Currently, a “multiple-hurdle” approach based on a combination of different antimicrobial interventions, including heat, is being utilized during meat processing to control the burden of spoilage and pathogenic bacteria. Our recent study suggests that U.S. beef cattle harbor Escherichia coli that possess the locus of heat resistance (LHR).
KEYWORDS: Escherichia coli, extreme heat resistance, locus of heat resistance, red meat, whole-genome sequencing
ABSTRACT
Microbial resistance to processing treatments poses a food safety concern, as treatment tolerant pathogens can emerge. Occasional foodborne outbreaks caused by pathogenic Escherichia coli have led to human and economic losses. Therefore, this study screened for the extreme heat resistance (XHR) phenotype as well as one known genetic marker, the locus of heat resistance (LHR), in 4,123 E. coli isolates from diverse meat animals at different processing stages. The prevalences of XHR and LHR among the meat-borne E. coli were found to be 10.3% and 11.4%, respectively, with 19% agreement between the two. Finished meat products showed the highest LHR prevalence (24.3%) compared to other processing stages (0 to 0.6%). None of the LHR+ E. coli in this study would be considered pathogens based on screening for virulence genes. Four high-quality genomes were generated by whole-genome sequencing of representative LHR+ isolates. Nine horizontally acquired LHRs were identified and characterized, four plasmid-borne and five chromosomal. Nine newly identified LHRs belong to ClpK1 LHR or ClpK2 LHR variants sharing 61 to 68% nucleotide sequence identity, while one LHR appears to be a hybrid. Our observations suggest positive correlation between the number of LHR regions present in isolates and the extent of heat resistance. The isolate exhibiting the highest degree of heat resistance possessed four LHRs belonging to three different variant groups. Maintenance of as many as four LHRs in a single genome emphasizes the benefits of the LHR in bacterial physiology and stress response.
IMPORTANCE Currently, a “multiple-hurdle” approach based on a combination of different antimicrobial interventions, including heat, is being utilized during meat processing to control the burden of spoilage and pathogenic bacteria. Our recent study (M. Guragain, G. E. Smith, D. A. King, and J. M. Bosilevac, J Food Prot 83:1438–1443, 2020, https://doi.org/10.4315/JFP-20-103) suggests that U.S. beef cattle harbor Escherichia coli that possess the locus of heat resistance (LHR). LHR seemingly contributes to the global stress tolerance in bacteria and hence poses a food safety concern. Therefore, it is important to understand the distribution of the LHRs among meat-borne bacteria identified at different stages of different meat processing systems. Complete genome sequencing and comparative analysis of selected heat-resistant bacteria provide a clearer understanding of stress and heat resistance mechanisms. Further, sequencing data may offer a platform to gain further insights into the genetic background that provides optimal bacterial tolerance against heat and other processing treatments.
INTRODUCTION
Escherichia coli is one of the top five pathogens in the United States contributing to domestically acquired foodborne illnesses requiring hospitalizations. During the last 5 years in the United States, meat-borne E. coli infections have caused multistate outbreaks with almost 300 reported cases. Almost all of these cases are caused by Shiga toxin-producing E. coli (STEC), a pathotype that commonly colonizes ruminant animals, such as cattle and sheep (1), but can be found among other meat animal types (2). Meat animals serve as a reservoir of these pathogens that can potentially get introduced at any point in the meat production system (as reviewed in reference 3). Currently, a “multiple-hurdle” approach based on a combination of different antimicrobial interventions is used to ensure meat safety (4–6). Existing interventions include hot water wash (80° to 85°C), organic (lactic, acetic, and citric) acids, peroxyacetic acid, and halogen-based compounds (acidified sodium chlorite, chlorine dioxide, sodium hypochlorite, and bromine) (7–10). Despite this, reports of meat-borne E. coli outbreaks continue to emerge.
Evidence suggests the role of a novel 15- to 19-kb genetic element called the locus of heat resistance (LHR) in E. coli stress tolerance. The LHR consists of 16 to 19 open reading frames (ORFs) putatively grouped into three functional motifs contributing to tolerance against heat shock, envelope stress, and oxidative stress. The heat shock motif possesses genes encoding the ATP-dependent protease ClpK and small heat shock proteins that are known to have roles in heat resistance. Two variants of ClpK have been reported; ClpK1 is associated with a 15-kb LHR, while ClpK2 is associated with a 19-kb LHR. The two ClpK2 LHRs that have been characterized to date are plasmid borne in nature and shown to have a complementary role to ClpK1 in the heat resistance phenotype (11–14). In some instances, the LHR is colocalized with genes related to multidrug resistance, biofilm formation, and an antibacterial peptide together with a self-immunity protein (11). E. coli strains carrying the LHR have increased resistance toward heat (60°C for 5 to 20 min) (11, 15) and antimicrobials (bleach, peroxyacetic acid, and hydrogen peroxide) (16). Further, the LHR-mediated extreme heat resistance (XHR) is transferable to both pathogenic and nonpathogenic E. coli via conjugative transfer (11).
The LHR is prevalent among many Enterobacteriaceae, including E. coli. Bioinformatic analysis of all E. coli in the NCBI genome assemblies and NCBI whole-genome shotgun sequences predicted LHR to be present in ∼2% of E. coli, none of which carry any virulence factors (12). High prevalences of the LHR and XHR phenotype were reported among E. coli isolates from Canadian beef (27.2%, n = 55) (12, 17) and Danish raw milk cheese (36.7%, n = 253) (18). In a subsequent study of Canadian beef E. coli isolates, prevalence of the LHR and XHR was found to be 4.3% (n = 92) (15). In another study, the LHR was found in 59% (n = 70) of E. coli isolated from chlorinated sewage water (19). These studies reported a lack of virulence factors in the majority of LHR+ E. coli. On the other hand, 0.49% of human clinical E. coli isolates (n = 613) (20, 21) and 2% of extended-spectrum beta-lactamase-producing E. coli isolates from hospital settings (n = 115) (22) carry the LHR with XHR phenotype. A recent study identified STEC of serogroups O26, O45, O145, and O157 that possessed the LHR and demonstrated moderate heat resistance (23). This indicates that LHR-mediated stress-tolerant enteric pathogens can be isolated and that more are likely to emerge.
Our recent study showed that LHR+ E. coli and XHR E. coli are present in beef and cull cattle types harvested in multiple geographical regions of the United States (24). Our current study aims to expand this investigation by examining a large and diverse collection of meat-borne E. coli isolated from various meat sources at different stages of processing, with the objective of establishing the incidence of the LHR and XHR phenotype E. coli isolated from red meat sources. These isolates have been recovered during previous studies of red meat animal sources (beef, veal, pork, and lamb) at different processing stages and archived at the U.S. Meat Animal Research Center. Further, we aimed to gain better insight into LHR-mediated heat resistance by comparative genomic analysis of the LHR+ isolates.
RESULTS AND DISCUSSION
E. coli isolate collection at USMARC.
E. coli isolates at the U.S. Meat Animal Research Center (USMARC) were collected from beef, veal, lamb, and pork through numerous studies focused on sources and control of pathogen contamination present during red meat animal harvest and processing (Table 1). Strains selected from the collection used in this study provided isolates from diverse meat animals and processing systems as summarized in Table 1. Beef and veal were represented by isolates from ground beef (25), beef trim intended for grinding (26), and veal carcasses during processing (27). Lamb was represented by STEC that were isolated from three stages of sheep harvest (28). Pork was represented by STEC previously isolated from pork carcasses at three steps of harvest process (29, 30). Nonpathogenic E. coli was not the target of the studies the strains were drawn from; therefore, generic E. coli from ground beef (31) and pork chops (32) that had been isolated during studies of antimicrobial resistance were added. Further, generic fecal E. coli from USMARC-raised pigs was isolated for use in this study to more fully represent the swine/pork continuum. Isolates of E. coli from beef cattle feces have been described previously in regard to XHR and LHR (18) and are not included here.
