TGFβ is essential for the generation of anti-tumor Th9 cells, while it causes resistance against anti-tumor immunity. Pei et al. identify BFAR as an E3 ligase of TGFβR1, through which it coordinates TGFβ signaling to modulate Th9-mediated cancer immunotherapy.
Abstract
TGFβ is essential for the generation of anti-tumor Th9 cells; on the other hand, it causes resistance against anti-tumor immunity. Despite recent progress, the underlying mechanism reconciling the double-edged effect of TGFβ signaling in Th9-mediated cancer immunotherapy remains elusive. Here, we find that TGFβ-induced down-regulation of bifunctional apoptosis regulator (BFAR) represents the key mechanism preventing the sustained activation of TGFβ signaling and thus impairing Th9 inducibility. Mechanistically, BFAR mediates K63-linked ubiquitination of TGFβR1 at K268, which is critical to activate TGFβ signaling. Thus, BFAR deficiency or K268R knock-in mutation suppresses TGFβR1 ubiquitination and Th9 differentiation, thereby inhibiting Th9-mediated cancer immunotherapy. More interestingly, BFAR-overexpressed Th9 cells exhibit promising therapeutic efficacy to curtail tumor growth and metastasis and promote the sensitivity of anti–PD-1–mediated checkpoint immunotherapy. Thus, our findings establish BFAR as a key TGFβ-regulated gene to fine-tune TGFβ signaling that causes Th9 induction insensitivity, and they highlight the translational potential of BFAR in promoting Th9-mediated cancer immunotherapy.
Introduction
TGFβ is known to be induced by a variety of tumors, and increased expression of this cytokine is associated with the induction of immunosuppressive profiles and contributes to poor prognosis in many cancers (Bruna et al., 2007; Calon et al., 2012; Calon et al., 2015; Delvenne et al., 2004; Tauriello et al., 2018). In addition, TGFβ has also been shown to act as a chief mechanism for the resistance of anti–programmed cell death ligand 1 (anti–PD-L1) or anti–CTL-associated antigen-4 (anti–CTLA-4)–mediated immunotherapy for the metastasis of urothelial cancer, colon cancer, or prostate cancer via diverse mechanisms (Jiao et al., 2019; Mariathasan et al., 2018; Tauriello et al., 2018). Therefore, blocking TGFβ reprograms the immune system to an anti-tumor active state, thereby exhibiting promising effects to suppress the growth and metastasis of multiple types of tumors with or without checkpoint immunotherapy (Arteaga, 2006; Biswas et al., 2017; Jiao et al., 2019; Mariathasan et al., 2018; Tauriello et al., 2018). However, the mechanism controlling TGFβ-mediated in vivo immunosuppression under tumor conditions is still debated.
TGFβ is also known to be essential for the induction of T helper type 9 (Th9) cells (Dardalhon et al., 2008; Elyaman et al., 2012; Nakatsukasa et al., 2015; Veldhoen et al., 2008; Wang et al., 2016), a subset of CD4+ effector T cells exerting robust anti-tumor activities (Chauhan et al., 2019; Lu et al., 2012; Lu et al., 2018; Purwar et al., 2012). Compared with Th1 and Th17 cells, Th9 cells are less exhausted, exhibit cytolytic activity as strong as that of Th1 cells, and persist as long as “stem cell–like” Th17 cells in vivo (Lu et al., 2018). Therefore, Th9 cells greatly suppress the growth and metastasis of various solid tumors more efficiently than Th1 or Th17 cells, thus enabling Th9 cells as powerful effector T cells for cancer adoptive cell therapy (ACT; Chauhan et al., 2019; Lu et al., 2012; Lu et al., 2018; Purwar et al., 2012; Rivera Vargas et al., 2017a). Given that TGFβ is a master cytokine for the induction of Th9 cell differentiation, speculation that TGFβ may promote the generation of IL-9–producing Th9 cells, which should antagonize tumor growth and metastasis, is reasonable. However, this speculation is contradicted by the observed phenomenon that TGFβ dictates an immunosuppressive profile to facilitate tumor growth and metastasis. Hitherto, whether and how TGFβ modulates in vivo Th9 cell differentiation and thus affects anti-tumor immunity remain unknown. We therefore conducted this study to investigate Th9 inducibility under tumor conditions to explore a novel approach for enhancing the therapeutic efficacy of Th9-based cancer immunotherapy.
Results
Elevated TGFβ impairs Th9 induction in gastrointestinal cancer (GIC) patients
Th9-mediated ACT has been proved to be a promising IL-9–dependent therapeutic strategy against solid tumors (Lu et al., 2012; Lu et al., 2018; Purwar et al., 2012; Rivera Vargas et al., 2017b). We observed that direct blocking of endogenous IL-9 markedly promoted tumor growth, whereas CD4+ T cell depletion abolished the pro-tumor effect of anti–IL-9 antibody in immune-competent C57BL/6 mice that were inoculated with B16 melanoma cells (Fig. 1, A–D), suggesting that endogenous IL-9 derived from self Th9 cells, instead of that from adoptively transferred cells, is also essential for Th9-mediated anti-tumor immunity. To investigate the effect of TGFβ on IL-9 production in vivo, we generated a mouse tumor model and analyzed the serum cytokine levels at different time points. As expected, the serum TGFβ gradually increased along with B16 melanoma growth. However, with elevated TGFβ levels, the serum IL-9 did not increase but decreased, and it was negatively correlated with TGFβ in the tumor-bearing mice (Fig. 1 E).
Figure 1.
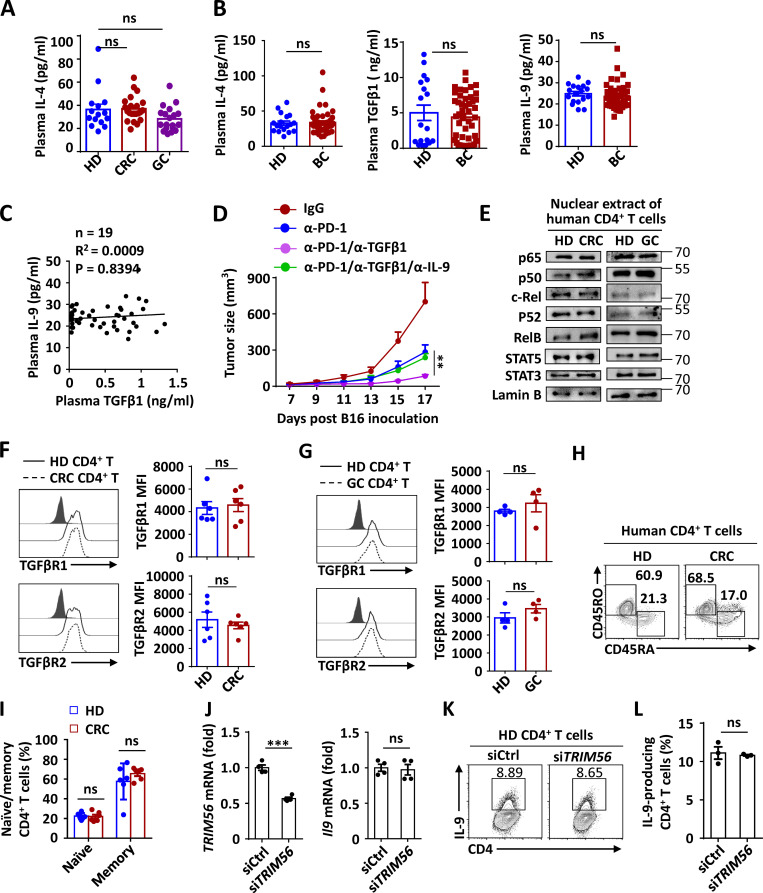
Elevated TGFβ impairs Th9 induction in GIC patients. (A–D) Tumor growth (A) and weight (B) of C57BL/6 mice that were s.c. injected with B16 melanoma cells and then treated with anti-CD4 (α-CD4; 100 µg/mouse), anti–IL-9 (α-IL-9; 100 µg/mouse; n = 6), or control antibody (IgG; n = 6) every 3 d starting from day 0. (E) ELISA measurements of serous IL-9 and TGFβ1 levels in the mice that were uninoculated (UI) or injected s.c. with B16 melanoma cells at the indicated time points, the changing trend of serous TGFβ1 in UI and tumor-bearing mice, and correlation analysis of serous TGFβ1 and IL-9 cytokine levels in the tumor-bearing mice. The results were plotted and analyzed with the linear regression t test. (F and G) ELISA measurements of plasma TGFβ1 and IL-9 levels in HDs (n = 26), CRC patients (n = 21), or GC patients (n = 19), and correlation analysis of plasma TGFβ1 and IL-9 cytokine levels of indicated cancer patients and HDs. The results were plotted and analyzed with the linear regression t test. (H and I) Flow cytometric analysis of Th9 inducibility (H) and the corresponding statistical analysis (I) of the CD4+ T cells from HDs (n = 20) and indicated patients with cancer (CRC, n = 16; GC, n = 33) and differentiated under Th9 conditions for 3 d. (J and K) Flow cytometric analysis of Th9 inducibility (J) and the corresponding statistical analysis (K) of HD-derived CD4+ T cells that were left untreated (NT) or that were pretreated with TGFβ1 (5 ng ml−1) for 12 h and then stimulated under Th9 condition for 3 d. (L) Tumor growth of C57BL/6 mice that were s.c. injected with B16-F10 cells and then i.v. administered control IgG or anti–PD-1 (α-PD-1) or α-PD-1 plus α-TGFβ. The antibodies were i.v. injected on days 7, 10, and 13 after tumor inoculation. n = 6 mice/group. (M–O) Heat map of average concentrations for a panel of Th cell–related cytokines in the serum (M) and flow cytometric analysis of tumor-infiltrating IL-9–producing Th9 cells (N and O) in tumor-bearing mice (n = 6) treated with α-PD-1 or with α-PD-1 plus α-TGFβ on day 17, as described in L. Each panel is representative of three independent experiments, and each circle represents one mouse in B, D, E, and O and one human individual in F, G, I, and K. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Considering that TGFβ was also reported to be induced in many GICs (Achyut and Yang, 2011; Calon et al., 2012; Calon et al., 2015; Ishimoto et al., 2017), we collected blood samples from colorectal cancer (CRC) and gastric cancer (GC) patients and examined the relationship between TGFβ and IL-9 under tumor conditions. Compared with healthy donors (HDs), TGFβ in the plasma of patients with CRC or GC was significantly increased (Fig. 1 F), whereas no significant change of IL-4 (Fig. S1 A), another cytokine essential for IL-9 induction in CD4+ T cells, was observed. In agreement with the mouse tumor data, plasma IL-9 of these cancer patients was also decreased and inversely correlated with TGFβ levels (Fig. 1, F and G). Nonetheless, these results were not phenocopied in breast cancer patients, in which comparable levels of plasma TGFβ were detected between the HDs and cancer patients (Fig. S1, B and C). These intriguing results prompted us to speculate that IL-9 inducibility was specifically impaired in the CD4+ T cells of TGFβ-elevated GIC patients. Compared with HD-derived cells, GIC patient–derived CD4+ T cells were, indeed, insensitive to induction of IL-9–producing cells under ex vivo Th9 differentiation conditions (Fig. 1, H and I). Furthermore, we confirmed that the Th9 induction was greatly impaired upon TGFβ pretreatment in HD-derived CD4+ T cells under in vitro Th9 differentiation conditions (Fig. 1, J and K).
Figure S1.
TGFβ1 negatively correlates with IL-9 production without affecting the surface expression of TGFβR1 and TGFβR2. (A and B) ELISA measurements of plasma IL-4, TGFβ, and IL-9 levels in HDs and CRC, GC, or breast cancer (BC) patients. (C) Correlation of plasma TGFβ1 and IL-9 cytokine levels in BC patients and HDs. The results were plotted and analyzed with the linear regression t test. (D) Tumor growth of C57BL/6 mice that were s.c. injected with B16-F10 cells and then i.v. administered control IgG; with α-PD-1; with α-PD-1 plus α-TGFβ1; or with α-PD-1 and α-TGFβ1 plus α-IL-9. The α-PD-1 or α-TGFβ1 was i.v. injected on days 7, 10, and 13 after tumor inoculation, and α-IL-9 was injected on days 10, 13, and 16 after tumor inoculation. n = 5 mice/group. (E) Immunoblot analysis of NF-κB and STAT proteins or actin and lamin B (loading controls) in cytoplasmic extracts (CEs) and nuclear extracts (NEs) of human CD4+ T cells derived from the indicated CRC or GC patients and HDs cultured under Th9 condition for 3 d. (F and G) Flow cytometric analysis of surface TGFβR1 and TGFβR2 expression in CD4+ T cells derived from the indicated CRC or GC patients and HDs cultured under Th9 condition for 3 d. Data are presented as representative plots (left panels) and summary graphs (right panels). MFI, mean fluorescence intensity. (H and I) Flow cytometric analysis of the frequencies of the naive or memory CD4+ T cells from the peripheral blood of HDs and CRC patients. (J) qPCR analysis of TRIM56 and IL9 mRNA expression in human HD-derived CD4+ T cells transduced with control siRNA or TRIM56-siRNA. (K and L) Flow cytometric analysis of the frequencies of IL-9–producing Th9 cells in human HD-derived CD4+ T cells were transduced with control siRNA or TRIM56-siRNA under Th9 condition for 3 d. Each panel is representative of two independent experiments, and each circle represents one human individual in A–C. Student’s t test was used. Bars, mean; error bars, SEM; ***, P < 0.001; ns, not significant.
