Abstract
Digital image analysis (DIA) is impacted by the quality of tissue staining. This study examined the influence of preanalytical variables—staining protocol design, reagent quality, section attributes, and instrumentation—on the performance of automated DIA software. Our hypotheses were that (1) staining intensity is impacted by subtle differences in protocol design, reagent quality, and section composition and that (2) identically programmed and loaded stainers will produce equivalent immunohistochemical (IHC) staining. We tested these propositions by using 1 hematoxylin and eosin stainer to process 13 formalin-fixed, paraffin-embedded (FFPE) mouse tissues and by using 3 identically programmed and loaded immunostainers to process 5 FFPE mouse tissues for 4 cell biomarkers. Digital images of stained sections acquired with a commercial whole slide scanner were analyzed by customizable algorithms incorporated into commercially available DIA software. Staining intensity as viewed qualitatively by an observer and/or quantitatively by DIA was affected by staining conditions and tissue attributes. Intrarun and inter-run IHC staining intensities were equivalent for each tissue when processed on a given stainer but varied measurably across stainers. Our data indicate that staining quality must be monitored for each method and stainer to ensure that preanalytical factors do not impact digital pathology data quality.
Keywords: digital pathology, histology process validation, image analysis, immunohistochemistry, preanalytical factors, precision, reproducibility
Introduction
Digital pathology is a rapidly evolving discipline in which digital image analysis (DIA) is used for such purposes as improving diagnostic sensitivity and rapidly extracting quantitative information from cytological and histological specimens. Common DIA tasks include measuring object counts or dimensions (eg, morphometry and stereology) and quantifying the distribution and intensities of various molecular markers (eg, immunohistochemical [IHC] and in situ hybridization [ISH] analyses).1,2 The importance of digital pathology data sets in answering basic experimental questions has been adapted to toxicology as a vital means for understanding the pathogenesis and mechanisms by which test articles may damage cell and tissue structures and compromise organ functions.3,4 Digital pathology data are also gaining in importance as an end point for optimizing the outcome of tissue evaluations in clinical practice5,6 including clinical trials of novel therapeutic candidates.7
Although DIA has considerable utility in evaluating stained tissue sections, effective application of this tool requires an understanding of various factors that impact the quality and validity of the resulting data. Preanalytical parameters such as collection conditions, fixation protocols, and histology processing procedures are known to substantially affect both the accuracy and precision of digital pathology data—and generally are not well-standardized across laboratories or across different types of studies for a single facility despite the existence of Good Laboratory Practice (GLP) guidelines and standard operating procedures (SOPs).8,9 Traditionally, pathologists have been trained to assess tissue sections effectively regardless of artifacts such as color differences (due to inconsistent section thickness or uneven staining) and structural defects (folds and tears). However, automated and reproducible DIA of tissue sections may be hindered or impossible in the face of such confounding variables when utilizing turn-key DIA algorithms in typical no-cost “shareware” products or when DIA expertise of technical staff is insufficient to effectively operate more advanced deep learning-based (“trainable”) platforms.
Prior work in our GLP-compliant contract histology laboratory has assessed the significance of many preanalytical factors associated with histology processing of banked human tissues. Key parameters in this regard include the protocol design, reagent quality, section thickness, staining instrument, and even the facility location; the influence of such variables applies to both routine methods (eg, hematoxylin and eosin [H&E]) and molecular techniques (eg, IHC).10-12 The comparative importance of these variables remains unclear with respect to which one(s) might be most critical to ensure the quality of digital pathology data acquired for nonclinical tissue specimens. The current study performed this comparison for several variables: H&E and IHC staining protocol designs, instrumentation, reagent quality, and section thickness. Our first hypothesis was that staining intensity for both conventional H&E and common IHC protocols is altered measurably by subtle differences in protocol design, reagent quality, and section features (composition and cell organization). Our second hypothesis was that performing an IHC procedure on different, properly maintained, automated instruments using an identical staining protocol and the same reagents will essentially eliminate variability in staining quality among instruments. Our data confirmed the first hypothesis by demonstrating that qualitative visualization of staining intensity by an observer and quantitative analysis of optical density (OD, a measure of staining intensity13) as calculated by DIA both are affected by the staining conditions and tissue attributes. With respect to the second hypothesis, the intensity of IHC labeling was equivalent for each tissue type within (“intrarun”) and between (“inter-run”) staining runs when processed on a given instrument, but intensities across identically programmed and loaded, automated immunostainers varied to some degree. Taken together, these data show that quality control (QC) documentation will be needed for each method and also for each stainer to be used for preparing tissue sections dedicated to DIA.
Materials and Methods
Animals, Tissues, Processing, and Sectioning
Tissues for H&E staining were obtained from wild-type adult C57BL/6 mice (purchased from Harlan Laboratories Inc by Premier Laboratory and bred in house at Michigan State University). All animals were used in accordance with protocols approved in advance by the Institutional Animal Care and Use Committee at their respective facilities and were maintained according to relevant federal and state laws as well as current guidelines for animal treatment.14 Mice were group-housed (N = 5/cage) in filter-capped microisolator cages and given pelleted chow and filter-purified tap water ad libitum during acclimatization (5 or more days). Constant environmental conditions were maintained (light–dark cycle, 12 hours each; temperature, 22 ± 2 °C; relative humidity, 40% ± 20%).
Mice were euthanized humanely with carbon dioxide to achieve unconsciousness followed by cervical dislocation. Selected tissues were collected immediately and fixed by immersion in neutral buffered 10% formalin for approximately 72 hours (for H&E) or 48 hours (for IHC) at room temperature (RT). Fixed tissues were processed routinely into paraffin on a Sakura Tissue-Tek VIP 6 tissue processor (Sakura) using a conventional processing protocol of 30 minutes per station, according to conventional practices for vertebrate tissues.15,16 A battery of 13 formalin-fixed, paraffin-embedded (FFPE) tissues was slated for H&E staining, while a limited list of 5 FFPE tissues was designated for IHC staining (Supplemental Figure 1).
Hematoxylin and Eosin Staining
Tissues
The organ list included tissues with high (pancreas, spleen); medium (cecum, colon, esophagus, kidney, jejunum, lung, preputial gland, salivary gland, tongue, urinary bladder); and low (skeletal muscle) numbers of nuclei. All tissues were embedded in 1 block. Mineralized tissues were avoided since an assessment of the impact of decalcification methods on staining intensity was outside the scope of this study. A total of 100 serial 4-µm thick sections were cut from a block prepared by Premier Laboratory; in addition, 9 slides were cut from the same block at various thicknesses ranging from 2 to 10 µm in 1-µm increments. All sections were mounted in the center of a positively charged, coated slide (Tanner Scientific). All slides were air-dried overnight at RT and baked at 60 °C for 30 minutes prior to H&E staining.
Hematoxylin and eosin stainer
All slides were processed on a Sakura Tissue-Tek Prisma Automated Slide Stainer (Sakura) at RT. Stainer settings for various protocol options are given in Table 1. Coverslips were applied over stained sections using a Sakura Tissue-Tek Glas cover-slipper (Sakura).
Table 1.
