set S2). When parsing antigen profiles of the infecting strain, we found that those individuals infected with EtpA-expressing ETEC exhibited significant increases in both ALS IgA (P = 0.005) and IgG (P = 0.02) responses in the week following infection relative to those infected with EtpA-negative strains (Fig. 2). As anticipated, we also observed significant increases in ALS immunoreactivity to the CssB subunit of CS6 that correlated with the production of CS6 by the infecting strain (Fig. S3).
FIG 2.
ALS responses to EtpA or EatA. Shown are microarray data for IgA (top) and IgG responses to EtpA (left) and the passenger domain of EatA (EatAp) (right) on days 2 and 7 following hospitalization. Data in each graph are segregated according to antigen expression in the infecting strain (negative or positive). P values reflect Kruskal-Wallis values, with post hoc analysis using Dunn’s test adjusted for multiple comparisons for between-group analysis.
EatA and EtpA are immunogenic in young children.
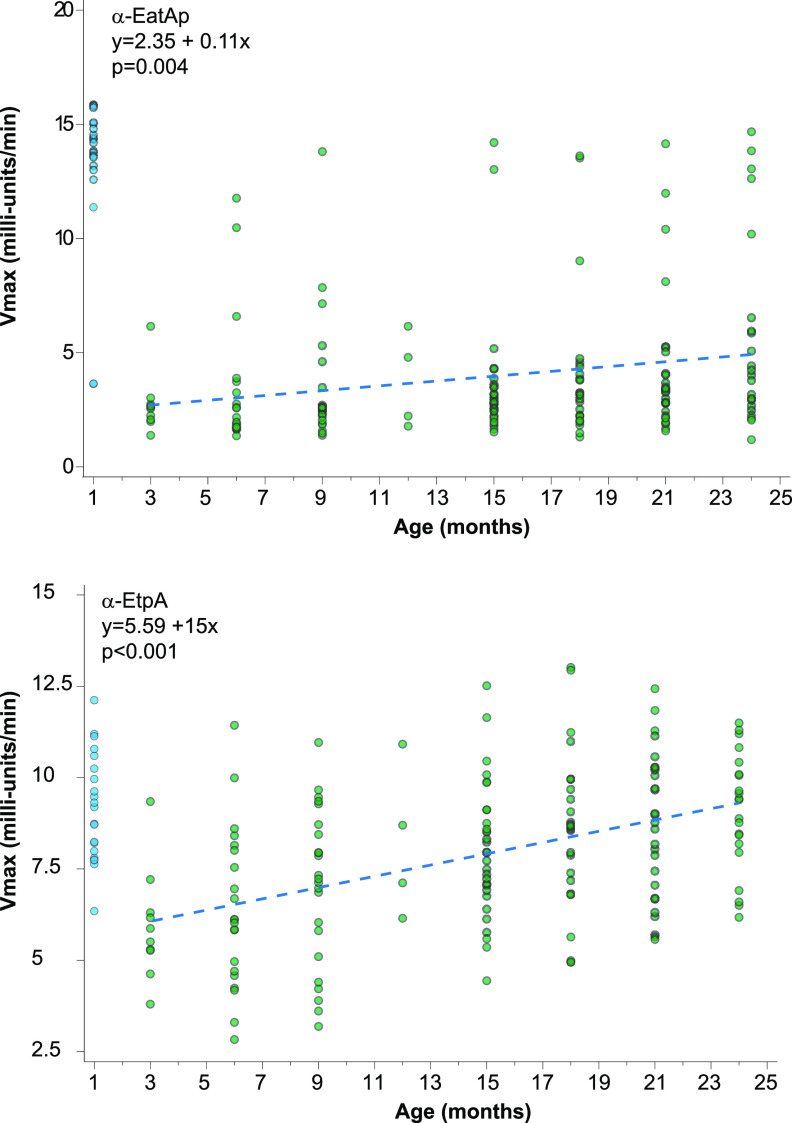
Data from recent controlled human infection model studies (7, 18) as well as earlier data from patients with natural ETEC infections (22) indicate that adults develop robust immune responses to noncanonical antigens, including EtpA and EatA. However, in areas of endemicity, young children are the population most severely impacted by ETEC, with incidence declining after 24 months of age, presumably as protection develops after infection. Therefore, we examined sera from a cohort of Bangladeshi children monitored from birth through 2 years of age (10) to profile the development of antibody responses to EatA and EtpA over time (Fig. 3). During the first month of life, the majority of children were observed to have elevated IgG responses to both EatA and EtpA, presumably reflecting the passive transfer of maternal antibodies (31). As anticipated, responses to both antigens decreased by 3 months of age, while mean responses to each antigen increased significantly through 24 months of age, likely reflecting early childhood infections with strains expressing EtpA and EatA.
FIG 3.
