The results demonstrated that PVDs are essential for the broad-spectrum antibacterial activities of strain YL-1 against both Gram-positive and Gram-negative bacteria under low-iron conditions. Our findings also highlight the effect of exogenous iron on the production of PVD and the importance of this bacterial product in bacterial interactions. As a biocontrol agent, PVDs can directly inhibit the proliferation of the tested bacteria in addition to participating in iron competition.
KEYWORDS: Pseudomonas chlororaphis, pyoverdine, low-iron, antibacterial, Xanthomonas oryzae pv. oryzae
ABSTRACT
Pseudomonas chlororaphis YL-1 has extensive antimicrobial activities against phytopathogens, and its genome harbors a pyoverdine (PVD) biosynthesis gene cluster. The alternative sigma factor PvdS in Pseudomonas aeruginosa PAO1 acts as a critical regulator in response to iron starvation. The assembly of the PVD backbone starts with peptide synthetase enzyme PvdL. PvdF catalyzes formylation of l-OH-Orn to produce l-N5-hydroxyornithine. Here, we describe the characterization of PVD production in YL-1 and its antimicrobial activity in comparison with that of its PVD-deficient ΔpvdS, ΔpvdF, and ΔpvdL mutants, which were obtained using a sacB-based site-specific mutagenesis strategy. Using in vitro methods, we examined the effect of exogenous iron under low-iron conditions and an iron-chelating agent under iron-sufficient conditions on PVD production, antibacterial activity, and the relative expression of the PVD transcription factor gene pvdS in YL-1. We found that strain YL-1, the ΔpvdF mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain produced visible PVDs and demonstrated a wide range of inhibitory effects against Gram-negative and Gram-positive bacteria in vitro under low-iron conditions and that with the increase of iron, its PVD production and antibacterial activity were reduced. The antibacterial compounds produced by strain YL-1 under low-iron conditions were PVDs based on liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Moreover, the antibacterial activity observed in vitro was correlated with in vivo control efficacies of strain YL-1 against rice bacterial leaf blight (BLB) disease caused by Xanthomonas oryzae pv. oryzae. Collectively, PVDs are responsible for the antibacterial activities of strain YL-1 under both natural and induced low-iron conditions.
IMPORTANCE The results demonstrated that PVDs are essential for the broad-spectrum antibacterial activities of strain YL-1 against both Gram-positive and Gram-negative bacteria under low-iron conditions. Our findings also highlight the effect of exogenous iron on the production of PVD and the importance of this bacterial product in bacterial interactions. As a biocontrol agent, PVDs can directly inhibit the proliferation of the tested bacteria in addition to participating in iron competition.
INTRODUCTION
Iron is an essential micronutrient for plants and associated microorganisms acquired from soil supplies (1, 2). Even so, soil supplies in most natural environments are inadequate due to iron's insolubility at neutral pH (3). Bacteria and fungi have, consequently, developed an efficient ferric ion-chelating agent, or siderophore, to scavenge iron from the extracellular environment and import it to maintain adequate intracellular levels of iron (1).
Among the siderophore-producing microorganisms are fluorescent pseudomonads of the genus Pseudomonas, Gram-negative bacteria highly studied due to their ubiquity and metabolic diversity. In fact, considerable research has been dedicated to the mechanism underlying the iron stress response exhibited by these bacteria (4, 5). Specifically, the siderophores were produced by fluorescent pseudomonads under conditions of iron deficiency, the most complex and common of which are pyoverdines (PVDs) (2).
PVDs are fluorescent siderophores with a high affinity for iron (10−32 M) (6). More than 60 PVDs are described among Pseudomonas species, including plant growth-promoting P. fluorescens SBW25, the plant pathogen P. syringae, and the human pathogen P. aeruginosa (7). Structurally, PVDs are composed of three elements: a side chain, a variable peptide moiety, and a dihydroxyquinoline chromophore moiety that is responsible for their fluorescence. PVDs are highly variable among species, often unique to each strain, and generally 6 to 12 amino acids in length, ranging from partial to entire, linear, or cyclic forms (8).
Three distinct PVD types can be produced by the Gram-negative pathogen P. aeruginosa PAO1, PVD-I, PVD-II, and PVD-III, which are characterized by different peptide chains, with PVD-I being the best investigated (8). The alternative sigma factor PvdS acts as an extracytoplasmic function (ECF) iron sigma factor (3). The PVD-I backbone is assembled in the cytoplasm by four nonribosomal peptide synthetases (NRPSs): PvdL, PvdI, PvdJ, and PvdD (8). Because the composition of the peptide is strain specific, the biosynthesis also involves three other cytoplasmic enzymes, namely, PvdA, PvdF, and PvdH, which participate in the synthesis of atypical amino acids present in the peptide moiety of PVD-I. The cytoplasmic PVD-I precursor has an unformed chromophore and a myristic or myristoleic acid chain bound to the first residue of the peptide backbone (Glu). This fatty acid chain probably keeps the cytoplasmic precursors bound to the inner membrane. It is believed that the nonfluorescent PVD-I precursor is transported via an ATP-binding cassette, in which PvdE is a subunit, to the periplasm for further maturation by the five enzymes PvdM, PvdN, PvdO, PvdP, and PvdQ (9, 10). Functionally, PVDs are diverse and have been associated with mechanisms that promote the induction of systemic resistance (11), plant growth (12), virulence (13), antagonistic activity (7, 14–17), and iron acquisition (18). Previously, PVDs have been used as biocontrol agents against Vibrio species (14), Xanthomonas oryzae pv. oryzae (Xoo) (7), Piricularia oryzae (15), Botrytis cinerea (16), and Aspergillus fumigatus (17). Apart from this, the necessity for new antimicrobial agents to limit the emergence of drug resistance has increased interest in microbial iron uptake systems. Clearly, research on exploiting siderophore uptake mechanisms in a “Trojan horse” strategy, i.e., coupling antibiotic and siderophore to increase biological activity by enhancing antibiotic transport into the bacterium, warrants future exploring (19, 20).
Strain YL-1 was isolated from a soybean root system and identified as Pseudomonas chlororaphis. Prior studies have shown that YL-1 strain demonstrate extensive antibacterial activities against plant pathogens significant to agribusiness (21). By applying bioinformatics techniques to its genome sequence (GenBank accession number AWWJ00000000.1) (22), its genome was found to harbor the PVD biosynthesis gene cluster. To demonstrate the function of PVD biosynthesis genes and the influence of PVD on antimicrobial mechanisms, the present study employed in vitro bioassays and verified these findings in greenhouse pot experiments. Based on the type strain P. aeruginosa PAO1 (8), we selected three target proteins involved in PVD regulation and synthesis, i.e., ECF iron sigma factor PvdS, nonribosomal peptide synthetase of the PVD chromophore PvdL, and N5-hydroxyornithine transformylase PvdF. Our results confirmed that under in vitro and environmental low-iron conditions, strain YL-1 produced PVDs that were liable for its antibacterial activity. Furthermore, this activity may directly inhibit the growth of indicator bacteria through an unknown mode of action, notwithstanding iron competition.
RESULTS
Growth and PVD production of P. chlororaphis YL-1 and mutants.
As expected, no difference was observed in culture broth color or growth capacity between P. chlororaphis YL-1 and mutants cultured in liquid LB medium (Fig. 1a and c). Likewise, strain YL-1 and mutants did not exhibit any fluorescence under UV light (Fig. 1b). The optical density at 405 nm (OD405)/OD600 values suggested that these strains could not produce PVD in liquid LB medium (Fig. 1d).
FIG 1.
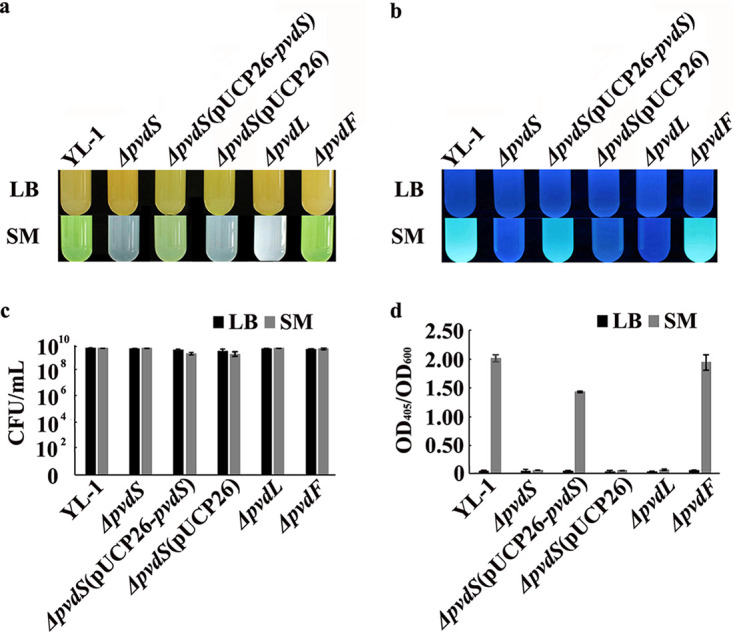
Comparison of the phenotypic differences between P. chlororaphis YL-1 and mutants cultured in liquid SM and LB medium. (a) Color of culture broth in visible light; (b) fluorescence under UV light; (c) cell number; (d) PVD production. P. chlororaphis YL-1 and mutants were cultured in liquid SM and LB medium, respectively, at 28°C and 200 rpm for 24 h. All liquid cultures were observed under visible light and UV light. The number of bacterial cells was determined by counting the numbers of CFU per ml. PVD production was measured as the OD405 of culture supernatants appropriately diluted in 0.1 M Tris-HCl (pH 8) and normalized to the OD600 of the corresponding cultures.