Thereafter, to validate the negative role of TGFβ in modulating in vivo Th9 induction, we examined IL-9 inducibility upon TGFβ blocking in tumor-bearing mice subjected to anti–PD-1 immunotherapy. Interestingly, blocking TGFβ at the early stage could enhance the therapeutic efficacy of anti–PD-1 antibody in eliminating tumor growth (Fig. 1 L), with dramatically increased serum levels of the Th9 signature cytokine IL-9 among all the Th-related cytokines examined at the late stage of tumor growth (Fig. 1 M). Expectedly, blocking TGFβ at the early stage also increased the infiltration of Th9 cells in a tumor microenvironment (TME; Fig. 1, N and O), and, accordingly, IL-9 neutralization at the late stage abolished the enhanced therapeutic efficacy caused by the TGFβ blocking at the early stage (Fig. S1 D). Collectively, these results suggested that the long-term exposure of TGFβ impairs the Th9 inducibility of CD4+ T cells under tumor conditions, thereby compromising anti-tumor immunity.
TGFβ inhibits the expression of TGFβR1 E3 ligase bifunctional apoptosis regulator (BFAR)
To dissect how TGFβ impairs in vivo Th9 cell differentiation, we examined the expression levels of transcription factors responsible for IL-9 induction (Goswami et al., 2012; Kaplan, 2017; Tamiya et al., 2013). The TGFβ-induced nuclear translocation of activated Smad2/3 was greatly inhibited in CD4+ T cells of GIC patients under Th9 conditions, as compared with that in HD-derived cells, while nuclear STAT6, NF-κB, and STAT proteins were comparable (Fig. 2 A and Fig. S1 E). Additionally, the surface expression of TGFβ receptors (TGFβRs) 1 and 2, primary receptors transducing the TGFβ signal to activate Smad2/3 (Budi et al., 2017), was comparable on CD4+ T cells of HDs and GIC patients (Fig. S1, F and G). However, TGFβR1 ubiquitination, an initial molecular event to activate downstream Smad2/3 (Budi et al., 2017; Kim and Baek, 2019), was dramatically inhibited in CD4+ T cells of GIC patients as compared with that in HD-derived cells upon TCR stimulation (Fig. 2 B). Moreover, pretreatment with TGFβ also sharply suppressed the TCR-induced ubiquitination of TGFβR1 in mouse primary CD4+ T cells (Fig. 2 C). A plausible explanation is that long-term exposure to elevated TGFβ may indirectly suppress TCR-induced TGFβR1 ubiquitination, thus impairing TGFβ-induced Smad2/3 activation under Th9 differentiation conditions.
Figure 2.
Elevated TGFβ in GIC patients inhibits the expression of TGFβR1 E3 ligase BFAR. (A) Immunoblot analysis of SMAD2/3 and STAT6 or actin and lamin B (loading controls) in cytoplasmic extracts (CE) and nuclear extracts (NE) of human CD4+ T cells derived from the indicated CRC or GC patients and HDs cultured under Th9 conditions for 3 d. (B and C) Endogenous ubiquitination of TGFβR1 in human CD4+ T cells isolated from peripheral blood of HDs and CRC or GC patients and then stimulated with anti-CD3 plus anti-CD28 (α-CD3/28; 3 µg ml−1) overnight (B) and in mouse primary CD4+ T cells that were left untreated (NT) or that were pretreated with TGFβ1 for 12 h and then stimulated with (+) or without (–) α-CD3/28 (3 µg ml−1) overnight (C), assessed by immunoblot (IB) analysis with anti-Ub or anti-TGFβR1 after immunoprecipitation (IP) with anti-TGFβR1 (top) and immunoblot analysis with input proteins and loading controls (below). (D) qPCR analysis showing the TGFβ1-induced alteration of the expression of genes encoding E3 ligases or deubiquitinases (DUBs) that have been reported to modulate TGFβR1 ubiquitination or are bound to TGFβR1 as detected by MS. (E) qPCR analysis of BFAR mRNA expression in CD4+ T cells of HDs and CRC or GC patients. (F and G) Flow cytometric analysis of Th9 inducibility (F) and the corresponding statistical analysis (G) of the naive or memory CD4+ T cells from HDs and CRC patients and differentiated under Th9 differentiation conditions for 3 d. (H) qPCR analysis of BFAR mRNA expression in naive or memory CD4+ T cells of HDs and CRC patients. (I) Correlation of the mRNA expression of BFAR in human CD4+ T cells with plasma TGFβ1 or IL-9 cytokine levels of CRC or GC patients and HDs. The results were plotted and analyzed with the linear regression t test. (J and K) Flow cytometric analysis (J) and the corresponding statistical analysis (K) of Th9 inducibility of HD-derived CD4+ T cells that were transfected with BFAR-specific or control siRNA, and CRC-derived CD4+ T cells that were reconstituted with empty vector (EV) or BFAR. (L and M) qPCR analysis of BFAR and IL9 mRNA expression in human HD-derived CD4+ T cells transduced with control siRNA or BFAR-siRNA or in CRC patient–derived CD4+ T cells with or without BFAR overexpression (OE) under Th9 condition for 3 d. Each panel is representative of three independent experiments, and each circle represents one human individual in E, G–I, and K. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; ***, P < 0.0001.
We thus performed an unbiased mass spectrometry (MS) screening and quantitative PCR (qPCR) analysis to identify potential ubiquitination-modulating enzymes that can bind to TGFβR1 (Table S1) and be modulated by TGFβ. Among the MS-identified and previously reported ubiquitination-modulating enzymes of TGFβR1 (Al-Salihi et al., 2012; Eichhorn et al., 2012; Kavsak et al., 2000; Lu et al., 2012; Suzuki et al., 2002; Wicks et al., 2005; Xie et al., 2013; Zhang et al., 2012), BFAR was the most significantly inhibited gene by TGFβ in mouse primary CD4+ T cells (Fig. 2 D). As expected, GIC patient–derived CD4+ T cells, especially memory T cells, showed decreased expression of BFAR and, accordingly, Th9 inducibility as compared with HD-derived cells (Fig. 2, E–H; and Fig. S1, H and I), which may be due to the fact that human memory CD4+ T cells are more sensitive to TGFβ stimulation than naive cells. In addition, BFAR expression in CD4+ T cells was inversely correlated with the levels of plasma TGFβ but positively correlated with plasma IL-9 in GIC patients (Fig. 2 I). Consistent with the correlation analysis, BFAR knockdown suppressed Th9 cell differentiation in HD-derived CD4+ T cells, whereas the overexpression of BFAR dramatically promoted the IL-9 inducibility of the CD4+ T cells from CRC patients (Fig. 2, J–M). By contrast, the knockdown of another MS-identified E3 ligase, namely TRIM56, associated with TGFβR1 and down-regulated by TGFβ, did not affect the Th9 differentiation of HD-derived CD4+ T cells (Fig. S1, J–L).
In contrast to the GIC cancer patients, TGFβ blocking significantly promoted BFAR expression in both splenic and intratumoral CD4+ T cells of tumor-bearing mice and thus enhanced the association of Smad2/3 with TGFβR1 and its activation in splenic CD4+ T cells under ex vivo Th9 conditions (Fig. 3, A–C). Next, we pretreated CD4+ T cells with TGFβ at a low/high dose corresponding to its average serum levels in tumor-bearing mice with/without TGFβ blocking (Fig. 3 A). As expected, pretreatment with high-dose TGFβ suppressed the association of Smad2/3 with TGFβR1 and its activation in mouse primary CD4+ T cells under Th9 conditions, and it accordingly compromised IL-9 inducibility as compared with that with low-dose TGFβ pretreatment (Fig. 3, D–F). Consistently, immunoblotting also confirmed that the expression BFAR protein was gradually down-regulated in mouse primary CD4+ T cells treated with the increased dose of TGFβ (Fig. 3 G). Moreover, TGFβ pretreatment compromised the IL-9 inducibility of mouse primary CD4+ T cells, and BFAR overexpression counteracted the inhibitory effect of the cytokine (Fig. 3 H), suggesting that the TGFβ-mediated insensitivity of Th9 induction is indeed dependent on its inhibitory effect on BFAR expression.
Figure 3.
High-level TGFβ-activated Smad2/3 suppresses BFAR transcription. (A) ELISA measurements of serum TGFβ1 levels in tumor-bearing mice treated with α-PD-1 or with α-PD-1 plus α-TGFβ. (B) qPCR analysis of the mRNA expression of BFAR in splenic and intratumoral CD4+ T cells of tumor-bearing mice treated with α-PD-1 or with α-PD-1 plus α-TGFβ. (C and D) Immunoblot analysis of the Smad2/3-TGFβR1 interaction and Smad2/3 phosphorylation in splenic CD4+ T cells of tumor-bearing mice treated with α-PD-1 or with α-PD-1 plus α-TGFβ (C) and in mouse primary CD4+ T cells that were pretreated with 0.5 ng ml−1 (low concentration) of TGFβ1 (Lo) or 10 ng ml−1 (high concentration) of TGFβ1 (Hi) for 12 h (D) and then stimulated under Th9 conditions for 3 d, assessed by immunoprecipitation (IP) with anti-TGFβR1 and immunoblotting with anti-TGFβR1 and anti-Smad2/3 (top) and immunoblot analysis of p-Smad2/3, TGFβR1, and Smad2/3 in lysates without IP (below). (E and F) Flow cytometric analysis of Th9 inducibility (E) and the corresponding statistical analysis (F) of naive CD4+ T cells that were pretreated with 0.5 ng ml−1 TGFβ1 (Low) or 10 ng ml−1 TGFβ1 (High) for 12 h and then stimulated under Th9 conditions for 3 d. (G) Immunoblot analysis of HA-BFAR in whole-cell lysates of CD4+ T cells reconstituted with HA-BFAR and then cultured under Th9 conditions with the indicated doses of TGFβ1. (H) qPCR analysis of BFAR and IL9 mRNA in human HD-derived CD4+ T cells left untreated (−) or pretreated with TGFβ1 (5 ng ml−1) for 12 h before reconstitution with an EV (−) or BFAR under Th9 conditions for 3 d. (I and J) Structure schema of the constructed luciferase reporter by using different truncated genomic sequences of BFAR genes (I), which were then used to test the luciferase (luc.) activity (J). (K) Luciferase activity of BFAR transcriptional activity in HEK293T cells that were transfected with a pBFAR-Luc2 (−70/+657)-driven luciferase reporter, together with EV or an expression vector encoding Ca-SMAD3. (L) ChIP-qPCR analysis of the binding activities of SMAD2/3 in the indicated regions of BFAR genes in human HD-derived CD4+ T cells left untreated (NT) or stimulated with TGFβ1 for 12 h. (M) Schematic representation showing a conserved SMAD2/3-binding motif located in the genomic region of human BFAR genes between +270 and +282. (N) Luciferase (Luc.) assay of BFAR transcriptional activity in HEK293T cells that were transfected with a WT pBFAR-Luc2-driven or mutant pBFAR-Luc2-driven luciferase reporter (as indicated in O), together with the EV (−) or an expression vector encoding Ca-SMAD3. Each panel is representative of two or three independent experiments. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; **, P < 0.01; and ***, P < 0.001; ns, not significant.
To figure out how TGFβ suppresses BFAR expression, we generated luciferase reporter plasmids by introducing distinct mouse BFAR genomic fragments into a pGL4-basic plasmid (Fig. 3 I), and we found that the genomic region between −70 and +657 contributed to the basal BFAR expression (Fig. 3 J). We also determined that this region between −70 and +657 effectively responded to constitutively activated (Ca)–Smad3 to suppress BFAR transcription (Fig. 3 K), and the chromatin immunoprecipitation (ChIP)–qPCR experiment further narrowed down the genomic region between +257 and +376 to be the DNA-binding site of TGFβ-activated Smad2/3 in human CD4+ T cells (Fig. 3 L). As expected, a conserved Smad2/3-binding motif was found in the genomic region between +270 and +282 (Fig. 3 M). Moreover, Ca-Smad3 failed to suppress BFAR transcription when five bases of the Smad2/3-binding motif were mutated (Fig. 3 N). In sum, these data demonstrated that long-term exposure of TGFβ suppresses BFAR transcription directly through activated SMAD2/3, leading to inhibited TGFβ downstream signaling, which eventually compromises Th9 differentiation under tumor conditions.
BFAR mediates K63-linked ubiquitination of TGFβR1
To investigate whether BFAR is an E3 ligase of TGFβR1, we first examined the association of these two proteins. The coimmunoprecipitation analysis revealed that BFAR was indeed bound to TGFβR1, both in HEK293T cells transfected with hemagglutinin (HA)-BFAR and Flag-TGFβR1 expression vectors and in primary CD4+ T cells transfected with HA-tagged BFAR, and TCR stimulation further enhanced the association of these two proteins (Fig. 4, A–D). In addition, the overexpression of full-length BFAR, but not a ring domain–deleted mutant (BFARΔR), specifically mediated the K63-linked ubiquitination, but not other types (K6, K9, K11, K27, K33, or K48 linked), of TGFβR1 (Fig. 4, E–G). An in vitro ubiquitination assay confirmed that the BFAR protein could directly add the polyubiquitin chains in the in vitro cell–free translated TGFβR1 protein (Fig. 4 H), indicating that BFAR is a direct E3 ligase of TGFβR1. To confirm the function of BFAR in mediating endogenous TGFβR1 ubiquitination, we generated conditional knockout (KO) mice to specifically delete BFAR in T cells (Fig. S2, A and B). The results showed that BFAR deficiency abolished TCR-induced endogenous TGFβR1 ubiquitination in mouse primary CD4+ T cells (Fig. 4 I). Under Th9 differentiation condition, BFAR overexpression dramatically promoted, whereas its deletion sharply inhibited, endogenous K63-linked ubiquitination of TGFβR1 (Fig. 4 J). Collectively, BFAR is an E3 ligase that directly mediates K63-linked ubiquitination of TGFβR1.