Variations for H&E Staining Protocol.a
| Protocol no. | Incubation times | |||
|---|---|---|---|---|
| Hematoxylin | Differentiation | Eosin | Alcohol | |
| 0 | 30 seconds | 10 minutes | 10 seconds | 50%—5 minutes |
| 1 | 30 seconds | 5 minutes | 30 seconds | 70%—5 minutes |
| 2 | 1 minute | 4 minutes | 30 seconds | 70%—3 minutes |
| 3 | 1 minute | 3 minutes | 30 seconds | 95%—2 minutes |
| 4 | 4 minutes | 1 minute | 1 minute | 95%—2 minutes |
| 5 | 6 minutes | 30 seconds | 3 minutes | 95%—30 seconds |
| 6 | 8 minutes | 30 seconds | 5 minutes | 95%—30 seconds |
| 7 | 15 minutes | 30 seconds | 15 minutes | 95%—30 seconds |
| 8 | 20 minutes | 0 second | 20 minutes | 0 second |
Abbreviation: H&E, hematoxylin and eosin.
a Protocol #4 (highlighted in pale blue) represents the conventional H&E staining method used in our laboratory.
Hematoxylin and eosin staining reagents
Staining runs combined both commercial reagents and solutions constituted in-house. All staining runs employed “off the shelf” hematoxylin-normal strength (Cat No. 812; Anatech) and eosin Y, alcoholic (Cat No. 832; Anatech). Both 0.5% aqueous ammonium hydroxide solution and 5% aqueous glacial acetic acid were made in-house from purchased reagents (procured from BDH VWR Analytical and Macron Fine Chemicals, respectively). Stains from the same lots were used throughout the study except for 2 different lots of expired hematoxylin (outdated by 6 weeks [Lot No. 5390] or 30 weeks [Lot No. 5261] at the time the study was initiated). Ammonium hydroxide and glacial acetic acid solutions were produced once in 5-gallon batches. All reagents were stored at RT until use.
Hematoxylin and eosin staining protocol variations
Four preanalytical differences during histology processing that might influence H&E staining quality were evaluated. These variables were the protocol design, staining precision, section thickness, and reagent quality.
For protocol design, staining quality for all 13 FFPE tissues was evaluated over a range of 8 designs in which incubation times for hematoxylin, glacial acetic acid, eosin Y, and alcohol were altered (Table 1). Protocol #4, which is the method used routinely to stain human and animal tissue sections in our laboratory, conforms to conventional best practice recommendations for H&E staining of vertebrate tissue sections.15,16
To examine precision (repeatability and reproducibility) in H&E staining quality, 1 slide per block was stained daily (Monday-Friday) for 4 months (from April 13 to July 27, 2016), for a total of 73 slides. All slides for each block were cut in advance and retained in a dust-free cabinet at RT until use. Staining reagents were changed biweekly in accordance with the laboratory SOP for H&E staining. This SOP was based on prior QC review of H&E-stained control slides (human and animal tissues) over time, which conforms to industry guidelines for H&E staining based on the volume of slides, types of staining solutions, and the mode (manual or automated) of staining.17
To assess section thickness and its influence on H&E staining quality, 9 slides with section thicknesses ranging from 2 to 10 µm were processed together in the same staining run. This portion of the project was performed only once since the outcome was obvious on both the slides and in the digital images by visual examination of the stained tissue sections.
To evaluate reagent quality on H&E staining, slides were stained with 2 lots of expired hematoxylin. Once weekly for 8 weeks, 1 slide per block was processed with hematoxylin batches that were either 6 weeks (Lot #5390) or 30 weeks (Lot #5261) past their effective use date at the beginning of the 8-week test period. These slides were processed in parallel with a hematoxylin batch (Lot #5822) that remained effective throughout the 8-week test period. Expired eosin was not considered for evaluation since the regular rotation (every 2 weeks) of this reagent as recommended by the manufacturer suggested that eosin age would be likely to impart clearly visible differences in staining intensity over the course of our study.
Immunohistochemical Staining
Tissues
The organ list—colon (with gut-associated lymphoid tissue [GALT]), kidney, liver, lymph node (mesenteric), and spleen—was dictated by the 4 antigens selected for evaluation (see “Immunohistochemical staining reagents and protocols” section). Specifically, the tissues were chosen to represent a range of anticipated levels of antigen expression and, therefore, staining intensities for each IHC procedure.
All tissues were embedded in a single block in random order. A total of 108 serial 4-µm-thick sections were cut from the block prepared by Premier Laboratory; in addition, 24 slides were cut at varying thicknesses ranging from 3 to 8 µm in 1-µm increments. All sections were placed in the center of a positively charged, coated slide (Tanner Scientific) to optimize adhesion. Slides were stored in a dust-free cabinet at RT for less than 1 week prior to initiating the various IHC staining runs, and all IHC runs were completed within 2 weeks of generating the sections. Intrarun and inter-run precision tests were examined over the course of 2 days at the end of this 2-week storage period. Due to the short time in storage and the robust retention of the chosen antigens during prior experiments in our facility, no analysis was performed for possible time-related epitope loss. Slides were air-dried overnight at RT and baked at 60 °C for 1 hour prior to IHC staining.
Immunohistochemical immunostainers
Three autostainers (Dako [Agilent Technologies]) designed for IHC and ISH multiplex procedures were employed. Two instruments were AutostainerPlus Link models (instrument nos. 0010 and 0083), while the third was an Autostainer Link 48 model (instrument no. 0151). Metadata for each IHC run (slide position on the stainer, reagents applied to each slide, reagent sequence, and incubation times, etc) were collected automatically according to built-in functions within the Dako stainer-specific software. As specified by institutional SOP for our laboratory, the metadata logs for the current IHC runs were evaluated to ensure that the stainer parameters fell within the required range of “normal” performance, after which the logs were employed to compile the data shown in various figures and tables for the current article and then retained on a facility-specific server using DakoLink software (version 4.0.3; Dako).
Immunohistochemical staining reagents and protocols
Well-characterized antibodies were selected to visualize 4 antigens (CD3, CD45, F4/80, and Ki-67) commonly expressed in mouse tissues. The specific IHC protocols for these antigens previously had been optimized in our laboratory and validated for use in animal toxicity studies using the specific reagents and IHC stainers employed in the current study. Primary antibodies were concentrates provided by several manufacturers (Table 2). The working solutions of the primary antibodies were prepared once in sufficient quantity so that the same IHC reagent solutions were used across all staining runs on all 3 autostainers, and all reagents for all staining runs were from single lots. This design was employed to eliminate subtle differences in solution preparation and lot-to-lot composition as variables. Staining runs were performed over several days. Sections were allocated among the stains and staining runs as shown in Supplemental Table 1.
Table 2.
Reagents for IHC Staining.
| Target | Primary antibody | Detection system | ||||||
|---|---|---|---|---|---|---|---|---|
| Host species | Antigen source | Clone no. | Concentration | Incubation time | Supplier | Cat no. | ||
| CD3 | Rabbit | Human | Polyclonal | 1.0 µg/mL | 30 minutes | Dako | A0452 | Envision+ rabbit HRP |
| CD45 | Rat | Mouse | 30-F11 | 0.2 µg/mL | 30 minutes | R&D Systems | MAB114 | Rabbit antirat secondary antibody with Envision+ rabbit HRP |
| F4/80 | Rat | Mouse | C1: A3-1 | 3.33 µg/mL | 60 minutes | Bio-Rad | MCA497 | Rabbit antirat secondary antibody with Envision+ rabbit HRP |
| Ki-67 | Rabbit | Human | SP6 | Not stated (titer = 1:120) | 60 minutes | Biocare Medical | CRM325 | Envision+ rabbit HRP |
Abbreviations: HRP, horseradish peroxidase; IHC, immunohistochemical.
