Abstract
The stability and high productivity of heterogeneous terpenoid production in Escherichia coli expression system is one of the most key issues for its large scale industrialization. In the current study on taking lycopene biosynthesis as an example, an integrated Escherichia coli system has been generated successfully, which resulted into stable and high lycopene production. In this process, two modules of mevalonate (MVA) pathway and one module of lycopene expression pathway were completely integrated in the chromosome. Firstly, the copy number and integrated position of three modules of heterologous pathways were rationally optimized. Later, a strain DH416 equipped with heterogeneous expression pathways through chromosomal integration was efficiently derived from parental strain DH411. The evolving DH416 strain efficiently produced the lycopene level of 1.22 g/L (49.9 mg/g DCW) in a 5 L fermenter with mean productivity of 61.0 mg/L/h. Additionally, the integrated strain showed more genetic stability than the plasmid systems after successive 21st passage.
Keywords: Escherichia coli, Lycopene biosynthesis, Rational model of expression pathways, Integrated MVA pathway, Fermentation
Abbreviations: MVA pathway, Mevalonate pathway
1. Introduction
The terpenoid family comprises of the largest class of organic products with over 80,000 characterized members [1]. Many terpenoids have substantial commercial values as perfumes, cosmetics, food additives, and pharmaceutical products [[2], [3], [4], [5], [6]]. Lycopene is a carotenoid C40 terpenoid with numerous therapeutic functions, such as anti-oxidative property, immunity enhancer, and effective against ocular and cardiovascular diseases [[7], [8], [9]]. In biological organism, all terpenoids are derived from the common C5 building blocks, such as isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP) are synthesized either from mevalonate (MVA) pathway or from 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway [[10], [11], [12]].
In the last two decades, several synthetic biological techniques have been developed to boost terpenoids yield in microbial host [11,[13], [14], [15]]. In this regard, due to the fast growth rate and availability of genetic tools, Escherichia coli has become one of the most commonly used hosts for synthetic production of terpenoids. Naturally, it utilizes the MEP pathway to produce terpenoids in smaller proportions, but it's not enough to meet the growing demand [11]. So, there are multiple engineering techniques that commonly exploit at the genomic level in Escherichia coli to achieve a high yield of terpenoids, either by enhancing the endogenous MEP pathway or by introducing heterogeneous MVA pathway to develop IPP and DMAPP pools. In 2003, the heterogeneous MVA pathway was also employed by Martin and his coworkers for significant production of sesquiterpene amorpha-4,11-diene in Escherichia coli. Previously, this same approach enabled a low-cost and environment friendly conversion of renewable acetyl-CoA into valuable terpenoid [11]. This genomic approach had proven to be more beneficial in comparison to the enhancement of endogenous MEP pathway due to certain aspects. Firstly, the heterogeneous MVA pathway bypassed the MEP pathway, which was regulated by unknown physiological stimuli in Escherichia coli [11]. Secondly, there was only one precursor involved in the MVA pathway; whereas, the MEP pathway encountered the metabolic problem of an imbalanced supply of precursors such as pyruvate and glyceraldehyde 3-phosphate (G3P) [16]. Thirdly, the MVA pathway depends upon cofactor availability to generate one lycopene molecule [17,18]. The demand of redox partners is quite higher for the MEP pathway along with hydroxymethylbutenyl 4-diphosphate (HMBPP) synthase, HMBPP reductase and DXP reductase, each requiring 1 NADPH for the accomplishment of a particular step. On the other hand, the MVA pathway needs 2 NADPH for the production of mevalonate by 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase. It demonstrates that three steps of the MEP pathway rely heavily on NADPH's availability for proper functioning, which is mainly supplied by pentose phosphate pathway. In the MVA pathway, only one step is regulated in this way. Therefore, the requirement of NADPH and ATP in the MVA pathway is quite lesser than the MEP pathway. The MVA pathway produces 46 mol NADH to generate one mole of terpenoids compared to 6 mol NADH produced as a result of MEP pathway. So, the former pathway ensures the supply of enough NADH molecules through the oxidation of pyruvate to acetyl-CoA, while consuming glucose as major carbon source [[18], [19], [20], [21], [22]]. Upon comparing the both pathways' attributes, the heterologous MVA pathway has shown better results of terpenoids production in Escherichia coli. (see Table S1, Supporting Information), such as isoprene [[23], [24], [25]], isopentenol [26,27], sabinene [28], pinene [[29], [30], [31]], limonene [32], geraniol [33,34], amorpha-4,11-diene [11,[35], [36], [37], [38]], bisabolane [39], farnesene [40], Farnesol [41], valerena-1,10-diene [42], β-caryophyllene [43,44], and lycopene [45,46]. Among these reported works, some effective strategies were implemented to enhance the terpenoid level, including the reduced formation of fermentation byproducts [23], analysis of targeted proteins [26,37,40], fusion of proteins [29], enzyme mutant [30], and controlled conversion of acetic acid [44]. All these outstanding results (Table S1 Supporting Information) demonstrate effectiveness of MVA pathway in Escherichia coli. Shen and his colleague conducted various scientific researches to get optimum lycopene production by overcoming genetic instability of plasmid-based expression systems through promoter engineering of integrated MVA pathway in Escherichia coli [47]. Thus the heterogeneous expression pathways were more stable, and had constant copy number. However, the lycopene level (529.45 mg/L) was lower than Zhou et al.‘s report (1.44 g/L) based on plasmid systems in a fed-batch culture mode [46,48]. This lower lycopene level might have been due to the imbalanced heterogeneous expressing pathways.
In the current study, the main focus is to rationally generate a stable integrated strain with higher lycopene productivity. In our previous work, one copy of PT7 (T7 promoter)-MVA pathway and PT7lac (T7lac promoter)-lycopene expression pathway was chromosomally integrated into Escherichia coli, and T7 RNA polymerase gene-tetR was integrated into the araB site, generating the strain DH411. Thus, the T7 RNA polymerase was under the control of araC-PBAD, and l-arabinose could be used as inductor to produce lycopene. However, its specific lycopene production was 0.66 mg/g DCW (dry cell weight), which was considered very low [49]. In order to improve the lower productivity, many previous studies have shown that chromosomal positioning has a notable impact on gene transcription to get the desired level of product [50,51]. Hence, in this study, a rational strategy is developed to engineer unique model of expression pathway which is employed to determine the optimum copy number and to select better chromosomal integrated position. The final Escherichia coli strain DH416 achieves the mean lycopene productivity of 61.0 mg/L/h after fed-batch culture. In addition, it has relatively better stability of lycopene productivity than the strain carrying plasmid systems.
2. Materials and methods
2.1. Cloning strains, primers and chemicals
The strains and plasmids used in this study are described in Table 1. E. coli DH5α, DH1 and DMT were used to construct plasmids. E. coli DMT was exploited for the elimination of the methylated plasmid in vivo. The manufacturing of all primers were executed by GENEWIZ Co. (Suzhou, China) and described in Table S2. Rapid DNA Ligation Kit (Lot 00225679), PrimeSTAR HS DNA polymerase, Restriction enzymes (Lot 00213114), Gel Extraction Kit (Lot 00071311), CloneExpress MultiS One Step Cloning Kit, and Plasmid Miniprep Kit (Lot 01915KA1) were purchased from Thermo Fisher Scientific Inc. (MA, USA), Vazyme Biotech Co. (Nanjing, China), TaKaRa Biotechnology Co. (Dalian, China) CloneExpress MultiS One Step Cloning Kit, Corning Inc. (Wujiang, China), or CloneExpress MultiS One Step Cloning Kit. All operations were commenced as per the manufacturer's instructions.