TABLE 1.
Number of E. coli isolates by meat production systems and processing stages
| Meat production system | No. found in: |
Total | Source of strain or reference(s) | ||||
|---|---|---|---|---|---|---|---|
| Fecesb | Hide, pelt, or skinc | Preintervention carcassd | Final carcasse | Finished productf | |||
| Beef | 0 | 0 | 0 | 0 | 1,548 | 1,548 | 25, 26, 31 |
| Pathogenic | 901a | 901 | |||||
| Nonpathogenic | 647 | 647 | |||||
| Veal | 0 | 0 | 948 | 0 | 46 | 994 | 27 |
| Pathogenic | 763 | 26a | 789 | ||||
| Nonpathogenic | 185 | 20 | 205 | ||||
| Sheep | 0 | 14 | 10 | 487 | 0 | 511 | 28 |
| Pathogenic | 14 | 10 | 484 | 508 | |||
| Nonpathogenic | 0 | 0 | 3 | 3 | |||
| Pork | 243 | 475 | 33 | 9 | 310 | 1,070 | 29, 32, this study |
| Pathogenic | 21 | 448 | 33 | 9 | 1a | 512 | |
| Nonpathogenic | 222 | 27 | 0 | 0 | 309 | 558 | |
| Total | 243 | 489 | 991 | 496 | 1,904 | 4,123 | |
| Pathogenic | 21 | 462 | 806 | 493 | 928a | 2,710 | |
| Nonpathogenic | 222 | 27 | 185 | 3 | 976 | 1,413 | |
Isolates possessing stx and/or eae genes are considered to have a pathogenic background.
Fecal samples of preharvest and harvest animals.
Skin/hide of preharvest and exsanguinated animals before antimicrobial interventions.
Carcass after exsanguinated animals were skinned and eviscerated but before any antimicrobial interventions.
Carcass after antimicrobial interventions and chilling.
Final carcass further handled and fabricated to produce consumer-ready products.
To ensure proper classification, all E. coli isolates were screened for the Shiga toxin (stx) and intimin (eae) virulence genes. Isolates were considered nonpathogenic background strains when neither of the virulence factors were present. One or both virulence factors were present in 65.8% of the isolates (Table 1). Out of a total of 4,123 isolates, 14.1%, 36.2%, 15.5%, and 34.2% were grouped as enterohemorrhagic E. coli (EHEC: stx+ eae+), Shiga-toxigenic E. coli (STEC: stx+ eae−), enteropathogenic E. coli (EPEC: stx− eae+), and nonpathogenic E. coli, respectively (Table 2). In some cases, the earlier studies characterized the pathoserotypes of the isolates and identified stx subtypes. However, since not all strains had been characterized in detail, we limited the definition of pathogens to these two genes. Further, in cases such as STEC isolated from pork processing, where most stx genes were the stx2e subtype, which is not commonly associated with human disease (33), severe STEC disease may still result (34). Thus, all STEC strains are considered pathogens for the sake of the current study.
TABLE 2.
| Pathotyped | Total no. of isolates (%)e | No. of isolates (%) |
|||
|---|---|---|---|---|---|
| LHR+f | LHR−f | XHR+f | XHR−f | ||
| EHEC | 582 (14.1) | 0 (0) | 582 (100) | 39 (6.7) | 543 (93.3) |
| STEC | 1,491 (36.2) | 3 (0.2) | 1,488 (99.8) | 81 (5.4) | 1,410 (94.6) |
| EPEC | 640 (15.5) | 0 (0) | 640 (100) | 55 (8.6) | 585 (91.4) |
| EC | 1,410 (34.2) | 467g (33) | 943g (67.9) | 176 (12.5) | 1,234g (87.5) |
| Total | 4,123 | 470 (11.4) | 3,653 (88.6) | 351 (8.5) | 3,772 (91.5) |
Values represent the number of E. coli isolates of each type that screened positive.
Presence of at least one region of locus of heat resistance (LHR) during four-plex PCR is considered LHR positive.
E. coli isolates surviving 60°C for 20 min are considered extremely heat resistant (XHR).
E. coli isolates possessing the stx and/or eae gene are considered to have a pathogenic background. Types of E. coli: enterohemorrhagic E. coli (EHEC; stx+ eae+), Shiga toxigenic E. coli (STEC; stx+ eae−), enteropathogenic E. coli (EPEC; stx− eae+), and nonpathogenic background E. coli (EC).
Numbers in parentheses are the percentage prevalence of each pathotype among total 4,123 isolates.
Numbers in parentheses are the percentage prevalence that screened positive among each pathotype.
The prevalence between pathogenic and nonpathogenic E. coli within a column is significantly different (P < 0.0001) as determined by Fisher’s exact test.
Prevalence of LHR in meat-borne E. coli.
In order to determine the prevalence of the molecular determinant of extreme heat resistance, the E. coli isolates (n = 4,123) were screened for the LHR using a multiplex PCR assay as previously described (24). Overall prevalence of the LHR among meat animal and meat-borne E. coli in the sample collection was found to be 11.4% (Table 3). The prevalence of LHR in meat processing environments was previously reported to be 4.3% and 27.2% (12, 17). The differing sources and sites of collection in the previous reports may have had an influence on the number isolates found to possess the LHR. The majority of the LHR-positive population we identified (82.8%) possessed an intact LHR based on the presence of all four PCR products in our assay that identifies the 5′-, 3′-, and two internal regions of the LHR (data not shown). The additional heat resistance-imparting factor, ClpK2, was found present in 6.5% of the isolates (data not shown). The prevalence of LHR was highest among isolates from beef (20.8%), followed by those from pork (13.4%), and least among isolates recovered from veal (1.0%) and sheep (0.4%). The prevalence of LHR also varied greatly among processing stages. Isolates recovered from finished meat products showed the highest LHR prevalence (24.5%) compared to isolates from earlier processing stages and animal feces (0 to 0.9%) (Table 3).
TABLE 3.
| Source of isolate | % found inh: |
Totalc | ||||
|---|---|---|---|---|---|---|
| Feces | Hide, pelt, or skin | Preintervention carcass | Final carcass | Finished product | ||
| Beef | NA | NA | NA | NA | 20.5 | 20.5Pe |
| Veal | NA | NA | 0.6 | NA | 2.2 | 0.6QR |
| Sheep | NA | 0 | 0 | 0.4 | NA | 0.4QR |
| Pork | 0 | 0 | 0 | 0 | 46.1g | 13.45S |
| Totald | 0Vf | 0V | 0.6V | 0.4V | 24.3Z | 11.4 |
Prevalence is calculated as the percentage of E. coli isolates that screened positive.
Presence of at least one region of locus of heat resistance (LHR) during four-plex PCR is considered LHR positive.
Prevalence of LHR in meat animal types.
Prevalence of LHR in meat processing stages.
LHR prevalence among meat animal types in a column with the same subscript letter are not significantly different (P > 0.05).
XHR prevalences among meat processing stages in a row with the same subscript letter are not significantly different (P > 0.05).
XHR prevalence in finished pork product is significantly different (P < 0.0001) compared to those from other meat processing stages of pig.
NA, data not available due to lack of samples.
