Water-based epidemiology (WBE) is a valuable early warning tool for tracking the circulation of the virus among the population, including not only symptomatic patients, but also asymptomatic, presymptomatic, and misdiagnosed carriers, which represent a high proportion of the infected population. In the specific case of Barcelona, wastewater surveillance anticipated by several weeks not only the original COVID-19 pandemic wave, but also the onset of the second wave.
KEYWORDS: SARS-CoV-2, COVID-19, epidemiology, surveillance, early warning, sewage
ABSTRACT
Two large wastewater treatment plants (WWTP), covering around 2.7 million inhabitants, which represents around 85% of the metropolitan area of Barcelona, were sampled before, during, and after the implementation of a complete lockdown. Five one-step reverse transcriptase quantitative PCR (RT-qPCR) assays, targeting the polymerase (IP2 and IP4), the envelope (E), and the nucleoprotein (N1 and N2) genome regions, were employed for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA detection in 24-h composite wastewater samples concentrated by polyethylene glycol (PEG) precipitation. SARS-CoV-2 was detected in a sewage sample collected 41 days ahead of the declaration of the first COVID-19 case. The evolution of SARS-CoV-2 genome copies in wastewater evidenced the validity of water-based epidemiology (WBE) to anticipate COVID-19 outbreaks, to evaluate the impact of control measures, and even to estimate the burden of shedders, including presymptomatic, asymptomatic, symptomatic, and undiagnosed cases. For the latter objective, a model was applied for the estimation of the total number of shedders, evidencing a high proportion of asymptomatic infected individuals. In this way, an infection prevalence of 2.0 to 6.5% was figured. On the other hand, proportions of around 0.12% and 0.09% of the total population were determined to be required for positive detection in the two WWTPs. At the end of the lockdown, SARS-CoV-2 RNA apparently disappeared in the WWTPs but could still be detected in grab samples from four urban sewers. Sewer monitoring allowed for location of specific hot spots of COVID-19, enabling the rapid adoption of appropriate mitigation measures.
IMPORTANCE Water-based epidemiology (WBE) is a valuable early warning tool for tracking the circulation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) among the population, including not only symptomatic patients but also asymptomatic, presymptomatic, and misdiagnosed carriers, which represent a high proportion of the infected population. In the specific case of Barcelona, wastewater surveillance anticipated by several weeks not only the original COVID-19 pandemic wave but also the onset of the second wave. In addition, SARS-CoV-2 occurrence in wastewater evidenced the efficacy of the adopted lockdown measures on the circulation of the virus. Health authorities profited from WBE to complement other inputs and adopt rapid and adequate measures to mitigate the effects of the pandemic. For example, sentinel surveillance of specific sewers helped to locate COVID-19 hot spots and to conduct massive numbers of RT-PCR tests among the population.
INTRODUCTION
Despite COVID-19 being a respiratory disease, the prolonged shedding of large amounts of coronavirus genomes in the feces (1, 2), which ultimately reach wastewater (3, 4), has been reported. Hence, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surveillance in sewage is considered a sensitive tool to monitor the spread of the virus among the population (5–7). There is no epidemiological evidence that sewage could be a transmission route for SARS-CoV-2, through contamination of bathing areas or irrigation waters, because very few studies report culture of infectious virus from stool (8). In fact, Zang and coworkers reported that SARS-CoV-2 released into the intestinal lumen is inactivated by human colonic fluid, and hence infectious virus is seldom recovered from the stool specimens of COVID-19 patients (9). In addition, the specific infectivity of SARS-CoV-2 in respiratory samples has been reported to be around 1 infectious unit per 106 or 107 genome copies (10, 11). Hence, infectious SARS-CoV-2 is unlikely to be present in wastewater.
