Summary
This protocol describes the embedding and processing of Drosophila pupae in paraffin to monitor tissue changes during development. Although multiple methods are available to evaluate developmental changes in Drosophila embryos, imaging detailed changes during metamorphosis is challenging as the animal is enclosed in the cuticle, rendering it inaccessible to whole mount imaging. Here, we present a protocol that focuses on developmental clearance of the larval salivary glands in Drosophila pupae that can be extended to examine other tissues/stages for similar purposes.
For complete details on the use and execution of this protocol, please refer to Velentzas et al. (2018).
Subject areas: Cell Biology, Developmental biology, Microscopy, Model Organisms
Graphical abstract

Highlights
-
•
Paraffin embedding and processing allows monitoring of tissue changes
-
•
Histology can be used to monitor tissue changes at different stages of development
-
•
Clearance of Drosophila larval salivary glands can be evaluated through histology
This protocol describes the embedding and processing of Drosophila pupae in paraffin to monitor tissue changes during development. Although multiple methods are available to evaluate developmental changes in Drosophila embryos, imaging detailed changes during metamorphosis is challenging as the animal is enclosed in the cuticle, rendering it inaccessible to whole mount imaging. Here, we present a protocol that focuses on developmental clearance of the larval salivary glands in Drosophila pupae that can be extended to examine other tissues/stages for similar purposes.
Before you begin
Drosophila melanogaster is used as a model organism for the study of many biological processes. Here, we describe a protocol for the study of the developmental clearance of the larval salivary glands during metamorphosis. This protocol can also be used to study the developmental changes of other tissues during the different stages of development and adulthood. The researcher should have a good knowledge of Drosophila morphology (Chyb and Gompel, 2013) and genetics (Greenspan, 2004). It is also critical to be able to distinguish the different pupal stages during Drosophila metamorphosis (Bainbridge and Bownes, 1981). This protocol is modified from Restifo and White (1991).
Fly crosses
Timing: approximately 1–2 weeks
-
1.
Collect virgin female animals of the appropriate genotype.
-
2.
In a vial containing fly food (Table 1), add ~8 virgin female and ~4 male adult animals of the appropriate genotype.
CRITICAL: Set-up the appropriate crosses for all the genetic controls needed.
-
3.
Incubate the vial with the flies in a 25°C incubator with a 16 h day/8 h night cycle.
-
4.
Let the animals lay eggs for 24 h.
-
5.
Remove the flies and add them to a new vial with fly food and incubate both vials in a 25°C incubator with a 16 h day/8 h night cycle.
-
6.
Repeat steps 4 and 5 at least five times.
Note: White pre-pupae will start forming on the side of the vial approximately five days after the eggs are laid.
Table 1.
.Composition of the Fly Food
| Reagent | Final concentration (%) | Amount |
|---|---|---|
| Agar | 0.65% | 6.5 g |
| Brewer’s Yeast | 6.3% | 63 g |
| Cornmeal | 6% | 60 g |
| Molasses | 6% | 60 mL |
| Propionic Acid | 0.2% | 2 mL |
| Phosphoric Acid | 0.2% | 2 mL |
| Tegosept | 0.13% | 1.3 g |
| Water | n/a | to 1 L |
| Total | n/a | 1 L |
Preparing fixative solution
Timing: approximately 20 min
-
7.In a 500 mL Erlenmeyer Flask, add:
-
a.200 mL of 100% Ethanol.
-
b.27.5 mL of 37% Formaldehyde.
-
c.12.5 mL of Glacial acetic acid.
-
d.10 mL of 25% Glutaraldehyde.
-
a.
-
8.
Mix the solution, on a magnetic stirrer at 20°C–25°C, for 5 min.
-
9.
Aliquot solution (Table 2) into 50 mL tubes.
-
10.
Store at 4°C.
CRITICAL: The contents of this solution are toxic, corrosive or flammable and should be handled carefully, while wearing the appropriate protective gear.
Note: The solution is stable for at least one month at 4°C.
Table 2.
.Composition of fixative solution
| Reagent | Final concentration (%) | Amount |
|---|---|---|
| Alcohol, 100% | 80% | 200 mL |
| Formaldehyde, 37% | 4% | 27.5 mL |
| Acetic Acid, Glacial | 5% | 12.5 mL |
| Glutaraldehyde 25% Aqueous Solution | 1% | 10 mL |
| Total | n/a | 250 mL |
Preparing Weigert’s Hematoxylin stain solution
Timing: approximately 10 min
-
11.In a 500 mL bottle for light-sensitive solutions, add:
-
a.150 mL of Weigert's Iron Hematoxylin solution A.
