This study shows that halolysin R4 from Haloferax mediterranei provides its host antagonistic and defensive activities against other haloarchaea, which expands our knowledge of the traditional function of haloarchaeal extracellular proteases. Haloarchaeal extracellular serine proteases have been previously discussed as growth phase-dependent proteins, whereas our study reports constitutive expression of halolysin R4.
KEYWORDS: Haloarchaea, halolysin, halocin, protease, bacteriocin, antagonisms, peptide antibiotics
ABSTRACT
Halolysins, which are subtilisin-like serine proteases of haloarchaea, are usually secreted into the extracellular matrix via the twin-arginine translocation pathway. A small number of activated molecules can greatly affect cell growth owing to their proteolytic activity. It is, however, unclear as to whether this proteolysis-based growth inhibition by halolysins conveys antagonistic or defensive effects against other resident and potentially competitive microorganisms. Here, we report that halolysin R4 (HlyR4), encoded by the hlyR4 gene, is the key enzyme in the initial steps of extracellular protein utilization in Haloferax mediterranei (Hfx. mediterranei). HlyR4 shows significant antagonistic activity against other haloarchaeal strains. Deletion of hlyR4 completely halts the inhibition activity of Hfx. mediterranei toward other haloarchaea, while correspondingly, complementation of hlyR4 almost completely restores the inhibition activity. Furthermore, Hfx. mediterranei strains containing hlyR4 showed a certain amount of resistance to halocins and halolysins in a hypersaline environment, and this function of hlyR4 is reproducible in Haloarcula hispanica. The versatility of HlyR4 enables its encoding organism to outcompete other haloarchaea living in the same hypersaline environment. Intriguingly, unlike the growth phase-dependent halolysins SptA and Nep, it is likely that HlyR4 may be secreted independently of growth phase. This study provides a new peptide antibiotic candidate in haloarchaea, as well as new insight into a better understanding of the ecological roles of halolysins.
IMPORTANCE This study shows that halolysin R4 from Haloferax mediterranei provides its host antagonistic and defensive activities against other haloarchaea, which expands our knowledge of the traditional function of haloarchaeal extracellular proteases. Haloarchaeal extracellular serine proteases have been previously discussed as growth phase-dependent proteins, whereas our study reports constitutive expression of halolysin R4. This work also clearly reveals a hidden diversity of extracellular proteases from haloarchaea. Studies on multifunctional halolysins reveal that they play an important ecological role in shaping microbial community composition and provide a new perspective toward understanding the intricate interactions between haloarchaeal cells in hypersaline environments. HlyR4 can lyse competing cells living in the same environment, and the cell debris may probably be utilized as nutrients, which may constitute an important part of nutrient cycling in extremely hypersaline environments.
INTRODUCTION
Halophilic archaea (Haloarchaea) live in hypersaline environments (3 to 5 M NaCl), such as salt lakes, salt ponds, and marine salterns, which are unfavorable to many other cohabiting microorganisms. They are a group of extremophilic microorganisms with unique physiological and biochemical properties (1, 2). Notably, they produce many biomolecules that remain active and stable under high salt concentrations, rendering them useful for various biotechnological and industrial applications (3). Halocins, a group of polypeptide biomolecules produced by numerous haloarchaea, are believed to be an effective weapon for competing with other residents in the same environment, which often has a limited supply of nutrients and space (4–6).
Halocin H4 (HalH4), a bacteriocin-like polypeptide from Haloferax mediterranei (Hfx. mediterranei) ATCC 33500 (7), is capable of killing sensitive haloarchaeal cells by targeting the cell wall or cytoplasmic membrane, thereby causing membrane leakage (4, 8). This compromises the integrity of the cell membrane and will eventually lead to cell swelling and lysis. In addition, sublethal concentrations of a halocin(s) can significantly promote DNA uptake by changing the cell surface structure of Hfx. mediterranei (9).
It has been reported that individual haloarchaeal species have evolved specific mechanisms to survive in the presence of an endogenous halocin(s). One such example is the halC8 gene from Halobacterium sp. strain AS7092, which encodes halocin C8 and its immune peptide HalI. HalI, which is localized both on the cell membrane and within the cell, likely binds to HalC8, thereby sequestering the halocin and inhibiting its activity (10). Many haloarchaea secrete proteolytic enzymes called halolysins to degrade halocins or other proteins in a hypersaline environment. In hypersaline environments where high quantities of halocin-producing strains have been isolated, it has been observed that a number of these strains produce high levels of extracellular proteases (11–13). The products created from such proteolysis, such as oligopeptides, dipeptides, and amino acid intermediates, are taken up as nutrients and fed through central metabolic pathways (14). A similar proteolytic mechanism is present in bacteria as well. Pseudoalteromonas sp. strain CF6-2 can secrete pseudoalterin, an extracellular metalloprotease, to kill Gram-positive bacteria by degrading their peptidoglycan and can subsequently utilize the degradation products as nutrients (15).
All of the extracellular proteases isolated from haloarchaea to date have been categorized as subtilisin-like serine proteases. Most of these proteases appear closely related to the subfamily S8A (COG1404) and have thus been designated halolysins (14, 16, 17). Several halolysins, e.g., 172P1 from Natrialba asiatica (18), HlyR4 from Hfx. mediterranei (18), SptA from Natrinema sp. strain J7-2 (19), and Nep from Natrialba magadii (20), have been characterized successfully. Several halolysins from different species are likely to share a secretory strategy; namely, the twin-arginine translocation (Tat) pathway (21, 22).
Many archaeal envelopes contain a protein coat or sheath composed of one or more exposed surface layer (S-layer) proteins (14). These S-layer proteins contribute to the cell’s structural integrity and protect the lipid membrane (23). Halolysins secreted by certain species of archaea have the natural ability to degrade these S-layer proteins or peptides (22), along with many proteins secreted by other haloarchaeal organisms. This property suggests that halolysin producers may possess an inhibition activity against other haloarchaeal cells and/or a resistance to proteinaceous antagonistic substances such as halocins or halolysins.
In this study, gene knockout and complementation of the hlyR4 gene, encoding HlyR4 in Hfx. mediterranei, and heterologous expression in Haloarcula hispanica (Har. hispanica) clearly demonstrated that HlyR4 is an extracellular serine protease. However, different from other reported extracellular serine proteases, it is constitutively expressed, as demonstrated here by reverse transcription analysis. Additionally, our results further indicated that HlyR4 is the key enzyme in the initial steps of extracellular protein utilization. In Hfx. mediterranei, HlyR4, rather than HalH4, is the main antimicrobial peptide acting against dozens of haloarchaeal strains from different genera. This paper presents experimental data that clearly demonstrate that the proteolytic activity of HlyR4 confers its encoding organism significant defensive and antagonistic effects toward other haloarchaeal strains.
RESULTS
Characterization of halolysin R4 and proteolytic activity of the producer.
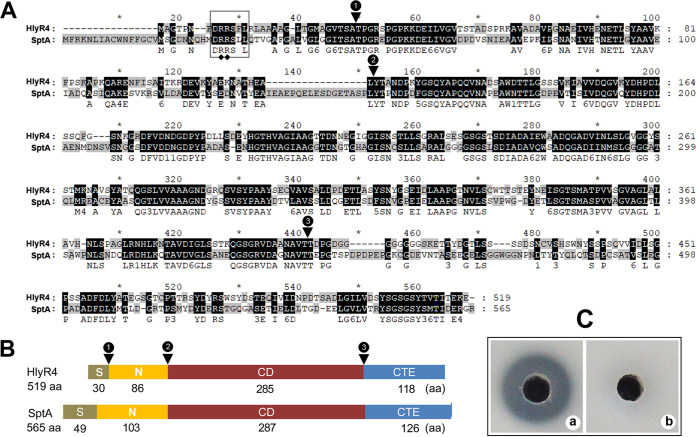
Halolysin R4 (HlyR4) (GenBank accession no. BAA10958.1) encoded by the hlyR4 gene (HFX_2419) located on the chromosome of Hfx. mediterranei ATCC 33500 consists of 519 amino acids (aa) and belongs to the Peptidase_S8 subtilase family in the Pfam protein domain database. HlyR4 is a serine protease with a calculated molecular mass of 53.5 kDa and a relatively low isoelectric point of 4.3 (24). Amino acid sequence alignment showed that HlyR4 (519 aa) is closely related to SptA (565 aa) (GenBank accession no. AAX19896.1) from Natrinema sp. J7-2, with a high sequence similarity value of 53.3% (query cover, 98%; E-value, 2e−169) (Fig. 1A). A typical twin-arginine motif (RR) of the Tat signal motif-like region (DRRSFL) was identified in the N-terminal signal peptide of HlyR4 (Fig. 1A).
FIG 1.
Schematic representation of the primary structure of halolysin R4 (HlyR4) compared with SptA. (A) Alignment of the amino acid sequences of HlyR4 (GenBank accession no. BAA10958.1) and SptA (AAX19896.1). The Tat motif (DRRSLL) in the signal peptide is shown in a box. The representative twin-arginine structure is marked by filled diamonds. Conjunction points between the signal peptide (S), the N-terminal propeptide (N), the catalytic domain (CD), and the C-terminal extension (CTE) are marked by filled triangles. (B) Schematic diagram of HlyR4 and SptA showing the length of the S, N, CD, and CTE. (C) Proteolytic activity of HlyR4 was assessed by dripping 100 μl of untreated (a) and PMSF-treated (b) supernatants of Hfx. mediterranei ATCC 33500 into the hole of a skim milk agar plate. The final concentration of the PMSF in supernatants was 60 μg ml−1 (b).
Both HlyR4 and SptA share the same protein structural domains, consisting of a signal peptide (S), an N-terminal propeptide (N), a catalytic domain (CD), and a C-terminal extension (CTE) (Fig. 1B).
Proteolytic activity detection assays on skim milk agar plates showed that the HlyR4-producing strain (Hfx. mediterranei ATCC 33500) (Fig. 1C) and other wild-type haloarchaeal strains, i.e., Haloarchaeobius sp. strain FL94, Halococcus saccharolyticus CGMCC 1.6994, Natrialba sp. J7, and Haloarchaeobius sp. strain FL176 (see Fig. S1 in the supplemental material), can degrade the protein in skim milk, indicated by a clear region surrounding the colony. Phenylmethylsulfonyl fluoride (PMSF) (10 μg ml−1) can completely inhibit this protease activity, indicating that the cell suspension of Hfx. mediterranei strain ATCC 33500 contains a typical serine protease, which is consistent with the production of the reported extracellular serine protease HlyR4 (18, 24).
Knockout and complementation of genes encoding halolysin and halocin.
Although some halocins exhibit significant inhibition activity against other haloarchaeal strains, it has been reported that HalH4, encoded by halH4, fails to show any antimicrobial activity against some strains from the genera Halorubrum and Halobacterium (9, 25). Whether halolysin R4, encoded by hlyR4, exerts antimicrobial activity against other haloarchaeal strains has not been explored. Based on the suicide plasmid pHFX and the overlapping PCR approach, the hlyR4 gene knockout plasmid pHFX-UDR4 was constructed and verified via restriction enzyme digestion analysis (Fig. S2a). Prior to transformation of Hfx. mediterranei strains EPS (deficient in the exopolysaccharide synthesis gene cluster) (26) and EPSH (the halH4 deletion mutant of strain EPS) (9), the DNA fragment inserted into pHFX was verified by DNA sequencing (data not shown). By utilizing the pop-in and pop-out gene knockout strategy, we obtained Hfx. mediterranei strains EPSR (the hlyR4 deletion mutant of strain EPS) and EPSHR (the hlyR4 deletion mutant of strain EPSH).