Table 1.
Strains and plasmids used in this study.
| Name | Description | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 λ−thi-1 gyrA96 relA1 | Takara |
| E. coli DH1 | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Our laboratory |
| E. coli DMT | lacZΔM15Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk− mk+) phoA supE44 thi-1 gyrA96 relA1 tonA | TRANSGEN |
| Strains used for lycopene production | ||
| DH411 | DH06 Δ23th:: T7lac GC (counter-clockwise) Δ64th:: T7 S1 Δ57th:: T7 ES araB::T7RNAP-tetA | [49] |
| DH412 | DH411 Δ8th:: T7 GC (counter-clockwise) | This study |
| DH422 | DH411 Δ44th:: T7 GC | This study |
| DH432 | DH411 Δ58th:: T7 GC | This study |
| DH442 | DH411 ΔlpxM:: T7 GC (counter-clockwise) | This study |
| DH414 | DH412 ΔlpxM:: T7 GC (counter-clockwise) | This study |
| DH416 | DH414 Δ58th:: T7 S1 | This study |
| D411E | DH411/pCC1E | This study |
| D411S | DH411/pCC1S | This study |
| D411G | DH411/pCC1G | This study |
| D411ES | DH411/pCC1ES | This study |
| D411EG | DH411/pCC1EG | This study |
| D411SG | DH411/pCC1SG | This study |
| D412SS | DH412/pCC1SS | This study |
| D412SG | DH412/pCC1SG | This study |
| D422SG | DH422/pCC1SG | This study |
| D432SG | DH432/pCC1SG | This study |
| D442SG | DH442/pCC1SG | This study |
| D414S | DH414/pCC1S | This study |
| D414SS | DH414/pCC1SS | This study |
| D414SG | DH414/pCC1SG | This study |
| Plasmids | ||
| pET3b | pMB1 origin, T7 promoter; AmpR | Novagen |
| Pete | pET3b derived, carrying T7 ES; AmpR | Our laboratory |
| pETS | pET3b derived, carrying T7 S1; AmpR | [49] |
| pETG | pET3b derived, carrying T7 GC; AmpR | [49] |
| pCC1FOS | oriV, ori2, repE, parA, parB, parC, cos, loxP, lacZ, redF; CmR | Epicentre |
| pCC1E | pCC1FOS derived, carrying T7 ES; CmR | This study |
| pCC1S | pCC1FOS derived, carrying T7 S1; CmR | [49] |
| pCC1G | pCC1FOS derived, carrying T7 GC; CmR | This study |
| pCC1ES | pCC1FOS derived, carrying T7 ES and T7 S1; CmR | This study |
| pCC1EG | pCC1FOS derived, carrying T7 ES and T7 GC; CmR | This study |
| pCC1SS | pCC1FOS derived, carrying T7 S1 and T7 S1; CmR | [49] |
| pCC1SG | pCC1FOS derived, carrying T7 S1 and T7 GC; CmR | [49] |
| Plasmid used for integration | ||
| Helper plasmid | ||
| pCNA | pKOBEG derived, carrying I-CreI endonuclease gene at Nhe I site; AmpR | [52] |
| pSNA | pKOBEG derived, carrying I-SceI endonuclease gene at Nhe I site; AmpR | [52] |
| pSNK | pKOBEG derived, carrying I-SceI endonuclease gene at Nhe I site; KanR | [52] |
| Landing pad plasmid | ||
| pBDC | p15A origin, sacB cassette, I-CreI sites; CmR | [58] |
| pBDK | p15A origin, sacB cassette, I-CreI sites; KanR | [58] |
| pBDC-8ri | pBDC derived, carrying eighth homologous region; CmR | [52] |
| pBDC-44i | pBDC derived, carrying 44th homologous region; CmR | [52] |
| pBDC-58i | pBDC derived, carrying 58th homologous region; CmR | [52] |
| pBDC-Li | pBDC derived, carrying lpxM homologous region; CmR | [49] |
| Donor plasmid | ||
| pKI | pMB1 origin, I-SceI sites; KanR | [49] |
| pETIS | pET3b derived, I-SceI sites; AmpR | [49] |
| pKILE | pMB1 origin, T7 ES, I-SceI sites; KanR, carrying lpxM homologous region | [49] |
| pKIS | pKIS derived, carrying T7 S1; KanR | [49] |
| pETISG | pETIS derived, carrying T7 GC; AmpR | [49] |
| pKLIG | pMB1 origin, T7 GC, I-SceI sites; KanR, carrying lpxM homologous region | This study |
2.2. Culture conditions
Luria Broth (LB) (10 g/L tryptone, 5 g/L yeast extract and 10 g/L NaCl) medium or agar plates medium was used for cell inoculation. LBS agar plates (LB agar plates without NaCl but containing 6% sucrose) were used for sacB counter selection of the landing pad plasmids [52]. Cells were grown at 30 °C and helper plasmid curing was done at 42 °C. For lycopene production, cells were cultured at 220 rpm at 30 °C in test tube with 5 mL FMGAT medium (15 g/L tryptone, 12 g/L yeast extract, 5 g/L NaH2PO4·2H2O, 7 g/L K2HPO4·3H2O, 2.5 g/L NaCl, 5 g/L Tween 80, 10 g/L glycerol, 0.5 g/L MgSO4, 2 g/L glucose, and 2 g/L l-arabinose), and original inoculum concentration was 0.05 (OD600nm) [49]. Ampicillin (50 μg/ml), chloramphenicol (25 μg/ml), and kanamycin (25 μg/ml) were added for plasmid maintenance.
2.3. Fermentation of strain DH416
To achieve the seed culture, two colonies of strain DH416 were grown overnight at 30 °C in each 240 mL of LB medium respectively until the OD600nm reached to 3.5. The 120 mL overnight culture from two bottles was together inoculated into 5 L bioreactor (Shanghai Guoqiang Bioengineering Equipment Co., Ltd, Shanghai, China) having initial volume of 3 L medium for fed-batch fermentation. The medium comprises of glucose (180 g), tryptone (45 g), yeast extract (36 g), NaH2PO4·2H2O (9 g), K2HPO4·3H2O (21 g), NaCl (7.5 g), Tween 80 (15 g), MgSO4 (2.2 g), and defoamer (5 g). At initial stage of fermentation, the temperature was maintained at 37 °C while the gas flow was 3 L/h, and pH was adjusted at 6.9–7.1 by using NH4OH. Moreover, the continuous agitation was adjusted at 500 rpm, which was required to achieve dissolved oxygen tension above 20%. After 7.5 h, 12 g l-arabinose was added into the system, and the temperature was changed to 33 °C. Then, 0.234 g glucose was added into the system within time period of 120 s. The 6 g l-arabinose was added into the system at 12 h of the fermentation [53].
2.4. Genes
The ES cassette catalyzes the conversion from acetyl-CoA to mevalonic acid (MVA upper pathway), including genes mvaE and mvaS amplified from genome of Enterococcus faecalis (GenBank: AF290092.1). The S1 cassette catalyzes the conversion from mevalonic acid to IPP and DMAPP (MVA lower pathway), consists of codon-optimized genes fni, mvk, mvaK2, and mvd1 from Streptococcus pneumoniae which synthesized by GENEWIZ, Inc (see Table S3, Supporting Information) [49]. The GC cassette catalyzes the transformation from IPP and DMAPP to lycopene (Lycopene expression pathway), comprise of codon-optimized idsA from Archaeoglobus fulgidus (see Table S3, Supporting Information) and crtI and crtB from genome of Pantoea agglomerans [52].
