Abstract
IDH1 and IDH2 mutations (IDH1/2Mut) are recognized as recurrent genetic alterations in acute myeloid leukemia (AML) and associated with both clinical impact and therapeutic opportunity due to the recent development of specific IDH1/2Mut inhibitors. In T-cell acute lymphoblastic leukemia (T-ALL), their incidence and prognostic implications remain poorly reported. Our targeted next-generation sequencing approach allowed comprehensive assessment of genotype across the entire IDH1 and IDH2 locus in 1085 consecutive unselected and newly diagnosed patients with T-ALL and identified 4% of, virtually exclusive (47 of 49 patients), IDH1/2Mut. Mutational patterns of IDH1/2Mut in T-ALL present some specific features compared to AML. Whereas IDH2R140Q mutation was frequent in T-ALL (25 of 51 mutations), the IDH2R172 AML hotspot was absent. IDH2 mutations were associated with older age, an immature phenotype, more frequent RAS gain-of-function mutations and epigenetic regulator loss-of-function alterations (DNMT3A and TET2). IDH2 mutations, contrary to IDH1 mutations, appeared to be an independent prognostic factor in multivariate analysis with the NOTCH1/FBXW7/RAS/PTEN classifier. IDH2Mut were significantly associated with a high cumulative incidence of relapse and very dismal outcome, suggesting that IDH2-mutated T-ALL cases should be identified at diagnosis in order to benefit from therapeutic intensification and/or specific IDH2 inhibitors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-021-01068-4.
Keywords: IDH1, IDH2, T-ALL
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is aggressive neoplasms resulting from the proliferation of T-lymphoid progenitors blocked at thymic stages of differentiation and account for 15% and 25% of pediatric and adult ALLs, respectively [1]. T-ALL is associated with a wide range of acquired genetic abnormalities that contribute to developmental arrest and abnormal proliferation [2]. Although intensive treatment protocols have markedly improved the outcomes of children with T-ALL, cure rates remain below 60% for adults and 85% for children [3–5]. The prognosis is particularly poor in relapsing patients, justifying the development of novel targeted therapies [6, 7]. For example, alterations affecting epigenetic factors may offer novel targeted therapeutic approaches in high-risk T-ALL [8].
Whole-genome sequencing of AML identified acquired mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) [9]. These paralogous genes encode two enzymes with distinct localizations (cytoplasmic for IDH1 and mitochondrial for IDH2). Both catabolize the conversion of isocitrate to α-ketoglutarate (α-KG). Gain-of-function IDH1/2 mutations (IDH1/2Mut) confer a neomorphic activity on the encoded enzymes, leading to the conversion of α-KG to 2-hydroglutatarate (2-HG) in a NAD phosphate-dependent manner [10]. Accumulation of the oncometabolite 2-HG induces multiple cellular alterations, including chromatin methylation and cellular differentiation, by inhibiting α-KG-dependent enzymes related to DNA methylation, such as Tet oncogene family members (TET2, TET3) [11]. IDH1/2Mut have been reported in 10 to 20% of AML cases, when they are predominantly located in the active site of the enzyme (IDH1R132, IDH2R140Q and IDH2R172). IDH1/2Mut in AML are associated with prognostic impact influenced by the genetic context [12, 13]. Importantly, specific drugs targeting mutant IDH1 or IDH2 have recently shown promise in IDH1/2Mut refractory or relapsed AML patients [14, 15].
In T-ALL, IDH1/2Mut have been partially explored and their prognostic impact poorly reported [16, 17]. We now provide the first comprehensive analysis and oncogenetic landscape of IDH1/2Mut in a cohort of 1085 T-ALL patients, when the nearly 4% of IDH1/2Mut are associated with extremely poor prognosis, specifically in IDH2-mutated cases.
