LETTER
Severe acute respiratory coronavirus 2 (SARS-CoV-2) was first detected as the causative agent for an outbreak of viral pneumonia in 2019 in Wuhan, China. The World Health Organization subsequently named the illness caused by SARS-CoV-2 as coronavirus disease 19 (COVID-19) and declared COVID-19 a world pandemic in March 2020 (1, 2). Due to the highly transmissible nature of COVID-19, rapid and accurate assays for SARS-CoV-2 remain the cornerstone for clinical management and effective isolation of symptomatic patients (2). Several diagnostic tests have been developed and commercialized for the detection of COVID-19 in response to increasing demand for testing (3). Although viral culture is the primary method for isolation of SARS-CoV-2, isolation of virus requires days and needs specialized facilities with a biosafety level-3 laboratory. Nucleic acid amplification tests (NAAT), such as RT-PCR, reduce the time frame to detection of viral nucleic acid down to minutes or hours and have excellent specificity and sensitivity (4), and are considered the gold standard for detecting COVID-19 by the Centers for Disease Control and Prevention (5). However, implementation is burdensome due to high costs and, as a result, this methodology is not broadly available (4). In contrast, rapid antigen tests can be used widely in clinical laboratories and point of care testing (POCT) settings.
The BD Veritor SARS-CoV-2 chromatographic immunoassay test (Becton, Dickinson, Sparks, MD, USA) detects SARS-CoV-2 nucleocapsid antigen from nasal samples with results available within 15 min (6). This antigen test received emergency use authorization (EUA) by the United States Food and Drug Administration (FDA) in July 2020 to be used in symptomatic patients within 5 days of symptom onset. The Veritor antigen test was implemented at our institution due to the high demand for testing and the limited allotment assigned for each of the NAAT tests in use. The short turnaround time (TAT) and the less restricted availability of antigen kits made it a potentially suitable alternative for diagnosis and management of patients receiving care at our hospital system. According to our ordering algorithm, the Veritor antigen test with reflex to RT-PCR test was indicated for patients with COVID-19 exposure and ≤5 days from symptom onset (fever/flu-like symptoms, unexplained shortness of breath, or new loss of taste).
The aim of this study was to evaluate the performance of the BD Veritor SARS-CoV-2 chromatographic immunoassay antigen test against RT-PCR as the reference method for the diagnosis of COVID-19 in symptomatic patients with high pretest probability and who were tested by both tests according to the ordering algorithm required at our health care system.
Between 20 October and 3 December 2020, a total of 1,384 patients meeting the criteria for Veritor antigen and reflex RT-PCR (median age 46.8, ranging from 1 to 98 years, 57.8% female) were tested. Paired nasal and nasopharyngeal samples were collected in the same encounter. Nasal samples for the antigen test were collected using flocked swabs (Becton, Dickinson, Sparks, MD, USA) and tested at the site of collection within 1 h of collection according to the manufacturer’s instructions (7). Briefly, the swabs were added to extraction buffer tubes and mixed for at least 15 to 30 s. The extraction buffer/specimen mixture was then added to the sample well of the test cartridge to initiate the testing. After the assays proceeded for 15 min, the test cartridges were inserted into the Veritor analyzer to obtain results.
For each patient tested by the Veritor antigen test, a nasopharyngeal sample was obtained at the same time, placed in 3 ml of viral transport medium (Remel, Lenexa, KS, USA), and submitted for RT-PCR testing using the Simplexa COVID-19 Direct EUA RT-PCR (Diasorin Molecular LLC, Cypress, CA, USA). The Simplexa COVID-19 direct assay targets two regions within the SARS-CoV-2 genome, the S gene encoding the spike protein (SP) and the ORF1ab genes encoding well-conserved nonstructural proteins of SARS-CoV-2. This assay was performed according to the manufacturer’s instructions (3). After swirling, the swab was discarded and 50 μl of sample and 50 μl of reaction mix were separately loaded into direct real-time PCR amplification-disc wells. The disc was then loaded onto the LIAISON MDX instrument (DiaSorin Molecular) and allowed to react for a 75-minute run. Extraction and amplification controls were used to detect PCR failure and/or inhibition. Positive- and negative-control samples were included in each run. After the assay’s completion, the instrument’s software (LIAISON MDX Studio-SW Version 2.1.0.4) automatically calculated and displayed results. Samples with median threshold cycle (CT) values of <40 (for one or both targets) were reported as positive for SARS-CoV-2. Sensitivity, specificity, and likelihood ratios were calculated along with the 95% confidence intervals (95% CI). Cycle times for each target sequence were examined and found to be nonparametric. Therefore, Wilcoxon rank sum testing was used to compare cycle times for S gene and ORF1ab targets between Simplexa RT-PCR positive/the Veritor test positive and Simplexa RT-PCR positive/the Veritor test negative samples. We held P < 0.05 to represent a statistically significant difference. As the sample size was fixed due to clinically available specimens, formal sample size calculations were not performed. Statistics were calculated with Stata 11.2.
