In this study, we comprehensively analyzed multispecific antibody kinetics of different immunoglobulins in hospitalized patients with acute severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Three hundred fifty-four blood samples longitudinally obtained from 81 IgG-seroconverting progressed coronavirus disease 2019 (CoVID-19) patients were quantified for spike 1 (S1), S2, and nucleocapsid protein (NCP)-specific IgM, IgA, IgG, and total Ig antibodies using a microarray, 11 different enzyme-linked immunosorbent assays (ELISAs)/chemiluminescence immunoassays (CLIAs), and 1 rapid test by seven manufacturers.
KEYWORDS: SARS, coronavirus, SARS-CoV-2, antibodies, immunoassay, microarray, IgA, ELISA
ABSTRACT
In this study, we comprehensively analyzed multispecific antibody kinetics of different immunoglobulins in hospitalized patients with acute severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Three hundred fifty-four blood samples longitudinally obtained from 81 IgG-seroconverting progressed coronavirus disease 2019 (CoVID-19) patients were quantified for spike 1 (S1), S2, and nucleocapsid protein (NCP)-specific IgM, IgA, IgG, and total Ig antibodies using a microarray, 11 different enzyme-linked immunosorbent assays (ELISAs)/chemiluminescence immunoassays (CLIAs), and 1 rapid test by seven manufacturers. The assays’ specificity was assessed in 130 non-CoVID-19 pneumonia patients. Using the microarray, NCP-specific IgA and IgG antibodies continuously displayed higher detection rates during acute CoVID-19 than S1- and S2-specific ones. S1-specific IgG antibodies, however, reached higher peak values. Until the 26th day post-symptom onset, all patients developed IgG responses against S1, S2, and NCP. Although detection rates by ELISAs/CLIAs generally resembled those of the microarray, corresponding to the target antigen, sensitivities and specificities varied among all tests. Notably, patients with more severe CoVID-19 displayed higher IgG and IgA levels, but this difference was mainly observed with S1-specific immunoassays. In patients with high SARS-CoV-2 levels in the lower respiratory tract, we observed high detection rates of IgG and total Ig immunoassays with a particular rise of S1-specific IgG antibodies when viral concentrations in the tracheal aspirate subsequently declined over time. In summary, our study demonstrates that differences in sensitivity among commercial immunoassays during acute SARS-CoV-2 infection are only partly related to the target antigen. Importantly, our data indicate that NCP-specific IgA and IgG antibodies are detected earlier, while higher S1-specific IgA antibody levels occur in severely ill patients.
INTRODUCTION
Since December 2019, the newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is causing a massive pandemic with currently over one million deaths worldwide and devastating effects on global health care systems and the world’s economy (1, 2). PCR-based techniques enabled the early identification of the virus and immediately became the backbone of laboratory diagnosis and contact tracing (3). Shortly after, a wide range of in-house and commercial antibody assays became available (4–11).
Meanwhile, antibody tests are widely applied in seroprevalence studies, aiming to evaluate the speed of SARS-CoV-2 spread on a population level (12–14). However, individuals with asymptomatic or clinically mild infections often display low antibody titers whose correct identification depends on the respective immunoassay’s sensitivity (15–18). Notably, antibody positivity in commercial immunoassays alone may not reflect functionally broad and long-lasting immunity against reinfections, limiting the epidemiological significance of serological antibody testing (6, 19).
In contrast, antibody assays may have considerable potential in diagnosing severe and symptomatic infections for three main reasons. First, patients with progressed coronavirus disease 2019 (CoVID-19) and pneumonia may display low virus concentrations in the upper respiratory tract, possibly causing false-negative PCR results from nasopharyngeal swabs (20–22). Second, the simultaneous measurement of different immunoglobulin (Ig) classes (IgM, IgA, and IgG) may help evaluate the infection stage and assess the cumulative risk of upcoming complications (23). Third, clinically severe CoVID-19 is associated with earlier seroconversion and higher antibody concentrations, posing a different diagnostic setting than mild or asymptomatic infection (4, 16).
However, recent studies comparing immunoassays with the SARS-CoV-2 nucleocapsid protein (NCP) and the spike (S) protein as target antigens indicate possible differences for sensitivity in early SARS-CoV-2 infection (24–27). Therefore, we longitudinally characterized NCP-, S1-, and S2-specific IgM, IgA, and IgG antibody kinetics in hospitalized patients with acute SARS-CoV-2 infection using a single commercial microarray and compared those kinetics with the detection rates of 12 other commercial immunoassays. Notably, we specifically included only seroconverting patients (in an early infection stage) who tested IgG negative at hospital admission.
MATERIALS AND METHODS
Patients and controls.
The study included 354 serum/plasma samples prospectively obtained from 81 patients (27 female, 54 male; median age, 73 years; range, 22 to 94) with PCR-confirmed, symptomatic SARS-CoV-2 infection (diagnosed by positive PCR from nasopharyngeal swab/respiratory specimen samples), hospitalized between the beginning of March and the end of April 2020. Out of a total of 218 patients who were hospitalized during this period, these 81 patients were included explicitly in the study since they fulfilled all of the following four inclusion criteria: (i) at hospital admission, these 81 patients tested negative for anti-SARS-CoV-2 IgG using the Euroimmun SARS-CoV-2 IgG enzyme-linked immunosorbent assay (ELISA) (the routine IgG assay at our center), (ii) from these 81 patients, a minimum of 2 prospectively collected serum/plasma samples with a minimum interval of 2 days were available, (iii) these 81 patients never received any convalescent plasma treatment during or before hospitalization, and (iv) these patients did not display any underlying immunosuppression, either by any disease or by treatment. The other 137 patients were excluded since they did not fulfill one or more inclusion criteria. The 81 study patients reported one or more of the following initial symptoms: loss of smell and taste, fever, cough, headache, and general weakness. Reasons for hospitalization were dyspnea and hypoxemia, medical observation of patients of high-risk groups, or the need for isolation of patients in nursing homes. The clinical severity of CoVID-19 was classified using the following WHO criteria: mild, symptomatic patients without evidence of pneumonia or hypoxia; moderate, clinical signs of pneumonia but no signs of severe pneumonia, including SpO2 of ≥90%; severe, clinical signs of pneumonia plus one of the following: respiratory rate of >30 breaths/min, severe respiratory distress, or SpO2 of <90%; critical, acute respiratory distress syndrome (ARDS) requiring intubation at the intensive care unit (ICU); and deceased.