Basic IHC protocol
A similar sequence of steps was adopted for all 4 procedures. Briefly, slides bearing FFPE sections were dried overnight, baked at 60 °C for 1 hour, deparaffinized in xylene, rinsed in alcohol, rehydrated in water, and equilibrated in wash buffer (TRIS-buffered saline with 0.05% Tween 20; Dako, Cat No. K8007). All IHC staining was performed at RT.
For CD3, CD45, and Ki-67, heat-induced epitope retrieval (HIER) was performed in a Dako PT Link Pre-treatment Module prior to loading slides on an autostainer. Depending on the IHC protocol, HIER employed either EnVision FLEX Target Retrieval Solution (TRS), low pH (about pH 6.1; Dako, Cat No. K8005), or EnVision FLEX TRS, high pH (approximately pH 9; Dako, Cat No. K8004). The PT Link was programmed to preheat to 80 °C and increase the temperature to 95 °C for 20 minutes after slides were added. Slides were then cooled and rinsed in wash buffer prior to loading on an autostainer.
Common steps in the autostainer included incubations in 3.0% hydrogen peroxide (5 minutes); serum-free protein block (Dako, Cat No. X0909, 5 minutes—not employed for Ki-67); primary antibody (as described below); EnVision+ antirabbit labeled polymer-horseradish peroxidase (HRP; Dako, Cat No. K4003, 30 minutes); Liquid DAB+ Substrate Chromogen system (Dako, Cat No. K3468, 5 minutes, where deposition of DAB [3,3′-diaminobenzidene] yields a brown product). After IHC staining, slides were removed from the autostainer, manually rinsed in tap water, and counterstained for 5 minutes in a modified Harris hematoxylin (Dako, Cat No. S3301). Slides were again rinsed in tap water, and wash buffer was used as the hematoxylin bluing reagent. The slides were then dehydrated in absolute alcohol solutions, cleared in xylene, and coverslipped.
CD3 protocol
This marker detects the T-cell receptor complex on the membranes of T-lymphocytes. Detection of CD3 was carried out (after HIER with TRS, high pH) using a rabbit polyclonal antihuman CD3 antibody (Dako, Cat No. A0452). Anti-CD3 primary antibody was applied for 30 minutes.
CD45 protocol
This marker is a receptor protein tyrosine phosphatase common to the membranes of all leukocytes except plasma cells. Detection of CD45 was carried out (after HIER with TRS, low pH) using a rat monoclonal antimouse CD45 antibody (clone 30-F11; R&D Systems, Cat No. MAB114). Anti-CD45 primary antibody was applied for 30 minutes followed by rabbit secondary antirat immunoglobulin G (IgG) antibody (1.25 µg/mL for 30 minutes; Abcam, Cat No. ab102248) prior to incubation with EnVision+ antirabbit labeled polymer HRP.
F4/80 protocol
This marker detects the F4/80 antigen that is expressed on the membranes of macrophages in many tissues. Detection of F4/80 was carried out using a rat monoclonal antimouse F4/80 antibody (clone Cl: A3-1; Bio-Rad, Cat No. MCA497). Instead of HIER prior to loading on the autostainer, a proteolytic-induced epitope retrieval step using proteinase K (undeclared number of enzyme units per mL in a prediluted solution, pH 7.5; Dako, Cat No. S3020, 2 minutes) was included in the autostainer program between the hydrogen peroxide and serum-free protein block steps. Anti-F4/80 primary antibody was applied for 60 minutes followed by rabbit secondary antirat IgG antibody (1.66 µg/mL for 30 minutes; Abcam Cat No. ab102248) prior to incubation with EnVision+ antirabbit labeled polymer HRP.
Ki-67 protocol
This marker detects an undefined epitope of the Ki-67 nuclear protein that is expressed highly in proliferating cells. Detection of Ki-67 was carried out (after HIER with TRS, low pH) using a rabbit monoclonal antihuman primary antibody (clone SP6; Biocare Medical, Cat No. CRM325). Anti-Ki-67 primary antibody was applied for 60 minutes.
Staining combinations
Two IHC procedures, 1 long (due to inclusion of a secondary antibody incubation) and 1 short (which lacked the secondary antibody step), were paired on each staining run. The pairing for 1 run was CD45 (long) with Ki-67 (short), while the grouping for the second run was F4/80 (long) and CD3 (short). The 2 long procedures (CD45 and F4/80) were performed separately since their secondary antibodies were applied at different concentrations.
Immunohistochemical staining protocol variations
Intrarun and inter-run precision in staining intensity was evaluated for each antibody on all 3 immunostainers using 3 runs per autostainer with 3 slides per run (for a total of 27 slides per antibody).
The impact of section thickness on IHC staining intensity was examined for all 4 antigens on 1 immunostainer (the Autostainer Link 48 model). For this purpose, multiple slides per block with section thicknesses ranging from 3 to 8 µm in 1-µm increments were processed together in the same staining run, with 1 slide per thickness dedicated to each antibody; an additional slide per thickness was stained with H&E in parallel for comparison. Again, this part of the study was done only once to mirror the design used for the H&E arm of the study. All IHC runs for all 3 instruments were performed by the same operator to remove technician-related variability as a potential confounding factor.
Semiquantitative Histopathologic Evaluation of Immunohistochemical Staining
Stained slides from colon (CD45 and Ki-67), liver (F4/80), lymph node (all biomarkers except F4/80), and spleen (all 4 biomarkers) were reviewed by an American College of Veterinary Pathologists (ACVP) board-certified veterinary anatomic pathologist (JSF). The specific tissue–biomarker combinations were chosen based on the known expression and distribution of the antigens in these organs. Each section was assigned a score from 0 to 5 based on DAB staining intensity (Supplemental Table 2) using a masked (“blinded”) histopathologic evaluation strategy.
Digital Image Analysis
Whole-slide images were acquired at 20× magnification using Leica Aperio scanners. An Aperio XT unit (scanner console version 101.0.0.23, halogen light source) was employed for H&E-stained sections, and an AT2 high-volume system (scanner console version 102.0.7.5, light-emitting diode light source) was used for IHC-stained sections. All scanners were calibrated prior to use according to the manufacturer’s instructions by capturing presnap images and setting line and area gains to correct for light reflection from the slide surface; importantly, the calibration step for all instruments was performed more frequently (weekly or more often) than is recommended by the manufacturer. Images were subjected to automated analysis using programmable algorithms in HALO software (versions 2.0 and 3.1, respectively, for the separate H&E and IHC arms of the study; Indica Labs).
Quantitative image analysis was used to determine the mean OD (staining intensity averaged across the entire section) for each stain (H&E and IHC) for each stained tissue section. Mean OD was calculated by adding the OD of all cells in each tissue and dividing by the total number of cells in that tissue. All algorithms were assessed for color accuracy visually by comparing the output overlay images to the original images on 3-color (red–green–blue [RGB]) monitors (model HP-2311x; Hewlitt-Packard) set at the factory default settings.
For H&E-stained sections, the OD of each tissue was analyzed separately using a modified area quantification algorithm. This algorithm defines the total stained area and the mean OD for hematoxylin- and eosin-colored pixels separately.