Culture broth of strain YL-1, the ΔpvdF mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain incubated in liquid succinic acid medium (SM) were yellow-green in visible light and exhibited fluorescence under UV light; however, culture broth of the ΔpvdS, ΔpvdS(pUCP26), and ΔpvdL mutants were white and did not exhibit fluorescence under UV light (Fig. 1a and b). The growth capacity of all the mutants was comparable to that of the wild-type strain (Fig. 1c); however, in comparison to the wild type, the OD405/OD600 values of the ΔpvdS, ΔpvdS(pUCP26), and ΔpvdL mutants decreased by 96.93%, 97.41%, and 96.69%, respectively (Fig. 1d). PVD production of the ΔpvdS(pUCP26-pvdS) complemented strain was partially rescued, demonstrated by an OD405/OD600 value of 1.44. No difference was observed in PVD production between strain YL-1 and its ΔpvdF mutant incubated in liquid SM, with OD405/OD600 values of 2.03 and 2.02, respectively (Fig. 1d). In addition, culture broth of strain YL-1 incubated in deferrated SM was also yellow-green in visible light and exhibited fluorescence under UV light (see Fig. S1 in the supplemental material), with an OD405/OD600 value of 2.89.
Antibacterial activity of P. chlororaphis YL-1 and mutants.
The antibacterial activities of strain YL-1 and mutants precultured in liquid LB medium were determined on LB plates. Strain YL-1 and mutants produced clear inhibitory zones with no observable difference in anti-Xoo and anti-Xanthomonas oryzae pv. oryzicola (anti-Xooc) activities (Fig. S2a). Inhibitory zones against Xoo ranged from 33.8 ± 1.8 mm to 37.0 ± 1.4 mm, and those against Xooc ranged from 30.5 ± 1.3 mm to 35.4 ± 1.1 mm in diameter (Table 1). There was a slight inhibitory effect against Bacillus megaterium and Erwinia amylovora by strain YL-1 and the mutants on LB plates (Table 1).
TABLE 1.
Antibacterial activity of P. chlororaphis YL-1 and mutants on LB plates and 1/2 TSA platesa
| Treatment | Inhibition zone diam (mm) |
|||||||
|---|---|---|---|---|---|---|---|---|
| LB |
1/2 TSA |
|||||||
| Xoo | Bacillus megaterium | Erwinia amylovora | Xooc | Xoo | Bacillus megaterium | Erwinia amylovora | Xooc | |
| YL-1 | 36.1 ± 1.1 A | 6.2 ± 0.5 A | 6.1 ± 0.2 A | 33.1 ± 1.7 A | 37.1 ± 1.2 A | 39.7 ± 1.0 A | 35.7 ± 1.3 A | 29.1 ± 1.1 A |
| YL-1 ΔpvdS | 37.0 ± 1.4 A | 6.5 ± 0.3 A | 6.1 ± 0.4 A | 33.3 ± 1.2 A | 7.0 ± 1.4 B | 6.0 ± 1.2 B | 9.5 ± 1.4 B | 6.0 ± 1.2 B |
| YL-1 ΔpvdS(pUCP26-pvdS) | 36.8 ± 1.2 A | 6.2 ± 0.4 A | 6.0 ± 0.3 A | 35.4 ± 1.1 A | 29.1 ± 1.1 A | 30.2 ± 2.0 A | 31.3 ± 1.2 A | 25.2 ± 1.3 A |
| YL-1 ΔpvdS(pUCP26) | 34.5 ± 1.3 A | 6.0 ± 0.5 A | 6.0 ± 0.5 A | 31.7 ± 1.4 A | 7.2 ± 1.5 B | 7.9 ± 1.5 B | 9.1 ± 1.5 B | 6.1 ± 1.1 B |
| YL-1 ΔpvdL | 33.8 ± 1.8 A | 6.0 ± 0.7 A | 6.1 ± 0.1 A | 30.5 ± 1.3 A | 8.3 ± 1.4 B | 10.8 ± 1.7 B | 6.5 ± 1.1 B | 6.1 ± 1.2 B |
| YL-1 ΔpvdF | 34.1 ± 1.2 A | 6.1 ± 0.6 A | 6.0 ± 0.2 A | 34.1 ± 1.5 A | 30.7 ± 1.3 A | 36.7 ± 1.3 A | 29.1 ± 1.5 A | 25.4 ± 1.5 A |
| Water | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
Values in columns followed by the same letter were not significantly different according to Fisher's protected LSD test (P = 0.05).
Strain YL-1 could produce PVD on both SM and 1/2 Trypticase soy agar (TSA) plates at 28°C (Fig. S3), which suggests that both SM and 1/2 TSA are low-iron media. Because most indicator bacteria used in the study can grow on 1/2 TSA plates, but not on low-iron SM, the antagonistic assays of P. chlororaphis YL-1 and mutants precultured in liquid SM were performed on 1/2 TSA plates. Interestingly, when grown in 1/2 TSA medium, strain YL-1, the ΔpvdF mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain precultured in liquid SM showed an apparent inhibitory effect for Xoo, whereas the ΔpvdS, ΔpvdS(pUCP26), and ΔpvdL mutants demonstrated significant reductions in anti-Xoo activity (Fig. S2b). In addition, strain YL-1, the ΔpvdF mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain also inhibited the growth of B. megaterium strain 329, E. amylovora strain 2029, and Xooc strain RS11, whereas the ΔpvdS, ΔpvdS(pUCP26), and ΔpvdL mutants nearly lost antibacterial activity (Table 1), which is consistent with the results for Xoo. These results suggest that strain YL-1 and its mutants secreted different secondary metabolites when cultured on LB plates compared with 1/2 TSA plates. The antibacterial activities of strain YL-1 and PVD-deficient mutants did not differ when grown on LB plates, indicating that the main antibacterial secondary metabolite secreted by strain YL-1 and mutants is not PVD. Because strain YL-1 and mutants were unable to produce PVD on LB medium (Fig. 1), the deletion of the PVD synthesis gene did not produce measurable differences in antibacterial activity. However, the antibacterial activities of the ΔpvdS, ΔpvdS(pUCP26), and ΔpvdL mutants had decreased significantly, and the three mutants cannot produce PVD (Fig. 1; Fig. S2b) when grown in 1/2 TSA plates, suggesting that few, if any, potential antibacterial secondary metabolites may be produced in 1/2 TSA medium. The main antibacterial secondary metabolite secreted by strain YL-1, the ΔpvdF mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain in the 1/2 TSA plates was PVD.
Biocontrol efficacy of P. chlororaphis YL-1 and mutants on BLB in pot experiments.
To determine whether P. chlororaphis YL-1 and mutants precultured in liquid SM or LB medium could control Xoo-induced BLB in rice, two pot experiments were conducted in June (Fig. 2a) and July (Fig. 2b) 2019. Strain YL-1 could control BLB effectively, with a lesion length of 5.44 cm when precultured in liquid SM and with a lesion length of 4.92 cm when precultured in LB medium. No difference in lesion lengths was observed upon treatment with the ΔpvdF mutant and the ΔpvdS(pUCP26-pvdS) complemented strain, with lesion lengths of 5.81 cm and 6.97 cm, respectively, when precultured in liquid SM and 5.50 cm and 7.65 cm, respectively, when precultured in liquid LB medium. Treatment with the ΔpvdS, ΔpvdS(pUCP26), and ΔpvdL mutants had little effect on BLB, whether preculture was in liquid SM or LB medium. The result of the second pot experiment was roughly the same as that of the first. No significant difference (P < 0.05) in the incidence of BLB was observed upon treatment with liquid SM, liquid LB medium, and sterile water, with lesion lengths of 12.05 to 12.88 cm in the first pot experiment (Fig. S4) and 7.84 to 8.20 cm in the second pot experiment. These data suggest that strain YL-1, the ΔpvdF mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain could effectively control BLB in a natural low-iron environment.
FIG 2.
Pot experiments of the suppression of rice BLB disease by strain YL-1 and mutants. BLB-susceptible rice cultivar Jingang 30 (JG30) was inoculated with 50 ml culture broth of strain YL-1 and mutants (1 × 108 CFU/ml) after being infected with Xoo using scissors dipped in a dilution of culture broth (1 × 108 CFU/ml) of Xoo. Treatment with distilled liquid SM or LB medium was used as the control. Sterile water was used as the blank treatment. The pot experiments were conducted over a period of 2 months. (a) The first field trial was performed in June 2019. (b) The second field trial was performed in July 2019.
Effects of exogenous iron or DSX on antibacterial activity and PVD production of strain YL-1.
The production of PVD decreased sharply with an increase in exogenous iron (FeCl3) concentration in liquid SM; the OD405/OD600 value of culture broth ranged from 2.03 ± 0.06 to 0.19 ± 0.02 when supplemented with a final concentration of 2 μM exogenous iron in liquid SM. Strain YL-1 showed limited production of PVD when the final concentration of exogenous iron reached 5 μM, with an OD405/OD600 value of 0.08 ± 0.01 (Table 2). The effect of exogenous iron on anti-Xoo activity was tested in 1/2 TSA plates. As expected, the anti-Xoo activity of strain YL-1 decreased with an increase in exogenous iron concentration. Inhibitory zones of strain YL-1 were 37.1 ± 1.2, 27.3 ± 1.5, 23.6 ± 1.7, 22.4 ± 1.1, and 21.1 ± 1.3 mm when the final concentrations of exogenous iron in 1/2 TSA plates were 0, 2, 5, 10, and 50 μM, respectively (Fig. S5). It was also noted that no visible effects of supplementation of either deferasirox (DSX) or exogenous iron were observed on the growth of either Xoo or strain YL-1 (data not shown).
TABLE 2.
Effect of exogenous iron or DSX on PVD production in strain YL-1a
| Concn of Fe3+ added to liquid SM (μM) | OD405/OD600 | Concn of DSX added to liquid LB medium (μM) | OD405/OD600 |
|---|---|---|---|
| 0 | 2.03 ± 0.06 A | 0 | 0.08 ± 0.01 C |
| 2 | 0.19 ± 0.02 AB | 5 | 0.08 ± 0.01 C |
| 5 | 0.08 ± 0.01 B | 25 | 0.11 ± 0.01 B |
| 10 | 0.07 ± 0.02 B | 50 | 0.14 ± 0.00 B |
| 50 | 0.06 ± 0.02 B | 200 | 0.22 ± 0.02 A |
Values in columns followed by the same letter were not significantly different according to Fisher's protected LSD test (P = 0.05).