During their hospital stay, the patients prospectively provided serum/plasma, nasopharyngeal swab, and/or tracheal aspirate samples (intubated patients) according to a protocol approved by the local ethics committee (EK 2283/2019). Informed written consent was obtained from each patient.
Serum samples from 130 non-SARS-CoV-2-infected, hospitalized patients with pneumonia (46 treated at an ICU) served as controls (59 female, 71 male; median age, 61 years; range, 1 to 93). Detailed information on these controls and how they were selected is given in supplemental material and methods.
PCR.
PCR analyses for SARS-CoV-2-specific RNA were performed as described previously. Information on the protocol is given in supplemental material and methods.
Antibody microarray.
Temporal antibody profiles of different immunoglobulin classes against S1, S2, and NCP were first quantified using a commercial, miniaturized, 96-well protein microarray, the SARS-CoV-2 IgM, IgA, and IgG ViraChip assay (Viramed, Planegg, Germany) according to the manufacturer’s instructions. In brief, this assay uses the purified spike proteins S1 and S2 and NCP as antigens fixed on a nitrocellulose membrane of the same microwell. Therefore, multispecific antibody responses against these antigens can be simultaneously detected in a single test run and differentiated by three immunoglobulin classes (IgM, IgA, or IgG). The antibodies are quantified by a chromogen/substrate reaction with the relative intensity (RI) of the specific colorimetric signal (correlated with a calibrator signal) given as ViraChip units. As shown in Fig. S1 in the supplemental material, we determined the cutoff levels for IgM, IgA, and IgG antibodies against S1, S2, and NCP, respectively, in the non-SARS-CoV-2-infected controls (mean plus two times the standard deviation, rounded to the nearest 10). The quantitative antibody measurement was performed on a ViraChip scanner using ViraChip software.
Additional immunoassays.
The 354 serum/plasma samples were further assessed using the (i) Euroimmun SARS-CoV-2 IgA and (ii) IgG ELISAs, and the (iii) Euroimmun NCP IgM and (iv) NCP IgG ELISAs (all by Euroimmun, Lübeck, Germany); the (v) Wantai SARS-CoV-2 IgM ELISA, (vi) Total Ab ELISA, and (vii) rapid test (all by Wantai, Bejing, China); (viii) the Liaison SARS-CoV-2 IgG chemiluminescent assay (CLIA; DiaSorin, Saluggia, Italy); (ix) the Platelia SARS-CoV-2 Total Ab assay (Bio-Rad, Hercules, USA); (x) the Elecsys anti-SARS-CoV-2 electrochemiluminescence immunoassay (ECLIA; Roche, Basel, Switzerland); and (xi) the COVID-19 ELISA IgM+IgA and (xii) COVID-19 ELISA IgG (both Vircell, Valencia, Spain). The tests were performed according to the manufacturers’ instructions using the recommended cutoffs (Table S1). The Wantai rapid test was performed and interpreted as described previously (28). Detailed information on these assays, including the respective target antigens, cutoff values, and covered immunoglobulin classes, are shown in Table S1. For the assessment of the immunoassays’ detection rates, borderline results were considered negative. In a previous study, we showed that although certain ELISAs used in this study are intended for semiquantitative assessment, stepwise serum/plasma dilution provided robust and accurate antibody quantification (28). More information on how samples were diluted is given in supplemental material and methods.
Neutralization assay.
One hundred thirty-eight samples from 20 patients of our study cohort had been assessed for neutralizing antibodies using an in-house neutralization assay (NT) in a previous study (29). In brief, duplicates of serial 2-fold dilutions of heat-inactivated serum samples were incubated with 50 to 100 TCID50 (50% tissue culture infective dose) of infectious SARS-CoV-2 (GISAID/EPI_ISL_438123/hCoV-19/Austria/CeMM0360/2020) for 1 h at 37°C. The mixture was then added to Vero E6 cells (ECACC), and incubation was continued for 2 to 3 days. NT titers were expressed as the reciprocal of the serum dilution that protected against virus-induced cytopathic effect (CPE). NT titers ≥10 were considered positive. For the present study, these quantitative NT titers were correlated with the microarray’s commercial results and the evaluated ELISAs and CLIAs.
Statistical analyses.
The relationship of neutralizing antibody titers with antibody concentrations assessed by the microarray and the ELISAs/CLIAs was analyzed using Spearman correlation. Differences in antibody levels relating to antigen specificity and antibody concentrations among patients with different disease severity were compared using the Wilcoxon rank-sum test and GraphPad Prism 8.3.1. software (GraphPad Software, San Diego, USA). A P value of <0.05 was considered statistically significant.
RESULTS
Patient characteristics.
The study included 354 serum/plasma samples from 81 hospitalized patients with PCR-confirmed SARS-CoV-2 infection, selected explicitly for IgG negativity at hospital admission. On average, patients were hospitalized at day 8 postonset of symptoms (range, days 1 to 20). The median concentration of SARS-CoV-2 RNA from nasopharyngeal swabs at hospital admission was 2.99 × 105 copies/ml (range, undetectable to 3.49 × 109). Kinetics of virus concentration in nasopharyngeal swabs and tracheal aspirates related to the clinical severity are shown in Fig. S2 in the supplemental material.
The 354 serum/plasma samples comprised a median of 4 serum/plasma samples per patient (range, 2 to 17), collected with a median interval of 2 days (range, 2 to 12), ranging from the 1st to the 26th day post-symptom onset. Table S2 displays the sample number per 2-day interval (using one sample per patient per interval step). Of note, 3 patients (4%) showed mild, 37 (45%) moderate, 11 (14%) severe, and 11 (14%) critical disease severity, while 19 (23%) patients deceased.
Specificity of immunoassays.