Methods
Patient’s protocol and clinical trials
Diagnostic peripheral blood or bone marrow samples from 1085 adults and children with T-ALL were analyzed after informed consent was obtained at diagnosis according to the Declaration of Helsinki. Among the 1085 T-ALL analyzed, 215 adult patients aged from 16–59 years were included in the GRAALL03/05 trials (details provide in supplementary) which were registered at clinicaltrials.gov (GRAALL-2003, #NCT00222027; GRAALL-2005, #NCT00327678). and 261 pediatric patients aged from 1 to 19 years were treated in 10 French pediatric hematology departments, members of the FRALLE study group, according to the FRALLE 2000 T guidelines (Additional file 2: Fig. S5 and Additional file 1: Table S3).
Gene mutation screening
A custom capture Nextera XT gene panel (Illumina, San Diego, CA) targeting all coding exons and their adjacent splice junctions of 80 genes was designed, based on available evidence in hematological neoplasms (Additional file 1: Table S1). DNA Libraries were prepared using Nextera Rapid Capture Enrichment protocol and underwent 2 × 150 bp paired-end sequencing on Illumina MiSeq sequencing system with MiSeq Reagent Kit v2 (Illumina). Briefly, sequence reads were filtered and mapped to the human genome (GRCh37/hg19) using in-house software (Polyweb, Institut Imagine, Paris). Annotated variants were selected after filtering out calls according to the following criteria: (1) coverage < 30×, < 10 alternative reads or variant allelic fraction (VAF) < 7%; (2) polymorphisms described in dbSNP, 1000Genomes, EVS, Gnomad and EXAC with a calculated mean population frequency > 0.1%. Non-filtered variants were annotated using somatic database COSMIC (version 78) and ProteinPaint (St Jude Children’s Research Hospital – Pediatric Cancer data portal). Lollipop plots were generated with ProteinPaint (https://pecan.stjude.org/#/proteinpaint).
Immunophenotypic and molecular characterization of T-ALL samples
Peripheral blood or bone marrow T-ALL samples were analyzed for immunophenotype, fusion transcripts (SIL-TAL1, CALM-AF10), oncogenic transcripts (HOXA9, TLX1 and TLX3) and T-cell receptor (TCR) recombination and NOTCH1/FBXW7/RAS/PTEN mutations, as previously described [4, 18, 19].
Minimal residual disease assessment
Immunoglobulin/T-cell receptor (Ig/TCR) gene rearrangement-based Minimal Residual Disease (MRD) evaluation was centrally assessed for patients who reached complete remission after the first induction cycle, on BM samples after induction (MRD1). MRD was centrally assessed by real-time quantitative allele-specific oligonucleotide PCR and interpreted according to EuroMRD group guidelines [20–22].
Statistical analysis
Comparisons for categorical and continues variables between IDH1Mut or IDH2Mut and IDHWT subgroups were performed with Fisher's exact test and Mann–Whitney test, respectively. Overall survival (OS) was calculated from the date of diagnosis to the last follow-up date censoring patients alive. The cumulative incidence of relapse (CIR) was calculated from the complete remission date to the date of relapse censoring patients alive without relapse at the last follow-up date. Relapse and death in complete remission were considered as competitive events. Univariate and multivariate analyses assessing the impact of categorical and continuous variables were performed with a Cox model. Proportional-hazards assumption was checked before conducting multivariate analyses. In univariate and multivariate analyses, age and log10(WBC) were considered as continuous variables. All analyses were stratified on the trial. Variables with a p value less than 0.1 in univariate analysis were included in the multivariable models. Statistical analyses were performed with STATA software (STATA 12.0 Corporation, College Station, TX). All p-values were two-sided, with p < 0.05 denoting statistical significance. Circos plots were generated using R software.