2.5. Plasmid construction
The backbone of pCC1E (fragment CC1) was amplified from plasmid pCC1FOS with the primers RP5 and FP3. The fragment ESCC containing T7 ES flanked by 15–20 bp homologous region of fragment CC1 was amplified from plasmid pETE with the primers BI5 and BI3. The fragment GCCC was obtained by the same method but its template was plasmid pETG [49]. Two fragments, CC1 and ESCC (or GCCC) were homologous assembled in vitro using One Step Cloning Kit (Vazyme Biotech Co.) to create the plasmid pCC1E (or pCC1G), and identified by PCR using the primers FP-0 and RP-1.
The fragment ESX containing T7 ES was achieved by PCR using primers BI5 and LP3 and pETE as a template. The fragments XS1 (XGC) were obtained by PCR using the same primers LP5 and BI3, and pETS (pETG) [49], as a template respectively. Three PCR products, CC1, ESX, and (XS1 or XGC) were assembled in vitro to create the plasmids pCC1ES and pCC1EG respectively.
The fragment KIL containing pBM1 ori, I-SceI endonuclease (Intron-encoded endonuclease) recognition sites, lpxM (Lipid A biosynthesis myristoyltransferase) non-essential region (GenBank: CP001637.1/1,937,085–1,938,013 bp), and kan gene was achieved by PCR using primers PKR5 and PKL3, and pKILE as a template. Two PCR products, KIL and GCCC were assembled in vitro to create the plasmid pKILG.
2.6. Integration method
The integration of expression pathway was performed according to the previous report [52]. In the first step, the helper plasmid produced I-CreI endonuclease (Homing endonuclease) and Red recombinases. The landing pad plasmid released the target DNA fragment containing cat gene, homologous regions, and landing pad regions digested by I-CreI endonuclease to replace the nonessential region. In the second step, the helper plasmid produced I-SceI endonuclease and Red recombinases. The chromosome DSB (double-strand break) was produced by excision of the cat gene (catalase) and then repaired with the heterogeneous expression pathway flanked by landing pad regions, which was released from the donor plasmid.
2.7. Analytical methods of lycopene
The collection of cells and detection of lycopene contents were carried out by following the previous studies [45,46,49,54]. The Escherichia coli cells were harvested by centrifugation, and then washed once with dd H2O (double-distilled water). The lycopene accumulated in cells were extracted with acetone in dark at 55 °C for 15 min. The samples were centrifuged at 11,340×g for 10 min. The lycopene content was determined by measuring the absorbance at 474 nm using an UV-2100 spectrophotometer (Ultraviolet-2100 spectrophotometer) (UNICO co., Shanghai, China). Lycopene purchased from analytical SIGMA-ALDRICH was used as the standard. Three independent experiments were performed with representation of mean ± standard deviation. The coefficient of 0.37 g DCW/OD600nm was used to calculate the specific lycopene production [49].
2.8. RNA isolation and qRT-PCR analysis
Mostly, the expression levels related to gfp (green fluorescent protein) mRNA was quantified through qRT-PCR technique in various BRY strains (experimental E. coli strain). The isolation of total RNA was achieved by an RNeasy Mini kit with on-column DNase I digestion (Qiagen) and RNA later (Ambion) stabilization solution. There would be a reverse transcription of total RNA by exploiting a tetro cDNA synthesis kit (Bioline) accompanied with oligonucleotides specific for both genes, termed as bglB and gfp. Concurrently, the Mx3000P qPCR system and (Agilent) and Brilliant III Ultra-Fast SYBR Green QPCR master mix (Agilent) employed cDNA as template in qPCR. However, the determination of relative expression levels of target gfp gene by taking bglB (beta-glucosidase) as reference gene is accomplished by normalizing reaction threshold cycle (CT) values. CT values for un-induced, no isopropyl -D1-thiogalactopyranoside is chosen as calibrators for examining the results by the relative quantification method (2−ΔΔCT) [55]. Three separate biological replicates were created by repeating each reaction three times in order to obtain mean and standard deviation values.
3. Results and discussion
3.1. Constructing of the model of expression pathway
In order to biosynthesize lycopene from acetyl-CoA in vivo, nine heterologous genes were simplified as three elements ES, S1, and GC cassette corresponding to “MVA upper pathway”, “MVA lower pathway”, and “Lycopene expression pathway” (Fig. 1). This rational model directs the pathway optimization design to construct the integrated heterogeneous expression pathway unlikely to plasmid based expression system (Fig. 2A). Thus, in present study relating to optimization of heterogeneous pathway, a series of sub-models were generated (Fig. 2B). To verify which heterogeneous expression pathway needed to be overproduced initially, the strain DH411 was transformed with the single copy plasmid pCC1FOS carrying ES cassette, S1 cassette, or GC cassette respectively, which in turn generating D411E (DH411/pCC1E), D411S (DH411/pCC1S), and D411G (DH411/pCC1G) strains (Fig. 2B). However, no obvious improvement in specific lycopene production was analyzed (Fig. 3A). It indicated that the next integrated set up comprised of more than one expression pathway for accelerated overexpression while comparing to initial design. So, the two of the three pathways were constructed on the backbone of plasmid pCC1FOS, generating pCC1ES, pCC1EG, and pCC1SG plasmids respectively. The strains D411ES, D411EG, and D411SG were generated by transforming the plasmids pCC1ES, pCC1EG, and pCC1SG into DH411 strain correspondingly. Based on the above mentioned results, D411SG was selected as the best integrated strain because of its highest specific lycopene production approximately up to 19.66 mg/g DCW, which is nearly 30-folds higher than starting strain DH411 (Fig. 3A). This result indicated that both MVA lower pathway and lycopene expression pathway needed to be overexpressed to amplify the final product yield same as in previous studies [49]. Moreover, this result showed that low copy of heterologous expression pathways was sufficient for lycopene production in integrated strain. So, the acquaintance of copy number of heterologous expression pathways is necessary to design efficient expression pathway.
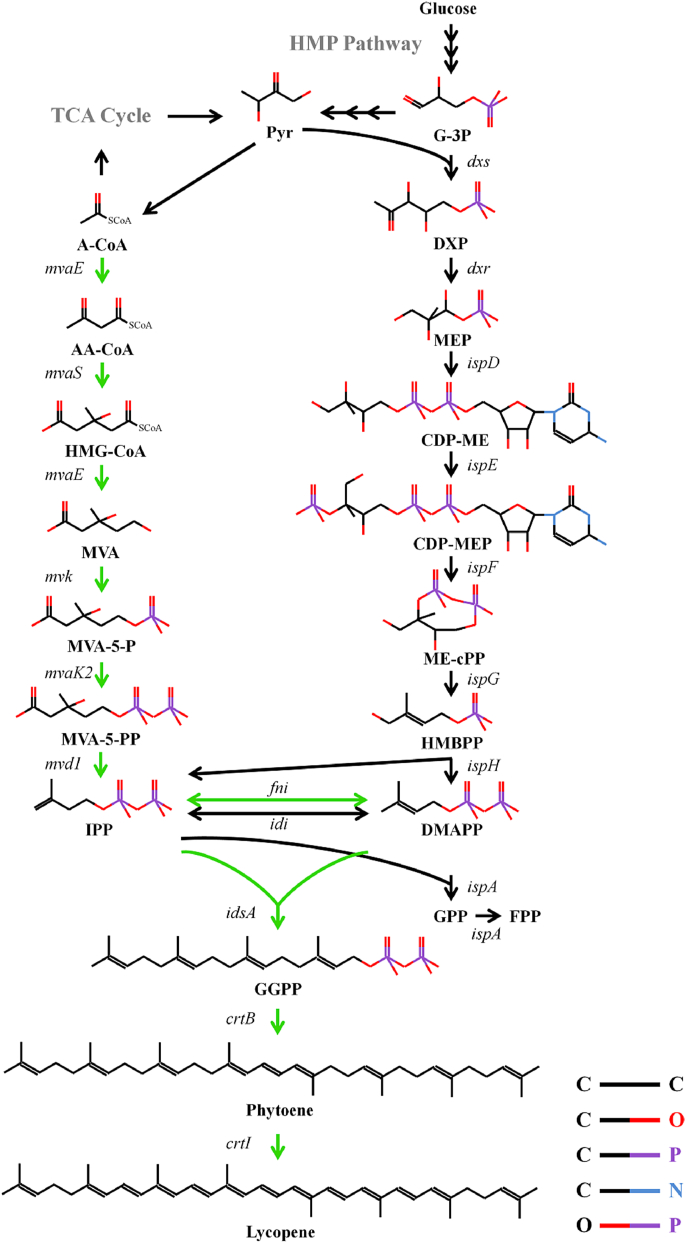
Fig. 1.