For IHC-stained sections, each tissue (5/slide) was manually annotated using separate layers to distinguish between distinct specimen types. All tissue folds and histological artifacts were traced using a negative pen tool to eliminate these regions from the analysis. Subsequently, an in-built cytonuclear algorithm was used to analyze all sections, where the algorithm measures the OD of each cell and classifies the cell as negative, weak positive, positive, or strong positive; measures the total tissue area; and calculates the total number of positive cells. Minor adjustments to the algorithm were made for each IHC stain, the chief of which were presetting the color values (RGB) for DAB and designating the cellular location of the labeled biomarker (nuclear or membrane) (Supplemental Table 3).
Statistical Analysis
The majority of current data are presented as graphs (descriptions). Where warranted, formal statistical analysis was performed using JMP Pro statistical software (version 15.1; SAS Institute) according to the manufacturer instructions. A P value ≤ 0.05 was used to indicate a significant difference.
For H&E-stained sections, a Student’s t test was employed to evaluate reproducibility in staining intensity over time (precision) as well as the influence of section thickness, reagent immersion times, and use of expired reagents on the OD of stained tissues.
For IHC-stained sections, nested variance component models18 were used to analyze the intrarun, inter-run, and instrument-related variability as well as the impact of section thickness on the relative numbers of positive cells and average OD. Variance components were expressed as a percentage of the coefficient of variation (%CV, calculated as the standard deviation divided by the mean) and summarized across the 20 tissues (5 tissues × 4 antibodies).
Results
Data are presented here separately for the H&E and IHC staining arms of this study. The reason for this organization is to emphasize the respective features evaluated by the designs of these 2 arms.
Impact of Preanalytical Variables on H&E Staining
Staining protocol design
The cumulative impact of altering the lengths of these steps across the various H&E staining protocols (Table 1) was detectable visually by examining the tissue section on the slide or the digitized image and was confirmed by the OD value yielded by DIA of the digitized image (Figure 1). As anticipated, protocol #4 (which uses mid-range conditions between the extremes of protocols #0 and #8 and represents an industry-standard design for H&E staining15,16) offered the best balance of staining intensity for both hematoxylin and eosin across tissue sections with cell populations possessing widely different numbers of nuclei and amounts of cytoplasm. The intensity of H&E staining increased as the incubation times in hematoxylin and eosin rose and the immersion time in differentiating solutions fell (Figure 1). For example, protocol #0 afforded much lighter staining intensity due to the very brief stays in hematoxylin (30 seconds) and eosin (10 seconds) relative to the extended periods for the differentiation (10 minutes) and alcohol (5 minutes) steps. In contrast, protocol #8 produced intense H&E staining due to the extended time in hematoxylin and eosin (20 minutes for each) and the absence (0 minutes) of the differentiation and alcohol steps. When comparing the entire range of tested conditions (using protocol #4 as the optimal procedure), hematoxylin OD was significantly degraded for protocols 0 to 3 while eosin OD was significantly impacted for protocols 1 and 5 to 8 (Supplemental Table 4). Depending on the study objective, the choice of an H&E staining option other than protocol #4 might provide improved visualization of certain tissue features for DIA. An example of this adjustment is boosting the intensity of eosin staining for skeletal muscle fibers by using protocol #5 (Figure 1B) or #6 (Figure 1A).
Figure 1.
Impact of the staining protocol design (reagent times) on staining intensities. Panel A depicts representative images of 4-µm thick H&E-stained sections of prototypic tissues characterized by different nuclear-to-cytoplasmic ratios in key cell populations showing the effect of changing the incubation lengths for staining and differentiation steps. Panel B shows shifts in measurements of staining intensity for both hematoxylin and eosin as the protocol design is altered. Protocol number 4 reflects the standard H&E method for vertebrate tissues.15,16 Protocol numbers 0 to 3 have shorter staining times and longer differentiation and alcohol steps, while protocol numbers 5 to 8 have longer staining times and shorter differentiation and alcohol steps. (For details on each protocol, see Table 1.). H&E indicates hematoxylin and eosin; OD, optical density.
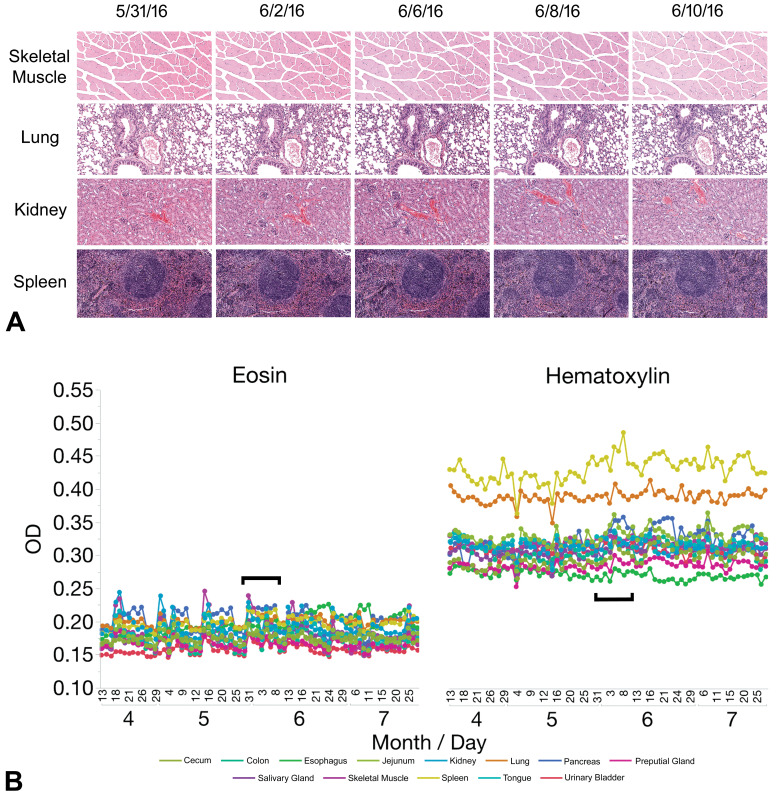
Staining precision
The consistency of H&E staining generally was comparable over time for a given tissue as assessed qualitatively in tissue sections (the chosen standard for assessing stain quality in the current study) or when viewing digitized images of the same sections (Figure 2A). However, more variability in the quality of staining could be appreciated for some tissues via fluctuations in OD values for hematoxylin and especially eosin over time (Figure 2B). For nearly all tissues, staining intensity was more stable (ie, characterized by a generally flatter curve) throughout the study for hematoxylin than for eosin. Divergence in hematoxylin OD was modest as shown by coefficients of variation (CVs) below 5.0% for all tissue types except pancreas and spleen, which had CVs of 7.0% and 5.7%, respectively (Supplemental Table 5). By comparison, eosin OD varied by approximately 2-fold from peak to trough over a 2-week cycle (Figure 2B and Supplemental Figure 2). The CVs for eosin staining intensity were less than 9.0% for all tissues except skeletal muscle, which had a CV of 12.8%. Peak staining intensity for eosin coincided with addition of fresh reagents to the H&E stainer.
Figure 2.
Precision (repeatability and reproducibility) of H&E staining varies over time for visual and OD-based assessments. Panel A shows a 2-week subset of representative images for H&E-stained sections of prototypic tissues characterized by different nuclear-to-cytoplasmic ratios in key cell populations taken at the middle of the 4-month study. Panel B shows the OD (a quantitative measure of staining intensity) of hematoxylin and eosin separately for each staining run, indicated by month and day. Note that the OD for each stain is fairly consistent over time for a given tissue but may vary among tissues (as indicated by the higher hematoxylin OD readings for lung and spleen). Black brackets represent the 2-week period during which the images were captured. H&E indicates hematoxylin and eosin; OD, optical density.