-
b.150 mL of Weigert's Iron Hematoxylin solution B.
-
a.
-
12.
Mix by inverting the bottle a few times.
-
13.
Store at 20°C–25°C in an appropriate safety cabinet.
CRITICAL: The contents of this solution are toxic, corrosive or flammable and should be handled carefully while wearing the appropriate protective gear.
Note: You can re-use the stain several times.
Preparing Pollak Trichrome stain solution
Timing: approximately 5–24 h
-
14.In a 400 mL beaker, add:
-
a.150 mL of 100% Ethanol.
-
b.150 mL of Milli-Q Water.
-
c.3 mL of Glacial acetic acid.
-
a.
-
15.
Mix the solution on a magnetic stirrer at 20°C–25°C for 2 min.
-
16.
Split the solution into four 250 mL beakers, adding approximately 75 mL of solution in each beaker.
-
17.To the solution in the first beaker, add:
-
a.0.5 g Acid Fuchsin.
-
b.1 g Ponceau Xylidine (2R).
-
a.
-
18.To the solution in the second beaker, add:
-
a.0.45 g Light Green SF Yellowish.
-
a.
-
19.To the solution in the third beaker, add:
-
a.0.75 g Orange G.
-
b.1.5 g Phosphotungstic Acid.
-
a.
-
20.To the solution in the fourth beaker, add:
-
a.1.5 g Phosphomolybdic acid hydrate.
-
a.
-
21.
Mix all the solutions on a magnetic stirrer at 20°C–25°C until all the powders are dissolved (1–24 h).
-
22.
Once completely dissolved, combine all four solutions in a 400 mL beaker.
-
23.
Mix the solution for 2 min.
-
24.
Filter solution through a 0.2 μm filter.
-
25.
Pour the solution into a 500 mL bottle for light-sensitive solutions.
-
26.
Store at 20°C–25°C in an appropriate safety cabinet.
CRITICAL: The contents of this solution are toxic, corrosive or flammable and should be handled carefully, while wearing the appropriate protective gear.
Note: The stain can be re-used several times.
Note: Some of the powder stains may not completely dissolve even after 24 h of mixing. Hence, the solution needs to be filtered.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Agar | MoorAgar | 41004 |
| Brewer’s yeast | Fisher Scientific | ICN90331225 |
| Cornmeal | Fisher Scientific | ICN90141125 |
| Molasses | Thomsen Food Services | 02625 |
| Propionic acid | Fisher Scientific | A258 |
| Phosphoric Acid | Fisher Scientific | A260 |
| Tegosept | Genesee | 20-259 |
| Weigert's Iron Hematoxylin A | Electron Microscopy Sciences | 26044-05 |
| Weigert's Iron Hematoxylin B | Electron Microscopy Sciences | 26044-15 |
| Acid Fuchsin | Electron Microscopy Sciences | 10035 |
| Orange G | Electron Microscopy Sciences | 19070 |
| Phosphotungstic Acid, Crystal | Electron Microscopy Sciences | 19500 |
| Ponceau Xylidine (2R) | Sigma-Aldrich | P2395 |
| Light Green SF Yellowish | Sigma-Aldrich | L1886 |
| Phosphomolybdic acid hydrate | Sigma-Aldrich | 221856 |
| Glutaraldehyde 25% Aqueous Solution | Electron Microscopy Sciences | 16220 |
| Formaldehyde, 37% by Weight | Fisher Scientific | F79-500 |
| Acetic Acid, Glacial | Fisher Scientific | BP1185-500 |
| Alcohol, 100%, HistoPrep™ | Fisher Scientific | HC-800-1GAL |
| Xylene, HistoPrep™ | Fisher Scientific | HC-700-1GAL |
| Paraplast® X-tra embedding agent for histology | McCormick Scientific | 39503002 |
| Permount™ Mounting Medium | Fisher Scientific | SP15-500 |
| DPBS, no calcium, no magnesium | Thermo Scientific | 14190144 |
| Experimental models: Organisms/strains | ||
| D. melanogaster, Canton-S | Bloomington Drosophila Stock Center | 64349 |
| D. melanogaster, w; ;Fkh-GAL4 | Eric Baehrecke | N/A |
| D. melanogaster, w; UAS-p35 | (Hay et al., 1994) | N/A |
| D. melanogaster, w; ;Atg13Δ74 | (Chang and Neufeld, 2009) | N/A |
| Software and algorithms | ||