The heterogeneous and endogenous pathways involved in lycopene production in E. coli. Green arrows: the heterogeneous MVA pathway and lycopene expression pathway. Black arrows: the endogenous pathway. The heterogeneous MVA pathway is from acetyl-CoA to isopentenyl diphosphate and dimethylallyl diphosphate, while the endogenous MEP pathway comes from pyruvate and glyceraldehyde 3-phosphate. Intermediates: A-CoA, acetyl-CoA; AA-CoA, acetoacetyl-CoA; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; MVA, mevalonate; MVA-5-P, mevalonate 5-phosphate; MVA-5-PP, mevalonate pyrophosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; Pyr, pyruvate; G3P, glyceraldehyde 3-phosphate; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2-C-methyl-d-erythritol 4-phosphate; CDP-ME, 4-diphosphocytidyl-2-C-methyl-D-erythritol; CDP-MEP, 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate; ME-cPP, 2-C-methyl-D-erythritol 2,4-cyclopyrophosphate; HMBPP, 1-hydroxy-2-methyl-2-(E)-butenyl 4-pyrophosphate. Heterogeneous pathway genes: mvaE, acetoacetyl-CoA thiolase/HMG-CoA reductase gene; mvaS, HMG-CoA synthase gene; mvk, mevalonate kinase gene; mvaK2, phosphomevalonate kinase gene; mvd1, mevalonate pyrophosphate decarboxylase gene; fni, IPP isomerase gene; idsA, bifunctional short chain isoprenyl diphosphate synthase gene; crtB, phytoene synthase gene; crtI, phytoene desaturase gene.
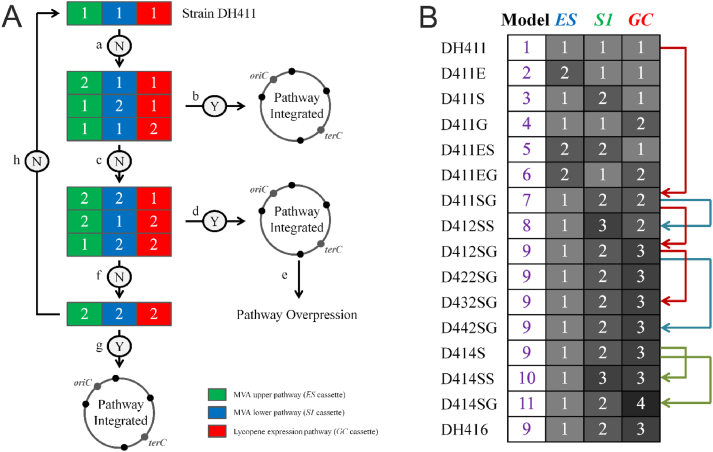
Fig. 2.
The rational model of heterogenous expression pathway. A) The project of the model. “N” indicates the lycopene yield isn't enhanced after genome's modification. “Y” demonstrates the improvement in lycopene yield. “a, b, c …” designates the various steps in rational model of heterogeneous expression pathway. B) The results of the model: The sub-model was based on the copy number of heterogeneous expression pathway. ES: mvaE-mvaS from E. faecalis V583; S1: fni-mvk-mvaK2-mvd1 from S. pneumoniae; GC: gps from A. fulgidus and crtI-crtB from P. agglomerans. Red line: the lycopene production increased; Blue line: the growth ability increased; Green line: the lycopene yield dropped.
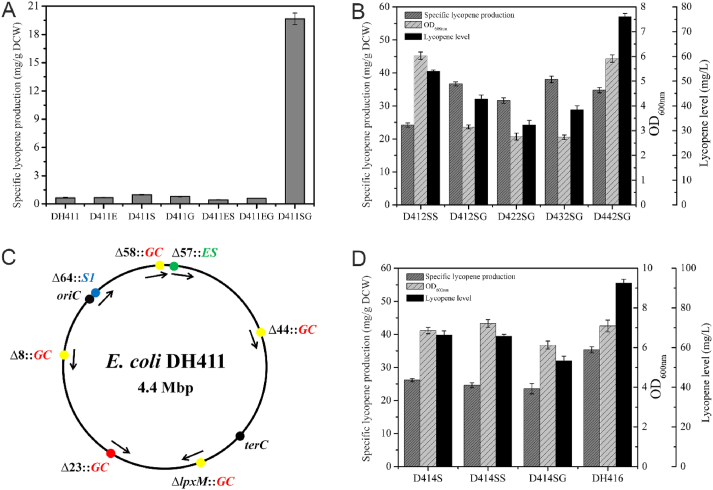
Fig. 3.
The specific lycopene production, growth ability, lycopene level of strains, and integrated positions of lycopene expression pathway. A) The specific lycopene production of Escherichia coliDH411, D411E, D411S, D411G, D411ES, D411EG, and D411SG. B) The specific lycopene production, growth ability, and lycopene level of D412SS, D412SG, D422SG, D432SG, and D442SG. C) Four positions (8th nonessential region, 44th nonessential region, 58th nonessential region, and lpxM site) were chosen for integration of lycopene expression pathway respectively. D) The specific lycopene production, growth ability, and lycopene level of D414S, D414SS, D414SG, and DH416.
As shown in Fig. 2B, the model of expression pathway was constructed according to the copy number of heterologous expression pathways. After construction of pathway, the best productive strain up to now is D411SG (DH411/pCC1SG) which carries overexpressing MVA lower pathway and lycopene expression pathway and belongs to 7th sub-model (Fig. 2B).
To test whether MVA lower pathway or lycopene expression pathway needed to be further overexpressed. The DH412 strain was designed which has one GC cassette at the position 8th nonessential region (GenBank: CP001637.1/413,909–430,258 bp) more than DH411strain and followed by transformation of the plasmids pCC1SS and pCC1SG into DH412 strain relatively. This transformation consequences into two evolve strains D412SS (DH412/pCC1SS) (8th sub-model) and D412SG (DH412/pCC1SG) (9th sub-model) which both had one copy of S1 cassette and GC cassette more than D411SG, respectively (Fig. 2B). Although the D412SS strain has better growth ability at OD600nm which is 6.03, but on other hand the D412SG strain achieved the better lycopene production of 36.68 mg/g DCW, which is 1.87-fold and 1.52-fold higher than D411SG and D412SS, respectively (Fig. 3B). Thus, the 9th sub-model (D412SG strain) is declared as the best productive sub-model among all previous eight sub models, consisting upon 1 copy of MVA upper pathway, 2 copies of MVA lower pathway, and 3 copies of lycopene expressing pathway.
