Mycobacterium avium subsp. paratuberculosis (MAP) is the causative agent of Johne's disease in ruminants, which has important health consequences for dairy cattle and enormous economic consequences for the dairy industry. Understanding which genes in this bacterium are correlated with key disease phenotypes can lead to functional experiments targeting these genes and ultimately lead to improved control strategies.
KEYWORDS: bacterial pan-GWAS, Mycobacterium avium subsp. paratuberculosis, Johne’s disease, population genomics, MAP, evolutionary genomics, pan-GWAS
ABSTRACT
Mycobacterium avium subsp. paratuberculosis (MAP) is the causative agent of Johne's disease in ruminants, which has important health consequences for dairy cattle. The Regional Dairy Quality Management Alliance (RDQMA) project is a multistate research program involving MAP isolates taken from three intensively studied commercial dairy farms in the northeastern United States, which emphasized longitudinal data collection of both MAP isolates and animal health in three regional dairy herds for a period of about 7 years. This paper reports the results of a pan-GWAS analysis involving 318 MAP isolates and dairy cow Johne’s disease phenotypes, taken from these three farms. Based on our highly curated accessory gene count, the pan-GWAS analysis identified several MAP genes associated with bovine Johne’s disease phenotypes scored from these three farms, with some of the genes having functions suggestive of possible cause/effect relationships with these phenotypes. This paper reports a pangenomic comparative analysis between MAP and Mycobacterium tuberculosis, assessing functional Gene Ontology category enrichments between these taxa. Finally, we also provide a population genomic perspective on the effectiveness of herd isolation, involving closed dairy farms, in preventing MAP interfarm cross infection on a microgeographic scale.
IMPORTANCE Mycobacterium avium subsp. paratuberculosis (MAP) is the causative agent of Johne's disease in ruminants, which has important health consequences for dairy cattle and enormous economic consequences for the dairy industry. Understanding which genes in this bacterium are correlated with key disease phenotypes can lead to functional experiments targeting these genes and ultimately lead to improved control strategies. This study represents a rare example of a prolonged longitudinal study of dairy cattle where the disease was measured and the bacteria were isolated from the same cows. The genome sequences of over 300 MAP isolates were analyzed for genes that were correlated with a wide range of Johne’s disease phenotypes. A number of genes were identified that were significantly associated with several aspects of the disease and suggestive of further experimental follow-up.
INTRODUCTION
Mycobacterium avium subsp. paratuberculosis (MAP) is the causative agent of Johne's disease in ruminants, a chronic granulomatous enteritis with an incubation period of several years (1, 2), and has important economic and health consequences for dairy and beef cattle, sheep, goats, and farmed deer throughout the world (3–5). Its greatest economic impact is in dairy cattle, with decreased milk production, weight loss, and premature culling costing the U.S. dairy industry approximately $250 million annually (6). MAP-containing milk may be of particular concern because the bacterium has been suggested as a cause of Crohn's disease in humans (7).
The Regional Dairy Quality Management Alliance (RDQMA) project is a multistate research program in collaboration with the USDA Agricultural Research Service (ARS), Cornell University, Pennsylvania State University, University of Pennsylvania, and University of Vermont. As part of this project, several epidemiological studies involving MAP isolates taken from three intensively studied commercial dairy farms in the northeastern United States have been conducted: farm A in New York State, farm B in Pennsylvania, and farm C in Vermont (8, 9). The project emphasized longitudinal data collection of both MAP isolates and animal health in these three dairy herds for a period of about 7 years. MLSSR (multilocus short-sequence-repeat) genotyping of a sample of isolates taken from these farms has been undertaken (8, 9) in an attempt to address various epidemiological questions.
The first genome sequence for MAP, strain K-10, was characterized in 2005 by Li et al. (10). There were 27 annotated genome sequences of MAP isolates from bovine publicly available on NCBI at the time of writing, with another 23 taken from several other host species. Comparative genomics studies have been conducted involving MAP from cattle compared to MAP from a number of different hosts, including sheep (11), camel (12), bison (13), human (14, 15), and other domestic and wild animals (16). In addition to these comparative genomic studies, phylogeographic analyses have also been conducted using whole-genome sequencing (WGS) data from Canadian isolates (17) and more globally (18). No pangenomic (the entire gene repertoire of a species including the core plus the accessory genome) study of MAP and/or pangenomic comparison to any other Mycobacterium species have been undertaken, and no comparative or population genomic study of MAP strains taken from these RDQMA farms have been undertaken.
Population genetics of MAP has been investigated at various geographic levels, involving an assortment of genetic techniques (17, 19–21). One of the more recent studies involved WGS data at an interprovincial and intraprovincial level in Canada (17). This study rejected the hypothesis of interprovincial panmixis but not so for intraprovincial data. Despite rejecting the strict hypothesis of panmixis, they did, however, find that most major clades were found in all provinces, suggesting a good deal of genotype mixing. The proposed explanation for this is that cattle movement is a major driver of MAP transmission at the herd level, further supported by the distinct lack of clustering within the more microgeographic area of southern Alberta. One of the main difficulties in evaluating the role of cattle movement in spreading MAP is the availability of detailed records on this subject. Sohal et al. (21), in a study of MAP in Quebec dairy herds using PCR, targeted interspersed repetitive units/variable-number tandem repeats (MIRU-VNTR) and found evidence for interherd genetic exchange, suggesting the explanation lies with a variety of possibilities, ranging from environmental means of transmission to interherd cattle movement.
Closing farms, indeed quarantining farms, is a strategy that is used to contain the spread of Johne’s disease (22) and is widely regarded as one of the most effective means of controlling the disease (23), with the tacit assumption that closing the farms would prevent interfarm spread of the bacteria. However, bacterial genetic evidence that would support that tacit assumption is lacking. The reason such a question is relevant is that although MAP is widely regarded as an obligate parasite and the vast majority of pathogen transmission is assumed to occur from animal-to-animal contact, studies have indicated that it can survive in environmental feces anywhere from 16 (24) to 55 weeks (25). Another study has shown experimentally that MAP can survive within freshwater amoebae and that strains can be found on farms within amoebae isolated from the cattle environment (26), suggesting infected amoebae are a reservoir and vector for the transmission of MAP. There is ample evidence for MAP in surface water sources (27–29). Environmental aerosols have even been suggested as a possible exposure source, since viable MAP has been recovered from air samples collected over rivers that drain livestock pastures (30). MAP can survive chlorine disinfection treatment used for treating municipal water sources (31) and has been detected in drinking water systems (32–35). Thus, although the tacit assumption is that herd isolation is an effective means to prevent the spread of the disease, there are reasons for doubt on this as well, and we are not aware of a case where this might have been even partially addressed with the level of precision that WGS data could provide and not on a microgeographic scale. Ideally, a thorough test of this question would involve closed and not closed farms at progressively greater distances, but having any perspective at all on this in terms of nearby farms would be of value.
An important aspect of the RDQMA project is the concomitance of MAP strain isolation, longitudinal MAP infection data, and a precisely documented dairy herd over a period of at least 7 years. The phenotypic data gathered on the RDQMA cows and their health status over these 7 years is considerable and includes such variables as milk production, clinical disease status, MAP enzyme-linked immunosorbent assay (ELISA) optical density (OD) level, fecal shedding, and postmortem feces or tissue infection status. Genes from MAP isolates sampled from these dairy cows could be correlated with these phenotypes and/or their relative severity and, therefore, might represent genes worth exploring for more causal explanations associated with the disease. Genome-wide association studies (GWAS), over the course of the last decade, have resulted in important advances in the understanding of complex traits and have identified hundreds of relevant genetic variants in humans (36, 37). GWAS analysis in bacteria was suggested over a decade ago (38), with discussions over the course of the last few years (39–41) resulting most recently with the development of several methods (42, 43). The purpose of GWAS is to identify statistically significant associations that indicate the presence of a causal relationship between genotype and phenotype, which, in microbes, because of their smaller genome sizes and ability to manipulate some of these genomes in the laboratory, may facilitate the confirmation of candidate loci. Methods for bacteria have been developed that will accommodate single-nucleotide polymorphisms (SNPs) (TreeWAS [43]) or the gene presence/absence characteristics typical of the bacterial accessory genome (pan-GWAS [42]). These new GWAS methods have been used to identify bacterial genetic associations to both subtle (e.g., antibiotic resistance) and more complex phenotypes, such as host association or invasiveness (43). Clearly, host immunity cannot be ignored when considering a comprehensive picture of the more complex phenotypes, but just as clearly, the genetic makeup of the pathogen also cannot be ignored. To our knowledge, employing these bacterial-GWAS approaches in an attempt to identify gene associations with disease phenotypes in dairy cattle, or indeed any agricultural animal, has not been undertaken and represents a useful first step to identifying pathogen-host causal relationships. Longitudinal biannual MAP strain isolation from the RDQMA dairy cattle hosts, concomitant with close documentation of these animals, including numerous and repeated phenotypic measurements over a course of 7 years, provides a unique opportunity to apply bacterial GWAS procedures in an attempt to associate MAP dairy cattle disease phenotypes with MAP gene polymorphisms and gene content.