| AxioVision | Zeiss | N/A |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| GraphPad Prism 8 | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| McPherson-Vannas Micro Dissecting Spring Scissors | Roboz Surgical | RS-5600 |
| Dumont #5 Forceps | Fine Science Tools | 11251-30 |
| BD Clay Adams™ Nutator Mixer | BD Diagnostics | 421105 |
| Incubator | VWR | 1510E |
| Incubator with day/night cycle | Percival Scientific | I-36VL |
| Slide warmer | Fisher Scientific | 12-594 |
| Microtome | American Optics Corp | 820 “Spencer’’ |
| Zeiss AxioImager Z1 | Zeiss | N/A |
| Superfrost™ Plus Microscope Slides | Fisher Scientific | 12-550-15 |
| VWR VistaVision™ Cover Glasses, No. 11/2, 22 × 50 mm | VWR | 16004-336 |
| Concavity Microscope Slide | Thermo Scientific | 1527-006 |
| Glass Scintillation vials, 20 mL | DWK Life Sciences | 74500-20 |
| 35 × 10 mm Dish, Non-treated | Thermo Scientific | 171099 |
| 3" × 5" Unruled Index Cards White | W.B. Mason | N/A |
| WHEATON® Complete (Dish, Cover, Rack, Handle) | DWK Life Sciences | 900200 |
| Cardboard Slide Tray | Fisher Scientific | 12-587-10 |
| Microscope Slide Box | Fisher Scientific | 03-446 |
Materials and equipment
Nutator or a rotator.
Incubator at 42°C.
Incubator at 56°C.
Incubator at 25°C with a 16 h day/8 h night cycle.
Slide warmer at 56°C.
Slide warmer at 42°C.
Microtome.
Brightfield microscope with a 5× objective and a color camera.
Step-by-step method details
Collection and fixation of Drosophila pupae
Timing: approximately 2–5 days
The steps below describe how to collect and fix Drosophila pupae of the appropriate developmental stage, to investigate the role of different genes in the clearance of the larval salivary glands during metamorphosis.
-
1.
Collect white prepupae (WPP) from the side wall of a culture vial (Figure 1A) using a pair of forceps and place them in a 35 mm petri dish lined with a piece of wet tissue paper (Figure 1B).
Note: New WPP form on the side wall of the culture vial every hour and can be collected multiple times each day.
-
2.
Incubate the WPP- containing petri dish in a 25°C incubator with a 16 h day/8 h night cycle for exactly 24 h.
CRITICAL: After 24 h of incubation, the animals should have transitioned from the pre-pupal to the pupal stage. Do not use animals that are still in the pre-pupal stage (troubleshooting 1).
-
3.
Using a pair of forceps, submerge a pupa in a concave slide containing PBS (Figure 1C and Methods video S1).
Figure 1.
.Collecting and preparing pupae for fixation
(A) White prepupa (WPP) (circle).
(B) Collected WPP in a petri dish lined with wet tissue paper.
(C) 24 h APF pupa (arrows indicate the position of the cuts).
(D) 24 h APF pupa after the cuts have been made (arrows point to the cut sites).
-
4.
Using a pair of dissecting scissors incise the pupal case first at the anterior end of the pupa and then at the posterior end (Figure 1D, Methods video S1).
-
5.
Puncture the cuticle at the posterior end of the developing animal using the tip of the dissecting scissors or a needle (Figure 1D and Methods video S1).
CRITICAL: During the process, be careful not to severely damage the developing animal inside the pupal case. When puncturing the cuticle or when cutting the pupal case at the posterior end of the animal, a small amount of tissue might leak out of the cut. These pupae can still be used unless they have sustained excessive damage (troubleshooting 2).
-
6.
Transfer the pupae to an Eppendorf tube with 500 μL of fixative solution.
-
7.
Incubate the pupae in fixative at 4°C for 48 h.
Pause point: After step 7 the animals can be stored in the fixative solution for at least two weeks without any noticeable variation.