At the time of this study, Spain ranked in fourth place in absolute number of cases worldwide and topped the list in Europe regarding the number of cases and deaths per 1 million inhabitants, with Barcelona being the second-most-affected area in the country (https://www.worldometers.info/coronavirus/). The first case in Barcelona (actually, the first in continental Spain) was reported on 15 February 2020. A complete lockdown was implemented in Spain on 15 March that gradually came to an end between 25 May and 21 June. The total number of reported cases in metropolitan Barcelona at the end of the lockdown in May 2020 was over 29,000 (https://salutweb.gencat.cat/ca/inici/nota-premsa/index.html?id=385948#googtrans(ca%Cen).
Two large wastewater treatment plants (WWTPs), WWTP1 (capacity, 525 million liters per day [MLD]) and WWTP2 (capacity, 420 MLD) cover around 2.7 million inhabitants, representing around 85% of the densely populated metropolitan area of Barcelona. The present extended study describes the evolution of the occurrence of SARS-CoV-2 RNA in these large WWTPs before, during, and after the lockdown, evidencing the validity of water-based epidemiology (WBE) to (i) anticipate COVID-19 outbreaks, (ii) evaluate the impact of the control measures, and (iii) estimate the burden of infected patients, including presymptomatic, asymptomatic, symptomatic, and undiagnosed cases.
RESULTS AND DISCUSSION
Time-evolution of SARS-CoV-2 RNA in wastewater during the pandemic.
The evolution of SARS-CoV-2 genome copies in sewage from the two main WWTPs in the metropolitan area of Barcelona is shown in Fig. 1. In WWTP1, maximum genome copy numbers of SARS-CoV-2 were detected in the initial sample collected on 13 April. A progressive decrease was observed thereafter. This decrease was observed employing polymerase (IP2 and IP4) targets (Fig. 1A) and confirmed with envelope (E) and nucleoprotein (N1 [Fig. 1B and C] and N2 [see Fig. S1 in the supplemental material]) targets. On 18 May, the genomes disappeared, although residual levels could be again detected on 25 May employing the N1 target.
FIG 1.
Evolution of SARS-CoV-2 genomes in two large Barcelona wastewater treatment plants (WWTP). (A and D) Detection of the RNA-dependent RNA polymerase gene (IP2 and IP4 primers). (B and E) Detection of the envelope protein gene (E primers). (C and F) Detection of the nucleoprotein gene (N1 primers). The absence of values on a given date is due to the unavailability of aliquots to assay. Dashed lines depict limits of detection. Red, orange, and green arrows indicate phase 1, phase 2, and phase 3 of the deconfinement, respectively (Table S1).
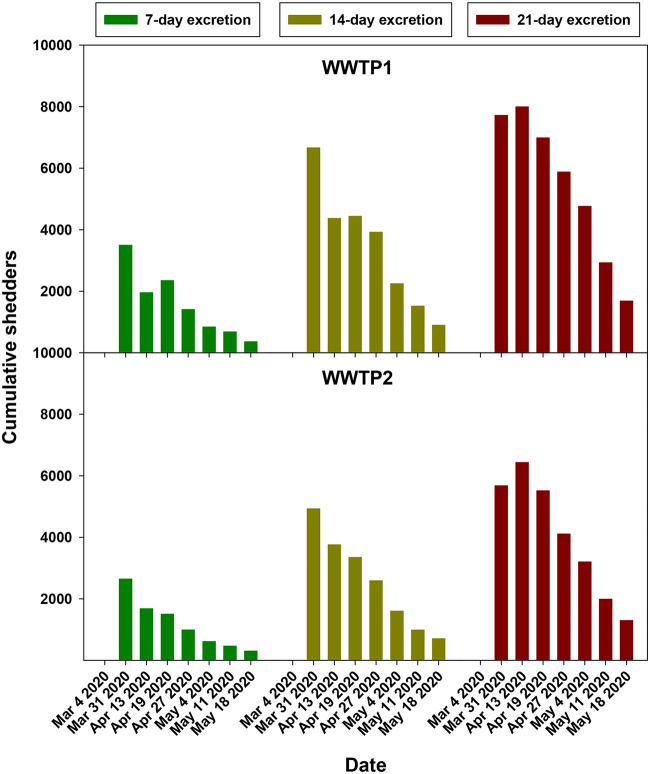
For WWTP2, samples from December 2019 to May 2020 were available, which opened the possibility to better analyze the dynamics of genome copy numbers in sewage. The analysis of archival samples revealed the increasing occurrence of SARS-CoV-2 genomes in samples from 15 January to 4 March employing the IP2, IP4, and E targets (Fig. 1D and E). Genome copy numbers peaked between 4 March and 4 May independently of the target used (Fig. 1D to F). Of note, SARS-CoV-2 was detected in sewage 41 days (15 January) ahead of the declaration of the first COVID-19 case (25 February), clearly evidencing the validity of wastewater surveillance to anticipate cases in the population. Again, as for WWTP1, genomes became undetectable on 18 May (Fig. 1D and E) except when employing the N1 target, whose signal completely disappeared on 25 May (Fig. 1F). The progressive decline in genome copy numbers in both WWTPs paralleled the diminution in the estimated cumulative number of shedders, based on the actual number of reported symptomatic cases and figured for 7-day, 14-day, and 21-day excretion periods before the sampling date (Fig. 2). This genome copy decay evidences the effect of the lockdown measures on the spread of the infection.