Similar to previous reports (11, 12, 19–22), the prevalences of LHR and virulence factors appear to have an inverse relationship. The majority of the LHR+ E. coli (99.4%) in our study did not carry virulence factors stx (Shiga toxin) and eae (intimin). Only 0.1% of virulent isolates were found to be LHR+ (Table 2). Together with predicted LHR domains, these findings suggest that the major role of the LHR is tolerance to environmental stresses rather than a pathogenicity advantage. E. coli strains lacking the virulence factors associated with EHEC, STEC, and EPEC are common nonpathogens harbored by all mammals and are merely an indicator of fecal contamination. Nevertheless, LHR detection in human clinical isolates and EHEC pathogenic serogroups (20, 21, 23), combined with LHR-mediated transfer of heat resistance to pathogens (11, 15), clearly emphasizes the likelihood that treatment-tolerant foodborne pathogenic E. coli can pose a serious public health threat.
Since nonpathogenic E. coli had the highest prevalence of the LHR, this explains the higher prevalence of the LHR among the beef and pork isolates, as these were the collections with the highest proportions of this type of E. coli (see Table S1 in the supplemental material). On the other hand, despite similar numbers of pathogenic and nonpathogenic isolates, finished meat products showed the highest prevalence of the LHR. Among pork isolates, which were represented across the processing continuum from animal feces to consumer-ready meat, the prevalence of the LHR was highest in those recovered from finished products (46.1%) while entirely absent from isolates found at earlier processing steps. A similar observation in a previous study of raw milk cheese E. coli isolates suggested that the LHR might be selected for or enriched during processing treatments (18). However, there is not enough current evidence to either conclude or suggest an alternative environmental source of LHR+ E. coli in finished products.
Prevalence of the XHR phenotype among meat-borne E. coli isolates.
E. coli isolates from various steps during animal harvest and meat processing were screened for the extreme heat resistance phenotype. We employed a previously validated XHR screening method that recovered the XHR/LHR type strain AW1.7 from samples and distinguished it from other heat-sensitive isolates (18). A total of 426 isolates (10.3%) survived 60°C for 20 min (Table 4). The prevalence of XHR was highest among E. coli isolated from beef (13.6%), followed by pork (10.6%), and veal (9.2%), and was lowest among lamb (2.3%). XHR E. coli was most prevalent in feces (17.3%) and least prevalent on final carcasses (1.8%) compared to other processing stages (7.3% to 12.6%). Only 90 (21.1%) of XHR isolates possessed the LHR, among which, the LHR was intact in 85 isolates. This is in agreement with our recent observation that only 9% (13/140) of XHR E. coli tested positive for LHR (24), indicating the need for investigation into non-LHR-mediated extreme heat resistance. Alternately, this incongruity may be the result of influences of water bath versus heating block phenotyping methods (23). In contrast to previous reports where strains possessing intact LHR and/or its variant always show the XHR phenotype (11, 12, 15, 22), 81% of LHR isolates in our study did not show the XHR phenotype (data not shown). This challenges the use of LHR alone as a reliable marker for the prediction of the XHR phenotype. Mutational analysis has shown that LHR transcription requires the presence of the two-component system EvgAS in the genome (13). More investigation of the cellular genetic background required for optimal LHR expression will lead to a better understanding of LHR-mediated resistance.
TABLE 4.
| Source of isolate | % found inh: |
Totalc | ||||
|---|---|---|---|---|---|---|
| Feces | Skin | Preintervention carcass | Final carcass | Finished product | ||
| Beef | NA | NA | NA | NA | 13.6 | 13.6Pe |
| Veal | NA | NA | 9.5 | NA | 2.2 | 9.2QS |
| Sheep | NA | 0 | 40g | 1.6 | NA | 2.3R |
| Pork | 17.3 | 7.6 | 18.2 | 11.1 | 9 | 10.6QS |
| Totald | 17.3Vf | 7.3WX | 10.1WXZ | 1.8Y | 12.6WZ | 10.3 |
Prevalence is calculated as percentage of E. coli isolates that screened positive.
E. coli isolates surviving 60°C for 20 min are considered extreme heat resistant (XHR).
Prevalence of XHR phenotype in meat animal types.
Prevalence of XHR phenotype in meat processing stages.
XHR prevalence among meat animal types in a column with the same subscript letter are not significantly different (P > 0.05).
XHR prevalence among meat processing stages in a row with the same subscript letter are not significantly different (P > 0.05).
XHR prevalence in finished lamb product is significantly different (P < 0.0001) compared to those from other meat processing stages of lamb.
NA, data not available due to lack of samples.
Two LHR variants are known to have complementary roles in the XHR phenotype (11), and an intact LHR has been shown to impart maximum heat resistance (13). Similarly, in our study, one LHR− ClpK2+ isolate, but none with less than 3 LHR fragments, showed the XHR phenotype (data not shown). Overall prevalences of the XHR phenotype in virulent and nonvirulent isolates were similar in our study (Table 2).
ClpK2-mediated heat resistance.
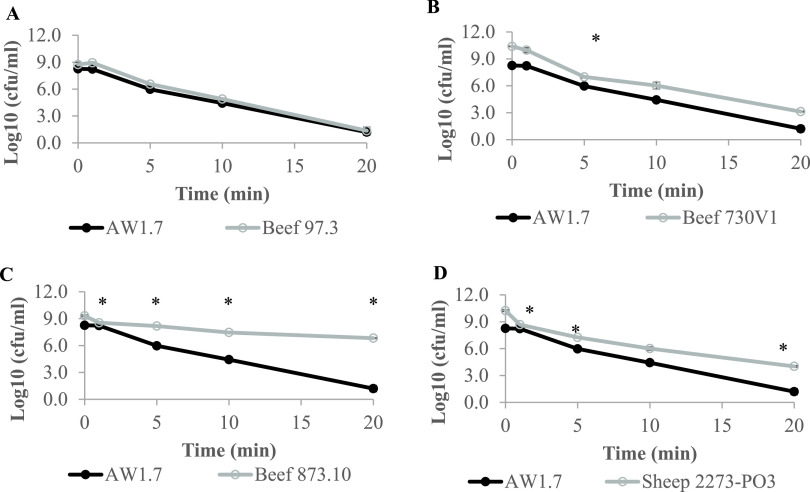
In addition to the LHR-detecting multiplex PCR, a ClpK2-specific PCR was performed to detect the second variant of the LHR (22). ClpK2 was prevalent in 6.5% of the isolates. While most ClpK2 isolates (91%) detected were in an intact LHR background, one was detected in an LHR− background (data not shown). LHR variants have been shown to have complementary roles in heat resistance, and the presence of both variants conferred a slight but significantly higher degree of heat resistance compared to the ClpK1 variant alone (11, 22). To determine if this was the case with the isolates identified here, a refined heat sensitivity measurement was performed with four XHR isolates. The isolates were selected based on the presence or absence of ClpK2 and various lengths of the LHR. Heat treatment of the prototype strain AW1.7 (intact LHR, ClpK2−) at 60°C for 1 min, 5 min, 10 min, and 20 min reduced cell counts by 2.3 log CFU/ml, 3.8 log CFU/ml, and 7.1 log CFU/ml, respectively (Fig. 1). Isolates Beef 97.3 (intact LHR, CLpK2−) (Fig. 1A) and Beef 730V1 (nonintact LHR, CLpK2−) (Fig. 1B) showed heat resistance profiles comparable to that of AW1.7. Isolate Beef 873.10 (intact LHR, ClpK2+) (Fig. 1C) showed remarkably high heat resistance, with only a 2.5 log (CFU/ml) reduction after 20m of heat treatment. Isolate Sheep 2273PO3 (intact LHR, ClpK2+) (Fig. 1D) showed an overall heat resistance profile comparable to that of AW1.7, with slightly increased survival at 20 min (6.2 log CFU/ml). While our findings clearly show a positive correlation between degree of heat resistance and presence of both LHR variants, a discrepancy in the extent of heat resistance between two ClpK2 strains led us to investigate the genomes of these isolates.