The complete hlyR4 with its native promoter was amplified and inserted into the shuttle plasmid pWL502 (27), resulting in the ΔhlyR4 complementation plasmid pWR4 (Fig. S2b). Plasmid pWR4 was introduced into strains EPSR and EPSHR, resulting in strains EPSR-R4 and EPSHR-R4, respectively.
Eight Hfx. mediterranei strains, i.e., DF50, EPS, EPSH, EPSR, EPSHR, EPSR-R4, EPSHR-R4, and EPSH-H4, were subsequently verified by PCR (Fig. S3a). The results clearly showed that this series of eight gene manipulation strains was constructed successfully.
Importance of HlyR4 in the initial steps of extracellular protein utilization.
Previous sequencing of the hlyR4 gene encoding HlyR4 led to its proposal as an extracellular serine protease-encoding gene (24, 28). There are probably several genes involved in the utilization of extracellular proteins. Deletion of hlyR4 from strains EPS and EPSH, resulting in strains EPSR and EPSHR, respectively, was accompanied by a loss of proteolytic activity on a skim milk agar plate. This indicated that proteolytic activity is dependent on the activation of hlyR4 (Fig. S3b). When hlyR4 was introduced back into the hlyR4-deficient strain, i.e., strain EPSR to strain EPSR-R4 and strain EPSHR to strain EPSHR-R4, proteolytic activity was restored (Fig. S3b). Furthermore, we found that hlyR4 was the only gene annotated as an extracellular serine protease-encoding gene within the Hfx. mediterranei ATCC 33500 genome (GCA_000306765.2). Based on this genome annotation and our proteolytic activity tests, we can assume that HlyR4 is the key enzyme in the initial steps of extracellular protein utilization in Hfx. mediterranei ATCC 33500, the original wild-type strain in this study (Fig. S3b).
Effect of halH4 on inhibiting other haloarchaeal strains.
If one microbe can inhibit the growth of another on an agar plate, a clear inhibition zone will occur around the colony. If the tested strain exhibits halocin production, it will be inhibiting the indicator strain contained within the medium. The diameter of the inhibition zone around the colony on the indicator agar plate reflects the level of antagonistic activity. Inhibition zones were clearly present around the spots of the tested strain EPS when spotted on an indicator agar plate, i.e., strain Halorubrum sp. strain G16-1 (Fig. 2A, left) and strain Haloarcula hispanica ATCC 33960 (Fig. 2A, right). However, deletion of halH4 from strain EPS, resulting in strain EPSH, yielded no reduction in the diameter of the inhibition zone against indicator strains such as Halorubrum sp. strains G16-1 (Fig. 2A) and LN72 (Fig. 2B), as well as Haloarcula hispanica ATCC 33960 (Fig. 2A). Furthermore, when halH4 was introduced back into strain EPSH through transformation using pWH4, resulting in strain EPSH-H4, the diameter of the inhibition zone still remained unchanged (Fig. 2). The inhibition activities of strains EPS, EPSH, and EPSH-H4 unequivocally showed that deletion or complementation of halH4 barely had any effect on the strains’ antagonistic activities against the indicator strains from the genera Halorubrum and Haloarcula (Fig. 2).
FIG 2.
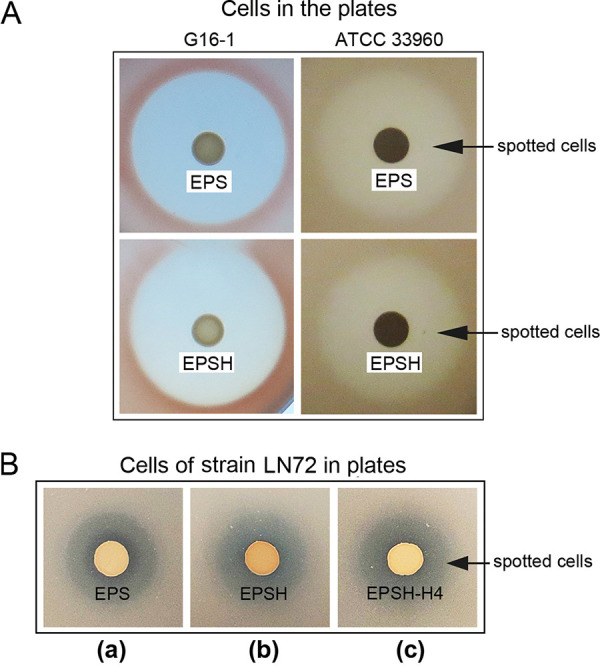
Contribution of halH4 in Hfx. mediterranei strain EPS to inhibition activity against other haloarchaeal strains. (A) Growth of Haloferax mediterranei strain EPS (strain EPS) and halH4-deficient Hfx. mediterranei strain EPS (strain EPSH) on indicator plates of Halorubrum sp. G16-1 and Haloarcula hispanica ATCC 33960. (B) Growth of strain EPS (a), strain EPSH (b), and halH4 complementation EPSH strain (strain EPSH-H4) (c) on Halorubrum sp. LN72.
The inhibition profiles of strains EPS and EPSH were compared against those of other haloarchaeal strains from the genera Natronomonas, Halorubrum, Halorubellus, Haloparvum, Halobaculum, Halobacterium, Haloarcula, and Haloarchaeobius (Table 1). Not all haloarchaeal strains were inhibited by strain EPS or EPSH, and this demonstrates the specificity of halocins in haloarchaea. For strains showing a clear inhibition zone, deletion of halH4 did not lead to a reduction in the diameter of the inhibition zone (Table 1), further supporting that halocin H4 bears no antagonistic activities against these strains.
TABLE 1.
Antagonistic activity against wild-type haloarchaeal strains by Hfx. mediterranei strains
| Indicator strain | Accession no. (16S rRNA gene) | Diam (mean ± SD) of inhibition zone (mm)a |
|||
|---|---|---|---|---|---|
| EPS | EPSH | EPSR | EPSR-R4 | ||
| Natronomonas sp. LN108 | MN826835 | 17.3 ± 0.5 | 16.3 ± 0.5 | 0 | 16.3 ± 0.5 |
| Saliphagus sp. LR7 | MG097860 | 0 | 0 | 0 | 0 |
| Natrialba sp. J3 | MN826721 | 0 | 0 | 0 | 0 |
| Natrialba sp. J7 | MN826722 | 0 | 0 | 0 | 0 |
| Haloterrigena salifodinae ZY19 | MG097861 | 0 | 0 | 0 | 0 |
| Haloterrigena sp. LN16 | MN826832 | 0 | 0 | 0 | 0 |
| Haloterrigena sp. J68 | MN826723 | 0 | 0 | 0 | 0 |
| Halostagnicola sp. GSM22 | MN856152 | 0 | 0 | 0 | 0 |
| Halorubrum sp. J88 | MN826725 | 0 | 0 | 0 | 0 |
| Halorubrum sp. LN72 | MN829452 | 19.7 ± 1.8 | 19.7 ± 2.1 | 0 | 19.0 ± 1.4 |
| Halorubrum sp. LN60 | MN826834 | 19.0 ± 0.8 | 18.7 ± 0.5 | 0 | 18.7 ± 0.5 |
| Halorubrum sp. LN11 | MN826831 | 10.6 ± 0.5 | 10.3 ± 0.5 | 0 | 11.0 ± 0.8 |
| Halorubrum sp. LN10 | MN826830 | 16.3 ± 0.5 | 16.3 ± 0.5 | 0 | 16.7 ± 0.5 |
| Halorubrum sp. F4 | MG097853 | 10.7 ± 0.5 | 10.0 ± 0.8 | 0 | 10.0 ± 0.8 |
| Halorubrum sp. G16-1 | MH106556 | 27.0 ± 1.4 | 26.7 ± 0.9 | 0 | 26.7 ± 1.2 |
| Halorubrum sp. LN27 | MN829451 | 23.3 ± 0.5 | 23.0 ± 0.0 | 20.7 ± 0.5 | 23.0 ± 0.9 |
| Halorubrum sp. G105 | MN826838 | 16.7 ± 0.9 | 16.7 ± 0.5 | 7.3 ± 0.5 | 16.0 ± 0.0 |
| Halorubrum sp. Y13 | MN826840 | 0 | 0 | 0 | 0 |
| Halorubrum sp. F18 | MN826839 | 8.7 ± 0.5 | 8.3 ± 0.5 | 7.3 ± 0.5 | 8.7 ± 0.5 |
| Halorubellus sp. FL87 | MN833415 | 17.3 ± 0.5 | 17.0 ± 0.8 | 18.3 ± 0.5 | 17.3 ± 0.5 |
| Halorhabdus sp. FL145 | MN098869 | 0 | 0 | 0 | 0 |
| Halopenitus sp. J523 | MN826728 | 0 | 0 | 0 | 0 |
| Haloparvum sedimenti DYS4 | KP202831 | 24.3 ± 1.2 | 23.3 ± 0.5 | 0 | 23.7 ± 0.5 |
| Haloparvum sedimenti Y2 | KP276581 | 19.7 ± 0.5 | 20.0 ± 0.0 | 0 | 20.7 ± 0.5 |
| Halohasta sp. FL93 | MN833416 | 0 | 0 | 0 | 0 |
| Haloferax sp. Q22 | KJ644210 | 0 | 0 | 0 | 0 |
| Halococcus salsus ZJ1 | MG097854 | 0 | 0 | 0 | 0 |
| Halococcus saccharolyticus CGMCC 1.6994 | AB663370 | 0 | 0 | 0 | 0 |
| Halobaculum roseum D90 | KX376701 | 23.0 ± 2.2 | 22.7 ± 1.9 | 23.0 ± 1.4 | 22.7 ± 1.2 |
| Halobacterium sp. FL45 | MN833413 | 26.3 ± 0.9 | 25.3 ± 0.5 | 0 | 25.3 ± 0.9 |
| Haloarcula sp. LN39 | MN826833 | 17.6 ± 0.5 | 18.0 ± 0.0 | 0 | 18.3 ± 0.5 |
| Haloarcula sp. LN121 | MN826836 | 9.7 ± 1.7 | 9.7 ± 1.2 | 0 | 10.0 ± 1.4 |
| Haloarchaeobius sp. FL94 | MN833417 | 17.0 ± 0.8 | 17.3 ± 0.5 | 9.0 ± 1.4 | 17.7 ± 0.9 |
| Haloarchaeobius sp. FL176 | MN833418 | 16.7 ± 1.7 | 16.3 ± 1.2 | 9.0 ± 1.4 | 17.0 ± 0.8 |
| Halalkalicoccus subterraneus GSM28 | MG097856 | 0 | 0 | 0 | 0 |
| Haladaptatus paucihalophilus CGMCC 1.8953 | NR_043744 | 0 | 0 | 0 | 0 |
Zero indicates no inhibition effect; the diameter of the lawn for each strain on the indicator plate was about 6 mm. Strain EPS, Hfx. mediterranei strain DF50 ΔEPS; strain EPSH, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4; strain EPSR, Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4; strain EPSR-R4, Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4::hlyR4. These data were derived from three biological replicates.
