LETTER
The pneumococcal serogroup 15 comprises four capsular polysaccharide serotypes (15A, 15B, 15C, 15F) that collectively account for an impactful disease burden (1, 2). These four serotypes are defined by Quellung reaction typing, employing specific serologic factor sera to visualize the capsule (3) (Table 1). Each of these four serotypes shares an identical pentasaccharide repeat unit that forms an identical linear structure in types 15A and 15F, and a distinct branched structure in types 15B and 15C (4). The two distinct repeat oligosaccharide polymerase genes, wzy15BC and wzy15AF (4), provide the basic structural difference between these two pairs of highly related serotypes (Fig. 1). The difference between serotypes 15B and 15C is dictated by O-acetylation of the polysaccharide in serotype 15B by the wciZ-encoded enzyme, with the two serotypes frequently interconverting due to reversible frameshifting of a TA repeat within wciZ (5). Similarly, the wciZ gene in serotype 15A contains multiple inactivating mutations compared to its functional counterpart in serotype 15F (4, 6). At the end of the cps15F operon is another putative O-acetyltransferase gene, wcjE (4), that potentially contributes to the serologic difference between 15A and 15F strains. The wzy primary target, combined with the wciZ secondary target, distinguishes these four serotypes in our whole-genome sequence-based pneumococcal bioinformatic pipeline (1). Note that the incomplete rhamnose biosynthetic apparatus (rmlB and rmlD) and the glf gene that encodes UDP-galactopyranose serve no apparent functions in serotype 15F (Fig. 1), since serogroup 15 capsules do not contain rhamnose or galactofuranose (7, 8).
TABLE 1.
Unique serogroup 15 factor reactivity of new serotype 15D employing CDC Streptococcus lab typing antisera and SSIa typing antisera
| Pneumococcal strain | CDC or SSI 15 poola | CDC factors 15bfb | CDC factors 15dec | CDC factor 15gd | CDC or SSI factor 15ee | CDCor SSI factor 15hf | SSI factor 15bg | SSI factor 15cg |
|---|---|---|---|---|---|---|---|---|
| 15A strain 389/39 | + | − | + | + | − | − | − | + |
| 15F strain 688/63 | + | + | − | − | − | − | + | + |
| 15B strain 7904/39 | + | + | + | − | + | + | + | − |
| 15C strain 553/62 | + | − | + | − | + | − | − | − |
| 15D invasive US 84245 | + | + | − | + | − | − | + | + |
| 15D carriage Mozambique MZ877 | + | + | − | + | − | − | + | + |
Pooled antiserum prepared against serotypes 15A, 15F, 15B, and 15C. Identical results were obtained using CDC and commercial antiserum from Serum Staten Institut (SSI, Copenhagen Denmark) where indicated. For CDC purposes, pooled antiserum, factor 15bf, factor 15de, and factor 15g routinely used to resolve serogroup 15 strains into its 4 different serotypes (and now can be used for identifying 5 different serotypes with the inclusion of 15D). The reagents were prepared as described (3).
Factor 15bf consists of antiserum prepared against serotype 15F strain followed by absorbing with serotype 15A strain.
Factor 15de consists of antiserum prepared against serotype 15C strain followed by absorbing with serotype 15F strain.
Factor 15g consists of antiserum prepared against serotype 15A strain followed by absorbing with serotype 15F strain and serotype 15C strain. We highlight this result since it is the only serologic difference shown in the table between serotypes 15F and 15D.
Factor 15e consists of antiserum prepared against serotype 15C strain followed by absorbing with serotype 15A strain.
Factor 15h consists of antiserum prepared against serotype 15B strain followed by absorbing with serotype 15F strain and 15C strain.
Factors 15b and 15c were obtained from SSI (Serum Staten Institut).
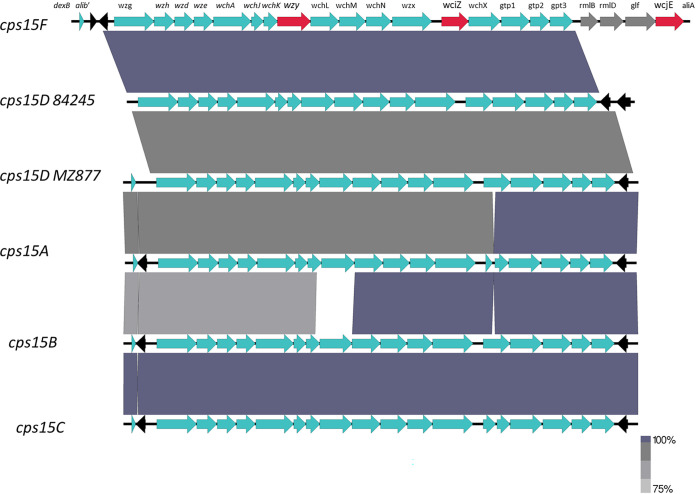
FIG 1.