This paper has several interrelated purposes. First, we provide a population genomic analysis of MAP isolates derived from three isolated, closed dairy farms for the purpose of providing some indication of the effectiveness of herd isolation in preventing MAP interfarm cross infection on a geographic scale of approximately one hundred to several hundred kilometers. Second, we present a pangenomic analysis of MAP along with a comparative pangenomic analysis to Mycobacterium tuberculosis (Mtb), the only other species of Mycobacterium for which there are sufficient publicly available annotated genome sequences to undertake any Mycobacterium interspecific functional characterization and comparison. Finally, we perform a bacterial GWAS analysis that evaluates genes correlated with various disease-related phenotypes of the MAP-infected dairy cattle of the RDQMA project.
RESULTS
Genome statistics and population genomics.
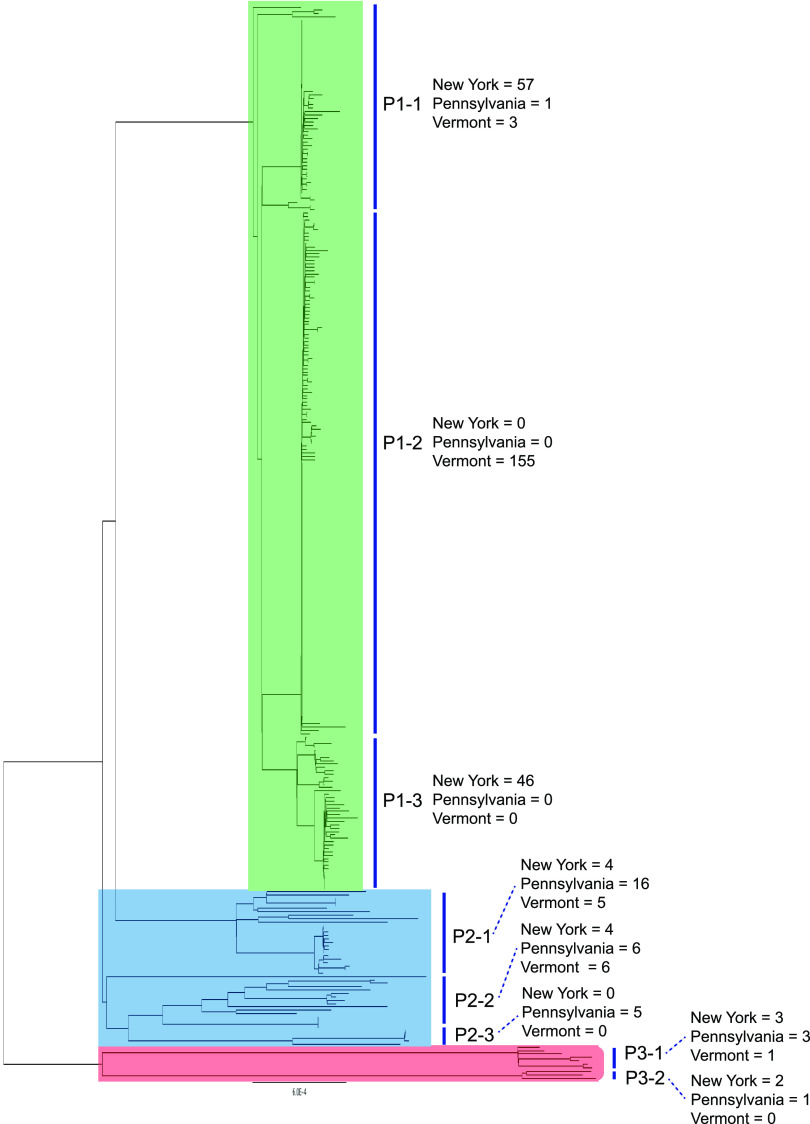
RealPhy produced an alignment that was 4,707,296 bp in length, and Phi detected no evidence of recombination (P = 0.21); from this, 1,418 SNPs were extracted, including 955 transitions (281C→T; T→C145; A→G145; 384G→A) and 464 transversions (C→A57; A→C61; G→T72; T→G42; G→C102; C→G95; A→T18; T→A17; Ts/Tv = 2.06). The resulting phylogenetic tree, based on these SNP data combined with the BAPS and HierBAPS analyses, provided strong evidence for genetically divergent clusters and nested population genetic structure (Fig. 1). The population structure analysis delineated the 318 isolates into three major populations, shaded green (P1), blue (P2), and red (P3) in Fig. 1. These three populations were substructured into three (P1-1, P1-2, and P1-3), three (P2-1, P2-2, and P2-3), and two (P3-1 and P3-2) subpopulations, respectively.
FIG 1.
Maximum likelihood phylogeny showing relationships among isolates. Three major populations are shaded green (P1), blue (P2), and red (P3). Substructuring within each population is shown with black vertical bars.
The vast majority of the Vermont isolates were included in a clade with the shortest branch lengths and comprised a single BAPS group (P1; highlighted in green in Fig. 1). Three nested or subpopulation groups arising from the HierBAPs analysis were evident within this P1 group, including one clade/subpopulation that was comprised exclusively of Vermont isolates and included 91% of all Vermont isolates in our analysis. The other two subpopulations within this P1 group included the majority of the New York isolates (89%), one of which was comprised of exclusively New York isolates (P1 to P3) and the other nearly so. Other clades in the tree had much longer branch lengths. Two of these clades comprised the BAPS-blue P2 group and included the majority of the Pennsylvania isolates, as well as several from each of New York and Vermont and with three subpopulations within. The third BAPS group (P3) (Fig. 1, red) consisted of highly divergent isolates and a mixture from all three states. Pennsylvania isolates on the whole had the greatest diversity.
Pangenomics: core and accessory genomes.
The core, accessory, and pangenome sizes were estimated from the 318 assembled MAP genomes. The homologous gene clustering of Panaroo detected 4,421 core gene clusters and 97 dispensable gene clusters, representing about 2.1% of a typical, complete, closed MAP genome. Roary, with paralog splitting mode off, identified 4,346 core gene clusters and 316 dispensable gene clusters, about 7% of a typical genome size and over three times higher than Panaroo. Roary with paralog splitting mode on (default setting) identified 4,306 core gene clusters and 541 dispensable gene clusters, about 12% of complete genome and over five times higher than the Panaroo finding. To explore the higher Roary dispensable genome estimation, we built a BLAST database that contained nucleotide sequences for all 318 genomes and a complete reference genome (strain MAP4; see Table S1 in the supplemental material). Searches against this database using putative dispensable genes as query sequences frequently revealed them to be core genes. Often, the query sequence had experienced truncation at the end of a sequence contig and/or frameshift.
The core genome consisted of 551 hypotheticals, about 12.5%, and the 97 genes representing the MAP accessory genome consisted of 20 genes annotated as hypotheticals; the vast majority of all hypotheticals were conserved hypotheticals (apparent orthologs across other species of Mycobacterium). Annotated genes in the accessory genome with known or suspected roles in Mycobacterium species pathogenesis and/or host adaptation included (among others) phenolphthiocerol synthesis polyketide synthase type 1 Pks15/1 (involved in fatty acid biosynthesis), KasB (involved in fatty acid biosynthesis), PPE family proteins (possible host endothelial-cell invasion and/or intracellular survival), PE family immunomodulator PE5 (evasion of host immune system), resuscitation-promoting factor RpfA (stimulates resuscitation of dormant cells), MCE family protein (mammalian cell entry protein), and MMPL family transporter (exports large, hydrophobic substrates essential for the cell envelope). Several of these genes were present as multiple copies; a complete list of accessory genes appears in Table S2.
Genome-wide GO comparisons between MAP and Mtb.