Role of hlyR4 in inhibition activity toward other haloarchaeal strains.
Most haloarchaeal cells have S-layer proteins on the cell surface (29), which may suggest that an extracellular protease-producing strain can inhibit some other haloarchaeal strains. Deletion of hlyR4 from strain EPS, resulting in strain EPSR, led to a loss of proteolytic activity, as indicated on skim milk plates (Fig. 3, upper row). Complementation of hlyR4, resulting in strain EPSR-R4, restored these proteolytic activities (Fig. 3, upper row). The loss and regain of proteolytic activity correspond to the loss and regain of inhibition activity in strains EPSR and EPSR-R4 against Halorubrum sp. strain LN72 (Fig. 3, bottom row), suggesting that the inhibition activity of Hfx. mediterranei against haloarchaeal strains is causatively associated with the proteolytic activity of HlyR4.
FIG 3.
Proteolytic and antagonistic activity of hlyR4-related Hfx. mediterranei strains. The proteolytic activities of EPS, EPSR, and EPSR-R4 were detected on skim milk nutrient agar plates (upper row). Haloferax mediterranei strains EPS, EPSR, and EPSR-R4 were also used to conduct the antagonistic assays to Halorubrum sp. LN72 (bottom row). Strain EPS, the eps deletion mutant of Haloferax mediterranei DF50; strain EPSR, the hlyR4 deletion mutant of Hfx. mediterranei strain EPS; strain EPSR-R4, the hlyR4 complementation strain of strain EPSR.
Furthermore, strains EPS, EPSR, and EPSR-R4 were used to comprehensively screen other haloarchaeal strains available from our laboratory (Table 1). Many strains from the genera Natronomonas, Halorubrum, Haloparvum, Halobacterium, and Haloarcula showed inhibition profiles similar to that of Halorubrum sp. strain LN72, which exhibited almost complete loss or restoration of inhibition for strains EPSR and EPSR-R4 proportional to the presence or absence of hlyR4. However, different inhibition profiles were discovered for the following: (i) Halorubrum sp. strains G105 and LN185, Haloarcula sp. strain H4, Haloarchaeobius sp. strain FL94, and Haloarchaeobius sp. strain FL176, in which a reduced inhibition zone surrounding Hfx. mediterranei was observed in the absence of hlyR4; (ii) Halorubrum sp. strains LN27 and FL87 and Halobaculum roseum D90, in which the size of the inhibition zone exhibited no significant reduction in the absence of hlyR4; (iii) strains from the genera Saliphagus, Natrialba, Haloterrigena, Halostagnicola, Halorhabdus, Halopenitus, Halohasta, Haloferax, Halococcus, Halalkalicoccus, and Haladaptatus, in which Hfx. mediterranei strains had no inhibition activity regardless of the absence or presence of hlyR4 (Table 1).
Although one type of halolysin cannot inhibit all haloarchaeal strains, halolysins may provide their encoding organisms with an ability to inhibit the growth of some competing haloarchaea. Other extracellular protease-producing strains, such as Haloarchaeobius sp. FL94 and FL176, Halococcus saccharolyticus CGMCC 1.6994, and Natrialba sp. J7, have been preserved in our laboratory, and those organisms shown to display proteolytic activity (Fig. S1) were assessed to determine their inhibition activities against other haloarchaeal strains (Table 2). The results showed that the extracellular protease-producing strains may inhibit some other haloarchaeal strains, especially those belonging to the genera Halorubrum and Haloarcula (Table 2), which illustrates that the inhibition spectrum of these halolysin-producing strains is not narrow. The inhibition activities might be attributable to the production of halolysins. If so, the spectrum of inhibition activities also indicates that extracellular proteases produced by haloarchaea vary among strains and species (Table 2). The antimicrobial spectra of these four extracellular protease-producing strains preserved in our laboratory are very different from one another, which illustrates that halolysin activities and specificities probably vary among species or strains.
TABLE 2.
Antagonistic activity of extracellular protease-producing strains
| Indicator strain | Accession no. (16S rRNA gene) | Diam (mean ± SD) of inhibition zone (mm)a |
|||||
|---|---|---|---|---|---|---|---|
| FL94 | FL176 | J7 | 1.6994 | ZJ1 | LN10 | ||
| Haloarcula sp. LN121 | MN826836 | 0 | 0 | 0 | 0 | 0 | 0 |
| Haloarcula sp. H4 | MN893883 | 15.0 ± 1.7 | 21.7 ± 2.8 | 23.7 ± 2.0 | 0 | 0 | 0 |
| Natronomonas sp. LN108 | MN826835 | 0 | 0 | 8.0 ± 0.0 | 0 | 0 | 0 |
| Halorubrum sp. LN10 | MN826830 | 7.3 ± 1.2 | 0 | 7.0 ± 1.0 | 8.7 ± 2.1 | 0 | 0 |
| Halorubrum sp. LN187 | MN826837 | 9.0 ± 1.0 | 0 | 0 | 0 | 0 | 0 |
| Halorubrum sp. LN60 | MN826834 | 7.7 ± 1.5 | 0 | 11.7 ± 2.0 | 29.7 ± 1.5 | 0 | 0 |
| Halorubrum sp. LN11 | MN826831 | 0 | 0 | 8.0 ± 1.0 | 0 | 0 | 0 |
| Halorubrum sp. FL23 | MT573937 | 18.5 ± 0.7 | 27.5 ± 0.7 | 9.1 ± 0.7 | 0 | 0 | 0 |
| Halorubrum sp. FL32 | MT573938 | 12.0 ± 0.5 | 28.5 ± 0.7 | 8.3 ± 1.0 | 0 | 0 | 0 |
| Halorubrum sp. Y13 | MN826840 | 0 | 0 | 0 | 0 | 0 | 0 |
| Halorubrum sp. LN185 | MN893884 | 0 | 0 | 7.0 ± 1.0 | 0 | 0 | 0 |
| Haloferax sp. Q22 | KJ644210 | 0 | 8.7 ± 1.5 | 21.7 ± 3.0 | 0 | 0 | 0 |
| Halobaculum roseum D90 | KX376701 | 8.7 ± 1.5 | 8.0 ± 1.0 | 24.7 ± 2.1 | 0 | 0 | 0 |
| Haloarchaeobius sp. FL94 | MN833417 | 0 | 0 | 0 | 0 | 0 | 0 |
| Halalkalicoccus subterraneus GSM28 | MG097856 | 0 | 0 | 10.7 ± 0.5 | 0 | 0 | 0 |
| Halococcus salsus ZJ1 | MG097854 | 0 | 0 | 0 | 0 | 0 | 0 |
| Halorhabdus sp. FL145 | MN098869 | 0 | 0 | 0 | 0 | 0 | 0 |
Zero indicates no inhibition activity; the diameter of the lawn for each strain on the indicator plate was about 6 mm, and the diameters (mean value ± SD) of their inhibition zones are shown. FL94, Haloarchaeobius sp. FL94; FL176, Haloarchaeobius sp. FL176; J7, Natrialba sp. J7; 1.6994, Halococcus saccharolyticus CGMCC 1.6994 (see Table 5); ZJ1, Halococcus salsus ZJ1; LN10, Halorubrum sp. LN10. Strains FL94, FL176, J7, and 1.6994 are four extracellular protease-producing haloarchaeal strains (see Fig. S1 in the supplemental material). Strains ZJ1 and LN10 were used as negative controls. These data were derived from three biological replicates.
The interaction between these gene manipulation strains of Hfx. mediterranei provides a clear picture of the inhibition activity of HlyR4. The hlyR4-positive strains, i.e., EPS, EPSH, EPSH-H4, EPSR-R4, and EPSHR-R4, all displayed significant inhibition activities against the hlyR4-negative strains, i.e., EPSR and EPSHR (Table 3 and Fig. S4). In addition, Haloarcula hispanica strain DF60 (pyrF-deficient Har. hispanica ATCC 33960) did not exert any inhibition effect on the indicator agar plates in which the cells of strain EPS, EPSR, or EPSR-R4 were embedded (Fig. 4a to c). When hlyR4 was introduced into cells of strain DF60, resulting in Har. hispanica strain DF60-R4 (Fig. S5), a clear inhibition zone was present on the indicator agar plate containing cells of strain EPSR (Fig. 4e), while no inhibition zone was present on the indicator agar plates containing cells of strain EPS or EPSR-R4 (Fig. 4d and f), both of which were hlyR4 positive.
TABLE 3.
Interaction between different gene manipulation Hfx. mediterranei strainsa
| Cells spotted on the plate | Cells in the plates |
||||||
|---|---|---|---|---|---|---|---|
| EPS | EPSH | EPSH-H4 | EPSR | EPSHR | EPSR-R4 | EPSHR-R4 | |
| EPS | – | – | – | + | + | – | – |
| EPSH | – | – | – | + | + | – | – |
| EPSH-H4 | – | – | – | + | + | – | – |
| EPSR | – | – | – | – | – | – | – |
| EPSHR | – | – | – | – | – | – | – |
| EPSR-R4 | – | – | – | + | + | – | – |
| EPSHR-R4 | – | – | – | + | + | – | – |
Strains shown at the top of the columns are the indicator strains, while strains in the first column are the tested strains dropped onto the indicator agar plates. Strain EPS, Hfx. mediterranei strain DF50 ΔEPS; strain EPSH, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4; strain EPSH-H4, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4::halH4; strain EPSR, Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4; strain EPSHR, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4 ΔhlyR4; strain EPSR-R4, Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4::hlyR4; strain EPSHR-R4, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4 ΔhlyR4::hlyR4. These results were obtained through three biological replicates.
FIG 4.
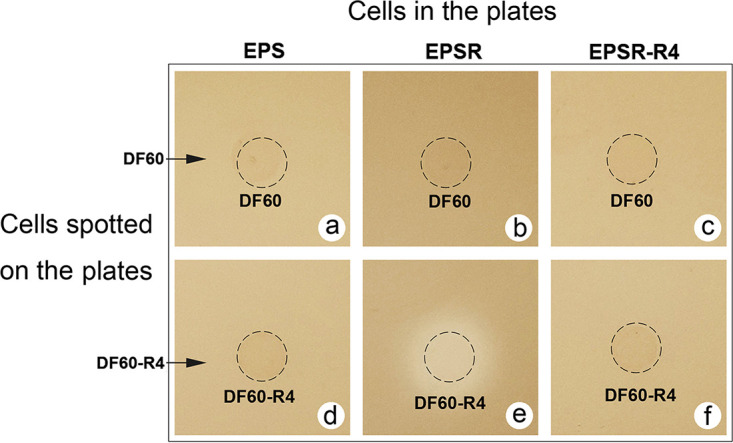
Effect of hlyR4 in Haloarcula hispanica strain DF60 against Hfx. mediterranei strain EPS and its derivatives. Cell-free supernatants of Haloarcula hispanica strain DF60 (a, b, and c) and Haloarcula hispanica strain DF60 containing hlyR4 (d, e, and f) were dropped onto the indicator plates of Hfx. mediterranei strain EPS (a and d), Hfx. mediterranei strain EPSR (b and e), and Hfx. mediterranei strain EPSR-R4 (c and f), respectively.