FIG 1.
Heat resistance of meat-borne E. coli isolates carrying LHR. Isolates were arbitrarily selected based on the presence or absence of ClpK2. Overnight cultures were exposed to 60°C for 1 min, 5 min, 10 min, and 20 min. The data presented are representative of at least two independent experiments, each with three technical replicates. Data indicate the mean ± standard deviations of three independent technical replicates. *, P < 0.01 (one-way ANOVA of log CFU/ml reduction at each time point).
Comparative genetics and phylogenetic analysis of LHR.
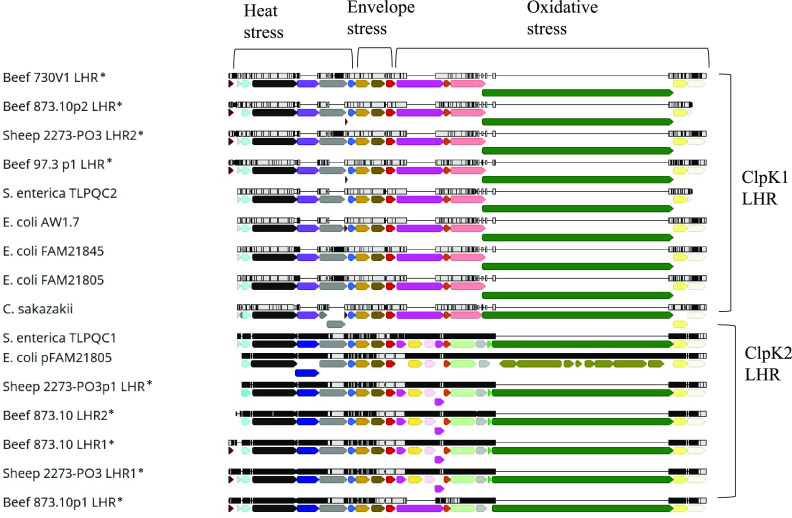
The four isolates identified above (Beef 730V1, Beef 97.3, Beef 873.10, and Sheep 2273PO3) were sequenced with the PacBio sequel system to examine the genomic differences underlying the varying degree of their heat resistance. Mapping of these genomes against a reference library of LHR sequences that were phenotypically and genotypically characterized in earlier studies (11, 13, 14, 35, 36) identified 9 LHR regions (Table 5) among the four isolates. Coordinates of LHR genes were derived from PGAP annotation (37, 38) and further confirmed using Prokka annotation (39). All LHRs identified in this study were flanked by mobile genetic elements and possess higher G+C content than the rest of the genome, indicating their horizontal acquisition. Each LHR harbors the core genes necessary for extreme heat resistance as previously identified (13) and appears to maintain genetic synteny (Fig. 2).
TABLE 5.
Loci of heat resistance among meat-borne E. coli isolates
| Organisma | LHRb | Location | Coordinates | G+C content (%) | Length (bp)c | ORF no. | LHR variantd |
|---|---|---|---|---|---|---|---|
| Beef 873.10 | 873.10 LHR1 | Chromosome | 22754–51098 | 57.7 | 28,345 | 28 | ClpK2 |
| 873.10 LHR2 | Chromosome | 2186098–2207888 | 59.8 | 21,791 | 22 | ClpK2 | |
| 873.10p1 LHR | Plasmid | 35177–52973 | 62.3 | 17,797 | 17 | Hybrid | |
| 873.10p2 LHR | Plasmid | 65948–79550 | 61.7 | 13,603 | 15 | ClpK1 | |
| Sheep 2273PO3 | 2273PO3 LHR1 | Chromosome | 2941510–2921761 | 61.3 | 19,750 | 22 | ClpK2 |
| 2273PO3 LHR2 | Chromosome | 3923433–3938938 | 62.1 | 15,506 | 17 | ClpK1 | |
| 2273PO3p1 LHR | Plasmid | 55553–36612 | 61.4 | 18,942 | 20 | ClpK2 | |
| Beef 730V1 | 730V1 LHR | Chromosome | 3826448–3841903 | 62.2 | 15,456 | 17 | ClpK1 |
| Beef 97.3 | 97.3p1 LHR | Plasmid | 139116–124551 | 61.8 | 14,566 | 18 | ClpK1 |
All isolates possess evgAS (a global regulator of LHR) (13); ClpA, ClpB, and ClpX (Clp chaperones); and ClpP (Clp protease).
Loci of heat resistance were identified by mapping the genomes against previously characterized LHR regions.
Length of LHR regions between flanking mobile elements.
The LHR variant is determined based on its homology to one of the two known variants of LHR, ClpK1 LHR and ClpK2 LHR.
FIG 2.
Multiple sequence analysis of LHR regions. The core LHR regions were manually extracted and aligned using MAFFT. The corresponding genes are represented by the same color. The alignment graph above each LHR shows identical nucleotides in gray and single nucleotide substitutions in black highlights. ORFs are denoted by the following colors: light gray, DNA binding protein; turquoise, sHsp20; black, ClpK; purple, hypothetical protein 2; dark blue, ClsA; dark gray, FtsH; bright blue, Hsp20; light brown, YfdX1; brown, YfdX2; red, HdeD; dark pink, hypothetical protein 4; gold, Msc; orange, Trx; pink, KefB; dark green, PsiE; green, Dgc; yellow, HtpX; white, DegP; olive green, microcin genes; light pink, hypothetical protein 5; gray, PAS domain protein; mint green, CPA; and dark red, small hypothetical proteins (1 and 3). The reference LHR sequences and corresponding NCBI accession numbers are E. coli FAM21805 (KY646173.1), E. coli FAM21845 (CP017220.1), E. coli AW1.7 (LDYJ01000141.1), Salmonella enterica serovar Senftenberg ATCC 43845 TLPQC1 and TLPQC2 (CP016838.1), Cronobacter sakazakii ATCC 29544 (CP011047.1), and E. coli pFAM21805 (KY416992.1). *, LHR identified in this study.