Anti-EtpA or anti-EatA IgG responses increase with age. Shown are representative kinetic ELISA data for serum IgG samples obtained from children aged 1 to 24 months enrolled in a birth cohort study. Scatterplots of anti-EtpA and anti-EatA IgG plotted against data for children aged 3 to 24 months with regression lines from linear repeated-measures models overlaid (dotted lines) demonstrate significant increases over time in the responses to the passenger domain of EatA (EatAp) (top) and EtpA (bottom). See Fig. S4 in the supplemental material for additional plots.
Anti-EtpA or -EatA responses relative to symptomatic diarrhea.
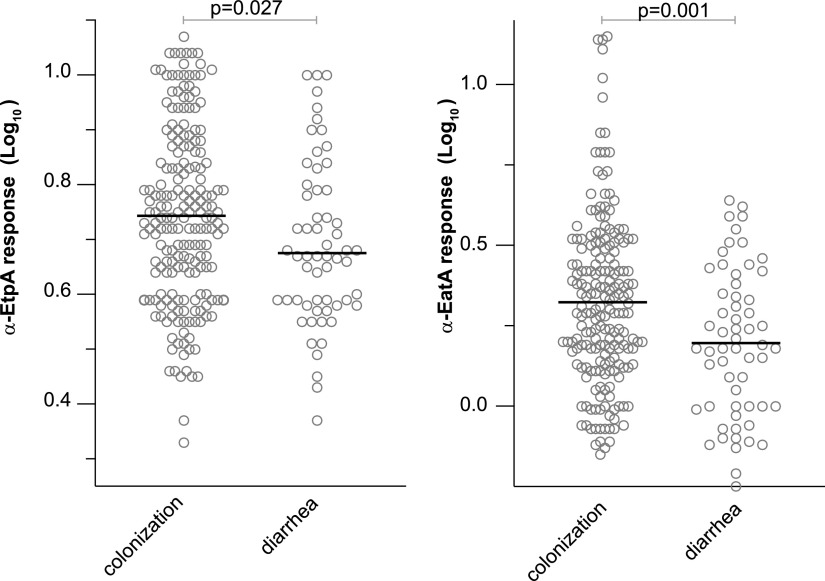
Immunological correlates of protection against ETEC are currently unknown (32). The majority of clinical studies to date have examined the impact of prior infection with strains producing particular colonization factors and/or LT (9) as well as antibody acquisition on the subsequent risk of infection with similar strains (33, 34). We hypothesized that because EtpA and EatA are relatively common antigens in the ETEC pathovar (19), higher antibody responses to these antigens may be associated with subsequent protection against symptomatic infection. After excluding antibody responses at 1 month of age, we examined the IgG antibody responses to EtpA and EatA preceding the detection of ETEC in either symptomatic or asymptomatic children between 4 and 24 months of age. Interestingly, we observed elevated responses to both antigens prior to the detection of ETEC in asymptomatic children relative to symptomatic cases (Fig. 4; Fig. S5), perhaps reflecting the overall mitigating impact of prior exposure on the development of diarrheal illness.
FIG 4.
Serum IgG responses preceding asymptomatic ETEC colonization and diarrhea. Shown are peak serum IgG responses for EtpA (left) or the EatA passenger domain (right) preceding either asymptomatic colonization or diarrheal illness with ETEC. Data shown are log10-transformed IgG antibody responses determined by a kinetic ELISA. Bars represent mean values. P values reflect Student’s t testing.
Association of eatA and etpA with virulence.
Although both EatA, a mucin-degrading serine protease, and the EtpA blood group A lectin are secreted by a diverse population of ETEC strains (19) and contribute to virulence phenotypes in vitro as well as in small-animal models of ETEC infection (23, 26, 35, 36), the role played by these antigens in human infections has yet to be explored in detail. To explore the association of eatA and etpA with symptomatic ETEC infection, we examined isolates collected in a birth cohort study in which stool specimens were collected at monthly intervals from asymptomatic children (asymptomatic colonization) or during surveillance for diarrhea (symptomatic infection) (10). Notably, the presence of etpA or eatA significantly increased the odds of having symptomatic diarrhea (unadjusted odds ratios of 2.1 and 3.1, respectively) (Table 1). Similarly, after adjusting for age, we observed significant associations between the presence of either EtpA (adjusted odds ratio of 1.98; P = 0.007) or EatA (adjusted odds ratio of 2.91; P < 0.001) and the development of diarrheal disease.
TABLE 1.