Results and discussion
Incidence of IDH1 and IDH2 mutations in 1085 T-ALL
A total of 51 (4%) mutations, mainly clonal, in either IDH1 or IDH2 were apparent in 49 cases (Fig. 1a and Additional file 1: Table S2, Additional file 2: Figs. S2, S3). IDH1 mutations were identified in 19 T-ALL cases (2%) and IDH2 mutations in 32 cases (3%). IDH1/2Mut were mutually exclusive except in 2 cases. The IDH2R140Q mutation was the most prevalent mutation affecting IDH2 (n = 25, 78%). We identified 7 IDH1 mutations located in the R132 hotspot (37% of IDH1 mutations), 3 cases with IDH1R132C mutation, 2 with IDH1R132S, 1 with IDH1R132H and IDH1R132G mutation. The most common IDH2 mutations in AML occur at R140 followed by residue IDH2R172. The latter mutation is virtually the only IDH mutation found in angio-immunoblastic T cell lymphoma, reported in about 30% of cases (Additional file 2: Fig. S1) [23]. IDH2R172 mutation has also been rarely and inconsistently described in peripheral T-cell lymphoma not otherwise specified (NOS) with T-follicular helper (TFH) phenotype [24, 25]. In striking contrast, IDH2R172 was not reported in our series of T-ALL. IDH1R132, the most frequent IDH1 mutation reported in our cohort, has recently been recognized to cooperate with NOTCH1 activation in a T-ALL mouse model [26]. These results highlight the specific consequence associated with IDH1/2Mut subtype during immature T-cell development.
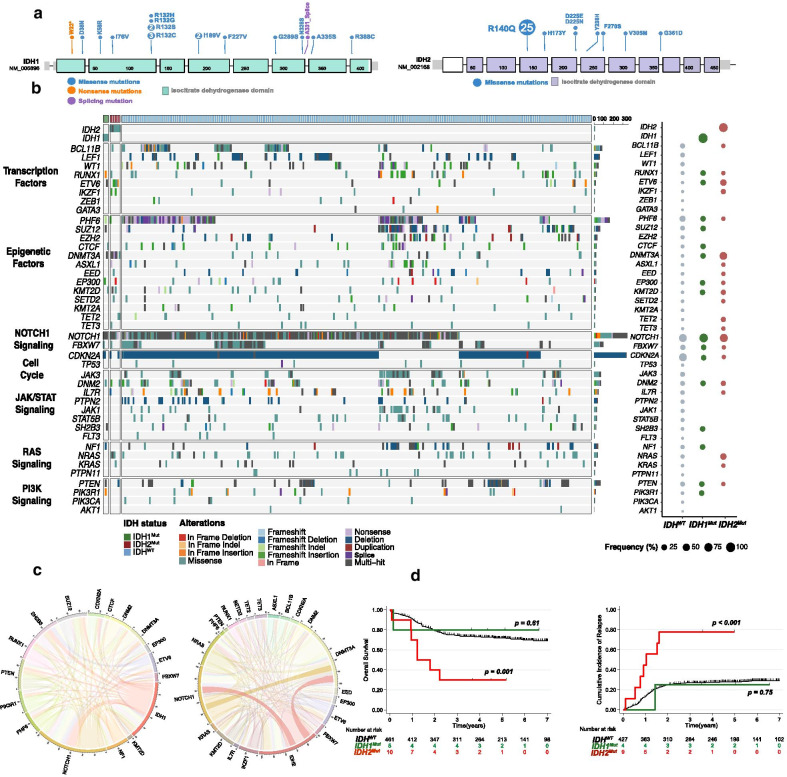
Fig. 1.
IDH1 and IDH2 mutations in the GRAALL03/05 and FRALLE2000 studies. a Lollipop plots indicating the observed mutations for each IDH gene and their consequences. b Oncoplot depicting the genetic anomalies observed in IDH1/2-Mutated or Wild type T-ALL cases of the GRAALL03/05 and FRALLE2000 studies. Genes are classified by functional groups. The right panel indicates the overall frequency of alterations per gene. c The circos plots depict the co-occurrences in genetic lesions observed in IDH1 (left panel) and IDH2 mutated T-ALL (right panel). d Clinical impact of IDH1 and IDH2 mutations in the GRAALL0305 and FRALLE2000 studies. Overall survival (left panel) and cumulative incidence of relapse (right panel). The red curve represents the IDH2-mutated patients, the green curve the IDH1-mutated patients and the black curve the IDHWt patients
Clinico-biological characteristics of IDH1/2Mut in GRAALL and FRALLE-treated T-ALLs
We then investigated the clinical characteristics linked to IDH1/2Mut in a subset of 476 patients, including 215 adults enrolled in the GRAALL-2003/2005 trials and 261 children enrolled in the FRALLE-2000 trial (Table 1 and Supplemental Methods). The incidence of IDH1/2Mut in this cohort was 3% (15/476). IDH1 mutations were detected in 5 patients (4 adult and 1 pediatric case), and IDH2 mutations were identified in 10 (6 adult and 4 pediatric cases) (Additional file 2: Fig. S2). IDH2R140Q was the most frequent mutation (n = 7, 70%) and was most prevalent in adults' patients (n = 6/7, 86%). Overall, IDH1/2Mut were observed in 5% of adults and 2% of children (p = 0.1).