3.2. Effect of integrated position of lycopene expression pathway
To further improve the lycopene production, the GC cassette was integrated into the 44th region (GenBank: CP001637.1/3,033,115–3,048,652 bp), 58th region (GenBank: CP001637.1/4,184,313–4,200,717 bp), and lpxM site (Fig. 3C) of DH411 strain, correspondingly, for the generation of DH422 (DH411 Δ44th: T7 GC), DH432 (DH411 Δ58th: T7 GC) and DH442 (DH411 ΔlpxM: T7 GC) strains. Then transformation of plasmid pCC1SG into DH422, DH432, and DH442 strains, resulted into three strains D422SG, D432SG, and D442SG, respectively. All latterly productions belonged to 9th sub-model (Fig. 2B), however, the integrated position of one copy of GC cassette was different from D412SG. As shown in Fig. 3B, strain D432SG had the best lycopene production which was 38.05 mg/g DCW. However, the D442SG strain had the best growth ability, nearly 1.8, 2.1, and 2.2-folds more than D412SG, D422SG, and D432SG strains, but its lycopene yield and growth ability was lower than the other strains of 9th sub-model. The lycopene level of D442SG (DH442/pCC1SG) strain was significantly increased by 80.7% from D412SG (76.04 mg/L of D442SG vs 42.81 mg/L of D412SG) because of development of growth ability after integration of GC cassette into lpxM site, while the lycopene yield of both D412SG and D442SG strains were similar (Fig. 3B).
In addition, the effect of integrated positions (the 44th region, 58th region and lpxM) were not limited by the heterogeneous pathways. The growth ability or lycopene level was increased about 1-fold after integration of ES cassette at lpxM site [49]. Moreover, the integration of ES cassette at 58th region successfully achieved the highest lycopene production [49]. Aforementioned results indicated the 58th region and lpxM site were superior integrated positions. Therefore, we suspected that the enhancement of growth ability was due to the deletion of nonessential genes or regions. Recently, Gyorgy Posfa and his co-worker developed the E. coli MDS42 strain, has a chromosome that is 14.3% smaller than that of its parental E. coli strain MG1655. This precise deletion of nonessential genes imparts favorable characteristics in MDS42 strain in the shape of robust growth and higher cell density under appropriate fermentation conditions, as well as increased transformation efficiency relative to MG1655 [56]. Consequently, the present study demonstrated that qRT-PCR is an applicable, simple and reproducible tool to study gene expression of lycopene pathway. The effect of chromosomal positions (8th, 44th, 58th, lpxM) on gene expression in E. coli was quite profound. Four chromosomal integrated lycopene producing strains D412SG, D422SG, D432SG and D442SG were studied under qRT-PCR technique to determine their relative gene transcription level as shown in Table 2.
Table 2.
Relative level of lycopene expression pathway, 16S as control.
| Strains | D412SG | D422SG | D432SG | D442SG |
|---|---|---|---|---|
| Relative gene transcription level | 1.727 | 1.903 | 1.647 | 1.741 |
This phenomenon of lycopene production through optimum positioning integration and its further effect on transcriptional and protein level should be explored in the future work.
3.3. Construction of an integrated lycopene production strain and its fed-batch fermentation
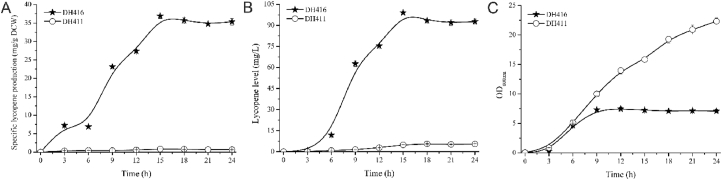
According to the result of chromosomal multiple position integration strategy, based on strain DH412, the lpxM site was chosen for integration of GC cassette, which in turn develop another strain, named as DH414 (DH412 ΔlpxM: T7 GC) and followed by plasmid transformation which were pCC1SS and pCC1SG plasmids to form D414SS (DH414/pCC1SS) (10th sub-model) and D414SG (DH414/pCC1SG) (11th sub-model) (Fig. 2B). After developing eleven sub-models (strains), the further increase in either MVA lower pathway or lycopene expression pathway to generate other sub-model (strain) didn't enhance the specific lycopene production that shown in (Fig. 3D). It denoted that the 9th sub-model was the best one while comparing with other eleven sub-models. The S1 cassette was integrated at 58th region to make DH416 strain. After 24 h of shaking at test tube scale, the specific lycopene production and lycopene level of integrated DH416 strain was reported as 35.34 mg/g DCW (Figs. 3D) and 92.66 mg/L (Fig. 4A) which was 53.6 and 17.0 fold higher than the starting strain DH411, correspondingly (Fig. 4). Moreover, DH416 had displayed specific lycopene production and lycopene level of about 36.92 mg/g DCW and 99.03 mg/L, respectively (Fig. 4A and B), with the mean productivity of 6.60 mg/L/h at 15 h.
Fig. 4.
Comparison of the growth ability, lycopene level, and specific lycopene production between DH416 and DH411 strains. The fermentation was carried out in 5 mL FMGAT medium at 220 rpm at 30 °C in test tubes.
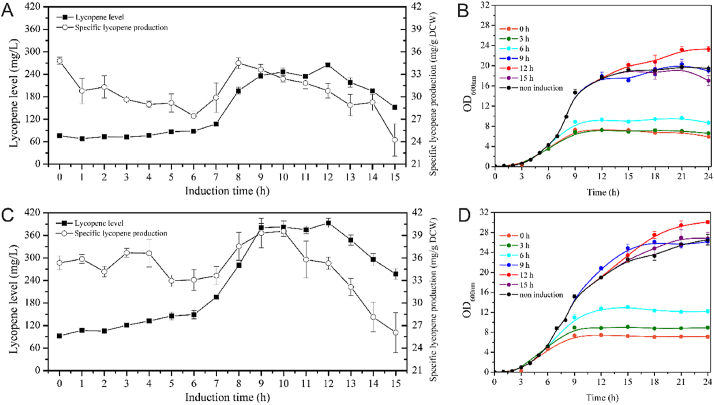
Next, the addition time of inducer l-arabinose was optimized. Although the lycopene production of D442SG strain at 12 h (30.80 mg/g DCW) was 11.5% lesser than the self-inducible condition (Fig. 5A), the lycopene level at 12 h was 3.5-fold higher than self-induction condition due to its better growth ability (Fig. 5B). The most suitable addition time of l-arabinose in DH416 strain was at 12 h with the lycopene level of 393.24 mg/L (see Fig. S1, Supporting Information), which was 4.2-fold higher than the self-inducible system (Figs. 5C) and 1.5-fold more than the D442SG strain under the same culture condition (Fig. 5A,C). As shown in Fig. 5D, the induction time had a significant impact on growth ability. The exponential stage was from 3 h to 15 h. After delaying the induction time points from 6 h to 9 h, the growth ability was enhanced dramatically from 12.10 (OD600nm) to 25.42 (OD600nm). This means that the growth is mostly sensitive to induction at the exponential stage. The best addition time of l-arabinose of both DH416 and D442SG was at 12 h. The selection of this same best induction time is mainly due to exploitation of same chassis parental strain and similar heterologous pathways.
Fig. 5.
Induction time as a single variable. A) Lycopene productivity of D442SG strain B) Growth curves of D442SG strain C) Lycopene productivity of DH416 strain D) Growth curves of DH416 strain.