Noncanonical antigen analysis of Bangladeshi birth cohort samplesa
| Antigen and toxin(s) | Presence of diarrhea | No. of samples (%) with antigen status |
Unadjusted odds ratio |
Age-adjusted odds ratio |
|||
|---|---|---|---|---|---|---|---|
| Negative | Positive | Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value | ||
| EtpA | |||||||
| All | − | 110 (40.6) | 161 (59.4) | 2.10 (1.28, 3.45) | 0.003 | 1.98 (1.21, 3.29) | 0.007 |
| + | 27 (24.5) | 83 (75.5) | |||||
| ST or ST/LT | − | 55 (34.4) | 105 (65.6) | 1.57 (0.89, 2.77) | 0.116 | 1.51 (0.86, 2.68) | 0.156 |
| + | 24 (25) | 72 (75) | |||||
| LT only | − | 55 (49.5) | 56 (50.5) | 3.60 (0.95, 13.61) | 0.047 | 2.59 (0.78, 10.79) | 0.144 |
| + | 3 (21.4) | 11 (78.6) | |||||
| EatA | |||||||
| All | − | 152 (56.1) | 119 (43.9) | 3.11 (1.93, 5.01) | <0.001 | 2.91 (1.81, 4.75) | <0.001 |
| + | 32 (29.1) | 78 (70.9) | |||||
| ST or ST/LT | − | 67 (41.9) | 93 (58.1) | 2.05 (1.18, 3.56) | 0.011 | 1.91 (1.1, 3.38) | 0.024 |
| + | 25 (26) | 71 (74) | |||||
| LT only | − | 85 (76.6) | 26 (23.4) | 3.27 (1.05, 10.18) | 0.051 | 2.36 (0.74, 7.46) | 0.142 |
| + | 7 (50) | 7 (50) | |||||
P values for unadjusted odds ratios were obtained from simple chi-square or Fisher’s exact tests, and P values for age-adjusted odds ratios were obtained from logistic regressions that included age as a covariate.
The eatA gene (21) and the etpBAC locus (20), encoding the two-partner secretion system responsible for EtpA secretion, were originally identified on the p948 plasmid of ETEC strain H10407, which also harbors the gene for STh (37), and our previous studies suggested that both loci are more commonly associated with ST-producing strains (19). Importantly, large epidemiological studies have demonstrated an association between ST- or ST/LT-producing ETEC and more severe disease than with LT-only-producing ETEC (38, 39). Similarly, we again found an association between ST-producing ETEC and symptomatic diarrhea, where 59.0% of colonizing ETEC isolates encode STh or STp (estH or estP positive), compared to 87.3% of diarrhea-associated isolates (adjusted odds ratio, 4.66 [95% confidence interval {CI}, 2.62, 8.85]; P < 0.001]). We therefore asked whether the eatA or etpA associations with virulence were independent of ST. The presence of either gene was associated with a higher risk of diarrheal illness independent of ST, although only the presence of eatA was significantly associated with illness adjusted for age. Collectively, however, these data suggest that these more recently discovered noncanonical antigens, now often referred to as “accessory” virulence factors, appear to be more frequently identified in ETEC strains causing acute diarrheal illness.
DISCUSSION
ETEC strains were initially discovered in patients presenting with severe diarrheal illness that mimicked clinical cholera (40–42). Following seminal discoveries of the heat-labile (LT) and heat-stable (ST) toxins that define ETEC and the initial characterization of plasmid-encoded colonization factor (CF) antigens, a canonical approach to vaccine development focused on LT and CFs emerged. However, subsequent studies have revealed that the molecular pathogenesis of ETEC likely involves a number of other plasmid-encoded as well as chromosomally-encoded features that may potentially expand the repertoire of target “noncanonical” antigens for use in ETEC vaccine development.
The intent of the protein microarray studies presented here was to facilitate the identification of novel, surface-expressed, immunogenic proteins conserved within the ETEC pathovar for investigation as feasible antigenic targets in future iterations of ETEC vaccines. To encompass as many relevant antigens as possible (“open aperture”), we selected any predicted surface-expressed protein encoded by more than 40% of ETEC strains but absent in commensal E. coli strains. Notably, because all of the known colonization factors, the principal targets for ETEC vaccines to date, failed to meet this minimal conservation cutoff, they were used to complement the conserved features on the array. In general, this expanded open-aperture assessment of immune responses to natural ETEC infections appears to reaffirm previous observations in human volunteer studies (7) demonstrating that there are relatively few conserved immunogenic targets in the potential repertoire of ETEC surface molecules.
Among the more recently discovered surface immunoreactive (7, 18, 43) antigens conserved within the ETEC pathovar (18, 19) are two high-molecular-weight secreted proteins, EtpA and EatA. Our understanding of the contribution of these antigens to ETEC virulence continues to evolve. Recent studies have revealed that the secreted 110-kDa passenger domain of the EatA autotransporter protein functions as a mucin-degrading enzyme capable of dissolving the MUC2 matrix that covers the surface of enterocytes, the target for ETEC binding and toxin delivery (23). EtpA, secreted by a two-partner secretion mechanism that requires both the EtpB outer membrane pore and EtpC, a glycosyltransferase (20), functions as an adhesin by bridging the bacteria (36) and GalNAc-containing host cell glycans present on enterocytes (44). However, despite an emerging understanding of the molecular function of these molecules, very little is known about their actual contribution to disease in human hosts.
