Based on our results, we observed a drastic reduction in gut microbiota diversity of children treated with antibiotics, which also affecting the abundance of Bifidobacterium, a bacterial genus commonly found in the infant gut. MIC experiments revealed that more than 98% of bifidobacterial strains tested were shown to be inhibited by the AMC antibiotic.
KEYWORDS: bifidobacteria, antibiotics, comparative genomics
ABSTRACT
Amoxicillin-clavulanic acid (AMC) is one of the most frequently prescribed antibiotic formulations in the Western world. Extensive oral use of this antimicrobial combination influences the gut microbiota. One of the most abundant early colonizers of the human gut microbiota is represented by different taxa of the Bifidobacterium genus, which include many members that are considered to bestow beneficial effects upon their host. In the current study, we investigated the impact of AMC administration on the gut microbiota composition, comparing the gut microbiota of 23 children that had undergone AMC antibiotic therapy to that of 19 children that had not been treated with antibiotics during the preceding 6 months. Moreover, we evaluated AMC sensitivity by MIC test of 261 bifidobacterial strains, including reference strains for the currently recognized 64 bifidobacterial (sub)species, as well as 197 bifidobacterial isolates of human origin. These assessments allowed the identification of four bifidobacterial strains that exhibit a high level of AMC insensitivity, which were subjected to genomic and transcriptomic analyses to identify the putative genetic determinants responsible for this AMC insensitivity. Furthermore, we investigated the ecological role of AMC-resistant bifidobacterial strains by in vitro batch cultures.
IMPORTANCE Based on our results, we observed a drastic reduction in gut microbiota diversity of children treated with antibiotics, which also affected the abundance of Bifidobacterium, a bacterial genus commonly found in the infant gut. MIC experiments revealed that more than 98% of bifidobacterial strains tested were shown to be inhibited by the AMC antibiotic. Isolation of four insensitive strains and sequencing of their genomes revealed the identity of possible genes involved in AMC resistance mechanisms. Moreover, gut-simulating in vitro experiments revealed that one strain, i.e., Bifidobacterium breve PRL2020, is able to persist in the presence of a complex microbiota combined with AMC antibiotic.
INTRODUCTION
The human gastrointestinal tract (GIT) harbors a complex and dynamic population of microorganisms, i.e., the gut microbiota, whose coordinated actions are believed to be important to support a healthy human physiology (1, 2). Microbes colonize the neonatal gut immediately following birth, and this early life event is believed to represent a crucial opportunity for microbiota modulation, which in turn may influence host health in later life (3). During a human life, the gut microbiota may frequently be exposed to various antibiotics, causing alterations in the associated microbial communities, with both immediate effects and potential indirect, long-term effects on host health (4, 5). In this context, early antibiotic exposure has a major impact on infant health through diversity reduction and compositional alteration of microbial communities in this ecosystem (6, 7).
Amoxicillin-clavulanic acid (AMC) is one of the most frequently prescribed antibiotics, especially in infants and adolescents (8). The combination of these two compounds is commonly used for treatment of different pediatric infectious diseases, including acute otitis media, sinusitis, pneumonia, urinary tract infections, and skin and soft tissue infections (9). Despite the extensive use of AMC, especially for pediatric patients, little is known about its impact on the gut microbiota.
The most prevalent and abundant early colonizers of the human infant gut encompass members of the Bifidobacterium genus (10–12). In recent years, the scientific interest in bifidobacteria has been growing because they are believed to have an impact on important immunomodulatory activities in the host at the very early stages of life and to influence the physiology of the gut ecosystem by their metabolic activities (11, 13). In vitro susceptibility assessments have demonstrated that Bifidobacterium species are generally sensitive to amoxicillin (14–16). Nonetheless, only limited knowledge is available pertaining to the effects of amoxicillin on its own or when combined with clavulanic acid on bifidobacterial communities residing in the human gut (17).
In the current study, we assessed the microbiota composition of children that had undergone AMC antibiotic treatment and compared this to that of children who had not been administered any antibiotics in the previous 6 months. Further analyses were carried out in order to investigate AMC sensitivity/resistance of a large collection of bifidobacterial isolates from humans, using the MIC assay. These analyses allowed the identification of two strains belonging to Bifidobacterium breve and two isolates representing Bifidobacterium longum subsp. longum strains that were shown to exhibit a relatively high level of AMC resistance. Genome sequencing coupled with transcriptomic analyses allowed the identification of genes that appear to be responsible for this high level of AMC insensitivity. Finally, we investigated the effect of AMC and AMC-insensitive bifidobacteria on the human gut microbiota through in vitro coculture experiments, obtaining insights into the ecological role played by such bifidobacterial strains.
RESULTS AND DISCUSSION
Microbial composition of gut microbiota of children taking AMC.
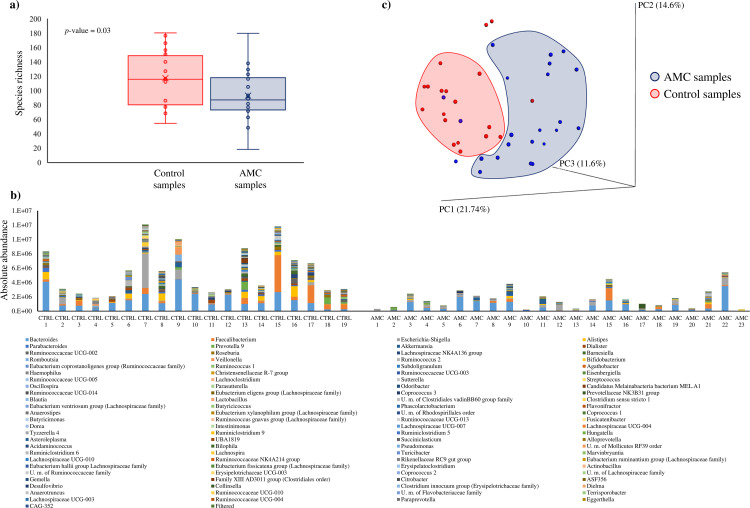
In order to assess the differences between the gut microbiota composition of 23 children undergoing AMC therapy for respiratory tract infection (AMC group) compared to that of 19 healthy, age-matched individuals (control [CTRL] group) who had not been taking any antibiotic during the preceding 6 months (see Table S1 in the supplemental material), 16S rRNA gene-based microbial profiling analyses were carried out on fecal samples as described previously (18). The analyses resulted in a total of 2,274,698 reads with an average of 54,159 ± 11,080 reads per sample. Statistical whisker plot analyses revealed a notable difference between the two groups analyzed (Fig. 1). Specifically, the 16S rRNA gene-based analysis showed a higher diversity of the CTRL group microbiota compared to that of the AMC group (P value = 0.03), demonstrating that the microbiota composition is significantly influenced by the antibiotic therapy to which individuals were subjected (Fig. 1a).
FIG 1.
Evaluation of the microbiota composition of amoxicillin-clavulanic acid (AMC) and control (CTRL) samples. (a) Whisker plot based on observed OTUs identified from AMC and CTRL samples. The x axis represents the different groups, while the y axis indicates the number of observed operational taxonomic units (OTUs). The boxes represent 50% of the data set, distributed between the first and the third quartiles. The median divides the boxes into the interquartile range, while “X” represents the mean. (b) Microbiota composition of AMC and CTRL samples based on 16S rRNA profiling normalized with a quantitative microbiome profiling approach employing flow cytometric enumeration of microbial cells for each sample. The x axis represents the different analyzed samples, while the y axis indicates normalized OTUs for each sample. (c) Beta diversity in AMC and CTRL samples. The predicted PCoA results encompassing all 43 analyzed samples are reported and the different clusters are represented by different colors.