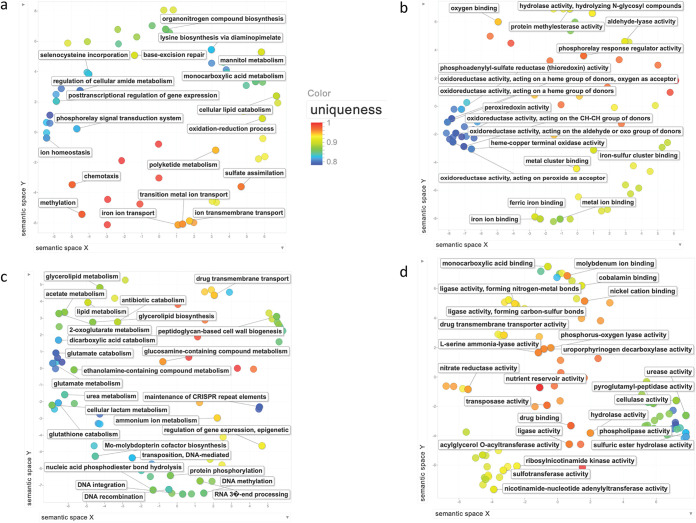
Earlier comparative genomic studies involving MAP have compared a single or very few genomes of the species to Mtb and to other Mycobacterium species. To our knowledge, a genome-wide Gene Ontology (GO) comparison has not been conducted, between MAP and any other species, using many multiple-genome sequences. The advantage of conducting such a comparison is that it can bring to light molecular features that differ interspecifically, both because it allows proportional statistical comparisons and because it includes an approximation of the gene repertoire of the accessory genome of each taxon. Our GO comparisons between MAP and Mtb revealed a total of 101 Biological Process terms that were enriched in MAP relative to Mtb, 28 Cellular Component terms, and 71 Molecular Function terms (Fig. 2a and b). GO categories that were underrepresented in MAP compared to Mtb included 106 Biological Process, 9 Cellular Component, and 116 Molecular Function (Fig. 2c and d). For MAP enrichments, terms related to ion homeostasis and iron transport, as well as numerous terms involving oxidoreductase and peroxiredoxin activity (Fig. 2a and b), were among those of prominence. MAP underrepresentations reflected the metabolic diversity of Mtb, including some GO terms with zero genes represented for MAP (further details are provided in Discussion). A complete listing of the GO results appears in Table S3 in the supplemental material.
FIG 2.
MAP enriched and underrepresented GO terms compared to Mtb, summarized and visualized as a REVIGO scatterplot. (a) Enriched Biological Process; (b) enriched Molecular Function; (c) underrepresented Biological Process; (d) underrepresented Molecular Function. Each circle represents a cluster of related GO terms, with a single term chosen by REVIGO as the cluster representative. Clusters are plotted according to semantic similarities to other GO terms (adjoining circles are most closely related). We have arbitrarily chosen to label a number of circles with their respective GO term in an attempt to reflect the overall range and emphasis. “Uniqueness” (the negative of average similarity of a term to all other terms) measures the degree to which the term is an outlier compared semantically to the whole list; blue and turquoise dots are among the least unique and, therefore, are of greatest average similarity to the set as a whole.
Pan-GWAS.
The results of our Panaroo curated set of accessory genes, and the following on Scoary analysis, identified several genes significantly associated with the presence or absence of measured traits (Table 1; for a complete list of measured traits, see Table 2; see Table S4 for a complete set of Scoary results and Table S5 for additional details and descriptions of the measured phenotypes). A few of these genes were significantly associated with more than one trait. The TreeWas analysis did not identify any SNPs significantly associated with measured bovine phenotypes.
TABLE 1.
Genes correlated with different RDQMA cow phenotypes
| Phenotype/characteristica | Positive state | Alternative state |
|---|---|---|
| Source | Type 1 polyketide_synthase MAP_RS09140 | C-hypotheticalb |
| Environment | 0 | 0 |
| LactIndex | 0 | 0 |
| SeroStatus | 0 | PPE32; MAP_RS07720 |
| ODcat | 0 | 0 |
| STpos | 0 | MCE family protein; MAP_RS09405; type 1 polyketide synthase MAP_RS09140 |
| AnteFC | Nonribosomal peptide synthase (NRPS); MAP_RS07215 | GTP-binding translation elongation factor TypA; MAP_RS13345 |
| FFS < 3 | mmps3; MAP_RS09845; type 1 polyketide_synthase MAP_RS09140 | SDR family NAD(P)-dependent oxidoreductase; MAP_RS06960 |
| FecalShedGroup | 0 | PPE32; MAP_RS07720; dimodular nonribosomal peptide synthase (dhbF); MAP_RS07215 |
| Prog | 0 | 0 |
See Table 2 for definitions of parameters.
C-hypothetical refers to conserved hypothetical (conserved across lineages other than MAP).
TABLE 2.
Dairy cow phenotypes or characteristics measured from the RDQMA farms and included in our GWAS analyses
| Phenotype/characteristic | Positive state (1) | Alternative state (0) | Na |
|---|---|---|---|
| Source | Cow was homebred | Cow was purchased | 304 |
| Environment | Isolate was sampled from cow’s environment | Isolate was sampled directly from cow, including fecal and tissue samples | 314 |
| LactIndex | Milk yield of cow decreased over lifetime compared to herd mates | Milk yield of cow stayed constant or improved over lifetime compared to herd mates | 56 |
| SeroStatus | Cow had at least one positive ELISA | Cow had no positive ELISA | 229 |
| ODcat | ∼Highest 10% OD values in ELISA of cow serum | ∼0–90th percentile of OD values in ELISA of cow serum | 229 |
| STpos | Cow was MAP culture positive in tissue (with or without additional positive feces or serum samples) | Cow was only MAP culture positive in feces | 160 |
| AnteFC | Cow had at least one MAP-positive fecal culture during lifetime (ante mortem) | Only samples taken from the cow’s carcass after culling/slaughter (post mortem) were MAP culture positive | 229 |
| FFS < 3 | Cow had first positive MAP fecal sample before the age of 3 yr | Cow had first positive MAP fecal sample at age of 3 yr or older | 229 |
| FecalShedGroup | Had at least one sample with >50 CFU/tube (no tissue), high fecal to super shedder | Negative samples or moderate fecal shedder (<50 CFU/tube) | 314 |
| Prog | Progressors show an increase in CFU number over time (fecal or tissue) | Nonprogressors show no increase in CFU number (fecal or tissue) | 131 |
N refers to the number of measurements made for the corresponding phenotype and for which a MAP isolate was also sequenced.
DISCUSSION
Population genomics.
A good deal of information now exists on population genetic variation in various bacterial pathogens, but the vast majority of this is on a much broader geographic scale than what we report here for MAP. It has been suggested that the pathogens that would pose the greatest challenge for disease management were those with the greatest evolutionary potential, with, for example, high mutation rates, large effective population sizes, and a high level of gene flow (44). However, there is growing concern that pathogens with a more clonal (or partially clonal) mode of reproduction can cause important outbreaks, particularly in agro‐ecosystems (45–47). Many agro‐ecosystems are characterized by the general uniformity of the host, the overall environment, and the associated agricultural practices.
Our results provide evidence for geographic population genetic structure in MAP on a microgeographic scale: three farms from adjacent states in the northeastern United States. Sampling of the cows for this study commenced in February, March, and November 2004 on the New York, Pennsylvania, and Vermont farms, respectively, and continued for about 7 years. During the study, the farms remained closed and did not purchase animals. The Pennsylvania farm was constituted from several herds 5 to 8 years prior to the start of the study. The Vermont farm was at this same location for 42 years prior to the onset of the study. For a short while it included a number of cows (about 20) from a neighboring dairy due to a barn fire at this neighboring dairy. This happened approximately 2 years into the RDQMA study. The New York farm was a closed farm for years before the start of the study, remained a closed farm throughout the study, and was in this same location for about 150 years. It is unclear where or how the original infections on each of these farms occurred. For the Pennsylvania farm, the most parsimonious conclusion would be that it arose from isolates associated with the diverse founder population of this farm. The long branch lengths typical of this population provide support to multiple lines of infection, likely arising from different lineages infecting different cows associated with the original founding of the farm. Mutation rates of MAP are estimated to be anywhere between 0.125 substitutions per genome per year to <0.3 substitutions per genome per year (48); assuming the median between these two estimates, with the minimal SNP differences associated with isolates in the Vermont clade, this would support a recent infection some time within a span of 10 years, perhaps arising from infection at the time of the neighbor’s barn fire. In addition, the Vermont clade provides some suggestion that there were two waves of infection, one very recent and the other closer to the 10-year interval. For New York, there also appears to be two waves of infection (P1-1 and P1-3; Fig. 1), and, based on SNP differences, both of these groups would be somewhat older than the Vermont infection, approximately 15 years. The pronounced MAP Pennsylvania diversity suggests multiple sources of infection arising from the multiple herds drawn upon for its original composition.
