LETTER
At-home respiratory specimen collection for pathogen testing enables community sampling. Furthermore, it requires neither a health care worker’s time nor personal protective equipment, and symptomatic individuals can continue to self-isolate. However, questions remain as to whether unsupervised upper respiratory specimen collection by individuals in their homes reliably produce specimens that are of high enough quality for pathogen testing. From October 2019 through May 2020, the Seattle Flu Study (1, 2) and the greater Seattle Coronavirus Assessment Network (SCAN; scanpublichealth.org) screened 16,785 midturbinate swabs that were self-collected by participants at home for respiratory pathogens. The at-home kits contained a flocked, midturbinate swab (Copan 56380CS01 or 56750CS01), either adult or pediatric, a tube of universal transport media (UTM), and instructions on how to self-collect a specimen or collect a specimen for a child and return it to the lab (2). Of the kits distributed to individuals in the Seattle metropolitan area, most resulted in swabs returned appropriately according to the instructions in the kit, but 138/16,785 (0.8%) kits were returned to the lab with the swab handle in the UTM tube rather than the swab itself. The swab handle is nontapered, hard plastic with decreased surface area compared with the flocked end of the swab (Fig. 1A). We were puzzled by this phenomenon and sought to evaluate whether handle-collected specimens were comparable to flocked swabs themselves for molecular pathogen detection. We also assessed demographic covariates associated with errors in swab collection.
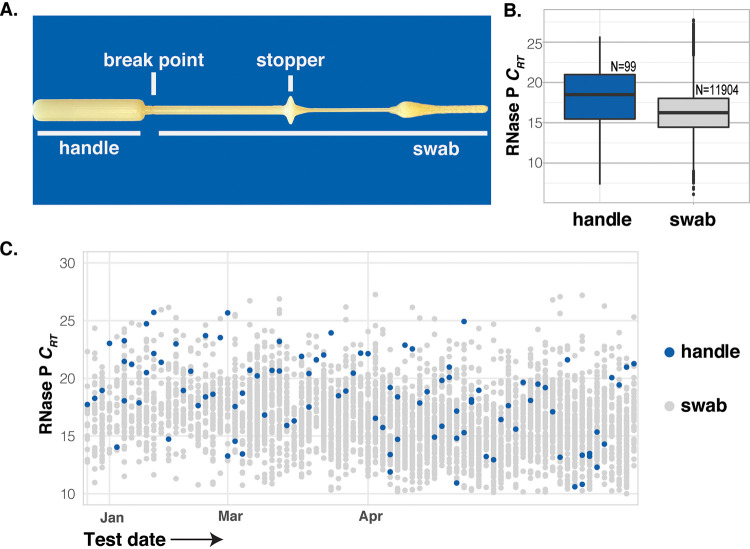
FIG 1.
(A) A midturbinate swab (Copan 56380CS01), underlined handle or swab, was placed in UTM by participants. (B) Crt values from all samples with RNase P detected; dashed line indicates detection limit. (C) Crt values for human RNase P among batches of specimens (arranged on the x axis by date) where at least one handle specimen was used.
Of the 16,782 specimens, 12,006 were analyzed for the presence of 24 respiratory pathogens using our TaqMan-based detection panel, including 99 of the 138 specimens collected with the handle (Table 1). Samples collected after 1 January 2020 were additionally tested for the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using a separate reverse transcriptase PCR (RT-PCR) assay. As a quality-control metric to determine if a sufficient nasal specimen was collected for each sample, both assay platforms measured the amount of human RNase P. Specimens with RNase P relative cycle threshold (Crt) of >28 were considered to be a failed collection. The failure rate for all properly collected specimens was 2.0% (238/12,142). We expected a high failure rate for the handle-collected specimens, but only 2.9% (3/102) failed this quality-control metric, a nonsignificant difference (P = 0.46, Fisher’s exact test). The Crt values for human marker RNase P for handle-collected specimens were higher than those for properly collected specimens (Fig. 1B), with a mean Crt value of 16.32 (95% confidence interval [CI], 16.27 to 16.37) for swabs and 18.19 (95% CI, 17.43 to 18.96) for handles (P < 0.01). However, the Crt from handle-collected specimens generally fell within the same range and well below the failure threshold (Fig. 1C), showing that the handles were indeed collecting human cells. In addition, we identified multiple respiratory pathogens, including SARS-CoV-2, at similar rates of detection with both swabs and swab handles (P = 0.52) (Table 1).
TABLE 1.