FIG 2.
Cumulated SARS-CoV-2 shedders associated with WWTP1 and WWTP2, determined by estimating fecal excretion periods of 7, 14, and 21 days and based on the actual number of reported symptomatic cases.
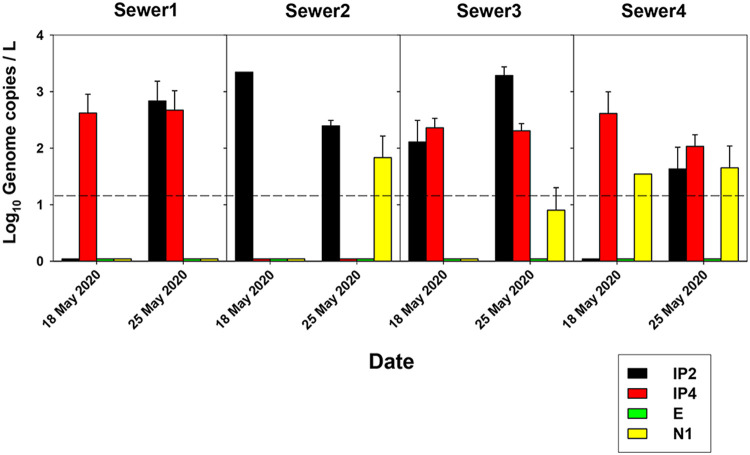
On 25 May, phase 1 of the gradual deconfinement was implemented (Fig. 1, Table S1). However, despite the apparent disappearance of SARS-CoV-2 RNA in the WWTPs around 18 to 25 May, the analysis of grab samples, collected at 8 to 9 a.m. on 18 and 25 May from four urban sewers, revealed the occurrence of virus genomes (Fig. 3), indicating that the virus was still circulating in the population. A higher dilution factor applies in the WWTPs than in the sewers, which together with possible differences applying between grab and composite samples, as well as bowel habits (12), could explain why the WWTP samples were negative for the virus, while genome copies were still detected in the sewer samples. Sewer analysis may provide the most relevant information for the specific localization of areas where COVID-19 cases reappear, enabling an immediate response to prevent spread of the outbreak. Nevertheless, it represents a more laborious and costly approach than surveillance through WWTP monitoring.
FIG 3.
SARS-CoV-2 genome copy levels in grab samples from four urban sewers, detected with primers for IP2, IP4, E, N1, and N2. Sewer1 drains into WWTP1, while sewer2, sewer3, and sewer4 drain into WWTP2.