Despite MAP being widely regarded as an obligate parasite, there is ample evidence the pathogen can live and survive in water and soil environments, with one study finding that MAP remained culturable in lake water microcosms for 632 days and persisted to 841 days, with sediment cores adjacent to river reservoirs showing evidence of MAP consistent with deposition over 50 years (27). Much of this study of MAP presence in waterways is associated with examinations of the potential linkage of MAP with Crohn’s disease (27, 49, 50). Despite MAP prevalence and hardiness in environmental samples, it is widely considered that the majority of bovine MAP infections arise from animal-animal contact. At the same time, the extremely high prevalence of MAP infection rates of dairy herds in the United States, estimated at somewhere between 68 and 90% (estimates for 2007 [51]), concomitant with the environmental presence and persistence of the pathogen, suggest interfarm environmental infection cannot be discarded. Thus, although the quarantine or closing of infected farms is a logical and, indeed, critical step to prevent interfarm infection, its overall effectiveness has to be regarded with a degree of uncertainty. A corollary issue is what might be the safe distance of one farm from another infected farm, given environmental transmission possibilities, such as interconnecting waterways. In our case, the distance from the Vermont-New York farms is about 130 km, and there are various interconnecting waterways; the distance between the Pennsylvania and New York farms is 657 km. Our results support a clear population genetic distinction of MAP isolates between New York and Vermont. This distinction included a total of 74 isolates in our set arising from environmental samples, and none of these showed any evidence of interfarm exchange. Ideally, one would prefer to have farms closer to both New York and Vermont to evaluate this thoroughly, but this cursory picture of herd closures, separated by about 130 km, suggests that at least over the 10-year span of this sample collection, it was effective. This is in sharp contrast to a study involving dairy cattle in three regions of southern Alberta (17), which showed no evidence of genotypic clustering involving farms sampled at distances of at least 500 km but with no reported history of closure.
Comparative genomics, pangenomics, and pan-GWAS.
We present a genome-wide GO comparison involving many multiple MAP isolates to another species of Mycobacterium, in this case Mtb. Several enriched GO terms and groups of terms from Biological Process and Molecular Function were evident that are related to the nature of MAP host survival and pathogenesis. Of particular note, several terms were related to ion homeostasis and iron transport, and numerous terms involved oxidoreductase and peroxiredoxin activity (Fig. 2a and b). The MAP evolutionary emphasis toward oxidoreductase activity is likely a reflection of the range of oxic to microoxic environments in which the pathogen can be found. Peroxiredoxins are a family of peroxidase enzymes that play dominant roles in regulating peroxide, one of the toxins produced as a by-product of using oxygen for respiration, and its enrichment here may reflect the more varied ways in which oxygen is made use of in MAP. MAP is the one species of pathogenic Mycobacterium that cannot produce mycobactin, a siderophore used by other members of the genus Mycobacterium to shuttle free extracellular iron ions into the cytoplasm of mycobacterial cells. To compensate for this, MAP has evolved other means of iron acquisition (52), and this is reflected in some of these GO enrichments. Enrichments in Cellular Component were dominated by terms referring to organelles, possibly reflecting MAP-specific membrane vesicles (MVs) (53, 54). Many bacterial pathogens produce and utilize MVs as a means of exporting various factors, such as toxins, lipids, polysaccharides, peptidoglycans, lipoproteins, and quorum-sensing molecules, across the bacterial cell envelope and into the host cells. MVs in Mtb have been shown to be involved in iron acquisition (55), TLR2-dependent immune modulation (56), and inhibition of T-cell activation (57). Mycobacterium avium produces MVs within phagosomes that carry products involved in modulation of host immune defenses and intracellular survival (54). MVs in MAP are not well studied, but these enrichments in GO terms, such as intracellular membrane bounded organelle (GO:0043231) and transmembrane transporter complex (GO:1902495), indicate a MAP evolutionary emphasis toward these structures and functions, suggesting they warrant further investigation. Underrepresentations included many terms related to the specific host adaptation and pathogenesis of Mtb compared to MAP (Fig. 2c and d). Of particular note were numerous terms emphasizing the metabolic diversity of Mtb (e.g., urea, acetate, lipid, hexose, glutamate, ammonium ion, glycerolipid, and glucosamine metabolism) as well as terms related to the drug tolerance of Mtb (e.g., drug transmembrane transport, antibiotic catabolism, and beta-lactamase activity). Several underrepresented GO categories had zero genes represented for MAP (e.g., urease activity and nickel cation binding); the reverse was not apparent.
Mycobacterium species are generally thought to have low or even nonexistent incidence of homologous recombination; indeed, our evaluation of recombination in our 318 MAP genome sequences yielded no significant evidence for core gene recombination. Mycobacterium species are, in fact, thought to be highly clonal; however, recent pangenomic analyses of Mtb provide evidence for an accessory genome comprising from 14% (58) to at least 21% of a typical Mtb genome (59). The former estimate was based on 36 complete genomes and involved the program PGAP (60). The latter was based on a diverse set of 1,595 WGS genomes and employed CD-hit clustering (61) followed by sensitivity analysis to demarcate the core and accessory genomes. Similarly, recent pangenomic studies of Mycobacterium bovis, a species very closely related to Mtb, report an accessory genome of around 20% of a typical genome size (62, 63). The latter two assessments were determined with Sybil (64) and Get_Homologues v2.0 (65), respectively. All of these approaches do nothing to accommodate for problematic issues associated with inflation of accessory count due to gene fragmentation at the ends of contigs. Tonkin-Hill et al. (66), in their Panaroo paper, indicate that the majority of errors leading to inflated accessory counts are due to genes being fragmented during assembly, although other factors, such as contamination, diverse gene families, and misassemblies, accumulate over the population, and all of these issues end up contributing to important consequences when analyzing the gene repertoire of a bacterial species. Tonkin-Hill et al. are not alone in pointing to annotation errors and fragmented assemblies as important sources of inflated gene numbers in draft genome assemblies (67, 68); indeed, our initial manual evaluation of the gene fragmentation issue involving our 318 MAP sequences largely concurred with the Panaroo assessment in significantly reducing the gene repertoire of the accessory genome, although the specifics differed somewhat (102 genes versus 97). Although we are confident in this assessment, it should be noted that this repertoire of genes should not be regarded as representative of the species as a whole. Our goal was to determine the most accurate assessment of accessory gene content that we could, since our purpose was to associate the presence/absence of genes with phenotypes. The restricted geographic range, the infection history, and strong population structure of our isolates would act to limit diversity and in turn limit the size of the accessory genome. We would expect the MAP species accessory genome to be somewhat larger than what we report for these 318 isolates. With the advent of pan-GWAS analysis tools and population genomic analyses of many sequences, from different hosts or environments, associating gene presence/absence on an accurate basis with the traits of interest is important and powerful, but only if it is done with considerable accuracy. We encourage a more curated approach to these assessments in the future.
The resulting MAP accessory genome of our 318 isolates was relatively small, but we did find a number of loci that were significantly associated with phenotypes. We also feel it is worth noting that because of the correction for common ancestry that Scoary incorporates, the bar of significance for associating a gene with a trait is set high. The analysis incorporates information on the phylogenetic structure of the sample in making the associations and, in the process, shifts emphasis to evolutionary transitions as the unit of importance rather than members of a clade. Such convergent evolution of genes with traits has long been regarded as one of our best estimates of adaptation and gene-function relationships in comparative biology (69), but like most good estimations in science, it is, by nature, conservative.
Several annotated loci were significantly correlated with traits. One of these, mmps3, was correlated with cows having their first positive fecal sample at less than 3 years old (FFS<3). mmps3 is a transport accessory protein, and other proteins in this family are known to be critical to Mtb virulence, specifically, mmps4, mmps5, and their interaction with mmpL4, through their transport of siderophores (70). Type 1 polyketide synthase was correlated with the positive state for FFS <3 as well as the alternative state of STpos (cow was only MAP culture positive in feces, i.e., not tissue). Type 1 polyketide synthases are multidomain enzymes that produce polyketides, which are secondary metabolites, many of which have antimicrobial or immunosuppressive properties. Another multicopy gene, MCE family protein (mammalian cell entry protein), was also correlated with the alternative state of STpos. MCE genes are organized into MCE operons of 6 to 10 open reading frames (ORFs), and MAP is unusual among the mycobacteria in possessing 8 such operons; Mtb has 4 (see MCE review in reference 71). Two of the MCE operons in MAP are duplicated, mce5 and mce7. The specific MCE locus in this case was RS09430 and is in one of the copies of MCE operon 7, locus mce7F (71); this operon is absent from Mtb. The functions for all of these MCE genes and all of these operons remains uncertain, but several have been implicated as important in invasion and survival within macrophages (71), while others function as a type of ABC-transporter system involved in import of fatty acids and the export of lipid virulence factors (71). Most of this functional work has been completed on Mtb. It is somewhat counterintuitive that a mammalian cell entry protein should be correlated, in our case, with a phenotype indicating the absence of tissue entry, but unfortunately not enough is known about the various functions of the genes in this complex family to formulate much of an explanation in that regard, particularly when this MCE operon is not found in Mtb. Nonetheless, the fact that an MCE family protein is correlated, either positively or negatively, with tissue entry suggests something worth experimentally investigating for this particular member (RS09430). MCE family genes were among the most common accessory genes in our set of isolates, totaling 9, further suggesting that their presence or absence could be playing a role in phenotypes not measured in our analysis.