Table 1 provides the comparative test results for antigen and RT-PCR testing for the 1,384 paired samples. Overall concordance was seen in 1,330/1,384 specimens (96.1%, 95% CI 95.0 to 97.1%). Utilizing Simplexa RT-PCR testing as the reference method, the Veritor antigen test sensitivity was 66.4% (95% CI 57.0 to 74.9%) and specificity was 98.8% (95% CI 98.1 to 99.3%). The likelihood ratio negative for the antigen test was 0.34 (95% CI 0.26 to 0.44) and the likelihood ratio positive was 56.1 (95% CI 33.4 to 94.3). Using Simplexa RT-PCR as a reference standard, the sample prevalence of disease was 116/1,384 (8.4%, 95% CI 7.0 to 10.0%).
TABLE 1.
Performance of the Veritor antigen test compared with the Simplexa RT-PCR as a reference standard
| Veritor antigen test result | No. of specimens by Simplexa RT-PCR result: |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 77 | 15 | 92 |
| Negative | 39 | 1,253 | 1,292 |
| Total | 116 | 1,268 | 1,384 |
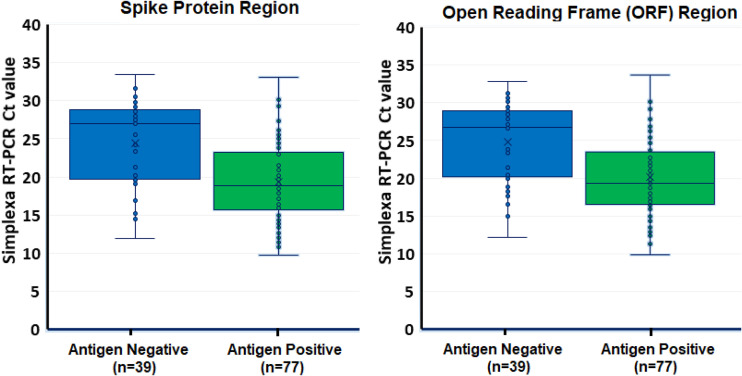
The CT value of the Simplexa RT-PCR among the 77 concordant positive samples was 19.1 for the S gene target (range: 10.6 to 33.1) and 19.8 for the ORF1ab target (range: 11.2 to 33.7), whereas the median CT for the 39 discordant (positive by the Simplexa RT-PCR and negative by the Veritor test) was 24.5 for the S gene target (range: 11.9 to 33.5) and 24.9 for the ORF1ab target (range: 11.2 to 32.8) (Fig. 1). For both the S gene and the ORFlab targets, these differences in Simplexa RT-PCR cycle times were significantly different in the Veritor positive samples (concordant) versus Veritor negative (discordant), P < 0.05.
FIG 1.
Comparison of the results of the Simplexa RT-PCR threshold cycle (CT) for both spike protein and open reading frame regions and the Veritor antigen test positive and negative samples.
Young et al. evaluated the Veritor test and reported an overall percent agreement of 97.4% for 251 participants with COVID-19 symptoms (6), consistent with results obtained in the current study. However, our study results showed a lower sensitivity than that reported by the manufacturer (66.4% versus 84%, respectively), probably due to differences in patient populations and different local prevalence rates (8.4% prevalence in our study versus 15.1% prevalence for Young et al. [6] and 13.7% prevalence for the Veritor validation study by the manufacturer [7]). It should be noted that most of the rapid antigen tests for detecting COVID-19 evaluated have demonstrated a lack of sensitivity, with reported sensitivities ranging from 30.2 to 93.9% (8–13).
The analytical performance of rapid antigen tests depends on the viral load of the samples (12). When we compared the concordant and discordant results between the Veritor antigen test and the Simplexa RT-PCR, the samples negative by antigen tests but positive by RT-PCR had higher CT values, indicating a lower viral load. These results suggest that the Veritor test might not detect SARS-CoV-2 in samples with low viral RNA, although the range of CT values overlapped with the range observed in positive samples (Fig. 1).
One of the limitations of this study is that different sample types were used, i.e., nasal swab for the antigen test and nasopharyngeal swab for the RT-PCR. This difference may also contribute to the lower sensitivity observed with the antigen test. However, in each case, the used sample types were those recommended for each test by the manufacturer and validated by our laboratory. In addition, the manufacturer also used nasal swabs compared to nasopharyngeal swabs for the RT-PCR for their validation data in the EUA submission to the FDA. Therefore, our study is comparable to that reported by the manufacturer to assess the clinical performance of the antigen test.
Overall, the Veritor test is rapid, easy to use, and can be performed as a POCT by nonlaboratory personnel, but requires a reader that needs to be replaced after 3,000 tests. In our institution, the average TAT is 29 min for the Veritor antigen test and 9 h for the Simplexa RT-PCR. The difference in TAT is because the antigen test is performed at the site of collection throughout our health care system, while the samples for Simplexa RT-PCR are sent to the laboratory for batched testing on first and second shift only.