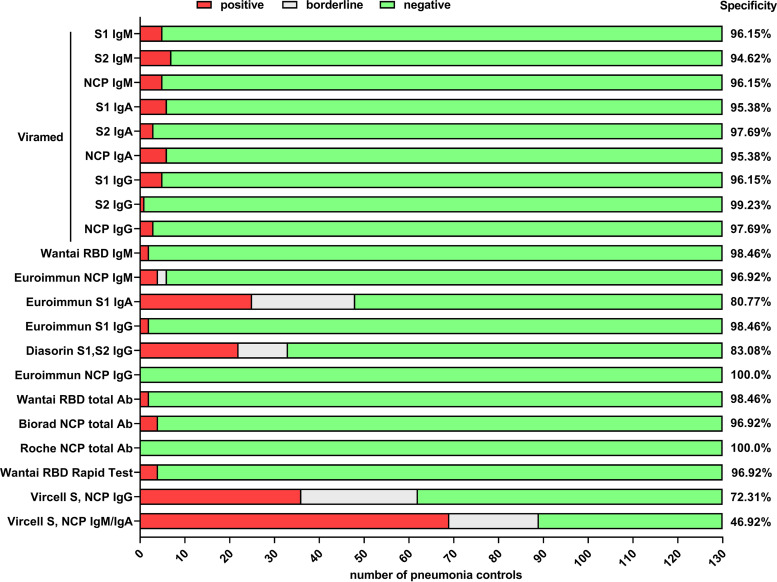
Test specificities of the evaluated immunoassays determined with 130 samples from non-SARS-CoV-2-infected pneumonia controls (see supplemental material and methods) are shown in Fig. 1. Notably, we found significant differences in individual assays’ specificities, ranging from 46.92% to 100%. The antibody levels in controls are shown in Fig. S3.
FIG 1.
Comparison of the assays’ specificity in 130 hospitalized patients with pneumonia without SARS-CoV-2 infection. The assays’ specificity is given as true-negativity rate, counting borderline results as negative.
Antibody kinetics assessed by microarray.
Using a microarray, we first analyzed the temporal profiles of S1-, S2-, and NCP-specific IgM, IgA, and IgG antibodies in the course of acute SARS-CoV-2 infection in hospitalized patients by a single test.
Regarding IgM antibody responses (Fig. 2A), we observed continuously higher detection rates within 2-day intervals postonset of symptoms for S1- and NCP-specific antibodies than for anti-S2-IgM antibodies. In contrast to IgM antibodies, as shown in Fig. 2B, IgA antibodies were earlier detected, particularly in the very early phase of the infection (until the 12th day postonset of symptoms). When we analyzed IgA and IgG antibody kinetics with respect to the target antigen, we observed that detection rates were continuously higher for NCP-specific antibodies over time than those directed against S1 and S2 (Fig. 2B and C).
FIG 2.
Detection rates by a commercial microarray (Viramed) in 354 blood samples longitudinally obtained from 81 hospitalized IgG-seroconverting patients with acute SARS-CoV-2 infection, grouped by a 2-day interval (one sample per patient per interval step). (A) S1-, S2-, and NCP-specific IgM antibodies; (B) S1-, S2-, and NCP-specific IgA antibodies; (C) S1-, S2-, and NCP-specific IgG antibodies.
Fig. S4 shows antibody levels quantified by the microarray. Of note, although NCP-specific IgG antibodies were detected earlier than S-specific antibodies, the median S1-specific IgG antibody units at the end of the observational period were significantly higher than NCP-specific IgG (P = 0.04; median RI, 393 for S1 versus 250 for NCP).
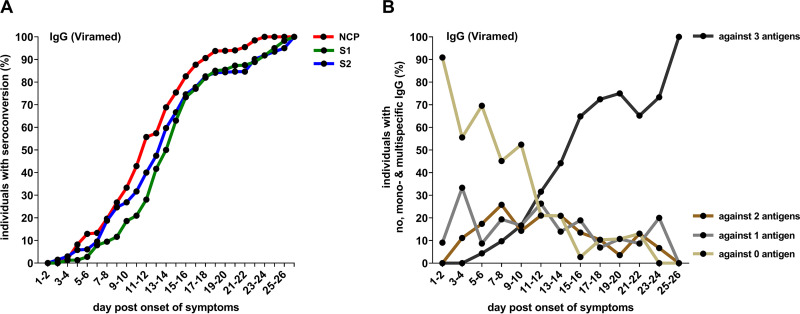
Corresponding to antibody detection rates within 2-day intervals postonset of symptoms, the NCP-specific IgG antibodies’ cumulative seroconversion rate was higher than that of S2- and S1-specific IgG antibodies (Fig. 3A). Of note, during acute SARS-CoV-2 infection, an increasing percentage of patients cumulatively developed IgG antibodies against more than one antigen (Fig. 3B) until at the end of the observational period, at which point all patients had detectable IgG responses against S1, S2, and NCP (Fig. 3B).
FIG 3.
(A) Cumulative microarray (Viramed) detection rates of IgG antibodies in 354 blood samples longitudinally obtained from 81 hospitalized patients with acute SARS-CoV-2 infection, grouped by a 2-day interval (one sample per patient per interval step). (B) Percentage of hospitalized patients with acute SARS-CoV-2 infection with detectable IgG responses (microarray, Viramed) against no, one, two, or all three target antigens (S1, S2, and NCP).
Detection rates of the 12 other immunoassays during acute SARS-CoV-2 infection.
With the microarray results as a basis, we evaluated the sensitivity of 12 immunoassays, grouped by Ig classes covered by the assays (IgM, IgA, IgG, and total Ig) and with respect to the assays’ respective target antigens (Fig. 4). Data from immunoassays with a specificity lower than 80% are presented as supplemental data.
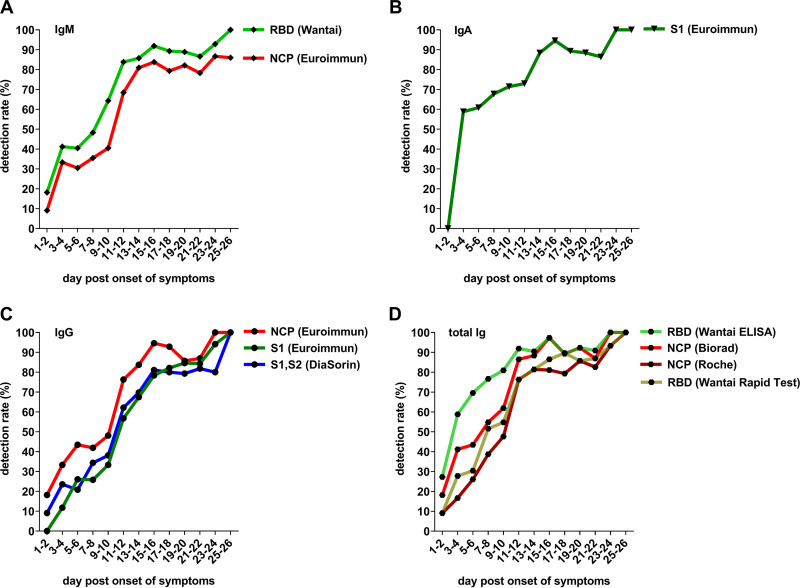
FIG 4.
