The long-lasting global COVID-19 pandemic demands timely genomic investigation of SARS-CoV-2 viruses. Here, we report a simple and efficient workflow for whole-genome sequencing utilizing one-step reverse transcription-PCR (RT-PCR) amplification on a microfluidic platform, followed by MiSeq amplicon sequencing.
KEYWORDS: COVID-19, next-generation sequencing, SARS-CoV-2, acute respiratory illness, emerging infectious viral disease, genomes, pandemic
ABSTRACT
The long-lasting global COVID-19 pandemic demands timely genomic investigation of SARS-CoV-2 viruses. Here, we report a simple and efficient workflow for whole-genome sequencing utilizing one-step reverse transcription-PCR (RT-PCR) amplification on a microfluidic platform, followed by MiSeq amplicon sequencing. The method uses Fluidigm integrated fluidic circuit (IFC) and instruments to amplify 48 samples with 39 pairs of primers, including 35 custom-designed primer pairs and four additional primer pairs from the ARTIC network protocol v3. Application of this method on RNA samples from both viral isolates and clinical specimens demonstrates robustness and efficiency in obtaining the full genome sequence of SARS-CoV-2.
INTRODUCTION
Severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) (family Coronaviridae, genus Betacoronavirus) is responsible for the global pandemic of coronavirus disease 2019 (COVID-19) (1–4). Since its emergence in Wuhan, China in November/December 2019, the disease has rapidly spread worldwide. As of 10 March 2021, there have been over 117 million confirmed cases and over 2.6 million deaths in 192 countries or regions (https://coronavirus.jhu.edu/) (5–8). In addition to the continuously growing number of SARS-CoV-2 infections, increased complexity and diversity of disease symptoms are also observed. Despite the large number of whole-genome sequences of SARS-CoV-2 available in GenBank and other public sources, the unprecedented scale of the viral transmission and the emergence of clinically significant variant strains demand more genomic data to be produced using cost-effective and quality-consistent methodology (9–12).
Targeted, whole-genome amplification and next-generation sequencing (NGS) techniques have been used in sequencing SARS-CoV-2 genomes (8, 10, 13). In contrast to the high-throughput capacity of NGS, using a conventional multiplexing reverse transcription and PCR (RT-PCR) procedures to amplify the large 30-Kb viral genomic RNA of SARS-CoV-2 is technically challenging, requires advanced primer and pooling design, demands experienced hands-on skills, and risks contamination and human error from manual manipulation. A rapid and streamlined approach with fewer manual steps to obtain whole-genome amplicons suitable for NGS is desired.
In this study, we utilize an integrated microfluidic nucleic acid amplification system (14), custom primer design, and a one-step RT-PCR program to amplify whole SARS-CoV-2 genomes. After NGS of the amplicons, an in-house developed bioinformatics pipeline is used to rapidly obtain genome sequences with graphic summaries for data and results visualization.
MATERIALS AND METHODS
SARS-CoV-2 RNA samples and real-time RT-PCR.
SARS-CoV-2 RNA samples used in this study include RNA extracted from viral isolate R4717 in the US Army Medical Research Institute of Infectious Disease (USAMRIID) and RNA extracted from deidentified clinical respiratory specimens. A one-step RT-qPCR method targeting RNA-dependent RNA polymerase (RdRp) was used to quantify SARS-CoV-2 RNA. Serially diluted in vitro transcripts (IVT), corresponding to nucleotide region 15431 to 15530 of NC_045512.2 for strain Wuhan-Hu-1, were prepared and used as real-time RT-PCR standards for quantification of genome equivalent copy number (GE) of SARS-CoV-2 RNA. Real-time RT-PCR was performed using the protocol from Carman et al. (15) with the SuperScript III one-step RT-PCR system and Platinum Taq polymerase. The QuantStudio 7 Flex real-time PCR system and software (Thermo Fisher Scientific, Inc.) were used for data acquisition.
Whole-genome RT-PCR amplification and Illumina sequencing.
For whole-genome RT-PCR amplification, 35 primer pairs covering the 29,903-bp SARS-CoV-2 reference genome (NC_045512.2) were custom designed by Fluidigm Corporation (South San Francisco, CA) (Table 1). The RT-PCR products are approximately 1 kb for all amplicons. One-step RT-PCR amplification was performed using the Fluidigm Access Array (AA) nucleic acids amplification system (Fluidigm Corporation, CA) and the SuperScript III one-step RT-qPCR System with Platinum Taq High Fidelity (Thermo Fisher Scientific, Inc.). Four additional pairs of primers were selected from the ARTIC network protocol v3 (https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye) and added to the primer panel to fill the gap/dip regions (Table 1).
TABLE 1.