Our findings demonstrate that the lower sensitivity of the Veritor test compared to RT-PCR led to false-negative results in 39 patients and therefore negative results in symptomatic patients require confirmation by RT-PCR where available. Furthermore, despite the high specificity of the antigen test (98.8%) found in this study and high pretest probability (exposure plus symptoms) of patients tested, 15 patients had a false-positive antigen result. This rate may be acceptable in outpatient settings where the patient with a positive result would be told to self-isolate, but could be too high for patients that will be admitted, as they could potentially be admitted to a COVID unit unnecessarily.
The need for confirmation of negative results in symptomatic patients reduces the clinical utility of this test, particularly in areas of low prevalence. But the high overall correlation of test results (96.1%) for symptomatic patients suggests that this antigen test can be a useful tool in areas where RT-PCR is not available or the TAT of RT-PCR is prolonged, especially given its rapid TAT and the possibility of its use in POCT locations.
ACKNOWLEDGMENT
We declare no competing interests.
REFERENCES
- 1.World Health Organization. 2020. WHO announces COVID-19 outbreak a pandemic. WHO Regional Office for Europe, Copenhagen, Denmark. https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/news/news/2020/3/who-announces-covid-19-outbreak-a-pandemic.
- 2.Nagura-Ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, Horiuchi M, Kato K, Imoto Y, Iwata M, Mimura S, Ito T, Tamura K, Kato Y. 2020. Clinical evaluation of self-collected saliva by quantitative reverse transcription-PCR (RT-qPCR), direct RT-qPCR, reverse transcription-loop-mediated isothermal amplification, and a rapid antigen test to diagnose COVID-19. J Clin Microbiol 58:e01438-20. 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordi L, Piralla A, Lalle E, Giardina F, Colavita F, Tallarita M, Sberna G, Novazzi F, Meschi S, Castilletti C, Brisci A, Minnucci G, Tettamanzi V, Baldanti F, Capobianchi MR. 2020. Rapid and sensitive detection of SARS-CoV-2 RNA using the Simplexa COVID-19 direct assay. J Clin Virol 128:104416. 10.1016/j.jcv.2020.104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, Chan RC, Tsang DN. 2020. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol 129:104500. 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2020. Interim guidance for antigen testing for SARS-CoV-2. CDC, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html.
- 6.Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, Griego-Fullbright C, Burgard C, Fernandez C, Eckert K, Andrews JC, Ren H, Allen J, Ackerman R, Cooper CK. 2020. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS antigen point-of-care test. J Clin Microbiol 59129:e02338-20. 10.1128/JCM.02338-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becton, Dickinson and Company. 2020. BD Veritor System for Rapid Detection of SARS-CoV-2, package insert EB. Becton, Dickinson and Company, Sparks-Glencoe, MD. https://www.fda.gov/media/139755/download. [Google Scholar]
- 8.Cerutti F, Burdino E, Milia MG, Allice T, Gregori G, Bruzzone B, Ghisetti V. 2020. Urgent need of rapid tests for SARS CoV-2 antigen detection: evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J Clin Virol 132:104654. 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenollar F, Bouam A, Ballouche M, Fuster L, Prudent E, Colson P, Tissot-Dupont H, Million M, Drancourt M, Raoult D, Fournier PE. 2020. Evaluation of the Panbio Covid-19 rapid antigen detection test device for the screening of patients with Covid-19. J Clin Microbiol 59:e02589-20. 10.1128/JCM.02589-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liotti FM, Menchinelli G, Lalle E, Palucci I, Marchetti S, Colavita F, La SM, Sberna G, Bordi L, Sanguinetti M, Cattani P, Capobianchi MR, Posteraro B. 2020. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clin Microbiol Infect 27:487–488. 10.1016/j.cmi.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porte L, Legarraga P, Vollrath V, Aguilera X, Munita JM, Araos R, Pizarro G, Vial P, Iruretagoyena M, Dittrich S, Weitzel T. 2020. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis 99:328–333. 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scohy A, Anantharajah A, Bodeus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. 2020. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol 129:104455. 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prince-Guerra JL, Almendares O, Nolen LD, Gunn JKL, Dale AP, Buono SA, Deutsch-Feldman M, Suppiah S, Hao L, Zeng Y, Stevens VA, Knipe K, Pompey J, Atherstone C, Bui DP, Powell T, Tamin A, Harcourt JL, Shewmaker PL, Medrzycki M, Wong P, Jain S, Tejada-Strop A, Rogers S, Emery B, Wang H, Petway M, Bohannon C, Folster JM, MacNeil A, Salerno R, Kuhnert-Tallman W, Tate JE, Thornburg NJ, Kirking HL, Sheiban K, Kudrna J, Cullen T, Komatsu KK, Villanueva JM, Rose DA, Neatherlin JC, Anderson M, Rota PA, Honein MA, Bower WA. 2021. Evaluation of Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection at two community-based testing sites—Pima County, Arizona, November 3–17, 2020. MMWR Morb Mortal Wkly Rep 70:100–105. 10.15585/mmwr.mm7003e3. [DOI] [PMC free article] [PubMed] [Google Scholar]