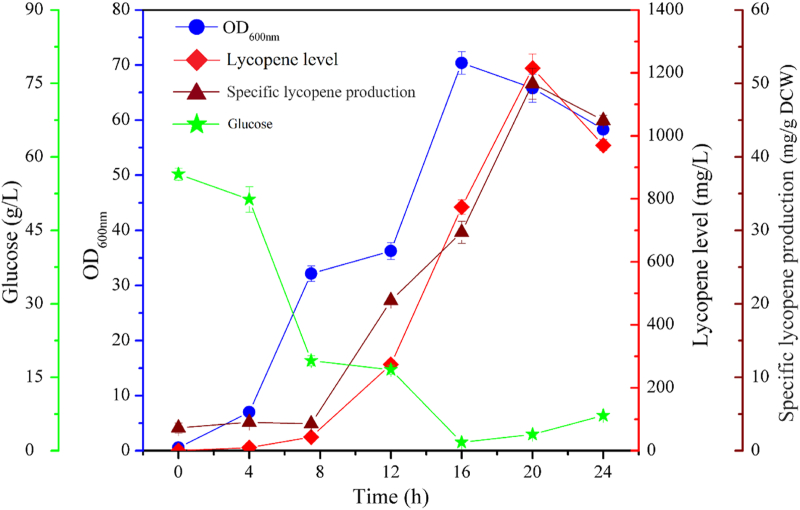
For further verification of lycopene level, DH416 strain was needed to be cultured in a 5 L fermenter. The lycopene level of 1.22 g/L (49.9 mg/g DCW) was successfully achieved after 20 h culture with mean productivity of 61.0 mg/L/h (Fig. 6 and see Fig. S1, Supporting Information). The lycopene level of DH416 was 2.3-fold higher than reported in Shen et al.‘s integrated strain [48], and it was 84.7% of Zhu et al.‘s findings (1.44 g/L) in a 2.5 L fermenter, and an approximately same percentage for some previous noted work, which results was about (1.23 g/L) in a 100 L fermenter [46]. However, the mean productivity of lycopene was about 2.0-fold and 10.0-fold higher than Zhu et al.‘s and Shen et al.‘s experimental outcomes [46,48]. This result supports the aim of our work to increase the lycopene production through suitable strain engineering. In the present work, the experimental strain has displayed vast potential in form of higher lycopene yield and optimum growth. Thus, these features seem ideal to proceed the tested strain for commercial production.
Fig. 6.
The time course of lycopene production of DH416 strain in 5 L fermenter. Lycopene level (◆),specific lycopene production (▲), growth ability (●), and glucose concentration (★). Induction of l-arabinose was carried out at 7.5 h and 12 h respectively.
Previous report utilized the pSC101 ori plasmid carrying the MVA lower pathway and regulate through different promoters [48]. Later, an effective effort has been applied to integrate the best combination at genomic level in the desire host. Nevertheless, the change in copy number of MVA lower pathway may cause the diversity of gene dosage which cause imbalancing of heterogeneous pathways. To confirm the best copy number of heterogeneous pathways, the model of expression pathway employed in this work is single copy plasmid pCC1FOS, so that the copy number of pathways didn't change. In addition, multiple position were selected which regulated the transcriptional level of integrated pathways [50]. This regulation can be achieved by high copy number of heterogeneous pathway which in certain case depending upon the utilization of chemically induced chromosomal evolution (CIChE) [47,57]. The best example to simplify the concept of chemically induced chromosomal evolution (CIChE) is by emergence of multidrug-resistant phenomenon through residual multiple antibiotic-resistant genes [48]. Whereas, the strains constructed in this study have not faced this kind of problem. Thus, the newly developed method shall be considered efficient while producing natural products which are the vital metabolites of multiple heterogenous expression pathways through chromosomal integrated strains.
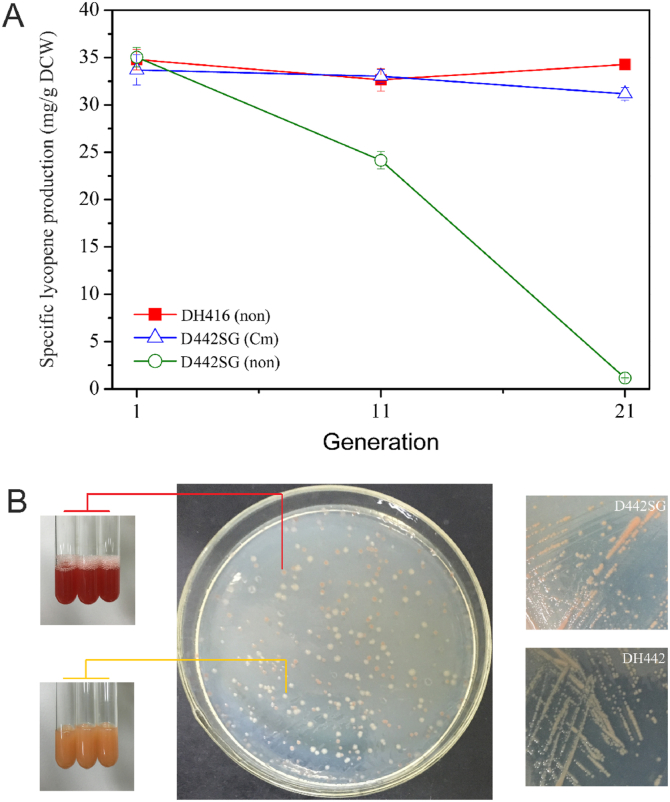
3.4. Qualitative analysis of lycopene synthetic stability between strains DH416 and D442SG
The consecutive cultures of recombinant engineer DH416 strain were carried out precisely. (see Fig. S2, Supporting Information). Strain DH416 could steadily synthetize lycopene after Generation 21st (Gen 21st) (Fig. 7A). The D442SG was chosen as control group as it contained a single copy plasmid pCC1SG plus the copy number of its heterologous expression pathway was same with DH416, and its lycopene production and growth ability was quite similar to DH416 strain. The D442SG strain could steadily synthetize lycopene after Gen 21st where Cm (chloramphenicol) was added during the successive generation (Fig. 7A). However, lycopene production dropped suddenly to 3.3% after Gen 21st without addition of Cm (Fig. 7A). Therefore, it is speculated that the loss of lycopene productivity was majorly due to segregational instability. As shown in Fig. 7B, two distinct colonies having different color types: orange ones and light yellow ones, were generated. The orange colonies preserved lycopene synthetic ability with Cm resistance while the light yellow ones had extremely low lycopene yield with Cm sensitive. Hence, it indicated that the light yellow colonies were mutated into DH442. Although the orange colonies were around 30%, the growth ability of DH442 strain was about 3-fold higher as compared to D442SG strain and the latter strain was resulting into lower yield at Gen 21st (cultured without Cm).
Fig. 7.
Biosynthesis stability of strains DH416 and D442SG. (A) The specific lycopene production of strains DH416 and D442SG after the Generation 21st. “non” means cultured without antibiotics. (B) The colonies of the Generation 21st of D442SG (cultured without Cm).
Comparing with the colonies of D442SG of Gen 21st (cultured without Cm), the DH442 and D442SG strains were plated on LB agar plates, relatively. The colonies of D442SG were orange, which were similar to the orange colonies of Gen 21st of D442SG strain (culture with Cm), whereas DH442 colonies were light yellow which was similar with the light yellow colonies of Gen 21st of D442SG strain (cultured without Cm) (Fig. 7B).