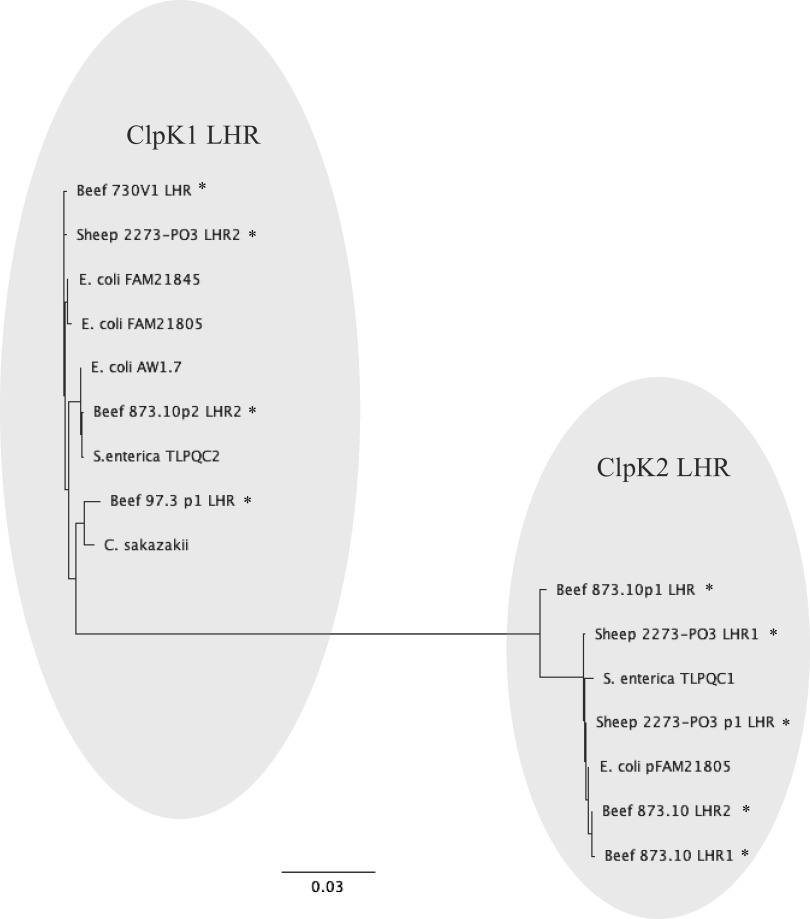
Phylogenetic analysis yielded two clades, and each clade distinctly included one of the two variants of reference LHRs, ClpK1 LHR or ClpK2 LHR (Fig. 3). ClpK1 LHR and ClpK2 LHR shared 61 to 68% nucleotide identity. The reference sequence pFAM21805 LHR (4) showed lower nucleotide identity with other LHRs (38 to 62%) due to the presence of an additional 11-kb microcin cluster (11). LHRs in both clades possess 12 common genes encoding DNA binding protein, small heat shock proteins sHSP20 and sHSP, ATP-dependent Clp protease ClpK, cell division protein FtsH, putative chaperone proteins YfdX1 and YfdX2, acid stress resistance membrane protein HdeD, thioredoxin reductase TRX2, phosphate-starvation-inducible protein PsiE, zinc-dependent protease HtpX, and periplasmic serine protease DegP. Most of the corresponding genes within a clade shared high levels of nucleotide identity (>90%). Between clades, the lowest level of identity is observed among HtpX (72%) and PsiE (76%). Unique to the ClpK1 LHR were highly conserved genes (96 to 100% identity) encoding two hypothetical proteins (2 and 4) and a glutathione-regulated potassium-efflux system protein, KefB. On the other hand, only ClpK2 LHRs possessed 6 highly conserved genes (>99% identity) encoding cardiolipin synthase (Cls), mechanosensitive channel (Msc), hypothetical protein 5, cation:proton antiporter (CPA), PAS-domain containing protein (PAS), and diguanylate cyclase (DGC). Cls was located at the position corresponding to hypothetical protein 2 of ClpK1 LHR. Msc and hypothetical protein 5 were found to be inserted in the hypothetical gene 4 of ClpK1 LHR (Fig. 2).
FIG 3.
Bayesian inference phylogenetic tree of LHR nucleotide sequences. The phylogenetic tree was constructed using MRBAYES utilizing generalized time reversible model with discrete gamma distribution (GTR+G). Ten newly identified LHR regions and seven genetically and phenotypically characterized LHR regions were reincluded in the MAFFT alignment, which was then used for phylogenetic analysis. *, LHR identified in this study.
The ClpK1 LHR was located chromosomally in isolates Beef 730V1 (730V1 LHR) and Sheep 2273PO3 (2273PO3 LHR2) and on the plasmids of isolates Beef 873.10 (873.10p2 LHR) and Beef 97.3 (97.3p1 LHR). The four newly identified ClpK1 LHRs within the first clade shared high nucleotide identity (92 to 100%) with each other as well as with previously described ClpK1 LHR, approximately 15 kb in size with a G+C content of 62% (11, 12, 14). Genes encoding FtsH in 873.10p2 LHR and 97.3p1 LHR, however, were shorter and shared only 41 to 43% identity with that of other ClpK1 LHR. In p873.10(2) LHR, the highly conserved gene degP, with a yet unidentified role in heat resistance, was severely truncated.
The second clade consisted of the five newly identified ClpK2 LHRs which shared high nucleotide identity with a previously described ClpK2 LHR, approximately 19 kb in size with a G+C content of 61 to 62% (11, 14). ClpK2 LHR was present in only two isolates, Beef 873.10 and Sheep 2273PO3, which is in agreement with the PCR results for the clpK2 gene. The presence of both ClpK LHR variants in these isolates correlated with their greater heat resistance as suggested by complementary functions (11). Chromosomes of the isolate Beef 873.10 harbored two (873.10 LHR1 and 873.10 LHR2) and Sheep 2273PO3 harbored one (2273PO3 LHR1) ClpK2 LHR. In addition, two more ClpK2 LHRs were also located on plasmids of these isolates (873.10p1 and 2273PO3p1). Four ClpK2 LHRs (873.10 LHR1, 873.10 LHR2, 2273PO3 LHR1, and 2273PO3p1 LHR) were >98% identical in their nucleotide sequences among themselves and to a previously identified TLPQC1 in Salmonella enterica serovar Senftenberg 775W (14). Some of the newly identified LHRs carried other genes in addition to core LHR genes. Additional genes include five genes encoding a phosphate transfer system and three genes encoding a 6-phospho beta-glucosidase system in 873.10 LHR1 and a DNA binding transcriptional regulator, DmlR, and a leucine efflux protein, LeuE, in 873.10 LHR2. Thirteen corresponding genes in these ClpK2 LHRs were nearly identical (>99%) to one another. Despite lower overall LHR sequence identity, corresponding individual genes in the reference sequence pFAM21805 LHR showed similarly high nucleotide identity with ClpK2 LHRs. Out of five ClpK2 LHR identified in this study, 873.10p1 LHR shared only 87% (versus >98%) sequence identity with other ClpK2 LHRs. Unlike all other ClpK2 LHRs, it lacked genes encoding Msc and hypothetical protein 5 and instead possessed a 1,146-bp gene encoding hypothetical protein 2 similar to that in the ClpK1 LHR. The gene trx2 shared relatively lower homology with that of other ClpK2 LHRs. Further, trx2 of 873.10p1 LHR shared higher identity with its ClpK1 LHR variant. This suggests possible genetic exchange/rearrangements between ClpK1 LHR and ClpK2 LHR 873.10p1 giving rise to a hybrid LHR.
Observations from this study indicate a correlation between the number of LHR regions present in an isolate and the extent of the heat-resistant phenotype (Beef 873.10 [4 LHRs] > Sheep 2273PO3 [3 LHRs] > Beef 730V1 [1 LHR] = Beef 97.3 [1 LHR]) and lay a foundation to investigate the stress tolerance potential of a hybrid LHR.
Conclusion.
This study assesses XHR and the LHR in meat-borne E. coli in the United States. A higher prevalence of LHR+ E. coli was observed in finished meat products and demands further studies directed toward identifying the source(s) of LHR+ E. coli in the meat processing continuum. The genomes of meat-borne LHR+ E. coli were characterized to better understand these LHR regions. Our analysis highlighted the unique features of LHR variants. We identified a novel hybrid LHR, which shared genes between two known LHR variants. Our study suggests that the comparatively higher heat resistance of some LHR isolates is positively correlated with the quantity of LHRs present in the isolate. Maintenance of as many as four LHRs in a single genome emphasizes the importance of LHR in bacterial physiology/survival. The findings of this work, therefore, will provide a platform for future investigations aimed at understanding LHR-mediated global stress tolerance and its molecular mechanisms.
MATERIALS AND METHODS
Samples and processing.