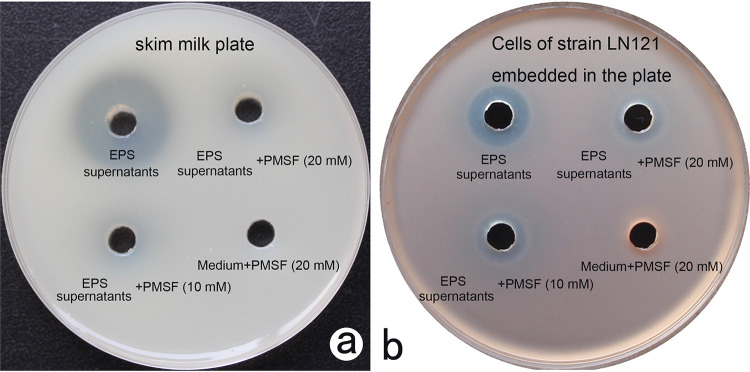
Supernatants of strain EPS harboring HlyR4 exhibited significant proteolytic activity on the skim milk plate, while the proteolytic activity disappeared in the presence of PMSF at 10 mM or 20 mM (Fig. 5a). The disappearance of the proteolytic activity was accompanied by a sharp decline in inhibition activity, although the inhibition zone did not disappear entirely (Fig. 5b). Cells in the presence of PMSF at a concentration of either 10 mM or 20 mM did not display any proteolytic activity on the skim milk plates or against the lawn cells of strain LN121 (Fig. 5).
FIG 5.
Correlation of proteolytic activity and antagonistic activity. (a and b) Proteolytic activity was detected using skim milk agar plates (a), while antagonistic activity was tested using strain LN121 indicator plates made by mixing the cells of strain LN121 with solid medium and pouring the mixture into the petri dishes (b). Supernatants of strain EPS under stationary phase were collected for activity tests. Phenylmethylsulfonyl fluoride (PMSF), an inhibitor of serine protease, was used to eliminate the proteolytic activities. The concentration of PMSF in the supernatants was 10 mM or 20 mM, while supernatants without PMSF and liquid AS-168 medium supplemented with PMSF (20 mM) were used as controls. LN121, Haloarcula sp. LN121; medium, liquid AS-168 medium.
Taken together, these data provide a strong indication that hlyR4 confers an inhibition activity on its encoding organism.
Inhibition effects of extracellular protease producers on other haloarchaeal strains living in the same habitat.
Due to the salt-tolerant proteolytic activity of halolysins, cell debris after cell lysis can be reused by other microorganisms living in hypersaline environments, which contributes substantially to the cycling of nutrients in this harsh niche ecosystem. Extracellular protease-producing strains are always found in hypersaline environments (13). Halorubrum sp. strains FL23 and FL32, as well as Haloarchaeobius sp. strains FL94 and FL176, were both isolated from the same deposit sample (data not shown). Haloarchaeobius sp. strains FL94 and FL176 are both extracellular protease producers (Fig. S1). Strains FL94 and FL176 exhibited significant inhibition activity against strains FL23 and FL32 (Table 2), likely through bacteriolysis. When the supernatants of strains FL94 and FL176 were mixed with an inhibitor of serine protease, PMSF, at a final concentration of 10 mM, the proteolytic activities vanished on the skim milk plate (Fig. S6a), which was accompanied by the disappearance of its inhibition activity on the indicator plate of Halorubrum sp. FL23 (Fig. S6b). It is proposed that extracellular proteases might possibly be involved in this process.
Correlation of hlyR4 with resistance against extracellular halocins and proteases.
As the proverb goes, offense is the best defense. The protein-hydrolyzing property of HlyR4 may protect its encoding organism from attacks by other antimicrobial peptides, i.e., halocins and exogenous halolysins. Haloferax sp. strain Q22, lacking in proteolytic activity (Fig. S7A, right half), showed a clear inhibition effect on the indicator agar plate of Halorubrum sp. strain LN10 (Fig. 6A, upper row; Fig. S7B). The antimicrobial substances in the strain Q22 cell-free supernatants were protease K and heat sensitive (Fig. S7B and C), and the molecular weight may exceed 50 kDa (Fig. S7D). From these data, we concluded that these antimicrobial substances were proteinaceous and may well be halocins. Strain Q22 exhibited extensive inhibition activity against other haloarchaeal strains due to the production of a halocin(s) (Table S1). Haloarchaeobius sp. strain FL94, which exhibited a clear proteolytic activity, also displayed an inhibition effect on the indicator agar plate containing Halorubrum sp. strain LN10 (Fig. 6A, bottom row). Strains Q22 and FL94 can significantly inhibit the growth of strain EPSR (deficient in hlyR4), but they did not exhibit any inhibition effect on the indicator agar plates in which the hlyR4-positive strain EPS or strain EPSR-R4 served as the lawn cells (Fig. 6B).
FIG 6.
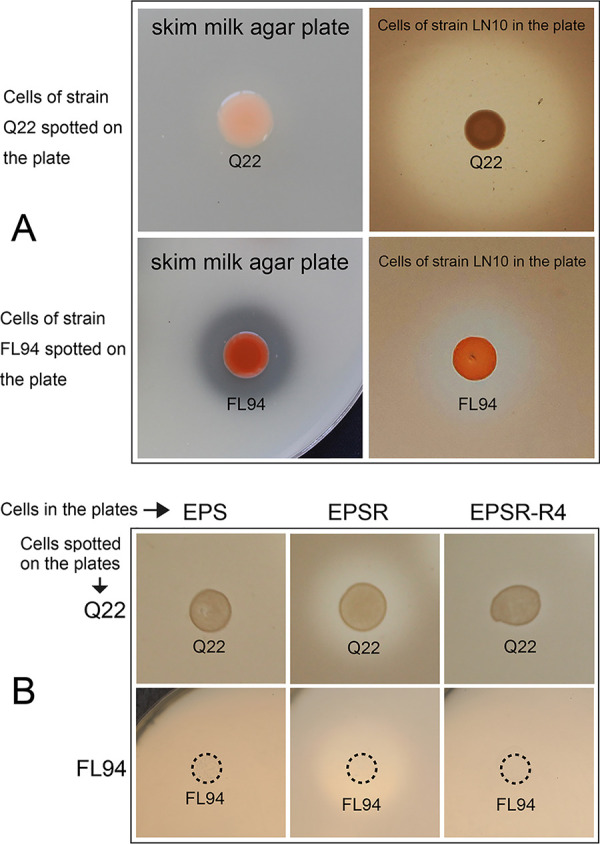
Defensive effect of hlyR4 against heterologous halocin(s) and extracellular protease(s) from haloarchaea. (A) Proteolytic and antagonistic activity of Haloferax sp. strain Q22 and Haloarchaeobius sp. strain FL94 on skim milk nutrient agar plates and on a Halorubrum sp. strain LN10 indicator plate, respectively. (B) Growth of Haloferax sp. strain Q22 and Haloarchaeobius sp. strain FL94 on indicator plates containing Hfx. mediterranei strain EPS, Hfx. mediterranei strain EPSR, and Hfx. mediterranei strain EPSR-R4, an hlyR4 complementation strain of the hlyR4 deletion mutant.
Moreover, the hlyR4-positive strains, i.e., EPS, EPSH, EPSH-H4, EPSR-R4, and EPSHR-R4, were resistant to attacks from either the hlyR4-carrying strains or hlyR4-deficient strains, i.e., EPSR or EPSHR (Table 3 and Fig. S4). In addition, strains EPS and EPSR-R4 displayed resistance to attacks from Har. hispanica strain DF60-R4, which harbors the plasmid pWR4 containing an exogenous hlyR4 (Fig. 4d and f).
Haloferax strain Q22, which exhibits both halocin(s) production and inhibition of many haloarchaeal strains, was used to determine the resistance of Hfx. mediterranei strains to halocins. Supernatants of Haloferax strain Q22 at 5- and 10-fold-concentrated solution showed significant inhibition of all Hfx. mediterranei strains (Table 4). When a 2.5-fold-concentrated solution (equal to 1/4 dilution in Table 4) was examined, only Hfx. mediterranei strain EPSHR-R4 (an hlyR4-positive strain) showed a distinct resistance to Haloferax strain Q22 supernatants (Table 4). When the concentration of the supernatant was decreased to a 1.25-fold-concentrated solution (equal to 1/8 dilution in Table 4), all hlyR4-positive strains, i.e., strains EPS, EPSH, and EPSR-R4, showed resistance to the assault from the halocin(s) in the supernatants (Table 4). Strains EPSR and EPSHR, the hlyR4-negative strains, were sensitive to concentrations of Haloferax strain Q22 supernatants with concentrations greater than the 1/16 dilution (Table 4). They could withstand a 1/32 dilution (equal to a 0.625-fold-concentrated solution) or a higher dilution of the originally concentrated Haloferax strain Q22 supernatants (10-fold-concentrated solution). When the dilution fold was increased, the concentration of the halocin(s) in the supernatants decreased sharply. Anything more than a 1/32 dilution of the Haloferax strain Q22 supernatants (10-fold-concentrated solution) displayed no inhibition effect on any Haloferax mediterranei strains constructed in this study (Table 4).
TABLE 4.
Defensive effects of the hlyR4-deficient Hfx. mediterranei mutants against halocin-containing supernatants
| Indicator plate (Hfx. mediterranei strain)a | Inhibition activity at indicated dilution of halocin-containing supernatantb |
|||||
|---|---|---|---|---|---|---|
| 1 | 1/2 | 1/4 | 1/8 | 1/16 | 1/32 | |
| EPS | ● | ● | ● | ○ | ○ | ○ |
| EPSH | ● | ● | ● | ○ | ○ | ○ |
| EPSHR | ● | ● | ● | ● | ● | ○ |
| EPSHR-R4 | ● | ● | ○ | ○ | ○ | ○ |
| EPSR | ● | ● | ● | ● | ● | ○ |
| EPSR-R4 | ● | ● | ● | ○ | ○ | ○ |
Strain EPS, Hfx. mediterranei strain DF50 ΔEPS; strain EPSH, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4; strain EPSHR, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4 ΔhlyR4; strain EPSHR-R4, Hfx. mediterranei strain DF50 ΔEPS ΔhalH4 ΔhlyR4::hlyR4; strain EPSR, Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4; strain EPSR-R4, Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4::hlyR4.
The dilution shown as 1 represents the 10-fold-concentrated halocin-containing supernatants produced by Haloferax sp. Q22 at late stationary phase, used for dilution. 1/2, 1/4, 1/8, 1/16, and 1/32 represent 2-, 4-, 8-, 16-, and 32-fold dilutions, respectively. Filled circle, significant inhibition activity; empty circle, no inhibition activity. These results were obtained through three biological replicates.
The results indicated that hlyR4 is correlated with the resistance of homologous or heterologous proteinaceous antagonistic substances, e.g., halocin(s) and extracellular proteases.
Competitive advantage conferred by hlyR4 on its encoding organism.