In order to explore the absolute bacterial abundance of the analyzed fecal samples, we used a quantitative microbiome profiling approach based on flow cytometric analyses for the enumeration of microbial cells present in each fecal sample assayed. This analysis allowed the identification of the microbial load of the 42 fecal samples, which was then used to normalize the 16S rRNA sequencing data. This procedure therefore generated absolute abundance data for each profiled taxon, as previously described (19). Notably, the absolute bacterial abundances for the two groups were shown to be significantly different (average absolute bacterial abundances of 2,448,888 ± 2,875,900 and 5,438,290 ± 3,345,390 in AMC and CTRL groups, respectively; P value < 0.001), revealing that the microbial abundance of the AMC samples was significantly reduced compared to that of the CTRL group (Fig. 1b). Interestingly, these analyses revealed differences in the composition at the genus level of the AMC group compared to the CTRL group microbiota, probably due to the intensive selective pressure imposed by the antibiotic therapy to which the AMC group children had been subjected. Specifically, comparison of the gut microbiota compositions of AMC and CTRL groups based on Student’s t test and linear discriminant analysis effect size (LEfSe) statistical analysis revealed significantly higher absolute abundance of 25 genera, such as Dialister (P value = 0.0001; false-discovery rate [FDR] = 0.0001), Roseburia (P value = 0.001; FDR = 0.001) Ruminococcus 1 (P value = 0.013; FDR = 0.025), Agathobacter (P value = 0.008; FDR = 0.019), and Odoribacter (P value = 0.009; FDR = 0.02), in CTRL samples (Fig. 1b; see also Tables S2 and S3 in the supplemental material). This finding is consistent with those of other studies that had previously assessed the microbiota of individuals treated with antibiotics (20–23). Interestingly, the comparison of the gut microbiota between the AMC and the CTRL groups also showed a 1.8-fold decrease in absolute abundance of the genus Bifidobacterium, revealing a possible correlation between the use of the antibiotic and the decrease of this bacterial genus. Bifidobacteria are widely considered to represent a positive biomarker for a healthy gut status and are characteristic of the infant’s microbiota (13, 24–26). Although no statistically significant differences were observed, the decrease in members of this genus is consistent with previous data that also showed a decline in bifidobacterial abundance following antibiotic exposure (5, 6).
In order to evaluate the interindividual differences between AMC and CTRL samples, we analyzed the beta diversity based on the weighted UniFrac distance metric and represented the obtained results through principal coordinate analysis (PCoA) (Fig. 1c). Interestingly, the PCoA analysis showed that most of the samples were grouped as two different clusters corresponding to AMC or CTRL samples, respectively, thus underlining the distinct microbiota composition of individuals that had or had not been treated with AMC (Fig. 1c). Moreover, a permutational analysis of variance (PERMANOVA) statistical analysis on the PCoA data revealed a significant division between the two analyzed groups (P value = 0.001, pseudo-F = 4.91, adonis P value = 0.001, R2 = 0.11). Furthermore, we performed a PCoA based on the host age (see Fig. S1 in the supplemental material), which revealed that there was no correlation between host age and gut microbiota profile (PERMANOVA P value = 0.541, pseudo-F = 0.97, adonis P value = 0.06, R2 = 0.04). These findings corroborate the fact that microbiota profiles differ considerably between samples, yet in the case of children that had been treated with antibiotics it was clear that such samples had generally suffered from bacterial depletion compared to control samples, underlining the expected detrimental effect of AMC on the microbiota composition.
Isolation of novel bifidobacterial strains and assessment of AMC susceptibility.
As previously reported, bifidobacterial species are commonly present in the infant microbiota and are generally considered to represent a positive microbial biomarker for gut health (13, 24–26). The possible correlation between the use of the antibiotic and the decrease in abundance of the Bifidobacterium genus (see above) prompted us to investigate the AMC sensitivity of the species belonging to this genus. For the purpose of evaluating the AMC sensitivity of different members of the Bifidobacterium genus, we performed MIC assays involving 237 different strains, including 63 type strains of currently recognized bifidobacterial (sub)species and 174 different strains belonging to four Bifidobacterium species, i.e., Bifidobacterium adolescentis, Bifidobacterium bifidum, Bifidobacterium breve, and Bifidobacterium longum (see Table S4 in the supplemental material), representing common colonizers of the human gut (27–31). Moreover, in order to isolate bifidobacterial strains with AMC insensitivity, we applied a bifidobacterial isolation protocol on fecal samples belonging to individuals subjected to AMC therapy (Table S4). The analysis (32, 33) described in Materials and Methods allowed the isolation of 24 novel strains (Table 1), of which 18 were shown to belong to the B. longum species, three to the B. breve species, and three to the B. pseudocatenulatum species. All tested strains showed a unimodal distribution of AMC MIC (MICAMC) breakpoint values, ranging from 0.125 μg/ml to 32 μg/ml (see Fig. S2 and Table S4 in the supplemental material). Of the strains, 96.5% exhibited a MIC of ≤1 μg/ml, five strains displayed a breakpoint value equal to 2 μg/ml, and, notably, four bifidobacterial strains showed a MIC equal to or higher than 4 μg/ml (Fig. S2 and Table S4). Interestingly, the newly isolated strain B. breve PRL2020 displayed the highest MICAMC value, i.e., 32 μg/ml (Table 1), which was 11-fold higher than the average bifidobacterial MICAMC. Moreover, B. breve strain M1D showed a MIC value of 16 μg/ml, which was 5-fold higher than the Bifidobacterium MIC average (Table 1). This antibiotic insensitivity is unusual for B. breve species, which in general exhibit a high sensitivity to various antibiotics, including AMC (34). Furthermore, two isolates belonging to the B. longum species, i.e., B. longum subsp. longum 1898B and B. longum subsp. longum 39B, were shown to exhibit MIC values of 8 μg/ml and 4 μg/ml (Table 1), 8- and 4-fold higher, respectively, than the average MIC value of other B. longum strains. These findings suggest that different levels of resistance/insensitivity to AMC reflect strain-specific characteristics rather than being a species-specific feature. Notably, the MICAMC breakpoints identified in 98.5% of the bifidobacterial strains tested were very low compared to MICAMC values previously identified for other members of the human gut microbiota, such as Escherichia coli, Citrobacter spp., Bacteroides spp., and Parabacteroides spp., which showed MICAMC breakpoint values higher than 8 μg/ml (35, 36). Interestingly, these data indicate that AMC resistance/insensitivity in bifidobacteria does not appear to follow a vertical route of evolution but may have been acquired through horizontal gene transfer (HGT), in a similar way to that described for other gut commensal microorganisms (37). Finally, we evaluated the susceptibility to the antibiotic amoxicillin alone for the four strains of Bifidobacterium insensitive to AMC. Interestingly, each strain showed higher MIC values, i.e., B. breve PRL2020 showed a MIC value of 64 μg/ml, B. breve strain M1D exhibited a MIC breakpoint of 32 μg/ml, and B. longum subsp. longum 1898B and B. longum subsp. longum 39B presented MIC values of 8 μg/ml. These data confirm the resistance of these strains to amoxicillin, both as the sole antibiotic and in combination with clavulanic acid.
TABLE 1.