E. coli isolates from the collection at the U.S. Meat Animal Research Center were used in this study. These isolates (n = 3,880) were previously collected from various steps (skin/hide, n = 489; preintervention carcass, n = 991; final carcass, n = 496; finished product, n = 1,904) during animal harvest in commercial meat processing plants. Due to concerns regarding anonymity of the participating commercial locations, the various anitmicrobials used in each plant to reduce contamination were only described in general terms. In short, the isolates were recovered from processors using single or sequential interventions consisting of hot (85°C) water, organic acids, and chlorine/bromine compounds. Difco tryptic soy broth (TSB; Beckton Dickinson, USA)-grown overnight cultures had been archived at −80°C with 17% glycerol as a cryoprotectant. Frozen cultures were revived on Difco tryptic soy agar (TSA; Beckton Dickinson, USA) plates following incubation at 37°C for 15 to 22 h. A single colony from each plate was utilized for the further studies. Pig fecal isolates (n = 243) were collected from rectal swabs of 5 arbitrarily selected pigs per each of 14 pens located within one enclosed swine feeding facility. Swabs were streaked for isolation onto the surface of CHROMagar E. coli medium (Paris, France) and incubated at 37°C for 18 to 24 h. Representative colonies (2 to 6 per sample) were selected for the study. Presumptive colonies were confirmed using a previously described PCR method for E. coli (40).
LHR detection assay.
A four-plex PCR assay was used to detect LHR as previously described (24). Briefly, oligonucleotides designed to target 5′, 3′, and interior regions of a 14-kb LHR were used to differentially amplify various regions across the LHR. Oligonucleotide primer pairs used were 1266F/1373R (AATGCAGGCGGTGATGAAGA/CGCTGATTGCCCATCAACAG), 4295F/4505R (CGAGGGAGAATTCCAGTCCG/GGCACTACGCTAATCCTGCT), 7069F/7404R (CTCATTGGATGCTTCGCTGC/ACGGAAACCATTGAGGCGAT), and 14160F/14699R (CCTGGCATTGTTTTCTGGCC/GGCTGTTCGATGACGCATTC). Isolates with all 4 amplicons present were considered to possess intact LHR. To detect the second variant of LHR, E. coli isolates were screened for the presence of marker gene ClpK2 using primers ClpK2F/ClpK2R (ACGATCACTATCGCCAACTG/AGTATTTATCCAGCTCGGGCGTG) as previously described (22). All amplicons were resolved on 1.5% agarose gels, electrophoresed in 1× sodium borate buffer (Faster Better Media LLC, USA), and stained with ethidium bromide for visualization. Prevalences were compared using Fisher’s exact test.
Heat killing assay.
XHR E. coli was identified in a previously validated high-throughput 96-well assay format (18). Briefly, each isolate was inoculated into Difco buffered peptone water (BPW; Beckton Dickinson, USA) and incubated statically at 37°C for 16 h. The cultures were diluted 1:10 into fresh BPW and further incubated at 37°C for 16 h to remove the influence of carbohydrates and buffers present in TSB, which would otherwise affect the heat-resistant phenotype. Then, 0.5 ml of each BPW cultured isolate was transferred to a thin-walled plastic test tube for heat treatments. Isolates were exposed to 60°C for 20 m in an aluminum heat block and then transferred and rapidly cooled in an ice-water bath for 45 s, followed by recovery at room temperature for 1 h. At the end of 1 h, an equal volume of 2× Difco MacConkey broth (Beckton Dickinson, USA) was added to each tube, and the mixture was incubated overnight at 37°C. Any heat-treated isolate showing growth, as evident by yellow coloration of medium, was considered an XHR E. coli. LHR+ XHR strain AW1.7 was included as a positive control in each series of heat treatments. Prevalences were compared using Fisher’s exact test.
Selected LHR isolates were further characterized for differences in their extent of heat resistance. Briefly, overnight cultures were grown in Luria Bertani (LB) broth (Beckton Dickinson, Sparks MD) at 37°C, 150 rpm. The optical densities at 600 nm (OD600nm) of overnight cultures were normalized to 0.2 before use. A 3-ml portion of each normalized culture was incubated in a 60°C water bath for 0, 1, 5, 10, and 20 min. At the end of each time point, 100 μl of the culture was immediately transferred to 900 μl BPW. Serial dilutions were then plated on 3M Petrifilm E. coli/coliform count plates (St. Paul, MN, USA) and incubated at 37°C for 18 to 24 h. Fold reductions in bacterial population were calculated by the log reduction in CFU/ml of untreated versus treated cells for each time point. Two independent experiments, each with three biologic replicates were carried out for each isolate.
Whole-genome sequencing and assembly.
Overnight cultures of the bacteria were diluted 1:40 times in LB, followed by incubation at 37°C at 190 rpm for 3 h. Genomic DNA was extracted and purified using Qiagen Genomic-tip 100/G columns and the Qiagen genomic DNA buffer set (Qiagen, Valencia, CA) using the manufacturer’s recommended protocol. Genomic DNA (gDNA) was sheared to an average size of 20 kb using g-TUBE (Covaris Inc, Woburn MA). Sequencing libraries were prepared using SMRTbell template prep kit 1.0 (Pacific Biosciences, Menlo Park, CA) and size selected for 20-kb fragments using BluePippin High Pass Plus cassettes (Sage Science, Inc., Beverly MA). Single-molecule real-time (SMRT) sequencing was performed on a PacBio Sequel system (Pacific Biosciences, Menlo Park, CA) with chemistry 3.0. De novo assembly, circularization, and plasmid identification were done using microbial assembly application within the PacBio SMRT analysis pipeline 8.0. The origin of replication was determined by using Ori-Finder 2.0 (41) and then was reset to position 1 of the chromosome. Replicon regions were identified by in silico analysis of the plasmid sequences with PlasmidFinder 2.1 (42). For contigs that were not circularized with SMRTLink microbial assembly, 50 kb sections from each end of the contigs were extracted and mapped to each other using the LASTZ aligner in Geneious Prime. Overlapping ends were trimmed and concatenated to form a complete chromosome. The manually closed chromosome was reoriented and mapped against error-corrected PacBio and Illumina reads. Consistent read depth coverage of the mapped reads over the chromosome indicated that it was correctly closed.
For the resulting assemblies, mean coverage and mean subread length ranged from 85× to 359× and 13,561 bp to 16,097 bp, respectively. The assembled genomes were analyzed using Geneious Prime (Biomatters Ltd., New Zealand). For error correction of the PacBio assemblies and detection of smaller plasmids, short reads were generated with an Illumina MiSeq 300-cycle kit 2 (paired-end sequencing). Genomic DNA was sheared to an average size of 350 bp using a Covaris microtube (Covaris, Inc, Woburn, MA). An Illumina (San Diego, CA) sequencing library was prepared using a TruSeq DNA PCR free LP kit. Paired read sequencing was performed on a MiSeq platform using kit 2 (300 cycles) (Illumina). Quality trimming and adapter clipping of raw reads was done with Trimmomatic 0.39 (43). Trimmed ends were mapped against PacBio assemblies using Pilon 1.23 (44). Unused reads were de novo assembled for smaller plasmids using the Geneious assembler.
Genome analysis.
LHR regions and the flanking mobile elements were located and identified by high-sensitivity mapping of full-length genomes against the reference sequence library using Geneious Mapper. Regions in de novo contigs showing at least 80% overlap identity (>99.9% confidence) with reference LHR sequences upon pairwise alignment over the full length were determined to be LHR. The reference sequence library consisted of seven genetically and phenotypically characterized LHR nucleotide sequences from earlier studies. The complete genomes were annotated with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (37, 38), and nucleotide sequences were submitted to GenBank. The coordinates of newly identified LHR regions and the flanking transposons were derived from PGAP annotations and further confirmed with rapid prokaryotic genome annotation software Prokka 1.14.6 (39). Transposons were further confirmed by in silico analysis using ISfinder (45).