However, PVD production increased gradually when DSX was added to liquid LB medium at final concentrations of 5, 25, 50, and 200 μM; the OD405/OD600 value ranged from 0.08 ± 0.01 to 0.22 ± 0.02 (Table 2). The effect of DSX on the anti-Xoo activity was also tested using LB plates. The inhibitory zones of strain YL-1 were 36.1 ± 1.1, 36.3 ± 1.8, 42.9 ± 1.1, 47.3 ± 1.2, and 61.5 ± 1.6 mm when the final concentrations of DSX in LB plates were 0, 5, 25, 50, and 200 μM, respectively (Fig. S6). PVD secreted by strain YL-1 was extremely sensitive to iron abundance in the culture medium, and the production of PVD was definitely related to the antibacterial activity of strain YL-1.
Expression of the pvdS gene and production of PVD at different time points.
Expression levels of pvdS of strain YL-1 cultured in liquid LB medium showed no significant difference (P < 0.05) from 3 h to 48 h. The expression levels of pvdS significantly increased (P < 0.05) when strain YL-1 was cultured in liquid SM. The relative quantification (RQ) value was increased to 6.28 at 3 h, reached a maximum of 12.10 at 9 h, and then decreased slowly (Fig. 3a). At 48 h, no difference was observed in the expression levels of pvdS when strain YL-1 was cultured in the four different media. Interestingly, the expression levels of pvdS increased gradually when DSX was added to liquid LB medium, with an RQ value of 2.15 at 9 h. Subsequently, the expression levels of pvdS decreased gradually until no difference was observed at 48 h when compared to that in LB medium. Moreover, the expression levels of pvdS decreased abruptly when exogenous iron was added to liquid SM. The RQ value was 1.09 at 3 h, and then the expression levels of pvdS increased gradually and showed no difference from those in liquid SM at 24 h, with an RQ value of 3.29 (Fig. 3a). These data suggest that it is beneficial to express the pvdS gene if strain YL-1 is cultured in low-iron medium, which leads to an increase in the production of PVD.
FIG 3.
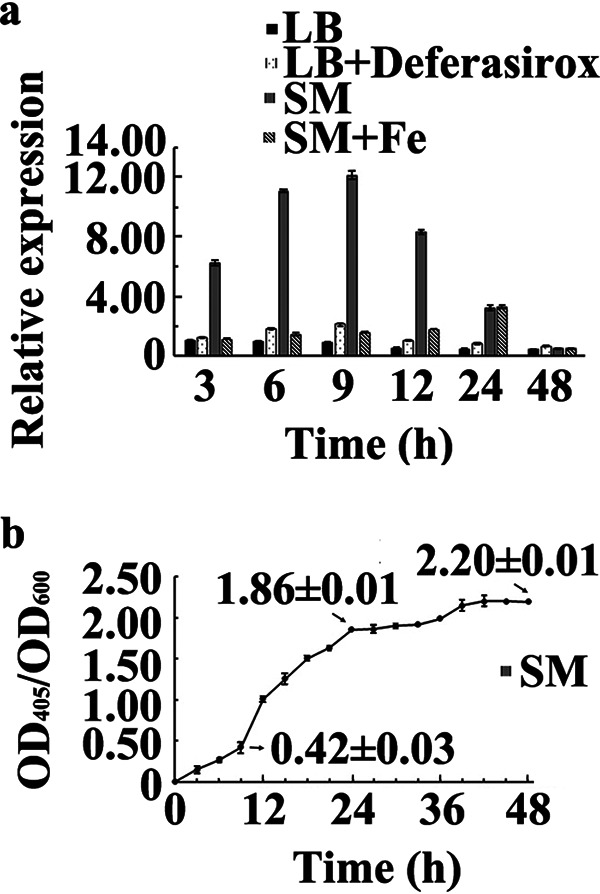
Expression of the pvdS gene (a) and PVD production (b) at different time points. (a) Strain YL-1 was precultured in four different liquid media: (i) LB medium, (ii) LB medium (200 μM DSX), (iii) SM, and (iv) SM (50 μM FeCl3). The total RNA at different time points was extracted and reverse transcribed. The 16S rRNA gene of strain YL-1 was used as an internal control for the real-time PCR assay. The expression of the pvdS gene in liquid LB medium for 3 h was used as the baseline. The relative quantification (RQ) values were calculated using the formula RQ = 2[ΔCT(pvdS in each treatment) − ΔCT(pvdS in LB, 3 h)]. (b) PVD production of strain YL-1 in liquid SM was measured by analyzing the OD405/OD600 at different time points.
Analysis of OD405/OD600 values showed that the PVD production of strain YL-1 incubated in liquid SM increased steadily from 0 to 9 h, increased rapidly from 9 to 24 h, and then increased gradually and finally stabilized at 48 h. The OD405/OD600 values were 0.42 ± 0.03, 1.86 ± 0.01, and 2.20 ± 0.01 when strain YL-1 was cultured in liquid SM at 9, 24, and 48 h, respectively (Fig. 3b).
Isolation and purification of PVDs.
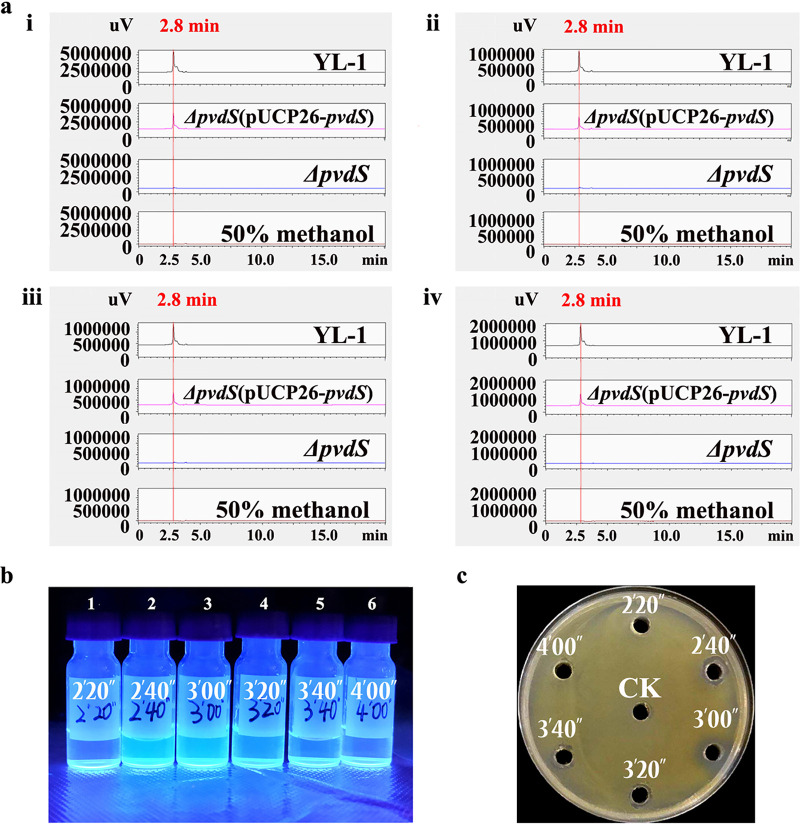
In this study, we used high-performance liquid chromatography (HPLC) carrying a diode array detector (DAD) to analyze the purified PVD products. It was found that comparable peaks produced by wild-type strain YL-1 (Fig. 4a, black line) and the ΔpvdS(pUCP26-pvdS) complemented strain (pink line) were almost unchanged under wavelengths of 225 to 400 nm simultaneously, with a retention time of 2.8 min. (Since every minute is divided into 10 parts in HPLC chromatograms, 2.8 min corresponds to 2′48″; hence, they are the same). The largest absorbance wavelength was 225 nm. No obvious peak was detected from the samples of the ΔpvdS mutant (blue line) and 50% methanol (red line) (Fig. 4a). Comparable peaks were also detected at a higher level in strain YL-1 than in the complemented strain (Fig. 4a). Fractions 2, 3, and 4, collected at 20 per s from a retention time of 2.33 min (2′20′′) to 3.33 min (3′20′′), exhibited fluorescence under UV light (Fig. 4b) and inhibited the growth of Xoo in vitro (Fig. 4c). The inhibitory zones of fractions 2, 3, and 4 were 8.7 ± 1.2 mm, 12.8 ± 1.3 mm, and 7.2 ± 1.1 mm, respectively. The results suggest that the primary antibacterial compounds produced by strain YL-1 in SM were PVDs.
FIG 4.
(a) Reversed-phase high-performance liquid chromatography (RP-HPLC) chromatograms. An overlay of the chromatograms at 225 nm (i), 300 nm (ii), 350 nm (iii), and 400 nm (iv) of the final purification step of the wild-type YL-1, the ΔpvdS mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain, using a C16 column (4.6 mm by 250 mm), is shown. Here, 50% methanol was used as a negative control. The gradient of the acetonitrile-water mobile phase was from 50% to 0% acetonitrile over 10 min at a flow rate of 1 ml/min. (b) Fractions of strain YL-1 per 20 s under UV light. (c) Anti-Xoo assay of fractions in 1/2 TSA medium. The diameter of the inhibitory zones was measured when Xoo covered the entire control plates. Fifty percent methanol was used as the control.