Bifidobacterium strains isolated from AMC samples and corresponding MICAMC values
| Species | Strain | MICAMC (μg/ml) |
|---|---|---|
| Bifidobacterium breve | PRL2020 | 32 |
| Bifidobacterium breve | M1D | 16 |
| Bifidobacterium breve | M5B | 1 |
| Bifidobacterium longum subsp. longum | 1898B | 8 |
| Bifidobacterium longum | 2195B | 1 |
| Bifidobacterium longum | 2196B | 1 |
| Bifidobacterium longum | 2197B | 1 |
| Bifidobacterium longum | 2198B | 0.5 |
| Bifidobacterium longum | 2199B | 1 |
| Bifidobacterium longum | 2202B | 0.125 |
| Bifidobacterium longum subsp. longum | 39B | 4 |
| Bifidobacterium longum | AD12C | 1 |
| Bifidobacterium longum | E2C | 1 |
| Bifidobacterium longum | E4F | 2 |
| Bifidobacterium longum | E6H | 1 |
| Bifidobacterium longum | F2A | 0.25 |
| Bifidobacterium longum | G7G | 0.25 |
| Bifidobacterium longum | G8F | 0.5 |
| Bifidobacterium longum | L5G | 2 |
| Bifidobacterium longum | MISS1F | 0.25 |
| Bifidobacterium longum | T3 | 2 |
| Bifidobacterium pseudocatenulatum | AN3D | 2 |
| Bifidobacterium pseudocatenulatum | L3G | 0.5 |
| Bifidobacterium pseudocatenulatum | M8H | 2 |
Comparative genomics and identification of putative resistance genes in AMC-insensitive Bifidobacterium strains.
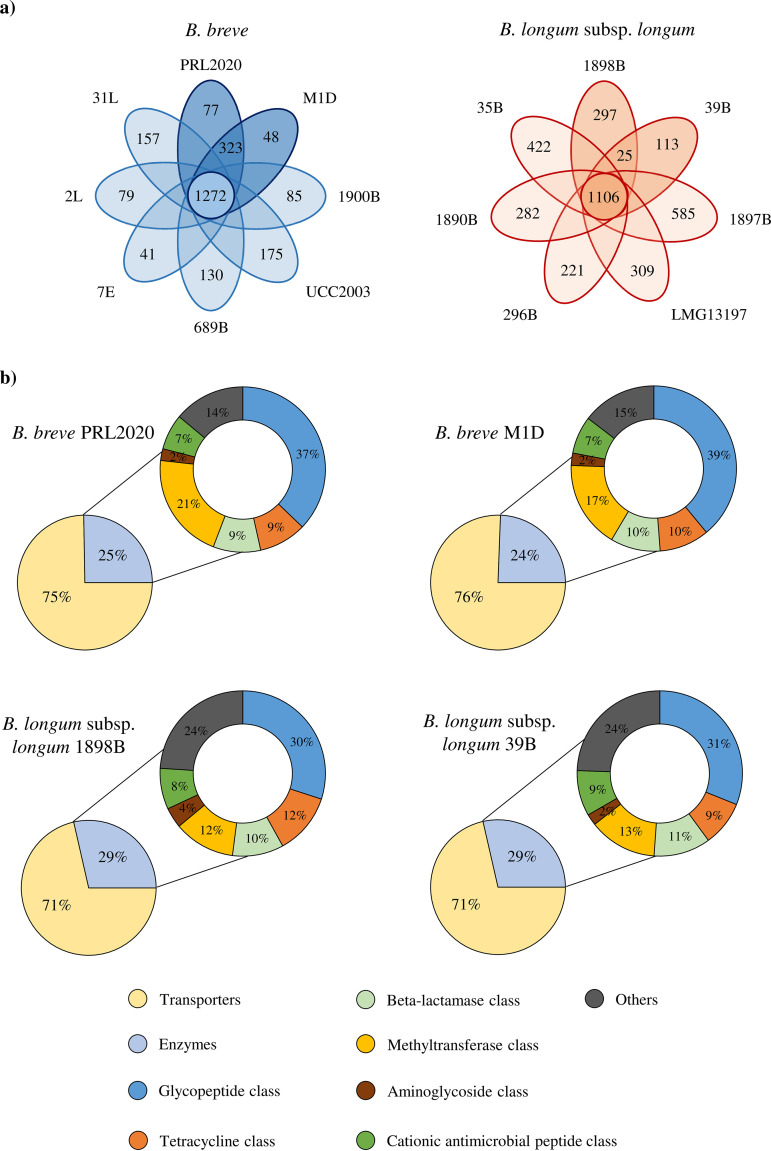
In order to identify the genetic features that are involved in AMC insensitivity in the four identified bifidobacterial strains, we sequenced, annotated and in silico analyzed the genomes of B. breve PRL2020, B. breve M1D, B. longum subsp. longum 39B, and B. longum subsp. longum 1898B, which were shown to elicit the highest MICAMC values (Table 1). The coverage depth of these four newly isolated Bifidobacterium chromosomes ranged from 72- to 388-fold, which upon assembly generated 1 to 25 contigs (Table 2). The number of predicted open reading frames (ORFs) ranged from 1,842 for B. longum subsp. longum 39B to 2,102 for B. breve PRL2020 (Table 2). We then evaluated the genetic similarities of B. breve PRL2020, B. breve M1D, B. longum subsp. longum 39B, and B. longum subsp. longum 1898B to other chromosome sequences that are publicly available for B. breve and B. longum subsp. longum through a comparative genomics analysis. Specifically, the genome sequences of B. breve PRL2020, B. breve M1D, B. longum subsp. longum 39B, and B. longum subsp. longum 1898B were compared with the chromosome sequences of six B. breve and five B. longum subsp. longum strains, including AMC-sensitive strains (see Table S5 in the supplemental material). In silico analysis revealed that 1,272 genes are commonly shared among the analyzed B. breve strains, representing the core genome of this taxon (Fig. 2a), whereas the core genome of the assessed B. longum subsp. longum strains was shown to be composed of 1,106 genes. Moreover, there are variable numbers of truly unique genes (TUGs) in each of the two investigated species, ranging between 175 for B. breve UCC2003 and 41 for B. breve 7E, and ranging from 585 for B. longum subsp. longum 1897B and 113 for B. longum subsp. longum 39B (Fig. 2a and Table S5). Interestingly, this analysis revealed the presence of 323 accessory genes shared between B. breve AMC-insensitive strains, i.e., B. breve PRL2020 and B. breve M1D, and absent in AMC-sensitive B. breve strains (Fig. 2a). Furthermore, the genomes of the AMC-resistant strains B. longum subsp. longum 39B and B. longum subsp. longum 1898B were shown to contain 25 genes that are shared between them but are absent in other analyzed B. longum strains (Fig. 2a). In addition, the genomes of the four identified AMC-insensitive Bifidobacterium strains were screened to identify putative antibiotic resistance (AR) genes, using the MEGARes database (38). This in silico analysis showed that the predicted resistome of B. breve PRL2020, B. breve M1D, B. longum subsp. longum 39B, and B. longum subsp. longum 1898B ranged from 167 genes in the case of B. breve M1D to 175 genes for B. longum subsp. longum 1898B (Fig. 2b). We included in our analyses both sequences coding for antibiotic-removing transporters and sequences specifying antibiotic-inactivating enzymes. The number of predicted antibiotic-removing transporters ranged from 109 for B. longum subsp. longum 39B to 127 genes for B. breve PRL2020 and B. breve M1D (Fig. 2b). Interestingly, the analyzed strains were shown to harbor genes putatively encoding beta-lactamases as part of their resistome; B. longum strains were predicted to encode five distinct beta-lactamases, whereas the assessed B. breve strains were predicted to specify four beta-lactamases. However, since these putative beta-lactamases are conserved among different strains of the same species, it appears that they are not involved in the observed AMC insensitivity phenotype for some strains.
TABLE 2.