For comparative analysis, nucleotide sequences for core LHR regions were extracted and aligned with MAFFT 7.450 (46) in Geneious Prime. The best fit model for the alignment as determined with jModelTest 2 (47, 48) using Akaike’s information criterion (49), was found to be a generalized time reversible model with discrete gamma distribution (GTR+G). These criteria were used to construct a Bayesian inference tree with MRBAYES 3.2.6 (50) in Geneious Prime. The Bayesian inference tree was supported with a maximum-likelihood tree generated using PhyML 3.3.20180621 (51). LHR variants were primarily determined based on the nucleotide identity between clpK genes; LHRs with clpK genes sharing ≥95% nucleotide identity were grouped as one variant. Further, the variant identity was confirmed by the length of LHR regions (∼15 kb versus ∼19 kb) and the presence or absence of highly conserved genes encoding cardiolipin synthase (clsA), diguanylate cyclase (DGC), and PAS-domain containing protein (PAS).
Data availability.
All of the genome sequences have been uploaded to NCBI under BioProject no. PRJNA663878. The GenBank accession numbers for the individual PGAP-annotated genomes are CP061749, CP061750, CP061751, CP061752, and CP061753 (Sheep 2273-PO3); CP061754, CP061755, CP061756, and CP061757 (Beef 873.10); CP061758, CP061759, CP061760, CP061761, CP061762, and CP061763 (Beef 97.3); and CP061764, CP061765, CP061766, and CP061767 (Beef 730V1).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by a grant from the Foundation for Meat and Poultry Research and Education (no. ARS-58-3040-7-010).
Thanks to Jody Gallagher for secretarial assistance, James Wells for providing pig fecal samples, Aaron Dickey for bioinformatics training, Kerry Brader and Julie Dyer for sequencing library preparation training, and Greg Smith, Bruce Jasch, Sydney Brodrick, Nirajan Bhattarai, and Kristen Kuhn for technical assistance.
Product names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of the name by USDA implies no approval of the product to the exclusion of others that may also be suitable.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Foodborne, Waterborne, and Environmental Diseases (DFWED). Reports of selected E. coli outbreak investigations. CDC, NCEZID, DFWED, Atlanta, GA. https://www.cdc.gov/ecoli/outbreaks.html.
- 2.Dutta TK, Roychoudhury P, Bandyopadhyay S, Wani SA, Hussain I. 2011. Detection & characterization of Shiga toxin producing Escherichia coli (STEC) & enteropathogenic Escherichia coli (EPEC) in poultry birds with diarrhoea. Indian J Med Res 133:541–545. [PMC free article] [PubMed] [Google Scholar]
- 3.Adam K, Brulisauer F. 2010. The application of food safety interventions in primary production of beef and lamb: a review. Int J Food Microbiol 141 Suppl 1(Suppl 1):S43–52. doi: 10.1016/j.ijfoodmicro.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 4.Bacon RT, Belk KE, Sofos JN, Clayton RP, Reagan JO, Smith GC. 2000. Microbial populations on animal hides and beef carcasses at different stages of slaughter in plants employing multiple-sequential interventions for decontamination. J Food Prot 63:1080–1086. doi: 10.4315/0362-028x-63.8.1080. [DOI] [PubMed] [Google Scholar]
- 5.Castillo A, Lucia LM, Goodson KJ, Savell JW, Acuff GR. 1999. Decontamination of beef carcass surface tissue by steam vacuuming alone and combined with hot water and lactic acid sprays. J Food Prot 62:146–151. doi: 10.4315/0362-028X-62.2.146. [DOI] [PubMed] [Google Scholar]
- 6.Kang DH, Koohmaraie M, Siragusa GR. 2001. Application of multiple antimicrobial interventions for microbial decontamination of commercial beef trim. J Food Protection 64:168–171. doi: 10.4315/0362-028X-64.2.168. [DOI] [PubMed] [Google Scholar]
- 7.Koohmaraie M, Arthur TM, Bosilevac JM, Guerini M, Shackelford SD, Wheeler TL. 2005. Post-harvest interventions to reduce/eliminate pathogens in beef. Meat Sci 71:79–91. doi: 10.1016/j.meatsci.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 8.U.S. FSIS. 2020. Safe and suitable ingredients used in production of meat, poultry, and egg products. FSIS Directive 7120.1. https://www.fsis.usda.gov/wps/portal/fsis/topics/regulations/directives/7000-series. Accessed 14 December 2020.
- 9.Schmidt JW, Wang R, Kalchayanand N, Wheeler TL, Koohmaraie M. 2012. Efficacy of hypobromous acid as a hide-on carcass antimicrobial intervention. J Food Prot 75:955–958. doi: 10.4315/0362-028X.JFP-11-433. [DOI] [PubMed] [Google Scholar]
- 10.Kocharunchitt C, Mellefont L, Bowman JP, Ross T. 2020. Application of chlorine dioxide and peroxyacetic acid during spray chilling as a potential antimicrobial intervention for beef carcasses. Food Microbiol 87:103355. doi: 10.1016/j.fm.2019.103355. [DOI] [PubMed] [Google Scholar]
- 11.Boll EJ, Marti R, Hasman H, Overballe-Petersen S, Stegger M, Ng K, Knochel S, Krogfelt KA, Hummerjohann J, Struve C. 2017. Turn up the heat: food and clinical Escherichia coli isolates feature two transferrable loci of heat resistance. Front Microbiol 8:579. doi: 10.3389/fmicb.2017.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer RG, Zheng J, Garcia-Hernandez R, Ruan L, Ganzle MG, McMullen LM. 2015. Genetic determinants of heat resistance in Escherichia coli. Front Microbiol 6:932. doi: 10.3389/fmicb.2015.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer R, Nguyen O, Ou Q, McMullen L, Ganzle MG. 2017. Functional analysis of genes comprising the locus of heat resistance in Escherichia coli. Appl Environ Microbiol 83:e01400-17. doi: 10.1128/AEM.01400-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen SV, Harhay GP, Bono JL, Smith TP, Harhay DM. 2017. Genome sequence of the thermotolerant foodborne pathogen Salmonella enterica serovar Senftenberg ATCC 43845 and phylogenetic analysis of loci encoding increased protein quality control mechanisms. mSystems 2:e00190-16. doi: 10.1128/mSystems.00190-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer RG, Walker BD, Yang X, McMullen LM, Ganzle MG. 2017. The locus of heat resistance (LHR) mediates heat resistance in Salmonella enterica, Escherichia coli and Enterobacter cloacae. Food Microbiol 64:96–103. doi: 10.1016/j.fm.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Fang Y, Zhi S, Simpson DJ, Gill A, McMullen LM, Neumann NF, Gänzle MG. 2019. The locus of heat resistance confers resistance to chlorine and other oxidizing chemicals in Escherichia coli. Appl Environ Microbiol 86:e02123-19. doi: 10.1128/AEM.02123-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dlusskaya EA, McMullen LM, Ganzle MG. 2011. Characterization of an extremely heat-resistant Escherichia coli obtained from a beef processing facility. J Appl Microbiol 110:840–849. doi: 10.1111/j.1365-2672.2011.04943.x. [DOI] [PubMed] [Google Scholar]
- 18.Marti R, Muniesa M, Schmid M, Ahrens CH, Naskova J, Hummerjohann J. 2016. Short communication: heat-resistant Escherichia coli as potential persistent reservoir of extended-spectrum beta-lactamases and Shiga toxin-encoding phages in dairy. J Dairy Sci 99:8622–8632. doi: 10.3168/jds.2016-11076. [DOI] [PubMed] [Google Scholar]