Other proteins of note correlated with cow phenotypes included DhbF_7 and PPE32. DhbF (dimodular nonribosomal peptide synthetase) was correlated with the alternative state of FecalShedGroup (negative or moderate fecal shedder); it is an example of a modular nonribosomal peptide synthetase (NRPS), which synthesize nonribosomal peptides, independent of mRNA. NRPS can be nonmodular or modular (72). DhbF proteins are modular, but our MAP genomes also harbored other NRPS annotated as nonmodular. Each NRPS can synthesize only one type of peptide, but these are involved in a wide range of functions, including toxins, siderophores, antibiotics, and pigments (72). Our MAP sequences harbored 10 copies of DhbF; 7 of them were core and they were highly divergent. dhbF_7 codes for a peptide that is the longest of these, at 4,038 amino acids in length, and it was a high-copy accessory gene (present in 95% of the isolates) in our analysis. It is not clear what peptide this NRPS is responsible for synthesizing, but a large proportion of the cows with MAP that produced this protein were low fecal shedders.
PPE32 is a member of the large PPE family of proteins present in Mycobacterium spp. In our analysis, one of the traits it was associated with was FecalShedGroup, and, like DhbF, it was associated with the negative or moderate fecal shedder state. Some PPE proteins of Mtb have been shown to elicit a Th1 cell response in humans and protect the host from pulmonary infection (73). Perhaps a similar phenomenon is at play here, with bovine Th1 cells recognizing these particular antigens, keeping the infection largely under control and the host thereby not attaining the supershedder phenotype. A similar line of logic may apply to the association of PPE32 with the absence of a positive SeroStatus measurement, or, in other words, an absence of a positive ELISA, which measures humoral immune response. The current perspective on MAP infection is that the bacteria can induce both the cellular (Th1) and the humoral (Th2) immune response, but early during infection the cellular (Th1) response dominates, which leads to inhibition of the humoral (Th2) response and effective control of MAP replication, resulting in limited bacterial shedding (74). A negative ELISA would reflect a stage of infection that was under Th1 control, and this could have been stimulated by this particular PPE32. Pathogenic mycobacteria have a type VII secretion system, ESX-5, which is a pathway for export of PE and PPE proteins. This particular PPE locus (MAP_1515) is located in one of the ESX-5 operons that also includes mycosin-5 and triacylglycerol lipase. It is a high-copy-number accessory gene, but cows in our analysis without this particular PPE32 locus were positive for FecalShedGroup (supershedders) and serostatus (positive ELISA), suggesting they have skipped Th1 response or at least had an earlier shift to Th2 immune control. Alignments and phylogenetic analyses of all PPE sequences taken from our MAP genomes indicate that this particular PPE is the most variable of all, with the vast majority of this variability occurring in the C-terminal end of the protein sequence. This PPE is a member of the PPE-SVP subfamily (sublineage IV [75]), and comparisons with other subspecies of Mycobacterium avium suggest that the MAP ancestral state was a peptide of 376 residues typical of these other subspecies and that subsequent deletions in this region resulted in various sequences of truncated length. The majority (although not all) of sequences associated with the alternative states for FecalShedGroup and serostatus were of the complete length.
Conclusions.
Our population genomics analysis of closed farms on a highly regional scale provided evidence for strong population subdivision and, in turn, genetic evidence in support of the strategy of quarantining or closing farms as an effective tool to stop interfarm spread of MAP, despite the existence of numerous other possible avenues of environmental transmission. Our pan-GWAS study involving MAP and Johne’s disease phenotypes identified a few loci worthy of further experimental consideration. Several of these genes appear to be linked to phenotypes suggestive of potentially important host immune responses. A better understanding of the interaction between MAP and the host immune system is germane to the longstanding and continuing efforts to develop an effective Johne’s vaccine. Our pan-GWAS identification of genes correlated with Johne’s phenotypes identifies specific loci for detailed follow-up causality experimentation involving both host immunity and MAP pathogenesis. Our study has highlighted the potential of pan-GWAS analysis in attempting gene-trait associations, even with complex phenotypes such as those measured here. Because of our microgeographic focus involving these largely closed farms, our study considered only a very small proportion of MAP genomic diversity, yet, even for this largely clonal organism, we identified a small but not insignificant accessory genome. This, in turn, suggests that a much broader survey of MAP genomic diversity would identify a larger accessory genome, and one that could differ in composition between areas. Such gene content variation between regions ultimately could be important in identifying and treating differences in the disease.
MATERIALS AND METHODS
Culturing, DNA extraction, and sequencing.
The original isolation and culture of MAP strains from the dairy cattle of these three RDQMA farms (Petersburgh, New York; Martinsburgh, Pennsylvania; and Florence, Vermont) are described elsewhere (76). For this study, strains were subcultured from glycerol stocks arising from these original isolations, in 4 ml 7H9 medium (7H9 broth, 10% Hardy Diagnostic Middlebrook oleic acid-albumin-dextrose-catalase, 0.05% Tween 80, 2 mg/liter Allied Monitor mycobactin J, 0.01% cycloheximide; BD Difco), and grown standing in closed-cap 14-ml cell culture tubes at 37°C for 8 to 12 weeks. Cultures were checked for contamination by plating an aliquot on BD chocolate agar and incubating overnight at 37°C. From the original collection of isolates derived from the RDQMA farms, a total of 337 isolates were successfully cultured and sequenced.
The Epicentre Masterpure Gram-positive DNA purification kit was used to extract genomic DNA, with some minor modifications to the kit protocol. Approximately 3 ml of culture resuspended in 150 μl TE buffer was heated at 80°C for 20 min. The samples were then treated with double the recommended volume of Ready-Lyse lysozyme (2 μl) for 2 h at 37°C and incubated in a rotating incubator with proteinase K lysis solution for 20 min at 65°C. Protein precipitation, RNase digest, and isopropanyl precipitation were performed as described in the kit protocol. DNA was resuspended in low-EDTA TE and prepared for sequencing using the Nextera XT Library Prep kit. Multiplexed libraries of 48 strains each were sequenced on the Illumina HiSeq 2500 with 2 × 100-bp Rapid Run paired-end reads. The resulting MAP sequences were first examined for possible evidence of multiple infections by identifying mixed SNPs. This was accomplished using the vSNP pipeline, https://github.com/USDA-VS/vSNP, which has been implemented for SNP calling and phylogenetics in recent studies of Mycobacterium spp. (77). Briefly, the Illumina sequence reads for each isolate were mapped to the reference genome MAP K-10 using the Burrows Wheeler Aligner (BWA [78]) and Genome Analysis Toolkit (GATK) (79–81) according to GATK best practices. Integrated Genomics Viewer was used to visually validate SNPs; MAP sequences with mixed nucleotide calls at any position were removed from further analysis, and this amounted to 6.0% of our original set of isolates, leaving us with 318, distributed between the three farms as the following: New York, 116; Pennsylvania, 32; Vermont, 170.
Population genomics and phylogenetics.
The Illumina sequence reads, for all 318 samples judged to be unique cultures, were quality controlled and assembled using the A5 pipeline (82). N50 ranged from 59,204 to 95,174, with an average of 78,878. Metadata and full assembly metrics for all isolates are shown in Table S1 in the supplemental material. For all genome assemblies, open reading frames were located and annotated using Prokka (83). For the Prokka annotation, a custom Mycobacterium database was built using available genome data at ref_seq (NCBI). We further refined the annotation using the updated complete MAP genome of strain K-10 (84) and BLASTn. An E value cutoff of 10−e5 was employed, and only the top hit was retained. Prokka uses Prodigal (85) to locate open reading frames. We pretrained the Prodigal model using a closed reference genome obtained from NCBI (see Table S1 for sequence details). To delineate the pangenome, amino acid sequences from all genomes were delineated into clusters with putative shared homology using the recently developed pangenome pipeline Panaroo (66). The pipeline generates initial gene clusters using a greedy incremental clustering approach that processes sequences based on length (as implemented in CD-HIT). Next, neighborhood information is generated by constructing a graph of the pangenome where nodes are gene clusters and edges connect genes adjacent on contigs. The graph and neighborhood information is then used to identify and merge fragmented or mistranslated genes and identify genes missed by the gene-calling algorithm. For comparison, we also used the established pipeline Roary (86). This pipeline first collapses redundant gene sequences and then uses the Markov cluster (MCL) algorithm of Enright et al. (87) to assign sequences to clusters with putative shared homology, with this shared homology being based on a BLASTp search between all pairs of protein sequences using a sequence identity threshold of 95%. We ran the pipeline with the paralog splitting mode both on and off.
To generate a core genomic alignment, we mapped our assemblies to six complete MAP reference genomes using the software REALPHY (88). The approach cuts contigs into 50-bp pieces, which are then mapped to each reference separately. A final nonredundant alignment is produced by merging each separate alignment. The approach reduces bias that could be introduced by using only a single reference. The reference genomes were obtained from NCBI (see Table S1 for details). The alignment was assessed for recombination using the pairwise homoplasy index (PHI) as implemented in Phi (89). Phi is a compatibility method that examines pairs of aligned nucleotide sites for homoplasy. SNPs were extracted from the final alignment and used to build a core phylogeny using IQ-TREE (90). This approach is applicable to SNP alignments, as it allows for ascertainment bias correction (ASC). The general time-reversible model with gamma and four rate categories was employed (GTR+G4).