Genetic features of AMC-resistant Bifidobacterium strains
| Strain | Genome length (bp) | No. of contigs | No. of ORFs | No. of tRNAs | No. of rRNA loci | Accession no. |
|---|---|---|---|---|---|---|
| B. breve PRL2020 | 2,427,222 | 6 | 2,102 | 54 | 3 | JACZEM000000000 |
| B. breve M1D | 2,421,612 | 7 | 2,053 | 54 | 3 | JACZEL000000000 |
| B. longum subsp. longum 1898B | 2,464,874 | 9 | 2,065 | 56 | 4 | JACZEK000000000 |
| B. longum subsp. longum 39B | 2,287,000 | 25 | 1,842 | 56 | 1 | JACZEJ000000000 |
FIG 2.
Comparative genomic and resistome analyses of AMC-resistant Bifidobacterium strains. (a) Venn diagrams. The numbers in the central circles represent the numbers of genes in the core genomes of the B. breve and B. longum subsp. longum strains analyzed. The numbers in the overlapping sections represent the numbers of shared genes between AMC-resistant strains, while the numbers in the ovals depict the number of TUGs for each strain. (b) Predicted resistomes of the AMC-resistant Bifidobacterium strains.
Moreover, further analysis of the in silico resistome data and comparison between genes found in the insensitive strains but absent from the sensitive strains allowed the identification of 11 predicted AR genes that are present only in the B. breve AMC-insensitive strains (see Table S6 in the supplemental material). Eight of these identified genes were predicted to encode transporters, whereas two genes seemed to encode enzymes that provide protection against glycopeptide antibiotics (39). Finally, one gene was predicted to encode a glycosyl hydrolase involved in the modification of the lipopolysaccharide (LPS) core and lipid A region with ethanolamine and addition of amino-arabinose to the 4′ phosphate of lipid A (40). Furthermore, carrying out the same analysis for members of the B. longum subsp. longum taxon, we identified just one gene uniquely shared between AMC-resistant B. longum subsp. longum strains, predicted to encode a putative AR transporter (Table S6). These identified genes are indeed credible genetic candidates responsible for the high MICAMC breakpoint values identified in B. breve PRL2020, B. breve M1D, B. longum subsp. longum 39B, and B. longum subsp. longum 1898B.
Transcriptomic analysis of Bifidobacterium breve PRL2020.
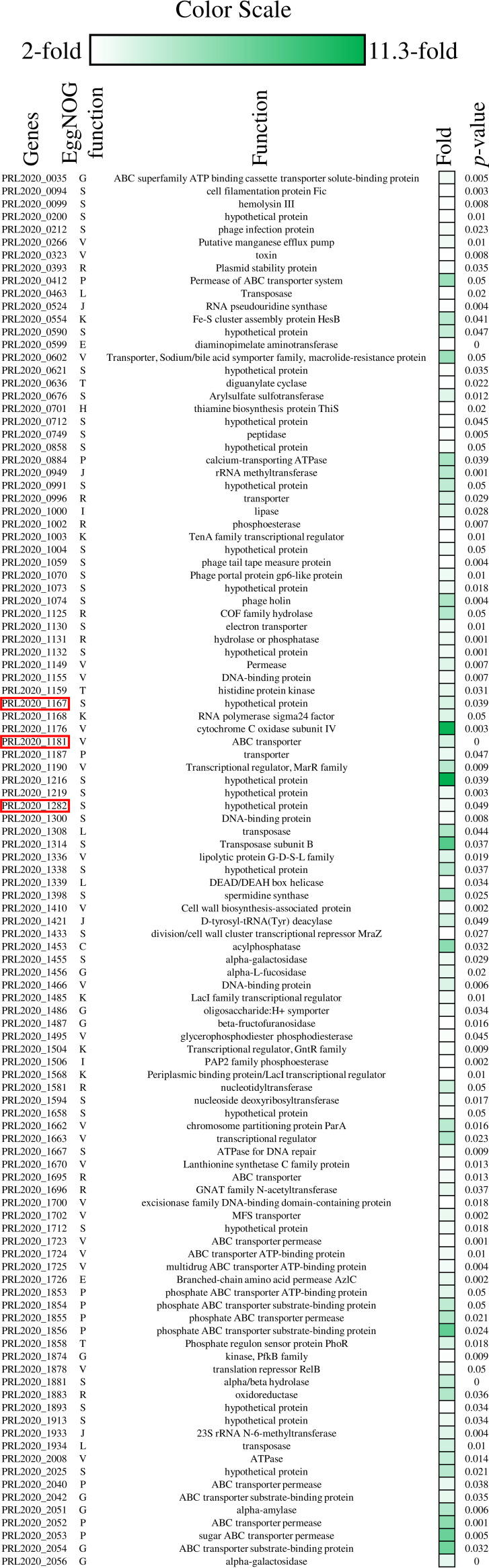
In order to investigate if and how the presence of AMC in the growth medium modulates the transcription of genes putatively involved in the AR described above, we explored the transcriptome of B. breve PRL2020 when cultivated in the presence or absence of AMC in De Man, Rogosa, and Sharpe (MRS) medium. This analysis showed significant modulation of 163 genes (>2-fold induction; P value ≤ 0.05), 109 of which were upregulated in the MRSAMC medium, while 52 genes were downregulated compared to the reference condition (Fig. 3). Functional assignment of the upregulated and downregulated transcribed genes based on the eggNOG database (41) revealed that the overexpressed transcripts corresponded to genes involved in defensive mechanisms and in the metabolism and transport of inorganic ions (20.2% and 9.2%, respectively) (Fig. 3). Conversely, downregulated transcripts corresponded to genes involved in the transport and metabolism of carbohydrates and nucleotides (13.7% and 25.5%, respectively). Interestingly, among the upregulated genes, one particular ORF, associated with locus tag PRL2020_1181 and encoding a predicted ABC transporter, exhibited 3.6-fold-increased transcription (P value = 0.0001; FDR = 0.002) (Fig. 3). This gene had been identified in the resistome of strain B. breve PRL2020, being shared with other B. breve AMC-resistant strains but not present in B. breve AMC-sensitive strains (Table S6). These findings suggest that this ABC transporter-encoding gene is involved in the observed insensitivity of B. breve PRL2020 to the AMC antibiotic. In addition, two TUGs observed in the genome of strain B. breve PRL2020, i.e., ORFs PRL2020_1167 and PRL2020_1282, were shown to be transcriptionally induced in the presence of AMC, exhibiting an upregulation of 3.4-fold (P value = 0.039; FDR = 0.04) and 2.7-fold (P value = 0.049; FDR = 0.044), respectively (Fig. 3), and in both cases encoding hypothetical proteins. These data indicate that these unique genes are involved in the high resistance of B. breve PRL2020 to AMC antibiotic.
FIG 3.
Transcriptional modulation of B. breve PRL2020 genes in the presence of AMC. The heatmap displays the subset of significantly upregulated encoding genes. Red boxes highlight the ORFs PRL2020_1181, PRL2020_1167, and PRL2020_1282, which are putatively involved in the AMC resistance mechanism. The color scale (green) at the top of the figure indicates increased transcription levels compared to those of the reference samples. The EggNOG letter for each significant upregulated gene is reported. Each letter stands for a function, as follows: S, function unknown; L, replication, recombination, and repair; R, general function prediction only; G, carbohydrate transport and metabolism; J, translation, ribosomal structure, and biogenesis; E, amino acid transport and metabolism; K, transcription; P, inorganic ion transport and metabolism; C, energy production and conversion; V, defense mechanisms; T, signal transduction mechanisms; H, coenzyme transport and metabolism; and I, lipid transport and metabolism.