Primers for RT-PCR amplification of SARS-CoV-2 genome using Fluidigm systema
| Forward primer name | Forward primer sequence | Reverse primer name | Reverse primer sequence | Amplicon length (bp) | From | To |
|---|---|---|---|---|---|---|
| nCoVF1 | TTCCCAGGTAACAAACCAACCAA | nCoVR1 | AGGTGTCTGCAATTCATAGCTCTT | 1011 | 17 | 1027 |
| nCoVF2 | CCGAACAACTGGACTTTATTGACAC | nCoVR2 | GCCTTCTGTAAAACACGCACAGA | 1048 | 912 | 1959 |
| nCoVF3 | ACTGAGTCCTCTTTATGCATTTG | nCoVR3 | AGCATCTGCCACAACACA | 1048 | 1852 | 2899 |
| nCoVF32 | AGAAGAAACTGGCCTACTCATGC | nCoVR32 | ACATTGGCTGCATTAACAACCAC | 1017 | 2937 | 3953 |
| nCoVF33 | TTTGGAATTTGGTGCCACTTCTG | nCoVR33 | CCTCTTGAACAACATCACCCACT | 1004 | 3642 | 4645 |
| nCoVF4 | AGAGTTTGTGTAGATACTGTTCG | nCoVR4 | TATAGGAACCAGCAAGTGAGATG | 1048 | 3752 | 4799 |
| nCoVF5 | GTTTCTGTTTCTTCACCTGATGC | nCoVR5 | TGGTGCTGACATCATAACAAAAG | 1008 | 4688 | 5695 |
| nCoVF6 | CCTTGTACGTGTGGTAAACAAGC | nCoVR6 | GCTAAACCATGAGTAGCAAGGGT | 1028 | 5621 | 6648 |
| nCoVF7 | TACAGAAGAGGTTGGCCACACAG | nCoVR7 | TGTACATTCGACTCTTGTTGCTCT | 1015 | 6523 | 7537 |
| nCoVF8 | ATGTGCATGTTGTAGACGGTTGT | nCoVR8 | GCAGCACTACGTATTTGTTTTCGT | 1009 | 7455 | 8463 |
| nCoVF9 | GCAGGTAGCAAAAAGTCACAACA | nCoVR9 | AGATGCTGATATGTCCAAAGCAC | 1036 | 8368 | 9403 |
| nCoVF10 | GGAGTTTTCTGTGGTGTAGATGC | nCoVR10 | AGGTGTCTTAGGATTGGCTGTAT | 1041 | 9311 | 10351 |
| nCoVF11 | TCTAAAGTTGCGTAGTGATGTGCT | nCoVR11 | TCCAGTTTGAGCAGAAAGAGGTC | 994 | 9835 | 10828 |
| nCoVF12 | GTTTGTTCGCATTCAACCAGGAC | nCoVR12 | ACACTCTCCTAGCACCATCATCA | 1031 | 10360 | 11390 |
| nCoVF34 | ATATGCCTGCTAGTTGGGTGATG | nCoVR34 | CTGCATCACGGTCAAATTCAGAT | 1026 | 11726 | 12751 |
| nCoVF35 | GCCTCAGAGTTTAGTTCCCTTCC | nCoVR35 | ATTAGTGATTGGTTGTCCCCCAC | 1047 | 12598 | 13644 |
| nCoVF13 | AAGCTGGTAATGCAACAGAAGTG | nCoVR13 | TTTCGCATGGCATCACAGAATTG | 1011 | 13023 | 14033 |
| nCoVF14 | TGTAGAAAACCCAGATATATTACGC | nCoVR14 | ATTTGTCTAGGTTGTTGACGATG | 1007 | 13935 | 14941 |
| nCoVF15 | TTGATTGTTACGATGGTGGCTGT | nCoVR15 | AGGTACACATAATCATCACCCTG | 1048 | 14879 | 15926 |
| nCoVF16 | TGTCTGAAGCAAAATGTTGGACT | nCoVR16 | CAACAGCATCACCATAGTCACCT | 1044 | 15821 | 16864 |
| nCoV-2019_54_LEFT* | TGAGTTAACAGGACACATGTTAGACA | nCoV-2019_56_RIGHT* | ACACTATGCGAGCAGAAGGGTA | 1033 | 16119 | 17152 |
| nCoV-2019_55_LEFT* | ACTCAACTTTACTTAGGAGGTATGAGCT | nCoV-2019_57_RIGHT* | GTAATTGAGCAGGGTCGCCAAT | 1035 | 16417 | 17452 |
| nCoVF17 | ACCTAGACCACCACTTAACCGAA | nCoVR17 | CAGCTTTTCTCCAAGCAGGGTTA | 1016 | 16749 | 17764 |
| nCoV-2019_56_LEFT* | ACCTAGACCACCACTTAACCGA | nCoV-2019_59_RIGHT* | AAGAGTCCTGTTACATTTTCAGCTTG | 1313 | 16749 | 18062 |
| nCoV-2019_58_LEFT* | TGATTTGAGTGTTGTCAATGCCAGA | nCoV-2019_60_RIGHT* | GGTACCAACAGCTTCTCTAGTAGC | 966 | 17382 | 18348 |
| nCoVF18 | GCTTAAAGCACATAAAGACAAATCA | nCoVR18 | GTGCGCTCAGGTCCTATTTT | 1041 | 17616 | 18656 |
| nCoVF19 | GTCTTATGGGCACATGGCTTTGA | nCoVR19 | AGCCACATTTTCTAAACTCTGAAGTC | 1050 | 18589 | 19638 |
| nCoVF20 | ACATGATGATCTCAGCTGGCTTT | nCoVR20 | TCACTTTGACAACCTTAGAAACTACA | 1047 | 19535 | 20581 |