Note: Collect at least 20 animals from each genotype/condition. The collection and fixation process can last several days. Animals of the same genotype/condition collected at different times/days can be added to the same tube.
Embedding Drosophila pupae in paraffin
Timing: approximately 2 days
These steps describe the process of embedding the Drosophila pupae in paraffin.
-
8.
Discard the fixative solution and add 500 μL of 80% ethanol to the pupae.
-
9.
Rotate the pupae with ethanol on a nutator at 20°C–25°C for 15 min.
-
10.
Discard 80% ethanol and add 500 μL of 85% ethanol.
-
11.
Rotate on a nutator at 20°C–25°C for 20 min.
-
12.
Discard 85% ethanol and add 500 μL of 90% ethanol.
-
13.
Rotate on a nutator at 20°C–25°C for 25 min.
-
14.
Discard 90% ethanol and add 500 μL of 100% ethanol.
-
15.
Rotate on a nutator at 20°C–25°C for 30 min.
-
16.
Discard 100% ethanol and add 500 μL of fresh 100% ethanol.
-
17.
Rotate on a nutator at 20°C–25°C for 30 min.
-
18.
Repeat steps 16 and 17 two additional times.
-
19.
Discard the 100% ethanol from the tube and add 500 μL of 100% xylene.
-
20.
Rotate on a nutator at 20°C–25°C for 30 min.
-
21.
Discard 100% xylene and add 500 μL of fresh 100% xylene.
-
22.
Rotate on a nutator at 20°C–25°C for 30 min.
-
23.
Repeat steps 21 and 22 two additional times.
-
24.In a 20 mL scintillation vial add:
-
a.~1.8 g of paraffin chips, Paraplast X-tra (not melted).
-
b.2 mL of 100% xylene.
-
a.
-
25.
Transfer the pupae into a scintillation vial.
Note: Add a maximum of 20 animals per vial. If more animals have been collected from one genotype/condition, split them into an appropriate number of vials.
Note: Remember to melt enough paraffin in a 56°C incubator which will be used the next day.
-
26.
Incubate the pupae in a 42°C incubator for 16–20 h.
-
27.
Discard the xylene/paraffin solution from the vial using a glass pipette.
-
28.
Add an adequate amount of melted paraffin, enough to cover all of the pupae.
-
29.
Incubate the vial containing the pupae in a 56°C incubator for 2 h.
-
30.
Replace melted paraffin with fresh melted paraffin and incubate in a 56°C incubator for 2 h.
-
31.
Repeat step 30 two additional times.
Pause point: The procedure can be paused for at least one week.
-
32.
Fill the vial with melted paraffin.
-
33.
Incubate the vial in a 56°C incubator for at least 30 min.
-
34.
Make a mold using a 3" × 5" white unruled index card (Figure 2A and Methods video S2).
Figure 2.
.Orientation of the processed pupae into the molds for embedding in paraffin
(A) Paper mold.
(B) Animals oriented in the molds and labeled.
-
35.
Cover the slide warmer with aluminum foil and pre-heat it to 56°C.
-
36.
Pre-heat the mold on the slide warmer.
-
37.
Quickly pour the melted paraffin containing the pupae into the mold.
-
38.
Orient the pupae, using a toothpick, with their ventral side facing down (spiracles pointing up) and ensure that there is an adequate amount of space between them. This step is to be performed quickly before the paraffin begins to harden (~3 min) (Figure 2B and Methods video S3 troubleshooting 3).
Note: You can add a small piece of paper (i.e., a tag) with the genotype/condition corresponding to each mold. You can also position up to five animals, side by side, as shown in Figure 2B.
-
39.
Switch off the slide warmer and leave the molds containing the pupae to cool for 16–20 h.
-
40.
Store the paraffin blocks with the embedded pupae at 20°C–25°C.
Pause point: The paraffin blocks can be stored at 20°C–25°C indefinitely.
Sectioning of the embedded Drosophila pupae
Timing: approximately 1 day
Here we describe how to prepare the embedded animals for sectioning with a microtome. We also describe the key steps in the sectioning process.
-
41.
Peel away the paper mold from the hardened paraffin block (Figure 3A).
-
42.
Cut the paraffin block into smaller, individual pyramid shaped blocks (Figure 3B).
-
43.
Mount a block on a microtome chuck by melting the paraffin on the bottom of the block and then letting it solidify on the microtome chuck (Figure 3C).