Section thickness
As expected, a visually obvious escalation in H&E staining intensity was evident as section thickness rose (Figure 3A). The OD for both hematoxylin and eosin surged in parallel (Figure 3B), although the extent of the effect depended on the stain. For hematoxylin, the OD was comparable for section thicknesses from 3 to 8 µm. In contrast, for eosin the staining intensity was more sensitive to variations in section thickness as indicated by significant differences in OD relative to a given baseline (assessed here as the eosin OD for a 4-µm thick section) for all thicknesses except 5 µm (Supplemental Table 6). The rate of the thickness-related OD increase was dependent on both the stain and the tissue; the OD in nucleus-rich tissues (eg, lung, spleen) was influenced more by hematoxylin, while the OD in cytoplasm- or connective tissue-rich tissues (eg, esophagus, skeletal muscle, tongue) was impacted more by eosin (Figure 3B). The optimal thickness to produce both suitable staining (as visualized by the pathologist) and a narrow range of OD variability (as assessed by CV values) for DIA appeared to be 4 to 6 µm across all tissues; this matches well with the usual staining thicknesses (4 µm for all tissues except bone and skin, which are cut at 5 µm) employed at Premier Laboratory for animal tissues from nonclinical studies. That said, DIA limited to a subset of tissues might be served better in some cases by using a different section thickness, such as 7 to 8 µm for cytoplasm- or connective tissue-rich tissues (Figure 3). Such adjustments are optional since conventional image analysis algorithms are capable of correctly assessing OD in sections of standard thickness.
Figure 3.
Staining intensity for H&E rises as section thickness increases. Panel A shows representative images of H&E-stained sections of various thicknesses for prototypic tissues characterized by different nuclear-to-cytoplasmic ratios in key cell populations. Panel B illustrates the progressive rise in OD ( a quantitative measure of staining intensity) for both hematoxylin and eosin as section thickness increases from 2 to 10 µm. Note that the eosin staining intensity is affected more substantially with increasing section thickness, especially in cytoplasm- and connective tissue-rich tissues like colon, esophagus, skeletal muscle, and tongue. H&E indicates hematoxylin and eosin; OD, optical density.
Reagent quality
Use of outdated hematoxylin did not produce a visible decline in H&E staining quality as assessed in digitized images of tissue sections (Figure 4A) but yielded a modest but measurable reduction in OD compared to sections stained with unexpired hematoxylin (Figure 4B). The extent of the OD decrease was greater for reagents that had been expired for a longer time. The eosin OD was not impacted systematically since the eosin lot employed for this experiment had not lapsed. That said, the intensity of eosin staining did vary over time as indicated by the clear decline in eosin OD for cytoplasm-rich tissue sections over the 2 weeks between replacement of used (but unexpired) eosin with fresh reagent (Figure 2B and Supplemental Figure 2).
Figure 4.
Reagent quality has a measurable impact on H&E staining intensity. Panel A demonstrates that sections stained with expired hematoxylin are still interpretable by visual examination, while panel B confirms that the older hematoxylin lots yield reduced OD (a quantitative measure of staining intensity). The same eosin lot was used throughout this experiment. Status of hematoxylin lots (on the start date for the study): 5261, expired for 30 weeks (left); 5390, expired for 6 weeks (middle); 5822, unexpired (right). H&E indicates hematoxylin and eosin; OD, optical density.
Impact of Preanalytical Variables in IHC Staining
Staining precision
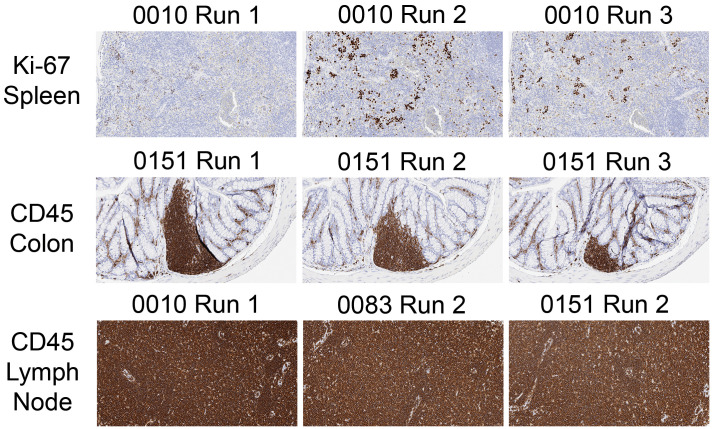
For each IHC method, staining precision (reproducibility) was assessed in 2 fashions. First, the intrarun and inter-run consistencies were assessed for each autostainer individually. Subsequently, the variation in staining among the 3 instruments was compared.
For a given instrument, data typically were equivalent both within a run and between runs (Figure 5). For a given stainer, the dispersion of DIA data depended on the biomarker and the tissue type but also on the end point under evaluation. For example, positive cell counts for all 4 stains were clustered tightly in colon and kidney for all 3 stainers (Figure 6), while average OD values were closely packed only for kidney (Figure 7). Despite the general consistency of mean OD readings for a given instrument across several runs (Figure 5A), in some cases analysis of data from each run in isolation showed that considerable variation existed across runs for all 3 autostainers. This fact was illustrated most clearly on all 3 stainers for CD45 in colon as well as Ki-67 in colon, lymph node, and to a lesser degree spleen (Figure 5B). An unexpected finding with respect to these inter-run differences was that the same pattern of variation from run #1 to run #3 occurred for all 3 stainers (Figure 5B); importantly, each run of a given IHC method was performed in parallel on all 3 instruments on the same day using identical reagent solutions. An explanation for this day-dependent, instrument-independent variability in IHC staining intensity was not apparent. Nested component variance models demonstrated that the contributions to staining intensity variations were 3.0% for the instrument and 4.6% for intrarun differences.
Figure 5.
Precision (repeatability and reproducibility) of IHC staining typically is similar within (intrarun) and between (inter-run) staining runs on a given instrument and across instruments. However, inter-run variability may occur depending on the biomarker and tissue (eg, CD45 and Ki67 in colon [panels A and B]). An unexpected result was that different instruments may share a similar pattern of inter-run variation (eg, CD45 and Ki67 in colon [panel B]). Each dot represents the average OD for deposition in 1 section. Instrument models: AutostainerPlus Link, Nos. 0010 and 0083; Autostainer Link 48, No. 0151. DAB indicates 3,3′-diaminobenzidene; IHC, immunohistochemical; OD, optical density.
Figure 6.
Precision (repeatability and reproducibility) of IHC staining depends principally on the biomarker and tissue rather than the stainer. Each dot represents the number of positive cells in 1 section. Instrument models: AutostainerPlus Link, Nos. 0010 and 0083; Autostainer Link 48, No. 0151. IHC indicates immunohistochemical.
Figure 7.
Precision (repeatability and reproducibility) of IHC staining for OD-based assessments generally is comparable across stainers, although subtle variations (eg, reduced average OD values for instrument 0151 for several biomarkers in colon, lymph node, and spleen) may be detected. These OD differences do not impact analysis of IHC data where the final interpretation is based on labeling of a specific tissue feature (eg, counts of positive cells; see Figure 6). IHC indicates immunohistochemical; OD, optical density.