| nCoVF21 | TTGGAGAAGCCGTAAAAACACAG | nCoVR21 | TTTATAGCCACGGAACCTCCAAG | 1045 | 20123 | 21167 |
| nCoVF22 | TTTAAGACAGTGGTTGCCTACGG | nCoVR22 | GGACTGGGTCTTCGAATCTAAAGT | 1001 | 20910 | 21910 |
| nCoVF23 | TAAGGGGTACTGCTGTTATGTCTT | nCoVR23 | TCAAGTGCACAGTCTACAGCATC | 1025 | 21419 | 22443 |
| nCoVF24 | GGGTTATCTTCAACCTAGGACTT | nCoVR24 | ACATCCTGATAAAGAACAGCAAC | 1044 | 22363 | 23406 |
| nCoVF25 | ACAATTTGGCAGAGACATTGCTG | nCoVR25 | AAACCTATAAGCCATTTGCATAGC | 1030 | 23251 | 24280 |
| nCoVF26 | CGGGTACAATCACTTCTGGTTGG | nCoVR26 | ACTATGGCAATCAAGCCAGCTAT | 1048 | 24198 | 25245 |
| nCoVF27 | TTCAAAAAGAAATTGACCGCCTC | nCoVR27 | CCGTCGATTGTGTGAATTTGGAC | 1047 | 25098 | 26144 |
| nCoVF28 | ACATGTTACCTTCTTCATCTACA | nCoVR28 | GACTGTATGCAGCAAAACCTG | 1045 | 26070 | 27114 |
| nCoVF29 | GTGACATCAAGGACCTGCCTAAA | nCoVR29 | ATAGGACACGGGTCATCAACTACAT | 1012 | 26998 | 28009 |
| nCoVF30 | CTGTAGCTGCATTTCACCAAGAA | nCoVR30 | CAAGCTGGTTCAATCTGTCAAGC | 1037 | 27928 | 28964 |
| nCoVF31 | GAACTTCTCCTGCTAGAATGGC | nCoVR31 | TCACATGGGGATAGCACTAC | 961 | 28884 | 29844 |
The primers were custom designed by Fluidigm, with four additional pairs (marked with *) from the ARTIC network protocol v3 (https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye).
SARS-CoV-2 R4717 RNA was serially diluted to the concentrations of 106, 105, 104, 103, 102, 10, and 1 GE/μl (genomic equivalence per microliter). For Fluidigm RT-PCR amplification, each sample well in the integrated fluidic circuit (IFC) chip contained 1.45 μl of RNA sample and 3.55 μl of sample mixture solution, consisting of 3 μl of 2× reaction mix, 0.2 μl of dimethyl sulfoxide (DMSO), 0.05 μl of RNaseOUT, 0.05 μl of SuperScript III RT/Platinum Taq High Fidelity enzyme mix, and 0.25 μl of 20× AA loading reagent. Each well of primer mix contained 10 μl of mixture containing one pair of primers at the final concentration of 4 μM for each primer and 1.25 mM MgSO4 in AA loading reagent. The primed IFC chip was loaded into the Fluidigm FC1 cycler and amplified using the following cycling conditions: 50°C for 30 min (reverse transcription), 94°C for 2 min, 35 cycles of 94°C for 30 s, 53°C for 30 s, 68°C for 90 s, and final extension at 68°C for 7 min, and then kept at 4°C. The RT-PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter, CA, USA) and then analyzed by using the Agilent TapeStation 4200 system and High Sensitivity DNA D5000 kit (Agilent Technologies, CA, USA) to determine quality and quantity of the amplicons.
The NGS libraries were prepared using Nextera DNA Flex library prep (currently Illumina DNA Prep) kit (Illumina, CA, USA). DNA was fragmentized by tagmentation for 15 min followed by indexing and library amplification, with 6 cycles used for 9 to 25 ng of amplicon or 12 cycles used for 1 to 9 ng of amplicon. The libraries were quantified by use of the Agilent TapeStation and DNA D5000 kit and then pooled with equal molar ratio for each library. Pooled libraries were denatured and diluted to a final concentration of 13.5 pM and then sequenced using the Illumina MiSeq system and reagent kit v3 (600 cycles).