For plasmids system, antibiotics were necessary to maintain their existence in most of the cases. Tyo et al. found that plasmid-based polyhydroxybutyrate (PHB) productivity drops to zero in five doublings without antibiotics [47]. For existence of antibiotics, 70% of the cells lost PHB productivity after 40 generations with maintenance of selection marker due to allele segregation [47]. In our study, different from the multi-copy plasmids (pBR and p15A) used in Tyo et al.‘s study, the target plasmid was single copy plasmid pCC1SG, and the result showed that about 67.6 ± 5.4% colonies of D442SG strain lost its plasmid after Gen 21st. Although, the lycopene yield of D442SG strain didn't decline significantly after Gen 21st of LB medium containing Cm (Fig. 7A), while the several colonies turned into light yellow color (see Fig. S3, Supporting Information). It is assumed that allele segregation of plasmid pCC1SG occurred which would be investigated in future work. On contrary to this, the color of all the DH416 colonies didn't change. Ten days of consecutive culture time was allowed in this study which considered as being longer than a fermentation period in industry, indicating that the strain DH416 could steadily synthesize lycopene.
4. Conclusions
To construct an integrated lycopene production strain, the conducted study described a rational model including optimization of copy number and selection of better integrated position of heterogeneous expression pathways. Moreover, the ninth sub-model of integrated strain, consisting upon one copy of MVA upper pathway, two copies of MVA lower pathway, and three copies of lycopene expression pathway was comprehended as the best performer in terms of lycopene productivity. The integrated strain DH416 has the lycopene level of 1.22 g/L (49.9 mg/g DCW) with mean productivity of 61.0 mg/L/h in a 5 L fermenter. In addition, it showed a better intrinsic stability than the strain carrying plasmid systems.
CRediT authorship contribution statement
Muhammad Hammad Hussain: Methodology, Validation, Investigation, Data curation, Visualization, Writing – original draft. Qi Hong: Methodology, Validation, Investigation, Data curation. Waqas Qamar Zaman: Data curation, Formal analysis, Writing – review & editing. Ali Mohsin: Formal analysis, Writing – review & editing. Yanlong Wei: Methodology, Investigation, Visualization, Writing – original draft, Writing – review & editing. Ning Zhang: Methodology, Validation, Investigation, Data curation. Hongqing Fang: Methodology, Validation. Zejian Wang: Methodology, Validation, Writing – review & editing. Haifeng Hang: Supervision, Funding acquisition, Project administration, Writing – review & editing. Yingping Zhuang: Supervision, Validation. Meijin Guo: Supervision, Funding acquisition, Project administration, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by grants from the National Key Research and Development Program of China (2018YFA0900300 and 2019YFA090141) and the National Science and Technology Major Project (2017ZX07402003).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supporting Information is available from the Wiley Online Library or from the author.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2021.04.001.
Contributor Information
Haifeng Hang, Email: hanghaifeng@ecust.edu.cn.
Meijin Guo, Email: guo_mj@ecust.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Pemberton T.A., Chen M., Harris G.G. Exploring the influence of domain architecture on the catalytic function of diterpene synthases. Biochemistry. 2017;56:2010–2023. doi: 10.1021/acs.biochem.7b00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao X., Wei L.J., Lin J.Y., Hua Q. Enhancing linalool production by engineering oleaginous yeast Yarrowia lipolytica. Bioresour Technol. 2017;245:1641–1644. doi: 10.1016/j.biortech.2017.06.105. [DOI] [PubMed] [Google Scholar]
- 3.Park S.Y., Binkley R.M., Kim W.J., Lee M.H., Lee S.Y. Metabolic engineering of Escherichia coli for high-level astaxanthin production with high productivity. Metab Eng. 2018;49:105–115. doi: 10.1016/j.ymben.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Bian G., Yuan Y., Tao H. Production of taxadiene by engineering of mevalonate pathway in Escherichia coli and endophytic fungus Alternaria alternata TPF6. Biotechnol J. 2017;12:1600697. doi: 10.1002/biot.201600697. [DOI] [PubMed] [Google Scholar]
- 5.Vranova E., Coman D., Gruissem W. Structure and dynamics of the isoprenoid pathway network. Mol Plant. 2012;5:318–333. doi: 10.1093/mp/sss015. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C., Seow V.Y., Chen X., Too H.P. Multidimensional heuristic process for high-yield production of astaxanthin and fragrance molecules in Escherichia coli. Nat Commun. 2018;9:1858. doi: 10.1038/s41467-018-04211-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng H.M., Koutsidis G., Lodge J.K. Tomato and lycopene supplementation and cardiovascular risk factors: a systematic review and meta-analysis. Atherosclerosis. 2017;257:100–108. doi: 10.1016/j.atherosclerosis.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Dherani M., Murthy G.V., Gupta S.K. Blood levels of vitamin C, carotenoids and retinol are inversely associated with cataract in a North Indian population. Invest Ophthalmol Vis Sci. 2008;49:3328–3335. doi: 10.1167/iovs.07-1202. [DOI] [PubMed] [Google Scholar]
- 9.Fuhrman B., Elis A., Aviram M. Hypocholesterolemic effect of lycopene and beta-carotene is related to suppression of cholesterol synthesis and augmentation of LDL receptor activity in macrophages. Biochem Biophys Res Commun. 1997;233:658–662. doi: 10.1006/bbrc.1997.6520. [DOI] [PubMed] [Google Scholar]
- 10.Kirby J., Keasling J.D. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 11.Martin V.J., Pitera D.J., Withers S.T., Newman J.D., Keasling J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 12.Yuan J., Ching C.B. Combinatorial engineering of mevalonate pathway for improved amorpha-4,11-diene production in budding yeast. Biotechnol Bioeng. 2014;111:608–617. doi: 10.1002/bit.25123. [DOI] [PubMed] [Google Scholar]
- 13.Ajikumar P.K., Xiao W.H., Tyo K.E. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paddon C.J., Westfall P.J., Pitera D.J. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 15.Ro D.K., Paradise E.M., Ouellet M. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 16.Li C., Ying L.Q., Zhang S.S. Modification of targets related to the Entner-Doudoroff/pentose phosphate pathway route for methyl-D-erythritol 4-phosphate-dependent carotenoid biosynthesis in Escherichia coli. Microb Cell Factories. 2015;14:117. doi: 10.1186/s12934-015-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evamaria Gruchattka O.H., Klamt S., Schütz V., Kayser O. In silico profiling of Escherichia coli and Saccharomyces cerevisiae as terpenoid factories. Microb Cell Factories. 2013;12:84. doi: 10.1186/1475-2859-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen H.J., Cheng B.Y., Zhang Y.M. Dynamic control of the mevalonate pathway expression for improved zeaxanthin production in Escherichia coli and comparative proteome analysis. Metab Eng. 2016;38:180–190. doi: 10.1016/j.ymben.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 19.Gruchattka E., Hädicke O., Schütz S. In silico profiling of Escherichia coli and Saccharomyces cerevisiae as terpenoid factories. Microb Cell Factories. 2013;12:1–18. doi: 10.1186/1475-2859-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng H., Wang Y., Hua Q. In silico analysis and experimental improvement of taxadiene heterologous biosynthesis in Escherichia coli. Biotechnol Bioproc Eng. 2011;16:205–215. doi: 10.1007/s12257-010-0329-z. [DOI] [Google Scholar]