Detection rates by 10 commercial immunoassays (6 ELISAs, 3 CLIAs, one rapid test; all with a specificity >80%) in the 354 blood samples from 81 hospitalized IgG-seroconverting patients with acute SARS-CoV-2 infection, grouped by a 2-day interval (one sample per patient per interval step). (A) IgM ELISAs; (B) IgA ELISA; (C) IgG ELISAs/CLIAs; (D) total Ig ELISAs/CLIAs and rapid tests. Borderline results were counted as negative.
As shown in Fig. 4A, the sensitivities of the Euroimmun NCP IgM and the Wantai SARS-CoV-2 IgM ELISA—with the receptor-binding domain (RBD) as antigen—were relatively similar over time. The RBD-specific Wantai IgM ELISA, however, continuously showed a higher sensitivity than the NCP-specific one by Euroimmun (Fig. 4A).
In agreement with the higher detection rates observed for IgA antibodies in the microarray, high sensitivity, especially in the early phase of the infection, was observed for the only commercial IgA ELISA, the SARS-CoV-2 IgA ELISA by Euroimmun (Fig. 4B).
The detection rates of the IgG ELISAs were similar to those of the microarray and were associated with the assays’ target antigen. Thus, higher detection rates were found for the NCP IgG ELISA by Euroimmun than for the S1-specific Euroimmun IgG ELISA and the Liaison SARS-CoV-2 IgG CLIA using S1 and S2 as antigens. Corresponding to the microarray results, all IgG immunoassays provided positive results at the end of the observational period (Fig. 4C).
Among the total Ig immunoassays, we observed a higher sensitivity for the RBD-specific Wantai total Ig ELISA than for the NCP-specific Platelia SARS-CoV-2 Total Ab assay and the Elecsys anti-SARS-CoV-2 ECLIA, especially during the early phase of the infection (Fig. 4D). Like the IgG immunoassays, all total Ig tests provided positive results at the end of the observational period (Fig. 4D).
Fig. S5 displays antibody levels measured by the commercial ELISAs and CLIAs over time. Fig. S6 shows all results from immunoassays with a specificity lower than 80%.
Correlation of the results by the microarray and ELISAs/CLIAs with neutralizing antibodies.
Next, we correlated the antibody levels assessed by the commercial immunoassays (microarray, ELISAs/CLIAs) with neutralizing antibody titers in 138 samples from 20 patients of our cohort. The results are shown in Fig. S7. Notably, although there was a statistically significant correlation between neutralizing antibody titers and antibody levels assessed by all evaluated commercial immunoassays, the correlation was stronger for those assays detecting IgG rather than IgM, IgA, and total Ig antibodies, as well as for antibody tests using S1 or RBD as the target antigens (Fig. S7 and S8).
Antibody kinetics in relation to viral load dynamics in the lower respiratory tract and performance of the immunoassays when PCR from nasopharyngeal swabs tested negative.
Immunoassays have considerable diagnostic potential in progressed CoVID-19, especially when PCR tests provide negative results from upper respiratory tract swabs. Therefore, we first analyzed the antibody kinetics assessed by the microarray and the ELISAs/CLIAs in relation to the virus concentration in the lower respiratory tract. From 20 patients, of whom all developed critical disease with ARDS and 9 patients deceased, subsequently collected tracheal aspirate samples were available (median number of samples per patient, 4; range, 2 to 6).
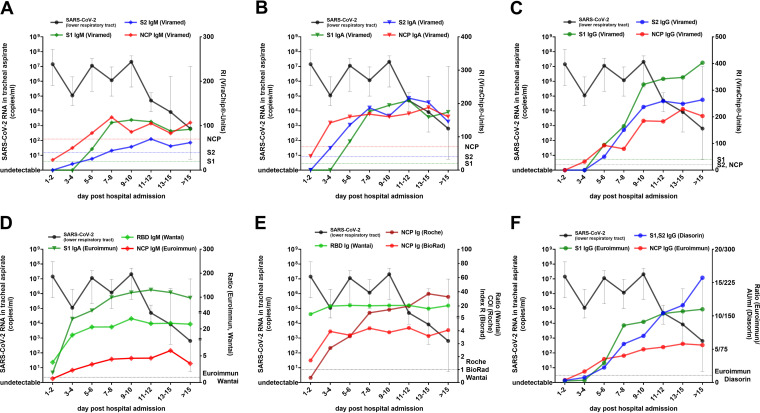
As shown in Fig. 5, the median virus concentration in tracheal aspirate samples decreased in these patients after the 11th day posthospital admission. Notably, the kinetics of SARS-CoV-2-specific IgM (Fig. 5A and D), IgA (Fig. 5B and E), and total Ig antibodies (Fig. 5E) were stable at an elevated level at this time point. In contrast, IgG antibody levels, especially those directed against S1, displayed a substantial increase immediately before viral concentrations in tracheal aspirate samples declined (Fig. 5C and F).
FIG 5.
Dynamics of median SARS-CoV-2 RNA concentration in tracheal aspirate samples (black line) and median antibody levels in 20 patients with critical CoVID-19 disease (of whom 9 patients deceased). Kinetics of SARS-CoV-2 S1-, S2-, and NCP-specific IgM (A), IgA (B), and IgG (C) antibodies as quantified by the microarray. Antibody levels as assessed by the anti-SARS-CoV-2 IgM and IgA (D), total Ig (E), and IgG ELISAs/CLIAs (F). Error bars indicate the interquartile range.