Reference genome mapping assembly.
The full SARS-CoV-2 reference genome, NC_045512.2, was downloaded from GenBank (16) and utilized for reference-based genome assembly using the WRAIR viral disease branch in-house bioinformatics pipeline ngs_mapper version 1.5.0 (https://github.com/VDBWRAIR/ngs_mapper). This pipeline incorporates a series of quality control and assembly processes for fastq sequence reads retrieved from the Illumina MiSeq and other platforms. These processes include filtration to drop poorly indexed reads, read trimming based on quality thresholds using Trimommatic (17), read mapping to the reference genome using the Burrows-Wheeler aligner with maximal exact matches (BWA-MEM) (18), read tagging, variant calling file (VCF) generation, read mapping visualizations, fastq statistic generation, and consensus sequence generation from the VCF using an in-house script basecaller.py. Basecaller.py takes into account the depth of coverage, the frequency of a variant, and the quality of bases to make a consensus base call.
A minimum Phred base quality score of 35 and a minimum depth of coverage of 10 were utilized as the configuration parameters for basecaller.py, meaning that any base with a Phred score below 35 was masked and not taken into account for consensus base calling.
Variants present at a frequency of 20% or higher were denoted in the consensus genome as ambiguous positions following the IUPAC code. The assembled genomes were further manually curated utilizing bam, consensus and variant calling files generated from the ngs_mapper pipeline. Geneious R10 software, integrated genome viewer (IGV) (19), and MEGA version 7 (20) were used for quality control of ambiguous calls, insertions, deletions, and primer-induced mutations. The genomes were processed to only include coding sequence regions by clipping the 5′ and 3′ untranslated regions. MEGA7 was used to align genomes using default parameters with multiple sequence comparison by log expectation (MUSCLE) (21).
Data availability.
Next-generation sequencing read data are available under NCBI BioProject no. PRJNA683873 (see also Table S1 in the supplemental material).
RESULTS
Correlation of Fluidigm RT-PCR yield with amount of input SARS-CoV-2 RNA.
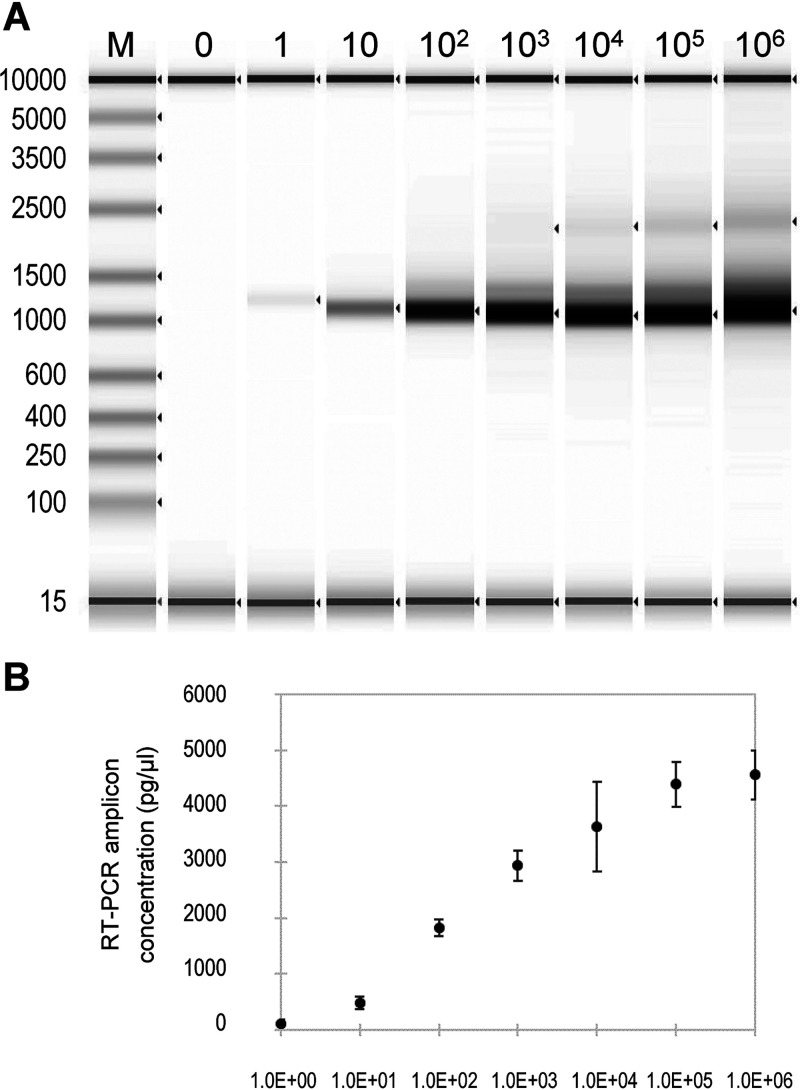
Fluidigm RT-PCR whole-genome amplification was done in the Access Array microfluidic chip using only 1.45 μl of RNA input for each sample and with a maximum capacity of 48 samples and 48 pairs of primers. In this study, a set of 35 primer pairs, i.e., nCOVF/R1-35 in Table 1, were designed and tested for genome RT-PCR amplification by using serial dilutions of purified RNA from SARS-CoV-2 isolate R4717. The experiment was done with four replicates processed in parallel. A single band of amplicons with expected sizes of approximately 1 kb was seen for all concentrations, with the band intensities correlated with copy numbers of SARS-CoV-2 in the serial dilutions. The correlation between the concentrations of yielded amplicons and the input SARS-CoV-2 genome copy numbers (Fig. 1) was significant, with a Pearson’s correlation P value of 3.38e−03.