Figure 4.
BFAR mediates K63-linked ubiquitination of TGFβR1 and downstream signaling activation. (A–D) Immunoblot analysis of the interaction of BFAR with TGFβR1 in HEK293T cells transfected with the indicated expression vectors or in HA-BFAR reconstituted (rec.) mouse CD4+ T cells or Jurkat T cells left unstimulated (−) or stimulated with (+) anti-CD3 plus anti-CD28 (α-CD3/28; 3 µg ml−1) overnight, assessed by immunoprecipitation (IP) with IgG, anti-Flag, anti-HA, or anti-TGFβR1 and by immunoblotting with input proteins in lysates and loading controls without immunoprecipitation. (E–H) Ubiquitination of TGFβR1 in HEK293T cells transfected with the expression vector encoding HA-BFAR, Flag-TGFβR1, and different types of Ub as indicated (E–G) or in vitro ubiquitination assay of TGFβR1 ubiquitination after a mixed reaction of Ub-charged E2 (UbcH5a), in vitro translated HA-TGFβR1, and with or without HA-BFAR proteins (H), assessed by immunoblot analysis with anti-HA, anti-Flag, anti-Ub, or anti-TGFβR1 or after IP with anti-Flag or by immunoblot analysis with input proteins without IP. (I and J) Endogenous ubiquitination of TGFβR1 in WT and BFAR-deficient or BFAR-overexpressed (OE) CD4+ T cells that were left unstimulated (−) or stimulated (+) with α-CD3/28 (3 µg ml−1) or under Th9 differentiation condition overnight, assessed by immunoblot analysis with anti-Ub, anti-K63 Ub (K63 Ub), or anti-TGFβR1 after IP with anti-TGFβR1 (top) and immunoblot analysis with input proteins and loading controls (below).BFAR-TKO, BFAR knock out in T cells. (K) Immunoblot analysis of the interaction of Smad2/3 and TGFβR1 in WT and BFAR-deficient CD4+ T cells that were stimulated with 3 µg ml−1 α-CD3/28 overnight and then left unstimulated (−) or stimulated (+) with 5 ng ml−1 TGFβ1 for 30 min, assessed by IP with anti-Smad2/3, immunoblot analysis with anti-TGFβR1 and anti-Smad2/3 (top), and immunoblot analysis of TGFβR1 and Smad2/3 in lysates without IP (below). (L) Immunoblot analysis of phosphorylated and total Smad2/3 in whole-cell lysates (WCLs) or Smad2/3 in cytoplasmic extract (CE) and nuclear extract (NE) fractions of WT and BFAR-deficient CD4+ T cells that were left untreated (−) or stimulated with 3 µg ml−1 α-CD3/28 overnight and then stimulated with TGFβ1 (5 ng ml−1) for the indicated time points. (M and N) Immunoblot analysis of phosphorylated and total Smad2/3 in WCLs and NE fractions of WT and BFAR-deficient CD4+ T cells that were stimulated under Th9 conditions for 3 d. PCNA, proliferating cell nuclear antigen. (O–Q) Immunoblot analysis of the nuclear NF-κB, STAT proteins, PU.1, and IRF4 in WT and BFAR-deficient Th9 cells (O) and the phosphorylated and total STAT6 or Smad2/3 in WCLs of WT and BFAR-deficient CD4+ T cells that were stimulated with TGFβ1 (5 ng ml−1) or IL-4 (10 ng ml−1) for the indicated time points (P and Q). Each panel is representative of three independent experiments.
Figure S2.
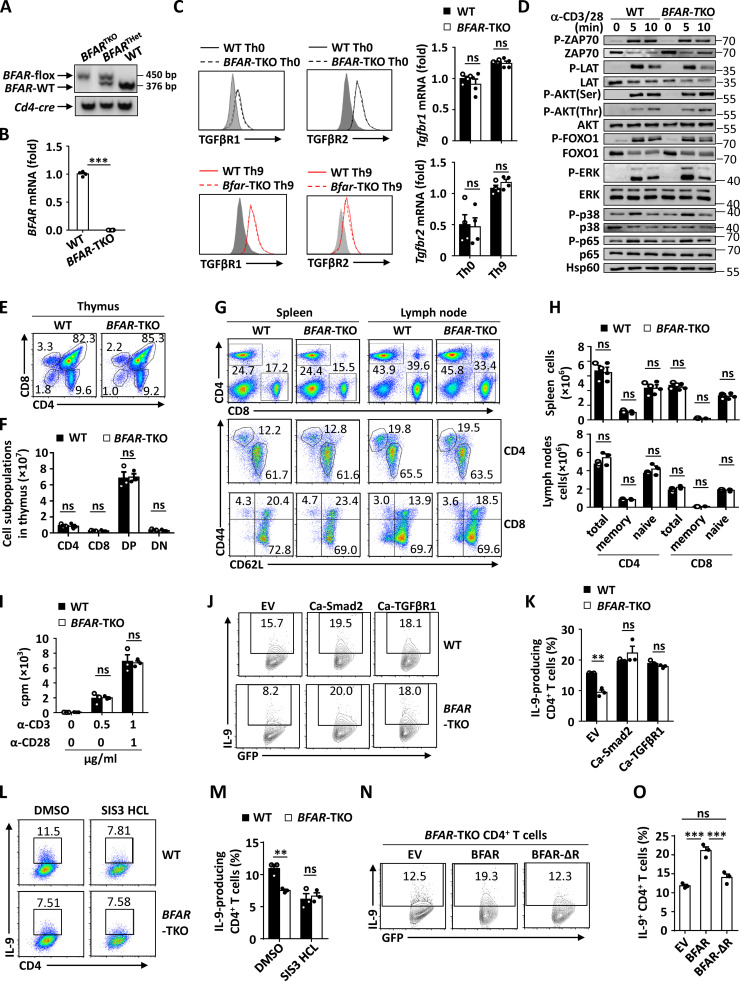
BFAR does not affect T cell development, proliferation, and TCR downstream signaling activation. (A) Genotyping PCR analysis of WT, T cell BFAR heterozygous (BFARTHet), and T cell BFAR homozygous KO (BFARTKO) mice. (B) qPCR analysis of BFAR mRNA expression in naive CD4+ T cells isolated from WT and BFAR-TKO mice. (C) Flow cytometric and qPCR analysis of TGFβR1 and TGFβR2 surface or mRNA expression in WT and BFAR-deficient naive CD4+ T cells cultured under Th0 or Th9 conditions for 3 d. (D) Immunoblot analysis of phosphorylated and total ZAP70, LAT, AKT, FOXO1, ERK, p38, and p65 in whole-cell lysates of WT and BFAR-deficient naive CD4+ T cells that were stimulated with anti-CD3 plus anti-CD28 (α-CD3/CD28; 1 µg ml−1) for the indicated time points. (E–H) Flow cytometric analysis of the frequencies of the indicated lymphoid immune cells in the thymus (E and F), spleen, and peripheral lymph nodes (G and H) of WT and BFAR-TKO mice. Data are presented as representative plots (E and G) and summary bar graphs (F and H). DP, double positive; DN, double negative. (I) Proliferation of WT and BFAR-deficient naive CD4+ T cells stimulated with anti-CD3 (α-CD3) and/or α-CD28 at the indicated concentrations for 72 h, assessed by [3H]thymidine incorporation. (J and K) Flow cytometric analysis of the frequencies of IL-9–producing Th9 cells in WT and BFAR-deficient naive CD4+ T cells that were reconstituted with EV, Ca-Smad2, or Ca-TGFβR1. Data are presented as representative plots (J) and summary graphs (K). (L–O) Flow cytometric analysis of the frequencies of IL-9–producing Th9 cells in WT and BFAR-deficient naive CD4+ T cells that were treated with DMSO or SIS3 HCl (Smad3 inhibitor; L and M) or in BFAR-deficient naive CD4+ T cells that were reconstituted with EV, BFAR, or Ring domain–deleted BFAR (BFAR-ΔR; N and O). Data are presented as representative plots (L and N) and summary graphs (M and O). Each panel is representative of three independent experiments. Student’s t test was used. Bars, mean; error bars, SEM; **, P < 0.01; and ***, P < 0.001; ns, not significant.
We then examined the TGFβR1 downstream signaling required for Th9 differentiation (Dardalhon et al., 2008; Tamiya et al., 2013; Veldhoen et al., 2008) upon BFAR deletion. The results revealed that BFAR deletion did not affect the TCR-induced activation of downstream signaling and the surface expression of TGFβR1 or TGFβR2 in mouse primary CD4+ T cells under Th0 or Th9 differentiation conditions (Fig. S2, C and D). However, after having been pretreated with anti-CD3 plus anti-CD28, the association of TGFβR1 with its downstream Smad2/3 was markedly suppressed, with or without TGFβ stimulation, in BFAR-deficient CD4+ T cells compared with WT cells (Fig. 4 K). Consequently, the loss of BFAR inhibited TGFβ-induced phosphorylation and the nuclear translocation of Smad2/3 in CD4+ T cells in the presence of TCR stimulation or under Th9 conditions (Fig. 4, L–N). However, BFAR deficiency affected neither nuclear translocation of NF-κB, STAT proteins, IFN regulatory factor 4 (IRF4), or PU.1 that are required for Il9 induction under Th9 conditions nor IL-4–induced STAT6 activation or TGFβ-induced Smad2/3 phosphorylation in CD4+ T cells without TCR pretreatment (Fig. 4, O–Q). These data suggested that BFAR-mediated Smad2/3 activation is dependent on its early-stage function in promoting TCR-induced TGFβR1 ubiquitination.
BFAR deficiency impairs Th9 differentiation and Th9-based cancer immunotherapy
Next, we found that BFAR deficiency affected neither the development, maturation, and activation of T cells nor the TCR-induced proliferation of CD4+ T cells (Fig. S2, E–I). As expected, the loss of BFAR dramatically inhibited Th9 cell differentiation (Fig. 5, A and B). In addition, introducing the constitutive activation of Smad2 or TGFβR1 in BFAR-deficient CD4+ T cells greatly enhanced Th9 differentiation and abolished the difference in Th9 inducibility between WT and BFAR-deficient CD4+ T cells (Fig. S2, J and K). By contrast, Smad3 inactivation by a selective inhibitor significantly suppressed the Th9 differentiation of WT CD4+ T cells and abolished the difference in Th9 inducibility between WT and BFAR-deficient CD4+ T cells (Fig. S2, L and M), suggesting that both Smad2 and Smad3 are essential for BFAR-mediated Th9 differentiation. Moreover, reconstitution with full-length BFAR, but not with E3 ligase functionally inactive BFARΔR, successfully rescued the Th9 inducibility in BFAR-deficient CD4+ T cells (Fig. S2, N and O). Therefore, these data established BFAR as an essential positive regulator of Th9 differentiation through modulating TGFβR1 and downstream signaling.
Figure 5.
BFAR deficiency impairs Th9 differentiation and Th9-based cancer immunotherapy. (A and B) Flow cytometric analysis of the frequencies of IL-9–producing WT and BFAR-deficient naive CD4+ T cells under Th0 or Th9 differentiation conditions for 3–4 d (A) and the corresponding statistical analysis (B). TKO, knock out in T cell. (C) RNA-sequencing (RNA-seq) analysis showing differentiated gene expression profiles in WT and BFAR-deficient naive CD4+ T cells differentiated under Th9 condition for 3 d, with representative Th9 signature genes indicated in the scatterplot. (D) qPCR analysis of the mRNA expression of Th9 signature genes in the WT and BFAR-deficient Th9 cells. (E–G) Tumor growth (E), tumor image (F), and tumor weight (G) in Rag1−/− mice that were s.c. inoculated with B16 melanoma cells and then adoptively transferred with WT or BFAR-deficient Th9 cells (n = 7 mice/group). (H) Flow cytometric analysis of tumor-infiltrating immune cells, as indicated in B16 melanoma-bearing mice on day 18, as described in E. Data are presented as representative plots (left panels) and summary graphs (right panels). (I) qPCR analysis of the mRNA expression of Il9 and Ccl20 in tumor-infiltrating CD45+CD11b−CD4+ T cells that were isolated from B16 melanoma-bearing mice on day 18, as described in E, and then stimulated with anti-CD3 (5 µg ml−1) for 24 h. (J) Tumor growth in Rag1−/− mice that were s.c. inoculated with B16 melanoma cells and received adoptive transferred of WT or BFAR-deficient Th9 cells and then i.v. injected with anti–IL-9 (α-IL-9) or control antibody (IgG) every 3 d starting from day 0 (n = 4 mice/group). (K and L) Tumor growth (K) and image (L) in C57BL/6 mice that were s.c. inoculated with B16-OVA melanoma cells and then adoptively transferred with WT (n = 8) or BFAR-deficient OT-II–specific Th9 cells (n = 6) and DCs at day 10 after tumor inoculation (arrow). Adjuvant cyclophosphamide was administered i.p. 1 d before Th9 cell transfer. Each panel is representative of three independent experiments, and each circle represents one mouse in G and H. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; **, P < 0.01; and ***, P < 0.001; ns, not significant.