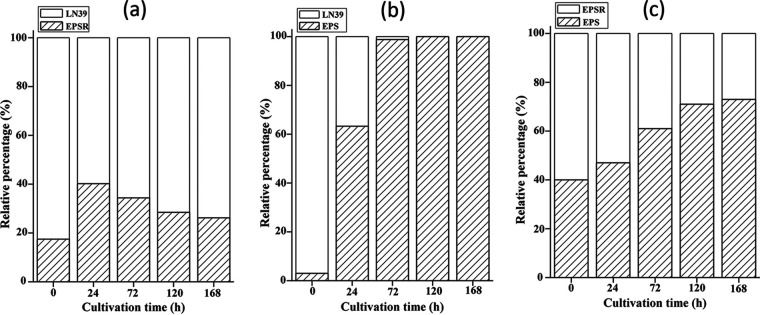
Protein hydrolysis is a common feature for all types of proteases. In contrast to bacterial extracellular serine proteases, halolysins secreted by haloarchaeal cells show bacteriostatic and defensive functions. These functions may convert to a competitive advantage for better survival in a harsh hypersaline environment. Strains EPS, EPSR, and LN39, exhibiting similar growth rates in the nutrient rich AS-168 medium (data not shown), were selected to investigate the competitive advantage supposedly attributable to the presence of hlyR4. When cells of strains EPSR and LN39 were mixed and inoculated into liquid AS-168 medium, the proportion of EPSR cells was about 17.5% at the start point (0 h) (Fig. 7a). After 168 h of cultivation, the proportion of EPSR cells was about 26.2%, which was slightly greater than that of EPSR cells from the beginning. The proportion of EPSR cells seems to have undergone a short stage of ascent, followed by a relatively long-term decline (Fig. 7a). When the EPSR cells were replaced by EPS cells, the proportion of EPS cells climbed rapidly from 3.0% (0 h) to 63.3% (24 h) after 24 h of cultivation (Fig. 7b). It took only 24 h for the abundance of EPS cells in the mixture to increase from the minority to the majority. Furthermore, the EPS cells were found to have entirely outcompeted the LN39 cells in the liquid system after less than 120 h of cultivation (Fig. 7b).
FIG 7.
Competitive advantage conferred by hlyR4. Pairwise competitive growth was tested between strains EPSR and LN39 (a), strains EPS and LN39 (b), and strains EPS and EPSR (c). Three sets of cell suspension were spread on agar plates for colony counting after 2 weeks of cultivation. Colony counting for strain EPSR cells on EPSR-LN39 plates and for strain EPS cells on EPS-LN39 plates was done directly, while EPS cells on EPS-EPSR plates were identified by PCR based on hlyR4. LN39, Haloarcula sp. LN39.
In the EPS and EPSR system, cells of EPS accounted for about 40.0% in the miscellaneous population at the beginning (0 h). The proportion of EPS cells increased steadily with cultivation time until reaching 73.0% abundance as the dominant group (Fig. 7c).
EPS cells were able to easily outcompete the growth of the EPSR and LN39 cells in liquid AS-168 medium within a very short time. However, a system containing EPSR and LN39 cells showed highly similar growth rates and abundances rather than clear transcendence (Fig. S8). It was obvious that hlyR4 provided its host with a significant competitive advantage against other halophiles.
Transcription activities of halH4 and hlyR4 in different growth phases.
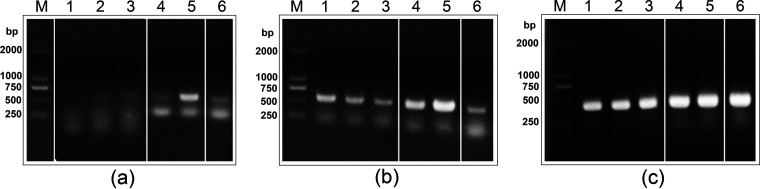
It has been experimentally determined that halH4 from Hfx. mediterranei is a growth phase-dependent gene (7). The sptA gene, encoding extracellular serine protease, SptA, in Natrinema sp. J7-2, has also been determined to be a growth phase-dependent expression gene (22). In order to determine if hlyR4 follows the same growth-dependent expression pattern, the trace amount of DNA contamination in the total RNA extraction was removed through digestion with RNase-free DNase I prior to conducting reverse transcription analysis (Fig. S9). The transcripts of halH4 were barely detectable before cultivation for 24 h and reached a peak value at cultivation for 96 h, followed by a gradual decline (Fig. 8), suggesting that halH4 is a growth phase-dependent expression gene. Cultivation times of 24 h and 96 h corresponded to strain EPS in the early logarithmic phase and late stationary phase, respectively (9). In contrast, the transcription of hlyR4 was steadily detected in all growth phases (Fig. 8), although protease activity in strain EPS supernatants was detected only after cultivation for 48 h (Fig. S10). The detection method used for checking the transcription activity of hlyR4 was reverse transcription-PCR, a technique more sensitive than what was used to detect protease activity. Therefore, the data illustrated that hlyR4 is constitutively expressed in all growth phases.
FIG 8.
Transcription activities of hlyR4 and halH4. (a and b) Transcription levels of halH4 (a) and hlyR4 (b) after incubation for 0 h (lane 1), 24 h (lane 2), 48 h (lane 3), 72 h (lane 4), 96 h (lane 5), and 120 h (lane 6) were determined by reverse transcription-PCR. (c) The 16S rRNA gene was used as a positive control. The molecular sizes of the standard DNA ladder (lane M) are shown on the left.
DISCUSSION
Haloarchaea thrive in environments with salt concentrations approaching saturation, such as natural brines, alkaline salt lakes, the Dead Sea, marine solar salterns, and rock salt deposits (30). Bacteriocins produced by Gram-negative bacteria such as Escherichia coli are ribosome-synthesized toxins (31). Halocins, bacteriocin-like proteinaceous antagonists, are produced by haloarchaea as a tool to compete against other strains for living space and nutrients (5, 32, 33). Kis-Papo and Oren (29) found that some halocin-producing strains isolated from natural brines failed to show any halocin activity even under exposure of highly concentrated supernatants (53-fold), which may be attributable to the production of haloarchaeal extracellular proteases (halolysins) (11–13). Our data have revealed that strains possessing an extracellular serine protease, halolysin R4 (HlyR4), are to a certain extent resistant to attack from halocins (Fig. 4 and 6). When HlyR4, a serine protease, was compared to SptA, the two were found to have similar gene structures and high amino acid sequence similarity values (Fig. 1), suggesting that they may possess similar protein properties.
It is well known that extracellular proteases such as halolysins have the ability to degrade proteins (13). Surface-layer (S-layer) proteins are widespread in almost all Archaea (34). In haloarchaea, S-layer proteins are present in the species Halobacterium salinarum, Haloarcula japonica, Haloferax volcanii, and Haloquadratum walsbyi but absent in the species Natronococcus occultus and Halococcus morrhuae (23). The naturally produced halolysin R4 (unconcentrated) can inhibit the growth of species not only in the genera Halobacterium and Haloarcula but also in species of the genera Natronomonas, Halorubrum, Haloparvum, and Haloarchaeobius as the main antagonistic substance (Table 1). This suggests that the cell envelopes of these species may possess typical S-layer proteins. In contrast to the species listed above, species from the genera Halococcus, Saliphagus, Natrialba, Haloterrigena, Halostagnicola, Halorhabdus, Halopenitus, Halohasta, Halalkalicoccus, and Haladaptatus exhibited significant resistance to unconcentrated halolysin R4 (Table 1), which may be attributable either to a lack of S-layer protein richness or possession of some unknown halolysin resistance mechanism. It has been reported that S-layer proteins are present on the cell envelope in the genus Haloferax (23), and, as such, halolysins may exert an inhibition effect on species in the genus Haloferax. Intriguingly, the Haloferax sp. strain Q22 showed resistance to halolysin R4 in the natural supernatants of Hfx. mediterranei strains (Table 1). There are several genes annotated through bioinformatic analysis as membrane-binding proteases in the genome of Haloferax volcanii and other haloarchaea (35), which may partially or largely contribute to their resistance toward halolysin R4.
Among these 11 strains from the genus Halorubrum used for screening antimicrobial activities (Table 1), there are three main categories. Halolysin R4 greatly inhibits seven strains, i.e., LN72, LN60, LN11, LN10, F4, G16-1, and G105, slightly inhibits strains LN27 and F18, and has no inhibition effect against strains Y13 and J88 (Table 1). The effects of halolysin R4 vary in strains even in the same genus, let alone from different genera. Thus, we can assume that the antagonistic activities in haloarchaea are probably much more complex than anticipated.
A unique and specific halocin resistance mechanism has been described for Halobacterium sp. strain AS7092, in which resistance is conferred by a single gene (halC8) encoding both halocin C8 (HalC8) and its immunity protein HalI (10). The halolysins, such as HlyR4, may exhibit nonspecific proteolysis, which serves to eliminate the attacking effects of halocins and halolysins that are either autosecreted (Table 3 and see Fig. S6 in the supplemental material) or secreted by other strains (Fig. 6). In addition, we have observed extracellular protease production in bacterial strains, indicating significant inhibition activity against several haloarchaeal strains (laboratory observation). This observation also supports the proposal that nonspecific proteolytic activity may provide a wide range of defensive mechanisms against many types of bacteriocin-like proteinaceous antagonists.
The synthesis of halolysins, e.g., Nep from Natrialba magadii and SptA from Natrinema sp. J7-2, is a well characterized indicator of entrance into the stationary phase; in other words, the synthesis of halolysins is growth phase dependent (22, 36). However, our data clearly show that hlyR4 is a constitutively expressed gene and that HlyR4 synthesis is most likely phase independent (Fig. 8 and Fig. S9). The features of halolysins varied between strains (Table 2), suggesting that they do not necessarily share a unified model. It was determined that out of the strains studied, Hfx. mediterranei ATCC 33500 was the only strain possessing both the halocin (HalH4)- and halolysin (HlyR4)-encoding genes, i.e., halH4 and hlyR4 (7, 24). Achieving a balance between antagonistic activities of the phase-dependent HalH4 and defensive effects of the phase-independent HlyR4 has resulted from long-term adaptive evolution in haloarchaea to improve their survival in hypersaline environments.
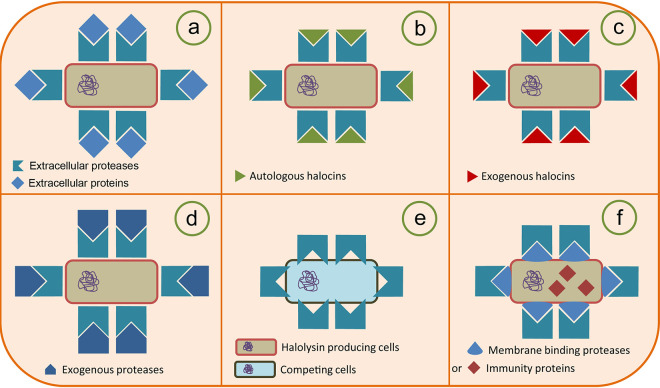
Here, we use HlyR4 as an example to summarize the multiple functions of halolysins and their proposed mechanisms of action (Fig. 9). It has been reported that HlyR4, a serine protease, has a primary proteolytic activity (Fig. 9a). Based on the findings in this study, we assume that HlyR4 can interact with autologous (Fig. 9b) or exogenous (Fig. 9c) halocins, thereby completely degrading them. It also exhibits resistance to attacks from halolysins from other haloarchaeal strains (Fig. 9d). HlyR4 can inhibit the growth of other haloarchaeal strains as a main or supplementary antagonistic substance (Fig. 9e). Some strains possess a resistance to halolysins owing to the proteolytic activity of endogenous halolysins or to membrane-binding proteases (MBPs) or immunity proteins (Fig. 9f) (35).