FIG 1.
Agilent TapeStation analysis of SARS-CoV-2 whole-genome RT-PCR amplification using the Fluidigm system. (A) TapeStation profile of RT-PCR products. From left to right, 0, 1, 10, 102, 103, 104, 105, and 106 copies of SARS-CoV-2 RNA from isolate R4717 were used. (B) SARS-CoV-2 amplicon concentrations for RT-PCR with serial dilutions of R4717 RNA as inputs. The concentration values were the average and standard deviation from four replicates.
Whole-genome coverage and alignment depth of SARS-CoV-2 sequence assembly.
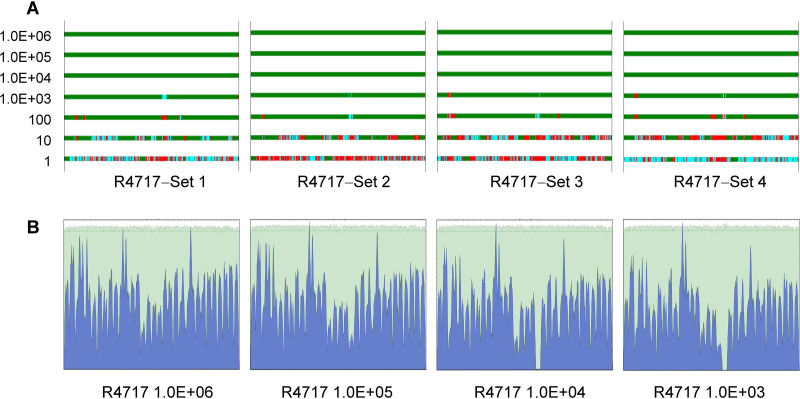
MiSeq data for the quadruplicated R4717 RNA serial dilutions described above were assembled using ngs_mapper pipeline with SARS-CoV-2 complete genome sequence NC_045512.2 as the mapping reference. As expected, genome assembly results correlated well with SARS-CoV-2 copy numbers in each sample. For reactions with 104 or higher SARS-CoV-2 RNA copies, complete genome sequences were readily obtained and, importantly, had uniform coverage depth across the genome with the peaks matching the regions of amplicon alignment (Fig. 2). For reactions with 103 copies of SARS-CoV-2 RNA, the assembled genome sequences were nearly complete, except for two small dip/gap at positions 1870 to 2500 and 16800 to 17700. The dip/gap regions were successfully filled by adding two extra primer pairs for each region to the panel of 35 primer pairs. These additional primers were selected from the ARTIC v3 protocol, paired to cover the two regions and added into four separate Fluidigm primer wells. In total, 39 pairs of primers were applied for whole-genome amplification in a single RT-PCR using Fluidigm nucleic acids amplification system.
FIG 2.
Genome coverage and sequence mapping alignment depth for MiSeq reads data. SARS-CoV-2 isolate R4717 RNA serial dilutions were subjected to Fluidigm RT-PCR amplification and MiSeq sequencing. Data were mapped using genome NC_045512.2 as the reference. (A) Genome coverage for 10× serial dilutions of R4717 RNA from 1.0E + 06 (top) to 1 (bottom) copies. Mapping depth was indicated in colors as follows: green, normal depth (depth ≥ 10); cyan, dip (depth 1 to 10); red, gap. Results for four replicates are shown. (B) Sequence mapping graphs for 106, 105, 104 and 103 copies of RNA from one set of serial dilutions.
SARS-CoV-2 genome sequencing of clinical respiratory specimens.