-
44.
Trim the blocks creating a truncated rectangular pyramid with only 0.5 mm of space surrounding the animals (Figure 3D and Methods video S4).
Figure 3.
.Preparation of embedded animals for sectioning
(A) Animals embedded in paraffin.
(B) Small piece of paraffin that contains a group of animals.
(C) Embedded animals mounted on microtome chucks.
(D) Trimmed block of paraffin with pupae.
CRITICAL: Make sure the shape is a truncated rectangular pyramid to obtain a straight ribbon when sectioning (Figure 3D and Methods video S4).
-
45.
Place a microscope slide onto a 42°C slide warmer and add distilled water on top of it (Methods video S5).
Note: Use a pencil or a chemical-resistant marker to label microscope slides.
-
46.
Section using a microtome at 7 μm thickness (troubleshooting 4).
-
47.
Float sections onto a microscope slide with water (Methods video S5).
-
48.
Let the sections expand for 1–2 min (Methods video S5).
-
49.
Remove the water from the slide using a paper towel along the side (Methods video S5).
-
50.
Leave the slides on the 42°C slide warmer for 16–20 h (Methods video S5).
Staining and image acquisition
Timing: approximately 4–6 h
In these steps we describe the process of staining the tissues on microscope slides.
-
51.
Place the microscope slides into a slide rack (Figure 4A).
-
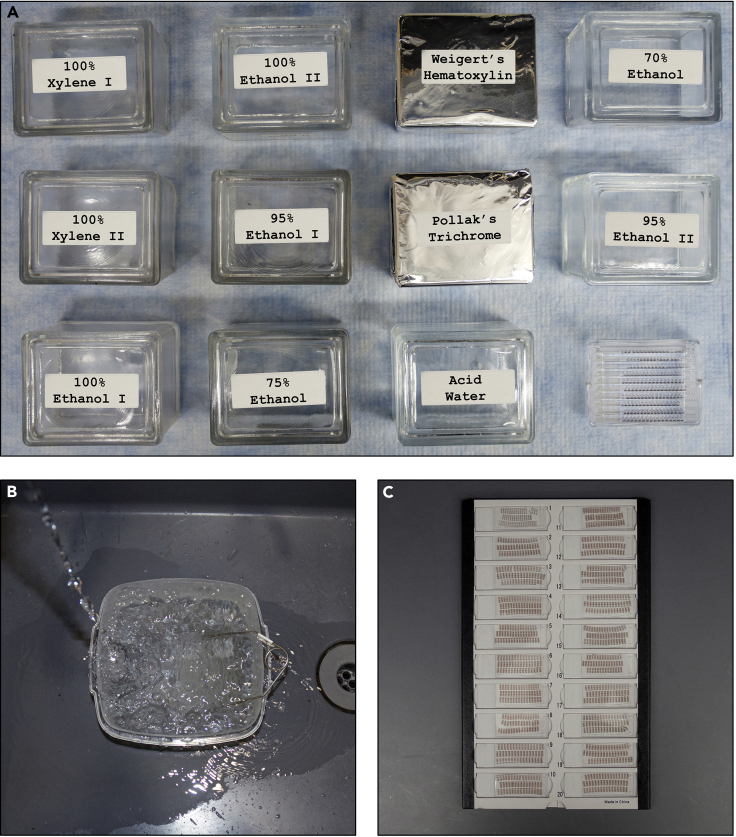
52.Rehydrate the tissue by incubating them sequentially in containers with the following solutions and time duration(s) under a chemical hood (Figure 4A):
-
a.100% Xylene I for 3 min.
-
b.100% Xylene II for 3 min.
-
c.100% Ethanol I for 3 min.
-
d.100% Ethanol II for 3 min.
-
e.95% Ethanol I for 3 min.
-
f.75% Ethanol for 3 min
-
g.Running tap water for 5 min (Figure 4B).
-
a.
CRITICAL: Use a large plastic container and do not let the stream of running water hit the slides (Figure 4B, troubleshooting 5).
-
53.Stain the tissue sections by incubating the slides containing the sections sequentially in containers with the following solutions and time duration(s):
-
a.Weigert’s Hematoxylin for 5 min.
-
b.Running tap water for 5 min (Figure 4B).
 CRITICAL: Use a large plastic container and do not let the stream of running water hit the slides (Figure 4B, troubleshooting 5).