Table 1.
Clinico-biological and outcome characteristics of adult and pediatric T-ALL (GRAALL and FRALLE protocols) according to IDH1/2 status
| Variable | IDH2Mut (n = 10) | p value2 | Overall (n = 476) | p value2 | IDH1Mut (n = 5) |
|---|---|---|---|---|---|
| Male | 7/10 (70%) | 0.72 | 357/476 (75%) | 0.34 | 5/5 (100%) |
| Age (y)1 | 47.6 (3.6–59.1) | 0.01 | 15.3 (1.1–59.1) | 0.26 | 21.6 (5.4–56.5) |
| WBC (G/L)1 | 9 (1–400) | 0.01 | 64 (0–980) | 0.60 | 80 (4–110) |
| CNS involvement | 1/10 (10%) | 0.99 | 51/474 (11%) | 0.99 | 0/5 (0%) |
| Immunophenotype | |||||
| ETP phenotype | 3/5 (60%) | 0.04 | 56/307 (18%) | 0.54 | 1/4 (25%) |
| Immature (IM0/δ/γ) | 5/7 (71%) | 0.006 | 89/419 (21%) | 0.99 | 1/5 (20%) |
| Cortical (IMB, preαβ) | 0/7 (0%) | 0.007 | 211/419 (50%) | 0.68 | 2/5 (40%) |
| Mature TCRαβ | 1/7 (14%) | 0.99 | 66/419 (16%) | 0.99 | 0/5 (0%) |
| Mature TCRγδ | 1/7 (14%) | 0.99 | 53/419 (13%) | 0.12 | 2/5 (40%) |
| Oncogenetic classification | |||||
| TLX1 | 0/8 (0%) | 0.60 | 54/415 (13%) | 0.99 | 0/5 (0%) |
| TLX3 | 1/8 (12%) | 0.99 | 72/415 (17%) | 0.21 | 2/5 (40%) |
| SIL-TAL1 | 0/8 (0%) | 0.61 | 57/415 (14%) | 0.99 | 0/5 (0%) |
| CALM-AF10 | 0/8 (0%) | 0.99 | 13/415 (3%) | 0.99 | 0/5 (0%) |
| High-risk classifier | 8/10 (80%) | 0.03 | 209/476 (44%) | 0.99 | 2/5 (40%) |
| Treatment response | |||||
| Rapid prednisone response | 3/10 (30%) | 0.12 | 259/467 (55%) | 0.66 | 2/5 (40%) |
| Complete Remission | 9/10 (90%) | 0.54 | 440/476 (92%) | 0.32 | 4/5 (80%) |
| MRD1 > 10–4 | 1/1 (100%) | 0.36 | 123/340 (36%) | 0.99 | 1/4 (25%) |
| Allo-HSCT | 2/10 (20%) | 0.99 | 101/456 (22%) | 0.99 | 1/5 (20%) |
| Outcome | |||||
| 4-year CIR (95% CI) | 78% (49;97) | < 0.0013 | 29% (25;33) | 0.753 | 25% (4;87) |
| 4-year OS (95% CI) | 30% (7;58) | 0.0013 | 71% (67;75) | 0.613 | 80% (20;97) |
| Univariate and multivariate analysis3 | ||||||
|---|---|---|---|---|---|---|
| Univariate | Multivariate | |||||
| CIR | SHR | 95%CI | p | SHR | 95%CI | p |
| Age | 1.01 | (0.98; 1.03) | 0.57 | - | - | - |
| CNS | 1.57 | (0.85; 2.59) | 0.08 | 1.33 | (0.80; 2.20) | 0.28 |
| Log(WBC) | 1.62 | (1.2; 2.18) | 0.002 | 1.63 | (1.20; 2.22) | 0.002 |
| Prednisone response | 0.67 | (0.47; 0.95) | 0.03 | 1.00 | (0.68; 1.46) | 0.99 |
| High-risk Classifier | 2.78 | (1.94; 3.99) | < 0.001 | 2.62 | (1.81; 3.79) | < 0.001 |
| IDH2Mut | 4.28 | (1.99; 9.23) | < 0.001 | 4.06 | (1.84; 8.96) | 0.001 |