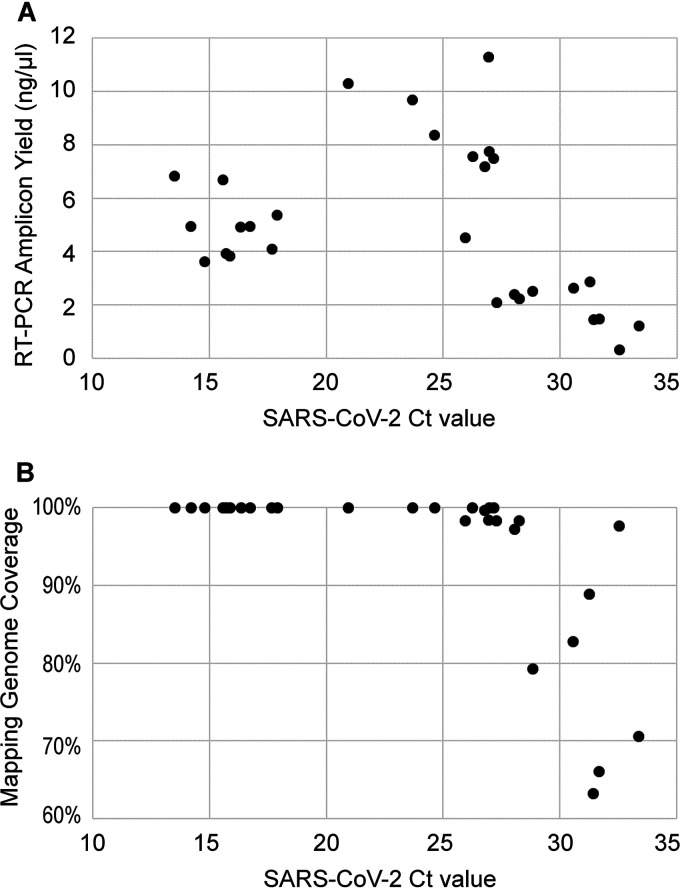
This method was subsequently used in genome sequencing of SARS-CoV-2 in RNA extracts purified from COVID-19-positive nasopharyngeal swabs. The set of 29 samples contained a wide range of titers, with RT-qPCR threshold cycle (CT) values from 13.5 to 33.4 (379 to 2.72 × 108 GE/μl) of SARS-CoV-2. The RT-PCR cDNA yield has significant correlation with viral titers in the CT range of 20 to 35, with P value of 1.26e−05. For the samples with CT values below 20, or exceedingly high concentrations of 1.0 × 107 or greater, RT-PCR yields were substantially lower than projected (Fig. 3). Importantly, this observation suggests that severe suppression of PCR could occur when samples of extremely high titer are used. Nevertheless, full or nearly complete genome coverage was obtained for all clinical specimens with titers above 1.0 × 104 GE/μl, or an approximate CT value of 29.
FIG 3.
SARS-CoV-2 whole-genome RT-PCR amplification using the Fluidigm system. Twenty nine RNA extracts from COVID-19-positive nasopharyngeal swabs were amplified and sequenced. RT-PCR yield was quantified using the Agilent TapeStation. Reference mapping coverage was determined by mapping MiSeq read data to reference genome sequence NC_045512.2. The represented coverage was for sequences with alignment depth of 10× or greater.
Comparison of assembled consensus sequences and nucleotide variations.
The Fluidigm one-step RT-PCR protocol was applied to SARS-CoV-2 RNA of highly varied titers. For SARS-CoV-2 isolate R4717 RNA 10× dilutions (Fig. 1), a total of 14 full genomes were assembled and curated. All the full genome consensus sequences were identical except for two replicates of 1.0 × 104 GE/μl. One sample had the ambiguous call Y (T or C) in the alignment nucleotide position 2105, while the other three replicates had a C. Another sample had a T in position 23260, while the other three had the ambiguous call Y. These were the only ambiguous positions found in the samples, showing low sample diversity at a variant frequency of 20% or higher. Taken together, this sequencing approach produces high-accuracy results.
DISCUSSION
Even before the disease was named COVID-19, the sequence was swiftly determined using next-generation sequencing technologies and SARS-CoV-2 was identified as the causative pathogen for the emerging acute respiratory disease (5, 22). The first sequence was made publicly available immediately, with a massive number of sequences subsequently generated and shared, which has greatly facilitated research and development (23–25). All the efforts in developing, improving, and sharing materials and/or methods have played an essential role in sequence-based investigations. All the known sequencing protocols use conventional PCR apparatuses with differences in design and selection of primers and reaction parameters. In this study, we applied a one-step RT-PCR protocol on a microfluidic platform (14) to establish a convenient workflow with throughput, speed, simplicity, consistency, and yield suitable for COVID-19 genome sequencing. The Fluidigm Access Array IFC, as well as the latest Fluidigm Juno model, holds 48 RNA samples (inlets) and 48 primer pairs (inlets). Steps for mixing sample with primers are obviated, which not only substantially reduces pipetting manipulation but also effectively mitigates the chance of sample-to-sample cross-contamination. In this report, we selected 39 primer pairs to obtain even genome coverage. Nine more individual pairs of primers can be easily added to the panel of 48 primer inlets to elevate coverage depth in regions that exhibit as dips/gaps and to quickly address emerging SARS-CoV-2 genetic divergence. Moreover, the total number of primer pairs can be further increased without difficulty by pooling together several compatible primer pairs and adding them into one primer inlet. Each sample is mixed with individual primer pairs in IFC microfluidic chambers for nanoliter (nl) RT-PCR amplifications in a simplex-independent reaction manner. In contrast, conventional PCR methods often need optimization of primer pooling and reaction parameters to circumvent primer-to-primer interference and to avoid highly variable yields among amplicons.