CRITICAL: Use a large plastic container and do not let the stream of running water hit the slides (Figure 4B, troubleshooting 5). -
c.Pollak’s Trichrome for 7 min.
-
d.Distilled water for 10 s to remove the excess of stain.
-
e.0.2% glacial acetic acid in distilled water for 10 s.
-
a.
-
54.Dehydrate the tissue sections by incubating in containers with the following solutions and time duration(s):
-
a.70% Ethanol 2–3 dips.
-
b.95% Ethanol II 2–3 dips.
-
c.95% Ethanol I for 3 min.
-
d.100% Ethanol I for 3 min.
-
e.100% Ethanol II for 3 min.
-
f.100% Xylene II for 3 min.
-
g.100% Xylene I for 3 min.
-
a.
-
55.
Dry the slides on a paper towel for 3 min.
-
56.
Use ~80 μL of Permount per slide to mount a 50 mm coverslip on the sections.
-
57.
Leave slides on a cardboard slide tray, at 20°C–25°C, for at least 2 h for the Permount to partially harden (Figure 4C).
-
58.
Using a brightfield microscope with a 5× objective, acquire representative images from each genotype/condition.
Note: Permount completely hardens after 24–48 h. Once Permount hardens, store slides in a slide box indefinitely.
Figure 4.
.Staining of the tissue sections
(A) Containers with the solutions used during the staining procedure.
(B) Plastic container with slides in the slide rack, under running water.
(C) Slides with stained tissues after a coverslip has been mounted.
Expected outcomes
In Figure 5 we describe the morphological changes to the larval salivary glands during metamorphosis. During the pre-pupal stages (~0–12 h after puparium formation, APF) the larval salivary glands are intact and appear tubular with a distinguishable lumen (Figures 5A and 5A’). Immediately after head eversion (~12–14 h APF), the larval salivary glands start to shrink (Figures 5B and 5B’), and by 24 h APF the glands disappear completely (Figures 5C and 5C’).
Figure 5.
.Tissue morphology of animals at different developmental stages
(A) Control animal 2–6 h APF (lu: lumen, sg: salivary gland, mg: midgut). (A’) Salivary gland tissue has been highlighted by digital removal of the surrounding tissues of (A) using image processing software.
(B) Control animal 12–14 h APF (sg: salivary gland, mg: midgut). (B’) Salivary gland tissue has been highlighted by digital removal of the surrounding tissues of (B).
(C) Control animal 24 h APF (mg: midgut). (C’) Clearance of salivary gland tissue has been highlighted by digital removal of the surrounding tissues of (C).
It has been shown that the clearance of the larval salivary glands requires both caspases and autophagy (Berry and Baehrecke, 2007). For example, inhibition of either caspases through the expression of the caspase inhibitor p35 (Hay et al., 1994) or autophagy through the mutation of Atg13, a gene that is required for autophagy initiation (Chang and Neufeld, 2009), leads to a partial salivary gland degradation phenotype with persistent cell fragments (Figure 6). It is also known that the larval midgut is degraded during metamorphosis and that this process requires autophagy (Denton et al., 2009). This is also seen in Figure 6C where the autophagy deficient animals fail to degrade the midgut (Figure 6C arrow).
Figure 6.
.Tissue morphology of animals after caspase or autophagy inhibition
(A) Animal with salivary gland-specific expression of p35 (w; +; fkh-GAL4/UAS-p35) 24 h APF (arrow indicates the midgut). (A’) Salivary gland tissue has been highlighted by digital removal of the surrounding tissues of (A).
(B) Quantification of data from (A).
(C) Mutant animal lacking Atg13 (w; +; Atg13Δ74) 24 h APF (arrow indicates the midgut). (C’) Salivary gland tissue has been highlighted by digital removal of the surrounding tissues of (C). (D) Quantification of data from (C).
Quantification and statistical analysis
For quantification, each animal is scored as either positive or negative for salivary gland cell fragments. For some genotypes, the gland cell fragments are easily distinguishable while not for others. We recommend looking at all the sections for each animal and score the animals for which there are gland fragments in multiple consecutive sections as positive. The data can be presented in a graph with the percentage of positive animals on the y-axis and the genotype/condition on the x-axis. For statistical analysis of the data, we use the software GraphPad Prism© to perform a chi-square test.