To confirm the essential role of BFAR in mediating Th9 differentiation, we examined IL-9 induction by using different doses of IL-4 or TGFβ alone or in combination or by using a different state of CD4+ T cells. The results showed that BFAR deficiency indeed greatly impaired IL-9 induction by using different doses of IL-4/TGFβ in naive CD4+ T cells or by using CD4+ T cells under memory state (Fig. S3, A and B). In addition, RNA sequencing, combined with qPCR analysis, confirmed that BFAR deletion suppressed the mRNA expression of Il9 and other Th9 signature genes, including Il1a, Ccl17, and Ccl20, under Th9 differentiation conditions (Fig. 5, C and D; and Fig. S3 C). Moreover, BFAR deficiency did not affect the proliferation and apoptosis of CD4+ T cells under Th9 conditions (Fig. S3, D–G), suggesting that the modulation of BFAR on Th9 differentiation is independent of its action on T cell proliferation and apoptosis. TGFβ-induced downstream signaling is also essential for the induction of Th17 and T regulatory (Treg) cell differentiation (Malhotra et al., 2010; Tone et al., 2008). However, BFAR deficiency did not affect the differentiation of Th1, Th2, Th17, and Treg cells (Fig. S4, A and B), suggesting that BFAR is a specific regulator for Th9 differentiation. In addition, high-dose TGFβ pretreatment that could impair Th9 differentiation (Fig. 3, E and F) failed to do so in the same way in Th17 and Treg cell differentiation (Fig. S4, C and D), and BFAR deletion also could not affect Th17 and Treg cell differentiation with different doses of TGFβ (Fig. S4, E–H). Therefore, BFAR-modulated TGFβ downstream signaling exhibits a unique function to specifically modulate Th9 differentiation.
Figure S3.
BFAR is critical for Th9 differentiation without affecting cellular proliferation and apoptosis. (A and B) Flow cytometric analysis of the frequencies of IL-9–producing Th9 cells in WT, BFAR-deficient naive (A), and WT and BFAR-deficient naive or memory CD4+ T cells (B) cultured with the indicated concentrations of IL-4 and/or TGFβ1 for 3 d. Med, medium. (C) qPCR analysis of the Ifng, Il4, Il17a, Foxp3, Il9, Il1a, Ccl17, and Ccl20 mRNA expression of the naive CD4+ T cells cultured under distinct differentiation conditions as indicated for 3 d. (D–G) Flow cytometric analysis of the proliferation (D and E) and apoptosis (F and G) in WT and BFAR-deficient naive CD4+ T cells or cultured under Th0 or Th9 conditions for 3 d. The proliferation ratio was assessed as CFSE dilution by FACS. Data are presented as representative plots (D and F) and summary graphs (E and G). Each panel is representative of two independent experiments. Student’s t test was used. Bars, mean; error bars, SEM; **, P < 0.01; ns, not significant.
Figure S4.
BFAR deficiency does not affect Th19 and Treg cell differentiation. (A and B) Flow cytometric analysis of the inducibility of Th1, Th2, Th17, or Treg cells of WT and BFAR-deficient naive CD4+ T cells under different differentiation conditions for 3 d. (C and D) Flow cytometric analysis of Th17 and Treg cell inducibility of mouse naive CD4+ T cells that were left pretreated with 0.5 ng ml−1 TGFβ1 (Lo) or 10 ng ml−1 TGFβ1 (Hi) for 12 h and then cultured under Th17 and Treg cell conditions for 3 d. (E–G) Flow cytometric analysis of the inducibility of Th17 and Treg cells of WT and BFAR-deficient naive CD4+ T cells under different differentiation conditions for 3 d with the indicated doses of TGFβ1. (H) ELISA measurements of supernatant TGFβ1 and IL-10 levels of Treg cells of WT and BFAR-deficient naive CD4+ T cells under Treg cell differentiation conditions for 3 d with the indicated doses of TGFβ1. Each panel is representative of two independent experiments. Student’s t test was used. Bars, mean; error bars, SEM; ns, not significant.
Th9 cells have been recognized as a promising CD4+ effector T cell subset to inhibit tumor growth (Chauhan et al., 2019; Lu et al., 2012; Lu et al., 2018; Purwar et al., 2012). Hence, we used an experimental melanoma mouse model to investigate whether BFAR deficiency impaired the anti-tumor response of Th9 cells. To avoid a potential nonspecific effect of CD8+ T cells, we adoptively transferred in vitro differentiated WT or BFAR-KO Th9 cells into Rag1-KO mice that were inoculated with B16 melanoma cells. As expected, mice that received BFAR-deficient Th9 cells showed increased growth of melanoma, with about twice as large tumor size on day 18, compared with that of mice receiving WT Th9 cells (Fig. 5, E–G). Moreover, the analysis of tumor-infiltrated immune cells indicated that Th9 cells and dendritic cells (DCs) were significantly inhibited (Fig. 5 H), which is inconsistent with the immune infiltration pattern in the TME after Th9 cell treatment (Lu et al., 2012). Accordingly, the expression of Th9 signature genes Il9 and Ccl20 was also significantly suppressed in the tumor-infiltrated CD4+ T cells of the mice that received BFAR-deficient Th9 cells (Fig. 5 I). Moreover, treatment with an IL-9 neutralizing antibody promoted tumor growth and abolished the difference in tumor growth between the mice that received WT Th9 cells and those that received BFAR-deficient Th9 cells (Fig. 5 J). Furthermore, BFAR deficiency also impaired Th9-mediated cancer immunotherapeutic efficacy against established B16-OVA melanoma in immune-competent C57BL/6 mice (Fig. 5, K and L). These data collectively confirmed the in vivo biological function of BFAR in mediating anti-tumor immunity in Th9 cells.
BFAR overexpression enhances Th9-mediated cancer immunotherapy
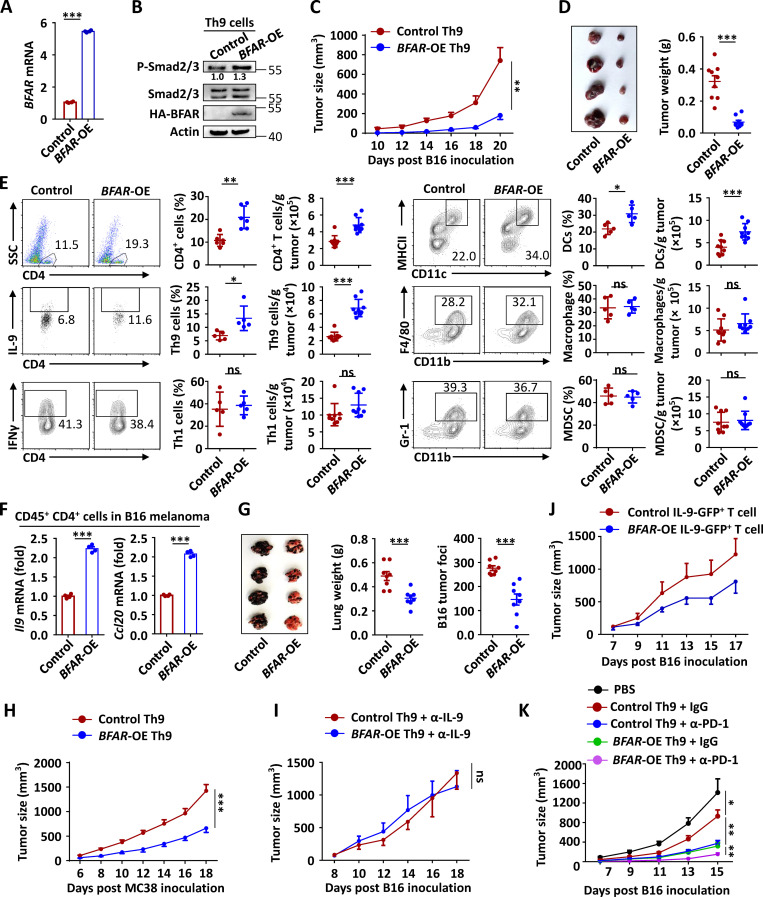
The essential role of BFAR in mediating Th9 differentiation prompted us to modulate this molecular target to enhance Th9-mediated cancer immunotherapy. To this end, we examined the anti-tumor response of BFAR-overexpressing Th9 cells through a cancer ACT model. Interestingly, BFAR overexpression promoted Smad2/3 activation under Th9 conditions (Fig. 6, A and B), and the overexpressed Th9 cells exhibited astonishing therapeutic efficacy to inhibit tumor growth as compared with WT control Th9 cells, reflected by remarkably smaller tumor size and almost no evident growth of melanoma at the initial 18-d observation time point (Fig. 6, C and D). Consistently, the analysis of tumor-infiltrated immune cells indicated that Th9 cells and DCs were specifically and significantly increased in the TME of the mice that received BFAR-overexpressing Th9 cells as compared with that of the mice receiving control Th9 cells (Fig. 6 E). Accordingly, the expression of Th9 signature genes Il9 and Ccl20 was also significantly promoted in the tumor-infiltrated CD4+ T cells of the mice that received BFAR-overexpressing Th9 cells (Fig. 6 F).
Figure 6.
BFAR overexpression enhances Th9-mediated cancer immunotherapy. (A and B) qPCR and immunoblot analysis of BFAR expression and Smad2/3 phosphorylation in CD4+ T cell reconstitution with an EV (−) or HA-BFAR and then cultured under Th9 conditions for 3 d. (C and D) Tumor growth (C), representative tumor images, and weight (D) of Rag1−/− mice that were s.c. inoculated with B16 melanoma cells and then adoptively transferred with control or BFAR-overexpressed (OE) Th9 cells (n = 8 mice/group). (E) Flow cytometric analysis of tumor-infiltrating immune cells, as indicated in B16 melanoma-bearing mice on day 20, as described in C. Data are presented as representative plots (left panels) and summary graphs (right panels). SSC, side scatter; MDSC, myeloid-derived suppressor cells. (F) qPCR analysis of the mRNA expression of Il9 and Ccl20 in tumor-infiltrating CD45+CD11b−CD4+ T cells that were isolated from B16 melanoma–bearing mice on day 20, as described in A, and then stimulated with anti-CD3 (5 µg ml−1) for 24 h. (G) Representative images, weight, and statistical analysis of the lung metastatic tumor foci on day 16 of C57BL/6 mice that were i.v. injected with B16F10-OVA melanoma cells and then adoptively transferred with control OT-II Th9 or BFAR-OE OT-II Th9 cells 1 d after tumor inoculation (n = 8 mice/group). (H and I) Tumor growth over time in Rag1−/− mice that were s.c. inoculated with MC38 tumor cells and then adoptively transferred with control or BFAR-OE Th9 cells (E; n = 6 mice/group) or that were s.c. inoculated with B16 melanoma cells, adoptively transferred with control or BFAR-OE Th9 cells, and then i.v. injected with anti–IL-9 (α-IL-9) every 3 d starting from day 0 (F; n = 4 mice/group). (J) Tumor growth of Rag1−/− mice that were s.c. inoculated with B16 melanoma cells and then adoptively transferred with control or BFAR-OE IL-9-GFP+ Th9 cells (n = 4 mice/group). (K) Tumor growth over time in Rag1−/− mice that were s.c. inoculated with B16-F10 melanoma cells, adoptively transferred with control or BFAR-OE Th9 cells, and then i.v. injected with control antibody (IgG) or α-PD-1 on days 7, 10, and 13 after tumor inoculation (n = 5–6 mice/group). Each panel is representative of three independent experiments, and each circle represents the data from one mouse. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; **, P < 0.01; and ***, P < 0.001; ns, not significant.
Compared with control Th9 cells, the overexpression of BFAR in Th9 cells also dramatically suppressed lung metastasis of i.v. injected B16 melanoma cells (Fig. 6 G). A parallel study revealed that the BFAR-overexpressing Th9 cells also displayed an enhanced anti-tumor response to suppress the growth of MC38 colon tumors (Fig. 6 H). Moreover, the neutralization of Th9 signature cytokine IL-9 abolished the difference in tumor growth in mice that were transferred with control or BFAR-overexpressing Th9 cells (Fig. 6 I), suggesting that the enhanced anti-tumor activity elicited by BFAR overexpression specifically depends on the Th9-mediated anti-tumor immune effect. By using newly generated IL-9–enhanced GFP (EGFP) mice, we collected the same amount of control and BFAR-overexpressing IL-9–producing CD4+ T cells for cancer ACT. The results revealed that although there was no statistically significant difference, BFAR-overexpressing IL-9–producing cells also exhibited an enhanced tendency for cancer immunotherapy (Fig. 6 J), suggesting that BFAR may also mediate the functional enhancement of Th9 cells, but its function for Th9-mediated cancer immunotherapy is still largely dependent on its role in promoting Th9 differentiation. Furthermore, the adoptive transfer of BFAR-overexpressing Th9 cells had a synergistic effect with anti–PD-1–mediated checkpoint immunotherapy in eliminating tumor growth (Fig. 6 K). These findings confirmed that BFAR is a promising molecular target for enhancing Th9-mediated cancer immunotherapy.