From 2 to 8 June, SARS-CoV-2 genomes reappeared in both studied WWTPs and increased thereafter. All the reverse transcriptase quantitative PCR (RT-qPCR) targets but the E target revealed this gradual raise (Fig. 1). Failure of the E target may be explained by the increasing circulation of viral variants with a recently described recurrent mutation affecting the probe-binding site (13). Throughout our study, five different RT-qPCR assays targeting different genome regions were employed for SARS-CoV-2 detection in order to increase the robustness of our data. From our own experience in this and other unpublished studies on the occurrence of SARS-CoV-2 in wastewater, only 10% of the samples are positive for the five RT-qPCR targets, indicating the need to employ more than one of these. In samples positive for all the five targets, the observed differences in quantification cycle (Cq) value did not translate into major differences in genome copy number in the corresponding standard curves (Fig. S1 and S2). However, since the N2 target provided some inconsistent results in comparison with the rest of the employed targets, for the sake of clarity, data generated with the N2 target are only shown in Fig. S3. Current RT-PCR assays employed for SARS-CoV-2 in WBE studies are diverse and demand harmonization as a step forward toward the development of standardized methodologies.
Phases 2 and 3 of the deconfinement were eventually applied on 8 and 21 June, respectively (Fig. 1); phase 3 was delayed due to the SARS-CoV-2 levels detected in sewage. Nevertheless, in early July, a huge outbreak was declared (over 300 cases confirmed in 2 weeks and around 10,000 cases in 14 weeks (https://canalsalut.gencat.cat/ca/inici/nota-premsa/index.html?id=387275#googtrans(ca%Cen) in a neighborhood whose sewers (Fig. 3, Sewer3 and Sewer4) drain into WWTP2, where genome copy numbers had started to increase around 3 to 4 weeks in advance (Fig. 1D and F).
Estimation of the total number of active shedders from SARS-CoV-2 RNA levels in wastewater.
WBE constitutes a valuable complementary tool for the surveillance of current infectious agents among the population (14, 15). In particular, WBE may contribute to comprehensive management of the spread of SARS-CoV-2 infection. Nevertheless, information is required to relate the detected genome copy numbers in wastewater to the numbers of infected individuals in the community, encompassing both symptomatic, presymptomatic, asymptomatic, and undiagnosed shedders.
A simple and intuitive model was elaborated based on the SARS-CoV-2 genome copy numbers per liter of sewage, wastewater flow at the sampling point during the sampling period (Table S2), and genome copy numbers shed in the feces of infected individuals. Wölfel and colleagues (11) reported SARS-CoV-2 shedding in stool based on RT-qPCR employing the E primers (16) (V. M. Corman, Charité Berlin, personal communication). Data generated with the E target were available from 13 April to 11 May and from 31 March to 11 May in samples from WWTP1 and WWTP2, respectively. The number of shedders, including symptomatic, presymptomatic, asymptomatic, and undiagnosed cases, was estimated following the model (Fig. 4). On 13 April, this estimation was 30,096 and 28,747 shedders, which accounted for a 2.0% and 2.4% prevalence, in WWTP1 and WWTP2, respectively. However, on 31 March, the estimation was 77,994 shedders and 6.5% prevalence in WWTP2. The applied model provided a sound estimation of the number of shedders in our setting. However, the simplicity of the model enables further refinements related to the percentage of shedders, which in our case was assumed to be 100%, and/or variations in virus load in feces of symptomatic, presymptomatic, and asymptomatic shedders when reliable data are available. An additional adjustment to the model is related to the threshold of genome copies in sewage to discern between periods of high and low stool excretion, which in our case was established to be 102.5 genome copies (gc)/liter. This value may vary depending on the WWTP type, the million liters per day capacity, or technical factors inherent to the SARS-CoV-2 detection pipeline, i.e., virus concentration, RNA extraction, and RT-qPCR efficiencies, and the reference material used in the standard curve, which all contribute to a degree of uncertainty. Additional sources of uncertainty are the limited number of assayed replicas; in our case, genome copies were determined in duplicate, while only a single value of the daily wastewater flow was available. Nevertheless, for influenza, a well-characterized respiratory infection with similar transmissibility and for which natural and/or vaccine-induced immunity exists, a 2018 CDC study determined that the percentage of the U.S. population sickened each season by flu was about 8%, with a range of between 3% and 11%. When asymptomatic cases were also considered, the estimate rose from 5% to 20% (17), which is not far from our estimate of COVID-19 in metropolitan Barcelona.