Since input RNA samples are partitioned into individual nl reaction chambers to cross-mix with individual primer pairs, microfluidic applications, including Fluidigm IFC, require sufficient genomic copies in order to achieve whole-genome amplification. In consequence, Fluidigm amplification-based whole-genome sequencing has limitations in sequencing low-titer samples. For RNA samples with 1,000 GE/μl or lower concentration, multiplex RT-PCR based methods or SARS-CoV-2 hybridization-based enrichment method might be a more suitable choice for whole-genome sequencing (26–29). The addition of a first-strand cDNA synthesis step prior to Fluidigm amplification, with a change of Fluidigm thermocycling program from RT-PCR to PCR, may help increase genome coverage for low-titer samples. Additionally, concentrating samples using the SpeedVac vacuum concentrator (Thermo Fisher Scientific) or similar devices and loading samples into multiple sample inlets are simple options to generate more amplicons from low-titer RNA samples. Many, if not all, COVID-19 specimens are tested with molecular tests and the CT values or equivalent titer scores are readily available for deciding whether one-step Fluidigm amplification-based genome sequencing protocol is appropriate.
Using this method, very few sequence assembly errors were observed throughout the tested SARS-CoV-2 sample genomes. These errors might be due to PCR, sequencing or basecalling algorithm errors, as well as due to normal fluctuations in the minor variant frequencies between the sample aliquots. When assembled sequences show potentially significant nucleotide alterations or indels, thorough examination of the data quality processing, primer trimming, curation of sequence assembly, and detailed laboratory records are needed. Whenever possible, running replicate samples, repeating the experiment entirely, or using other methods are important to validate genome variations and minor variants.
In conclusion, our study demonstrates a convenient SARS-CoV-2 whole-genome sequencing protocol by incorporating one-step RT-PCR amplification, microfluidic technology, and next-generation sequencing to achieve a simple and fast workflow with consistent and high-quality data. The performance of the protocol was verified using viral isolate RNA and tested by sequencing clinical respiratory samples of various viral titers.
Supplementary Material
ACKNOWLEDGMENTS
We thank James S. Hilaire, Nicole R. Nicholas, Tuan K. Nguyen, and April N. Griggs for their assistance in project management, sample tracking, storage, and retrieval.
Funding was received from the Global Emerging Infections Surveillance and Response System (GEIS), a division of the Armed Forces Health Surveillance Branch.
We declare no conflicts of interest.
The material has been reviewed by the authors’ respective institutions. There are no objections to its presentation and/or publication. The views expressed here are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the U.S. Government.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Hu B, Guo H, Zhou P, Shi ZL. 2021. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19:141–154. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hon KL, Leung KKY, Leung AKC, Sridhar S, Qian S, Lee SL, Colin AA. 2020. Overview: the history and pediatric perspectives of severe acute respiratory syndromes: novel or just like SARS. Pediatr Pulmonol 55:1584–1591. 10.1002/ppul.24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Habibzadeh P, Stoneman EK. 2020. The novel coronavirus: a bird's eye view. Int J Occup Environ Med 11:65–71. 10.15171/ijoem.2020.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gralinski LE, Menachery VD. 2020. Return of the coronavirus: 2019-nCoV. Viruses 12:135. 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang C, Horby PW, Hayden FG, Gao GF. 2020. A novel coronavirus outbreak of global health concern. Lancet 395:470–473. 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauver JR, Petrone ME, Hodcroft EB, Shioda K, Ehrlich HY, Watts AG, Vogels CBF, Brito AF, Alpert T, Muyombwe A, Razeq J, Downing R, Cheemarla NR, Wyllie AL, Kalinich CC, Ott IM, Quick J, Loman NJ, Neugebauer KM, Greninger AL, Jerome KR, Roychoudhury P, Xie H, Shrestha L, Huang ML, Pitzer VE, Iwasaki A, Omer SB, Khan K, Bogoch II, Martinello RA, Foxman EF, Landry ML, Neher RA, Ko AI, Grubaugh ND. 2020. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell 181:990–996.e5. 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worobey M, Pekar J, Larsen BB, Nelson MI, Hill V, Joy JB, Rambaut A, Suchard MA, Wertheim JO, Lemey P. 2020. The emergence of SARS-CoV-2 in Europe and North America. Science 370:564–570. 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockett RJ, Arnott A, Lam C, Sadsad R, Timms V, Gray KA, Eden JS, Chang S, Gall M, Draper J, Sim EM, Bachmann NL, Carter I, Basile K, Byun R, O'Sullivan MV, Chen SC, Maddocks S, Sorrell TC, Dwyer DE, Holmes EC, Kok J, Prokopenko M, Sintchenko V. 2020. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat Med 26:1398–1404. 10.1038/s41591-020-1000-7. [DOI] [PubMed] [Google Scholar]