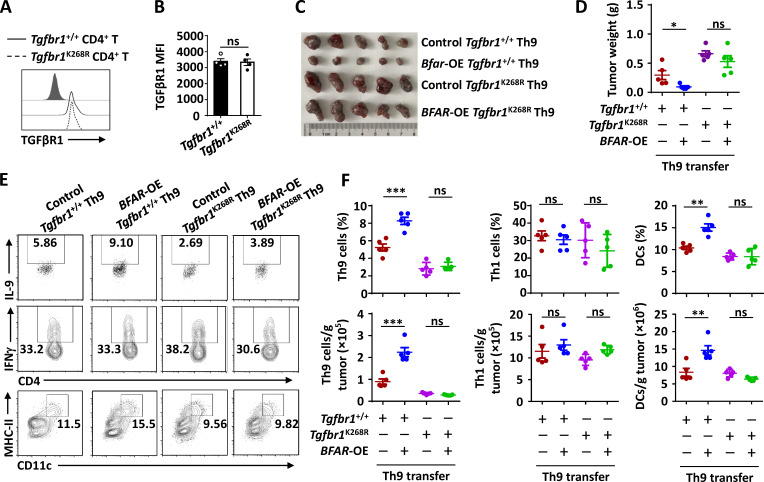
BFAR-induced TGFβR1 ubiquitination at K268 is essential for Th9-mediated therapy
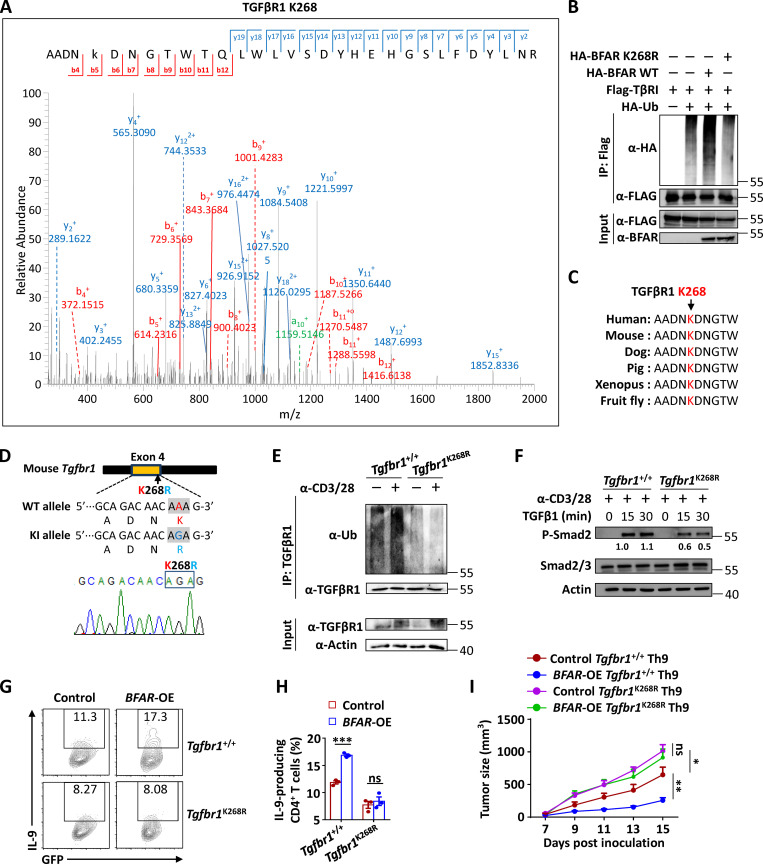
To validate whether BFAR-mediated Th9 differentiation and Th9-based cancer immunotherapy are dependent on their role in promoting TGFβR1 ubiquitination, we performed MS analysis of Flag-tagged TGFβR1 in the presence of BFAR overexpression and identified lysine 268 (K268) as a potential ubiquitination site targeted by BFAR (Fig. 7 A). BFAR failed to promote the ubiquitination of K268R point-mutated TGFβR1 (Fig. 7 B), indicating that K268 is the ubiquitination site targeted by BFAR. Interestingly, the K268 site is evolutionarily conserved in TGFβR1 proteins among different species (Fig. 7 C), suggesting the potential functional essence of this ubiquitination site.
Figure 7.
BFAR-induced TGFβR1 ubiquitination at K268 is essential for Th9-mediated cancer therapy. (A) MS showing the potential ubiquitination site of TGFβR1 at K268 after TGFβR1 immunoprecipitation (IP) in HEK293T cells transfected with TGFβR1, BFAR, and Ub. (B) Ubiquitination of TGFβR1 in HEK293T cells transfected with the indicated expression vectors, assessed by immunoblotting with anti-HA and anti-Flag after IP with anti-Flag or by immunoblotting with input proteins in lysates without IP. (C) Amino acid sequence alignment of TGFβR1 among the indicated species, with Lys268 (K268) highlighted in red. (D) The knock-in (KI) strategy (upper panel) and sequencing verification of the codon replacement (lower panel) for Tgfbr1-K268R by using the CRISPR/Cas9 technique. (E) Endogenous ubiquitination of TGFβR1 in Tgfbr1+/+ and Tgfbr1K268R CD4+ T cells that were left unstimulated (−) or stimulated (+) with anti-CD3 plus anti-CD28 (α-CD3/28; 3 µg ml−1) overnight, assessed by immunoblotting with anti-Ub and anti-TGFβR1 after IP with anti-TGFβR1 (top) and immunoblotting with input proteins and loading controls (below). (F) Immunoblot analysis of phosphorylated and total Smad2/3 in whole-cell lysates of Tgfbr1+/+ and Tgfbr1K268R CD4+ T cells that were stimulated with α-CD3/28 (3 µg ml−1) overnight and then stimulated with TGFβ1 (5 ng ml−1) for the indicated time points. (G and H) Flow cytometric analysis of the frequencies of IL-9–producing Tgfbr1+/+ and Tgfbr1K268R CD4+ T cells that were reconstituted with EV (control) or BFAR-overexpressing (OE) vector under Th9 differentiation conditions for 3–4 d (G) and the corresponding statistical analysis (H). (I) Tumor growth over time in Rag1−/− mice that were s.c. inoculated with B16 melanoma cells and then received adoptive transfer of Tgfbr1+/+ and Tgfbr1K268R Th9 cells with or without BFAR-OE (n = 5 mice/group). Each panel is representative of three independent experiments. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; **, P < 0.01; and ***, P < 0.001; ns, not significant.
Hence, we generated K268R knock-in mice with both copies of the mutant Tgfbr1 gene by using the CRISPR/Cas9 system and verified the mutations by genomic DNA sequencing to confirm the importance of endogenous TGFβR1 K268 ubiquitination (Fig. 7 D). K268R knock-in did not affect the surface expression of TGFβR1 in mouse primary CD4+ T cells (Fig. S5, A and B). However, this knock-in mutation abolished the TCR-induced ubiquitination of TGFβR1 and, as expected, greatly suppressed TGFβ-induced downstream Smad2 activation in CD4+ T cells in the presence of TCR (Fig. 7, E and F). Accordingly, the inducibility of Th9 differentiation was suppressed in K268R knock-in CD4+ T cells in comparison with WT cells, and BFAR overexpression failed to promote Th9 differentiation in the knock-in CD4+ T cells like it did in WT cells (Fig. 7, G and H). Consistent with the differentiation result, K268R knock-in suppressed the anti-tumor effect of adoptively transferred Th9 cells, and the knocked-in Th9 cells were not responsive to BFAR overexpression, thereby failing to improve the Th9-mediated therapeutic efficacy in eliminating tumor growth (Fig. 7 I; and Fig. S5, C–F). Together these findings confirmed the essential function of TGFβR1 K268 ubiquitination by BFAR in mediating Th9 differentiation and related cancer immunotherapy.
Figure S5.
TGFβR1 ubiquitination at K268 is critical for Th9-mediated cancer immunotherapy without affecting the surface TGFβR1 expression. (A and B) Flow cytometric analysis of surface TGFβR1 expression in Tgfbr1+/+ and Tgfbr1K268R naive CD4+ T cells. Data are presented as a representative plot (A) and a bar graph (B). MFI, mean fluorescence intensity. (C–F) Representative tumor images (C), weights (D), and tumor-infiltrating immune cells (E and F) of Rag1−/− mice that were s.c. inoculated with B16 melanoma cells and then adoptively transferred with Tgfbr1+/+ and Tgfbr1K268R Th9 cells with or without BFAR overexpression (OE). Each panel is representative of two independent experiments, and each circle represents the data from one mouse. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; **, P < 0.01; and ***, P < 0.001; ns, not significant.
BFAR-overexpressing Th9 suppresses human PDX tumor
To evaluate the clinical significance and translational potential of BFAR in treating human solid tumors, we generated a patient-derived xenograft (PDX) colorectal tumor model in immunodeficient nonobese diabetic/ShiLtJGpt-Prkdcem26Cd52Il2rgem26Cd22/Gpt (NCG) mice, which were then adoptively transferred with patient-derived DCs alone or with Th9 cells differentiated from the CD4+ T cells of human CRC patients. Interestingly, mice receiving Th9 cells with DCs greatly suppressed tumor growth as compared with the mice that were transferred with DCs alone; moreover, BFAR overexpression further enhanced the therapeutic efficacy of transferred Th9 cells, as reflected by a remarkably reduced tumor size (Fig. 8, A–C). Additional flow cytometric analysis revealed that HLA-DR+, a T cell activation marker (Capasso et al., 2019), or IL-9–producing CD4+ T cell infiltration was significantly increased in the TME of the mice transferred with BFAR-overexpressing Th9 cells (Fig. 8, D and E). Moreover, BFAR overexpression markedly promoted the expression of Th9-related genes, including IL9 and CCL20, but not the Th1 gene IFNG or the Treg cell gene FOXP3, in tumor-infiltrated CD4+ T cells (Fig. 8 F). Accordingly, the serum level of human IL-9, but not IFNγ, was significantly increased in the mice receiving human BFAR-overexpressing Th9 cells when compared with those receiving control Th9 cells (Fig. 8 G). These results further confirmed the therapeutic potential of BFAR in promoting Th9-mediated immunotherapy against human cancer.
Figure 8.
BFAR-overexpressed Th9 cells suppress human PDX tumor. (A–C) Tumor growth (A), tumor images (B), and weight (C) of human CRC PDX in NCG mice (n = 10 mice/group) that were adoptively transferred with CRC-derived DCs alone or DCs plus CRC-derived Th9 cells with or without BFAR overexpression (OE). (D and E) Flow cytometric analysis of the frequencies of tumor-infiltrating HLA-DR+CD4+ T cells and IL-9–producing Th9 cells in tumor-bearing mice on day 27 after tumor inoculation, as described in A. Data are presented as a representative plot (D) and summary graphs (E). (F) qPCR analysis of T cell–related gene expression in tumor-infiltrating CD45+CD11b−CD4+ T cells sorted from the tumor-bearing mice on day 27 after tumor inoculation, as described in A, and then stimulated with anti-CD3 plus anti-CD28 for 24 h. (G) ELISA measurements of serum human IL-9 and IFNγ levels in tumor-bearing mice on day 27 after tumor inoculation. ND, not detected. (H) Tumor-derived TGFβ impairs Th9 inducibility by antagonizing the expression of BFAR, which decreased the production of Th9 signature cytokine IL-9 and chemokine CCL20, leading to reduced recruitment of DCs from the circulation system. Consequently, T cell activation and Th9 differentiation were inhibited, thereby impairing the anti-tumor immune response. Each panel is representative of two independent experiments, and each circle represents the data from one mouse. Student’s t test was used. Bars, mean; error bars, SEM; *, P < 0.05; **, P < 0.01; and ***, P < 0.001; ns, not significant.
Discussion
TGFβ is a pleiotropic cytokine that plays a role in tumor biology and immune modulation (Batlle and Massagué, 2019; Budi et al., 2017; Travis and Sheppard, 2014). The biological function of this cytokine in directly regulating tumor progression, such as modulating epithelial-to-mesenchymal transition, stemness, and mobile features of cancer cells, has been well studied and characterized (Calon et al., 2012; Polyak and Weinberg, 2009; Su et al., 2020; Zeisberg et al., 2003). However, little is known about how TGFβ reprograms immune profiles to affect anti-tumor immune responses. It is widely known that, in vitro, TGFβ and IL-4 can act together to induce CD4+ T cell differentiation into Th9 cells with a robust anti-tumor activity (Nakatsukasa et al., 2015; Tamiya et al., 2013; Wang et al., 2016). Nevertheless, under in vivo tumor conditions, with or without immunotherapy, the role of TGFβ in modulating Th9 inducibility, thus affecting anti-tumor immunity, remains unexplored. In this study, we found that the GIC patient–derived CD4+ T cells exposed to TGFβ for a long time were insensitive to Th9 induction, which contradicts the in vitro data indicating that TGFβ is an essential cytokine for Th9 induction. A plausible explanation is that long-term exposure to TGFβ may indirectly impair the Th9 inducibility of CD4+ T cells through an unknown negative regulatory mechanism. We further identified BFAR as a direct E3 ligase to mediate the K63-linked ubiquitination of TGFβR1- and TGFβ-suppressed BFAR expression, thereby inhibiting TGFβR1 ubiquitination. Consequently, the activation of downstream Smad2/3 was impaired in tumor patient–derived CD4+ T cells that underwent long-term exposure to TGFβ when stimulated with TGFβ again under Th9 induction conditions. This fine-tuned negative regulatory mechanism was confirmed by the finding that BFAR deletion suppressed TGFβR1 ubiquitination, thus impairing Th9-mediated cancer immunotherapy, whereas BFAR overexpression exhibited an opposite effect. Therefore, even with elevated levels of TGFβ in tumor patients, these patients’ CD4+ T cells still cannot be efficiently induced into anti-tumor Th9 cells due to the inhibition of TGFβR1 E3 ligase BFAR. However, since suppressed BFAR expression and Th9 inducibility is only in CD4+ T memory cells of cancer patients, we also could not rule out the possibility of the ground state difference of CD4+ T memory cells between HDs and cancer patients in affecting their potential to become Th9 cells. Hence, our findings disclose a novel mechanism by which tumors resist the anti-tumor immunity to maintain their survival and metastasis.