Staining for a given IHC procedure typically was comparable for a given tissue for all 3 immunostainers when visually evaluating stained slides (Supplemental Table 7) and interpreting measurements of positive cell counts obtained by DIA (Figures 5A and 8A). In contrast, when using mean OD as a measure of staining intensity, values across all 3 stainers were similar for only 2 tissues (kidney, liver); for the other 3 tissues (colon, lymph node, spleen), the staining intensity imparted by the Autostainer Link 48 instrument (no. 0151) was decreased to a modest degree relative to the AutostainerPlus Link machines (nos. 0010 and 0083; Figure 8B). The reduction in staining intensity for the Autostainer Link 48 depended on the biomarker and tissue (Figure 7 and Supplemental Table 7). In the final analysis, little of the variation in DAB staining intensity—only 0.017% for the mean positive cell counts and 0.56% for the mean OD values—was attributable to inter-instrument variation. Instead, most of the variation in staining intensity was linked to the biomarker being detected (Figure 9) and the tissue type (Figures 5-7) and not the instrument (Supplemental Table 8). For intensely stained (“saturated”) tissues (eg, CD45-stained lymph node and spleen sections ≥ 6 µm in thickness; Figure 9), subtle differences in OD related to the tone of DAB staining could be discriminated by DIA (Figures 5 and 8) that were difficult to discriminate by eye (Supplemental Table 7). These subtle differences were not statistically significant and were deemed to be inconsequential to data interpretation.
Figure 8.
The choice of qualifying end point impacts the apparent equivalence of IHC procedures when done on different instruments. Comparisons of stainers based on measurements linked to a particular tissue feature (eg, labeled objects, such as positive cells [panel A]) show less variation for some tissues than do quantitative data based on staining intensity (OD) across the entire section (panel B). For a given instrument, each dot represents the average for all runs of all 4 biomarkers for that tissue. Instrument models: AutostainerPlus Link, Nos. 0010 and 0083; Autostainer Link 48, No. 0151. DAB indicates 3,3′-diaminobenzidene; IHC, immunohistochemical; OD, optical density.
Figure 9.
The staining intensity of a particular IHC method is influenced more by the biomarker than the choice of stainer. Data are shown as the CV (calculated as the SD divided by the mean) for the average OD within each staining run for each biomarker on a given instrument for all tissues. The box plots represent the distribution of the data, where the box defines the range encompassing values between the 25th and 75th quartiles, the line within the box is drawn at the median OD, and the whiskers demonstrate the expected variation in the data. Dots located over some boxes (eg, CD3 on instrument 0083) plot data that fell beyond the whiskers, which are indicative of staining runs with higher than expected variability. For Ki-67, the expanded ranges (longer boxes) on all 3 stainers denote that staining was more variable across runs for sections containing lymphoid tissue (see Figure 5). Instrument models: AutostainerPlus Link, Nos. 0010 and 0083; Autostainer Link 48, No. 0151. CV indicates coefficient of variation; DAB, 3,3′-diaminobenzidene; IHC, immunohistochemical; OD, optical density; SD, standard deviation.
Section thickness
In most tissues, staining intensity for a given IHC procedure depended on the labeled biomarker more than section thickness as indicated by no substantial difference in DAB deposition in digitized images (Figure 10A) and relatively flat curves for DAB OD (Figure 10B). The exceptions to this trend were that staining intensity (OD) in thinner sections (<5 µm thick) was lower for CD45 (pan-leukocyte marker) in lymphoid organs and F4/80 (macrophage marker) in spleen (Figure 10B) in the absence of substantive diminishment in DAB staining in sections of variable thickness (Figure 10A [for CD45]).
Figure 10.
Staining intensity for IHC methods in sections of different thicknesses may be comparable by visual inspection of tissue where specific tissue features are the subject of the evaluation (panel A, depicting CD45) but nonetheless have a quantitative difference in optical OD for some biomarkers in certain tissues as section thickness increases from 3 to 8 µm (panel B, showing lower OD for sections <5 µm thick for CD45 in lymph node and F4/80 in spleen). IHC indicates immunohistochemical; OD, optical density.
Operational considerations
While not a part of the original study design, observations in the course of the IHC arm revealed 2 other preanalytical factors that can impact the suitability of tissue sections for DIA. The first is the OD for sections from the same block, which may exhibit inter-run variation due to differences in the trimming plane (Figure 11). The extent of the difference will be greater for biomarkers expressed in only a few cells within the section. The second and more insidious source of variation is an inadvertent change in the IHC staining protocol, which can occur when separate IHC staining protocols are conducted in the same staining run on this staining platform. When 2 staining protocols of variable durations are done together on 1 instrument, the slides being stained for the shorter protocol will incubate in wash buffer between steps for a longer duration than if only 1 protocol was conducted at a time. This possibility was demonstrated clearly by the visual fading in Ki-67 staining intensity for sections processed in an autostainer run where increased time in buffer rinse steps was used as an accommodation for including CD45 (which included an extra incubation and buffer rinse to incorporate the secondary antibody prior to the polymer reagent) in the same run (Figure 12). Interestingly, the fading due to extended buffer rinses was method-dependent since CD3 staining remained robust despite having experienced similar extended buffer incubations (data not shown).
Figure 11.
Optical density as a measure of staining intensity must be verified against the tissue architecture prior to interpretation. This quality control step is necessary chiefly for methods in which staining intensity varies across runs based on the IHC protocol design (top row; see also Figures 9 and 12) or the variable presence of the cell population expressing the biomarker (middle row) but is not needed for sections where tissue composition is consistent across the entire section (bottom row). Images for each row show representative step sections for the same site of each tissue. IHC indicates immunohistochemical.
Figure 12.
Staining intensity may be impacted unexpectedly by inadvertent adjustments to the IHC protocol design. In this example, Ki-67-positive cells in 4-µm-thick step sections exhibit stronger staining when processed using a protocol in which the incubation in primary antibody is followed rapidly by application of the visualization reagent (ie, the standard protocol [column A]) relative to 4-µm thick step sections in which the primary antibody was followed by a 35-min incubation in buffer before addition of the visualization reagent (column B); the extended buffer incubation for Ki-67 corresponded to the time needed to accommodate another biomarker in the same staining run (CD45, which requires incubation with a secondary antibody prior to application of the visualization reagent). The decreased staining intensity associated with extended rinsing for some staining runs likely explains the higher variation in Ki-67 labeling across instruments (see Figure 9) relative to labeling provided by the other biomarkers tested in this study (CD3, CD45, F4/80). IHC indicates immunohistochemical.
Discussion
Digital pathology is a rapidly evolving field in which the conventional semiquantitative process of histopathologic diagnosis and severity grading is supplemented with quantitative data obtained by DIA. Acquisition of credible digital pathology data requires that tissue sections slated for DIA be of high quality. Section quality is determined by such preanalytical factors as sample collection, fixation, and histological processing. Our current data reinforce the fact that all aspects of the histological endeavor influence the appearance of tissue sections and thus their suitability for digital pathology. In this regard, several elements of our study provide insight for pathologists involved in designing, implementing, and interpreting digital pathology studies.