FIG 9.
Proposal for the new biological function of haloarchaeal extracellular protease by using halolysin R4 from Hfx. mediterranei (HlyR4) as an example. HlyR4, encoded by hlyR4, was initially reported as a subtilisin-like extracellular serine protease from Hfx. mediterranei. HlyR4, an extracellular protease with a primarily proteolytic activity, was secreted by Hfx. mediterranei into its surroundings to degrade proteinaceous complexes and recalcitrant substrates (e.g., protein, glycoprotein, and lipoprotein, etc.). These degradation products could then be utilized as nutrients for the secreting strain (a). Due to its proteolytic activity, the HlyR4-producing strain displayed resistance to autologous (b) or exogenous (c) halocins (proteinaceous substances). The strain was also resistant to the attack from exogenous proteases (d). The HlyR4-producing strain inhibited the growth of other haloarchaeal cells by hydrolyzing their cell surface glycoproteins (e). Strains producing HlyR4 possess some membrane-binding proteases (MBPs) on their cytomembrane. These MBPs can largely relieve the proteolytic effects from their own HlyR4, which may contribute to their survival in environments containing a certain concentration of proteases (f).
The interaction between Pseudoalteromonas sp. and Gram-positive bacteria has been proposed as a typical predator-prey interaction in bacteria (15). Pseudoalterin, an extracellular metalloprotease secreted by Pseudoalteromonas sp. strain CF62, can bind to and degrade the peptidoglycan peptide chains of the cell wall in Gram-positive bacteria, resulting in death of the target strain. These hydrolysates can then be utilized as nutrients for growth in the pseudoalterin producer (15). Here, the proteolytic activities were directly related to the inhibition activities not only in bacteria but also in haloarchaea (Fig. 3 and 5). The present study hints that halolysins, the extracellular proteases isolated from haloarchaea, may have the ability not only to lyse sensitive cells but also to utilize the hydrolysates from those lysed cells as nutrients for growth. Halolysins are a newly discovered variant of proteinaceous antagonists and have been reported to exhibit defensive responses to other proteinaceous antagonists in haloarchaea. Like the interaction between haloviruses and haloarchaeal cells (37, 38), halolysin-producing strains and cells that are sensitive to their effects may represent another predator-prey interaction in hypersaline environments. Notably, such interactions play important roles in the cycling of organic matter in hypersaline environments. Strains with extracellular protease production, such as FL94 and FL176, certainly inhibit the growth of other phylogenetically related strains such as FL23 and FL32 that are isolated from the same habitat. Moreover, in a mimic experiment, HlyR4 likely enabled the cells of its encoding organism to outcompete those of other haloarchaea living in the same hypersaline environment (Fig. 7).
MATERIALS AND METHODS
Strains, medium, and culture conditions.
The genetically engineered strains used in this study are listed in Table 5. Wild-type haloarchaeal strains used for the screening of inhibition spectra are listed in Table 1. Escherichia coli JM109 and E. coli JM110 were cultivated in Luria-Bertani (LB) medium (39) at 37°C. If necessary, ampicillin was added to a final concentration of 100 μg ml−1. Plasmids were shuttled in E. coli JM110 prior to haloarchaeal transformation in order to eliminate methylation patterns. Wild-type haloarchaeal strains and the hlyR4 complementation strains were cultured at 37°C in AS-168 medium containing 200 g NaCl, 20 g MgSO4·7H2O, 2 g KCl, 3 g trisodium citrate, 1.8 g sodium glutamate, 50 mg FeSO4·7H2O, 0.36 mg MnCl2·4H2O, 5 g Bacto Casamino Acids (Difco, USA), and 5 g yeast extract (Oxoid, England) (pH 7.2) per liter. The medium was autoclaved at 121°C for 20 min after preparation (40). For solid medium, 1.5% (for LB medium) or 2.0% (for AS-168 medium) (wt/vol) agar powders were added to the liquid medium before autoclaving.
TABLE 5.
Genetically engineered strains and plasmids used in this study
| Strain (abbreviation) or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Haloferax mediterranei ATCC 33500 | Wild-type haloarchaeal strain | CGMCCa |
| Hfx. mediterranei strain DF50 (DF50) | pyrF gene deletion mutant of Hfx. mediterranei ATCC 33500 | 41 |
| Hfx. mediterranei strain DF50 ΔEPS (EPS) | eps gene deletion mutant of Hfx. mediterranei DF50 | 26 |
| Hfx. mediterranei strain DF50 ΔEPS ΔhalH4 (EPSH) | halH4 deletion mutant of Hfx. mediterranei strain EPS | 9 |
| Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4 (EPSR) | hlyR4 deletion mutant of Hfx. mediterranei strain EPS | This study |
| Hfx. mediterranei strain DF50 ΔEPS ΔhlyR4::hlyR4 (EPSR-R4) | Strain EPSR harboring a recombinant plasmid, pWR4; hlyR4+ pyrF+ | This study |
| Hfx. mediterranei strain DF50 ΔEPS ΔhalH4 ΔhlyR4 (EPSHR) | hlyR4 and halH4 double deletion mutant of Hfx. mediterranei strain EPS | This study |
| Hfx. mediterranei strain DF50 ΔEPS ΔhalH4 ΔhlyR4::hlyR4 (EPSHR-R4) | Strain EPSHR harboring a recombinant plasmid, pWR4; hlyR4+ pyrF+ | This study |
| Hfx. mediterranei strain DF50 ΔEPS ΔhalH4::halH4 (EPSH-H4) | Strain EPSH harboring a recombinant plasmid, pWH4; halH4+ pyrF+ | 9 |
| Haloarcula hispanica ATCC 33960 | Wild-type haloarchaeal strain | CGMCC |
| Haloarcula hispanica strain DF60 (DF60) | ΔpyrF strain of Haloarcula hispanica ATCC 33960 | 45 |
| Haloarcula hispanica strain DF60-R4 (DF60-R4) | Haloarcula hispanica strain DF60 harboring a recombinant plasmid, pWR4; hlyR4+ pyrF+ | This study |
| Escherichia coli JM109 | Widely used host strain for molecular cloning; recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 | Novagen |
| E. coli JM110 | dam- and dcm-negative strain of E. coli JM109 | TaKaRa |
| Plasmids | ||
| pMD-18T | 2.7-kb cloning T-vector; Ampr | TaKaRa |
| pHFX | 4.0 kb, original plasmid for gene knockout lacking the origin of replication in haloarchaea; Ampr | 41 |
| pHFX-UDR4 | The upstream (509-bp) and downstream (541-bp) fragments of hlyR4 were linked and inserted into plasmid pHFX at the multiple cloning sites for gene knockout of hlyR4 | This study |
| pWL502 | 7.9-kb shuttle vector with pyrF gene; Ampr | 41 |
| pWR4 | 9.5-kb derivative of pWL502 containing hlyR4 and its native promoter | This study |
CGMCC, China General Microbiological Culture Collection Center.
Naming and description of Hfx. mediterranei derivatives.
Strain Hfx. mediterranei ATCC 33500 originated from the American Type Culture Collection (ATCC). Deletion of pyrF resulted in strain DF50. Without the gene cluster responsible for exopolysaccharide synthesis, strain DF50 was termed strain EPS. The halH4 deletion strain of EPS was named EPSH, and the hlyR4 deletion strain of EPS was named EPSR. The halH4 hlyR4 double deletion strain of EPS was termed strain EPSHR. Strain EPSR harboring the plasmid pWR4 for complementation of hlyR4 deficiency was termed strain EPSR-R4. Likewise, strain EPSHR harboring the plasmid pWR4 was termed strain EPSHR-R4, and strain EPSHR harboring the plasmid pWH4 was termed strain EPSH-H4 (9).
For cultivation of the pyrF-deficient Hfx. mediterranei strains, e.g., DF50, EPS, EPSH, EPSR, and EPSHR, and Haloarcula hispanica DF60, uracil was added to the AS-168 medium to a final concentration of 50 μg ml−1. For propagation of cells and supernatants, liquid cultivation techniques were performed in a 250-ml shake flask (180 rpm) for 120 h for haloarchaeal strains or 24 h for E. coli strains.
Plasmid construction, gene knockout, and complementation.
In order to construct the hlyR4 gene knockout plasmid, a 509-bp DNA fragment that was located immediately upstream of hlyR4 and a 541-bp DNA fragment that was located immediately downstream of hlyR4 were amplified with primer pairs UPR4F1/UPR4R1 and DWR4F2/DWR4R2, respectively (Table 6). These two DNA fragments were used as templates for overlapping extension PCR with primer pair UPR4F1/DWR4R2. The overlapping PCR product was digested with HindIII plus KpnI and then inserted into the pyrF-based integration plasmid pHFX (41), resulting in pHFX-UDR4 (Table 5). All oligonucleotides used in this study were prepared at a concentration of 10 μmol ml−1. Forward and reverse primers in quantities of 10 pmol each were used in 25-μl PCR mixtures.
TABLE 6.
Oligonucleotides used in this study
| Name | Sequence (5′–3′)a | Description |
|---|---|---|
| EPS16SF | GCGGACGATAACCTCGGGAAACTG | Internal reference for RT-PCR of halH4 and hlyR4 (393 bp) |
| EPS16SR | GCGGCTTTAGGCCCAATAATATCG | |
| H4F | ATTACACCGACTTTGCGCTC | For detection of halH4 (469 bp) |
| H4R | GCAACGTACACCATCTCGTC | |
| UPR4F1 | CGCAAGCTTCCGCGCGACCCTCGACGGCA | For amplification of upstream fragment of hlyR4 (509 bp) |
| UPR4R1 | CGCTTTCTTGGGGAGAGGGGCAAACATGACTCAGTGCACTACGATATTA | |
| DWR4F2 | CTGAGTCATGTTTGCCCCTCTCCCCAAGAAAGCGCGCTAGCCGTCTTCC | For amplification of downstream fragment of hlyR4 (541 bp) |
| DWR4R2 | CGCGGTACCGTCAGACCTCTCTCGGCACC | |
| R4DPF | CTACGCTGCAGTCAAGTTCC | For detection of hlyR4 (567 bp) |
| R4DPR | TTCATCGTCGAGGAGTAGCC | |
| 33500HlyF | GCGGGTACCGCAGACCGACGGAGCCGACG | For amplification of complete hlyR4 with its native promoter (1,700 bp) |
| 33500HlyR | GCGGGATCCTTATTCCTTTTCCGTGATGG |
Restriction sites HindIII (AAGCTT), KpnI (GGTACC), and BamHI (GGATCC) are in italics.
To construct the hlyR4-deficient mutants, pHFX-UDR4, the hlyR4 gene knockout plasmid, was introduced into strains EPS and EPSH through polyethylene glycol (PEG)-mediated transformation (42) after shuttling in E. coli JM110 (41). AS-168Y medium (AS-168 medium without yeast extract) was used to screen for successful transformants. Then, colonies grown on AS-168Y agar plates were transferred onto AS-168 agar plates supplemented with uracil and 5-fluoroorotic acid (250 μg ml−1). Subsequently, hlyR4 deletion mutants were confirmed by colony PCR with primers R4DPF/R4DPR (Table 6).