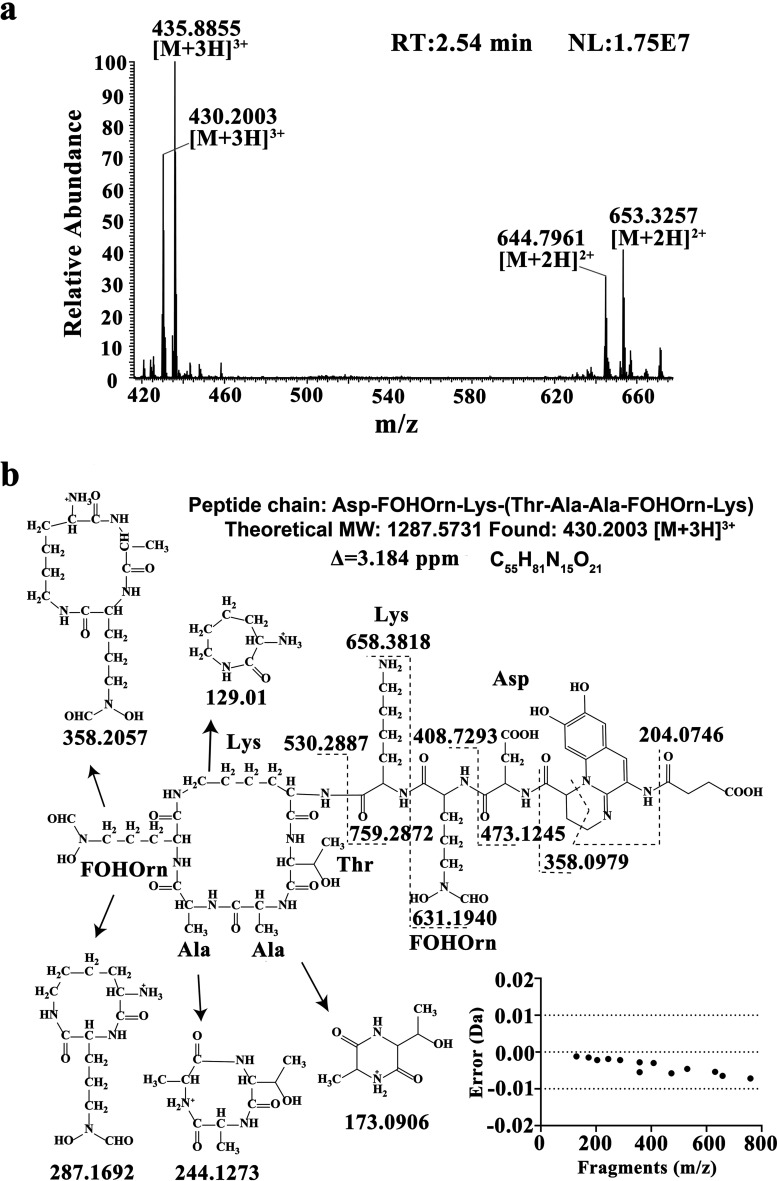
The fractions from 2.33 to 3.00 min described above were further analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). As shown in Fig. 5a, one of the quasi-molecular ions displayed from the main peak at 2.54 min was [M + 3H]3+ at 430.2003, corresponding to a molecular mass of 1,287.58 Da and a C55H81N15O21 molecular formula. As shown in Fig. 5b, the ions with m/z values of 204.07 and 358.10 are characteristic fragment ions of the quinoline ring of PVDs (23). The peptide chains of PVDs can be inferred from the presence of b and y ions in the MS/MS spectrum. As a result, the peptide chain of PVD in P. chlororaphis YL-1 was deduced to be Asp-FOHOrn-Lys-(Thr-Ala-Ala-FOHOrn-Lys) (Fig. 5b), which is consistent with those of PVD in P. fluorescens CHA0 as previously reported (24, 25). The mass errors (measured m/z minus theoretical m/z) of all characteristic fragment ions were less than 0.01 Da, which further supported this inference. At 2.54 min, another quasi-molecular ion was [M + 3H]3+ at m/z 435.89, leading to a 1,304.63-Da molecular mass (Fig. 5a), which also provided a characteristic fragment ion of the quinoline ring of PVDs (m/z 204.07).
FIG 5.
(a) MS spectrum at 2.54 min. The flow rate was maintained at 0.2 ml/min, and the mobile phase comprised water (A) and ACN (B) as follows: 0 min, 50% B; 0 to 10 min, 0% B; 10 to 20 min, 0% B; and 20 to 25 min, 50% B. The detecting wavelength was 225 nm, and the sample volume injected was 2 μl. The ESI source parameters were set as follows: positive ion mode, 3-kV spray voltage, 350°C capillary temperature, and nitrogen serving as both the sheath gas (35 U) and the auxiliary gas (10 U). (b) Proposed characteristic fragment ions of pyoverdines in P. chlororaphis YL-1. The matched b and y ions are indicated, as well as the corresponding mass error.
Antibacterial activity of PVD compounds.
The PVD compounds, extracted from 100 ml fermentation broth of strain YL-1 in SM, were dried to 2 g in a freeze dryer and dissolved in 10 ml 50% methanol (the technical concentration is 200 mg/ml). The results showed that 20 μl of 10-fold-diluted PVD compounds (20 mg/ml) could effectively inhibit the growth of four indicator bacteria in vitro. No difference in inhibitory effects was observed when antibacterial activities against the tested bacteria were assayed on LB and 1/2 TSA plates (Fig. S7).
Anti-Xoo activity of PVD compounds was examined on 1/2 TSA plates in the presence of various concentrations of exogenous iron. As shown in Fig. 6, the inhibitory zones against Xoo increased as the volume of PVD compounds increased on 1/2 TSA plates, with diameters of 14.9 ± 0.5, 22.5 ± 0.6, 27.5 ± 0.2, and 34.9 ± 0.3 mm when treated with 10, 25, 50, and 100 μl of PVD compounds, respectively. Specifically, when treated with the same volume of PVD compounds, the results demonstrated that decreases in inhibitory zone diameter correlated with increases in the iron content of the medium. Of course, the inhibitory activity upon treatment with a small volume of PVD compounds was substantially weaker than that upon treatment with a large volume at any concentration of exogenous iron.
FIG 6.
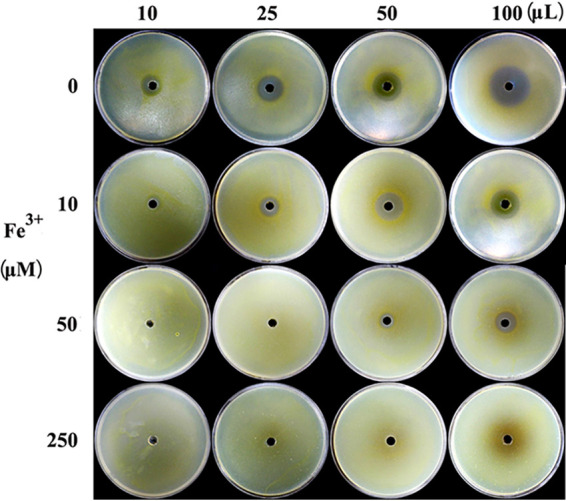
Anti-Xoo activity of PVD compounds on 1/2 TSA plates with different concentrations of exogenous iron. After spraying culture broth with Xoo, 10, 25, 50, and 100 μl of purified PVD were injected into the central hole of 1/2 TSA plates with increasing concentrations of exogenous iron (0, 10, 50, and 250 μM), respectively. Here, 50% methanol was used as the control. The diameters of the inhibitory zones were measured when Xoo covered the entire control plates containing increasing concentrations of exogenous iron.
It is noteworthy that the inhibitory zones against B. megaterium also increased on the deferrated SM as the amount of PVD compounds increased, with 21.9 ± 0.3, 26.8 ± 0.5, 29.2 ± 0.4, and 40.0 ± 0.5 mm, respectively, when treated with 10, 25, 50, and 100 μl of PVD compounds (Fig. 7). Moreover, no difference was observed in the inhibitory zones when tested on deferrated SM and standard SM plates simultaneously.
FIG 7.
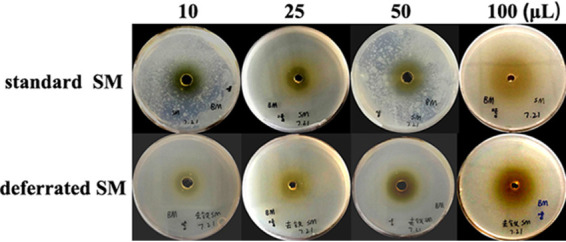
Inhibitory effect of PVD compounds on B. megaterium on standard SM plates (top) and the deferrated SM plates (bottom). Here, 50% methanol was used as the control. B. megaterium was precultured in liquid standard SM or the deferrated SM at 28°C for 24 h to the desired cell density of 2 × 108 CFU/ml and then evenly sprayed onto the surface of standard SM plates and deferrated SM plates for 1 s. Subsequently, 10, 25, 50, and 100 μl of PVD compounds were injected into the central hole of deferrated SM plates, respectively. The diameters of the inhibitory zones were measured when B. megaterium covered the entire control plates.
Collectively, these results suggest the following. (i) No obvious difference in inhibitory effects of PVDs was observed when antibacterial activities were tested on LB and 1/2 TSA plates, which does not depend on the indicator species. (ii) Under low-iron conditions (such as 1/2 TSA), the inhibitory activity of PVDs seems to be more dependent on exogenous iron concentrations. (iii) The ferric-PVD complex has no antibacterial activity. (iv) As an antibiotic, PVD may directly inhibit the growth of indicator bacteria with an unknown mechanism in ironless medium, notwithstanding iron competition.
DISCUSSION
PVD biosynthesis involves a complex pathway comprising cytosolic nonribosomal peptide synthetases. Most enzymes of structure involved in PVD chromophore maturation have been elucidated (10). In the Gram-negative pathogen P. aeruginosa PAO1, the alternative sigma factor PvdS acts as a critical regulator in response to iron starvation (26). The four most substantial PVD synthesis genes in P. aeruginosa strain PAO1, i.e., pvdL, pvdI, pvdJ, and pvdD, encode peptide synthetase enzymes (10). The pvdF gene is required for the transformylation step in the synthesis of PVD because PVD from P. aeruginosa PAO1 lacks the N5-hydroxyornithine (OHOrn) cyclic derivative 3-amino-1-hydroxy-2-piperidone, which is present in other PVDs (27). In this study, when the deletion of the pvdS and pvdL genes occurred, PVD synthesis was disrupted, as expected. However, PVD synthesis was not disrupted when the pvdF gene was deleted, i.e., the production of PVD and the antibacterial activity of the ΔpvdF mutant remained unchanged in comparison to that of the wild-type strain YL-1. Thus, we speculate that the derivative described above should be present in the PVD synthetic pathway of P. chlororaphis YL-1, and synthesis of the PVD may not require the transformylation of PvdF. Further investigation is needed to explore variations among the Pseudomonas strains.