The K63-linked ubiquitination is critical for the activation of signaling molecules (Wu and Karin, 2015; Yu et al., 2019; Zhu et al., 2020); TGFβR1 K63-linked ubiquitination is speculated to be an initial molecular event for the TGFβ-induced activation of downstream Smad2/3. Previous studies have identified several E3 ligases or deubiquitinases, such as SMURF1/2 and USP4, to modulate the K48-linked ubiquitination and/or degradation of TGFβR1 (Kavsak et al., 2000; Kim and Baek, 2019; Suzuki et al., 2002; Zhang et al., 2012). However, the direct E3 ubiquitin (Ub) ligase that mediates K63-linked ubiquitination of TGFβR1 is so far unknown. The present study not only identified BFAR as the first direct E3 ligase to mediate the K63-linked ubiquitination of TGFβR1 and downstream Smad2/3 activation but also found that K268 is the functional site where BFAR adds K63-linked polyubiquitination chains in TGFβR1. In addition, K268R mutation at the genetic level by CRISPR/Cas9-mediated knock-in also confirmed the biological function of K63-linked ubiquitination at K268 of TGFβR1 in mediating Th9 cell differentiation and Th9-based cancer immunotherapy.
Our research also provided genetic evidence that BFAR is critical for TGFβ-induced phosphorylation and the nuclear translocation of Smad2 and Smad3, thus specifically mediating Th9 differentiation without affecting Treg and Th17 cell differentiation. A plausible explanation is that the role of TGFβ in inducing the differentiation of different Th subsets depends on the synergistic activities of distinct transcription factors with Smad proteins (Massagué, 2012). For Th17 differentiation, Smad2 cooperates with STAT3 to induce IL-17A expression, but Smad3 plays an opposite role in IL-17A induction (Fu et al., 2017; Malhotra et al., 2010; Yoon et al., 2015). By contrast, Smad3 cooperates with NFAT to induce Foxp3 expression under Treg cell differentiation conditions, which is independent of the activity of Smad2 (Tone et al., 2008). However, for Th9 differentiation, Smad2 acts with Smad3 cooperating with IRF4 to induce IL-9 expression (Tamiya et al., 2013). The need for both Smad2 and Smad3 in BFAR-mediated Th9 differentiation was further confirmed by the reconstitution of Ca-Smad2 or the use of the selective inhibitor of Smad3. Moreover, an alternative possibility would be that BFAR recruitment to TGFβR1 following TCR stimulation is unique to the Th9 condition or that the nuclear translocation of Smads is independent of BFAR in the Th17/Treg cell condition.
In sum, our work demonstrated that tumor-derived TGFβ impairs Th9 inducibility of GIC patient–derived CD4+ T cells by antagonizing the expression of BFAR, a direct E3 ligase, to mediate the K63-linked ubiquitination of TGFβR1. Consequently, the production of Th9 signature cytokine IL-9 and chemokine CCL20 was decreased, leading to reduced recruitment of DCs for T cell activation and Th9 differentiation, thereby impairing the anti-tumor immune response. (Fig. 8 H). More interestingly, we identified BFAR as a promising molecular target that can be used to boost Th9 differentiation, thereby enhancing the therapeutic efficacy of Th9-mediated cancer immunotherapy.
Materials and methods
Human blood samples
Human peripheral blood from HDs and CRC or GC patients was obtained from Fudan University Shanghai Cancer Center. Informed consent was obtained from all study subjects before their inclusion in this study. The sample collection for this study was reviewed and approved by the ethics committee of Fudan University Shanghai Cancer Center and complied with all relevant ethical regulations.
Mice
BFAR floxed mice and Tgfbr1 K268R knock-ins were generated by the Shanghai Research Center for Model Organisms. T cell–specific BFAR conditional KO mice were generated by crossing Bfarfl/fl mice with mice expressing Cre under the control of the CD4 promoter (CD4-Cre). IL-9–internal ribosome entry site (IRES)-EGFP mice were generated by Biocytogen by inserting IRES-EGFP-SV-pA before the termination codon of the Il9 gene through CRISPR/Cas9. Rag1−/− and OT-II TCR transgenic mice with a C57BL/6 background were purchased from Shanghai Research Center for Model Organisms. NCG mice were purchased from GemPharmatech. In all of the experiments with gene KO or knock-in mice, WT littermate controls were used. All mice were bred and maintained under specific-pathogen-free conditions. All animal experiments were performed in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and were approved by the institutional biomedical research ethics committee of the Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences.
Plasmids, antibodies, and reagents
MSCV-PIG and pCL-Eco were kindly provided by Dr. Xing Chang (Westlake University, Hangzhou, China). pCMV-Ca-SMAD3 was kindly provided by Dr. Long Zhang (Zhejiang University, Hangzhou, China). pCDH-MCS-copGFP was kindly provided by Dr. Feng Yu (Jiangsu University, Zhenjiang, China). The cDNAs encoding Ca-SMAD2, BFAR, and TGFβR1 were cloned and constructed into PMX-IRES, pLVX-IRES-GFP, p3×FLAG-CMV, or pcDNA vector. The pMD2.G, psPAX2, and expression vectors encoding different HA-tagged types of Ub were as previously described (Zhu et al., 2020). For the generation of TGFβR1 K268R and Ca-TGFβR1 (TGFβR1 T204D) plasmids, point mutations were constructed by site-directed mutagenesis. All homemade and requested constructs were confirmed by DNA sequencing.
For Western blots, antibody for BFAR (LS-C482996) was from LSBio. Antibody for TGFβRII (06-318) was from Merck. Antibody for TGFβR1 (ab31013) was from Abcam. Antibodies for Hsp60 (H-1; sc-13115), lamin B (C-20; sc-6216), p65 (C-20; sc-372), p38 (H-147; sc-7149), Erk1 (K-23; sc-94), p-Erk (E-4; sc-7383), Ub (P4D1; sc-8017), p-STAT6 (sc-11762), STAT6 (M-20; sc-981), p105/p50 (c-19; sc-1190), p100/p52 (c-5; sc-7386), c-Rel (sc-71), RelB (c-19; sc-226), ZAP70 (1E7.2; sc-32760), LAT (FL-233; sc-7948), STAT1 (M-22; sc-592), IRF4 (M-17; sc-6059), and donkey anti-goat IgG (HRP; sc-2020) were from Santa Cruz Biotechnology. Antibodies for STAT3 (9132), STAT5 (9363), p-SMAD2/3 (8828), SMAD2/3 (8685), p-p65 (3033), p-p38 (9215), p-AKT (4060), p-AKT (2965), p-FOXO1 (9464), FOXO1 (2880), p-ZAP70 (2701), p-LAT (3584), K63-linkage specific polyubiquitin (5621), PU.1 (2258), and normal rabbit IgG (2729) were from Cell Signaling Technology. Antibodies for β-actin (A2228) and Flag (A8592) were from Sigma-Aldrich. Antibody for HA (2013819) was from Roche. Antibody for Alexa Fluor Plus 488–conjugated rabbit IgG (A11034) was from Thermo Fisher Scientific.
For the flow cytometric analysis, anti-mouse CD3ε (145-2C11), anti-mouse CD8 (53-6.7), anti-mouse CD4 (RM4-5), anti-mouse CD45 (30-F11), anti-mouse CD44 (IM7), anti-mouse CD62L (MEL-14), anti-Ki-67 (16A8), anti-mouse CD11b (M1/70), anti-mouse F4/80 (BM8), anti-mouse-Ly-6G/Ly6C (RB6-8C5), anti-mouse CD11c (N418), anti-mouse FOXP3 (FJK-16s), anti-mouse IFNγ (XMG1.2), anti-mouse IL-4 (11B11), anti–MHC-II (AF6-120.1), and anti-mouse IL-17a (eBio17B7) were purchased from eBioscience. Anti-human IL-9 (MH9A), anti-mouse IL-9 (RM9A4), anti-human CD45 (2D1), anti-human HLA-DR (L243), anti-human CD45RO, and anti-human CD4 (A161A1) were purchased from BioLegend. Anti-human CD45RA (clone HI100; 555489) was purchased from BD Pharmingen.
Hamster monoclonal anti-CD3ε (145-2C11; BE0001-1), hamster monoclonal anti-CD28 (27.51; BE0015-1), InVivoPlus anti-mouse IFNγ (BP0055), InVivoPlus anti-mouse IL-4 (BP0045), InVivoPlus anti-mouse CD4 (BP0003-1), InVivoPlus anti-mouse PD-1 (BP0033-2), InVivoPlus anti-mouse TGFβ (BE0057), and InVivoPlus anti-mouse IL-9 (BE0181) were purchased from Bio X Cell. FastStart Universal SYBR Green Master Mix (4913914001) was from Roche. Proteinase K (A300491) and DAPI dihydrochloride (A606584) were from Sangon Biotech. Klenow fragment DNA polymerase I (2140A) and the PrimeScript RT reagent kit (RR037A) were from Takara. The LIVE/DEAD Fixable Violet Dead Cell Stain Kit (L34963), the CellTrace CFSE Cell Proliferation Kit (L34955), TRIzol reagent (15596018), RNase A (8003089), and Lipofectamine 3000 (L3000015) were from Thermo Fisher Scientific. LipoFilter Liposomal Transfection Reagent (HB-LF10001) was from Hanbio. EZ-ChIP ChIP kit (17-371) and Immobilon Western Chemiluminescent HRP Substrate (WBKLS0500) were from Millipore. The dual-luciferase reporter assay system (E1960) and TNT quick coupled transcription/translation system (L1170) for in vitro protein expression were from Promega. UbcH5a/UBE2D1-Ub charged (human recombinant; E2-800) for in vitro ubiquitination assay was from Boston Biochem. The ClonExpress II one-step cloning kit (C112) and AxyPrep PCR cleanup kit (AP-PCR-250) were from Vazyme and Axygen, respectively. FBS (10270), 2-mercaptoethanol (21985023), penicillin-streptomycin (15140-122), and GlutaMAX Supplement (35050-061) were from Gibco. DMEM/high glucose (SH30243.01) and RPMI medium modified (SH30809.01) were from HyClone. Protease inhibitor cocktail (B14001) and phosphatase inhibitor cocktail (B15001) were from Bimake. N-ethylmaleimide (E3876) was from Sigma-Aldrich. Recombinant mouse IL-4 (CK74) and human IL-4 (CD03) were from Novoprotein. Recombinant human TGFβ1 (240-B-002), recombinant mouse IL-2 (402-ML-020), recombinant mouse IL-23 (1887-ML), and recombinant mouse IL-6 (406-ML-005) were from R&D Systems. Recombinant mouse IL-12 (210-12) and IL-1β (210-11B) proteins were from PeproTech. SIS3 HCL (S7959) was purchased from Selleck.
Cell line culture
Mouse melanoma cell lines B16-F10 and B16-OVA, colon adenocarcinoma cell line MC-38, human embryonic kidney HEK293T cells, and Plat-E cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. Human Jurkat T cells were cultured with RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were routinely tested for mycoplasma contamination using the Mycoplasma Detection Kit and were found to be negative.
Cytokine measurements
Cytokine levels were determined using a single-plex sandwich ELISA (Senxiong Biotech). The assay was performed according to the manufacturer’s instructions.
T cell culture and differentiation
Primary T cells were isolated from the spleen and lymph nodes of the 6–8-wk-old mice. Naive CD4+CD44loCD62Lhi T cells were purified by flow cytometry from enriched CD4+ T cells via negative selection with the MojoSort Mouse CD4+ T Cell Isolation Kit (480033; BioLegend) according to the manufacturer’s instructions. The sorted naive CD4+ T cells were stimulated with plate-bound anti-CD3 (5 µg ml−1) and anti-CD28 (5 µg ml−1) antibodies and polarized into effector CD4+ T lymphocyte subsets with anti–IFN-γ (10 µg ml−1) and anti–IL-4 (10 µg ml−1) for Th0 cells; or with IL-12 (20 ng ml−1) and anti–IL-4 (10 µg ml−1) for Th1 cells; or with IL-4 (20 ng ml−1) and anti–IFN-γ (10 µg ml−1) for Th2 cells; or with TGFβ1 (1, 2, or 10 ng ml−1), IL-4 (1, 5, or 20 ng ml−1), and anti–IFN-γ (10 µg ml−1) for Th9 cells; or with TGFβ1 (2 ng ml−1), IL-6 (25 ng ml−1), anti–IFN-γ (10 µg ml−1), and anti–IL-4 (10 µg ml−1) for Th17 cells; or with IL-1β (10 ng ml−1), IL-23 (20 ng ml−1), and IL-6 (10 ng ml−1) for Th17 cells; or with TGFβ1 (2 ng ml−1), anti–IFN-γ (10 µg ml−1), and anti–IL-4 (10 µg ml−1) for Treg cells. Cells were classically harvested on day 3 in T cell medium (RPMI 1640, 10% FBS, 1× antibiotics, 1× nonessential amino acids, and 50 µM β-mercaptoethanol), and cytokine expression was detected by flow cytometry, ELISA, or real-time PCR.
For in vitro human Th9 cell differentiation, CD4+ T cells were isolated by flow cytometry from the peripheral blood mononuclear cells of different donors with the MojoSort Human CD4+ T Cell Isolation Kit (480010; BioLegend), stimulated with plate-bound anti-CD3 (5 µg ml−1; Bio X Cell) and anti-CD28 (5 µg ml−1; Bio X Cell) antibodies, and polarized into Th9 cells with human TGFβ1 (10 ng ml−1) and IL-4 (20 ng ml−1).