FIG 4.
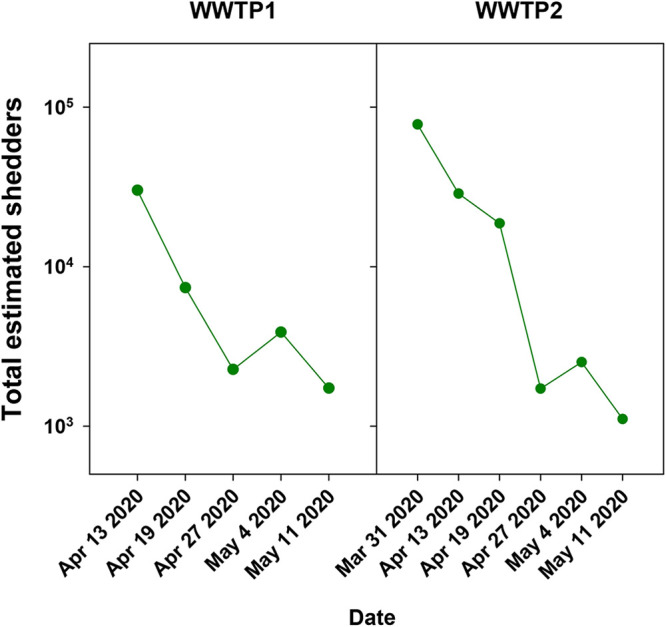
Estimation of the total number of SARS-CoV-2 infected shedders, including symptomatic, presymptomatic, asymptomatic, and undiagnosed cases. A model was developed based on the genome copies at the wastewater treatment plants detected during the first wave of the pandemic using the E target; the reported numbers of genome copies excreted in feces were also determined by employing the E target (11) and considering the actual daily wastewater flow.
Our data fall within the range of seroprevalence reported in the literature, taking into account the uncertainty of the seroprevalence assays, associated with the time of sample collection in the convalescence phase, the immuno-status, and/or the age of the patients and the determination kit employed. A study conducted in Spain based on the detection of antibodies directed to the S protein revealed an overall 5% seroprevalence (18), with substantial geographic variability, e.g., over 10% and 7% in the Madrid and Barcelona areas, respectively. Similarly, adjusted estimates of the persons seroreactive to SARS-CoV-2 spike protein antibodies in the San Francisco and New York City areas were 1% and 7%, respectively (19).
Hart and Halden (20) reported through computational analysis that, in worst-case conditions, a 0.88% prevalence is required for successful detection of SARS-CoV-2 in sewage, while Ahmed and coworkers reduced this requirement to a prevalence of only 0.025% (5). In the present study, the last positive RNA signal with the E target was observed on 11 March in both WWTP (Fig. 1). Applying our model, a proportion of around 0.12% and 0.09% of the total population (1,732 and 1,109 infected individuals, respectively) is required for positive detection in WWTP1 and WWTP2, respectively (Fig. 4).
Our SARS-CoV-2 early detection in sewage supports the idea that cases may have been present in the population before the first imported case was reported. COVID-19 cases may have been misclassified as influenza diagnoses in primary care, boosting community transmission before public health measures were taken (21). Most COVID-19 cases show mild influenza-like symptoms (22), and it has been suggested that some uncharacterized influenza cases may have masked some COVID-19 cases in the 2019–2020 season (21).
Our data reveal the significant proportion of presymptomatic and asymptomatic carriers that nevertheless shed SARS-CoV-2 and contribute to the spread of the virus (23, 24). The enormous burden in morbidity and mortality of COVID-19 calls for sentinel surveillance of SARS-CoV-2 in wastewater to enable rapid mitigation measures in pandemic waves and to evaluate the usefulness of lockdown and deconfinement measures. Presently, surveillance networks comprising 56 WWTP in Catalonia (Catalonian Health Authority, Catalonian Water Agency, and Catalonian Institute of Water Research, https://sarsaigua.icra.cat/) and 30 WWTP in Spain (VATar Project, Ministry of Health and Ministry of the Environment, https://www.miteco.gob.es/es/agua/temas/concesiones-y-autorizaciones/vertidos-de-aguas-residuales/alerta-temprana-covid19/default.aspx) have been implemented.