Open Prism and make a new project file. Select the new contingency data table option with outcome A being the number of animals negative for salivary gland cell fragments and outcome B the number of animals positive for salivary gland cell fragments. Rows 1 and 2 contain the experimental group and the appropriate control, respectively. In the respective cells enter the actual numbers of the animals and not the percentages since chi-square test is dependent on the sample size. Analyze the data by selecting chi-square test option to obtain the p-value. A difference is characterized as significant when the percentage of the positive animals is more than 50% and the p-value below 0.05.
Limitations
The resolution in paraffin sections is limited, and only substantial morphological changes can be observed. Genetic or other manipulations that induce minor morphological changes cannot be distinguished. Genetic tools, such as the GAL4/UAS system, often possess expression in multiple tissues. Hence, selection of the correct genetic tools that have specific expression patterns is critical for acquiring reliable results. Degradation of larval tissues and the development of the adult tissues are temporally regulated. This should be taken into consideration, as genetic mutations or manipulations can cause developmental arrest or delay making the observations unreliable.
Troubleshooting
Problem 1
The animal’s tissues seem to be that of a larva and not that of a pupa.
Potential solution
In step 2, after 24 h of incubation, the animals should have transitioned from the pre-pupal to the pupal stage and a clear definition of the three major body segments, head, thorax and abdomen should be visible (Figure 7A and Methods video S5). Animals that are still in the pre-pupal stage (Figure 7B and Methods video S5) should not be used.
Figure 7.
.Morphology of pupae 24 h APF
(A) A 24 h APF pupa with normal morphology.
(B) A 24 h APF pupa that has arrested development.
Problem 2
The morphology of the animal’s tissues seems highly irregular in sections.
Potential solution
During steps 4 and 5 the developing animal was probably damaged. Apply minimal pressure on the developing animal when cutting the pupal case and use a sharp needle to puncture the posterior end instead of the tip of the scissors. In addition, always make the cut at the anterior of the pupal case first, to release the pressure from the air inside the case.
Problem 3
The paraffin hardens before proper orientation of the animals inside the mold (step 38).
Potential solution
Transfer fewer animals per vial in step 25. Orienting the animals in the mold requires practice, begin by transferring only 5–10 animals until you become familiar with the process.
Problem 4
Cannot obtain a straight paraffin ribbon when sectioning (step 46).
Potential solution
Make sure that the block is properly aligned on the microtome and that during the trimming (step 44) you have created a truncated rectangular pyramid with equal space around the animals.
Problem 5
The sections on the slides look damaged after staining.
Potential solution
In steps 52(g) and 53(b) use a large plastic container and do not let the stream of running water hit the slides to avoid damage (Figure 4B).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Eric Baehrecke (eric.baehrecke.umassmed.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze [datasets/code].
Acknowledgments
This work was supported by National Institutes of Health (USA) grant GM079431 to E.H.B.
Author contributions
All experiments were performed by P.D.V., and P.D.V. and E.H.B. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.xpro.2021.100473.
Contributor Information
Panagiotis D. Velentzas, Email: panagiotis.velentzas@umassmed.edu.
Eric H. Baehrecke, Email: eric.baehrecke@umassmed.edu.
References
- Bainbridge S.P., Bownes M. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- Berry D.L., Baehrecke E.H. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.Y., Neufeld T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyb S., Gompel N. Academic Press; 2013. Atlas of Drosophila morphology: wild-type and classical mutants. [Google Scholar]
- Denton D., Shravage B., Simin R., Mills K., Berry D.L., Baehrecke E.H., Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr. Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan R.J. 2nd Edition. Cold Spring Harbor Laboratory Press; 2004. Fly pushing : the theory and practice of Drosophila genetics. [Google Scholar]
- Hay B.A., Wolff T., Rubin G.M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Restifo L.L., White K. Mutations in a steroid hormone-regulated gene disrupt the metamorphosis of the central nervous system in Drosophila. Dev. Biol. 1991;148:174–194. doi: 10.1016/0012-1606(91)90328-z. [DOI] [PubMed] [Google Scholar]
- Velentzas P.D., Zhang L., Das G., Chang T.K., Nelson C., Kobertz W.R., Baehrecke E.H. The proton-coupled monocarboxylate transporter hermes is necessary for autophagy during cell death. Dev. Cell. 2018;47:281–293.e4. doi: 10.1016/j.devcel.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze [datasets/code].