The present studies extend previous observations to a cohort of naturally infected children in Bangladesh (10) and suggest that these noncanonical antigens play critical roles in determining the outcome of ETEC infections. The finding that genes encoding these antigens are significantly associated with the development of symptomatic infection may have important implications for the interpretation of large-scale epidemiological studies that have employed a population-attributable fraction methodology in which ETEC strains detected in cases of diarrheal illness are compared to those of asymptomatically colonized controls (4). The present studies seem to suggest that additional characterization of ETEC beyond the pathovar-defining heat-labile or heat-stable toxins could be required to accurately assess the contribution of ETEC to the global burden of diarrheal disease.
As with a number of important enteric pathogens, true mechanistic correlates of immune responses directly responsible for protection (45) against ETEC remain undefined (32, 46). Although we observed high IgG serum antibody responses to both EtpA and EatA in children who were simply colonized with ETEC compared to those with diarrhea, suggesting that these antigens could afford some protection against symptomatic illness, these findings need to be interpreted cautiously. Both EtpA and EatA are relatively common antigens among strains circulating in Bangladesh; therefore, the identification of antibodies could simply reflect prior infection that mitigates infection through responses to other antigens.
Altogether, however, the findings reported here suggest that antigens that have not been part of traditional approaches to vaccine development may play important roles in virulence and in acquired immunity to ETEC. Further studies will clearly be needed to examine the efficacy of these more recently discovered antigens as protective immunogens and their role in complementing colonization factors and heat-labile toxin that have played a fundamental role in canonical approaches to vaccine development.
MATERIALS AND METHODS
Clinical samples used in this study.
Specimens used in these studies were obtained from archived studies on an ETEC birth cohort carried out in Mirpur in Dhaka city (10) as well as other studies (28) at the International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b). Frozen ETEC isolates were retrieved from storage (−80°C), and duplicate vials were shipped to Washington University for subsequent antigen detection.
Microbial genome analysis and bioinformatic antigen selection.
Genomes from 89 clinical ETEC isolates previously collected at icddr,b were used to identify conserved surface proteins. Sequence data for all 89 clinical isolates examined in this study are available in GenBank (47). Paired-end Illumina sequence data for each isolate were generated de novo, and contigs were binned using a previously described protocol (47). The ETEC genomes were compared using large-scale BLAST score ratio (LS-BSR) analysis as previously described (48–50). The predicted protein-encoding genes of each genome that had ≥90% nucleotide identity to each other were assigned to gene clusters using uclust (51). Representative sequences of each gene cluster were then compared to each genome using TBLASTN (52) with composition-based adjustment turned off, and the TBLASTN scores were used to generate a BSR value indicating the detection of each gene cluster in each of the genomes analyzed. The BSR value was determined by dividing the score of a gene compared to a genome by the score of the gene compared to its own sequence. The predicted protein function of each gene cluster was determined using an ergatis-based (53) in-house annotation pipeline (54). A total of 13,835 nonredundant putative genes (referred to here as “centroids”) were extracted from the 89 genomes.
All 13,835 centroids in this study were subjected to a reverse-vaccinology pipeline (illustrated in Fig. S1 in the supplemental material) (Institute for Genome Sciences, MD, USA) to identify molecules conserved in the ETEC pathovar that contained features that suggested that they were surface exposed. Briefly, subtractive analysis was first conducted by filtering centroids (BLASTx and BLASTn) against the genome contents of six E. coli commensal and laboratory strains, yielding 6,444 ETEC pathovar-specific centroids. These data were further refined by selecting centroids with a BSR (55) of ≥0.8 and present in at least 40% of the clinical isolates, yielding 316 conserved, virulence-linked genetic features for further analysis. BLASTx was next used to assign a putative function to these virulence-linked centroids. This analysis was coupled with results from pSORTv3.0 (56), SubLoc (57), and CELLO (58) to predict subcellular localization, altogether resulting in down-selection to 118 potential surface-expressed molecules. These features were complemented with all known and putative colonization factor subunits (n = 52), toxin subunits, and subdomains of novel antigens for inclusion on the microarrays (Data Set S1).
Microarray production.
Antigen-encoding regions selected for the microarrays were amplified by PCR using primers listed in Data Set S1 and constructed as previously described (7, 18, 59). Recombinant versions of select antigens, including EtpA, EatA, LT-A, LT-B, YghJ, STh, and EaeH, were also included on the arrays.
Microarray processing.
Microarrays were shipped to icddr,b, where they were rehydrated for 10 min with 100 μl array blocking buffer (catalog number 10485356; GVS). The E. coli lysate was reconstituted in a final volume of 20% in array blocking buffer. Archived antibody lymphocyte supernatant (ALS) samples prepared from the blood of ETEC patients as previously described (29) were diluted 1:2.5 in the resuspended lysate, followed by loading onto the microarrays, and incubated in the dark for 2 h at 25°C on a rotating platform. Microarrays were then washed three times with TBS-T (0.05% Tween in Tris-buffered saline [TBS] [pH 7.5]), followed by incubation for 5 min in TBS-T at 25°C. This process was repeated once with TBS, followed by a final wash in distilled water. Slides were dried by centrifugation (10 min at 500 × g) and then stored in desiccated boxes prior to shipping to the Felgner Laboratory, University of California, Irvine, for processing with anti-human IgA and anti-human IgG.