Then, we evaluated the immunoassays’ performance in 12 patients from our cohort who developed pneumonia before or during hospitalization and then displayed a negative PCR result from a nasopharyngeal swab. SARS-CoV-2 infection was verified in these patients by PCR positivity from a previously collected nasopharyngeal swab or a simultaneously obtained tracheal aspirate sample. Fig. 6 displays PCR and immunoassay results (including the detection rates) at the exact day when PCR from nasopharyngeal swabs initially tested negative in these patients (in the median, the 20th day post-symptom onset; range, days 9 to 31). Of note, the detection rates were higher than 90% for all evaluated IgM, IgA, IgG, and total Ig immunoassays.
FIG 6.
PCR and immunoassay results from 12 SARS-CoV-2-infected patients of the whole cohort who developed pneumonia and then displayed a negative PCR result from a nasopharyngeal swab (results from the initial day the nasopharyngeal swab tested negative; median, 20th day post-symptom onset; range, days 9 to 31). (A) Virus concentration by quantitative PCR in nasopharyngeal swab and tracheal aspirate samples; (B) microarray results (detection rates); (C) ELISA/CLIA results (detection rates); (D) results from the rapid test (detection rate).
Antibody levels in association with disease severity.
Finally, we compared antibody titers and kinetics assessed by all immunoassays (microarray, ELISAs, and CLIAs) among hospitalized patients with mild (n = 3) or moderate (n = 37) disease severity and patients who developed severe (n = 11) or critical (n = 11) CoVID-19 or deceased (n = 19). As shown in Fig. 7A, IgA and IgG immunoassays using S1 as the target antigen (microarray, Euroimmun ELISAs) yielded higher median antibody levels in patients with a more severe course of CoVID-19. The most significant differences in median values were observed for S1-specific IgA antibodies (P = 0.021 for microarray, median difference, 68.20 ViraChip units; P = 0.003 for ELISA, median difference, ratio 35.08).
FIG 7.
Longitudinal median levels of IgA, IgG, and total Ig antibodies directed against S1 (A) and NCP (B) among the hospitalized patients with mild (n = 3) or moderate (n = 37) disease severity and patients who displayed severe (n = 11) or critical (n = 11) courses of CoVID-19 or deceased (n = 19).
In contrast, differences according to disease severity were less prominent for NCP- and S2-specific immunoassays or for S1- and RBD-specific IgM and total Ig tests (Fig. 7B; Fig. S9).
DISCUSSION
In this study, we longitudinally analyzed SARS-CoV-2-specific antibody responses with a commercial microarray in hospitalized CoVID-19 patients. Thus, we provide comparable antibody data by a single test on the whole spectrum of IgM, IgA, and IgG, focusing at the same time on their respective specificities against the viral proteins S1, S2, and NCP. With the microarray results serving as background information, we compared detection rates of 12 other commercial immunoassays during acute SARS-CoV-2 infection.
A wide range of commercial SARS-CoV-2 immunoassays have become available, using different target antigens, covering different immunoglobulin classes, and differing in the respective test methods (e.g., ELISA and CLIA) (5, 8). There is also a variety of potential applications (e.g., to stage acute SARS-CoV-2 infection, to identify past infection independently of symptoms, and to characterize convalescent plasma quantitatively) (12, 13, 23). However, previous observations by others and ourselves propose particular potential for antibody assays in progressed CoVID-19 when PCR from nasopharyngeal swabs may yield negative results (4, 20, 21).
Therefore, we specifically analyzed antibody kinetics in relation to the dynamics of the virus concentration in the lower respiratory tract and performed a subanalysis of the immunoassays' performance in patients with severe disease in whom SARS-CoV-2 RNA from nasopharyngeal swabs became undetectable by PCR, while PCR of simultaneously collected tracheal aspirate samples tested positive. In this diagnostic setting, six commercial ELISAs/CLIAs and the only evaluated rapid test displayed a 100% detection rate. Furthermore, we observed that particularly S1-specific IgG antibodies increased when virus concentrations in the tracheal aspirate declined over time. These data confirm previous reports that antibody assays may indeed aid PCR in diagnosing and staging of severe SARS-CoV-2 infections, especially when patients present at a progressed stage of CoVID-19 (e.g., with pneumonia) and nasopharyngeal swabs test negative without deep sputum or tracheal aspirate being immediately available (20–22).
Furthermore, the microarray analysis revealed that NCP-specific IgA and IgG antibodies continuously displayed higher detection rates in hospitalized patients, although significantly higher antibody peak levels were found for S1-specific IgG antibodies. Notably, the microarray’s detection rates strongly reflected those of the NCP-specific IgG and total Ig ELISAs/CLIAs. In contrast, test sensitivities early after symptom onset (until the 10th day postonset of symptoms) were highest for the RBD-specific total Ig ELISA by Wantai and the S1-specific IgA ELISA by Euroimmun. Of note, the Wantai total Ig ELISA is the only evaluated ELISA with a double-antigen sandwich principle using RBD as the antigen, and the Euroimmun SARS-CoV-2 IgA ELISA has already been demonstrated to be highly sensitive (but with limited specificity) (4, 9, 30).
Similar to our findings, a previous longitudinal study demonstrated earlier detection of NCP-specific antibodies during acute SARS-CoV-2 infection using multiple commercial IgG and total Ig ELISAs/CLIAs (26). Other in-house and commercial immunoassays indicated higher sensitivity early after the infection for NCP-specific rather than for S1-specific IgG antibodies (24, 25, 27). Here, we quantitatively assessed antibody kinetics with a single microarray covering the antigen specificity of multiple immunoglobulin classes, including a relatively high number of samples per patient. Therefore, our data extend previous findings, demonstrating that anti-NCP IgG and IgA antibodies preceded those against S1 and S2. Our data further underline that, independently of the applied test, more robust antibody responses are directed against NCP in the early phase of the infection, possibly due to higher circulating NCP levels acting as a stronger early-phase immunogen than the spike protein.
However, we included only hospitalized patients who tested IgG negative at hospital admission and at the end of the observational period (the 26th day postonset of symptoms); all of those patients had developed detectable IgG antibody responses against all three antigens (S1, S2, and NCP), corresponding to 100% detection rates by all evaluated IgA, IgG, and total Ig ELISAs/CLIAs. This fact, together with our observation that in these IgG-seroconverting patients, the tests’ detection rates varied more widely during the early phase of the infection (until the 10th day post-symptoms onset), indicates that particular antibody tests may have excellent sensitivity in assessing past infections but may be less well suited to support PCR in the diagnosis and staging of acute infections. Therefore, immunoassays should be specifically selected for their diagnostic performance in the respective application area. Since individuals with mild or moderate symptoms display lower antibody levels than hospitalized patients, the test performances in our cohort might thus not directly apply to nonhospitalized individuals (4, 16, 31).