| OS | HR | 95%CI | p | HR | 95%CI | p |
| Age | 1.03 | (1.01; 1.05) | 0.001 | 1.04 | (1.02; 1.07) | < 0.001 |
| CNS | 2.00 | (1.28; 3.14) | 0.002 | 1.67 | (1.02; 1.07) | 0.03 |
| Log(WBC) | 1.99 | (1.48; 2.67) | < 0.001 | 2.00 | (1.46; 2.76) | < 0.001 |
| Prednisone response | 0.54 | (0.38; 0.76) | < 0.001 | 0.85 | (0.59; 1.24) | 0.41 |
| High-risk Classifier | 2.93 | (2.06; 4.17) | < 0.001 | 2.90 | (2.00; 4.19) | < 0.001 |
| IDH2Mut | 3.56 | (1.66; 7.65) | 0.001 | 1.98 | (0.86; 4.57) | 0.11 |
p-values < 0.05 are indicated in bold
MRD1 correspond to MRD evaluation after induction and was performed by allele-specific oligonucleotides polymerase chain reaction. T-cell receptor status and oncogenic were performed as described in supplemental methods. IDH1Mut and IDH2Mut were statistically compared to IDH1WT and IDH2WT patients, respectively
T-ALL: T-cell acute lymphoblastic leukemia; WBC, white blood count; CNS, central nervous system; ETP, early thymic precursor; High Risk classifier, NOTCH1/FBXW7-RAS/PTEN classifier as previously described [3, 4]; CR, complete remission; MRD, minimal residual disease; Allo-HSCT, allogenic hematopoietic stem cell transplantation; CIR, cumulative incidence of relapse; OS, overall survival; HR: hazard ratio, SHR: specific hazard ratio, CI: confidence interval
1Statistics presented: Median (Minimum–Maximum)
2Statistical tests performed: Fisher's exact test; Wilcoxon rank-sum test
3Univariate and multivariate Cox analyses stratified on protocol
IDH1 and IDH2 mutations are associated with both specific clinical and mutational profiles
Patients with IDH2Mut were significantly older than IDHWT (median 47.6 years vs 15.0, p = 0.01). IDH2Mut were associated with an immature immunophenotype (5/7, 71% vs 83/407, 20%, p = 0.006) and ETP-phenotype (3/5, 60% vs 52/298, 17%, p = 0.04). In line with this, IDH2Mut correlated positively with abnormalities known to be associated with an immature phenotype, including RAS (50% vs 11%, p = 0.02), ETV6 (40% vs 3%, p < 0.01), DNMT3A (70% vs 3%, p < 0.01), IKZF1 (20% vs 2%, p = 0.02) and TET2 (20% vs 2%, p = 0.04) mutations (Fig. 1b, c). IDH2Mut were mutually exclusive with SIL-TAL1 + cases, associated with a mature TCRαβ lineage. Interestingly, contrary to IDH2-mutated cases, IDH1Mut did not statistically differ from IDHWT patient regarding age, immunophenotype or mutational co-occurrence.