Hierarchal population structure was delineated using a Bayesian clustering/assignment approach, as implemented in the Bayesian Analysis of Population Structure (BAPS and hierBAPS) software (91). The approach first determines the optimal number of genetically distinct clusters (populations) (K) such that the genetic variation within clusters is minimized and the variation among them is maximized. Each isolate is then assigned to a population (structured hierarchically). More specifically, the posterior probability distribution of model parameters from a mixture model derived using Bayesian predictive classification theory are sampled using a Metropolis-Hastings implementation of Markov chain Monte Carlo (MCMC) probability distribution sampling. Model parameters represent SNP frequencies and individual/SNP population assignment probabilities. The core SNP alignment was used as the input.
Comparative genomics and bacterial pan-GWAS.
To determine what gene functional categories from MAP isolates were enriched compared to those of Mtb, we analyzed genome-wide amino acid sequences from our study isolates to genomes from 300 randomly selected Mtb isolates from ref_seq at NCBI (see Table S1 for genome sequence details). For our study isolates, we used genes for which called ORFs and annotation were in agreement between Prokka and Panaroo. Gene Ontology (GO) terms were assigned to genes from each genome using Interproscan (92) in default mode, and the output was used to assess term enrichment using Fisher exact tests as implemented in the find_enrichment.py script in GOATOOLS v0.5.9 (93). P values were adjusted to account for increased type I errors due to multiple-hypothesis testing following the false discovery rate (FDR) procedure of Benjamini and Hochberg (94). Tests were judged significant when the adjusted P value fell below the FDR threshold of 0.05.
Dairy cow phenotypes were evaluated and scored as part of earlier RDQMA studies of these three farms (8). The phenotypes, and a brief explanation of each, are presented in Table 2; more detailed descriptions are provided in Table S5.
To identify genes that were statistically associated with each phenotype, we used the software Scoary (42), which identifies the presence/absence of genes from the accessory genome that are significantly correlated with phenotypes. The method corrects for the possibility of false-positive gene-trait correlations due to population structure and isolate evolutionary history by implementing the pairwise comparisons algorithm (95, 96), which finds the maximum number of phylogenetic pairs of isolates that contrast in the state of both genotype and phenotype; it does this by evaluating how each gene-trait pair is distributed through the underlying phylogeny, which in our case was our core SNP phylogeny. By identifying the maximal number of contrasting pairs in the context of this phylogeny, it counts the minimum number of independent coemergence of a given gene-trait combination in the evolutionary history of the sample population. Enrichment of dispensable genes from isolates within each sampling location (New York, Pennsylvania, and Vermont) was determined using Fisher exact tests within Scoary (42). Gene presence/absence among isolates was determined using Panaroo.
In an attempt to identify SNPs that were statistically associated with phenotypes, we used the software TreeWAS. Similar to Scoary, TreeWAS is a phylogenetic method that measures the statistical associations between phenotype and genotype at all bacterial loci while correcting for the confounding effects of clonal population structure and homologous recombination (43). In addition to the binary coded phenotype data, the core SNP alignment and phylogeny were used as the input for the program.
Data availability.
Sequence read data associated with this project have been deposited at NCBI in the Sequence Read Archive database under the following BioProject accession number: PRJNA686527.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter Schweitzer and the Cornell Biotechnology Resource Center for genome sequencing of MAP isolates.
This work was funded by USDA-NIFA AFRI grant number 2014-67015-2240 as part of the joint USDA-NSF-NIH-BBSRC-BSF Ecology and Evolution of Infectious Diseases program.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Gollnick NS, Mitchell RM, Baumgart M, Janagama HK, Sreevatsan S, Schukken YH. 2007. Survival of Mycobacterium avium subsp. paratuberculosis in bovine monocyte-derived macrophages is not affected by host infection status but depends on the infecting bacterial genotype. Vet Immunol Immunopathol 120:93–105. 10.1016/j.vetimm.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 2.Benedictus A, Mitchell RM, Linde-Widmann M, Sweeney R, Fyock T, Schukken YH, Whitlock RH. 2008. Transmission parameters of Mycobacterium avium subspecies paratuberculosis infections in a dairy herd going through a control program. Prev Vet Med 83:215–227. 10.1016/j.prevetmed.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Wells SJ, Wagner BA. 2000. Herd-level risk factors for infection with Mycobacterium paratuberculosis in US dairies and association between familiarity of the herd manager with the disease or prior diagnosis of the disease in that herd and use of preventive measures. J Am Vet Med Assoc 216:1450–1457. 10.2460/javma.2000.216.1450. [DOI] [PubMed] [Google Scholar]
- 4.Motiwala AS, Amonsin A, Strother M, Manning EJ, Kapur V, Sreevatsan S. 2004. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis isolates recovered from wild animal species. J Clin Microbiol 42:1703–1712. 10.1128/jcm.42.4.1703-1712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Möbius P, Luyven G, Hotzel H, Köhler H. 2008. High genetic diversity among Mycobacterium avium subsp. paratuberculosis strains from German cattle herds shown by combination of IS900 restriction fragment length polymorphism analysis and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing. J Clin Microbiol 46:972–981. 10.1128/JCM.01801-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ott SL, Wells SJ, Wagner BA. 1999. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev Vet Med 40:179–192. 10.1016/S0167-5877(99)00037-9. [DOI] [PubMed] [Google Scholar]
- 7.Greenstein RJ. 2003. Is Crohn's disease caused by a mycobacterium? Comparisons with leprosy, tuberculosis, and Johne's disease. Lancet Infect Dis 3:507–514. 10.1016/S1473-3099(03)00724-2. [DOI] [PubMed] [Google Scholar]
- 8.Pradhan AK, Mitchell RM, Kramer AJ, Zurakowski MJ, Fyock TL, Whitlock RH, Smith JM, Hovingh E, Van Kessel JA, Karns JS, Schukken YH. 2011. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis in a longitudinal study of three dairy herds. J Clin Microbiol 49:893–901. 10.1128/JCM.01107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell RM, Beaver A, Knupfer E, Pradhan AK, Fyock T, Whitlock RH, Schukken YH. 2019. Elucidating transmission patterns of endemic Mycobacterium avium subsp. paratuberculosis using molecular epidemiology. Vet Sci 6:32. 10.3390/vetsci6010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, Banerji N, Kanjilal S, Kapur V. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc Natl Acad Sci U S A 102:12344–12349. 10.1073/pnas.0505662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bannantine JP, Wu CW, Hsu C, Zhou S, Schwartz DC, Bayles DO, Paustian ML, Alt DP, Sreevatsan S, Kapur V, Talaat AM. 2012. Genome sequencing of ovine isolates of Mycobacterium avium subspecies paratuberculosis offers insights into host association. BMC Genomics 13:89. 10.1186/1471-2164-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh P, Hsu C, Alyamani EJ, Shehata MM, Al-Dubaib MA, Al-Naeem A, Hashad M, Mahmoud OM, Alharbi KB, Al-Busadah K, Al-Swailem AM, Talaat AM. 2012. Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius). PLoS One 7:e31947. 10.1371/journal.pone.0031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohal JS, Arsenault J, Labrecque O, Fairbrother JH, Roy JP, Fecteau G, L'Homme Y. 2014. Genetic structure of Mycobacterium avium subsp. paratuberculosis population in cattle herds in Quebec as revealed by using a combination of multilocus genomic analyses. J Clin Microbiol 52:2764–2775. 10.1128/JCM.00386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynne JW, Bull TJ, Seemann T, Wynne JW, Bull TJ, Seemann T, Bulach DM, Wagner J, Kirkwood CD, Michalski WP. 2011. Exploring the zoonotic potential of Mycobacterium avium subspecies paratuberculosis through comparative genomics. PLoS One 6:e22171. 10.1371/journal.pone.0022171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timms VJ, Hassan KA, Mitchell HM, Neilan BA. 2015. Comparative genomics between human and animal associated subspecies of the Mycobacterium avium complex: a basis for pathogenicity. BMC Genomics 16:695. 10.1186/s12864-015-1889-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu CY, Wu CW, Talaat AM. 2011. Genome-wide sequence variation among Mycobacterium avium subspecies paratuberculosis isolates: a better understanding of Johne's disease transmission dynamics. Front Microbiol 2:236. 