To construct the hlyR4 complementation strains, hlyR4 was obtained from Hfx. mediterranei ATCC 33500 via PCR using primers 33500HlyF and 33500HlyR. The PCR product was purified with a DNA extraction kit (Oxygen, USA), digested with KpnI plus BamHI, and then inserted into the shuttle plasmid pWL502, resulting in plasmid pWR4 (27). Plasmid pWR4 was introduced into the hlyR4 deletion mutants, Hfx. mediterranei strains EPSR and EPSHR, and the Haloarcula hispanica strain DF60 via PEG-mediated transformation in order to verify the biological function of hlyR4 (42).
Proteolytic activity detection for Hfx. mediterranei and other haloarchaeal strains.
Skim milk agar plates were utilized in order to determine the proteolytic activity of the Hfx. mediterranei and other haloarchaeal strains before and after deletion of hlyR4. In order to prepare the skim milk plates, 10 ml 10% (wt/vol) skim milk solution (BD-Difco, USA) was heated in boiling water for 15 min. After the boiling period, the skim milk solution was rapidly added to 100 ml AS-168 medium with 2% (wt/vol) agar warmed at 55°C, and this solution was mixed evenly and then poured into petri dishes (20 ml per dish). All studied strains were grown and assayed on plates prepared using this method.
Cell suspensions were made from cells in the early stationary phase of growth from Hfx. mediterranei and other haloarchaeal strains in order to test for proteolytic activity. The optical density at 600 nm (OD600) of all these cell suspensions was adjusted to 2.0 with AS-168 medium. Cell suspensions (10 μl for each strain) were spotted onto skim milk agar plates to detect proteolytic activity levels after cultivation for 2 weeks.
It has been reported that halolysin R4 (also named strain R4) from Hfx. mediterranei ATCC 33500 is an extracellular serine protease (24). To verify whether this classification as a serine protease is correct, supernatants of Hfx. mediterranei ATCC 33500 collected by centrifugation at 12,000 rpm for 3 min were treated with the serine protease inhibitor PMSF (final concentration, 10 μg ml−1) at 37°C for 2 h. Treated supernatants (100 μl) were then used to conduct agar diffusion assays (10), while the PMSF solution alone was used as a control.
Inhibition activity assay.
Haloferax mediterranei strains, i.e., EPS, EPSH, EPSR, and EPSR-R4, were used as test strains for detection of hlyR4- and halH4-related inhibition activities against other haloarchaeal strains from different genera that had been previously isolated, identified, and preserved in our laboratory (Table 1). The testing procedure was modified from the method described by Naor et al. (25). Cell suspensions of Hfx. mediterranei strains were prepared as described above and spotted onto the indicator plates, which were prepared in the same manner as the skim milk agar plates but with cell suspensions of other haloarchaeal strains in the late logarithmic phase replacing the skim milk solution in the plate medium.
The interaction between any two Hfx. mediterranei strains, i.e., EPS, EPSH, EPSR, EPSHR, EPSHR-R4, EPSH-H4, and EPSR-R4, was investigated by using these strains in pairs, in which one, the indicator, was contained within the medium, while the other, the tester, was plated onto the medium.
In addition, six wild-type haloarchaeal strains, including four extracellular protease-producing strains and two extracellular protease-negative strains belonging to different genera, were used to investigate the universality of their inhibition activity against other haloarchaeal strains (Table 2). All of these strains were able to grow in the AS-168 medium and under the same cultivation conditions. The procedure was identical to that used for Hfx. mediterranei strains.
Correlation between proteolytic activity and inhibition effect.
Strain EPS derived from Hfx. mediterranei ATCC 33500 is an extracellular serine protease production strain (24). To determine the correlation between the proteolytic activity and the inhibition effect of the extracellular serine protease, the supernatants of strain EPS in the late logarithmic or stationary phase (OD600, 2.0 to 3.0) were collected through centrifugation. Cells of strain Haloarcula sp. strain LN121 in the logarithmic phase (OD600, 1.0) were used to construct the indicator plates. The strain LN121 indicator plates were generated by adding 1 ml cell suspension in the logarithmic phase to 100 ml AS-168 solid medium at 50°C and pouring the mixture into petri dishes. A description of the preparation of skim milk agar plates can be found in the previous section.
To detect the inhibition activity of haloarchaeal strains such as EPS and Haloarchaeobius sp. FL94 and FL176 after elimination of the activity of serine protease, phenylmethylsulfonyl fluoride (PMSF) was added to the supernatants of these strains at a final concentration of either 10 mM or 20 mM, while supernatants without PMSF and liquid medium with PMSF (20 mM) were used as controls. These supernatants and solution were poured in quantities of 100 μl into the holes on the skim milk and corresponding indicator plates (strain LN121 for strain EPS; strain FL23 for strains FL94 and FL176).
Defensive effect of the strains containing hlyR4.
To determine the type of the inhibition substances from Haloferax sp. strain Q22, streaking on skim milk agar plate was performed. Also, before these tests were carried out, cell-free supernatants of strain Q22 were treated with protease K (20 μg ml−1 for 1 h), heating (85°C for 1 h), and ultrafiltration (molecular weight cutoff [MWCO], 50 kDa) to probe the general properties of substances exhibiting inhibition activity in the suspensions. The procedure for constructing skim milk and indicator plates has already been described. Here, Halorubrum sp. strain LN10 was used to construct the indicator plate.
To investigate the resistance of strains possessing hlyR4 toward other proteinaceous antagonistic substances (halocins or halolysins), Haloferax sp. Q22 (Table 1) (see reference 9), a halocin-producing strain, and Haloarchaeobius sp. FL94 (Table 1), an extracellular protease-producing strain, were used as the testing strains, while the Hfx. mediterranei strains, i.e., EPS, EPSR, and EPSR-R4, were used as the indicator strains.
To explore the hlyR4-related resistance of Hfx. mediterranei strains toward halocin(s) produced by Haloferax sp. Q22, 10-fold-concentrated cell-free supernatants obtained from late-stationary-phase cultures of Haloferax sp. Q22 were used as the original stock to make serial dilutions of 1 (equal to 10-fold-concentrated solution), 1/2 (equal to 5-fold), 1/4 (equal to 2.5-fold), 1/8 (equal to 1.25-fold), 1/16 (equal to 0.625-fold), and 1/32 (equal to ∼0.313-fold) to test the resistance of Hfx. mediterranei strains. One-fold-concentrated solution indicates unconcentrated supernatants. Concentration fold levels below 1 indicate a dilution of the natural supernatants of Haloferax sp. Q22. Haloferax mediterranei strains were used to make the indicating agar plates in accordance with the procedure described previously.
Pairwise growth competition assays.
To determine the competitive advantage of the strains in liquid culture, pairwise competitive growth assays were performed as previously described (43). Strains EPS, EPSR, and Haloarcula sp. LN39 (wild halophiles) were cultivated in AS-168 broth supplemented with uracil. Cells in the logarithmic phase were selected for competitive growth. Cells of two strains (500 μl for each strain), i.e., EPS and Haloarcula sp. LN39, EPSR and Haloarcula sp. LN39, or EPS and EPSR, were mixed and inoculated into 100 ml AS-168 broth supplemented with uracil.
After cultivation in liquid medium for 0, 24, 72, 120, and 168 h, a culture from each time point was diluted in an appropriate dilution that depended on the cell concentration, and 150-μl aliquots of these different cell suspensions were plated on AS-168 plates supplemented with uracil. Cells on the plates were counted after 2 weeks of cultivation. Cells of EPS and EPSR are phenotypically distinguishable from those of Haloarcula sp. LN39, as the colonies show different coloration; thus, we conducted the colony counting directly for the plates with the EPS and Haloarcula sp. LN39 mixture or the EPSR and Haloarcula sp. LN39 mixture. Colonies of strains EPS or EPSR on the plates spread with the EPS and EPSR mixture were detected by PCR using hlyR4 as the marker gene, and the ratio was calculated based on 100 random colonies.
Reverse transcription of halH4 and hlyR4.
To analyze transcriptional levels of halH4 and hlyR4 during different growth phases, reverse transcription-PCR was conducted. The inoculum amount of Hfx. mediterranei strain EPS was 0.5% (vol/vol), using a late-stationary-phase cell suspension. Throughout the cultivation process, equal amounts of cells cultivated for 0 h, 24 h, 48 h, 72 h, 96 h, and 120 h were collected by centrifugation at 12,000 rpm for 3 min. Cells were resuspended with 500 μl RNase-free sterilized 2% (wt/vol) NaCl solution prepared with diethyl pyrocarbonate (DEPC)-treated water. Total RNA extraction was performed according to the procedures outlined in the Rnaiso Plus kit (TaKaRa, Japan). All the containers and instruments, including tips and Eppendorf tubes used for RNA extraction and reverse transcription, were treated with DEPC.
Prior to conducting reverse transcription-PCR, trace amounts of contaminant DNA were removed by RNase-free DNase I (TaKaRa, Japan). Then, the concentration and purity of total RNAs were assessed by measuring the absorbance at 260 nm (A260) and determining the A260/A280 ratio. In order to check for trace DNA contamination in the total RNA samples, primer pairs EPS16SF/EPS16SR, H4F/H4R, and R4DPF/R4DPR were utilized to amplify a partial sequence of the 16S rRNA gene, hlaH4, and hlyR4, respectively (Table 6). The reverse primers, EPS16SR, H4R, and R4DPR, were used to generate cDNAs through reverse transcription with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, USA) (Table 6). After reverse transcription, these cDNAs were used as PCR templates, and primer pairs EPS16SF/EPS16SR, H4F/H4R, and R4DPF/R4DPR (Table 6) were utilized to determine the presence of the halH4 and hlyR4 transcripts during different growth phases.
Detection of the proteolytic activity of strain EPS supernatants along with growth phase.
To detect proteolytic activity in supernatants of cells in different growth phases, cell suspensions of strain EPS in the logarithmic phase (1 ml, OD600 = 1.0) were inoculated into AS-168 medium supplemented with uracil (100 ml). Supernatants were collected at 24, 48, 96, 120, 144, and 168 h after cultivation by centrifugation. Azocasein was used as the substrate to quantify the serine protease activity through the release of a chromogenic product. The OD at 335 nm was then measured (44).
Data availability.
The almost complete 16S rRNA genes of the haloarchaeal strains used in this study have been deposited in the GenBank/EMBL/DDBJ. Accession numbers are listed in Tables 1 and 2.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dahe Zhao and Jian Zhou from the Institute of Microbiology, Chinese Academy of Sciences, for their technical assistance and Rong Gao and Libby Helfant from Rutgers, The State University of New Jersey, and Ping-Shin Lee from Anhui Normal University for their constructive comments and suggestions.