The secondary metabolites produced by a microorganism and its antimicrobial activities can vary depending on the culture medium used for growth. For example, the magnitude of the antagonistic activity of Lysobacter antibioticus is medium dependent, with potent antifungal activity on R2A medium and weak antifungal activity on potato-dextrose agar (PDA) medium (28). In this study, there was no difference in antibacterial activity between the wild-type strain and all the mutants on the iron-sufficient LB medium. Strain YL-1 and all the mutants produced clear inhibitory zones against Xoo and Xooc and had little inhibitory effect against B. megaterium and E. amylovora (see Fig. S2 in the supplemental material). As we know, neither P. chlororaphis YL-1 nor the PVD-deficient mutants were able to produce PVD on LB medium (Fig. 1), which indicated that another antibacterial compound(s) secreted by strain YL-1 and mutants is responsible for the activity against Xoo and Xooc. However, in the low-iron 1/2 TSA medium, the ΔpvdS, ΔpvdS(pUCP26), and ΔpvdL mutants were unable to produce PVD, and the antibacterial activity of these three mutants decreased considerably (Fig. 1; Fig. S2), indicating that production of other antibacterial substances did not occur when growth was in 1/2 TSA medium. The antibacterial activities of strain YL-1, the ΔpvdF mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain did not change because they can secrete PVD in a low-iron environment (Fig. 1). Moreover, when the wild-type strain YL-1, the ΔpvdS mutant, and the ΔpvdS(pUCP26-pvdS) complemented strain were cultured in SM to extract PVD, it was found that peaks produced by strain YL-1 and the ΔpvdS(pUCP26-pvdS) complemented strain were almost unchanged under different tested wavelengths, while no obvious peak produced by the ΔpvdS mutant was detected (Fig. 4a). Therefore, the data suggest that PVD is essential for the antibacterial activity of strain YL-1 under low-iron conditions and that the main antibacterial compound produced by strain YL-1 under low-iron conditions is PVD.
PVD, produced by many Pseudomonas strains, can inhibit the growth of fungi and oomycete pathogens and is required for pathogenesis in Caenorhabditis elegans and many mammalian infection models (13, 14, 29, 30). Previous reports on Pseudomonas-produced PVDs have focused exclusively on its effect on Gram-negative bacteria and fungi (7, 11, 15–17). However, in the present study, we found that the PVDs produced by strain YL-1 demonstrated antibacterial activity against both Gram-positive and Gram-negative bacteria. Therefore, we hypothesized that the PVDs secreted by YL-1 might be different from those secreted by other Pseudomonas strains, as reported previously. Since the first PVD structure was elucidated at the beginning of the 1980s (31), the numerous PVDs of fluorescent Pseudomonas could usually be differentiated from each other by two most practical and most used methods, i.e., (i) PVD isoelectrofocusing (PVD-IEF), and (ii) the determination of bacterial specificity of the PVD-mediated iron uptake. However, a few PVDs could not be differentiated because of identical IEF and iron uptake behaviors (25). Meyer et al. reported that determination of the molecular mass of PVDs by MS should be considered a third powerful siderotyping method (25). LC-MS/MS combines the resolution of LC with the detection specificity of MS/MS, overcoming the limitations of the conventional approaches and providing a direct and rapid methodology to resolve the structure of chemical compounds accumulated in the culture without prior purification or formation of derivatives (32). In this study, using LC-MS/MS, two key peptides were isolated and identified from PVD compounds as Asp-FOHOrn-Lys-(Thr-Ala-Ala-FOHOrn-Lys) with a molecular weight of 1,287.58, consistent with those of PVD in P. fluorescens CHA0 as previously reported (23, 24), and an unknown peptide with a molecular weight of 1,304.63, which could be important components of PVDs of P. chlororaphis YL-1. It was reported that the PVD from P. chlororaphis D-TR133, with the peptide chain Asp-FOHOrn-Lys-(Thr-Ala-Ala-FOHOrn-Ala), showed mutual acceptance with the PVD of P. fluorescens CHA0. This is explained by a structural similarity between the two PVDs except for the replacement of Lys by Ala in the C-terminal part of the molecules (33). Moreover, the main PVD of P. chlororaphis D-TR133 is accompanied by a minor one where Ala is replaced by Gly (33). The structural variations in the peptide chain of PVDs occur either between aromatic amino acids or between those with an alkyl chain. Therefore, we speculate that the unknown peptide produced by P. chlororaphis YL-1 may be a variant, possibly a cyclopeptide with a similar C terminus. Collectively, this research demonstrates that the key antibacterial compounds produced by strain YL-1 under low-iron conditions are PVDs.
It is well known that the in vitro antimicrobial activity of biocontrol bacteria does not necessarily translate into effective disease control in the field. For example, strains of Lysobacter capsici have strong antifungal activity against the pathogen Rhizoctonia solani in vitro. However, no significant or consistent suppression of R. solani damping-off disease was observed when the strains were introduced into the soil (28). Here, we describe P. chlororaphis YL-1, a plant growth-promoting rhizobacterium, demonstrating in vitro activity against Gram-positive and -negative bacteria, including Xoo, as well as against BLB in pot experiments, which is consistent with the results obtained using total culture broth and cell-free supernatants of Pseudomonas taiwanensis (7). Additionally, we observed that the biocontrol efficiencies in BLB for all strains precultured in the liquid LB medium were consistent with those precultured in liquid SM. These results suggest that P. chlororaphis YL-1 is a promising biocontrol bacterium; it produced PVD and played a role in the control of BLB in the natural low-iron environment. Additional studies on the biological characteristics of strain YL-1, such as colonization, survival ability, and product formulation, are needed to confirm the biocontrol effect of strain YL-1 in the field.
In 1952, it was observed that iron concentrations affected the production of fluorescein by P. aeruginosa (34). After that, the role of PVD in iron acquisition by fluorescent pseudomonads was established in the late 1970s (35). Based on these fundamentals and previous reports on other Pseudomonas spp., we sought to determine the effect of iron-dependent PVD production on antibacterial activity and relative expression of the transcription factor gene, pvdS, in strain YL-1. Consistent with previous Pseudomonas species work (7, 29, 36), our findings suggest that when grown under low-iron conditions, strain YL-1 produces PVD and displays antibacterial activity against the Gram-positive and -negative bacteria tested in this study. These findings were further substantiated using real-time PCR, which indicated that under low-iron conditions created by the SM medium, the expression of the pvdS gene was initially increased, with the expression peak at approximately 9 h. The production of PVD that resulted from the expression of the pvdS gene was monitored using OD values and showed that PVD production increased significantly from 9 h, reached the highest level at approximately 24 h, and subsequently stabilized.
The biosynthesis and secretion of PVD are regulated by many factors. The type VI secretion system (T6SS) is necessary for the secretion of PVD in P. taiwanensis (7). However, it has been reported that PvdRT-OpmQ and MdtABC-OpmB efflux systems are involved in PVD secretion in Pseudomonas putida KT2440 (37). In addition to the transcription factor PvdS, PVD biosynthesis is regulated by many other factors, such as ECF sigma factor Fpvl (38), quorum sensing via secreted signaling molecules such as PQS (39), Gac/Rsm signaling networks (40), the AlgZR two-component system (41), and intracellular signaling via secondary messengers (i.e., cyclic diguanylate monophosphate [c-di-GMP]) (42). Moreover, it has been demonstrated that cell aggregation is an essential cue in triggering the production of PVD and virulence in P. aeruginosa PAO1 (40). Therefore, the genetic regulation of PVD biosynthesis of strain YL-1 remains to be investigated to optimize the production of it as a potential biocontrol agent.
In conclusion, the findings of this research demonstrate that strain YL-1, when grown under low-iron conditions, could produce PVDs, which have a broad spectrum of inhibitory effects on both Gram-negative and Gram-positive bacteria, as demonstrated by using bacterial cells, culture broth, and purified compounds. Our findings highlight the effect of exogenous iron on the production of PVD and the importance of this product in bacterial interactions. PVD, as a biocontrol agent, can directly inhibit the proliferation of pathogenic bacteria in addition to participating in iron competition. PVD with a higher ability to chelate Fe3+ to form ferri-PVD complexes could not be utilized by pathogenic bacteria. Thus, PVD may represent a promising agent for application in bacterial disease control.
MATERIALS AND METHODS
Microorganisms, media, and culture conditions.
Bacterial strains used in the study are listed in Table 3. Pseudomonas strains and Escherichia coli were routinely cultured at 28°C and 37°C, respectively, on Luria-Bertani (or lysogeny broth) (LB) medium and preserved in 20% glycerol at −80°C in long-term storage. To obtain a large amount of PVD, P. chlororaphis YL-1 and its mutants were grown in natural low-iron succinic acid medium (SM) (43, 44). The iron chelator deferasirox (DSX), purchased from APExBIO (Shanghai, China), was added to LB medium (known as natural iron-sufficient medium) to achieve low-iron conditions. In contrast, iron-sufficient conditions were achieved by supplementing FeCl3 in either SM or 1/2 Trypticase soy agar (TSA). All chemicals used in this study were of analytical purity and purchased from Sangon (Shanghai, China) unless otherwise stated.