Noncanonical antigen ELISA.
Plates (384-well plates, product number 3540; Corning) were coated with a recombinant EatA passenger domain (rEatp) (10 μg/ml in carbonate buffer [15 mM Na2CO3, 35 mM NaHCO3, 0.2 g/liter NaN3 {pH 9.6}]) or recombinant EtpA (rEtpA) (1 μg/ml in carbonate buffer) and shipped to icddr,b, being maintained at 4°C prior to use. The enzyme-linked immunosorbent assay (ELISA) plates were manually washed three times with PBS-T (phosphate-buffered saline [PBS] with 0.05% Tween), including brief centrifugation for 30 s at 200 × g on a tabletop centrifuge between washes. Plates were rehydrated with 1% bovine serum albumin (BSA) in PBS-T overnight at 4°C. The following day, serum or plasma samples and plates were warmed to ambient temperature (∼25°C), and serum was diluted 1:200 in PBS-T with 1% BSA and briefly vortexed. Ten microliters of diluted serum was added to the plates, centrifuged as described above, sealed, and incubated at 37°C for 1 h. After incubation, plates were washed 3 times with PBS-T as described above. Ten microliters of horseradish peroxidase (HRP)-conjugated anti-human IgG (catalog number 309-035-006; Jackson ImmunoResearch Laboratories, West Grove, PA) was diluted 1:2,000 in 1% BSA in PBS-T, followed by incubation and washing as described above. ELISA plates were read using 10 μl of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (catalog number 50-76-00; Seracare, Milford, MA). A kinetic ELISA (60) was used to determine relative antibody concentrations (Eon, Gen5 Take3, v.2.00.18; BioTek). Each plate was analyzed separately, and Vmax titers were normalized to the mean negative-control value of the plate.
Strain characterization by PCR and immunoblotting.
Frozen glycerol stocks of ETEC strains maintained at −80°C were used to inoculate lysogeny broth (LB) for overnight growth at 37°C at 250 rpm. One microliter of the culture grown overnight was diluted in 100 μl of PBS, of which 1 μl was used as the DNA template in initial PCR screening with primers listed in Table S1. The thermocycler conditions for eatA and etpA were a denaturation step for 5 min at 95°C with 30 amplification cycles utilizing 95°C for 30 s, 52°C for 30 s, and 72°C for 2 min. The toxin multiplex assays (genes eltB, estH, and estP) were conducted as follows: 5 min at 95°C with 32 cycles of amplification using 94°C for 15 s, 55°C for 15 s, and, finally, 72°C for 30 s. Amplicons were visualized as described above using a 0.8% agarose gel with ethidium bromide. The H10407 strain (eatA, etpA, estH, estP, and eltB positive) was used as a positive control for the assays.
To adjudicate discordant results, PCR was performed using genomic DNA (gDNA) extraction with the Invitrogen PureLink Quick plasmid miniprep kit (catalog number K210010; Thermo Fisher, Waltham, MA). If toxin multiplex PCRs were negative, isolates were deemed to have lost their original plasmid during storage, transportation, or culture passage and subsequently excluded from the analysis.
Immunoblotting for EatA and EtpA was performed on tricarboxylic acid (TCA)-precipitated culture supernatants, as previously described (19), using affinity-purified polyclonal rabbit antibodies against the passenger domain of EatA (21) or EtpA (20) (dilutions of 1:1,000 and 1:5,000, respectively). Primary antibodies were detected using HRP-conjugated anti-rabbit IgG secondary antibody (1:5,000 dilution) (catalog number A16110; Invitrogen) for 1 h at room temperature. HRP was detected with the ECL Western blotting substrate (catalog number ABIN412579; Bio-Rad).
Statistical analysis.
Categorical outcomes were analyzed using chi-square tests, Fisher’s exact tests, or age-adjusted logistic regression analyses as appropriate. Serum data were analyzed using a linear repeated-measures model with a compound symmetry covariance structure. P values of <0.05 were considered significant. Analyses were conducted using SAS version 9.4 (SAS Institute Inc., Cary, NC, USA), SPSS v.24 (IBM, Armonk, NY, USA), or GraphPad Prism v9.0.0.
Ethics statement.
These studies were approved by the Research Review and Ethical Review Committee of the icddr,b and the Institutional Review Board of the Washington University School of Medicine in St. Louis.
Supplementary Material
ACKNOWLEDGMENTS
These studies were supported in part by funding from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under award numbers R01AI089894, R01AI126887 (J.M.F.), and K23AI30389 (F.M.K.) and from Department of Veterans Affairs award I01BX004825 (J.M.F.). The icddr,b is supported by the governments of Bangladesh, Canada, Sweden, and the United Kingdom.