Effect of resistant Bifidobacterium strains on the human gut microbiota in the presence of AMC.
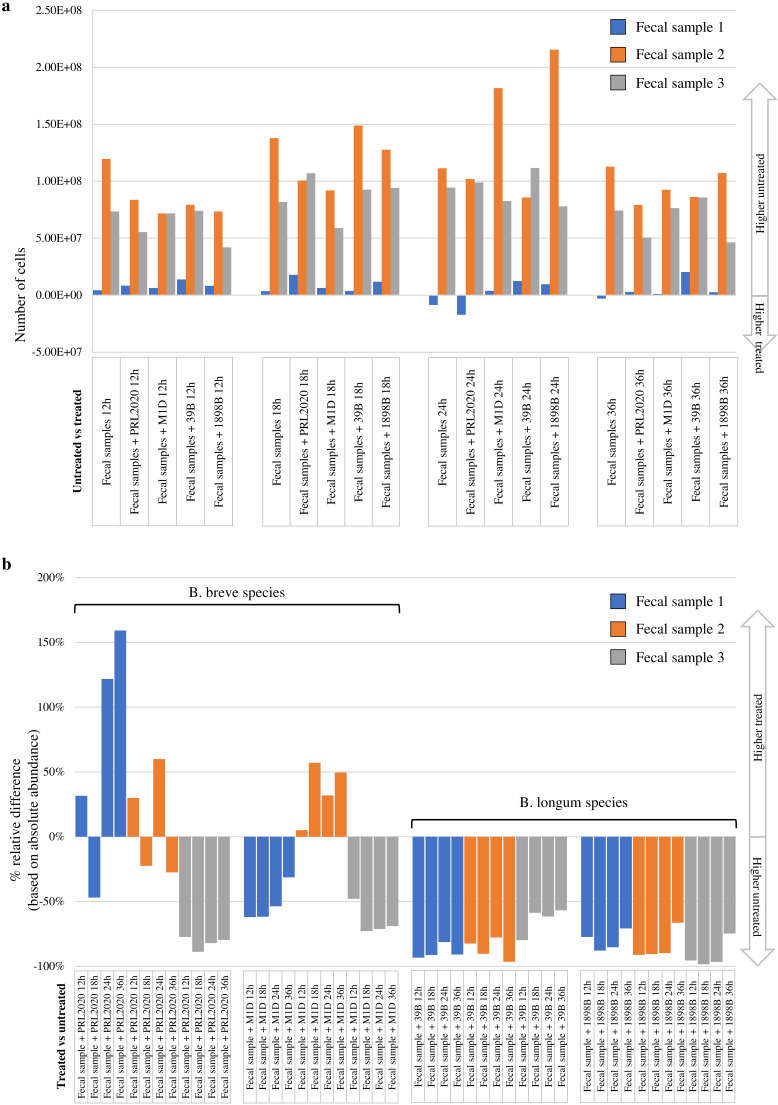
The GIT microbiota composition is known to be influenced by antibiotic compounds (4, 5). In particular, antibiotic exposure will alter the microbial composition in the infant gut community (6, 7). In order to evaluate the effects of AMC on the human gut microbiota in the presence of AMC-insensitive Bifidobacterium strains retrieved in this study, three different fresh infant fecal samples were used separately to perform coculture experiments. In detail, each fecal sample was used to test four different coculture conditions for each AMC-insensitive strain, i.e., (i) fecal sample, (ii) fecal sample with AMC-insensitive strain, (iii) fecal sample with AMC, and (iv) fecal sample with AMC and AMC-insensitive strain (see Materials and Methods) (4). The cocultures were monitored at four different time points, i.e., 12 h, 18 h, 24 h, and 36 h after the addition of inoculum. For each time point and for each cultivation condition tested, the changes in the microbiota composition were assessed by shallow shotgun metagenomics analysis. The analyses resulted in a total of 4,659,664 reads, with an average of 38,831 ± 21,806 reads per sample (see Table S7 in the supplemental material). In order to obtain a comprehensive biological interpretation of the analyzed batch culture microbiome complexity, we performed a quantitative microbiome profiling experiment based on flow cytometric analyses for the enumeration of microbial cells present in each coculture condition at each time point (19). Interestingly, comparison of each coculture experiment with its own control revealed a decrease of the number of microbial cells in 95% of the samples to which AMC was added (Fig. 4a; see also Table S8 in the supplemental material). Moreover, statistical analysis revealed a significant decrease (P value = 0.0001) of 1.94-fold in the number of cells in the AMC-treated group (average of 6.85 × 107 ± 4.32 × 107) compared to the control sample group (average of 1.33 × 108 ± 8.21 × 107).
FIG 4.
Growth experiments in fecal medium. (a) Comparison of the absolute abundances of the coculture experiments in MRSAMC and the reference condition. (b) Relative percentage difference between AMC-treated and untreated samples based on the absolute abundance of the bacterial species corresponding to the strain analyzed (B. breve for PRL2020 and M1D strains and B. longum for 39B and 1898B strains).
Focusing on the absolute abundance of the species corresponding to the identified AMC-resistant Bifidobacterium strains, the strains that seemed to increase in abundance following the AMC treatment were B. breve PRL2020 and B. breve M1D (Fig. 4b and Table S8). In fact, the fecal samples treated with AMC and cocultivated with PRL2020 or M1D showed an increase of abundance of B. breve species in 42% and 33% of the samples, respectively, compared to each untreated condition (Fig. 4b). Moreover, the B. breve PRL2020 strain displayed the greatest AMC resistance, with a decrease in abundance of <50% in only 33% of the samples, while B. breve M1D revealed a decrease in abundance of <50% in 50% of the samples (Fig. 4b). In contrast, B. longum subsp. longum 39B and B. longum subsp. longum 1898B showed a decrease in absolute abundance always greater than 50% (Fig. 4b). Furthermore, alignment analysis based on Bowtie 2 software revealed that all these reads classified as B. breve or B. longum align with each AMC-resistant Bifidobacterium strain’s reference genome (alignment identity of 99%). Therefore, these results may support the apparent improved ability of B. breve PRL2020 to grow in the presence of AMC antibiotic, making this strain an interesting candidate for the development of Bifidobacterium-containing probiotic products. Certainly, further in vitro experiments aimed at evaluating the ability of the strain to survive gastrointestinal passage and to achieve a high level of viability during industrial production and storage could promote the B. breve PRL 2020 strain as a putative novel health-promoting bacterium.
Conclusions.
Many pharmaceuticals are well known to influence the human gut microbiota. Since the combination of amoxicillin and clavulanic acid is one of the most frequently prescribed antibiotics in the Western world, especially during infancy and adolescence, we decided to explore the impact of this antibiotic formulation on the gut microbiota of 23 children and compare this to that of a control group who had not been treated with AMC. Interestingly, results indicated a drastic reduction of bacteria in AMC-treated children compared to the CTRL group, including a reduction in absolute abundance of the Bifidobacterium genus. We decided to perform MIC experiments with several strains belonging to our collection and novel strains that had been isolated from children treated with AMC. The determination of sensitivity/resistance of these intestinal Bifidobacterium strains showed that 98.5% of them are sensitive to this antibiotic. We isolated four strains that showed higher resistance, of which B. breve strain PRL2020 displayed the highest MICAMC value. We identified gene candidates responsible for the higher resistance of certain bifidobacterial strains to AMC compared to that of the majority of bifidobacteria, which appear to be highly sensitive to these antimicrobial molecules. In a follow-up study, we will further investigate the genetic determinants and molecular mechanism of the observed AMC resistance by applying gene inactivation protocols and/or the heterologous expression of the predicted AMC resistance genes in sensitive bifidobacterial strains. Finally, by simulating gut microbiota experiments it emerged that the PRL2020 strain is able to survive in the presence of a complex microbiota combined with AMC antibiotic, opening up the possibility, after verifying that horizontal gene transfer of the AR genes is unlikely to take place, of using this strain in a probiotic product when AMC therapy is prescribed.