TABLE 3.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristicsa,b | Source or reference (yr) |
|---|---|---|
| Strains | ||
| Pseudomonas chlororaphis | ||
| YL-1 | Wild-type strain, Ampr | Liu et al. (2015) (21) |
| YL-1 ΔpvdS | pvdS in-frame deletion mutant of strain YL-1, Ampr | Liu et al. (2019) (48) |
| YL-1 ΔpvdL | pvdL in-frame deletion mutant of strain YL-1, Ampr | This study |
| YL-1 ΔpvdF | pvdF in-frame deletion mutant of strain YL-1, Ampr | This study |
| YL-1 ΔpvdS(pUCP26) | Mutant strain ΔpvdS harboring plasmid pUCP | This study |
| YL-1 ΔpvdS(pUCP26-pvdS) | Mutant strain ΔpvdS harboring plasmid pUCP-pvdS | This study |
| Escherichia coli | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 | Sangon Corp. |
| S17-1λpir | (F−)RP4-2-Tc::Mu aphA::Tn7 recA λpir lysogen Smr Tpr | Simon et al. (1983) (47) |
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB+ lacIq lacZΔM15] | Promega Corp. |
| Plasmids | ||
| pUCP26 | Broad-host-range vector; Tcr | West et al. (1994) (49) |
| pEX18Gm | Suicide plasmid with a sacB gene, Gmr | Hoang et al. (1998) (46) |
| pEX18-pvdS | pEX18GM with two flanking fragments of pvdS | Liu et al. (2019) (48) |
| pUCP26-pvdS | Recombinant plasmid | This study |
| pEX18-pvdL | pEX18GM with two flanking fragments of pvdL | This study |
| pEX18-pvdF | pEX18GM with two flanking fragments of pvdF | This study |
| Primers | ||
| pvdS-F1 | 5′-GGGGTACCACTGCTGTGGCGGGATGTTC (KpnI) -3′ | Liu et al. (2019) (48) |
| pvdS-R1 | 5′-CCCAAGCTTCCGCGATTTTGACCAGGATC (HindIII) -3′ | Liu et al. (2019) (48) |
| pvdS-F2 | 5′-CCCAAGCTTCACCTACGCCCGCCACTGAT (HindIII) -3′ | Liu et al. (2019) (48) |
| pvdS-R2 | 5′-GCTCTAGAGGATGGCGTGAGGAGTGGAT (XbaI) -3′ | Liu et al. (2019) (48) |
| pvdL-F1 | 5′-CGGAATTCGGAAGAAAATCGGCACCACG (EcoRI) -3′ | This study |
| pvdL-R1 | 5′-CGGGGTACCGAGGAACTGGCGCAGTACAT (KpnI) -3′ | This study |
| pvdL-F2 | 5′-CGGGGTACCCTCTGATCCTGCTCTTCGGC (KpnI) -3′ | This study |
| pvdL-R2 | 5′-GCTCTAGAGCCTGCTTGCGATAGTGGTC (XbaI) -3′ | This study |
| pvdF-F1 | 5′-CGGGGTACCTCGCCCAGAAAACCACCTTG (KpnI) -3′ | This study |
| pvdF-R1 | 5′-CCCAAGCTTCGGAAGCGGGTTACATCACC (HindIII) -3′ | This study |
| pvdF-F2 | 5′-CCCAAGCTTCTTTGCCGCTGAGGGATTGG (HindIII) -3′ | This study |
| pvdF-R2 | 5′-CGGAATTCGGAAAACGCCGCAGGAACTC (EcoRI) -3′ | This study |
| PvdS FP | 5′- CAGGAATTCATGACGGAACAAGTATCCAC (EcoRI) -3′ | This study |
| PvdS RP | 5′- CACAAGCTTTCAGTGGCGGGCGTAGGTGT (HindIII) -3′ | This study |
| RT-pvdS-F | 5′-GCGAGATGCCGTTGATCAGC-3′ | This study |
| RT-pvdS-R | 5′-TACGAGCAGTTCACCATCCG-3′ | This study |
| RT-16S rRNA gene-F | 5′-ACGTCCTACGGGAGAAAGC-3′ | This study |
| RT-16S rRNA gene-R | 5′-CGTGTCTCAGTTCCAGTGTGA-3′ | This study |
Ampr , Gmr, Tcr, resistance to ampicillin, gentamicin, and tetracycline, respectively.
Restriction enzyme digestion sites in sequences are bold.
To evaluate the antibacterial activity of PVD compounds against Bacillus megaterium under ironless conditions, SM was treated with an equal volume of chloroform containing 1.5% 8-hydroxyquinoline to remove the traces of iron in the medium, and the traces of hydroxyquinoline were removed by washing three times with chloroform to obtain the deferrated SM (43, 44).
Generation of deletion mutants.
All primers used for PCR and cloning are listed in Table 3. The B. subtilis sacB gene, which encodes levansucrase, which is toxic to Gram-negative bacteria in the presence of sucrose, was considered a counterselection marker in the system (45). We selected a well-characterized pvdS gene as a representative example to generate an in-frame deletion mutant (see Fig. S8 in the supplemental material). First, the upstream and downstream homolog fragments of pvdS in wild-type strain YL-1 were cloned by using two pairs of primers (pvdS-F1 and pvdS-R1, pvdS-F2 and pvdS-R2). Then, a sacB-containing suicide vector, pEX18Gm (46), was used to create a recombinant plasmid, pEX18-pvdS. The recombinant plasmid was initially transformed into E. coli S17-1 (47) and then transformed into wild-type strain YL-1 via an optimized bacterial conjugal approach. Through a double-crossover homologous recombinant strategy, an in-frame deletion mutant of pvdS, described as the ΔpvdS mutant, was generated and validated (48). The ΔpvdL and ΔpvdF mutants were generated as described above (Fig. S8).
The pvdS gene was selected as a representative model to generate a complemented strain, because it encodes a sigma factor known to control PVD biosynthesis at the transcription level (2, 3). The ΔpvdS(pUCP26-pvdS) complemented strain was generated using the plasmid pUCP26 Escherichia-Pseudomonas shuttle vector, as previously described (49, 50). Employing primers PvdS FP and PvdS RP, the pvdS PCR product of strain YL-1 was confirmed by sequence analysis. Following double restriction enzyme digestion, the PCR product was ligated into the plasmid pUCP26 to generate the plasmid pUCP26-pvdS. pUCP26-pvdS was electroporated into E. coli JM109 cells for identity confirmation. After the sequencing confirmation, the plasmid was introduced into ΔpvdS mutant cells by electroporation. The ΔpvdS(pUCP26) mutant was generated via electroporation of the empty vector pUCP26 into the ΔpvdS mutant and used as a control. Gene replacements and insertions were verified by PCR amplification using specific primers and DNA sequencing.
Growth and PVD measurements.
To examine growth and PVD production under low-iron and iron-sufficient conditions, P. chlororaphis YL-1 and mutants (described above) were individually incubated in a 250-ml flask containing 100 ml liquid SM or LB medium at 28°C with shaking (200 rpm for 24 h). All liquid cultures were observed under visible light and UV light simultaneously. For growth measurements, the numbers of CFU per ml were determined by standard serial dilution (7). PVD production in liquid cultures was measured using the optical density according to the procedure described by Imperi et al. (51). P. chlororaphis YL-1 incubated in 100 ml liquid deferrated SM was used as a control. The experiment was repeated three times independently.
Antibacterial activity of strain YL-1 and mutants.
Antibacterial activities of YL-1 and PVD-deficient mutants were evaluated using a plate bioassay protocol as described previously (50). To determine activity in an iron-sufficient medium, the bacterial strains were incubated in liquid LB medium at 28°C with shaking (200 rpm for 24 h) and resuspended in sterile liquid LB medium to the desired cell density of 108 CFU/ml. Culture broth of the indicator bacterium, Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99 (7), was evenly sprayed onto the surface of LB plates for 1 s using sterile laryngeal spray (Taizhou Medical Instrument Co., Ltd., China). Thereafter, 50 μl culture broth (108 CFU/ml) of strain YL-1 or mutants was injected into a central hole (5 mm) of the LB plates and incubated at 28°C for 48 h. The inhibitory zone diameters of strain YL-1 and mutants were measured when Xoo covered the entire control plates. Distilled liquid LB medium was used as a control. Each treatment was replicated three times, and each experiment was repeated three times independently. Strain YL-1 and PVD-deficient mutants were evaluated against the phytopathogenic bacteria Erwinia amylovora 2029 (50), X. oryzae pv. oryzicola (Xooc) RS11 (52), and Gram-positive Bacillus megaterium 329 (52) in the same manner as for Xoo.
The antagonistic assays of P. chlororaphis YL-1 and mutants precultured in liquid SM were performed on 1/2 TSA plates. The experimental procedures were the same as those described above. Sterile liquid SM was used as the control.
Anti-Xoo activity of strain YL-1 and mutants in pot experiments.
For the pot experiments, BLB-susceptible rice cultivar Jingang 30, provided by the Institute of Food Crops, Jiangsu Academy of Agricultural Sciences, was used as the host plant (53). Four-leaf seedlings of rice were transplanted in plastic pots (diameter, 40 cm) and supplied with nutrition soil (Anhui Rongfeng Biotechnology Co., Ltd., China). The plants were grown in a greenhouse at the Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, with 12-h light/12-h dark cycles at 28°C and 75% relative humidity. In the rice tillering stage, 20 leaves of Jingang 30 rice per treatment were infected with Xoo using scissors dipped into a dilution of culture broth (1 × 108 CFU/ml) of Xoo (7). After 24 h, the rice was sprayed with 50 ml culture broth (1 × 108 CFU/ml) of P. chlororaphis YL-1 or mutants, which were precultured in liquid SM or LB medium. Treatment with sterile liquid SM or LB medium was used as a control; sterile water was used as the blank treatment. Treatments were maintained at constant humidity and temperature, like those described above, for 3 weeks, and the lesion lengths of BLB were recorded (7). Each treatment was replicated three times, and the experiment was repeated twice.
Effects of exogenous iron or DSX on anti-Xoo activity and PVD production of strain YL-1.
The effect of iron abundance on the antagonistic activity of YL1 was investigated by repeating the procedure used to assess antibacterial activity (see above) with modifications to the iron content of the culture medium. This was accomplished by supplementing SM with exogeneous iron in the form of FeCl3 and lowering iron availability in LB by the addition of DSX. In brief, strain YL-1 was first incubated in liquid SM containing FeCl3 at 28°C with shaking (200 rpm for 24 h). Thereafter, 50 μl of culture broth (108 CFU/ml) was injected into a central hole (5 mm) of 1/2 TSA plates containing FeCl3 and incubated at 28°C. FeCl3 was added to final concentrations of 2, 5, 10, and 50 μM in liquid SM and 1/2 TSA plates, respectively. Similarly, strain YL-1 was incubated in liquid LB medium mixed with DSX, and the anti-Xoo activity was tested on LB plates mixed with DSX to final concentrations of 5, 25, 50, and 200 μM. The production of PVD and anti-Xoo activity was measured as described above. Each treatment was replicated three times, and each experiment was repeated three times independently.
Real-time PCR assay.
The pvdS gene was selected as a representative example to investigate the effects of adding exogenous iron under low-iron conditions or DSX under iron-sufficient conditions on its relative expression in YL-1. Consequently, the total RNA of strain YL-1 precultured in liquid LB medium, LB (200 μM DSX), SM, and SM (50 μM FeCl3) was extracted for pvdS gene expression analysis.