J.M.F. is listed as the inventor on U.S. patent 8323668.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.GBD 2016 Diarrhoeal Disease Collaborators. 2018. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 18:1211–1228. 10.1016/S1473-3099(18)30362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson JD, IV, Bagamian KH, Muhib F, Amaya MP, Laytner LA, Wierzba T, Rheingans R. 2019. Burden of enterotoxigenic Escherichia coli and Shigella non-fatal diarrhoeal infections in 79 low-income and lower middle-income countries: a modelling analysis. Lancet Glob Health 7:e321–e330. 10.1016/S2214-109X(18)30483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosangadi D, Smith PG, Giersing BK. 2019. Considerations for using ETEC and Shigella disease burden estimates to guide vaccine development strategy. Vaccine 37:7372–7380. 10.1016/j.vaccine.2017.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanata CF, Black RE. 2018. Estimating the true burden of an enteric pathogen: enterotoxigenic Escherichia coli and Shigella spp. Lancet Infect Dis 18:1165–1166. 10.1016/S1473-3099(18)30546-2. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe K, Petri WA, Jr.. 2016. Environmental enteropathy: elusive but significant subclinical abnormalities in developing countries. EBioMedicine 10:25–32. 10.1016/j.ebiom.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosangadi D, Smith PG, Kaslow DC, Giersing BK, Who E, WHO ETEC & Shigella Vaccine Consultation Expert Group. 2019. WHO consultation on ETEC and Shigella burden of disease, Geneva, 6-7th April 2017: meeting report. Vaccine 37:7381–7390. 10.1016/j.vaccine.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty S, Randall A, Vickers TJ, Molina D, Harro CD, DeNearing B, Brubaker J, Sack DA, Bourgeois AL, Felgner PL, Liang X, Mani S, Wenzel H, Townsend RR, Gilmore PE, Darsley MJ, Rasko DA, Fleckenstein JM. 2018. Human experimental challenge with enterotoxigenic Escherichia coli elicits immune responses to canonical and novel antigens relevant to vaccine development. J Infect Dis 218:1436–1446. 10.1093/infdis/jiy312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine MM, Nalin DR, Hoover DL, Bergquist EJ, Hornick RB, Young CR. 1979. Immunity to enterotoxigenic Escherichia coli. Infect Immun 23:729–736. 10.1128/IAI.23.3.729-736.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinsland H, Valentiner-Branth P, Gjessing HK, Aaby P, Molbak K, Sommerfelt H. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362:286–291. 10.1016/S0140-6736(03)13971-2. [DOI] [PubMed] [Google Scholar]
- 10.Qadri F, Saha A, Ahmed T, Al Tarique A, Begum YA, Svennerholm AM. 2007. Disease burden due to enterotoxigenic Escherichia coli in the first 2 years of life in an urban community in Bangladesh. Infect Immun 75:3961–3968. 10.1128/IAI.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaastra W, Svennerholm AM. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 4:444–452. 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 12.Clements JD, Norton EB. 2018. The mucosal vaccine adjuvant LT(R192G/L211A) or dmLT. mSphere 3:e00215-18. 10.1128/mSphere.00215-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleckenstein JM, Kuhlmann FM. 2019. Enterotoxigenic Escherichia coli infections. Curr Infect Dis Rep 21:9. 10.1007/s11908-019-0665-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zegeye ED, Govasli ML, Sommerfelt H, Puntervoll P. 2019. Development of an enterotoxigenic Escherichia coli vaccine based on the heat-stable toxin. Hum Vaccin Immunother 15:1379–1388. 10.1080/21645515.2018.1496768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taxt AM, Diaz Y, Aasland R, Clements JD, Nataro JP, Sommerfelt H, Puntervoll P. 2016. Towards rational design of a toxoid vaccine against the heat-stable toxin of Escherichia coli. Infect Immun 84:1239–1249. 10.1128/IAI.01225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahl JW, Sistrunk JR, Baby NI, Begum Y, Luo Q, Sheikh A, Qadri F, Fleckenstein JM, Rasko DA. 2017. Insights into enterotoxigenic Escherichia coli diversity in Bangladesh utilizing genomic epidemiology. Sci Rep 7:3402. 10.1038/s41598-017-03631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mottram L, Chakraborty S, Cox E, Fleckenstein J. 2019. How genomics can be used to understand host susceptibility to enteric infection, aiding in the development of vaccines and immunotherapeutic interventions. Vaccine 37:4805–4810. 10.1016/j.vaccine.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakraborty S, Randall A, Vickers TJ, Molina D, Harro CD, De Nearing B, Brubaker J, Sack DA, Bourgeois AL, Felgner PL, Liang X, Mani S, Wenzel H, Townsend RR, Gilmore PE, Darsley MJ, Rasko DA, Fleckenstein JM. 2019. Interrogation of a live-attenuated enterotoxigenic Escherichia coli vaccine highlights features unique to wild-type infection. NPJ Vaccines 4:37. 