Alignment of cps operons from the four known serogroup 15 serotypes and from the two isolates expressing the new serotype 15D. The three genes indicated in red provide basis by which to resolve the five different serotypes shown. The gray open reading frames encode enzymes that have functions unrelated to serogroup 15 polysaccharide structure. The short black open reading frames are transposase gene remnants. The two serotype 15D strains were subjected to whole-genome sequencing as previously described (1, 2). Sequences encompassing the dexB 3′ end and the aliA 5′ end were extracted from these two strains and were aligned with published GenBank sequences for cps15F (CR931666), cps15A (CR931663), cps15B (CR931664), and 15C (CR931665) (see references 4 and 6). The extracted sequences and published serogroup 15 cps operons were subjected to Prokka to identify and annotate open reading frames (9). The resultant annotated sequences were analyzed by BLAST and the homolog cps locus regions were aligned into figures using EasyFig version 2.2.3 software (10). Strains 84245 and MZ877 were assigned BioSample accession numbers SAMN14150919 and SAMN17684515, respectively, under project number PRJNA697931.
We recently encountered two strains (invasive U.S. blood isolate 84245 recovered during 2018 and Mozambique carriage isolate MZ877 recovered during 2019) that our bioinformatics pipeline identified as serotype 15A due to an identical wzy15AF target combined with a divergent wciZ15F target sequence (92% sequence identity). Strains 84245 and MZ877 shared near-identity over the full-length wciZ gene (977/978 identical bp) but had only 2 to 3 conservative amino acid substitutions compared to the WciZ15F protein (not shown). Strains 84245 and MZ877 also lacked the wcjE gene present in cps15F (Fig. 1). Consistent with their unique cps operons, these two strains displayed unique reactivity with serogroup 15 serotyping factors (Table 1). To summarize, this new serotype, designated 15D, is potentially predicted by the unique combination of wzy15AF-encoded polymerase activity, wciZ-encoded O-acetyltransferase activity, and lack of a wcjE-encoded O-acetyltransferase.
Strain 84245 (BioSample accession no. SAMN14150919), described during US invasive pneumococcal disease surveillance (2), was predicted to have low-level penicillin resistance and macrolide resistance, while strain MZ877 (SAMN17684515) was predicted to have reduced penicillin susceptibility combined with co-trimoxazole-resistance. Strain 84245 had a new multilocus sequence type (ST15307) that is a single locus variant of ST9692, associated with serotype 15A strains recovered in Kenya, while MZ877 has the unrelated genotype ST10654, reported from 15A strains recovered in South Africa (https://pubmlst.org/organisms/streptococcus-pneumoniae). Our finding is conceivably important for future pneumococcal vaccine formulations. We were originally mistaken in assigning these two strains the serotype 15A. It is conceivable that other researchers have also mistakenly assigned serotype 15A or 15F to strains that actually express serotype 15D.
Data availability. Strains 84245 and MZ877 were assigned BioSample accession numbers SAMN14150919 and SAMN17684515, respectively, under project number PRJNA697931.
ACKNOWLEDGMENT
The opinions expressed by the authors do not necessarily reflect the opinions of the Centers for Disease Control and Prevention or the institutions with which the authors are affiliated.
REFERENCES
- 1.Metcalf BJ, Gertz RE, Jr., Gladstone RA, Walker H, Sherwood LK, Jackson D, Li Z, Law C, Hawkins PA, Chochua S, Sheth M, Rayamajhi N, Bentley SD, Kim L, Whitney CG, McGee L, Beall B, Active Bacterial Core surveillance Team. 2016. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin Microbiol Infect 22:e9–60.e29. 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metcalf BJ, Chochua S, Walker H, Tran T, Li Z, Varghese J, Snippes Vagnone PM, Lynfield R, McGee L, Li Y, Pilishvili T, Beall B. 2021. Invasive pneumococcal strain distributions and isolate clusters associated with persons experiencing homelessness during 2018. Clin Inf Dis 10.1093/cid/ciaa1680. [DOI] [PubMed] [Google Scholar]
- 3.Lund E, Henrichsen J. 1978. Laboratory diagnosis, serology and epidemiology of Streptococcus pneumoniae, p 1241–1262. In Methods in microbiology, Bergan T, Norris JR (ed), Academic Press, London, United Kingdom. [Google Scholar]
- 4.Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, Reeves PR, Bentley SD, Spratt BG. 2007. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol 189:7841–7855. 10.1128/JB.00836-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Selm S, van Cann LM, Kolkman MA, van der Zeijst BA, van Putten JP. 2003. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect Immun 71:6192–6198. 10.1128/iai.71.11.6192-6198.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aanensen DM, Mavroidi A, Bentley SD, Reeves PR, Spratt BG. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol 189:7856–7876. 10.1128/JB.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamerling JP. 2000. Pneumococcal polysaccharides: a chemical view, p 81–114. In Tomasz A (ed), Streptococcus pneumoniae molecular biology and mechanisms of disease. Mary Ann Liebert, Inc., Larchmont, NY. [Google Scholar]
- 8.Jones C, Lemercinier X. 2005. Full NMR assignment and revised structure for the capsular polysaccharide from Streptococcus pneumoniae type 15B. Carbohydr Res 340:403–409. 10.1016/j.carres.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 9.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]