At various time points, the total RNA was extracted using a total RNA miniprep purification kit (Sangon Biotech, Shanghai). The total RNA was reverse transcribed using PrimeScript RT master mix (TaKaRa, Japan). Quantitative real-time (RT) PCR was performed on a Roche LightCycler 2.0 system (USA) using Power TB Green PCR master mix (TaKaRa, Japan). The 16S rRNA gene was used as an internal control in the real-time PCR assay (54). Each 20-μl RT-PCR volume contained 1 ng cDNA, 5× TB Green mix, and 4 μM forward and reverse primers (Table 3). The threshold cycle (CT) values of pvdS used to determine transcript levels were normalized to those of the 16S rRNA gene according to the calculation ΔCT = CT(pvdS) − CT(16S rRNA gene). The expression of the pvdS gene in liquid LB medium for 3 h was used as the baseline. The relative quantification (RQ) values were calculated with the formula RQ = 2[ΔCT (pvdS in each treatment) − ΔCT (pvdS in LB, 3 h)]. When the RQ was equal to 1, transcript levels of each treatment, including the baseline, were equal. Three replicates of RT-PCR analysis for each reaction were performed and repeated independently. Additionally, PVD production of strain YL-1 in liquid SM was also measured at different time points. The experiment was repeated three times.
Purification and determination of PVD.
The purification of PVD was performed as described by Yin et al. (55), with some modifications. In brief, a 250-ml flask with 100 ml of P. chlororaphis YL-1, the ΔpvdS mutant, or the ΔpvdS(pUCP26-pvdS) complemented strain was incubated in SM at 28°C with shaking (200 rpm for 24 h). The culture supernatant was collected by centrifugation at 10,000 × g for 20 min at 4°C and filtered through 0.22-μm sterile membrane filters (Millex-GV; Millipore). The filtered culture supernatant was mixed with binding buffer (0.02 M Na2HPO4 and 1 M NaCl, pH 7.2) at a 1:1 ratio. The mixed solution (200 ml) was loaded onto a Cu-Sepharose column to purify the PVD. Finally, PVD was eluted using an elution buffer (0.02 M Na2HPO4 and 1 M NH4Cl, pH 7.2) and dried in a freeze dryer (LyoAlfa 15-85; Telstar, Spain).
The purified compounds were dissolved in 10 ml 50% methanol and analyzed by high-performance liquid chromatography (HPLC) (Shimadzu LC-6AD, Japan) using an RP-Amide C16 column (4.6 by 250 mm, 5 μM; Sigma-Aldrich). The acetonitrile (ACN)-water gradient of the HPLC mobile phase demonstrated 50% to 0% acetonitrile over 10 min at a flow rate of 1 ml/min and 5 μl per injection volume. Fractions of strain YL-1, collected every 20 s from retention time 2.0 min to 4.0 min, were monitored by UV light. Before the antibacterial assay, the fractions were first dried in a freeze dryer (LyoAlfa 15-85; Telstar, Spain), separately, and then dissolved in 50 μl 50% methanol. The anti-Xoo activity was evaluated as described above. Fifty percent methanol was used as a control.
The fraction from retention time 2.33 to 3.00 min was further separated and determined with a Thermo Fisher Scientific UHPLC-LTQ-Orbitrap mass spectrometer (LC-MS/MS) equipped with electrospray ionization (ESI). The HPLC column used was Hypersil GOLD C18 (2.1 by 100 mm, 3 μm; Thermo Fisher Scientific). The temperature of the column oven was set at 35°C. The flow rate was maintained at 0.2 ml/min, and the mobile phase comprised water (A) and ACN (B) as follows: 0 min, 50% B; 0 to 10 min, 0% B; 10 to 20 min, 0% B; and 20 to 25 min, 50% B. The detecting wavelength was 225 nm, and the sample volume injected was 2 μl. The ESI source parameters were set as follows: positive ion mode, 3-kV spray voltage, 350°C capillary temperature, and nitrogen serving as both the sheath gas (35 U) and the auxiliary gas (10 U). A data-dependent acquisition was applied. From a high-resolution (resolving power, 60,000) full scan of mass spectrometry (MS), the three most intense precursor ions were selected for fragmentation, using higher-energy C-trap dissociation (HCD). The normalized collision energy was set to a value of 45%. Accurate mass analyses were calibrated before the experiment according to the manufacturer’s guidelines.
Anti-Xoo activity of PVD compounds in different media.
The anti-Xoo activity of purified PVD compounds was assessed by plate assays as described above, which were divided into three parts. First, to test the efficacy of purified compounds under iron-sufficient and low-iron conditions, a Xoo suspension (2 × 108 CFU/ml) was evenly sprayed onto LB and 1/2 TSA plates for 1 s, and then 20 μl of PVD compounds (20 mg/ml) was injected into a central hole (5 mm) of each plate. Second, the effect of various levels of exogenous iron was evaluated by repeating the procedure using 1/2 TSA plates. A Xoo suspension (2 × 108 CFU/ml) was evenly sprayed onto the 1/2 TSA plates with different exogenous iron concentrations (0, 10, 50, and 250 μM) for 1 s, and then 10, 25, 50, and 100 μl of PVD compounds were injected into a central hole (5 mm) of each plate. Third, the effect of purified PVD compounds on the ironless medium was evaluated by repeating the procedure as described above. A suspension of B. megaterium (2 × 108 CFU/ml) was sprayed onto the deferrated SM and standard SM plates for 1 s, and 10, 25, 50, and 100 μl of PVD compound were injected into a central hole (5 mm) of each plate.
All plates were incubated at 28°C, and 50% methanol was used as a control. The inhibitory zone diameters were measured when the indicator bacteria covered the entire control plates. Each treatment was replicated three times, and each experiment was repeated three times independently.
Statistical analysis.
Data were processed by analysis of variance (ANOVA) using the SAS GLM procedure (SAS Institute, Inc., Cary, NC). A P of <0.05 was considered statistically significant. Significant means were further compared by using Fisher's protected least significant difference (PLSD).
Supplementary Material
ACKNOWLEDGMENTS
We thank Weiwei Zhang (Ningbo University) and Guoliang Qian (Nanjing Agricultural University) for optimizing the extraction method of pyoverdine and providing some essential materials. We also thank Kate Phillips for English proofreading.
This research was funded by the National Key R&D Program of China (grant no. 2017YFD0200400) and the National Natural Science Foundation of China (grant no. 31672076). This research was also partially funded by the Science and Technology Project of Jiangsu Province (grant no. BE2018359) and Suzhou Science and Technology Projects (grant no. SNG2018095). This research was partially supported by the USDA NIFA project MIS-401200.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Schalk IJ. 2008. Metal trafficking via siderophores in Gram-negative bacteria: specificities and characteristics of the pyoverdine pathway. J Inorg Biochem 102:1159–1169. doi: 10.1016/j.jinorgbio.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 2.Moon CD, Zhang XX, Matthijs S, Schäfer M, Budzikiewicz H, Rainey PB. 2008. Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol 8:7. doi: 10.1186/1471-2180-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Owen JG, Ackerley DF. 2011. Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a. BMC Microbiol 11:218. doi: 10.1186/1471-2180-11-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subashri R, Raman G, Sakthivel N. 2013. Biological control of pathogens and plant growth promotion potential of fluorescent pseudomonads, p 77–110. In Maheshwari D (ed), Bacteria in agrobiology: disease management. Springer, Berlin, Germany. doi: 10.1007/978-3-642-33639-3_4. [DOI] [Google Scholar]
- 5.Anokhina TO, Volkova OV, Puntus IF, Filonov AE, Kochetkov VV, Boronin AM. 2006. Plant growth-promoting Pseudomonas bearing catabolic plasmids: naphthalene degradation and effect on plants. Process Biochem 41:2417–2423. doi: 10.1016/j.procbio.2006.06.026. [DOI] [Google Scholar]
- 6.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. 1996. Pyoverdine is essential for virulence of Pseudomonas aeruginosa. Infect Immun 64:518–523. doi: 10.1128/IAI.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen WJ, Kuo TY, Hsieh FC, Chen PY, Wang CS, Shih YL, Lai YM, Liu JR, Yang YL, Shih MC. 2016. Involvement of type VI secretion system in secretion of iron chelator pyoverdine in Pseudomonas taiwanensis. Sci Rep 6:32950. doi: 10.1038/srep32950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schalk IJ, Guillon L. 2013. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ Microbiol 15:1661–1673. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 9.Pol NJ, Gudrun K, Carlos RR, Remco M, Hans R, Margot JS, Robbert HC, Wim JQ. 2014. PvdP is a tyrosinase that drives maturation of the pyoverdine chromophore in Pseudomonas aeruginosa. J Bacteriol 196:2681–2690. doi: 10.1128/JB.01376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poppe J, Reichelt J, Blankenfeldt W. 2018. Pseudomonas aeruginosa pyoverdine maturation enzyme PvdP has a noncanonical domain architecture and affords insight into a new subclass of tyrosinases. J Biol Chem 293:14926–14936. doi: 10.1074/jbc.RA118.002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leeman M, Van PJ, Den OF. 1995. Induction of systemic resistance against Fusarium wilt of radish by lipopolysaccharides of Pseudomonas fluorescens. Phytopathology 85:1021–1027. doi: 10.1094/Phyto-85-1021. [DOI] [Google Scholar]