10.1038/s41541-019-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhlmann FM, Martin J, Hazen TH, Vickers TJ, Pashos M, Okhuysen PC, Gomez-Duarte OG, Cebelinski E, Boxrud D, Del Canto F, Vidal R, Qadri F, Mitreva M, Rasko DA, Fleckenstein JM. 2019. Conservation and global distribution of non-canonical antigens in enterotoxigenic Escherichia coli. PLoS Negl Trop Dis 13:e0007825. 10.1371/journal.pntd.0007825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleckenstein JM, Roy K, Fischer JF, Burkitt M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect Immun 74:2245–2258. 10.1128/IAI.74.4.2245-2258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel SK, Dotson J, Allen KP, Fleckenstein JM. 2004. Identification and molecular characterization of EatA, an autotransporter protein of enterotoxigenic Escherichia coli. Infect Immun 72:1786–1794. 10.1128/iai.72.3.1786-1794.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Q, Qadri F, Kansal R, Rasko DA, Sheikh A, Fleckenstein JM. 2015. Conservation and immunogenicity of novel antigens in diverse isolates of enterotoxigenic Escherichia coli. PLoS Negl Trop Dis 9:e0003446. 10.1371/journal.pntd.0003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar P, Luo Q, Vickers TJ, Sheikh A, Lewis WG, Fleckenstein JM. 2014. EatA, an immunogenic protective antigen of enterotoxigenic Escherichia coli, degrades intestinal mucin. Infect Immun 82:500–508. 10.1128/IAI.01078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo Q, Vickers TJ, Fleckenstein JM. 2016. Immunogenicity and protective efficacy against enterotoxigenic Escherichia coli colonization following intradermal, sublingual, or oral vaccination with EtpA adhesin. Clin Vaccine Immunol 23:628–637. 10.1128/CVI.00248-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy K, Hamilton D, Ostmann MM, Fleckenstein JM. 2009. Vaccination with EtpA glycoprotein or flagellin protects against colonization with enterotoxigenic Escherichia coli in a murine model. Vaccine 27:4601–4608. 10.1016/j.vaccine.2009.05.076. [DOI] [PubMed] [Google Scholar]
- 26.Roy K, Hamilton D, Allen KP, Randolph MP, Fleckenstein JM. 2008. The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect Immun 76:2106–2112. 10.1128/IAI.01304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy SP, Rahman MM, Yu XD, Tuittila M, Knight SD, Zavialov AV. 2012. Crystal structure of enterotoxigenic Escherichia coli colonization factor CS6 reveals a novel type of functional assembly. Mol Microbiol 86:1100–1115. 10.1111/mmi.12044. [DOI] [PubMed] [Google Scholar]
- 28.Qadri F, Ahmed T, Ahmed F, Bhuiyan MS, Mostofa MG, Cassels FJ, Helander A, Svennerholm AM. 2007. Mucosal and systemic immune responses in patients with diarrhea due to CS6-expressing enterotoxigenic Escherichia coli. Infect Immun 75:2269–2274. 10.1128/IAI.01856-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang HS, Sack DA. 2001. Development of a novel in vitro assay (ALS assay) for evaluation of vaccine-induced antibody secretion from circulating mucosal lymphocytes. Clin Diagn Lab Immunol 8:482–488. 10.1128/CDLI.8.3.482-488.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forrest BD. 1988. Identification of an intestinal immune response using peripheral blood lymphocytes. Lancet i:81–83. 10.1016/S0140-6736(88)90284-X. [DOI] [PubMed] [Google Scholar]
- 31.Bhuiyan TR, Saha A, Lundgren A, Qadri F, Svennerholm AM. 2010. Immune responses to Helicobacter pylori infection in Bangladeshi children during their first two years of life and the association between maternal antibodies and onset of infection. J Infect Dis 202:1676–1684. 10.1086/657085. [DOI] [PubMed] [Google Scholar]
- 32.Holmgren J, Parashar UD, Plotkin S, Louis J, Ng SP, Desauziers E, Picot V, Saadatian-Elahi M. 2017. Correlates of protection for enteric vaccines. Vaccine 35:3355–3363. 10.1016/j.vaccine.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tobias J, Andersson K, Bialik A, Cohen D. 2008. Preexisting antibodies to homologous colonization factors and heat-labile toxin in serum, and the risk to develop enterotoxigenic Escherichia coli-associated diarrhea. Diagn Microbiol Infect Dis 60:229–231. 10.1016/j.diagmicrobio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Rao MR, Wierzba TF, Savarino SJ, Abu-Elyazeed R, El-Ghoreb N, Hall ER, Naficy A, Abdel-Messih I, Frenck RW, Jr, Svennerholm AM, Clemens JD. 2005. Serologic correlates of protection against enterotoxigenic Escherichia coli diarrhea. J Infect Dis 191:562–570. 10.1086/427662. [DOI] [PubMed] [Google Scholar]