First, DIA detected quantitative changes in staining intensity (in terms of OD, the amount of light that can pass through a stained structure) associated with all preanalytical factors explored during this study: staining protocol design (Figures 1 and 12); precision (Figures 2 and 6); reagent quality (eg, freshness, as in Figures 2, 4, and 5); section thickness (Figures 3 and 10); and staining instrumentation (Figure 8). This trend applied to both conventional H&E staining and for several IHC procedures when performed with commercial automated stainers and reagents. This universal applicability indicates that all these histological parameters should be scrutinized as potential sources of variability in section quality when DIA is a possible end point of interest. Systems to monitor all these factors are a natural consequence when histological laboratories process sections using well-written SOPs, especially if performed in compliance with GLP guidelines.
Second, our current data clearly emphasized that some preanalytical factors are more critical than others in terms of standardizing staining quality. The major preanalytical determinants of staining intensity for both H&E and IHC methods were the staining protocol design; section thickness, biomarker being detected (for IHC, Figures 5, 6, and 9); and the tissue (Figures 2, 5, and 6)—assuming that routine conventions were maintained for the remainder of the histology procedure (eg, standard section thickness, utilization of unexpired reagents). Sections having different compositions may be affected unequally when processing conditions are altered for particular stain components. For example, the staining intensity of cytoplasm-rich tissues like skeletal muscle may fluctuate to some degree based on the freshness of eosin (Figure 2 and Supplemental Figure 2). This fact suggests that several different tissues may need to be available so that appropriate method-specific control materials are available for QC monitoring. A corollary element that deserves consideration during histology QC is the structural homology of sections. A decrease or lack of the desired cell population or tissue feature by default will diminish staining relative to other sections, as we noted during the current study based on the variable presence of GALT in colon (Figure 11). This observation emphasizes that an observer needs to verify that section quality is comparable when interpreting digital pathology data.
Third, our present data indicated that each automated stainer must be monitored individually with respect to maintaining staining precision over time. Precision is an evaluation of the extent to which repeated measurements under the same conditions yield the same results. Two aspects of precision are repeatability, the degree to which performance by an instrument or operator varies over a short period when working under the same conditions, and reproducibility, which is the extent to which an entire multistep process (eg, an IHC protocol or DIA algorithm) can be duplicated. Our data for 3 identically programmed and loaded immunostainers show that intrarun and, to a lesser extent, inter-run variation is low for tissue step sections stained on a given instrument (Figures 5 and 6), but that measurable differences in quantification of staining intensity (as OD) exist between stainers (Figure 8). Such modest discrepancies will not impact interpretation of DIA data if the end point being examined ties staining to a particular structure (eg, labeled cells), whereas interpretation based only on the cumulative staining intensity (ie, positive pixels, wherever they occur in the section) may be impacted if 1 instrument consistently delivers a lighter stain (Figures 7 and 8). An ancillary concern for IHC procedures is that staining intensity may be impacted inadvertently when multiple procedures are included in the same staining run. Programmed run times for 2 studies with different steps may result in extended buffer incubations that may remove bound antibody, thereby leading to reduced chromogen deposition. This variable may be instrument-dependent and is likely method-dependent, with less robust antibody–antigen pairs being more susceptible to reduced staining from extended buffer rinses. This inference suggests that extended buffer rinsing caused fading for Ki-67 (Figure 12) but not the more robust CD3 method. Again, this possibility highlights that an experienced observer should provide QC for staining quality, keeping in mind that the staining intensity relative to historical control patterns may be as vital as comparison to concurrent controls.
Fourth, our current data demonstrate that preanalytical factors known to impact the quality of human tissues10,11 also apply similarly to tissues of nonclinical species like mice (as shown in this article) and presumably other test species as well. The average OD of H&E-stained sections from humans and mice are similar (Supplemental Table 9). This outcome was anticipated given that H&E is the gold standard for staining biological tissue sections, IHC methods are the bulwark of molecular biology and molecular pathology investigations for all species, and that staining protocols for these procedures are equivalent across species (except for the need for primary antibodies directed against species-specific antigens for IHC). Nonetheless, this confirmation of interspecies concordance deserves mention given that tissue structure19 and function vary across species.
Finally, our present data confirm that the choice to perform a conventional histopathologic evaluation, a digital pathology analysis, or both depends on the research question. For many tissues and stains in the current study, qualitative or semiquantitative visual review of a tissue section or digitized image of that section was equivalent to DIA in terms of detecting differences in staining quality (Figures 1, 3, 10 and Supplemental Table 7). This concordance suggests that in many cases a rapid review by an experienced histotechnologist will be suitable for QC and that a semiquantitative histopathology evaluation by a qualified pathologist will provide high-quality data acceptable for interpretation. That said, automated DIA clearly showed (and rapidly quantified) differences in staining intensity below the limit of visual discrimination by the human eye (Figures 2 and 4), indicating that instrument-based analysis is the most effective and appropriate approach to address some research questions.
Our study design did not collect data to formally address several other questions regarding how preanalytical variables might impact DIA, but our experience over many projects has provided insight regarding potential answers for them. For instance, what parameters intrinsic to a specific stainer (presence or absence of covertiles, slide numbers per run, slide positions, etc) might affect staining quality? Our work over the years with multiple stainers from several vendors with differing instrument designs (with or without covertiles, variable slide payloads and reagent application mechanisms) indicates that instruments have their own idiosyncrasies, but that most may be used without concern as long as systems are properly maintained and calibrated and a record of staining precision over time is generated to assure that the stainer is operating within normal parameters. In like manner, when performing large IHC projects, should slides be processed serially on a single instrument or might they be processed in parallel on several appropriately maintained and calibrated systems? Again, our experience with many platforms indicates that different representatives of a given stainer model exhibit generally equivalent performance, so multiple instruments and several runs may be employed to complete larger IHC studies. As a third example, if technical issues in processing (eg, breakage, staining or tissues artifacts) are detected during an IHC project, may the individual slides be rerun on the original stainer and then slotted into the preexisting slide set or must all slides in the set be rerun? Our current practice is to record the breakage, reprocess the individual slides, and—if the staining pattern for the control tissue exhibits the expected distribution of the target antigen—insert them into the slide set. By visual inspection (multiple histologists and pathologists), this approach does not seem to have affected the IHC data, but this premise has not been explored by DIA.
Two additional questions presented themselves during the analysis and interpretation phases of our current project, but our study data cannot answer them. First, could an approach be devised whereby a simple value inherent in (eg, OD) or calculated from (eg, %CV) a stainer’s metadata might serve consistently as an acceptable threshold for validating the data quality for a given staining run? The authors anticipate that performance reliability and reproducibility will be validated by formal analysis of staining precision rather than scrutiny of the OD and/or %CV. This supposition is based on the observable inter-run and intrarun variation in OD that may be observed in performing a given IHC method over time. For the second unanswered question, can DIA data for a given method be compared across studies over time that employ the same reagents and stainer or is data interpretation valid only within the context of a given study? In a qualitative sense, pathologists routinely make diagnoses across studies for a particular histology method based on its known pattern of staining. However, the authors posit that quantifiable stain attributes of tissue sections slated for DIA may vary substantially over time as reagent lots are replaced and environmental conditions within the facility fluctuate (eg, with the seasons11). Our current results do not address these questions, but our findings do provide an outline of key data categories and QC activities that will be essential considerations in investigating such topics.