MATERIALS AND METHODS
Sample collection.
For the purpose of this study, a total of 42 human fecal samples were collected and divided into two groups; the first one was represented by fecal samples obtained from 23 children who were undergoing AMC (amoxicillin:clavulanic acid ratio of 7:1) treatment from 7 to 10 days (average of 8.9 ± 1.3 days) due to respiratory tract infections, while the second group was represented by fecal samples from 19 healthy children (see Table S1 in the supplemental material). Collected samples, which consisted of approximately 10 g of fresh fecal material, were kept on ice, shipped under subzero conditions to the laboratory, and stored at −80°C until further processing.
Bacterial DNA extraction and 16S rRNA gene PCR amplification and sequencing.
Stool samples were subjected to DNA extraction using the QIAamp DNA stool minikit following the manufacturer’s instructions (Qiagen). Partial 16S rRNA gene sequences were amplified from extracted DNA using the primer pair Probio_Uni/Probio_Rev, targeting the V3 region of the 16S rRNA gene sequence (18). Illumina adapter overhang nucleotide sequences were added to these partial 16S rRNA gene-specific amplicons, which were further processed employing the 16S metagenomic sequencing library preparation protocol (part no. 15044223 rev. B; Illumina). Amplifications were carried out using a Verity thermocycler (Applied Biosystems). The integrity of the PCR amplicons was analyzed by electrophoresis on a 2200 TapeStation instrument (Agilent Technologies, USA). DNA products obtained following PCR-mediated amplification of the 16S rRNA gene sequences were purified by a magnetic purification step involving Agencourt AMPure XP DNA purification beads (Beckman Coulter Genomics GmbH, Bernried, Germany) in order to remove primer dimers. The DNA concentration of the amplified sequence library was determined by a fluorimetric Qubit quantification system (Life Technologies, USA). Amplicons were diluted to a concentration of 4 nM, and 5-μl quantities of each diluted DNA amplicon sample were mixed to prepare the pooled final library. Sequencing was performed using an Illumina MiSeq sequencer with MiSeq reagent kit v3 chemicals. Following sequencing, the FASTQ files were processed using a custom script based on the QIIME software suite (42). Paired-end read pairs were assembled to reconstruct the complete Probio_Uni/Probio_Rev amplicons. Quality control retained sequences with a length between 140 and 400 bp and a mean sequence quality score of >20, while sequences with homopolymers of >7 bp and mismatched primers were omitted. In order to calculate downstream diversity measures (alpha and beta diversity indices and UniFrac analysis), 16S rRNA operational taxonomic units (OTUs) were defined at 100% sequence homology using DADA2 (43); OTUs not encompassing at least 2 sequences derived from the same sample were removed. Notably, this approach allows highly distinctive taxonomic classification at single-nucleotide accuracy (43). All reads were classified to the lowest possible taxonomic rank using QIIME2 (42, 44) and a reference data set from the SILVA database. Biodiversity within a given sample (alpha diversity) was calculated based on the observed OTU index. Similarities between samples (beta diversity) were calculated by weighted UniFrac (45). The range of similarities is calculated between values 0 and 1. PCoA representations of beta diversity were performed using QIIME2 (42, 44).
Evaluation of cell density by flow cytometry assay.
For bacterial cell counting, 0.2 g of fecal sample was diluted in a physiological solution (phosphate-buffered saline [PBS]). Subsequently, bacterial cells were stained with 1 μl SYBR green I and incubated in the dark for at least 15 min before measurement. All count experiments were performed in triplicate using an Attune NxT flow cytometer (Invitrogen, Thermo Fisher Scientific) equipped with a blue laser set at 50 mW and tuned to an excitation wavelength of 488 nm. Multiparametric analyses were performed on both scattering signals (forward scatter [FSC] and side scatter [SSC]) and SYBR green I fluorescence was detected on the FL1 channel. Cell debris and eukaryotic cells were excluded from acquisition analysis by a sample-specific FL1 threshold. All data sets were statistically analyzed with Attune NxT flow cytometer software. Utilizing these cell counts to normalize the sequencing data into absolute abundance of each profiled taxon, we were able to perform quantitative microbiome profiling using a previously described method (19).
Isolation of novel Bifidobacterium strains.
In order to explore the AMC resistance of the Bifidobacterium genus, 24 novel strains were isolated from fecal samples of individuals that had been treated with AMC for various numbers of days (Table 1). One gram of feces from each collected fecal sample was mixed with 9 ml of PBS (pH 6.5). Serial dilutions and subsequent plating were performed using MRS agar supplemented with 50 μg/ml mupirocin (Delchimica, Italy), 0.05% (wt/vol) l-cysteine hydrochloride, and 8 μg/ml of AMC (Merck, Germany). Morphologically distinct colonies that developed on MRS plates were randomly picked and restreaked in order to isolate purified bacterial strains. All novel isolates were subjected to DNA isolation and characterized as previously described (46). The Bifidobacterium strains isolated in this study are listed in Table 1.
Amoxicillin-clavulanic acid susceptibility tests.
The MIC breakpoints (in μg/ml) of AMC were determined using the broth microdilution method (MDIL) (47). Microplates were incubated under anaerobic conditions for 48 h at 37°C. Cell density was monitored by optical density measurements at 600 nm (OD600) using a plate reader (BioTek, VT, USA). Furthermore, the same MIC analysis was performed for the antibiotic amoxicillin alone (European Standard, Merck, Germany). The MIC breakpoint represents the highest concentration of a given antibiotic to which a particular bacterial strain was shown to be resistant.
Genome sequencing and assemblies.
DNA samples extracted from AMC-resistant bifidobacterial isolates were subjected to genome sequencing using the MiSeq platform (Illumina, UK) at GenProbio srl (Parma, Italy) according to the supplier’s protocol (Illumina, UK). FASTQ files of the paired-end reads obtained from targeted genome sequencing of isolated strains were utilized as input for genome assemblies through the MEGAnnotator pipeline (48). SPAdes software was used for de novo assembly of each Bifidobacterium genome sequence (49, 50), while protein-encoding ORFs were predicted using Prodigal (51).
Comparative genomics.
In order to identify unique protein families encoded by newly isolated AMC-resistant Bifidobacterium strains, a PGAP analysis was performed (52). B. breve PRL2020 and B. breve M1D genomes were analyzed with six other B. breve genomes tested for AMC resistance (Table S5). Simultaneously, B. longum subsp. longum 39B and B. longum subsp. longum 1898B genomes were compared with five B. longum subsp. longum genomes used in the MICAMC analyses (Table S5). Each predicted proteome of a given Bifidobacterium genome was screened for orthologues against the proteome of every Bifidobacterium strain belonging to the same species by means of BLAST analyses (53) (E value cutoff of <1 × 10−4 and 50% identity across at least 80% of both protein sequences). Protein families shared between analyzed genomes allowed us to identify the core genomes of B. breve and B. longum subsp. longum.
Functional annotation of each protein of B. breve PRL2020 was performed employing the eggNOG database (41).
Prediction of antibiotic resistance genes.