MATERIALS AND METHODS
Wastewater samples.
Composite raw sewage samples corresponding to 24 h were collected weekly from two large wastewater treatment plants (WWTP1 and WWTP2) in the metropolitan area of Barcelona from 13 April, in the peak of the COVID-19 first wave, to 7 July. In addition, for WWTP2, frozen archival samples collected monthly from January to March 2020 were also assayed. Furthermore, grab samples were collected from sewer maintenance holes on 18 and 25 May at 8 to 9 a.m. The time of grab sample collection was selected according the bowel habits of the population (12).
Wastewater concentration.
Samples of wastewater (800 ml) were concentrated through precipitation with 20% polyethylene glycol 6000 and resuspended in 3 ml of phosphate-buffered saline (PBS), pH 7.4 (25). This procedure provided us with a mean recovery efficiency of 2.53% ± 0.17% of the attenuated porcine coronavirus PUR46-MAD strain of transmissible gastroenteritis virus (kindly provided by L. Enjuanes and I. Sola, National Center of Biotechnology, Cantoblanco, Madrid; 26).
Nucleic acid extraction and virus quantification.
Nucleic acid extraction was performed from 1 ml of the concentrate and eluted in 50 μl using the NucliSENS miniMAG extraction system (bioMérieux).
Five one-step RT-qPCR assays (RNA UltraSense one-step quantitative RT-PCR system; Invitrogen, Life Technologies) were employed targeting the RNA-dependent RNA polymerase (RdRp) gene, IP2 and IP4 fragments (Institut Pasteur, Paris; protocol, real-time RT-PCR assays for the detection of SARS-CoV-2 [2020]; https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2), the envelope protein (E) gene, E fragment (Charité, Berlin) (16), and the nucleoprotein (N) N1 and N2 fragments (CDC, Atlanta; CDC 2019-Novel Coronavirus [2019-nCoV] real-time RT-PCR diagnostic panel 2020; https://www.fda.gov/media/134922/download).
Standard curves were constructed using the Twist synthetic SARS-CoV-2 RNA control 2 (MN908947.3; Twist Bioscience). Figure S1 shows the average standard curves for each of the targets used.
Quality control and quality assurance to determine any potential contamination and/or inhibition were conducted using negative and positive controls, respectively. Positive controls consisted of the addition of two distilled water samples containing 5 × 103 copies of the Twist RNA, which were run in parallel in each RT-qPCR plate. Direct and 1/10 diluted replicas were assayed to ascertain assay inhibition. All quantitative assays were performed in duplicate; hence, the depicted genome copy numbers correspond to the mean of four values. Negative controls comprised five distilled water samples per run, two from the beginning of the assay, to control any potential contamination during the RNA extraction, and three in the RT-PCR, to control any potential contamination during nucleic acid amplification.
Estimation of SARS-CoV-2 shedders.
The number of symptomatic SARS-CoV-2 shedders was determined from the actual number of reported cases in the metropolitan Barcelona area (https://aquas.gencat.cat/ca/actualitat/ultimes-dades-coronavirus/mapa-per-municipis/). Since SARS-CoV-2 excretion in stool has been reported to be variable and long-lasting (11, 27), we calculated the cumulative number of symptomatic shedders at each given date considering all cases reported on this date and in each of the previous seven (1 to 7) days, 14 (1 to 14) days, and 21 (1 to 21) days.
The total number of SARS-CoV-2 shedders (S), including asymptomatic, presymptomatic, and undiagnosed virus carriers, was determined applying a model integrating the genome copy numbers per liter of sewage (gc/liter), the actual 24-h flow in liters corresponding to each assayed composite sample (F), and the mean genome copy numbers per gram (gc/g) shed per infected patient: S = gc/liter · F/(gc/g stool · g stool).