- 35.Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J Biol Chem 286:29771–29779. 10.1074/jbc.M111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy K, Hilliard GM, Hamilton DJ, Luo J, Ostmann MM, Fleckenstein JM. 2009. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457:594–598. 10.1038/nature07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SG, Henderson IR. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol 192:5822–5831. 10.1128/JB.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O’Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 39.Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, McCormick BJ, McGrath M, Olortegui MP, Samie A, Shakoor S, Mondal D, Lima IF, Hariraju D, Rayamajhi BB, Qureshi S, Kabir F, Yori PP, Mufamadi B, Amour C, Carreon JD, Richard SA, Lang D, Bessong P, Mduma E, Ahmed T, Lima AA, Mason CJ, Zaidi AK, Bhutta ZA, Kosek M, Guerrant RL, Gottlieb M, Miller M, Kang G, Houpt ER, MAL-ED Network Investigators. 2015. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 3:e564–e575. 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De SN, Bhattacharya K, Sarkar JK. 1956. A study of the pathogenicity of strains of Bacterium coli from acute and chronic enteritis. J Pathol Bacteriol 71:201–209. 10.1002/path.1700710126. [DOI] [PubMed] [Google Scholar]
- 41.Nalin DR, Bhattacharjee AK, Richardson SH. 1974. Cholera-like toxic effect of culture filtrates of Escherichia coli. J Infect Dis 130:595–607. 10.1093/infdis/130.6.595. [DOI] [PubMed] [Google Scholar]
- 42.Sack RB, Gorbach SL, Banwell JG, Jacobs B, Chatterjee BD, Mitra RC. 1971. Enterotoxigenic Escherichia coli isolated from patients with severe cholera-like disease. J Infect Dis 123:378–385. 10.1093/infdis/123.4.378. [DOI] [PubMed] [Google Scholar]
- 43.Roy K, Bartels S, Qadri F, Fleckenstein JM. 2010. Enterotoxigenic Escherichia coli elicits immune responses to multiple surface proteins. Infect Immun 78:3027–3035. 10.1128/IAI.00264-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar P, Kuhlmann FM, Chakraborty S, Bourgeois AL, Foulke-Abel J, Tumala B, Vickers TJ, Sack DA, DeNearing B, Harro CD, Wright WS, Gildersleeve JC, Ciorba MA, Santhanam S, Porter CK, Gutierrez RL, Prouty MG, Riddle MS, Polino A, Sheikh A, Donowitz M, Fleckenstein JM. 2018. Enterotoxigenic Escherichia coli-blood group A interactions intensify diarrheal severity. J Clin Invest 128:3298–3311. 10.1172/JCI97659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plotkin SA, Gilbert PB. 2012. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis 54:1615–1617. 10.1093/cid/cis238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mani S, Toapanta FR, McArthur MA, Qadri F, Svennerholm AM, Devriendt B, Phalipon A, Cohen D, Sztein MB. 2019. Role of antigen specific T and B cells in systemic and mucosal immune responses in ETEC and Shigella infections, and their potential to serve as correlates of protection in vaccine development. Vaccine 37:4787–4793. 10.1016/j.vaccine.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahl JW, Sistrunk JR, Fraser CM, Hine E, Baby N, Begum Y, Luo Q, Sheikh A, Qadri F, Fleckenstein JM, Rasko DA. 2015. Examination of the enterotoxigenic Escherichia coli population structure during human infection. mBio 6:e00501-15. 10.1128/mBio.00501-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahl JW, Caporaso JG, Rasko DA, Keim P. 2014. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ 2:e332. 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hazen TH, Sahl JW, Fraser CM, Donnenberg MS, Scheutz F, Rasko DA. 2013. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. Proc Natl Acad Sci U S A 110:12810–12815. 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. 10.1371/journal.pone.0054287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. 2006. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol 4:41. 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orvis J, Crabtree J, Galens K, Gussman A, Inman JM, Lee E, Nampally S, Riley D, Sundaram JP, Felix V, Whitty B, Mahurkar A, Wortman J, White O, Angiuoli SV. 2010. Ergatis: a Web interface and scalable software system for bioinformatics workflows. Bioinformatics 26:1488–1492. 10.1093/bioinformatics/btq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galens K, Orvis J, Daugherty S, Creasy HH, Angiuoli S, White O, Wortman J, Mahurkar A, Giglio MG. 2011. The IGS standard operating procedure for automated prokaryotic annotation. Stand Genomic Sci 4:244–251. 10.4056/sigs.1223234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasko DA, Myers GS, Ravel J. 2005. Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6:2. 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen H, Huang N, Sun Z. 2006. SubLoc: a server/client suite for protein subcellular location based on SOAP. Bioinformatics 22:376–377. 10.1093/bioinformatics/bti822. [DOI] [PubMed] [Google Scholar]
- 58.Yu CS, Lin CJ, Hwang JK. 2004. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci 13:1402–1406. 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A 102:547–552. 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsang VC, Wilson BC, Maddison SE. 1980. Kinetic studies of a quantitative single-tube enzyme-linked immunosorbent assay. Clin Chem 26:1255–1260. 10.1093/clinchem/26.9.1255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.