IDH2 mutations, but not IDH1, are associated with a poor prognosis in T-ALL
To investigate the prognostic value of IDH1/2Mut, survival analyses were performed on the 476 patient cohort. IDH1/2Mut cases did not differ significantly with regard to sex, white blood cell count (WBC) or central nervous system (CNS) involvement (Table1). Despite an initial good treatment response (IDH2Mut cases achieved 90% complete remission rate and IDH2Mut did not confer increased poor prednisone response), patients with IDH2Mut had an inferior outcome compared to IDH2Wt (Table1, Fig. 1d, Additional file 2: Fig. S4), with an increased cumulative incidence of relapse (CIR) (4y-CIR: 78% vs 29%; specific hazard ratio (SHR) 4.3, 95%CI (2.0–9.2); p < 0.001) and a shorter overall survival (OS) (4y-OS: 30% vs 71%; hazard ratio: 3.6, 95%CI (1.7–7.7); p = 0.001). In multivariate analysis considering variables associated with CIR and OS in univariate analyses as covariates, IDH2Mut predicted a trend for lower OS (HR: 1.98, 95%CI (0.86–4.57); p = 0.11) and statistically higher CIR (SHR, 4.06, 95%CI (1.84–8.96), p = 0.001) even after adjustment on the 4-gene NOTCH1/FBXW7/RAS/PTEN (NFRP) classifier which identified poor prognosis patients in both GRAALL and FRALLE trials [3, 4]. Conversely to IDH2Mut, IDH1Mut was not associated with poor prognostic impact in T-ALL (4y-CIR: 25% vs 29%, p = 0.75 and 4y-OS: 80% vs 71%, p = 0.61).
We provide the largest comprehensive analysis of IDH1 and IDH2 mutations in T-ALL and highlight for the first time both their clinical profile and, most importantly, the extremely poor prognosis impact associated with IDH2Mut. We describe the specific oncogenetic landscape of IDH1/2Mut and interestingly report that IDH2Mut T-ALL conversely to IDH1Mut were associated with an immature phenotype and alterations such as RAS mutations, transcription factors alterations (ETV6, IKZF1) and epigenetic regulators alterations (TET2, DNMT3A).
Recent studies have shed light on new prognostic factor in T-ALL allowing sharper prediction of the risk of relapse (e.g., NFRP classifier, level of MRD1, IKZF1 alterations) [3, 4, 27]. Despite this, a significant number of T-ALL relapses remain unpredicted, so new predictive markers are needed, given the extremely poor prognosis associated with T-ALL relapse. We therefore consider that IDH2Mut T-ALL cases should be identified at diagnosis to benefit from therapeutic intensification and/or specific IDH2Mut inhibitors [15].
Supplementary Information
Additional file 1. Supplemental Table 1: Custom capture Nextera XT gene panel. Supplemental Table 2: IDH1 and IDH2 mutations identified in 1085 patients with T-ALL. Supplemental Table 3: Chemotherapy in the FRALLE 2000 standard risk group T1 and high risk T2.
Additional file 2. Figure S1: Lollipop plots indicating the observed mutations for IDH1 and IDH2 in the present series confront with Cosmic-reported mutations for AML and AITL. Figure S2: Lollipop plots indicating the observed mutations for IDH1 and IDH2 affecting patients included in FRALLE and GRAALL protocol. Figure S3: Variant Allele Frequency (VAF) of individual IDH1 and IDH2 mutations observed in 1085 T-ALL. Figure S4: OS and CIR according to the IDH1 or IDH2Mut status in the two subgroups (FRALLE and GRALL 03/05). Figure S5: General design of FRALLE 2000 T guidelines.
Acknowledgements
The authors would like to thank all participants in the GRAALL-2003 and GRAALL-2005 study groups, the SFCE and the investigators of the 16 SFCE centers involved in collection and provision of data and patient samples, and V. Lheritier for collection of clinical data.