- 10.Oude Munnink BB, Nieuwenhuijse DF, Stein M, O'Toole Á, Haverkate M, Mollers M, Kamga SK, Schapendonk C, Pronk M, Lexmond P, van der Linden A, Bestebroer T, Chestakova I, Overmars RJ, van Nieuwkoop S, Molenkamp R, van der Eijk AA, GeurtsvanKessel C, Vennema H, Meijer A, Rambaut A, van Dissel J, Sikkema RS, Timen A, Koopmans M, Dutch-Covid-19 response team . 2020. Rapid SARS-CoV-2 whole-genome sequencing and analysis for informed public health decision-making in the Netherlands. Nat Med 26:1405–1410. 10.1038/s41591-020-0997-y. [DOI] [PubMed] [Google Scholar]
- 11.Licastro D, Rajasekharan S, Dal Monego S, Segat L, D’Agaro P, Marcello A. 2020. Isolation and full-length genome characterization of SARS-CoV-2 from COVID-19 cases in northern Italy. J Virol 94:e00543-20. 10.1128/JVI.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam MR, Hoque MN, Rahman MS, Alam A, Akther M, Puspo JA, Akter S, Sultana M, Crandall KA, Hossain MA. 2020. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci Rep 10:14004. 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefanelli P, Faggioni G, Lo Presti A, Fiore S, Marchi A, Benedetti E, Fabiani C, Anselmo A, Ciammaruconi A, Fortunato A, De Santis R, Fillo S, Capobianchi MR, Gismondo MR, Ciervo A, Rezza G, Castrucci MR, Lista F, on behalf of ISS Covid-19 study group . 2020. Whole genome and phylogenetic analysis of two SARS-CoV-2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill 25:2000305. 10.2807/1560-7917.ES.2020.25.13.2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M, Escudero-Ibarz L, Moody S, Zeng N, Clipson A, Huang Y, Xue X, Grigoropoulos NF, Barrans S, Worrillow L, Forshew T, Su J, Firth A, Martin H, Jack A, Brugger K, Du MQ. 2015. Somatic mutation screening using archival formalin-fixed, paraffin-embedded tissues by Fluidigm multiplex PCR and Illumina sequencing. J Mol Diagn 17:521–532. 10.1016/j.jmoldx.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eurosurveillance Editorial Team. 2020. Note from the editors: novel coronavirus (2019-nCoV). Euro Surveill 25:2001231. 10.2807/1560-7917.ES.2020.25.3.2001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corman VM, Muller MA, Costabel U, Timm J, Binger T, Meyer B, Kreher P, Lattwein E, Eschbach-Bludau M, Nitsche A, Bleicker T, Landt O, Schweiger B, Drexler JF, Osterhaus AD, Haagmans BL, Dittmer U, Bonin F, Wolff T, Drosten C. 2012. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill 17:20334. 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 24.Nalla AK, Casto AM, Huang MW, Perchetti GA, Sampoleo R, Shrestha L, Wei Y, Zhu H, Jerome KR, Greninger AL. 2020. Comparative performance of SARS-CoV-2 detection assays using seven different primer-probe sets and one assay kit. J Clin Microbiol 58:e00557-20. 10.1128/JCM.00557-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan M, Wu NC, Zhu X, Lee CD, So RTY, Lv H, Mok CKP, Wilson IA. 2020. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368:630–633. 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itokawa K, Sekizuka T, Hashino M, Tanaka R, Kuroda M. 2020. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. PLoS One 15:e0239403. 10.1371/journal.pone.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paden CR, Tao Y, Queen K, Zhang J, Li Y, Uehara A, Tong S. 2020. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:2401–2405. 10.3201/eid2610.201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasir JA, Kozak RA, Aftanas P, Raphenya AR, Smith KM, Maguire F, Maan H, Alruwaili M, Banerjee A, Mbareche H, Alcock BP, Knox NC, Mossman K, Wang B, Hiscox JA, McArthur AG, Mubareka S. 2020. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random hexamers, and bait capture. Viruses 12:895. 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Addetia A, Lin MJ, Peddu V, Roychoudhury P, Jerome KR, Greninger AL. 2020. Sensitive recovery of complete SARS-CoV-2 genomes from clinical samples by use of Swift Biosciences’ SARS-CoV-2 multiplex amplicon sequencing panel. J Clin Microbiol 59:e02226-20. 10.1128/JCM.02226-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing read data are available under NCBI BioProject no. PRJNA683873 (see also Table S1 in the supplemental material).