S.C. and H.X. conceived the project, S.S., R.W., H.F., and S.C. performed the study, S.C., S.S., R.W., and H.X. analyzed the data, and S.C. drafted the manuscript. S.C. and H.X. critically revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (grant no. 31460003 and 91751201), the Anhui Provincial Key Laboratory of the Conservation and Exploitation of Biological Resources (grant no. 591601), and the Excellent Young Talents Fund Project for Universities in Anhui Province (grant no. gxyqZD2017011).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Falb M, Muller K, Konigsmaier L, Oberwinkler T, Horn P, von Gronau S, Gonzalez O, Pfeiffer F, Bornberg-Bauer E, Oesterhelt D. 2008. Metabolism of halophilic archaea. Extremophiles 12:177–196. 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oren A. 2002. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol Ecol 39:1–7. 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 3.Singh A, Singh AK. 2017. Haloarchaea: worth exploring for their biotechnological potential. Biotechnol Lett 39:1793–1800. 10.1007/s10529-017-2434-y. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Xiang H, Liu J, Zhou M, Tan H. 2003. Purification and biological characterization of halocin C8, a novel peptide antibiotic from Halobacterium strain AS7092. Extremophiles 7:401–407. 10.1007/s00792-003-0335-6. [DOI] [PubMed] [Google Scholar]
- 5.Besse A, Peduzzi J, Rebuffat S, Carre-Mlouka A. 2015. Antimicrobial peptides and proteins in the face of extremes: lessons from archaeocins. Biochimie 118:344–355. 10.1016/j.biochi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Valera F, Juez G, Kushner DJ. 1982. Halocins: salt-dependent bacteriocins produced by extremely halophilic rods. Can J Microbiol 28:151–154. 10.1139/m82-019. [DOI] [Google Scholar]
- 7.Cheung J, Danna KJ, O'Connor EM, Price LB, Shand RF. 1997. Isolation, sequence, and expression of the gene encoding halocin H4, a bacteriocin from the halophilic archaeon Haloferax mediterranei R4. J Bacteriol 179:548–551. 10.1128/jb.179.2.548-551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meseguer I, Torreblanca M, Konishi T. 1995. Specific inhibition of the halobacterial Na+/H+ antiporter by halocin H6. J Biol Chem 270:6450–6455. 10.1074/jbc.270.12.6450. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Sun S, Korfanty GA, Liu J, Xiang H. 2019. A halocin promotes DNA uptake in Haloferax mediterranei. Front Microbiol 10:1960. 10.3389/fmicb.2019.01960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun C, Li Y, Mei S, Lu Q, Zhou L, Xiang H. 2005. A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Mol Microbiol 57:537–549. 10.1111/j.1365-2958.2005.04705.x. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Porro C, Martin S, Mellado E, Ventosa A. 2003. Diversity of moderately halophilic bacteria producing extracellular hydrolytic enzymes. J Appl Microbiol 94:295–300. 10.1046/j.1365-2672.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- 12.Rohban R, Amoozegar MA, Ventosa A. 2009. Screening and isolation of halophilic bacteria producing extracellular hydrolyses from Howz Soltan Lake, Iran. J Ind Microbiol Biotechnol 36:333–340. 10.1007/s10295-008-0500-0. [DOI] [PubMed] [Google Scholar]
- 13.Karray F, Ben Abdallah M, Kallel N, Hamza M, Fakhfakh M, Sayadi S. 2018. Extracellular hydrolytic enzymes produced by halophilic bacteria and archaea isolated from hypersaline lake. Mol Biol Rep 45:1297–1309. 10.1007/s11033-018-4286-5. [DOI] [PubMed] [Google Scholar]
- 14.De Castro RE, Maupin-Furlow JA, Gimenez MI, Herrera Seitz MK, Sanchez JJ. 2006. Haloarchaeal proteases and proteolytic systems. FEMS Microbiol Rev 30:17–35. 10.1111/j.1574-6976.2005.00003.x. [DOI] [PubMed] [Google Scholar]
- 15.Tang BL, Yang J, Chen XL, Wang P, Zhao HL, Su HN, Li CY, Yu Y, Zhong S, Wang L, Lidbury I, Ding H, Wang M, McMinn A, Zhang XY, Chen Y, Zhang YZ. 2020. A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria. Nat Commun 11:285. 10.1038/s41467-019-14133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Yin J, Mei S, Wang X, Tang XF, Tang B. 2018. Halolysin SptA, a serine protease, contributes to growth-phase transition of haloarchaeon Natrinema sp. J7-2, and its expression involves cooperative action of multiple cis-regulatory elements. Front Microbiol 9:1799. 10.3389/fmicb.2018.01799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siezen RJ, Leunissen JA. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci 6:501–523. 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamekura M, Seno Y, Holmes ML, Dyall-Smith ML. 1992. Molecular cloning and sequencing of the gene for a halophilic alkaline serine protease (halolysin) from an unidentified halophilic archaea strain (172P1) and expression of the gene in Haloferax volcanii. J Bacteriol 174:736–742. 10.1128/jb.174.3.736-742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi W, Tang XF, Huang Y, Gan F, Tang B, Shen P. 2006. An extracellular halophilic protease SptA from a halophilic archaeon Natrinema sp. J7: gene cloning, expression and characterization. Extremophiles 10:599–606. 10.1007/s00792-006-0003-8. [DOI] [PubMed] [Google Scholar]
- 20.De Castro RE, Ruiz DM, Gimenez MI, Silveyra MX, Paggi RA, Maupin-Furlow JA. 2008. Gene cloning and heterologous synthesis of a haloalkaliphilic extracellular protease of Natrialba magadii (Nep). Extremophiles 12:677–687. 10.1007/s00792-008-0174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz DM, Paggi RA, Gimenez MI, De Castro RE. 2012. Autocatalytic maturation of the Tat-dependent halophilic subtilase Nep produced by the archaeon Natrialba magadii. J Bacteriol 194:3700–3707. 10.1128/JB.06792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du X, Li M, Tang W, Zhang Y, Zhang L, Wang J, Li T, Tang B, Tang XF. 2015. Secretion of Tat-dependent halolysin SptA capable of autocatalytic activation and its relation to haloarchaeal growth. Mol Microbiol 96:548–565. 10.1111/mmi.12955. [DOI] [PubMed] [Google Scholar]
- 23.Albers SV, Meyer BH. 2011. The archaeal cell envelope. Nat Rev Microbiol 9:414–426. 10.1038/nrmicro2576. [DOI] [PubMed] [Google Scholar]
- 24.Kamekura M, Seno Y, Dyall-Smith M. 1996. Halolysin R4, a serine proteinase from the halophilic archaeon Haloferax mediterranei; gene cloning, expression and structural studies. Biochim Biophys Acta 1294:159–167. 10.1016/0167-4838(96)00016-7. [DOI] [PubMed] [Google Scholar]
- 25.Naor A, Yair Y, Gophna U. 2013. A halocin-H4 mutant Haloferax mediterranei strain retains the ability to inhibit growth of other halophilic archaea. Extremophiles 17:973–979. 10.1007/s00792-013-0579-8. [DOI] [PubMed] [Google Scholar]
- 26.Zhao D, Cai L, Wu J, Li M, Liu H, Han J, Zhou J, Xiang H. 2013. Improving polyhydroxyalkanoate production by knocking out the genes involved in exopolysaccharide biosynthesis in Haloferax mediterranei. Appl Microbiol Biotechnol 97:3027–3036. 10.1007/s00253-012-4415-3. [DOI] [PubMed] [Google Scholar]
- 27.Cai S, Cai L, Liu H, Liu X, Han J, Zhou J, Xiang H. 2012. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl Environ Microbiol 78:1946–1952. 10.1128/AEM.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamekura M, Seno Y. 1993. Partial sequence of the gene for a serine protease from a halophilic archaeum Haloferax mediterranei R4, and nucleotide sequences of 16S rRNA encoding genes from several halophilic archaea. Experientia 49:503–513. 10.1007/BF01955152. [DOI] [PubMed] [Google Scholar]
- 29.Kis-Papo T, Oren A. 2000. Halocins: are they involved in the competition between halobacteria in saltern ponds? Extremophiles 4:35–41. 10.1007/s007920050005. [DOI] [PubMed] [Google Scholar]
- 30.Stan-Lotter H, Fendrihan S. 2015. Halophilic archaea: life with desiccation, radiation and oligotrophy over geological times. Life (Basel) 5:1487–1496. 10.3390/life5031487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flaherty RA, Freed SD, Lee SW. 2014. The wide world of ribosomally encoded bacterial peptides. PLoS Pathog 10:e1004221. 10.1371/journal.ppat.1004221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley MA, Wertz JE. 2002. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol 56:117–137. 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 33.Salgaonkar BB, Mani K, Nair A, Gangadharan S, Braganca JM. 2012. Interspecific interactions among members of family Halobacteriaceae from natural solar salterns. Probiotics Antimicrob Proteins 4:98–107. 10.1007/s12602-012-9097-8. [DOI] [PubMed] [Google Scholar]
- 34.Sleytr UB, Schuster B, Egelseer EM, Pum D. 2014. S-layers: principles and applications. FEMS Microbiol Rev 38:823–864. 10.1111/1574-6976.12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gimenez MI, Cerletti M, De Castro RE. 2015. Archaeal membrane-associated proteases: insights on Haloferax volcanii and other haloarchaea. Front Microbiol 6:39. 10.3389/fmicb.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paggi RA, Madrid EA, D'Alessandro CP, Cerletti M, De Castro RE. 2010. Growth phase-dependent biosynthesis of Nep, a halolysin-like protease secreted by the alkaliphilic haloarchaeon Natrialba magadii. Lett Appl Microbiol 51:36–41. 10.1111/j.1472-765X.2010.02855.x. [DOI] [PubMed] [Google Scholar]
- 37.Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. 2012. Global network of specific virus-host interactions in hypersaline environments. Environ Microbiol 14:426–440. 10.1111/j.1462-2920.2011.02603.x. [DOI] [PubMed] [Google Scholar]
- 38.Motlagh AM, Bhattacharjee AS, Coutinho FH, Dutilh BE, Casjens SR, Goel RK. 2017. Insights of phage-host interaction in hypersaline ecosystem through metagenomics analyses. Front Microbiol 8:352. 10.3389/fmicb.2017.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch EF, Maniatis T (ed). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 40.Han J, Hou J, Zhang F, Ai G, Li M, Cai S, Liu H, Wang L, Wang Z, Zhang S, Cai L, Zhao D, Zhou J, Xiang H. 2013. Multiple propionyl coenzyme A-supplying pathways for production of the bioplastic poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in Haloferax mediterranei. Appl Environ Microbiol 79:2922–2931. 10.1128/AEM.03915-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Han J, Liu X, Zhou J, Xiang H. 2011. Development of pyrF-based gene knockout systems for genome-wide manipulation of the archaea Haloferax mediterranei and Haloarcula hispanica. J Genet Genomics 38:261–269. 10.1016/j.jgg.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. 1989. Transformation methods for halophilic archaebacteria. Can J Microbiol 35:148–152. 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- 43.Delmas S, Shunburne L, Ngo HP, Allers T. 2009. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet 5:e1000552. 10.1371/journal.pgen.1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bendicho S, Martí G, Hernández T, Martín O.. 2002. Determination of proteolytic activity in different milk systems. Food Chem 79:245–249. 10.1016/S0308-8146(02)00126-7. [DOI] [Google Scholar]
- 45.Li M, Wang R, Zhao D, Xiang H. 2014. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res 42:2483–2492. 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The almost complete 16S rRNA genes of the haloarchaeal strains used in this study have been deposited in the GenBank/EMBL/DDBJ. Accession numbers are listed in Tables 1 and 2.