- 21.Ward V.C.A., Chatzivasileiou A.O., Stephanopoulos G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol Lett. 2018;365:10. doi: 10.1093/femsle/fny079. [DOI] [PubMed] [Google Scholar]
- 22.Yadav V.G., Mey M.D., Lim C.G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng. 2012;14:233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J.H., Wang C., Jang H.J. Isoprene production by Escherichia coli through the exogenous mevalonate pathway with reduced formation of fermentation byproducts. Microb Cell Factories. 2016;15:214. doi: 10.1186/s12934-016-0612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Zhao G., Sun Y. Bio-isoprene production using exogenous MVA pathway and isoprene synthase in Escherichia coli. Bioresour Technol. 2012;104:642–647. doi: 10.1016/j.biortech.2011.10.042. [DOI] [PubMed] [Google Scholar]
- 25.Yang J.M., Xian M., Su S.Z. Enhancing production of bio-isoprene using hybrid MVA pathway and isoprene synthase in E-coli. PloS One. 2012;7 doi: 10.1371/journal.pone.0033509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George K.W., Chen A., Jain A. Correlation analysis of targeted proteins and metabolites to assess and engineer microbial isopentenol production. Biotechnol Bioeng. 2014;111:1648–1658. doi: 10.1002/bit.25226. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y.N., Liu Q., Li L.L. Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnol Biofuels. 2013;6:57. doi: 10.1186/1754-6834-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Liu Q., Cao Y. Microbial production of sabinene--a new terpene-based precursor of advanced biofuel. Microb Cell Factories. 2014;13:20. doi: 10.1186/1475-2859-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarria S., Wong B., Garcia Martin H., Keasling J.D., Peralta-Yahya P. Microbial synthesis of pinene. ACS Synth Biol. 2014;3:466–475. doi: 10.1021/sb4001382. [DOI] [PubMed] [Google Scholar]
- 30.Tashiro M., Kiyota H., Kawai-Noma S. Bacterial production of pinene by a laboratory-evolved pinene-synthase. ACS Synth Biol. 2016;5:1011–1020. doi: 10.1021/acssynbio.6b00140. [DOI] [PubMed] [Google Scholar]
- 31.Yang J., Nie Q., Ren M. Metabolic engineering of Escherichia coli for the biosynthesis of alpha-pinene. Biotechnol Biofuels. 2013;6:60. doi: 10.1186/1754-6834-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alonso-Gutierrez J., Chan R., Batth T.S. Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metab Eng. 2013;19:33–41. doi: 10.1016/j.ymben.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Zhou J., Wang C., Yang L., Choi E.S., Kim S.W. Geranyl diphosphate synthase: an important regulation point in balancing a recombinant monoterpene pathway in Escherichia coli. Enzym Microb Technol. 2015;68:50–55. doi: 10.1016/j.enzmictec.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J., Wang C., Yoon S.H. Engineering Escherichia coli for selective geraniol production with minimized endogenous dehydrogenation. J Biotechnol. 2014;169:42–50. doi: 10.1016/j.jbiotec.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 35.Anthony J.R., Anthony L.C., Nowroozi F. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab Eng. 2009;11:13–19. doi: 10.1016/j.ymben.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Newman J.D., Marshall J., Chang M. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol Bioeng. 2006;95:684–691. doi: 10.1002/bit.21017. [DOI] [PubMed] [Google Scholar]
- 37.Redding-Johanson A.M., Batth T.S., Chan R. Targeted proteomics for metabolic pathway optimization: application to terpene production. Metab Eng. 2011;13:194–203. doi: 10.1016/j.ymben.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 38.Tsuruta H., Paddon C.J., Eng D. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PloS One. 2009;4:e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peralta-Yahya P.P., Ouellet M., Chan R. Identification and microbial production of a terpene-based advanced biofuel. Nat Commun. 2011;2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu F., Zhong X., Hu M. In vitro reconstitution of mevalonate pathway and targeted engineering of farnesene overproduction in Escherichia coli. Biotechnol Bioeng. 2014;111:1396–1405. doi: 10.1002/bit.25198. [DOI] [PubMed] [Google Scholar]
- 41.Wang C., Yoon S.H., Shah A.A. Farnesol production from Escherichia coli by harnessing the exogenous mevalonate pathway. Biotechnol Bioeng. 2010;107:421–429. doi: 10.1002/bit.22831. [DOI] [PubMed] [Google Scholar]
- 42.Nybo S.E., Saunders J., McCormick S.P. Metabolic engineering of Escherichia coli for production of valerenadiene. J Biotechnol. 2017;262:60–66. doi: 10.1016/j.jbiotec.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Yang J., Li Z., Guo L., Du J., Bae H.J. Biosynthesis of β-caryophyllene, a novel terpene-based high-density biofuel precursor, using engineered Escherichia coli. Renew Energy. 2016;99:216–223. doi: 10.1016/j.renene.2016.06.061. [DOI] [Google Scholar]
- 44.Yang J., Nie Q. Engineering Escherichia coli to convert acetic acid to beta-caryophyllene. Microb Cell Factories. 2016;15:74. doi: 10.1186/s12934-016-0475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon S.H., Lee Y.M., Kim J.E. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol Bioeng. 2006;94:1025–1032. doi: 10.1002/bit.20912. [DOI] [PubMed] [Google Scholar]
- 46.Zhu F.Y., Lu L., Fu S. Targeted engineering and scale up of lycopene overproduction in Escherichia coli. Process Biochem. 2015;50:341–346. doi: 10.1016/j.procbio.2014.12.008. [DOI] [Google Scholar]
- 47.Tyo K.E., Ajikumar P.K., Stephanopoulos G. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol. 2009;27:760–765. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 48.Shen H.J., Hu J.J., Li X.R., Liu J.Z. Engineering of Escherichia coli for lycopene production through promoter engineering. Curr Pharmaceut Biotechnol. 2015;16:1094–1103. doi: 10.2174/1389201016666150731110536. [DOI] [PubMed] [Google Scholar]
- 49.Wei Y., Mohsin A., Hong Q., Guo M., Fang H. Enhanced production of biosynthesized lycopene via heterogenous MVA pathway based on chromosomal multiple position integration strategy plus plasmid systems in Escherichia coli. Bioresour Technol. 2018;250:382–389. doi: 10.1016/j.biortech.2017.11.035. [DOI] [PubMed] [Google Scholar]
- 50.Bryant J.A., Sellars L.E., Busby S.J., Lee D.J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014;42:11383–11392. doi: 10.1093/nar/gku828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gerganova V., Berger M., Zaldastanishvili E. Chromosomal position shift of a regulatory gene alters the bacterial phenotype. Nucleic Acids Res. 2015;43:8215–8226. doi: 10.1093/nar/gkv709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Y., Deng P., Mohsin A. An electroporation-free method based on Red recombineering for markerless deletion and genomic replacement in the Escherichia coli DH1 genome. PloS One. 2017;12 doi: 10.1371/journal.pone.0186891. e0186891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong Z.Q., Guo M.J., Chu J., Zhuang Y.P., Zhang S.L. On-line specific growth rate control for improving reduced glutathione production in Saccharomyces cerevisiae. Biotechnol Bioproc Eng. 2015;20:887–893. doi: 10.1007/s12257-015-0018-z. [DOI] [Google Scholar]
- 54.Kim S.K., Han G.H., Seong W. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab Eng. 2016;38:228–240. doi: 10.1016/j.ymben.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Posfai G., Plunkett G., Feher T. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y.Y., Shen H.J., Cui Y.Y. Chromosomal evolution of Escherichia coli for the efficient production of lycopene. BMC Biotechnol. 2013;13:6. doi: 10.1186/1472-6750-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu Y., Yang Y., Den P. Sci China Life Sci. 2016;59:1034. doi: 10.1007/s11427-016-5100-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.