The in silico proteomes of four Bifidobacterium genomes isolated in this study were screened for proteins with similarity to antibiotic resistance proteins acting through inactivation and/or removal of antibiotic molecules. The screening was carried out using the MEGARes database through BLASTP analysis (E value cutoff of 1 × 10−5) (38, 53, 54). The core database was obtained by nonredundant compilation of sequences contained in ResFinder, ARG-ANNOT, the Comprehensive Antibiotic Resistance Database (CARD), and the NCBI Lahey Clinic beta-lactamase archive (55–58). Following this, a manual examination of sequences with an E value below 1 × 10−5 was performed in order to detect distant homologs.
RNA extraction.
Aliquots of B. breve PRL2020 cells were grown to an optical density at 600 nm ranging from 0.6 to 0.8. The experiment was conducted in three biological replicates. Total RNA was isolated from bifidobacterial cultures grown in MRS or MRS with 32 μg/ml of AMC. RNA extraction was performed as previously described (59). Briefly, cultures were centrifuged at 4,000 rpm for 10 min at 4°C. Cells pellets were treated with TES buffer (Tris-EDTA-sodium dodecyl sulfate buffer) and lysozyme, followed by resuspension in 1 ml of QIAzol lysis reagent (Qiagen, UK), and placed in a sterile tube containing glass beads (Merck, Germany). The cells were lysed by shaking the mix on a Mini-BeadBeater-24 (BioSpec Products, USA) for 2 min followed by 2 min of static cooling; this step was repeated three times. The lysate was centrifuged at 12,000 × g for 15 min, and the upper phase was recovered. RNA samples were washed from proteins by means of chloroform, and, finally, samples were purified by means of the RNeasy minikit (Qiagen, UK), following the manufacturer’s instructions. Quality and integrity of the RNA were checked by 2200 TapeStation (Agilent Technologies, USA) analysis. RNA concentration was then determined by a fluorimetric Qubit quantification system (Life Technologies).
Transcriptome sequencing (RNAseq) analysis performed by the MiSeq Illumina platform.
Total RNA (1 μg) was treated by the TruSeq stranded total RNA protocol (Illumina, Inc., San Diego, CA), following the manufacturer’s instructions. Ribosomal depletion is included in the protocol. Quality and quantity of libraries were checked by 2200 TapeStation (Agilent Technologies) analysis and a fluorimetric Qubit quantification system (Life Technologies), respectively. Samples were loaded into a flow cell from the MiSeq v3 kit (600 cycles; Illumina) as instructed by the technical support guide. The reads were depleted of adapters, quality filtered (with overall quality, quality window, and length filters), and aligned to the Bifidobacterium reference genomes through Bowtie 2 software (60). Reads per kilobase per million (RPKM) values were evaluated by means of Artemis software (61).
In vitro simulation of the effect of AMC and Bifidobacterium strains on the human gut microbiota.
The effect of AMC antibiotic and four selected AMC-insensitive Bifidobacterium strains on the human gut microbiota was evaluated in vitro through anaerobic, pH- and temperature-controlled batch cultures. We evaluated four different conditions, and the growth medium used was based on the fecal medium described by Macfarlane et al. (62). Each batch culture condition was performed using three different fresh fecal samples from children (aged 4 years) who had not undergone antibiotic treatment for at least 3 months prior to sample collection and who had not consumed probiotic bacteria. The fresh fecal sample was previously analyzed in order to confirm the absence of B. breve and B. longum species DNA. We tested four different growth conditions for each strain. Briefly, the first condition consisted of 40 ml of growth medium supplemented with AMC (20 μM) in which an overnight culture of a particular Bifidobacterium strain and the fresh fecal sample were inoculated, each at 1% (vol/vol and wt/vol, respectively). The concentration of AMC used was based on another study in which different antibiotic and nonantibiotic compounds were tested for their impact on the gut microbiota (4). The second condition consisted of 40 ml of fecal medium inoculated with 1% (vol/vol) of an overnight culture of a Bifidobacterium strain and 1% (wt/vol) of the fresh fecal sample. The third batch culture was made of 40 ml of fecal medium supplemented with AMC at 20 μM and in which the fecal sample was inoculated at 1% (wt/vol). Finally, the fourth condition was composed of 40 ml of growth medium in which only the fecal sample was inoculated at 1% (wt/vol). For each experiment, an aliquot of culture was taken at four different time points, namely, 12 h, 18 h, 24 h, and 36 h after inoculum addition. Each aliquot was subjected to DNA extraction using the QIAamp DNA stool minikit following the manufacturer’s instructions (Qiagen, UK) for sequencing library preparation.
Shallow shotgun metagenomics and evaluation of cell density by flow cytometry assay of coculture experiments.
Extracted DNA was prepared for sequencing purposes following the Illumina Nextera XT protocol. Briefly, DNA samples were enzymatically fragmented, barcoded, and purified using magnetic beads. Then, samples were quantified using the fluorometric Qubit quantification system (Life Technologies), loaded on a 2200 TapeStation instrument (Agilent Technologies, USA), and normalized to 4 nM. Single-end sequencing was performed using an Illumina MiSeq sequencer with a flow cell from the MiSeq v3 kit (600 cycles; Illumina). For bacterial cell counting, the batch cultures were diluted in PBS physiological solution. Subsequently, bacterial cells were stained with 1 μl SYBR green I and incubated in the dark for at least 15 min before measurement. All count experiments were performed in triplicate as described previously (see above). All data sets were statistically analyzed with Attune NxT flow cytometer software. Utilizing these cell counts to normalize the sequencing data into absolute abundance of each profiled taxa, we were able to perform quantitative microbiome profiling using a previously described method (19).
Statistical analysis.
SPSS software (IBM, Italy) was used to perform statistical analysis between the AMC group and the healthy group, as well as to apply Student’s t test to the RNAseq. PCoA statistical analysis was performed using PERMANOVA and adonis tests.
Data availability.
Raw sequences of the 16S profiling experiments, shallow shotgun metagenomics experiments, and RNAseq experiments are accessible under BioProject accession number PRJNA663786. Newly isolated Bifidobacterium genomes were sequenced and deposited in DDBJ/ENA/GenBank under the accession numbers reported in Table 2.
Supplementary Material
ACKNOWLEDGMENTS
We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. D.V.S. is a member of APC Microbiome Ireland, which is funded by SFI through the Irish Government’s National Development Plan (grant numbers SFI/12/RC/2273-P1 and SFI/12/RC/2273-P2).