Detection rates of respiratory pathogens
| Pathogen | No. (%) of respiratory pathogens |
|
|---|---|---|
| Handle present | Swab present | |
| Adenovirus | 1 (1.0) | 77 (0.6) |
| Bocavirus | 0 (0.0) | 12 (0.1) |
| Enterovirus | 1 (1.0) | 29 (0.2) |
| Influenza A | 4 (4.0) | 394 (3.3) |
| Influenza B | 0 (0.0) | 250 (2.1) |
| Influenza C | 0 (0.0) | 0 (0.0) |
| Metapneumovirus | 0 (0.0) | 77 (0.6) |
| Parainfluenza | 1 (1.0) | 50 (0.4) |
| Parechovirus | 0 (0.0) | 0 (0.0) |
| Respiratory syncytial virus | 2 (2.0) | 116 (1.0) |
| Rhinovirus | 6 (6.0) | 620 (5.2) |
| SARS-CoV-2 | 1 (1.1) | 119 (1.1) |
| Seasonal coronavirus | 3 (3.0) | 350 (2.9) |
| Chlamydia pneumoniae | 0 (0.0) | 8 (0.1) |
| Mycoplasma pneumoniae | 0 (0.0) | 28 (0.2) |
| Streptococcus pneumoniae | 3 (3.0) | 275 (2.3) |
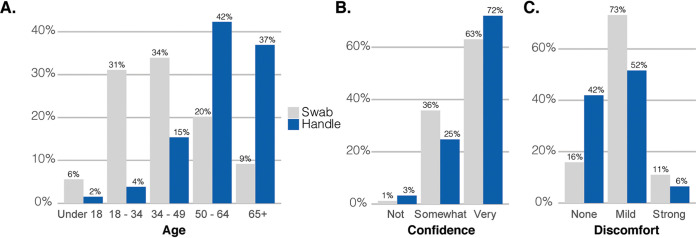
We examined the clinical data associated with the samples to determine which participants were more likely to collect a specimen with the handle. Participants who swabbed with the handle were more likely to be older (Fig. 2A), with a median age of 62 compared with 39 for those who followed the instructions (P < 0.01). There was no significant difference in handle use between men and women (P = 0.22) or across income brackets (supplemental material). Interestingly, participants who had erroneously used the handle were more confident that they had collected a quality specimen (Fig. 2B) (73% highly confident with the handle versus 62% with the swab, P = 0.02) and reported lower overall discomfort (Fig. 2C) (42% reported no discomfort with the handle versus 16% with the swab, P < 0.01). The greater reported comfort, combined with the larger size of the handles, suggests that these specimens were collected from the anterior nares rather than the midturbinate.
FIG 2.
(A) Age of participants. (B) Self-reported confidence in specimen collection. (C) Self-reported discomfort during specimen collection by which end of the swab was used.
We investigated unanticipated operator error in two large studies employing at-home midturbinate swab collection and determined that participants who used the plastic handle rather than flocked swab to collect their sample and submit it to a laboratory were able to collect an adequate nasal specimen for molecular detection of respiratory pathogens. Like other studies (3), these results suggest that the use of specialty swabs may result in only marginal increases in pathogen detection. They also suggest that even if participants do not closely adhere to instructions, they can still collect a sample that is sufficient for the molecular detection of respiratory pathogens, including influenza and SARS-CoV-2.
The Seattle Flu Study received approval by the University of Washington’s Institutional Review Board (UW IRB; STUDY00006181), and informed consent was obtained prior to study enrollment. Participants joined SCAN as part of public health surveillance.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Seattle Flu Study and SCAN participants for their invaluable contributions to this research, the entire Seattle Flu Study team for making this study possible, Katrina Van Raay for R code, and Kristen Huden for first spotting handles in UTM.
The Seattle Flu Study and SCAN are administered by the Brotman Baty Institute for Precision Medicine and funded by Gates Ventures, the private office of Bill Gates. The funder was not involved in the design of the study and does not have any ownership over the management and conduct of the study, the data, or the rights to publish. L.M.S. and J.S. are funded by 1RM1HG010461-01 from the NHGRI, and J.S. is an investigator of the Howard Hughes Medical Institute. REDCap at ITHS is supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1 TR002319.
H.Y.C. is a consultant for Merck and GlaxoSmithKline and receives research funding from Ellume, Cepheid, and Sanofi-Pasteur. J.S. is a consultant with Guardant Health, Maze Therapeutics, Camp4 Therapeutics, Nanostring, Phase Genomics, Adaptive Biotechnologies, and Stratos Genomics and has a research collaboration with Illumina. J.A.E. is a consultant with Sanofi Pasteur and Meissa Vaccines.
Seattle Flu Study principal investigators from Seattle, WA are as follows: Helen Y. Chu, Department of Medicine, University of Washington, Brotman Baty Institute for Precision Medicine; Michael Boeckh, Department of Medicine, University of Washington, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Brotman Baty Institute for Precision Medicine; Jeffrey S. Duchin, Public Health–Seattle King County; Janet A. Englund, Seattle Children’s Research Institute, Brotman Baty Institute for Precision Medicine; Michael Famulare, Institute for Disease Modeling; Barry R. Lutz, Department of Bioengineering, University of Washington, Brotman Baty Institute for Precision Medicine; Deborah A. Nickerson, Department of Genome Sciences, University of Washington, Brotman Baty Institute for Precision Medicine; Mark J. Rieder, Brotman Baty Institute for Precision Medicine; Lea M. Starita, Department of Genome Sciences, University of Washington, Brotman Baty Institute for Precision Medicine; Matthew Thompson, Department of Family Medicine, University of Washington; Jay Shendure, Department of Genome Sciences, University of Washington, Brotman Baty Institute for Precision Medicine, Howard Hughes Medical Institute; and Trevor Bedford, Vaccine and Infectious Disease Division, Fred Hutchinson Cancer Research Center, Department of Genome Sciences, University of Washington, Brotman Baty Institute for Precision Medicine.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JCM.02934-20.
Supplemental material is available online only.
REFERENCES
- 1.Chu HY, Englund JA, Starita LM, Famulare M, Brandstetter E, Nickerson DA, Rieder MJ, Adler A, Lacombe K, Kim AE, Graham C, Logue J, Wolf CR, Heimonen J, McCulloch DJ, Han PD, Sibley TR, Lee J, Ilcisin M, Fay K, Burstein R, Martin B, