Abstract
Novel coronavirus SARS-CoV-2 was recently outbreak worldwide causes severe acute respiratory syndrome along with gastrointestinal symptoms for some infected patients. Information on detail pathogenesis, host immune responses and responsible biological pathways are limited. Therefore, infection specific host gut responses and dietary supplements to neutralize immune inflammation demand extensive research. This study aimed to find differences in global co-expression protein-protein interaction sub-network and enriched biological processes in SARS-CoV and SARS-CoV-2 infected gut enterocytes cell line. Attempts have also been made to predict some dietary supplements to boost human health. The SARS-CoV and SARS-CoV-2 infected differential express proteins were integrated with the human protein interaction network and co-expression subnetworks were constructed. Common hubs of these sub-networks reshape central cellular pathways of metabolic processes, lipid localization, hypoxia response to decrease oxygen level and transport of bio-molecules. The major biological process enriched in the unique hub of SARS-CoV-2 significantly differ from SARS-CoV, related to interferon signaling, regulation of viral process and influenza-A enzymatic pathway. Predicted dietary supplements can improve SARS-CoV-2 infected person’'s health by boosting the host immunity/reducing inflammation. To the best of our knowledge this is the first report on co-expression network mediated biological process in human gut enterocytes to predict dietary supplements/compounds.
Keywords: SARS-CoV, SARS-CoV-2, RNA-Seq, Disease specific co-expression network, Dietary supplementary
Graphical abstract
1. Introduction
Severe acute respiratory syndrome (SARS) first emerged in 2003 caused by coronavirus SARS-CoV (Drosten et al., 2003). In late December 2019, a novel coronavirus (SARS-CoV-2) epidemic happened from China and on 30th January 2020 World Health Organization (WHO) declared COVID-19 as a pandemic (Zhu et al., 2020; Li et al., 2020). Coronaviruses (CoVs) are the single stranded RNA viruses that infects animals and humans causing respiratory, gastrointestinal and hepatic disease (Leibowitz and Weiss, 2013; Lamers et al., 2020). Till date, there have been seven human coronaviruses (HCoVs) identified, including HCoVs-NL63, HCoVs-229E, HCoVs-OC43, HCoVs-HKU1, SARS-CoV, MERS-CoV and novel SARS-CoV-2 (Ye et al., 2020). Despite some common clinical symptoms, SARS-CoV-2 has the highest pathogenicity with 106,125,682 confirmed cases and 2,320,497 deaths globally as of 10th February 2021 much more than SARS-CoV (8422 people infected in 26 countries, leading to 916 deaths) according to WHO. (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Along with their common clinical symptoms subset of patients showed severe gastrointestinal problems for SARS-CoV-2 (Lamers et al., 2020).
Although there are some reports of host responses on infected lung epithelial cells, less research has been done on human gut infection which is another important site for SARS-CoV-2 causing gastrointestinal problems. Early reports revealed that in SARS patients, there is a pulmonary infection and severe lung damage associated with elevated pro-inflammatory cytokines in serum (IL-6, IL-8, IFN-γ, IL-1β, TNF-α; Azkur et al., 2020; Prasad et al., 2020; Liang et al., 2020).
There are some recent reports on RNA-Seq expression for SARS-CoV and SARS-CoV-2 infected lung epithelial and gut enterocytes cell line to characterize the differentially expressed genes and their responsible metabolic pathways, however, lacking the global co-expression profile of intestinal cells (Lamers et al., 2020; Lieberman et al., 2020). Protein-protein interaction (PPI) network from differentially expressed datasets and their co-expression profile may provide a global picture of cellular processes that can be used as a target to improve diagnostic, prognostic and therapeutic for HCoVs (Dhal et al., 2014; Krishnamoorthy et al., 2021; Prasad et al., 2020).To boost the patient's health by neutralizing excessive immune response/inflammation proper dietary supplements/compounds also needed.
In this study a systematic approach was made to decipher the following objects 1) How SARS-CoV-2 specific human protein interaction co-expressed network differ from SARS-CoV in 60 h of gut enterocytes cells 2) What are different enriched biological processes are globally activated during SARS-CoV-2 and SARS-CoV infection 3) What are host immune responses and disease severity in the gut during SARS-CoV-2 infection 4) What are the topmost hub proteins according to their topological importance in the network and their metabolic pathways 5) What are probable dietary supplements/compounds that improve patients health and reduce disease severity during SARS-CoV-2 infection.
To the best of our knowledge, this is the first report on SARS-CoV and SARS-CoV-2 gut infection specific global PIN and host immune response during gastrointestinal tract infection as well as suggestions for dietary supplements/compounds based inflammation reduction to improve patient's health of COVID-19 infected patients.
2. Methods
2.1. Protein-protein interaction network and gene expression data
The human binary protein-protein interaction dataset (Human Protein Reference Database) was visualized by importing the network to Cytoscape 3.8.0 v-3.8.0 (Shannon et al., 2003) from Network Data Exchange (NDEx) (www.ndexbio.org) named here as HPRD static protein protein interaction network (HPRD PIN) (Supplementary Fig. S1).
The RNA-Seq FASTQ files of human gut enterocytes cell line infected with SARS-CoV and SARS-CoV-2 and their respective control gene expression datasets were downloaded from GEO database (Lamers et al., 2020). Details descriptions of all the collected datasets are given in Supplementary Table S1.
2.2. RNA-Seq data processing and identification of differentially expressed protein coding genes
SARS-CoV and SARS-CoV-2 infected and control RNA-Seq datasets were processed to determine the differentially expressed protein coding genes (DEPCGs) following the established protocol with some minute modifications (Contreras-López et al., 2018). Selected FASTQ files were trimmed using Trimmomatic v-0.36 (Bolger et al., 2014) with a minimum length 36 and slidingwindow 10:30. Trimmed sequenced were aligned using Hista2 v-2.2.9 (Kim et al., 2015) against human reference sequence (GRCh38.p13; released date 2019.02.28) and only the protein coding genes were extracted for further analysis. The DEPCGs were enlisted using DESeq2 (Love et al., 2014) and their sequence reads were normalized through EBSeq package of Bioconductor (Leng et al., 2013) in R v-3.6.2 (Contreras-López et al., 2018).
2.3. Determination of Pearson correlation coefficient (PCC) of DEPCGs and construction of human-SARS-CoV and human-SARS-CoV-2 co-expression networks
Each DEPCGs data was formatted uniformly and their correlation profiles were measured by calculating the Pearson Correlation Coefficient (PCC) between each gene pair based on their expression profile using psych package of R (Revelle, 2017). Now the infection specific DEPCGs data of SARS-CoV and SARS-CoV-2 were integrated with HPRD PIN and human-SARS-CoV and human-SARS-CoV-2 co-expression network were formed after removing the nodes which had less than 5 interactor partners. Nodes having greater than or equal to 5 interactor partners termed as a hub.
2.4. Common and unique hubs finding and disease specific sub-network construction
Common and unique hubs of human-SARS-CoV and human-SARS-CoV-2 co-expression network were determined by Venny v-2.1 (Oliveros, 2007). Now, SARS-CoV and SARS-CoV-2 infection specific co-expression subnetworks were constructed using the unique hubs of human-SARS-CoV and human-SARS-CoV-2 co-expression networks respectively. Statistical analyses for both of the networks were done by Cytoscape plugin NetworkAnalyzer v-4.4.6 (Assenov et al., 2008).
2.5. Functional group annotation and identification of enriched gene ontology of infection specific subnetworks
In this study, we identified the biological process (BP) represented by the unique hubs for infection specific co-expression subnetwork and dynamicity of common hubs for human-SARS-CoV, human-SARS-CoV-2 using CluGo functional analysis with medium network specificity and classification stringency (Bindea et al., 2009). Enriched gene ontology (GO) analysis of these hub proteins was determined with group P-value and P-value corrected with Bonferroni step down ≤0.05.
2.6. Identification of top 20 hub proteins from infection specific co-expression subnetworks and relevant pathway analysis
To identify the top 20 hubs from these co-expression subnetworks, we calculate four different topological parameters of all involved hubs from the infection specific unique SARS-CoV and SARS-CoV-2 co-expression subnetwork using degree centrality (DC), closeness centrality (CC), betweenness centrality (BC), eigenvector centrality (EC) values. Next, we also computed the median ranking score for each of those proteins instead of exploring individual score. The most significant pathway (P-value ≤0.05) represented by these top 20 proteins were predicted in the Reactome database (Jassal et al., 2020).
2.7. Dietary supplement/anti-inflammatory compounds-protein interaction analysis
Dietary supplement/anti-inflammatory compounds–target interaction information for the selected 20 hub proteins were collected from the Comparative Toxicogenomics Database (CTD) (Davis et al., 2019). The predicted dietary supplement/anti-inflammatory molecules for hub proteins through the protein-compound interaction databases were used for constructing the compound-protein network using STITCH database (Kuhn et al., 2008). The interactions in STITCH database is derived from three main sources, mainly by automated text-mining, high-throughput lab experiments and previous knowledge from databases with a high confidence score (0.7) (Prasad et al., 2020).
3. Results
3.1. RNA-Seq data and DEPCGs specific network for SARS-CoV and SARS-CoV-2
The cumulative host cell response because of SARS-CoV and SARS-CoV-2 infection can be conceptualized by the host-viral proteins protein interactions. Beside mild to severe respiratory symptom, infected patients also reported having gastrointestinal problems with novel SARS-CoV-2 (Bojkova et al., 2020; Lamers et al., 2020). To better understand the difference in the global molecular mechanism of host defense response against these two viral infections we have used the 60 h of SARS-CoV and SARS-CoV-2 post infected RNA-Seq data of human gut enterocytes cell line and utilize them for DEPCGS specific SARS-CoV and SARS-CoV-2 co-expression network based analysis. The RNA-Seq data were trimmed and aligned with the human reference genome to extract a total of 17198 protein-coding genes and by removing the proteins that do not have any read total of 16409 proteins were selected. From this set of proteins, we got 1058 and 1037 DEPCGs (cutoff value log2FC > 1 and adjusted p-value <0.01) for SARS-CoV and SARS-CoV-2 respectively. The correlations of each of the individual proteins with the rest of the enlisted proteins of these DEPCGs data sets were calculated by PCC. Finally, 8017 and 6877 correlation sets for differentially expressed protein-coding genes (DEPCGs-CR) were listed for SARS-CoV and SARS-CoV-2 respectively (Supplementary Table S2).
3.2. Human-SARS-CoV and SARS-CoV-2 co-expression networks and infection specific SARS-CoV and SARS-CoV-2 co-expression subnetworks
Condition-specific dynamic sub-network model allows us to identify the key regulatory protein concerning different infections (Dhal et al., 2014). To find the differences in host responses due to SARS-CoV and SARS-CoV-2 infection four dynamic networks were constructed. The static HPRD PIN consists of 37,039 interactions, in which 9465 proteins are interconnected like a circuit with a clustering coefficient of 0.106 and network density 0.001. The DEPCGs-CR sets of both SARS-CoV and SARS-CoV-2 were integrated with HPRD PIN and co-expression of all individual hubs and their interacting partners were quantified. We identified 899 and 834 hubs having 7814 and 6510 interactors for human-SARS-CoV (clustering coefficient 0.604 and network density 0.019) and human-SARS-CoV-2 (clustering coefficient 0.596 and network density 0.019) co-expression network, respectively (Supplementary Fig. S2; Supplementary Fig. S3; Table 1 ). Among them 436 hubs were common for both, 463 and 398 hubs were unique for human-SARS-CoV and human-SARS-CoV-2 respectively (Supplementary Fig. S4; Supplementary Table S3). Using these unique proteins we have constructed infection specific co-expression subnetwork of SARS-CoV and SARS-CoV-2 (Supplementary Fig. S5; Supplementary Fig.S6). Infection specific SARS-CoV co-expression subnetwork was consist of 463 hubs with 1762 interactors having a clustering coefficient of 0.0.588 and 0.018 network density, similarly, SARS-CoV-2 consists of 398 hubs with 1394 interactors having clustering coefficient 0.573 and 0.018 network density (Table 1).
Table 1.
Characteristics information of HPRD static network, human-SARS-CoV & human-SARS-CoV-2 co-expression network, and SARS-CoV & SARS-CoV-2 infection specific subnetwork.
| Network | No. of Nodes | No. of Edges | Clustering Coefficient | Network Density |
|---|---|---|---|---|
| HPRD (Release 9) | 9465 | 37,039 | 0.106 | 0.001 |
| Human-SARS-CoV co-expression network (SARS-CoV + HPRD) with Degree ≥5 | 899 | 7814 | 0.604 | 0.019 |
| SARS-CoV infection specific co-expression subnetwork (unique hubs of SARS-CoV + HPRD) | 463 | 1762 | 0.588 | 0.018 |
| Human-SARS-CoV-2 co-expression network (SARS-CoV-2 + HPRD) with Degree ≥5 | 834 | 6510 | 0.596 | 0.019 |
| SARS-CoV-2 infection specific co-expression subnetwork (unique hubs of SARS-CoV-2 + HPRD) | 398 | 1394 | 0.573 | 0.018 |
3.3. Functional annotation of common and infection specific unique hub proteins of SARS-CoV and SARS-CoV-2
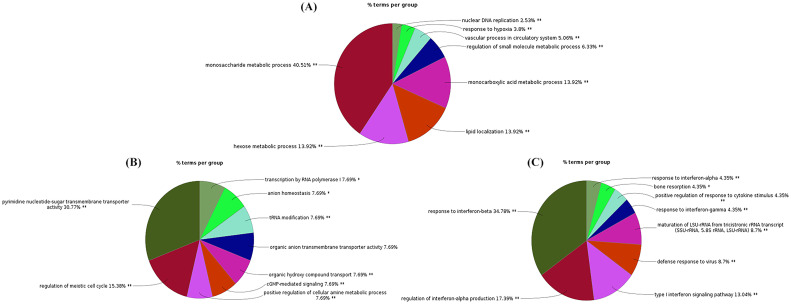
The Gene Ontology (GO) analysis was performed via CluGo plugin of Cytoscape for common and unique hub proteins to find out the main enriched GO biological processes they involved for. The major enriched biological processes were encoded by 436 common hubs were mainly the metabolic processes which represent 77.68% of total biological processes (GO:0062012, GO:0006082, GO:0072521, GO:0019693, GO:0019752, GO:0006631, GO:0032787, GO:0006163, GO:0009259, GO:0006090, GO:0009150), followed by lipid localization; 13.92% (GO:0010876), transport of different kinds of molecules; 5.06% (GO:0006869, GO:0006820, GO:0015850),hypoxia response to decreasing oxygen level; 3.8% (GO:0001666, GO:0036293, GO:0070482) and Nuclear DNA replication; 2.53% (GO:0033260) (Supplementary Table S4; Fig. 1A).
Fig. 1.
Pie diagram representation for the biological process of common and unique hub proteins of co-expression networks. Percentages of gene involved and enriched biological processes were represented in pie diagram. A) Enriched biological processes of common hub proteins B) SARS-CoV specific enriched biological processes C) SARS-CoV-2 specific enriched biological processes.
The unique hub proteins of SARS-CoV involved in the biological processes were transmembrane transporter which was 46.15% of the total biological processes (GO:0005338, GO:0008514, GO:0015165, GO:0015780, GO:0015850 and GO:0090481) followed by transcription by RNA polymerase I, tRNA modification and regulation of meiotic cell cycle; 25.38% (GO: 0006400, GO:0006360, GO:0040020 and GO:00051445) (Table 2 ; Fig. 1B). Interestingly unique hub proteins of SARS-CoV-2 involved in the human innate immune system through interferonalpha, beta and gamma response and regulation which was represented by 73.91% of the total responsible biological processes (GO:32479, GO:0032606, GO:0032607, GO:0032647, GO:0034340, GO:0034341, GO:0035455 and GO:0035456) followed by negative regulation of viral process and inflammatory cytokine; 13.05% (GO:0045069, GO:0050792, GO:0045071 and GO:0048525) (Table 3 ; Fig. 1C). So, the main biological processes for SARS-CoV-2 were related to interferon signaling, negative regulation of viral process and inflammatory cytokine biological process of immune response while SARS-CoV related to transmembrane transporter and cell cycle regulation.
Table 2.
Significantly enriched gene ontology biological process and associated hub proteins of SARS-CoV with their log2 Fold Change (down regulate highlighted with red color).
Table 3.
Significantly enriched gene ontology biological process and associated hub proteins of SARS-CoV-2 with their log2 Fold Change (down regulate highlighted with red color).
3.4. Identification of the most important proteins from infection specific unique network and their role on the infection
From the infection specific SARS-CoV and SARS-CoV-2 subnetwork we predict topologically important top 20 proteins based on their median ranking score. For SARS-CoV, these top 20 proteins were KAT5, CRELD1, SAT1, RGL2, UPP1, GBP2, ZNF215, JUNB, C5orf63, TAPBPL, PDE3A, FOS, SLC2A3, INTU, UTP14A, ZNF615, MAP1S, TTF1, CENPS and LOC105376526 (Supplementary Table S5). The most relevant pathways encoded by them were transmembrane transport and IL4 and IL13 signaling (Supplementary Table S6). For SARS-CoV-2,the top 20 proteins were ANKRD49, GGA1, NAGLU, SORL1, TRIM59, GAS2L3, PTPRH, DRD1, RHOV, VPS35L, ZNF581, SAMD9, TBC1D3, DUOXA1, IL18, LPAR2, OPN5, GSTM3 and SPATA12 responsible for interleukin signaling pathway of the immune system (Supplementary Table S7; Supplementary Table S8) and interestingly most of them reported to be involved induced during different kind of viral infection having some role on host defense response and inflammation (Menner et al., 2015; Filyk et al., 2020).
3.5. Dietary supplement/anti-inflammation compound-protein interaction analysis
The proposed network-based dietary supplements/anti-inflammatory compounds discovery depends on the hypothesis that the important hub proteins that functionally govern viral infection localized in the corresponding subnetwork would be the target for compounds or dietary supplements (Filyk et al., 2020). Using Comparative Toxicogenomics Database (CTD), we identified the possible dietary supplements/ant-inflammatory compounds which known to have possible interaction with the hub proteins. Based on the chemical-protein interaction results, we used STITCH database for the final categorization of the compound-protein interaction network based on the high interaction score. In total, we have identified 10 compounds that can interact with a few of the top 20 hub proteins of SARS-CoV (JUNB, PDE3A and FOS). Within these 10 compounds, 7 compounds (arachidonic acid, omega-3-fatty acid, EGCG, calcitrol, lactate, curcumin, and resveratrol) were dietary supplements/vitamin, 2 compounds (ginsenoside rh-1 and andrographolide) were anti-inflammatory/antioxidant and theophyline used against respiratory disease and anti-inflammation (Fig. 2 ). For SARS-CoV-2 we selected 3 dietary supplements/compounds which known to have interacted with the top 20 hub proteins of SARS-CoV-2 (DRD1, IL18 and LPAR2) (Fig. 3 ). Within them resveratrol and lactate used as antioxidant and dietary supplements respectively and theophylline used for respiratory disease and anti-inflammation.
Fig. 2.
Dietary supplement/compound-protein interaction of SARS-CoV. Using topological important top 20 proteins of SARS-CoV infection specific subnetwork and their related dietary supplements derived from Comparative Toxicogenomics Database.
Fig. 3.
Dietary supplement/compound-protein interaction of SARS-CoV-2. Using topological important top 20 proteins of SARS-CoV-2 infection specific subnetwork and their related dietary supplements derived from Comparative Toxicogenomics Database.
4. Discussion
Protein-protein interaction networks are static as they include all possible binary interactions without their expression profile. The integration of expression data with the PPI network allowed us to identify functionally important proteins (Dhal et al., 2014).
Some RNA-Seq expression data are available for SARS-CoV and SARS-CoV-2 infected on lung epithelial and gut enterocytes cell line to characterize the differentially expressed genes, however, lacking the entire interconnected expression condition of the cells (Lamers et al., 2020; Lieberman et al., 2020). Any biological response is a multi-protein activity that can be predicted through dynamic co-expression PIN (Dhal et al., 2014; Prasad et al., 2020). Our analysis provides infection specific PPI network of the human to provide a map of involved host proteins affected by the viral infection. Importance has been given in finding the difference of host defense response when gut enterocytes cell line infected with SARS-CoV and SARS-CoV-2 (ex-vivo). We consider the human gut enterocytes cell line as a prototype of the human gastrointestinal tract because SARS-CoV-2 infects gut enterocytes cell as a primary target causing gastrointestinal problem including diarrhea. An effort was also made to predict the new targets related to dietary supplements and anti-inflammatory molecules to boost human heaths and immunity to neutralize the viral responses.
In the present work, the differentially expressed protein coding genes of SARS-CoV and SARS-CoV-2 infection specific condition is determined. To measure whether DEPCGs are likely to be co-expressed, we use PCC in all the studied conditions and disease specific co-expression network (human-SARS-CoV and human-SARS-CoV-2) have been constructed. Compare with human-SARS-CoV co-expression network, human-SARS-CoV-2 have 65 fewer hub proteins and 1304 interactors with 0.008 higher clustering coefficient and same network density. So, host cell expression may be more specific and stringent during SARS-CoV-2 infection.
From human-SARS-CoV and human-SARS-CoV-2 co-expression network, we identified 436 common hubs while 463 and 398 hubs are unique respectively. The major enriched biological processes involved in the common hubs are related to metabolic processes, biosynthetic process, lipid localization, transport of different kinds of molecules, hypoxia and nuclear DNA replication. The metabolic process of the viral infected cell is high because energy and biomolecules (small molecules and lipid) may require for viral particle synthesis (Soliman et al., 2020). Those biological processes altered in the host cell during SARS-CoV-2 infection well indicated in the previous findings (Krishnamoorthy et al., 2021; Liu et al., 2020; Soliman et al., 2020). It is also reported that during different pathogenic infection those are the biological processes get changed causing diarrhea and hypoxia (Kuntumalla et al., 2011; Singh et al., 2007). So these biological processes may responsible for SARS-CoV-2 infected diarrhea, gastrointestinal problems and hypoxia.
The unique hub proteins of SARS-CoV involved mainly in transmembrane transporter, transcription by RNA polymerase I, tRNA modification, cGMP-mediated signaling, positive regulation of cellular amine metabolic process and regulation of meiotic cell cycle indicating the absence of any major immunological defense response in 60 h of SARS-CoV infection. According to the previous investigation, the combined induction of antibodies and virus-specific T cells provides optimal protective immunity against SARS-CoV (Liang et al., 2020). After infection SARS-CoV encodes multiple structural and non-structural proteins that antagonize innate IFN response, alteration of antigen-presenting cell function and impaired dendritic cell migration is the possible reason for the delayed adaptive immune response (Totura and Baric, 2012; Yoshikawa et al., 2009). For SARS-CoV infected patients IgM and IgG production level peaked at approx 1 and 2–4 months, respectively after symptoms (Liang et al., 2020). So, SARS-CoV activates different types of biological processes of the human gut enterocytes cell line to create a suitable environment for them but the immunological response still not activates at 60 h of post-infection, indicated in this study.
Interestingly unique hub proteins of SARS-CoV-2 involved in the human immune system through interferon alpha, beta and gamma response and regulation, type I interferon production, negative regulation of viral process and positive regulation of response to cytokine stimulus. Based on our findings, it can be hypothesized that the immunological response of SARS-CoV-2 includes innate and adaptive immune responses. Infection of enterocytes cell by SARS-CoV-2 induces a robust intrinsic immune response characterized by the production of type I IFNs results in the reduction of viral replication and a significant decrease in the production of infectious de novo virus particles. Production of cytokine is associated with the severity of SARS-CoV-2 patients, which is characterized by increased interleukins. Therefore, the body may have experienced a cytokine storm caused by excessive immunity in SARS-CoV-2 infected patients. It's already reported that type I and III IFNs induce an antiviral state thereby restricting SARS-CoV-2 replication in cells (Mantlo et al., 2020; Stanifer et al., 2020). On the other hand, at the later stages of the disease, the balance of the immune system becomes impaired, leading to inflammatory over-reactions and cytokine storm happens (Prasad et al., 2020). So, this study proposes the immune response in gut enterocytes cell is in line with the previously reported immune response of human against SARS-CoV-2 (Huang et al., 2020; Liang et al., 2020; Mehta et al., 2020).
According to the host response and symptoms (Reactome pathways based on the top 20 hub proteins of co-expression subnetworks) against SARS-CoV and SARS-CoV-2, we propose dietary supplements and compounds from compound-protein interaction analysis in STITCH database. For SARS-CoV and SARS-CoV-2 infected patients we predict dietary supplements (curcumin, arachidonic acid, omega-3-fatty acid, EGCG, calcitrol, lactate and resveratrol) and anti-inflammatory/antioxidant drugs (ginsenoside rh-1, andrographolide and theophyline) to neutralize the viral response (Supplementary Table S9) (Kahkhaie et al., 2019; Arreola et al., 2016; Malaguarnera, 2019; Perdigon et al., 2002; Allen and Diwari, 2019; Tallima and El Ridi, 2018).
All of the food supplements and compounds reduce the inflammation (Cytokine storm) of SARS-CoV-2 infected patients by inhibition IL-1, IL-6, IL-4, IL-12, IL-23, NF-κB and TNF-α. Along with it Omega-3 fatty acid, ECGC, Calcitrol and Andrographolide induce innate immune responses by regulating macrophage and monocyte (Gutiérrez et al., 2019; Hajian, 2014; Iddir et al., 2020; Prietl et al., 2013; Santos et al., 2019). Ginsenoside rh-1 suppress the production of inflammatory enzymes, such as inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) may be helpful against SARS inflammation (Kim et al., 2015). Andrographolide and Omega-3 fatty acid induce adaptive immune responses by regulating macrophage and antigen-specific antibody production through MAPK and PI3K pathways(Wang et al., 2010). Lastly, antioxidative agent EGCG can be beneficial as immune restorative properties by maintaining the balance of Th1/Th2 system (Hajian, 2014; Kuo et al., 2014).
Our suggested dietary supplements and compounds can improve SARS-CoV-2 infected as well as non-infected person's health and reduce mortality by boosting the host immunity, stress relieves, reducing the inflammation (cytokine storm) and damage reduction of infected cells.
5. Conclusions
In summary, our integrative interactome and network topology analyses showed that
-
1.
Host cell expression may be more specific and stringent during SARS-CoV-2 infection.
-
2.
The human-SARS-CoV and human-SARS-CoV-2 specific co-expression network, total 436 common hubs, while 463 and 398 unique hubs are identified respectively, may be designated as disease specific hub proteins.
-
3.
Major enriched biological processes for SARS-CoV-2 were related to interferon signaling, negative regulation of viral process and inflammatory cytokine response while SARS-CoV related to transmembrane transporter and cell cycle regulation.
-
4.
During 60 h of post-infection SARS-CoV-2 developed a strong cytokine and low INFs response while SARS-CoV response on host immunity not activated.
-
5.
During host gut infection the balance of the immune system becomes impaired, leading to inflammatory over-reactions, cytokine storm, and possible autoimmune responses happened.
-
6.
From the top 20 hub proteins JUNB, PDE3A, FOS of SARS-CoV and DRD1, IL18, LPAR2 of SARS-CoV-2 can be targeted to neutralize the inflammation of SARS-CoV and SARS-CoV-2.
-
7.
Curcumin, arachidonic acid, omega-3-fatty acid, EGCG, calcitrol, lactate and resveratrol ginsenoside rh-1, andrographolide, theophyline and dopamine may be used to neutralize the viral response by inhibiting the cytokine response based inflammation and activating the INF response.
The following is the supplementary data related to this article.
Supplementary material Supplementary Fig. S1: Human PPI network. Network was visualized through Cytoscape protein-protein interaction visualization tool. The hub proteins with ≥5 interacting partners in the human PPI network were represented. Supplementary Fig. S2: Human-SARS-CoV co-expression network. SARS-CoV co-expression data were merged with human static PIN in Cytoscape. Common hub proteins which present in the both SARS-CoV and SARS-CoV-2 co-expression network highlighted with purple. Supplementary Fig. S3: Human-SARS-CoV co-expression network. SARS-CoV co-expression data were merged with human static PIN in Cytoscape. Common hub proteins which present in the both SARS-CoV and SARS-CoV-2 co-expression network highlighted with purple. Supplementary Fig. S4: SARS-CoV infection specific subnetwork. From human-SARS-CoV co-expression PIN we select the infection specific unique hubs in Cytoscape and subnetwork was constructed. Supplementary Fig. S5: SARS-CoV-2 infection specific subnetwork. From human-SARS-CoV-2 co-expression PIN we select the infection specific unique hubs in Cytoscape and subnetwork was constructed. Supplementary Fig. S6: Venn diagram for unique and common hubs. According to the Venn diagram, 436 hubs were common for both human-SARS-CoV and human-SARS-CoV-2 co-expression network and 463 and 398 hubs are unique for SARS-CoV and SARS-CoV-2 respectively. Supplementary Table S1: GEO datasets used in this study. Supplementary Table S2: Step by step sequence processing information, identifying the protein coding genes and determination of the differentially expressed protein coding genes of GEO datasets used in this study Supplementary Table S3: Venn dataset of unique hubs of SARS-CoV, unique hubs of SARS-CoV-2, and Common hubs for human-SARS-CoV and human-SARS-Cov-2 co-expression PIN. Supplementary Table S4: Significantly enriched gene ontology biological process and associated common hub proteins. Supplementary Table S5: Topologically important top 20 hub proteins of SARS-CoV infection specific subnetwork based on their median ranking scores. Supplementary Table S6: Most significant and relevant pathways for top 20 proteins of SARS-CoV infection derived from Reactome database. Supplementary Table S7: Topologically important top 20 hub proteins of SARS-CoV-2 infection specific subnetwork based on their median ranking scores. Supplementary Table S8: Most significant and relevant pathways for top 20 proteins of SARS-CoV-2 infection derived from Reactome database. Supplementary Table S9: Dietary supplements/compounds related defense mechanism and literature survey against infected patients, which helps to improve the host immune response.
Funding
No one funded this research.
Competing interests
None declared.
Acknowledgements
The authors acknowledge the Science & Engineering Research Board (SERB), Ministry of Science and Technology (Grant No. EEQ/2018/000006) for the high performance computing workstation. We are grateful to Jadavpur University, Life Sciences and Biotechnology Department, for providing lab space to carry out the work.
References
- Allen S., Diwari T. Theophylline as a systemic anti-inflammatory agent: the need for its revival as a possible adjunctive treatment for “inflammaging.”. Biol. Eng. Med. 2019;4 doi: 10.15761/bem.1000162. [DOI] [Google Scholar]
- Arreola R., Alvarez-Herrera S., Pérez-Sánchez G., Becerril-Villanueva E., Cruz-Fuentes C., Flores-Gutierrez E.O., Garcés-Alvarez M.E., de la Cruz-Aguilera D.L., Medina-Rivero E., Hurtado-Alvarado G., Quintero-Fabián S., Pavón L. Immunomodulatory effects mediated by dopamine. J. Immunol. Res. 2016;2016:31. doi: 10.1155/2016/3160486. Article Id: 3160486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y., Ramírez F., Schelhorn S.-E., Lengauer T., Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- Azkur A.K., Akdis M., Azkur D., Sokolowska M., Veen W., Brüggen M., O’Mahony L., Gao Y., Nadeau K., Akdis C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75:1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-López O., Moyano T.C., Soto D.C., Gutiérrez R.A. Methods in Molecular Biology. Humana Press Inc; 2018. Step-by-step construction of gene co-expression networks from high-throughput Arabidopsis RNA sequencing data; pp. 275–301. [DOI] [PubMed] [Google Scholar]
- Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47:D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhal P.K., Barman R.K., Saha S., Das S. Dynamic modularity of host protein interaction networks in Salmonella Typhi infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- Filyk H.A., Sharon A.J., Fonseca N.M., Simister R.L., Yuen W., Hardman B.K., Robinson H.G., Seo J.H., Rocha-Pereira J., Welch I., others STAT1-dependent tolerance of intestinal viral infection. bioRxiv. 2020 doi: 10.1101/2020.02.13.936252. [DOI] [Google Scholar]
- Gutiérrez S., Svahn S.L., Johansson M.E. Effects of omega-3 fatty acids on immune cells. Int. J. Mol. Sci. 2019;20:5028. doi: 10.3390/ijms20205028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajian S. Positive effect of antioxidants on immune system. Immunopathol. Persa. 2014;1 [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., others Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iddir M., Brito A., Dingeo G., Fernandez Del Campo S.S., Samouda H., La Frano M.R., Bohn T. Strengthening the immune system and reducing inflammation and oxidative stress through diet and nutrition: considerations during the COVID-19 crisis. Nutrients. 2020;12:1562. doi: 10.3390/nu12061562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., Loney F., May B., Milacic M., Rothfels K., Sevilla C., Shamovsky V., Shorser S., Varusai T., Weiser J., Wu G., Stein L., Hermjakob H., D’Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahkhaie K.R., Mirhosseini A., Aliabadi A., Mohammadi A., Mousavi M.J., Haftcheshmeh S.M., Sathyapalan T., Sahebkar A. Curcumin: a modulator of inflammatory signaling pathways in the immune system. Inflammopharmacology. 2019;27:885–900. doi: 10.1007/s10787-019-00607-3. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy P., Raj A.S., Roy S., Kumar N.S., Kumar H. Comparative transcriptome analysis of SARS-CoV, MERS-CoV, and SARS-CoV-2 to identify potential pathways for drug repurposing. Comput. Biol. Med. 2021;128:104123. doi: 10.1016/j.compbiomed.2020.104123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntumalla S., Zhang Q., Braisted J.C., Fleischmann R.D., Peterson S.N., Donohue-Rolfe A., Tzipori S., Pieper R. In vivo versus in vitro protein abundance analysis of Shigella dysenteriae type 1 reveals changes in the expression of proteins involved in virulence, stress and energy metabolism. BMC Microbiol. 2011;11:1–13. doi: 10.1186/1471-2180-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.-L., Chen T.-S., Liou S.-Y., Hsieh C.-C. Immunomodulatory effects of EGCG fraction of green tea extract in innate and adaptive immunity via T regulatory cells in murine model. Immunopharmacol. Immunotoxicol. 2014;36:364–370. doi: 10.3109/08923973.2014.953637. [DOI] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., Van Der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Van Schayck J.P., Mykytyn A.Z., Duimel H.Q., Van Donselaar E., Riesebosch S., Kuijpers H.J.H., Schipper D., De Wetering W.J.V., De Graaf M., Koopmans M., Cuppen E., Peters P.J., Haagmans B.L., Clevers H. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;80(369):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J.L., Weiss S.R. San Francisco, School of Medicine (currently at Department of Microbiology, University of Pennsylvania, Philadelphia, PA 19104) Biochem. Biol. Coronaviruses. 2013;142:227. [Google Scholar]
- Leng N., Dawson J.A., Thomson J.A., Ruotti V., Rissman A.I., Smits B.M.G., Haag J.D., Gould M.N., Stewart R.M., Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–1043. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Yang Y., Ren L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect. Genet. Evol. 2020 doi: 10.1016/j.meegid.2020.104285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Wang M.-L., Chien C.-S., Yarmishyn A.A., Yang Y.-P., Lai W.-Y., Luo Y.-H., Lin Y.-T., Chen Y.-J., Chang P.-C., others Highlight of immune pathogenic response and hematopathologic effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 infection. Front. Immunol. 2020;11:1022. doi: 10.3389/fimmu.2020.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman N., Peddu V., Xie H., Shrestha L., Huang M.-L., Mears M., Cajimat M., Bente D., Shi P.-Y., Bovier F., Roychoudhury P., Jerome K., Moscona A., Porotto M., Greninger A. In vivo antiviral host response to SARS-CoV-2 by viral load, sex, and age. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.06.22.165225. 2020.06.22.165225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Chen S., Liu M., Nie H., Lu H. Comorbid chronic diseases are strongly correlated with disease severity among COVID-19 patients: a systematic review and meta-analysis. Aging Dis. 2020 doi: 10.14336/AD.2020.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaguarnera L. Influence of resveratrol on the immune response. Nutrients. 2019;11:946. doi: 10.3390/nu11050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantlo E., Bukreyeva N., Maruyama J., Paessler S., Huang C. Antiviral activities of type I interferons to SARS-CoV-2 infection. Antivir. Res. 2020;179:104811. doi: 10.1016/j.antiviral.2020.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menner A.J., Rauch K.S., Aichele P., Pircher H., Schachtrup C., Schachtrup K. Id3 controls cell death of 2B4 + virus-specific CD8 + T cells in chronic viral infection. J. Immunol. 2015;195:2103–2114. doi: 10.4049/jimmunol.1402607. [DOI] [PubMed] [Google Scholar]
- Oliveros J.C. 2007. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. [Google Scholar]
- Perdigon G., Galdeano C.M., Valdez J.C., Medici M. Interaction of lactic acid bacteria with the gut immune system. Eur. J. Clin. Nutr. 2002;56:S21–S26. doi: 10.1038/sj.ejcn.1601658. [DOI] [PubMed] [Google Scholar]
- Prasad K., Khatoon F., Rashid S., Ali N., AlAsmari A.F., Ahmed M.Z., Alqahtani A.S., Alqahtani M.S., Kumar V. Targeting hub genes and pathways of innate immune response in COVID-19: a network biology perspective. Int. J. Biol. Macromol. 2020;163:1–8. doi: 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prietl B., Treiber G., Pieber T.R., Amrein K. Vitamin D and immune function. Nutrients. 2013;5:2502–2521. doi: 10.3390/nu5072502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle W.R. 2017. psych: Procedures for Personality and Psychological Research. [Google Scholar]
- Santos N., Pereira-Nunes A., Baltazar F., Granja S. Lactate as a regulator of cancer inflammation and immunity. Immunometabolism. 2019;1 [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Malik B.K., Sharma D.K. Choke point analysis of metabolic pathways in E. histolytica: a computational approach for drug target identification. Bioinformation. 2007;2:68. doi: 10.6026/97320630002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman S., Faris M.E., Ratemi Z., Halwani R. Switching host metabolism as an approach to dampen SARS-CoV-2 infection. Ann. Nutr. Metab. 2020;76:297–303. doi: 10.1159/000510508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanifer M.L., Kee C., Cortese M., Zumaran C.M., Triana S., Mukenhirn M., Kraeusslich H.-G., Alexandrov T., Bartenschlager R., Boulant S. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallima H., El Ridi R. Arachidonic acid: physiological roles and potential health benefits--a review. J. Adv. Res. 2018;11:33–41. doi: 10.1016/j.jare.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura A.L., Baric R.S. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang J., Dong S., Liu C., Italiani P., Sun S., Xu J., Boraschi D., Ma S., Qu D. Immunomodulatory activity of andrographolide on macrophage activation and specific antibody response. Acta Pharmacol. Sin. 2010;31:191–201. doi: 10.1038/aps.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020 doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa T., Hill T., Li K., Peters C.J., Tseng C.-T.K. Severe acute respiratory syndrome (SARS) coronavirus-induced lung epithelial cytokines exacerbate SARS pathogenesis by modulating intrinsic functions of monocyte-derived macrophages and dendritic cells. J. Virol. 2009;83:3039–3048. doi: 10.1128/JVI.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., others A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Supplementary Fig. S1: Human PPI network. Network was visualized through Cytoscape protein-protein interaction visualization tool. The hub proteins with ≥5 interacting partners in the human PPI network were represented. Supplementary Fig. S2: Human-SARS-CoV co-expression network. SARS-CoV co-expression data were merged with human static PIN in Cytoscape. Common hub proteins which present in the both SARS-CoV and SARS-CoV-2 co-expression network highlighted with purple. Supplementary Fig. S3: Human-SARS-CoV co-expression network. SARS-CoV co-expression data were merged with human static PIN in Cytoscape. Common hub proteins which present in the both SARS-CoV and SARS-CoV-2 co-expression network highlighted with purple. Supplementary Fig. S4: SARS-CoV infection specific subnetwork. From human-SARS-CoV co-expression PIN we select the infection specific unique hubs in Cytoscape and subnetwork was constructed. Supplementary Fig. S5: SARS-CoV-2 infection specific subnetwork. From human-SARS-CoV-2 co-expression PIN we select the infection specific unique hubs in Cytoscape and subnetwork was constructed. Supplementary Fig. S6: Venn diagram for unique and common hubs. According to the Venn diagram, 436 hubs were common for both human-SARS-CoV and human-SARS-CoV-2 co-expression network and 463 and 398 hubs are unique for SARS-CoV and SARS-CoV-2 respectively. Supplementary Table S1: GEO datasets used in this study. Supplementary Table S2: Step by step sequence processing information, identifying the protein coding genes and determination of the differentially expressed protein coding genes of GEO datasets used in this study Supplementary Table S3: Venn dataset of unique hubs of SARS-CoV, unique hubs of SARS-CoV-2, and Common hubs for human-SARS-CoV and human-SARS-Cov-2 co-expression PIN. Supplementary Table S4: Significantly enriched gene ontology biological process and associated common hub proteins. Supplementary Table S5: Topologically important top 20 hub proteins of SARS-CoV infection specific subnetwork based on their median ranking scores. Supplementary Table S6: Most significant and relevant pathways for top 20 proteins of SARS-CoV infection derived from Reactome database. Supplementary Table S7: Topologically important top 20 hub proteins of SARS-CoV-2 infection specific subnetwork based on their median ranking scores. Supplementary Table S8: Most significant and relevant pathways for top 20 proteins of SARS-CoV-2 infection derived from Reactome database. Supplementary Table S9: Dietary supplements/compounds related defense mechanism and literature survey against infected patients, which helps to improve the host immune response.