Abbreviations
- IDH1/2Mut
IDH1-IDH2 Mutations
- AML
Acute myeloid leukemia
- T-ALL
T-cell acute lymphoblastic leukemia
- NFRP
NOTCH1/FBXW7/RAS/PTEN
- CNS
Central nervous system
- WBC
White blood cell count
- NOS
Not otherwise specified
- TFH
T-Follicular helper
- ETP
Early thymic precursor
- MRD
Minimal residual disease
Authors' contributions
N.B, V.A and M.S conceived the study and oversaw the project; M.S, A.S, C.B, ME.D, E.L, C.G, N.G, JM.C, I.A, V.G, F.H, S.D, N.I, H.D, A.B, A.P, N.B provided study materials or patients; M.S, C.B, A.S, E.M, G.P.A and V.A performed molecular analyses; M.S, A.S, C.B, V.A. collected and assembled data; N.B and M.S performed statistical analysis; M.S, A.S, C.B, V.A, N.B, G.P.A analyzed and interpreted data; M.S, N.B, A.S, C.B, E.M, V.A wrote the manuscript. All authors read and approved the final manuscript.
Funding
The GRAALL was supported by grants P0200701 and P030425/AOM03081 from the Programme Hospitalier de Recherche Clinique, Ministère de l’Emploi et de la Solidarité, France and the Swiss Federal Government in Switzerland. Samples were collected and processed by the AP-HP “Direction de Recherche Clinique” Tumor Bank at Necker-Enfants Malades. MS was supported by « Action Leucémie» and « Soutien pour la formation à la recherche translationnelle en cancérologie dans le cadre du Plan cancer 2009–2013». This work was supported by grants to Necker laboratory from the “Association Laurette Fugain”, Association pour la Recherche contre le Cancer (Equipe labellisée), Institut National du Cancer PRT-K 18–071 and the Fédération Leucémie espoir and Horizon Hemato.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Studies were conducted in accordance with the Declaration of Helsinki and approved by local and multicenter research ethical committees.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med. 2015;373:1541–1552. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 2.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129:1113–1123. doi: 10.1182/blood-2016-10-706465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petit A, Trinquand A, Chevret S, Ballerini P, Cayuela J-M, Grardel N, et al. Oncogenetic mutations combined with MRD improve outcome prediction in pediatric T-cell acute lymphoblastic leukemia. Blood. 2018;131:289–300. doi: 10.1182/blood-2017-04-778829. [DOI] [PubMed] [Google Scholar]
- 4.Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengliné E, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol. 2013;31:4333–4342. doi: 10.1200/JCO.2012.48.5292. [DOI] [PubMed] [Google Scholar]
- 5.Gökbuget N, Kneba M, Raff T, Trautmann H, Bartram C-R, Arnold R, et al. Adult patients with acute lymphoblastic leukemia and molecular failure display a poor prognosis and are candidates for stem cell transplantation and targeted therapies. Blood. 2012;120:1868–1876. doi: 10.1182/blood-2011-09-377713. [DOI] [PubMed] [Google Scholar]
- 6.Gökbuget N, Stanze D, Beck J, Diedrich H, Horst H-A, Hüttmann A, et al. Outcome of relapsed adult lymphoblastic leukemia depends on response to salvage chemotherapy, prognostic factors, and performance of stem cell transplantation. Blood. 2012;120:2032–2041. doi: 10.1182/blood-2011-12-399287. [DOI] [PubMed] [Google Scholar]
- 7.Desjonquères A, Chevallier P, Thomas X, Huguet F, Leguay T, Bernard M, et al. Acute lymphoblastic leukemia relapsing after first-line pediatric-inspired therapy: a retrospective GRAALL study. Blood Cancer J. 2016;6:e504. doi: 10.1038/bcj.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Meulen J, Van Roy N, Van Vlierberghe P, Speleman F. The epigenetic landscape of T-cell acute lymphoblastic leukemia. Int J Biochem Cell Biol. 2014;53:547–557. doi: 10.1016/j.biocel.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boissel N, Nibourel O, Renneville A, Gardin C, Reman O, Contentin N, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28:3717–3723. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- 13.DiNardo CD, Ravandi F, Agresta S, Konopleva M, Takahashi K, Kadia T, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90:732–736. doi: 10.1002/ajh.