10.3389/fmicb.2011.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahlstrom C, Barkema HW, Stevenson K, Zadoks RN, Biek R, Kao R, Trewby H, Haupstein D, Kelton DF, Fecteau G, Labrecque O, Keefe GP, McKenna SL, Tahlan K, De Buck J. 2016. Genome-wide diversity and phylogeography of Mycobacterium avium subsp. paratuberculosis in Canadian dairy cattle. PLoS One 11:e0149017. 10.1371/journal.pone.0149017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bryant JM, Thibault VC, Smith DG, McLuckie J, Heron I, Sevilla IA, Biet F, Harris SR, Maskell DJ, Bentley SD, Parkhill J, Stevenson K. 2016. Phylogenomic exploration of the relationships between strains of Mycobacterium avium subspecies paratuberculosis. BMC Genomics 17:79. 10.1186/s12864-015-2234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amonsin A, Li LL, Zhang Q, Bannantine JP, Motiwala AS, Sreevatsan S, Kapur V. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J Clin Microbiol 42:1694–1702. 10.1128/jcm.42.4.1694-1702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thibault VC, Grayon M, Boschiroli ML, Hubbans C, Overduin P, Stevenson K, Gutierrez MC, Supply P, Biet F. 2007. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: comparison with IS900 and IS1245 restriction fragment length polymorphism typing. J Clin Microbiol 45:2404–2410. 10.1128/JCM.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohal JS, Sheoran N, Narayanasamy K, Brahmachari V, Singh S, Subodh S. 2009. Genomic analysis of local isolate of Mycobacterium avium subspecies paratuberculosis. Vet Microbiol 134:375–382. 10.1016/j.vetmic.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 22.Whittington R, Donat K, Weber MF, Kelton D, Nielsen SS, Eisenberg S, Arrigoni N, Juste R, Sáez JL, Dhand N, Santi A, Michel A, Barkema H, Kralik P, Kostoulas P, Citer L, Griffin F, Barwell R, Moreira MAS, Slana I, Koehler H, Singh SV, Yoo HS, Chávez-Gris G, Goodridge A, Ocepek M, Garrido J, Stevenson K, Collins M, Alonso B, Cirone K, Paolicchi F, Gavey L, Rahman MT, de Marchin E, Van Praet W, Bauman C, Fecteau G, McKenna S, Salgado M, Fernández-Silva J, Dziedzinska R, Echeverría G, Seppänen J, Thibault V, Fridriksdottir V, Derakhshandeh A, Haghkhah M, Ruocco L, Kawaji S, Momotani E, Heuer C, et al. 2019. Control of paratuberculosis: who, why and how. A review of 48 countries. BMC Vet Res 15:198. 10.1186/s12917-019-1943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna SL, Keefe GP, Tiwari A, VanLeeuwen J, Barkema HW. 2006. Johne's disease in Canada part II: disease impacts, risk factors, and control programs for dairy producers. Can Vet J 47:1089–1099. [PMC free article] [PubMed] [Google Scholar]
- 24.Eppleston J, Begg DJ, Dhand NK, Watt B, Whittington RJ. 2014. Environmental survival of Mycobacterium avium subsp. paratuberculosis in different climatic zones of eastern Australia. Appl Environ Microbiol 80:2337–2342. 10.1128/AEM.03630-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittington RJ, Marshall DJ, Nicholls PJ, Marsh IB, Reddacliff LA. 2004. Survival and dormancy of Mycobacterium avium subsp. paratuberculosis in the environment. Appl Environ Microbiol 70:2989–3004. 10.1128/aem.70.5.2989-3004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samba-Louaka A, Robino E, Cochard T, Samba-Louaka A, Robino E, Cochard T, Branger M, Delafont V, Aucher W, Wambeke W, Bannantine JP, Biet F, Héchard Y. 2018. Environmental Mycobacterium avium subsp. paratuberculosis hosted by free-living amoebae. Front Cell Infect Microbiol 8:28. 10.3389/fcimb.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickup RW, Rhodes G, Arnott S, Sidi-Boumedine K, Bull TJ, Weightman A, Hurley M, Hermon-Taylor J. 2005. Mycobacterium avium subsp. paratuberculosis in the catchment area and water of the River Taff in South Wales, United Kingdom, and its potential relationship to clustering of Crohn's disease cases in the city of Cardiff. Appl Environ Microbiol 71:2130–2139. 10.1128/AEM.71.4.2130-2139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whan L, Ball HJ, Grant IR, Rowe MT. 2005. Occurrence of Mycobacterium avium subsp. paratuberculosis in untreated water in Northern Ireland. Appl Environ Microbiol 71:7107–7112. 10.1128/AEM.71.11.7107-7112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickup RW, Rhodes G, Bull TJ, Arnott S, Sidi-Boumedine K, Hurley M, Hermon-Taylor J. 2006. Mycobacterium avium subsp. paratuberculosis in lake catchments, in river water abstracted for domestic use, and in effluent from domestic sewage treatment works: diverse opportunities for environmental cycling and human exposure. Appl Environ Microbiol 72:4067–4077. 10.1128/AEM.02490-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes G, Richardson H, Hermon-Taylor J, Weightman A, Higham A, Pickup R. 2014. Mycobacterium avium subspecies paratuberculosis: human exposure through environmental and domestic aerosols. Pathogens 3:577–595. 10.3390/pathogens3030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor RH, Falkinham JO, III, Norton CD, LeChevallier MW. 2000. Chlorine, chloramine, chlorine dioxide, and ozone susceptibility of Mycobacterium avium. Appl Environ Microbiol 66:1702–1705. 10.1128/aem.66.4.1702-1705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beumer A, King D, Donohue M, Mistry J, Covert T, Pfaller S. 2010. Detection of Mycobacterium avium subsp. paratuberculosis in drinking water and biofilms by quantitative PCR. Appl Environ Microbiol 76:7367–7370. 10.1128/AEM.00730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aboagye G, Rowe MT. 2011. Occurrence of Mycobacterium avium subsp. paratuberculosis in raw water and water treatment operations for the production of potable water. Water Res 45:3271–3278. 10.1016/j.watres.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 34.Klanicova B, Seda J, Slana I, Slany M, Pavlik I. 2013. The tracing of mycobacteria in drinking water supply systems by culture, conventional, and real time PCRs. Curr Microbiol 67:725–731. 10.1007/s00284-013-0427-1. [DOI] [PubMed] [Google Scholar]
- 35.Donohue MJ, Vesper S, Mistry J, Donohue JM. 2019. Impact of chlorine and chloramine on the detection and quantification of Legionella pneumophila and Mycobacterium species. Appl Environ Microbiol 85:e01942-19. 10.1128/AEM.01942-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Büning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, International IBD Genetics Consortium (IIBDGC), et al. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, Amin N, Buchkovich ML, Croteau-Chonka DC, Day FR, Duan Y, Fall T, Fehrmann R, Ferreira T, Jackson AU, Karjalainen J, Lo KS, Locke AE, Mägi R, Mihailov E, Porcu E, Randall JC, Scherag A, Vinkhuyzen AA, Westra HJ, Winkler TW, Workalemahu T, Zhao JH, Absher D, Albrecht E, Anderson D, Baron J, Beekman M, Demirkan A, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Fraser RM, Goel A, Gong J, Justice AE, Kanoni S, Kleber ME, Kristiansson K, Lim U, et al. 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46:1173–1186. 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falush D, Bowden R. 2006. Genome-wide association mapping in bacteria? Trends Microbiol 14:353–355. 10.1016/j.tim.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Read TD, Massey RC. 2014. Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: a new direction for bacteriology. Genome Med 6:109. 10.1186/s13073-014-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen PE, Shapiro BJ. 2015. The advent of genome-wide association studies for bacteria. Curr Opin Microbiol 25:17–24. 10.1016/j.mib.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Power RA, Parkhill J, de Oliveira T. 2017. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet 18:41–50. 10.1038/nrg.2016.132. [DOI] [PubMed] [Google Scholar]
- 42.Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. 2016. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol 17:238. 10.1186/s13059-016-1108-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins C, Didelot X. 2018. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLoS Comput Biol 14:e1005958. 10.1371/journal.pcbi.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonald BA, Linde C. 2002. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379. 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- 45.Pennisi E. 2010. Armed and dangerous. Science 327:804–805. 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- 46.Tibayrenc M, Ayala FJ. 2012. Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci U S A 109:E3305–E3313. 10.1073/pnas.1212452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonald BA, Stukenbrock EH. 2016. Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Philos Trans R Soc B 371:20160026. 10.1098/rstb.2016.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson K. 2015. Genetic diversity of Mycobacterium avium subspecies paratuberculosis and the influence of strain type on infection and pathogenesis: a review. Vet Res 46:64. 10.1186/s13567-015-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNees AL, Markesich D, Zayyani NR, Graham DY. 2015. Mycobacterium paratuberculosis as a cause of Crohn's disease. Expert Rev Gastroenterol Hepatol 9:1523–1534. 10.1586/17474124.2015.1093931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graham DY, Naser SA, Offman E, Kassir N, Hardi R, Welton T, Rydzewska G, Stepien B, Arlukowicz T, Wos A, Fehrmann C, Anderson P, Bibliowicz A, McLean P, Fathi R, Harris MS, Kalfus IN. 2019. RHB-104, a fixed-dose, oral antibiotic combination against Mycobacterium avium paratuberculosis (MAP) infection, is effective in moderately to severely active Crohn's disease. Am J Gastroenterol 114:S376–S377. 10.14309/01.ajg.0000592108.53051.68. [DOI] [Google Scholar]