Part of this research was conducted using the high-performance computing (HPC) facility of the University of Parma.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Young VB. 2012. The intestinal microbiota in health and disease. Curr Opin Gastroenterol 28:63–69. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A. 2018. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francino MP. 2015. Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Front Microbiol 6:1543. doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, Shirakawa T, Sonomoto K, Nakayama J. 2009. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol 56:80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 7.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. 2012. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goossens H, Ferech M, Vander Stichele R, Elseviers M, ESAC Project Group. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579–587. doi: 10.1016/S0140-6736(05)70799-6. [DOI] [PubMed] [Google Scholar]
- 9.Klein JO. 2003. Amoxicillin/clavulanate for infections in infants and children: past, present and future. Pediatr Infect Dis J 22:S139–S148. doi: 10.1097/00006454-200308001-00005. [DOI] [PubMed] [Google Scholar]
- 10.de Muinck EJ, Trosvik P. 2018. Individuality and convergence of the infant gut microbiota during the first year of life. Nat Commun 9:2233. doi: 10.1038/s41467-018-04641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, van Sinderen D, Ventura M. 2018. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci 75:103–118. doi: 10.1007/s00018-017-2672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker WA. 2013. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab 63(Suppl 2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 13.Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. 2018. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol 26:339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Delgado S, Florez AB, Mayo B. 2005. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr Microbiol 50:202–207. doi: 10.1007/s00284-004-4431-3. [DOI] [PubMed] [Google Scholar]
- 15.Masco L, Van Hoorde K, De Brandt E, Swings J, Huys G. 2006. Antimicrobial susceptibility of Bifidobacterium strains from humans, animals and probiotic products. J Antimicrob Chemother 58:85–94. doi: 10.1093/jac/dkl197. [DOI] [PubMed] [Google Scholar]
- 16.Moubareck C, Gavini F, Vaugien L, Butel MJ, Doucet-Populaire F. 2005. Antimicrobial susceptibility of bifidobacteria. J Antimicrob Chemother 55:38–44. doi: 10.1093/jac/dkh495. [DOI] [PubMed] [Google Scholar]
- 17.Mangin I, Bouhnik Y, Bisetti N, Decaris B. 1999. Molecular monitoring of human intestinal Bifidobacterium strain diversity. Res Microbiol 150:343–350. doi: 10.1016/s0923-2508(99)80060-6. [DOI] [PubMed] [Google Scholar]
- 18.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized Ion Torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandeputte D, Kathagen G, D'Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, Vermeire S, Falony G, Raes J. 2017. Quantitative microbiome profiling links gut community variation to microbial load. Nature 551:507–511. doi: 10.1038/nature24460. [DOI] [PubMed] [Google Scholar]
- 20.Young VB, Schmidt TM. 2004. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol 42:1203–1206. doi: 10.1128/jcm.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinosa-Gongora C, Jessen LR, Kieler IN, Damborg P, Bjornvad CR, Gudeta DD, Pires Dos Santos T, Sablier-Gallis F, Sayah-Jeanne S, Corbel T, Neviere A, Hugon P, Saint-Lu N, de Gunzburg J, Guardabassi L. 2020. Impact of oral amoxicillin and amoxicillin/clavulanic acid treatment on bacterial diversity and beta-lactam resistance in the canine faecal microbiota. J Antimicrob Chemother 75:351–361. doi: 10.1093/jac/dkz458. [DOI] [PubMed] [Google Scholar]
- 22.MacPherson CW, Mathieu O, Tremblay J, Champagne J, Nantel A, Girard SA, Tompkins TA. 2018. Gut bacterial microbiota and its resistome rapidly recover to basal state levels after short-term amoxicillin-clavulanic acid treatment in healthy adults. Sci Rep 8:11192. doi: 10.1038/s41598-018-29229-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangin I, Leveque C, Magne F, Suau A, Pochart P. 2012. Long-term changes in human colonic Bifidobacterium populations induced by a 5-day oral amoxicillin-clavulanic acid treatment. PLoS One 7:e50257. doi: 10.1371/journal.pone.0050257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ventura M, Turroni F, Lugli GA, van Sinderen D. 2014. Bifidobacteria and humans: our special friends, from ecological to genomics perspectives. J Sci Food Agric 94:163–168. doi: 10.1002/jsfa.6356. [DOI] [PubMed] [Google Scholar]
- 25.Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sanchez B, Margolles A. 2017. Bifidobacteria and their health-promoting effects, p 73–98. In Britton R, Cani P (ed), Bugs as drugs. ASM Press, Washington, DC. doi: 10.1128/microbiolspec.BAD-0010-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duranti S, Gaiani F, Mancabelli L, Milani C, Grandi A, Bolchi A, Santoni A, Lugli GA, Ferrario C, Mangifesta M, Viappiani A, Bertoni S, Vivo V, Serafini F, Barbaro MR, Fugazza A, Barbara G, Gioiosa L, Palanza P, Cantoni AM, de’Angelis GL, Barocelli E, de'Angelis N, van Sinderen D, Ventura M, Turroni F. 2016. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol 92:fiw191. doi: 10.1093/femsec/fiw191. [DOI] [PubMed] [Google Scholar]
- 27.Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sanchez B, Margolles A, van Sinderen D, Ventura M. 2016. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep 6:23971. doi: 10.1038/srep23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 29.Turroni F, Duranti S, Milani C, Lugli GA, van Sinderen D, Ventura M. 2019. Bifidobacterium bifidum: a key member of the early human gut microbiota. Microorganisms 7:544. doi: 10.3390/microorganisms7110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bottacini F, O’Connell Motherway M, Kuczynski J, O’Connell KJ, Serafini F, Duranti S, Milani C, Turroni F, Lugli GA, Zomer A, Zhurina D, Riedel C, Ventura M, van Sinderen D. 2014. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Callaghan A, Bottacini F, O’Connell Motherway M, van Sinderen D. 2015. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics 16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10:1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrario C, Milani C, Mancabelli L, Lugli GA, Turroni F, Duranti S, Mangifesta M, Viappiani A, Sinderen D, Ventura M. 2015. A genome-based identification approach for members of the genus Bifidobacterium. FEMS Microbiol Ecol 91:fiv009. doi: 10.1093/femsec/fiv009. [DOI] [PubMed] [Google Scholar]
- 34.Duranti S, Lugli GA, Mancabelli L, Turroni F, Milani C, Mangifesta M, Ferrario C, Anzalone R, Viappiani A, van Sinderen D, Ventura M. 2017. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl Environ Microbiol 83:e02894-16. doi: 10.1128/AEM.02894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duytschaever G, Huys G, Boulanger L, De Boeck K, Vandamme P. 2013. Amoxicillin-clavulanic acid resistance in fecal Enterobacteriaceae from patients with cystic fibrosis and healthy siblings. J Cyst Fibros 12:780–783. doi: 10.1016/j.jcf.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Nakano V, Nascimento e Silva A, Merino VR, Wexler HM, Avila-Campos MJ. 2011. Antimicrobial resistance and prevalence of resistance genes in intestinal Bacteroidales strains. Clinics (Sao Paulo) 66:543–547. doi: 10.1590/s1807-59322011000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huddleston JR. 2014. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect Drug Resist 7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lakin SM, Dean C, Noyes NR, Dettenwanger A, Ross AS, Doster E, Rovira P, Abdo Z, Jones KL, Ruiz J, Belk KE, Morley PS, Boucher C. 2017. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res 45:D574–D580. doi: 10.1093/nar/gkw1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gudeta DD, Moodley A, Bortolaia V, Guardabassi L. 2014. vanO, a new glycopeptide resistance operon in environmental Rhodococcus equi isolates. Antimicrob Agents Chemother 58:1768–1770. doi: 10.1128/AAC.01880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 41.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldon T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M. 2009. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J 3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 47.European Food Safety Authority. 2012. Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10:2740. [Google Scholar]
- 48.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 49.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: Pan-Genomes Analysis Pipeline. Bioinformatics 28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 54.Mancino W, Lugli GA, Sinderen DV, Ventura M, Turroni F. 2019. Mobilome and resistome reconstruction from genomes belonging to members of the Bifidobacterium genus. Microorganisms 7:638. doi: 10.3390/microorganisms7120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milani C, Duranti S, Napoli S, Alessandri G, Mancabelli L, Anzalone R, Longhi G, Viappiani A, Mangifesta M, Lugli GA, Bernasconi S, Ossiprandi MC, van Sinderen D, Ventura M, Turroni F. 2019. Colonization of the human gut by bovine bacteria present in Parmesan cheese. Nat Commun 10:1286. doi: 10.1038/s41467-019-09303-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macfarlane GT, Macfarlane S, Gibson GR. 1998. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35:180–187. doi: 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences of the 16S profiling experiments, shallow shotgun metagenomics experiments, and RNAseq experiments are accessible under BioProject accession number PRJNA663786. Newly isolated Bifidobacterium genomes were sequenced and deposited in DDBJ/ENA/GenBank under the accession numbers reported in Table 2.