Conclusion
In summary, our findings confirm that many preanalytical factors encountered during routine histological processing will impact staining quality of tissue sections. The most critical variables to control across all procedures are the staining protocol design, reagent quality, section thickness, and tissue homology (ie, the presence of the cells or tissue feature of interest). The choice of biomarker and tissue battery also will impact staining intensity for IHC procedures, but these factors actually represent key elements of the research question rather than preanalytical variables per se. Our data show that staining quality will be comparable within and between runs on a given automated stainer, but that staining intensity will diverge for sections processed on different stainers even if they are programmed and loaded identically. Taken together, these data indicate that QC will be needed on each histological procedure and each stainer to be employed in preparing sections slated for DIA. More specifically, our data clearly demonstrate that “turn-key” algorithms in DIA software must be sufficiently robust to accommodate variation intrinsic to histology processing and strongly suggest that operators utilizing DIA “shareware” have the ability to troubleshoot unusual data patterns in DIA data sets.
Supplemental Material
Supplemental Material, sj-docx-1-tpx-10.1177_0192623320970534 for Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis by Elizabeth A. Chlipala, Mark Butters, Miles Brous, Jessica S. Fortin, Roni Archuletta, Karen Copeland and Brad Bolon in Toxicologic Pathology
Supplemental Material, sj-tif-1-tpx-10.1177_0192623320970534 for Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis by Elizabeth A. Chlipala, Mark Butters, Miles Brous, Jessica S. Fortin, Roni Archuletta, Karen Copeland and Brad Bolon in Toxicologic Pathology
Supplemental Material, sj-tif-2-tpx-10.1177_0192623320970534 for Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis by Elizabeth A. Chlipala, Mark Butters, Miles Brous, Jessica S. Fortin, Roni Archuletta, Karen Copeland and Brad Bolon in Toxicologic Pathology
Acknowledgments
The authors would like to thank Adam Smith of Indica Labs for image analysis guidance and the use of HALO Image Analysis software. Ada Feldman of Anatech, Ltd generously supplied the expired hematoxylin-normal strength reagents. Beth Mahler, the journal’s Illustrations Editor, was instrumental in helping to optimize the figures.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Elizabeth A. Chlipala  https://orcid.org/0000-0002-2194-7645
https://orcid.org/0000-0002-2194-7645
Mark Butters  https://orcid.org/0000-0002-6307-2496
https://orcid.org/0000-0002-6307-2496
Jessica S. Fortin  https://orcid.org/0000-0002-1007-9360
https://orcid.org/0000-0002-1007-9360
Karen Copeland  https://orcid.org/0000-0002-6455-3283
https://orcid.org/0000-0002-6455-3283
Brad Bolon  https://orcid.org/0000-0002-6065-1492
https://orcid.org/0000-0002-6065-1492
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Aeffner F, Zarella MD, Buchbinder N, et al. Introduction to digital image analysis in whole-slide imaging: a white paper from the Digital Pathology Association. J Pathol Inform. 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Webster JD, Dunstan RW. Whole-slide imaging and automated image analysis: considerations and opportunities in the practice of pathology. Vet Pathol. 2014;51(1):211–223. [DOI] [PubMed] [Google Scholar]
- 3. Potts SJ. Digital pathology in drug discovery and development: multisite integration. Drug Discov Today. 2009;14(19-20):935–941. [DOI] [PubMed] [Google Scholar]
- 4. Gauthier BE, Gervais F, Hamm G, O’Shea D, Piton A, Schumacher VL. Toxicologic Pathology Forum: opinion on integrating innovative digital pathology tools in the regulatory framework. Toxicol Pathol. 2019;47(4):436–443. [DOI] [PubMed] [Google Scholar]
- 5. Griffin J, Treanor D. Digital pathology in clinical use: where are we now and what is holding us back? Histopathology. 2017;70(1):134–145. [DOI] [PubMed] [Google Scholar]
- 6. Rizzardi AE, Johnson AT, Vogel RI, et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn Pathol. 2012;7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pell R, Oien K, Robinson M, et al. The use of digital pathology and image analysis in clinical trials. J Pathol Clin Res. 2019;5(2):81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunstan RW, Wharton KA, Jr, Quigley C, Lowe A. The use of immunohistochemistry for biomarker assessment—can it compete with other technologies? Toxicol Pathol. 2011;39(6):988–1002. [DOI] [PubMed] [Google Scholar]
- 9. Potts SJ, Young GD, Voelker FA. The role and impact of quantitative discovery pathology. Drug Discov Today. 2010;15(21-22):943–950. [DOI] [PubMed] [Google Scholar]
- 10. Chlipala E, Bendzinski CM, Chu K, et al. Optical density-based image analysis method for the evaluation of hematoxylin and eosin staining precision. J Histotechnol. 2020;43(1):29–37. [DOI] [PubMed] [Google Scholar]
- 11. Chlipala EA, Bendzinski CM, Dorner C, et al. An image analysis solution for quantification and determination of immunohistochemistry staining reproducibility. App Immunohistochem Mol Morphol. 2020;28(6):428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janardhan KS, Jensen H, Clayton NP, Herbert RA. Immunohistochemistry in investigative and toxicologic pathology. Toxicol Pathol. 2018;46(5):488–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Der Laak JA, Pahlplatz MM, Hanselaar AG, de Wilde PC. Hue-saturation-density (HSD) model for stain recognition in digital images from transmitted light microscopy. Cytometry. 2000;39(4):275–284. [PubMed] [Google Scholar]
- 14. NRC-ILAS (National Research Council, Institute of Laboratory Animal Sciences). Guide for the Care and Use of Laboratory Animals. 8th ed. National Academy Press; 2011. [Google Scholar]
- 15. Carson FL, Hladik C. Histotechnology: A Self-Instructional Text. 3rd ed. American Society for Clinical Pathology Press; 2009. [Google Scholar]
- 16. Gamble M. The hematoxylins and eosin. In: Bancroft JD, Gamble M, eds. Theory and Practice of Histological Techniques. 6th ed. Elsevier; 2011. [Google Scholar]
- 17. NSH (National Society for Histotechnology). Guidelines for hematoxylin & eosin staining. Published 2001. Accessed September 15, 2020. http://nsh.org/sites/default/files/Guidelines_For_Hematoxylin_and_Eosin_Staining.pdf.
- 18. NIST/SEMATECH (U.S. National Institute of Standards and Technology / Semiconductor Manufacturing Technology Consortium). Comparisons based on data from more than two processes. e-Handbook of Statistical Methods. Published 2003. Updated October 30, 2013. Accessed September 15, 2020. https://www.itl.nist.gov/div898/handbook/prc/section4/prc4.htm.
- 19. Treuting PM, Dintzis SM, Montine KS, eds. Comparative Anatomy and Histology: A Mouse, Rat, and Human Atlas. 2nd ed. Academic Press (Elsevier); 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-docx-1-tpx-10.1177_0192623320970534 for Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis by Elizabeth A. Chlipala, Mark Butters, Miles Brous, Jessica S. Fortin, Roni Archuletta, Karen Copeland and Brad Bolon in Toxicologic Pathology
Supplemental Material, sj-tif-1-tpx-10.1177_0192623320970534 for Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis by Elizabeth A. Chlipala, Mark Butters, Miles Brous, Jessica S. Fortin, Roni Archuletta, Karen Copeland and Brad Bolon in Toxicologic Pathology
Supplemental Material, sj-tif-2-tpx-10.1177_0192623320970534 for Impact of Preanalytical Factors During Histology Processing on Section Suitability for Digital Image Analysis by Elizabeth A. Chlipala, Mark Butters, Miles Brous, Jessica S. Fortin, Roni Archuletta, Karen Copeland and Brad Bolon in Toxicologic Pathology