- 51.Lombard JE, Gardner IA, Jafarzadeh SR, Fossler CP, Harris B, Capsel RT, Wagner BA, Johnson WO. 2013. Herd-level prevalence of Mycobacterium avium subsp. paratuberculosis infection in United States dairy herds in 2007. Prev Vet Med 108:234–238. 10.1016/j.prevetmed.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Moolji J, Dufort A, Staffa A, Domenech P, Reed MB, Behr MA. 2015. Iron acquisition in Mycobacterium avium subsp. paratuberculosis. J Bacteriol 198:857–866. 10.1128/JB.00922-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin WS. 2016. The isolation and proteomic analysis of Mycobacterium avium subspecies paratuberculosis membrane vesicles. M.S. thesis. Department of Molecular and Cellular Biology, University of Guelph, Ontario, Canada. http://hdl.handle.net/10214/9744. [Google Scholar]
- 54.Chiplunkar SS, Silva CA, Bermudez LE, Danelishvili L. 2019. Characterization of membrane vesicles released by Mycobacterium avium in response to environment mimicking the macrophage phagosome. Future Microbiol 14:293–313. 10.2217/fmb-2018-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prados-Rosales R, Weinrick BC, Piqué DG, Jacobs WR, Jr, Casadevall A, Rodriguez GM. 2014. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol 196:1250–1256. 10.1128/JB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, Jimenez J, Glatman-Freedman A, Jacobs WR, Jr, Porcelli SA, Casadevall A. 2011. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Investig 121:1471–1483. 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Athman JJ, Sande OJ, Groft SG, Reba SM, Nagy N, Wearsch PA, Richardson ET, Rojas R, Boom WH, Shukla S, Harding CV. 2017. Mycobacterium tuberculosis membrane vesicles inhibit T cell activation. J Immunol 198:2028–2037. 10.4049/jimmunol.1601199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang T, Zhong J, Zhang J, Yang T, Zhong J, Zhang J, Li C, Yu X, Xiao J, Jia X, Ding N, Ma G, Wang G, Yue L, Liang Q, Sheng Y, Sun Y, Huang H, Chen F. 2018. Pan-genomic study of Mycobacterium tuberculosis reflecting the primary/secondary genes, generality/individuality, and the interconversion through copy number variations. Front Microbiol 9:1886. 10.3389/fmicb.2018.01886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kavvas ES, Catoiu E, Mih N, Yurkovich JT, Seif Y, Dillon N, Heckmann D, Anand A, Yang L, Nizet V, Monk JM, Palsson BO. 2018. Machine learning and structural analysis of Mycobacterium tuberculosis pan-genome identifies genetic signatures of antibiotic resistance. Nat Commun 9:4306. 10.1038/s41467-018-06634-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 62.Zimpel CK, Brandão PE, de Souza Filho AF, de Souza RF, Ikuta CY, Ferreira Neto JS, Camargo NCS, Heinemann MB, Guimarães AMS. 2017. Complete genome sequencing of Mycobacterium bovis SP38 and comparative genomics of Mycobacterium bovis and M. tuberculosis strains. Front Microbiol 8:2389. 10.3389/fmicb.2017.02389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patané JS, Martins J, Beatriz Castelão A, Patané JS, Martins J, Beatriz Castelão A, Nishibe C, Montera L, Bigi F, Zumárraga MJ, Cataldi AA, Fonseca Junior A, Roxo E, Luiza A, Osório AR, Jorge KS, Thacker TC, Almeida NF, Araújo FR, Setubal JC. 2017. Patterns and processes of Mycobacterium bovis evolution revealed by phylogenomic analyses. Genome Biol Evol 9:521–535. 10.1093/gbe/evx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riley DR, Angiuoli SV, Crabtree J, Dunning Hotopp JC, Tettelin H. 2012. Using Sybil for interactive comparative genomics of microbes on the web. Bioinformatics 28:160–166. 10.1093/bioinformatics/btr652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, Gladstone RA, Lo S, Beaudoin C, Floto RA, Frost SDW, Corander J, Bentley SD, Parkhill J. 2020. Producing polished prokaryotic pangenomes with the panaroo pipeline. Genome Biol 21:180. 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Denton JF, Lugo-Martinez J, Tucker AE, Schrider DR, Warren WC, Hahn MW. 2014. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput Biol 10:e1003998. 10.1371/journal.pcbi.1003998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salzberg SL. 2019. Next-generation genome annotation: we still struggle to get it right. Genome Biol 20:92. 10.1186/s13059-019-1715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ridley M. 1983. The explanation of organic diversity: the comparative method and adaptations for mating. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 70.Wells RM, Jones CM, Xi Z, Speer A, Danilchanka O, Doornbos KS, Sun P, Wu F, Tian C, Niederweis M. 2013. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog 9:e1003120. 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hemati Z, Derakhshandeh A, Haghkhah M, Chaubey KK, Gupta S, Singh M, Singh SV, Dhama K. 2019. Mammalian cell entry operons; novel and major subset candidates for diagnostics with special reference to Mycobacterium avium subspecies paratuberculosis infection. Vet Q 39:65–75. 10.1080/01652176.2019.1641764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martínez-Núñez MA, López VEL. 2016. Nonribosomal peptides synthetases and their applications in industry. Sustain Chem Process 4:13. 10.1186/s40508-016-0057-6. [DOI] [Google Scholar]
- 73.Sayes F, Pawlik A, Frigui W, Sayes F, Pawlik A, Frigui W, Gröschel MI, Crommelynck S, Fayolle C, Cia F, Bancroft GJ, Bottai D, Leclerc C, Brosch R, Majlessi L. 2016. CD4+ T cells recognizing PE/PPE antigens directly or via cross reactivity are protective against pulmonary Mycobacterium tuberculosis infection. PLoS Pathog 12:e1005770. 10.1371/journal.ppat.1005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ganusov VV, Klinkenberg D, Bakker D, Koets AP. 2015. Evaluating contribution of the cellular and humoral immune responses to the control of shedding of Mycobacterium avium spp. paratuberculosis in cattle. Vet Res 46:62. 10.1186/s13567-015-0204-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fishbein S, van Wyk N, Warren RM, Sampson SL. 2015. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol 96:901–916. 10.1111/mmi.12981. [DOI] [PubMed] [Google Scholar]
- 76.Pradhan AK, Van Kessel JS, Karns JS, Wolfgang DR, Hovingh E, Nelen KA, Smith JM, Whitlock RH, Fyock T, Ladely S, Fedorka-Cray PJ, Schukken YH. 2009. Dynamics of endemic infectious diseases of animal and human importance on three dairy herds in the northeastern United States. J Dairy Sci 92:1811–1825. 10.3168/jds.2008-1486. [DOI] [PubMed] [Google Scholar]
- 77.Salvador LCM, O'Brien DJ, Cosgrove MK, Stuber TP, Schooley AM, Crispell J, Church SV, Gröhn YT, Robbe-Austerman S, Kao RR. 2019. Disease management at the wildlife-livestock interface: using whole-genome sequencing to study the role of elk in Mycobacterium bovis transmission in Michigan, USA. Mol Ecol 28:2192–2205. 10.1111/mec.15061. [DOI] [PubMed] [Google Scholar]
- 78.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.10.33. 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 83.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 84.Wynne JW, Seemann T, Bulach DM, Coutts SA, Talaat AM, Michalski WP. 2010. Resequencing the Mycobacterium avium subsp. paratuberculosis K10 genome: improved annotation and revised genome sequence. J Bacteriol 192:6319–6320. 10.1128/JB.00972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. 2014. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol 31:1077–1088. 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bruen TC, Philippe H, Bryant D. 2006. A simple and robust statistical test for detecting the presence of recombination. Genetics 172:2665–2681. 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30:1224–1228. 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klopfenstein DV, Zhang L, Pedersen BS, Ramírez F, Warwick Vesztrocy A, Naldi A, Mungall CJ, Yunes JM, Botvinnik O, Weigel M, Dampier W, Dessimoz C, Flick P, Tang H. 2018. GOATOOLS: a Python library for Gene Ontology analyses. Sci Rep 8:10872. 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 95.Read AF, Nee S. 1995. Inference from binary comparative data. J Theor Biol 173:99–108. 10.1006/jtbi.1995.0047. [DOI] [Google Scholar]
- 96.Maddison WP. 2000. Testing character correlation using pairwise comparisons on a phylogeny. J Theor Biol 202:195–204. 10.1006/jtbi.1999.1050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence read data associated with this project have been deposited at NCBI in the Sequence Read Archive database under the following BioProject accession number: PRJNA686527.