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiNardo CD, Stein EM, de Botton S, Roboz GJ, Altman JK, Mims AS, et al. Durable remissions with ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N Engl J Med. 2018;378:2386–2398. doi: 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 15.Stein EM, DiNardo CD, Pollyea DA, Fathi AT, Roboz GJ, Altman JK, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 2017;130:722–731. doi: 10.1182/blood-2017-04-779405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208:2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, Hadler M, Paietta E, Tallman MS, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122:74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asnafi V, Beldjord K, Boulanger E, Comba B, Le Tutour P, Estienne M-H, et al. Analysis of TCR, pT alpha, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood. 2003;101:2693–2703. doi: 10.1182/blood-2002-08-2438. [DOI] [PubMed] [Google Scholar]
- 19.Bond J, Marchand T, Touzart A, Cieslak A, Trinquand A, Sutton L, et al. An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. Haematologica Haematologica. 2016;101:732–740. doi: 10.3324/haematol.2015.141218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dongen JJM, Lhermitte L, Böttcher S, Almeida J, van der Velden VHJ, Flores-Montero J, et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia. 2012;26:1908–1975. doi: 10.1038/leu.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pongers-Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E, et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia. 1998;12:2006–2014. doi: 10.1038/sj.leu.2401246. [DOI] [PubMed] [Google Scholar]
- 22.van der Velden VHJ, Cazzaniga G, Schrauder A, Hancock J, Bader P, Panzer-Grumayer ER, et al. Analysis of minimal residual disease by Ig/TCR gene rearrangements: guidelines for interpretation of real-time quantitative PCR data. Leukemia. 2007;21:604–611. doi: 10.1038/sj.leu.2404586. [DOI] [PubMed] [Google Scholar]
- 23.Lemonnier F, Cairns RA, Inoue S, Li WY, Dupuy A, Broutin S, et al. The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development. Proc Natl Acad Sci U S A. 2016;113:15084–15089. doi: 10.1073/pnas.1617929114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heavican TB, Bouska A, Yu J, Lone W, Amador C, Gong Q, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. 2019;133:1664–1676. doi: 10.1182/blood-2018-09-872549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watatani Y, Sato Y, Miyoshi H, Sakamoto K, Nishida K, Gion Y, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33:2867–2883. doi: 10.1038/s41375-019-0473-1. [DOI] [PubMed] [Google Scholar]
- 26.Hao Z, Cairns RA, Inoue S, Li WY, Sheng Y, Lemonnier F, et al. Idh1 mutations contribute to the development of T-cell malignancies in genetically engineered mice. Proc Natl Acad Sci U S A. 2016;113:1387–1392. doi: 10.1073/pnas.1525354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonin M, Lhermitte L, Dourthe M-E, Lengline E, Graux C, Grardel N, et al. IKZF1 alterations predict poor prognosis in adult and pediatric T-ALL. Blood. 2020 doi: 10.1182/blood.2020007959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental Table 1: Custom capture Nextera XT gene panel. Supplemental Table 2: IDH1 and IDH2 mutations identified in 1085 patients with T-ALL. Supplemental Table 3: Chemotherapy in the FRALLE 2000 standard risk group T1 and high risk T2.
Additional file 2. Figure S1: Lollipop plots indicating the observed mutations for IDH1 and IDH2 in the present series confront with Cosmic-reported mutations for AML and AITL. Figure S2: Lollipop plots indicating the observed mutations for IDH1 and IDH2 affecting patients included in FRALLE and GRAALL protocol. Figure S3: Variant Allele Frequency (VAF) of individual IDH1 and IDH2 mutations observed in 1085 T-ALL. Figure S4: OS and CIR according to the IDH1 or IDH2Mut status in the two subgroups (FRALLE and GRALL 03/05). Figure S5: General design of FRALLE 2000 T guidelines.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.