When the detection and seroconversion rates in our study are directly compared to those assessed in hospitalized patients with the same immunoassays as in previous studies, the following factors should be considered. Our study only included patients who were IgG negative at hospital admission. Furthermore, we considered all equivocal test results as negative, and since we investigated a relatively large sample number, we report the actual detection rates within 2-day intervals post-symptom onset rather than the cumulative seroconversion rate. These factors may have resulted in slightly lower detection and later seroconversion rates in our cohort than those previously observed (4, 26).
Compared with other studies using the same ELISAs/CLIAs, we observed a lower specificity for particular immunoassays (32, 33). Notably, we specifically included non-SARS-CoV-2-infected pneumonia patients as controls to assess false-positivity rates in a more realistic setting. Since cross-reactivity may be higher when humoral immune responses are upregulated against other pathogens, the lower specificities we observed are not surprising (7, 9, 30, 33). Moreover, similar to our findings, a strong effect of microbial infections other than SARS-CoV-2 on specific immunoassays’ specificity has been recently demonstrated (34).
Although our study cohort was relatively small for such a comparative analysis, we observed that hospitalized patients with a more severe course of CoVID-19 displayed higher antibody levels, which were mainly observed with IgG and especially IgA immunoassays (microarray and ELISAs) using S1 as the target antigen. Higher antibody levels in clinically more severe disease have already been demonstrated with multiple immunoassays, including S- and NCP-specific ELISAs/CLIAs, as well as neutralization assays (4, 15, 16, 18, 20, 31, 35). Thus, the question arises whether higher S1-specific IgA and IgG antibody titers, which we observed in more severe CoVID-19, have a functional origin or are a test-related phenomenon in our specific study cohort. On one hand, more robust IgA antibody responses have already been described in clinically more severe CoVID-19 cases, possibly indicating higher levels of virus replication in the respiratory tract (36, 37). On the other hand, comprehensive B-cell analyses revealed a strong humoral response against the spike protein in severely ill individuals, but NCP-specific responses even exceeded those against S (38). Further studies with higher patient numbers are needed and underway to comprehensively clarify the correlation of antibody levels of different immunoglobulin classes and against variable viral proteins with the clinical severity of CoVID-19.
Notably, the specific antibody kinetics (i.e., the respective signal strength) indicates that not all ELISAs/CLIAs included in this study were equally capable of quantifying high antibody concentrations, as indicated by a plateau of the measured antibody ratios/indices in individual assays (Fig. S5 in the supplemental material). Since most of the evaluated ELISAs/CLIAs are designed for semiquantitative rather than quantitative use, it is indeed possible that higher antibody concentrations in severely ill patients were mainly detected with those immunoassays with a more linear signal-concentration relationship at higher antibody concentrations. Nonetheless, since multiple immunoassays (microarray, ELISA) concordantly identified significantly higher IgA antibody values in patients with more severe CoVID-19, further studies with larger patient numbers should clarify to which extent SARS-CoV-2-specific IgA may indeed predict a more severe disease course.
Although our study has the limitation of a relatively small patient number, it nonetheless provides single-test microarray data on isotype- and antigen-specific differences in anti-SARS-CoV-2 antibody kinetics, partly affecting the sensitivity of commercial ELISAs/CLIAs. Finally, due to the large panel of additionally evaluated immunoassays, our observations significantly contribute to a comprehensive assessment of the commercial immunoassays’ longitudinal performance during acute SARS-CoV-2 infections in hospitalized patients.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hannah Griebler for the excellent technical support and her valuable input. We also thank Anja Eckerstorfer and Jutta Hutecek for their technical assistance.
We declare that we do not have a commercial or other association that might pose a conflict of interest.
Reagents for the NCP-specific IgM and IgG ELISAs by Euroimmun, the NCP-specific total Ab ELISA by Bio-Rad, and the NCP-specific total Ab ECLIA by Roche were provided by the manufacturers free of charge.
This study was funded by the Medical Scientific Fund of the mayor of the city of Vienna.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eurosurveillance Editorial Team. 2020. Note from the editors: World Health Organization declares novel coronavirus (2019-nCoV) sixth public health emergency of international concern. Euro Surveill 25:200131e. 10.2807/1560-7917.ES.2020.25.5.200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders D, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, Wang X, Yuan J, Li T, Li J, Qian S, Hong C, Wang F, Liu Y, Wang Z, He Q, Li Z, He B, Zhang T, Fu Y, Ge S, Liu L, Zhang J, Xia N, Zhang Z. 2020. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis 71:2027–2034. 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Ai J, Loeffelholz MJ, Tang YW, Zhang W. 2020. Meta-analysis of diagnostic performance of serology tests for COVID-19: impact of assay design and post-symptom-onset intervals. Emerg Microbes Infect 9:2200–2211. 10.1080/22221751.2020.1826362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.GeurtsvanKessel CH, Okba NMA, Igloi Z, Bogers S, Embregts CWE, Laksono BM, Leijten L, Rokx C, Rijnders B, Rahamat-Langendoen J, van den Akker JPC, van Kampen JJA, van der Eijk AA, van Binnendijk RS, Haagmans B, Koopmans M. 2020. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun 11:3436. 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken C, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 26:1478–1488. 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaaskelainen AJ, Kuivanen S, Kekalainen E, Ahava MJ, Loginov R, Kallio-Kokko H, Vapalahti O, Jarva H, Kurkela S, Lappalainen M. 2020. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 129:104512. 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaaskelainen AJ, Kekalainen E, Kallio-Kokko H, Mannonen L, Kortela E, Vapalahti O, Kurkela S, Lappalainen M. 2020. Evaluation of commercial and automated SARS-CoV-2 IgG and IgA ELISAs using coronavirus disease (COVID-19) patient samples. Euro Surveill 25:2000603. 10.2807/1560-7917.ES.2020.25.18.2000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, Jiang K, Asthagiri Arunkumar G, Jurczyszak D, Polanco J, Bermudez-Gonzalez M, Kleiner G, Aydillo T, Miorin L, Fierer D, Amarilis Lugo L, Milunka Kojic E, Stoever J, Liu STH, Cunningham-Rundles C, Felgner PL, Moran T, Garcia-Sastre A, Caplivski D, Cheng A, Kedzierska K, Vapalahti O, Hepojoki JM, Simon V, Krammer F. 2020. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med 26:1033–1036. 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krammer F, Simon V. 2020. Serology assays to manage COVID-19. Science 368:1060–1061. 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 12.Pollán M, Pérez-Gómez B, Pastor-Barriuso R, Oteo J, Hernán MA, Pérez-Olmeda M, Sanmartín JL, Fernández-García A, Cruz I, Fernández de Larrea N, Molina M, Rodríguez-Cabrera F, Martín M, Merino-Amador P, León Paniagua J, Muñoz-Montalvo JF, Blanco F, Yotti R, Blanco F, Gutiérrez Fernández R, Martín M, Mezcua Navarro S, Molina M, Muñoz-Montalvo JF, Salinero Hernández M, Sanmartín JL, Cuenca-Estrella M, Yotti R, León Paniagua J, Fernández de Larrea N, Fernández-Navarro P, Pastor-Barriuso R, Pérez-Gómez B, Pollán M, Avellón A, Fedele G, Fernández-García A, Oteo Iglesias J, Pérez Olmeda MT, Cruz I, Fernandez Martinez ME, Rodríguez-Cabrera FD, Hernán MA, Padrones Fernández S, Rumbao Aguirre JM, Navarro Marí JM, Palop Borrás B, Pérez Jiménez AB, Rodríguez-Iglesias M, Calvo Gascón AM, et al. 2020. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet 396:535–544., 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, Arnthorsson AO, Helgason D, Bjarnadottir K, Ingvarsson RF, Thorsteinsdottir B, Kristjansdottir S, Birgisdottir K, Kristinsdottir AM, Sigurdsson MI, Arnadottir GA, Ivarsdottir EV, Andresdottir M, Jonsson F, Agustsdottir AB, Berglund J, Eiriksdottir B, Fridriksdottir R, Gardarsdottir EE, Gottfredsson M, Gretarsdottir OS, Gudmundsdottir S, Gudmundsson KR, Gunnarsdottir TR, Gylfason A, Helgason A, Jensson BO, Jonasdottir A, Jonsson H, Kristjansson T, Kristinsson KG, Magnusdottir DN, Magnusson OT, Olafsdottir LB, Rognvaldsson S, Le Roux L, Sigmundsdottir G, Sigurdsson A, Sveinbjornsson G, Sveinsdottir KE, Sveinsdottir M, Thorarensen EA, Thorbjornsson B, Thordardottir M, Saemundsdottir J, et al. 2020. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 383:1724–1734. 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wajnberg A, Mansour M, Leven E, Bouvier NM, Patel G, Firpo-Betancourt A, Mendu R, Jhang J, Arinsburg S, Gitman M, Houldsworth J, Sordillo E, Paniz-Mondolfi A, Baine I, Simon V, Aberg J, Krammer F, Reich D, Cordon-Cardo C. 2020. Humoral response and PCR positivity in patients with COVID-19 in the New York City region, USA: an observational study. Lancet Microbe 10.1016/S2666-5247(20)30120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, Hu JL, Xu W, Zhang Y, Lv FJ, Su K, Zhang F, Gong J, Wu B, Liu XM, Li JJ, Qiu JF, Chen J, Huang AL. 2020. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 26:1200–1204. 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 16.Rijkers G, Murk JL, Wintermans B, van Looy B, van den Berge M, Veenemans J, Stohr J, Reusken C, van der Pol P, Reimerink J. 2020. Differences in antibody kinetics and functionality between severe and mild severe acute respiratory syndrome coronavirus 2 infections. J Infect Dis 222:1265–1269. 10.1093/infdis/jiaa463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orth-Holler D, Eigentler A, Stiasny K, Weseslindtner L, Most J. 2020. Kinetics of SARS-CoV-2 specific antibodies (IgM, IgA, IgG) in non-hospitalized patients four months following infection. J Infect 82:282–327. 10.1016/j.jinf.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marklund E, Leach S, Axelsson H, Nystrom K, Norder H, Bemark M, Angeletti D, Lundgren A, Nilsson S, Andersson LM, Yilmaz A, Lindh M, Liljeqvist JA, Gisslen M. 2020. Serum-IgG responses to SARS-CoV-2 after mild and severe COVID-19 infection and analysis of IgG non-responders. PLoS One 15:e0241104. 10.1371/journal.pone.0241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang AT, Garcia-Carreras B, Hitchings MDT, Yang B, Katzelnick LC, Rattigan SM, Borgert BA, Moreno CA, Solomon BD, Trimmer-Smith L, Etienne V, Rodriguez-Barraquer I, Lessler J, Salje H, Burke DS, Wesolowski A, Cummings DAT. 2020. A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association with severity. Nat Commun 11:4704. 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traugott MT, Hoepler W, Seitz T, Baumgartner S, Karolyi M, Pawelka E, Friese E, Neuhold S, Kelani H, Thalhammer F, Zoufaly A, Laferl H, Aberle JH, Wenisch C, Puchhammer-Stockl E, Stiasny K, Aberle SW, Weseslindtner L. 2021. Diagnosis of COVID-19 using multiple antibody assays in two cases with negative PCR results from nasopharyngeal swabs. Infection 49:171–175. 10.1007/s15010-020-01497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdivia A, Torres I, Huntley D, Alcaraz MJ, Albert E, Gonzalez C, Colomina J, Navarro D. 2020. Diagnostic significance of SARS-CoV-2 IgM positive/IgG negative antibody profile in symptomatic patients with suspected COVID-19 testing negative by RT-PCR. J Infect 82:e15–e16. 10.1016/j.jinf.2020.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang Q-W, Xu S-Y, Zhu H-D, Xu Y-C, Jin Q, Sharma L, Wang L, Wang J. 2020. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis 71:778–785. 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, Strich JR, Chertow DS, Davey RT, Cohen JI. 2020. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis 222:206–213. 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Algaissi A, Alfaleh MA, Hala S, Abujamel TS, Alamri SS, Almahboub SA, Alluhaybi KA, Hobani HI, Alsulaiman RM, AlHarbi RH, ElAssouli MA, Alhabbab RY, AlSaieedi AA, Abdulaal WH, Al-Somali AA, Alofi FS, Khogeer AA, Alkayyal AA, Mahmoud AB, Almontashiri NAM, Pain A, Hashem AM. 2020. SARS-CoV-2 S1 and N-based serological assays reveal rapid seroconversion and induction of specific antibody response in COVID-19 patients. Sci Rep 10:16561. 10.1038/s41598-020-73491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Elslande J, Decru B, Jonckheere S, Van Wijngaerden E, Houben E, Vandecandelaere P, Indevuyst C, Depypere M, Desmet S, Andre E, Van Ranst M, Lagrou K, Vermeersch P. 2020. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin Microbiol Infect 26:1557.e1–1557.e7. 10.1016/j.cmi.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turbett SE, Anahtar M, Dighe AS, Garcia Beltran W, Miller T, Scott H, Durbin SM, Bharadwaj M, Thomas J, Gogakos TS, Astudillo M, Lennerz J, Rosenberg ES, Branda JA. 2020. Evaluation of three commercial SARS-CoV-2 serologic assays and their performance in two-test algorithms. J Clin Microbiol 59:e01892-20. 10.1128/JCM.01892-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jungbauer C, Weseslindtner L, Weidner L, Gansdorfer S, Farcet MR, Gschaider-Reichhart E, Kreil TR. 2021. Characterization of 100 sequential SARS-CoV-2 convalescent plasma donations. Transfusion 61:12–16. 10.1111/trf.16119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koblischke M, Traugott MT, Medits I, Spitzer FS, Zoufaly A, Weseslindtner L, Simonitsch C, Seitz T, Hoepler W, Puchhammer-Stockl E, Aberle SW, Fodinger M, Bergthaler A, Kundi M, Heinz FX, Stiasny K, Aberle JH. 2020. Dynamics of CD4 T cell and antibody responses in COVID-19 patients with different disease severity. Front Med (Lausanne) 7:592629. 10.3389/fmed.2020.592629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traugott M, Aberle SW, Aberle JH, Griebler H, Karolyi M, Pawelka E, Puchhammer-Stockl E, Zoufaly A, Weseslindtner L. 2020. Performance of severe acute respiratory syndrome coronavirus 2 antibody assays in different stages of infection: comparison of commercial enzyme-linked immunosorbent assays and rapid tests. J Infect Dis 222:362–366. 10.1093/infdis/jiaa305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P, Liu L, Nair MS, Yin MT, Luo Y, Wang Q, Yuan T, Mori K, Solis AG, Yamashita M, Garg A, Purpura LJ, Laracy JC, Yu J, Joshua-Tor L, Sodroski J, Huang Y, Ho DD. 2020. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect 9:2091–2093. 10.1080/22221751.2020.1823890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naaber P, Hunt K, Pesukova J, Haljasmagi L, Rumm P, Peterson P, Hololejenko J, Eero I, Jogi P, Toompere K, Sepp E. 2020. Evaluation of SARS-CoV-2 IgG antibody response in PCR positive patients: comparison of nine tests in relation to clinical data. PLoS One 15:e0237548. 10.1371/journal.pone.0237548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manthei DM, Whalen JF, Schroeder LF, Sinay AM, Li SH, Valdez R, Giacherio DA, Gherasim C. 2020. Differences in performance characteristics among four high-throughput assays for the detection of antibodies against SARS-CoV-2 using a common set of patient samples. Am J Clin Pathol 155:267–279. 10.1093/ajcp/aqaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boukli N, Le Mene M, Schnuriger A, Cuervo NS, Laroche C, Morand-Joubert L, Gozlan J. 2020. High incidence of false-positive results in patients with acute infections other than COVID-19 by the Liaison SARS-CoV-2 commercial chemiluminescent microparticle immunoassay for detection of IgG anti-SARS-CoV-2 antibodies. J Clin Microbiol 58:e01352-20. 10.1128/JCM.01352-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grzelak L, Temmam S, Planchais C, Demeret C, Tondeur L, Huon C, Guivel-Benhassine F, Staropoli I, Chazal M, Dufloo J, Planas D, Buchrieser J, Rajah MM, Robinot R, Porrot F, Albert M, Chen K-Y, Crescenzo-Chaigne B, Donati F, Anna F, Souque P, Gransagne M, Bellalou J, Nowakowski M, Backovic M, Bouadma L, Le Fevre L, Le Hingrat Q, Descamps D, Pourbaix A, Laouénan C, Ghosn J, Yazdanpanah Y, Besombes C, Jolly N, Pellerin-Fernandes S, Cheny O, Ungeheuer M-N, Mellon G, Morel P, Rolland S, Rey FA, Behillil S, Enouf V, Lemaitre A, Créach M-A, Petres S, Escriou N, Charneau P, Fontanet A, et al. 2020. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci Transl Med 12:eabc3103. 10.1126/scitranslmed.abc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henss L, Scholz T, von Rhein C, Wieters I, Borgans F, Eberhardt FJ, Zacharowski K, Ciesek S, Rohde G, Vehreschild M, Stephan C, Wolf T, Hofmann-Winkler H, Scheiblauer H, Schnierle BS. 2020. Analysis of humoral immune responses in SARS-CoV-2 infected patients with severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis 223:56–61. 10.1093/infdis/jiaa680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, Tong X, Li Y, Gu B, Yan J, Liu Y, Shen H, Huang R, Wu C. 2020. A comprehensive, longitudinal analysis of humoral responses specific to four recombinant antigens of SARS-CoV-2 in severe and non-severe COVID-19 patients. PLoS Pathog 16:e1008796. 10.1371/journal.ppat.1008796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guthmiller JJ, Stovicek O, Wang J, Changrob S, Li L, Halfmann P, Zheng NY, Utset H, Stamper CT, Dugan HL, Miller WD, Huang M, Dai YN, Nelson CA, Hall PD, Jansen M, Shanmugarajah K, Donington JS, Krammer F, Fremont D, Joachimiak A, Kawaoka Y, Tesic V, Madariaga ML, Wilson PC. 2021. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike. mBio 12:e02940-20. 10.1128/mBio.02940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.