Lymphocyte staining and flow cytometry
For cell surface staining, cells were washed with staining buffer (2% FBS in PBS) and incubated with the indicated antibodies on ice for 30 min. Cells were washed two more times with staining buffer before being analyzed with a Beckman Coulter Gallios machine.
For intracellular cytokine staining, cells were stimulated for 4 h at 37°C with PMA (100 ng ml−1), ionomycin (500 ng ml−1), and GolgiPlug (1:1,000 dilution; BD Pharmingen), followed by staining with fixation/permeabilization buffer solution according to the manufacturer’s protocol (BD Biosciences). Intranuclear staining was performed with fixation/permeabilization buffer solution according to the manufacturer’s protocol (eBioscience). Stained cells were analyzed on a Beckman Coulter Gallios machine.
To determine cell viability, cells were stained with annexin V and propidium iodide (Annexin V Apoptosis Detection Kit I; BD Biosciences), and annexin V–negative cells were determined as viable cells. To analyze cell proliferation, naive CD4+ T cells were purified and labeled with CellTrace Violet reagent (Life Technologies) and cultured under Th9 conditions; 3 d later, analysis was completed by using the Gallios machine.
Gene knockdown or overexpression
For gene knockdown in human CD4+ T cells, siRNAs were transfected into human CD4+ T cells by using TransIT-TKO (Mirus Bio) according to the manufacturer’s instructions. 24 h after transfection, the CD4+ T cells were stimulated with plate-bound anti-CD3 and anti-CD28 antibodies, differentiated into Th9 cells as described above, and cultured for an additional 72 h before real-time qPCR and flow cytometric analysis. The siRNA sequences are listed in Table S2.
For BFAR overexpression in human CD4+ T cells, lentivirus was generated by transfecting HEK293T cells with pCDH-MCS-copGFP-HA-BFAR, pMD2, and psPAX2 plasmids. The 1.75 ml of lentiviral supernatant plus 0.75 ml of fresh complete RPMI medium were used to resuspend 106 human CD4+ T cells in 6-well plates, then spin-infected for 45 min at 1,800 g in the presence of polybrene (8 µg ml−1) to increase infection efficiency. The infected cells were cultured at 37°C for an additional 2–6 h before being resuspended in the indicated human Th9 cell differentiation medium.
For BFAR overexpression in mouse CD4+ T cell infection, naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 for ∼18 h and then infected with the supernatant containing fresh retrovirus produced by transfecting Plat-E cells with the indicated plasmids. The infected T cells were spun for 1.5 h at 1,800 g in the presence of polybrene (8 µg ml−1), and were cultured at 37°C for an additional 2–6 h before being resuspended in the mouse Th9 cell differentiation medium.
Murine tumor models
For induction of the tumor model, age- and sex-matched Rag1−/− mice or C57BL/6 mice (6–8 wk old) received a s.c. abdominal injection with 106 B16-F10 or MC-38 tumor cells. Tumor volumes were measured every 2 d with a caliper and calculated by using the equation: V = (minor tumor axis)2 × (major tumor axis) × π/6. To analyze the phenotype of tumor-infiltrating immune cells, mice were euthanized on days 15–20 after tumor inoculation. Tumors were dissected and weighed, and the infiltrated immune cells were collected for flow cytometric analysis. The tumor-infiltrating CD4+ T cells were sorted from the collected immune cells by flow cytometry and were examined for gene expression by qPCR analysis. The peripheral blood of the tumor-bearing mice was collected for cytokine measurement by ELISA or flow cytometric analysis. For the induction of metastatic B16-OVA lung melanoma, C57BL/6 mice were injected i.v. with 105 B16-OVA tumor cells and were sacrificed for enumeration of metastatic lung foci at day 18 after tumor cell injection.
Adoptive Th9 cell transfer therapy
For the B16-F10 s.c. tumor model, Rag1−/− or C57BL/6 mice received an i.v. injection of 5 × 106 Th9 cells with the indicated phenotype on the same day as tumor cell inoculation. For antibody neutralization in vivo, mice were i.v. injected with anti–IL-9 (100 µg/mouse, BE0181; Bio X Cell) or anti-TGFβ (100 µg/mouse, BE0057; Bio X Cell) or anti–PD-1 (100 µg/mouse, BP0033-2; Bio X Cell) or anti-CD4 (100 µg/mouse, BP0003-1; Bio X Cell) or control mouse IgG2a at the indicated time points. For the B16-OVA lung melanoma model, C57BL/6 mice were i.v. injected with 5 × 106 OVA-specific OT-II–Th9 cells with the indicated phenotype 1 d after tumor cell inoculation.
To evaluate the therapeutic effects of antigen-specific Th9 cells against established tumors, mice received a s.c. injection with 106 B16-OVA tumor cells. At day 10 after tumor injection, mice were adoptively transferred with 2.5 × 106 OVA-specific OT-II–Th9 cells, followed by i.v. injection of 2.5 × 105 OVA peptide-pulsed bone marrow–derived DCs. Cyclophosphamide (Sigma-Aldrich) was administered i.p. as a single dose at 200 mg kg−1 1 d before Th9 transfer as previously described (Lu et al., 2018). For examining the therapeutic effects of IL-9–producing CD4+ T cells, Rag1−/− mice received i.v. injection of 5 × 105 GFP+ cells, which were sorted from control or BFAR-overexpressed Th9 cells that differentiated from IL-9–EGFP CD4+ T cells on the same day as tumor cell inoculation.
For the treatment of the PDX model, immunodeficient NCG mice that received implants of human CRC tissues were purchased from GemPharmatech. 10 d later, the NCG mice were i.v. injected with in vitro matured CRC patient–derived DCs (105 cells/mouse) alone or with in vitro differentiated CRC patient–derived control or BFAR-overexpressed Th9 cells (5 × 106 cells/mouse). At day 27 after tumor implantation, tumors were dissected and weighed, and the infiltrated immune cells were collected for flow cytometric analysis or sorted by flow cytometry for qPCR analysis. The peripheral blood was collected for cytokine measurement through ELISA analysis.
Real-time qPCR
Total RNA was extracted using TRIzol reagent according to the manufacturer’s protocol. The cDNA was synthesized using the PrimeScript RT Reagent Kit (Takara). qPCR was performed in triplicate with SYBR Green Master Mix (Roche). The relative expression of genes was calculated by a standard curve method and normalized to the expression level of Actb. Gene-specific PCR primers are listed in Table S3.
RNA-sequencing analysis
WT and BFAR-deficient naive CD4+ T cells cultured under Th9-polarizing conditions for 72 h were applied for total RNA extraction with TRIzol (Invitrogen) and subjected to RNA-sequencing analysis. RNA sequencing was performed by BGI Tech Solutions. The raw transcriptomic reads were mapped to a reference genome (GRCm38/mm10) using Bowtie. Gene expression levels were quantified using the RSEM software package. Significantly affected genes were acquired by setting a fold change >1.5 and a false discovery rate threshold of 0.05. Differentially expressed genes were analyzed using the Ingenuity Pathway Analysis and DAVID bioinformatics platforms.
Luciferase reporter assay
The human BFAR conserved promoter and several exon regions (positions −920 to −160, positions −70 to +657, and positions +616 to +1,692) were amplified from genomic DNA by PCR, then cloned into the pGL4 basic luciferase reporter gene vector. BFAR promoter activity was assessed by a dual-luciferase assay system according to the manufacturer’s instructions (Promega). All samples were normalized for transfection efficiency by division of firefly luciferase activity by Renilla luciferase activity.
MS
For the identification of TGFβR1-binding E3 ligases or deubiquitinases, Flag-TGFβR1 expression plasmids were transfected into HEK293T cells. Cells were harvested 48 h after transfection, and the lysates were precleared with protein A/G–coupled agarose beads and then incubated with anti-Flag on a shaker at 4°C overnight. On the next day, the immunoprecipitated proteins were collected by incubation with protein A/G–coupled agarose beads on a shaker at 4°C for 4 h, then beads were washed with cell lysis buffer, eluted with glycine-Tris-HCl buffer (pH 2.5), and sent for processing with MS analysis of protein interactions in the National Facility for Protein Science in Shanghai, Zhangjiang Lab.
For the identification of TGFβR1 ubiquitination sites, Flag-TGFβR1, HA-Ub, and HA-BFAR expression plasmids were cotransfected into HEK293T cells. Cells were harvested 48 h after transfection, and the lysates were immunoprecipitated with anti-Flag antibody. After washing, the eluted samples were resolved with SDS-PAGE, followed by Coomassie Brilliant Blue staining. The sample of the TGFβR1 band was cut and sent for processing with MS analysis using a QE1 system at the National Facility for Protein Science in Shanghai, Zhangjiang Lab.
Immunoblot and ubiquitination assay
Jurkat T cells and CD4+ T cells were left unstimulated or stimulated for appropriate times by anti-CD3 plus anti-CD28 or by IL-4 or TGFβ1, and total or subcellular extracts were prepared for immunoprecipitation or immunoblot analysis with specific antibodies. The immunoblot intensity was quantified using ImageJ.
For the in vivo ubiquitination assay, CD4+ T cells that were left unstimulated or which were stimulated with anti-CD3 plus anti-CD28 or 293T cells that transfected with the desired plasmids were lysed with cell lysis radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA) containing protease inhibitor and N-ethylmaleimide. After saving some cell extracts for input analysis, the remaining cell extracts were added to SDS to a final concentration of 1% and then were boiled at 100°C for 5 min, which will dissociate TGFβR1-interacting proteins under such denaturing conditions. The boiled cell extracts were diluted with RIPA buffer until the SDS concentration was 0.1%, precleaned with protein A/B–coupled agarose beads, then incubated with specific immunoprecipitation antibody on a shaker under denaturing conditions (0.1% SDS) at 4°C overnight. The next day, the immunoprecipitated proteins were collected by incubation with protein A/B–coupled agarose beads on a shaker at 4°C for 2 h; washed with RIPA buffer containing protease inhibitors, PMSF, and N-ethylmaleimide; boiled at 100°C for 5 min; and then loaded to run SDS-PAGE. The immunoprecipitates were immunoblotted with anti-Ub or the indicated antibodies.
For the in vitro ubiquitination assay, TGFβR1 and BFAR proteins were expressed in vitro with the TNT Quick Coupled Transcription/Translation System (Promega). In vitro ubiquitination reactions were performed with a ubiquitination kit (Boston Biochem) according to the manufacturer’s instructions.
ChIP-qPCR assay
The ChIP assay procedure was modified from the manufacturer’s instructions (EZ-ChIP; EMD Millipore). Briefly, human CD4+ T cells (107 cells) were fixed with 1% formaldehyde (Sigma-Aldrich) at room temperature for 10 min in 10 ml medium, followed by quenching with 125 mM glycine. Nuclear extracts were sonicated with the Covaris E220 sonicator for 660 s. After preclearing with normal IgG for 1 h, the sonicated cell lysates were immunoprecipitated with the SMAD2/3 antibody overnight on a nutator at 4°C. On the next day, protein A/G magnetic beads were added, and cell lysates were incubated on a nutator for another 2 h. After washing with buffers, chromatin was eluted from the protein–DNA complex and digested with proteinase K and RNase A at 65°C overnight to reverse cross-links. The freed DNA was purified with the AxyPrep PCR cleanup kit (Axygen) and subjected to qPCR analysis using SYBR Green Master Mix. All the sequences of the primers used for ChIP-qPCR are shown in Table S1.
Statistical analysis
The data are shown as mean ± SEM, and, unless otherwise indicated, all the presented data are representative results of at least two or three independent repeats. Statistical analyses were performed using GraphPad Prism 7, and the statistics were analyzed by two-tailed Student’s t test or one-way or two-way ANOVA as indicated. Differences were considered to be significant at P ≤ 0.05 and are indicated by ∗; those at P ≤ 0.01 are indicated by ∗∗; and those at P ≤ 0.001 are indicated by ∗∗∗.
Online supplemental material
Fig. S1 shows that IL-9 neutralization at the late stage abolishes the enhanced therapeutic efficacy caused by TGFβ blocking and that activated NF-κB, STAT proteins, surface expression of TGFβR1/2 in Th9 cells, and the percentage of naive/memory CD4+ T cells are comparable between HDs and GIC patients. Fig. S2 shows that BFAR is dispensable for T cell homeostasis and proliferation but positively regulates Th9 differentiation through modulating TGFβR1 and downstream signaling. Fig. S3 shows that BFAR is critical for Th9 differentiation without influencing T cell proliferation and apoptosis. Fig. S4 shows that BFAR deletion could not affect Th17 and Treg cell differentiation. Fig. S5 shows that TGFβR1 ubiquitination at K268 is critical for Th9-mediated cancer immunotherapy without affecting the surface TGFβR1 expression. Table S1 lists TGFβR1-interacting E3 ligases and deubiquitinase proteins identified by MS. Table S2 lists sequences of siRNA for specific gene knockdown. Table S3 lists primers used for real-time qPCR.
Data availability
The RNA-sequencing data have been deposited in the Gene Expression Omnibus (accession no. GSE153133). All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
lists TGFβR1-interacting E3 ligase and deubiquitinase proteins identified by MS.
lists sequences of siRNA for specific gene knockdown.
lists primers used for real-time qPCR.
Acknowledgments
We thank Dr. Long Zhang (Zhejiang University, China), Dr. Feng Yu (Jiangsu University, China), and Dr. Xing Chang (Westlake University, China) for the generous gift of plasmids and cell lines.