- 12.Trapet P, Avoscan L, Klinguer A, Pateyron S, Citerne S, Chervin C, Mazurier S, Lemanceau P, Wendehenne D, Besson-Bard A. 2016. The Pseudomonas fluorescens siderophore pyoverdine weakens Arabidopsis thaliana defense in favor of growth in iron-deficient conditions. Plant Physiol 171:675–693. doi: 10.1104/pp.15.01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang D, Kirienko DR, Webster P, Fisher AL, Kirienko NV. 2018. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 9:804–817. doi: 10.1080/21505594.2018.1449508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang WW, Liang WK, Li CH. 2016. Inhibition of marine Vibrio sp. by pyoverdine from Pseudomonas aeruginosa PA1. J Hazard Mater 302:217–224. doi: 10.1016/j.jhazmat.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Xu YQ, Gao H, Tong GL, Zhu XD. 1999. Siderophore production and their activity against Piricularia oryzae by Pseudomonas JKD-2. Microbiol China 26:180–183. [Google Scholar]
- 16.Ran LX, Xiang ML, Zhou B, Bakker PAHM. 2005. Siderophores are the main determinants of fluorescent Pseudomonas strains in suppression of grey mould in Eucalyptus urophylla. Acta Phytopathol Sin 35:6–12. [Google Scholar]
- 17.Sass G, Nazik H, Penner J, Shah H, Ansari SR, Clemons KV, Groleau M-C, Dietl A-M, Visca P, Haas H, Déziel E, Stevens DA. 2017. Studies of Pseudomonas aeruginosa mutants indicate pyoverdine as the central factor in inhibition of Aspergillus fumigatus biofilm. J Bacteriol 200:e00345-17. doi: 10.1128/JB.00345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang D, Kirienko NV. 2017. High-throughput genetic screen reveals that early attachment and biofilm formation are necessary for full pyoverdine production by Pseudomonas aeruginosa. Front Microbiol 8:1707. doi: 10.3389/fmicb.2017.01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wencewicz TA, Möllmann U, Long TE, Miller MJ. 2009. Is drug release necessary for antimicrobial activity of siderophore-drug conjugates? Syntheses and biological studies of the naturally occurring salmycin “Trojan Horse” antibiotics and synthetic desferridanoxamine-antibiotic conjugates. Biometals 22:633–648. doi: 10.1007/s10534-009-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mislin GL, Schalk IJ. 2014. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 6:408–420. doi: 10.1039/c3mt00359k. [DOI] [PubMed] [Google Scholar]
- 21.Liu YZ, Lu S-E, Baird SM, Qiao JQ, Du Y. 2015. Cloning the genes of Pseudomonas chlororaphis YL-1 dedicated to antibacterial activities against microbial phytopathogens. Acta Phytopathol Sin 45:307–316. [Google Scholar]
- 22.Liu Y, Lu S-E, Baird SM, Qiao JQ, Du Y. 2014. Draft genome sequence of Pseudomonas chlororaphis YL-1, a biocontrol strain suppressing plant microbial pathogens. Genome Announc 2:e01225-13. doi: 10.1128/genomeA.01225-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budzikiewicz H, Schäfer M, Meyer JM. 2007. Siderotyping of fluorescent pseudomonads—problems in the determination of molecular masses by mass spectrometry. Mini-Rev Organic Chem 4:246–253. doi: 10.2174/157019307781369968. [DOI] [Google Scholar]
- 24.Wong-Lun-Sang S, Bernardini J, Hennard C, Kyslik P, Dell A, Abdallah MA. 1996. Bacterial siderophores: structure elucidation, 2D 1H and 13C NMR assignments of pyoverdins produced by Pseudomonas fluorescens CHA0. Tetrahedron Lett 37:3329–3332. doi: 10.1016/0040-4039(96)00569-2. [DOI] [Google Scholar]
- 25.Meyer JM, Gruffaz C, Raharinosy V, Bezverbnaya I, Schafer M, Budzikiewicz H. 2008. Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 21:259–271. doi: 10.1007/s10534-007-9115-6. [DOI] [PubMed] [Google Scholar]
- 26.Imperi F, Tiburzi F, Fimia GM, Visca P. 2010. Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ Microbiol 12:1630–1642. doi: 10.1111/j.1462-2920.2010.02210.x. [DOI] [PubMed] [Google Scholar]
- 27.Visca P, Imperi F, Lamont IL. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol 15:22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Gómez ER, Postma J, Raaijmakers JM, De Bruijn I. 2015. Diversity and activity of Lysobacter species from disease suppressive soils. Front Microbiol 6:1243. doi: 10.3389/fmicb.2015.01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torres-Rubio MG, Valencia-Plata SA, Bernal-Castillo J, Martinez-Nieto P. 2000. Isolation of Enterobacteria, Azotobacter sp. and Pseudomonas sp., producers of indole-3-acetic acid and siderophores, from Colombian rice rhizosphere. Rev Latinoam Microbiol 42:171–176. [Google Scholar]
- 30.Santoyo G, Valencia-Cantero E, Orozco-Mosqueda MDC, Pena-Cabriales JJ, Farias-Rodriguez R. 2010. Role of siderophores in antagonic activity of Pseudomonas fluorescens ZUM80 against plant fungi. Terra Latinoam 28:53–60. [Google Scholar]
- 31.Teintze M, Hossain MB, Barnes CL, Leong J, van der Helm D. 1981. Structure of ferric pseudobactin, a siderophore from a plant growth promoting Pseudomonas. Biochemistry 20:6446–6457. doi: 10.1021/bi00525a025. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Arjol I, Bassas-Galia M, Bermudo E, Garcia F, Manresa A. 2010. Identification of oxylipins with antifungal activity by LC–MS/MS from the supernatant of Pseudomonas 42A2. Chem Phys Lipids 163:341–346. doi: 10.1016/j.chemphyslip.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Barelmann I, Fernandez DU, Budzikiewicz H, Meyer JM. 2003. The pyoverdine from Pseudomonas chlororaphis D-TR133 showing mutual acceptance with the pyoverdine of Pseudomonas fluorescens CHA0. BioMetals 16:263–270. doi: 10.1023/A:1020615830765. [DOI] [PubMed] [Google Scholar]
- 34.Totter JR, Moseley FT. 1953. Influence of the concentration of iron on the production of fluorescin by Pseudomonas aeruginosa. J Bacteriol 65:45–47. doi: 10.1128/JB.65.1.45-47.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer JM, Hornsperger JM. 1978. Role of pyoverdine the iron binding fluorescent pigment of Pseudomonas fluorescens iron transport. J Gen Microbiol 107:329–331. doi: 10.1099/00221287-107-2-329. [DOI] [Google Scholar]
- 36.Wensing A, Braun SD, Büttner P, Expert D, Volksch B, Ullrich MS, Weingart H. 2010. Impact of siderophore production by Pseudomonas syringae pv. syringae 22d/93 on epiphytic fitness and biocontrol activity against Pseudomonas syringae pv. glycinea 1a/96. Appl Environ Microbiol 76:2704–2711. doi: 10.1128/AEM.02979-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henríquez T, Stein NV, Jung H. 2019. PvdRT-OpmQ and MdtABC-OpmB efflux systems are involved in pyoverdine secretion in Pseudomonas putida KT2440. Environ Microbiol Rep 11:98–106. doi: 10.1111/1758-2229.12708. [DOI] [PubMed] [Google Scholar]
- 38.Edgar RJ, Xu X, Shirley M, Konings AF, Martin LW, Ackerley DF, Lamont IL. 2014. Interactions between an anti-sigma protein and two sigma factors that regulate the pyoverdine signaling pathway in Pseudomonas aeruginosa. BMC Microbiol 14:287. doi: 10.1186/s12866-014-0287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang D, Turner KE, Kirienko NV. 2017. PqsA promotes pyoverdine production via biofilm formation. Pathogens 7:3. doi: 10.3390/pathogens7010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visaggio D, Pasqua M, Bonchi C, Kaever V, Visca P, Imperi F. 2015. Cell aggregation promotes pyoverdine-dependent iron uptake and virulence in Pseudomonas aeruginosa. Front Microbiol 6:902. doi: 10.3389/fmicb.2015.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Little AS, Okkotsu Y, Reinhart AA, Damron FH, Barbier M, Barrett B, Oglesby-Sherrouse AG, Goldberg JB, Cody WL, Schurr MJ, Vasil ML, Schurr MJ. 2018. Pseudomonas aeruginosa AlgR phosphorylation status differentially regulates pyocyanin and pyoverdine production. mBio 9:e02318-17. doi: 10.1128/mBio.02318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T, Cai Z, Shao X, Zhang W, Xie Y, Zhang Y, Hua C, Schuster SC, Yang L, Deng X. 2019. Pleiotropic effects of c-di-GMP content in Pseudomonas syringae. Appl Environ Microbiol 85:e00152-19. doi: 10.1128/AEM.00152-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bharucha UD, Patel KC, Trivedi UB. 2013. Antifungal activity of catecholate type siderophore produced by Bacillus sp. Int J Res Pharm Sci 4:528–531. [Google Scholar]
- 44.Meyer JM, Abdallah MA. 1978. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J Gen Microbiol 107:319–328. doi: 10.1099/00221287-107-2-319. [DOI] [Google Scholar]
- 45.Gay P, Coq DL, Steinmetz M, Ferrari E, Hoch JA. 1983. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol 153:1424–1431. doi: 10.1128/JB.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 47.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 48.Liu YZ, Zhang TT, Zhou YQ, Qiao JQ, Liu YF. 2019. Establishment of sacB-mediated genetic manipulation system of Pseudomonas chlororaphis YL-1. Chinese J Biol Control 35:622–629. [Google Scholar]
- 49.West SEH, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 50.Liu YZ, Baird SM, Qiao JQ, Du Y, Lu S-E. 2015. SecG is required for antibiotic activities of Pseudomonas sp. YL23 against Erwinia amylovora and Dickeya chrysanthemi. J Basic Microbiol 55:617–624. doi: 10.1002/jobm.201400491. [DOI] [PubMed] [Google Scholar]
- 51.Imperi F, Tiburzi F, Visca P. 2009. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 106:20440–20445. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu YZ, Qiao JQ, Liu YF, Liang XJ, Zhou YQ, Liu JB. 2019. Characterization of Lysobacter capsici strain NF87–2 and its biocontrol activities against phytopathogens. Eur J Plant Pathol 155:859–869. doi: 10.1007/s10658-019-01817-9. [DOI] [Google Scholar]
- 53.Fan YL, Chen XW, Wang CL, Zhu LH, Zhang Q, Zhao KJ. 2006. Mapping the rice bacterial blight resistance gene Xa 23 with RFLP markers and converting RFLP to STS marker. Acta Agron Sin 32:931–935. [Google Scholar]
- 54.Chen QZ, Tu H, Huang F, Wang YC, Dong WL, Wang WH, Li ZK, Wang F, Cui ZL. 2016. Impact of pnpR, a LysR-type regulator-encoding gene, on the cellular processes of Pseudomonas putida DLL-E4. FEMS Microbiol Lett 363:fnw110. doi: 10.1093/femsle/fnw110. [DOI] [PubMed] [Google Scholar]
- 55.Yin K, Zhang W, Chen L. 2014. Pyoverdine secreted by Pseudomonas aeruginosa as a biological recognition element for the fluorescent detection of furazolidone. Biosens Bioelectron 51:90–96. doi: 10.1016/j.bios.2013.07.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.