- 19.Zhi S, Banting G, Li Q, Edge TA, Topp E, Sokurenko M, Scott C, Braithwaite S, Ruecker NJ, Yasui Y, McAllister T, Chui L, Neumann NF. 2016. Evidence of naturalized stress-tolerant strains of Escherichia coli in municipal wastewater treatment plants. Appl Environ Microbiol 82:5505–5518. doi: 10.1128/AEM.00143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma A, Chui L. 2017. Identification of heat resistant Escherichia coli by qPCR for the locus of heat resistance. J Microbiol Methods 133:87–89. doi: 10.1016/j.mimet.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 21.Ma A, Glassman H, Chui L. 2020. Characterization of Escherichia coli possessing the locus of heat resistance isolated from human cases of acute gastroenteritis. Food Microbiol 88:103400. doi: 10.1016/j.fm.2019.103400. [DOI] [PubMed] [Google Scholar]
- 22.Boll EJ, Frimodt-Moller J, Olesen B, Krogfelt KA, Struve C. 2016. Heat resistance in extended-spectrum beta-lactamase-producing Escherichia coli may favor environmental survival in a hospital setting. Res Microbiol 167:345–349. doi: 10.1016/j.resmic.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 23.de Souza Figueiredo EE, Yang X, Zhang P, Reuter T, Stanford K. 2019. Comparison of heating block and water bath methods to determine heat resistance in Shiga-toxin producing Escherichia coli with and without the locus of heat resistance. J Microbiol Methods 164:105679. doi: 10.1016/j.mimet.2019.105679. [DOI] [PubMed] [Google Scholar]
- 24.Guragain M, Smith GE, King DA, Bosilevac JM. 2020. Prevalence of extreme heat-resistant Gram-negative bacteria carried by US cattle at harvest. J Food Prot 83:1438–1443. doi: 10.4315/JFP-20-103. [DOI] [PubMed] [Google Scholar]
- 25.Bosilevac JM, Koohmaraie M. 2011. Prevalence and characterization of non-O157 shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl Environ Microbiol 77:2103–2112. doi: 10.1128/AEM.02833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosilevac JM, Guerini MN, Brichta-Harhay DM, Arthur TM, Koohmaraie M. 2007. Microbiological characterization of imported and domestic boneless beef trim used for ground beef. J Food Prot 70:440–449. doi: 10.4315/0362-028x-70.2.440. [DOI] [PubMed] [Google Scholar]
- 27.Bosilevac JM, Wang R, Luedtke BE, Hinkley S, Wheeler TL, Koohmaraie M. 2017. Characterization of enterohemorrhagic Escherichia coli on veal hides and carcasses. J Food Prot 80:136–145. doi: 10.4315/0362-028X.JFP-16-247. [DOI] [PubMed] [Google Scholar]
- 28.Kalchayanand N, Arthur TM, Bosilevac JM, Brichta-Harhay DM, Guerini MN, Shackelford SD, Wheeler TL, Koohmaraie M. 2007. Microbiological characterization of lamb carcasses at commercial processing plants in the United States. J Food Prot 70:1811–1819. doi: 10.4315/0362-028x-70.8.1811. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt JW, Brichta-Harhay DM, Kalchayanand N, Bosilevac JM, Shackelford SD, Wheeler TL, Koohmaraie M. 2012. Prevalence, enumeration, serotypes, and antimicrobial resistance phenotypes of Salmonella enterica isolates from carcasses at two large United States pork processing plants. Appl Environ Microbiol 78:2716–2726. doi: 10.1128/AEM.07015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nastasijevic I, Schmidt JW, Boskovic M, Glisic M, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M, Bosilevac JM. 2020. Seasonal prevalence and characterization of Shiga toxin-producing Escherichia coli on pork carcasses for three steps of the harvest process at two commercial processing plants in the United States. Appl Environ Microbiol 87:e01711-20. doi: 10.1128/AEM.01711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vikram A, Miller E, Arthur TM, Bosilevac JM, Wheeler TL, Schmidt JW. 2018. Similar levels of antimicrobial resistance in U.S. food service ground beef products with and without a “raised without antibiotics” claim. J Food Prot 81:2007–2018. doi: 10.4315/0362-028X.JFP-18-299. [DOI] [PubMed] [Google Scholar]
- 32.Vikram A, Miller E, Arthur TM, Bosilevac JM, Wheeler TL, Schmidt JW. 2019. Food service pork chops from three U.S. regions harbor similar levels of antimicrobial resistance regardless of antibiotic use claims. J Food Prot 82:1667–1676. doi: 10.4315/0362-028X.JFP-19-139. [DOI] [PubMed] [Google Scholar]
- 33.Beutin L, Kruger U, Krause G, Miko A, Martin A, Strauch E. 2008. Evaluation of major types of Shiga toxin 2E-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans. Appl Environ Microbiol 74:4806–4816. doi: 10.1128/AEM.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fasel D, Mellmann A, Cernela N, Hachler H, Fruth A, Khanna N, Egli A, Beckmann C, Hirsch HH, Goldenberger D, Stephan R. 2014. Hemolytic uremic syndrome in a 65-Year-old male linked to a very unusual type of stx2e- and eae-harboring O51:H49 Shiga toxin-producing Escherichia coli. J Clin Microbiol 52:1301–1303. doi: 10.1128/JCM.03459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marti R, Schmid M, Kulli S, Schneeberger K, Naskova J, Knochel S, Ahrens CH, Hummerjohann J. 2017. Biofilm formation potential of heat-resistant Escherichia coli dairy isolates and the complete genome of multidrug-resistant, heat-resistant strain FAM21845. Appl Environ Microbiol 83:e00628-17. doi: 10.1128/AEM.00628-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gajdosova J, Benedikovicova K, Kamodyova N, Tothova L, Kaclikova E, Stuchlik S, Turna J, Drahovska H. 2011. Analysis of the DNA region mediating increased thermotolerance at 58°C in Cronobacter sp and other enterobacterial strains. Antonie Van Leeuwenhoek 100:279–289. doi: 10.1007/s10482-011-9585-y. [DOI] [PubMed] [Google Scholar]
- 37.Haft DH, DiCuccio M, Badretdin A, Brover V, Chetvernin V, O'Neill K, Li WJ, Chitsaz F, Derbyshire MK, Gonzales NR, Gwadz M, Lu F, Marchler GH, Song JS, Thanki N, Yamashita RA, Zheng C, Thibaud-Nissen F, Geer LY, Marchler-Bauer A, Pruitt KD. 2018. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res 46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt K, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 40.Horakova K, Mlejnkova H, Mlejnek P. 2008. Specific detection of Escherichia coli isolated from water samples using polymerase chain reaction targeting four genes: cytochrome bd complex, lactose permease, beta-D-glucuronidase, and beta-D-galactosidase. J Appl Microbiol 105:970–976. doi: 10.1111/j.1365-2672.2008.03838.x. [DOI] [PubMed] [Google Scholar]
- 41.Luo H, Zhang CT, Gao F. 2014. Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes. Front Microbiol 5:482. doi: 10.3389/fmicb.2014.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carattoli A, Hasman H. 2020. PlasmidFinder and in silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol 2075:285–294. doi: 10.1007/978-1-4939-9877-7_20. [DOI] [PubMed] [Google Scholar]
- 43.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–6. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 48.Santorum JM, Darriba D, Taboada GL, Posada D. 2014. jmodeltest.org: selection of nucleotide substitution models on the cloud. Bioinformatics 30:1310–1311. doi: 10.1093/bioinformatics/btu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akaike H. 1974. New look at statistical-model identification. IEEE Trans Automat Contr 19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 50.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 51.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the genome sequences have been uploaded to NCBI under BioProject no. PRJNA663878. The GenBank accession numbers for the individual PGAP-annotated genomes are CP061749, CP061750, CP061751, CP061752, and CP061753 (Sheep 2273-PO3); CP061754, CP061755, CP061756, and CP061757 (Beef 873.10); CP061758, CP061759, CP061760, CP061761, CP061762, and CP061763 (Beef 97.3); and CP061764, CP061765, CP061766, and CP061767 (Beef 730V1).