This research was supported by the grants from the National Key R&D Program of China (2018YFA0902703, 2018YFA0107201), the National Natural Science Foundation of China (82030041, 81770567, 81972176), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB39030300), the Program of Shanghai Academic/Technology Research Leader (20XD1424600), the Key Research Program of the Chinese Academy of Sciences (KFZD-SW-216), the Zhejiang Provincial Medical and Health Science Foundation (2021441200, 2020380312), and the Chinese Academy of Sciences Key Laboratory of Tissue Microenvironment and Tumor.
Author contributions: S. Pei designed and performed the experiments, prepared the figures, and wrote part of the manuscript; M. Huang, J. Huang, X. Zhu, H. Wang, K. Shen, Y. Luo, Q. Luo, Z. Wen, and D. Dai contributed to human sample collection; S. Romano, X. Deng, Y. Wang, S. Hao, J. Xu, T. Yu, Q. Zhu, J. Yuan, Z. Liu, and G. Hu contributed to the experiments; C. Peng contributed to mass spectrum analysis; Y. Xiao initiated, designed, and supervised this study, prepared the figures, and wrote the manuscript.
References
- Achyut, B.R., and Yang L.. 2011. Transforming growth factor-β in the gastrointestinal and hepatic tumor microenvironment. Gastroenterology. 141:1167–1178. 10.1053/j.gastro.2011.07.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Salihi, M.A., Herhaus L., Macartney T., and Sapkota G.P.. 2012. USP11 augments TGFβ signalling by deubiquitylating ALK5. Open Biol. 2:120063. 10.1098/rsob.120063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga, C.L. 2006. Inhibition of TGFβ signaling in cancer therapy. Curr. Opin. Genet. Dev. 16:30–37. 10.1016/j.gde.2005.12.009 [DOI] [PubMed] [Google Scholar]
- Batlle, E., and Massagué J.. 2019. Transforming growth factor-β signaling in immunity and cancer. Immunity. 50:924–940. 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S., Guix M., Rinehart C., Dugger T.C., Chytil A., Moses H.L., Freeman M.L., and Arteaga C.L.. 2017. Inhibition of TGF-β with neutralizing antibodies prevents radiation-induced acceleration of metastatic cancer progression. J. Clin. Invest. 127:1116. 10.1172/JCI93333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna, A., Darken R.S., Rojo F., Ocaña A., Peñuelas S., Arias A., Paris R., Tortosa A., Mora J., Baselga J., et al. 2007. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 11:147–160. 10.1016/j.ccr.2006.11.023 [DOI] [PubMed] [Google Scholar]
- Budi, E.H., Duan D., and Derynck R.. 2017. Transforming growth factor-β receptors and Smads: regulatory complexity and functional versatility. Trends Cell Biol. 27:658–672. 10.1016/j.tcb.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Calon, A., Espinet E., Palomo-Ponce S., Tauriello D.V., Iglesias M., Céspedes M.V., Sevillano M., Nadal C., Jung P., Zhang X.H., et al. 2012. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 22:571–584. 10.1016/j.ccr.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon, A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M., Sevillano M., Palomo-Ponce S., Tauriello D.V., Byrom D., et al. 2015. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 47:320–329. 10.1038/ng.3225 [DOI] [PubMed] [Google Scholar]
- Capasso, A., Lang J., Pitts T.M., Jordan K.R., Lieu C.H., Davis S.L., Diamond J.R., Kopetz S., Barbee J., Peterson J., et al. 2019. Characterization of immune responses to anti-PD-1 mono and combination immunotherapy in hematopoietic humanized mice implanted with tumor xenografts. J. Immunother. Cancer. 7:37. 10.1186/s40425-019-0518-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, S.R., Singhal P.G., Sharma U., Bandil K., Chakraborty K., and Bharadwaj M.. 2019. Th9 cytokines curb cervical cancer progression and immune evasion. Hum. Immunol. 80:1020–1025. 10.1016/j.humimm.2019.09.009 [DOI] [PubMed] [Google Scholar]
- Dardalhon, V., Awasthi A., Kwon H., Galileos G., Gao W., Sobel R.A., Mitsdoerffer M., Strom T.B., Elyaman W., Ho I.C., et al. 2008. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+ IL-10+ Foxp3− effector T cells. Nat. Immunol. 9:1347–1355. 10.1038/ni.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvenne, P., Hubert P., and Jacobs N.. 2004. Epithelial metaplasia: an inadequate environment for antitumour immunity? Trends Immunol. 25:169–173. 10.1016/j.it.2004.02.002 [DOI] [PubMed] [Google Scholar]
- Eichhorn, P.J., Rodón L., Gonzàlez-Juncà A., Dirac A., Gili M., Martínez-Sáez E., Aura C., Barba I., Peg V., Prat A., et al. 2012. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med. 18:429–435. 10.1038/nm.2619 [DOI] [PubMed] [Google Scholar]
- Elyaman, W., Bassil R., Bradshaw E.M., Orent W., Lahoud Y., Zhu B., Radtke F., Yagita H., and Khoury S.J.. 2012. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity. 36:623–634. 10.1016/j.immuni.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, G., Xu Q., Qiu Y., Jin X., Xu T., Dong S., Wang J., Ke Y., Hu H., Cao X., et al. 2017. Suppression of Th17 cell differentiation by misshapen/NIK-related kinase MINK1. J. Exp. Med. 214:1453–1469. 10.1084/jem.20161120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, R., Jabeen R., Yagi R., Pham D., Zhu J., Goenka S., and Kaplan M.H.. 2012. STAT6-dependent regulation of Th9 development. J. Immunol. 188:968–975. 10.4049/jimmunol.1102840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto, T., Miyake K., Nandi T., Yashiro M., Onishi N., Huang K.K., Lin S.J., Kalpana R., Tay S.T., Suzuki Y., et al. 2017. Activation of transforming growth factor β1 signaling in gastric cancer-associated fibroblasts increases their motility, via expression of rhomboid 5 homolog 2, and ability to induce invasiveness of gastric cancer cells. Gastroenterology. 153:191–204.e16. 10.1053/j.gastro.2017.03.046 [DOI] [PubMed] [Google Scholar]
- Jiao, S., Subudhi S.K., Aparicio A., Ge Z., Guan B., Miura Y., and Sharma P.. 2019. Differences in tumor microenvironment dictate T helper lineage polarization and response to immune checkpoint therapy. Cell. 179:1177–1190.e13. 10.1016/j.cell.2019.10.029 [DOI] [PubMed] [Google Scholar]
- Kaplan, M.H. 2017. The transcription factor network in Th9 cells. Semin. Immunopathol. 39:11–20. 10.1007/s00281-016-0600-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavsak, P., Rasmussen R.K., Causing C.G., Bonni S., Zhu H., Thomsen G.H., and Wrana J.L.. 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol. Cell. 6:1365–1375. 10.1016/S1097-2765(00)00134-9 [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., and Baek K.H.. 2019. TGF-β signaling pathway mediated by deubiquitinating enzymes. Cell. Mol. Life Sci. 76:653–665. 10.1007/s00018-018-2949-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Hong S., Li H., Park J., Hong B., Wang L., Zheng Y., Liu Z., Xu J., He J., et al. 2012. Th9 cells promote antitumor immune responses in vivo. J. Clin. Invest. 122:4160–4171. 10.1172/JCI65459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Wang Q., Xue G., Bi E., Ma X., Wang A., Qian J., Dong C., and Yi Q.. 2018. Th9 cells represent a unique subset of CD4+ T cells endowed with the ability to eradicate advanced tumors. Cancer Cell. 33:1048–1060.e7. 10.1016/j.ccell.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra, N., Robertson E., and Kang J.. 2010. SMAD2 is essential for TGFβ-mediated Th17 cell generation. J. Biol. Chem. 285:29044–29048. 10.1074/jbc.C110.156745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan, S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., Kadel E.E. III, Koeppen H., Astarita J.L., Cubas R., et al. 2018. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 554:544–548. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué, J. 2012. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13:616–630. 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa, H., Zhang D., Maruyama T., Chen H., Cui K., Ishikawa M., Deng L., Zanvit P., Tu E., Jin W., et al. 2015. The DNA-binding inhibitor Id3 regulates IL-9 production in CD4+ T cells. Nat. Immunol. 16:1077–1084. 10.1038/ni.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak, K., and Weinberg R.A.. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer. 9:265–273. 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- Purwar, R., Schlapbach C., Xiao S., Kang H.S., Elyaman W., Jiang X., Jetten A.M., Khoury S.J., Fuhlbrigge R.C., Kuchroo V.K., et al. 2012. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat. Med. 18:1248–1253. 10.1038/nm.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera Vargas, T., Cai Z., Shen Y., Dosset M., Benoit-Lizon I., Martin T., Roussey A., Flavell R.A., Ghiringhelli F., and Apetoh L.. 2017a. Selective degradation of PU.1 during autophagy represses the differentiation and antitumour activity of TH9 cells. Nat. Commun. 8:559. 10.1038/s41467-017-00468-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera Vargas, T., Humblin E., Végran F., Ghiringhelli F., and Apetoh L.. 2017b. TH9 cells in anti-tumor immunity. Semin. Immunopathol. 39:39–46. 10.1007/s00281-016-0599-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, J., Morgani S.M., David C.J., Wang Q., Er E.E., Huang Y.H., Basnet H., Zou Y., Shu W., Soni R.K., et al. 2020. TGF-β orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature. 577:566–571. 10.1038/s41586-019-1897-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, C., Murakami G., Fukuchi M., Shimanuki T., Shikauchi Y., Imamura T., and Miyazono K.. 2002. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 277:39919–39925. 10.1074/jbc.M201901200 [DOI] [PubMed] [Google Scholar]
- Tamiya, T., Ichiyama K., Kotani H., Fukaya T., Sekiya T., Shichita T., Honma K., Yui K., Matsuyama T., Nakao T., et al. 2013. Smad2/3 and IRF4 play a cooperative role in IL-9-producing T cell induction. J. Immunol. 191:2360–2371. 10.4049/jimmunol.1301276 [DOI] [PubMed] [Google Scholar]
- Tauriello, D.V.F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., Sevillano M., Ibiza S., Cañellas A., Hernando-Momblona X., et al. 2018. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 554:538–543. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- Tone, Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., and Tone M.. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202. 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Travis, M.A., and Sheppard D.. 2014. TGF-β activation and function in immunity. Annu. Rev. Immunol. 32:51–82. 10.1146/annurev-immunol-032713-120257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen, M., Uyttenhove C., van Snick J., Helmby H., Westendorf A., Buer J., Martin B., Wilhelm C., and Stockinger B.. 2008. Transforming growth factor-β ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 9:1341–1346. 10.1038/ni.1659 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Bi Y., Chen X., Li C., Li Y., Zhang Z., Wang J., Lu Y., Yu Q., Su H., et al. 2016. Histone deacetylase SIRT1 negatively regulates the differentiation of interleukin-9-producing CD4+ T cells. Immunity. 44:1337–1349. 10.1016/j.immuni.2016.05.009 [DOI] [PubMed] [Google Scholar]
- Wicks, S.J., Haros K., Maillard M., Song L., Cohen R.E., Dijke P.T., and Chantry A.. 2005. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-β signalling. Oncogene. 24:8080–8084. 10.1038/sj.onc.1208944 [DOI] [PubMed] [Google Scholar]
- Wu, X., and Karin M.. 2015. Emerging roles of Lys63-linked polyubiquitylation in immune responses. Immunol. Rev. 266:161–174. 10.1111/imr.12310 [DOI] [PubMed] [Google Scholar]
- Xie, Y., Avello M., Schirle M., McWhinnie E., Feng Y., Bric-Furlong E., Wilson C., Nathans R., Zhang J., Kirschner M.W., et al. 2013. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J. Biol. Chem. 288:2976–2985. 10.1074/jbc.M112.430066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J.H., Sudo K., Kuroda M., Kato M., Lee I.K., Han J.S., Nakae S., Imamura T., Kim J., Ju J.H., et al. 2015. Phosphorylation status determines the opposing functions of Smad2/Smad3 as STAT3 cofactors in TH17 differentiation. Nat. Commun. 6:7600. 10.1038/ncomms8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T., Gan S., Zhu Q., Dai D., Li N., Wang H., Chen X., Hou D., Wang Y., Pan Q., et al. 2019. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat. Commun. 10:4353. 10.1038/s41467-019-12384-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg, M., Hanai J., Sugimoto H., Mammoto T., Charytan D., Strutz F., and Kalluri R.. 2003. BMP-7 counteracts TGF-β1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat. Med. 9:964–968. 10.1038/nm888 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Zhou F., Drabsch Y., Gao R., Snaar-Jagalska B.E., Mickanin C., Huang H., Sheppard K.A., Porter J.A., Lu C.X., et al. 2012. USP4 is regulated by AKT phosphorylation and directly deubiquitylates TGF-β type I receptor. Nat. Cell Biol. 14:717–726. 10.1038/ncb2522 [DOI] [PubMed] [Google Scholar]
- Zhu, Q., Yu T., Gan S., Wang Y., Pei Y., Zhao Q., Pei S., Hao S., Yuan J., Xu J., et al. 2020. TRIM24 facilitates antiviral immunity through mediating K63-linked TRAF3 ubiquitination. J. Exp. Med. 217:e20192083. 10.1084/jem.20192083 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists TGFβR1-interacting E3 ligase and deubiquitinase proteins identified by MS.
lists sequences of siRNA for specific gene knockdown.
lists primers used for real-time qPCR.
Data Availability Statement
The RNA-sequencing data have been deposited in the Gene Expression Omnibus (accession no. GSE153133). All other data supporting the findings of this study are available from the corresponding authors on reasonable request.