Genome copy numbers in sewage were determined using the same E-targeted RT-qPCR assay developed at Charité, Berlin, employed for the quantification of the genomes present in stool (11, 16). The number of genomes excreted per patient, was determined using the product of the mean genome copy numbers excreted per gram of stool and the average daily wet weight (wt/wt) of feces. The latter amount was figured to be 380 g, based on an excretion of 30 g (wt/wt) per 5.5 kg of body weight, assuming an average weight of the Spanish population of 70 kg (https://www.mscbs.gob.es/estadEstudios/sanidadDatos/), which falls within the previously reported range (28, 29). The number of genome copies shed by patients has been reported to range from less than 103 gc/g, to over 107 gc/g, depending on the time course of the infection, with higher titers during the first 10 days post-symptom onset (11). Based on these data, we assumed a fecal excretion of 105.3 gc/g (average for the first 10 days) or 104.9 gc/g (average for the rest of the excretion period up to 21 days), depending on whether the number of genomes detected in sewage was higher or lower than 102.5 gc/liter (threshold established to discern between periods of high and low excretion), respectively.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by the REVEAL project, funded by SUEZ Spain.
We thank our colleagues from Aigües de Barcelona, Cetaqua and Labaqua, who provided insight and expertise for the research. We are indebted with the Metropolitan Area of Barcelona (AMB) for a fruitful collaboration. We are grateful to M. Jané-Checa and A. Martínez-Mateo of the Public Health Agency of Catalonia (ASPCAT) for providing epidemiological data and useful discussion. We are indebted to Luis Enjuanes and Isabel Sola (National Center of Biotechnology, Cantoblanco, Madrid) for providing us with the attenuated PUR46-MAD strain of transmissible gastroenteritis virus.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. 2020. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Wang S, Xue Y. 2020. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol 92:680–682. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. 2020. Presence of SARS-Coronavirus-2 in sewage. medRxiv 2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- 4.Lodder W, de Roda Husman AM. 2020. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol Hepatol 5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O'Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas KV, Mueller JF. 2020. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ 728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.La Rosa G, Iaconelli M, Mancini P, Bonanno Ferraro G, Veneri C, Bonadonna L, Lucentini L, Suffredini E. 2020. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ 736:139652. doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G. 2020. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res 181:115942–115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao F, Sun J, Xu Y, Li F, Huang X, Li H, Zhao J, Huang J, Zhao J. 2020. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis 26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zang R, Castro MFG, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ, Ding S. 2020. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. 2020. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brunink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 12.Heaton KW, Radvan J, Cripps H, Mountford RA, Braddon FE, Hughes AO. 1992. Defecation frequency and timing, and stool form in the general population: a prospective study. Gut 33:818–824. doi: 10.1136/gut.33.6.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artesi M, Bontems S, Göbbels P, Franckh M, Maes P, Boreux R, Meex C, Melin P, Hayette M-P, Bours V, Durkin K. 2020. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol 58:e01598-20. doi: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sims N, Kasprzyk-Hordern B. 2020. Future perspectives of wastewater-based epidemiology: monitoring infectious disease spread and resistance to the community level. Environ Int 139:105689. doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barras C. 2018. Going to waste. Nat Med 24:1484–1487. doi: 10.1038/s41591-018-0218-0. [DOI] [PubMed] [Google Scholar]
- 16.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokars JI, Olsen SJ, Reed C. 2018. Seasonal incidence of symptomatic influenza in the United States. Clin Infect Dis 66:1511–1518. doi: 10.1093/cid/cix1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollán M, Pérez-Gómez B, Pastor-Barriuso R, Oteo J, Hernán MA, Pérez-Olmeda M, Sanmartín JL, Fernández-García A, Cruz I, Fernández de Larrea N, Molina M, Rodríguez-Cabrera F, Martín M, Merino-Amador P, León Paniagua J, Muñoz-Montalvo JF, Blanco F, Yotti R, Blanco F, Gutiérrez Fernández R, Martín M, Mezcua Navarro S, Molina M, Muñoz-Montalvo JF, Salinero Hernández M, Sanmartín JL, Cuenca-Estrella M, Yotti R, León Paniagua J, Fernández de Larrea N, Fernández-Navarro P, Pastor-Barriuso R, Pérez-Gómez B, Pollán M, Avellón A, Fedele G, Fernández-García A, Oteo Iglesias J, Pérez Olmeda MT, Cruz I, Fernandez Martinez ME, Rodríguez-Cabrera FD, Hernán MA, Padrones Fernández S, Rumbao Aguirre JM, Navarro Marí JM, Palop Borrás B, Pérez Jiménez AB, Rodríguez-Iglesias M, Calvo Gascón AM, et al. 2020. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet 396:535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havers FP, Reed C, Lim T, Montgomery JM, Klena JD, Hall AJ, Fry AM, Cannon DL, Chiang CF, Gibbons A, Krapiunaya I, Morales-Betoulle M, Roguski K, Rasheed MAU, Freeman B, Lester S, Mills L, Carroll DS, Owen SM, Johnson JA, Semenova V, Blackmore C, Blog D, Chai SJ, Dunn A, Hand J, Jain S, Lindquist S, Lynfield R, Pritchard S, Sokol T, Sosa L, Turabelidze G, Watkins SM, Wiesman J, Williams RW, Yendell S, Schiffer J, Thornburg NJ. 2020. Seroprevalence of Antibodies to SARS-CoV-2 in 10 Sites in the United States, March 23-May 12, 2020. JAMA Intern Med 180:1576. doi: 10.1001/jamainternmed.2020.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hart OE, Halden RU. 2020. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci Total Environ 730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coma E, Mora N, Prats-Uribe A, Fina F, Prieto-Alhambra D, Medina-Peralta M. 2020. Excess cases of influenza suggest an earlier start to the coronavirus epidemic in Spain than official figures tell us: an analysis of primary care electronic medical records from over 6 million people from Catalonia. medRxiv 2020.04.09.20056259. [Google Scholar]
- 22.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, Ng OT, Marimuthu K, Ang LW, Mak TM, Lau SK, Anderson DE, Chan KS, Tan TY, Ng TY, Cui L, Said Z, Kurupatham L, Chen MI, Chan M, Vasoo S, Wang LF, Tan BH, Lin RTP, Lee VJM, Leo YS, Lye DC, Singapore 2019 Novel Coronavirus Outbreak Research Team. 2020. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA 323:1488–1494. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. 2020. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Kang H, Liu X, Tong Z. 2020. Asymptomatic cases with SARS-CoV-2 infection. J Med Virol 92:1401–1403. doi: 10.1002/jmv.25990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco A, Abid I, Al-Otaibi N, Perez-Rodriguez FJ, Fuentes C, Guix S, Pinto RM, Bosch A. 2019. Glass wool concentration optimization for the detection of enveloped and non-enveloped waterborne viruses. Food Environ Virol 11:184–192. doi: 10.1007/s12560-019-09378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez CM, Jimenez G, Laviada MD, Correa I, Sune C, Bullido M, Gebauer F, Smerdou C, Callebaut P, Escribano JM, Enjuanes L. 1990. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology 174:410–417. doi: 10.1016/0042-6822(90)90094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, Yin H, Xiao Q, Tang Y, Qu X, Kuang L, Fang X, Mishra N, Lu J, Shan H, Jiang G, Huang X. 2020. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penn R, Ward BJ, Strande L, Maurer M. 2018. Review of synthetic human faeces and faecal sludge for sanitation and wastewater research. Water Res 132:222–240. doi: 10.1016/j.watres.2017.12.063. [DOI] [PubMed] [Google Scholar]
- 29.Rose C, Parker A, Jefferson B, Cartmell E. 2015. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit Rev Environ Sci Technol 45:1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.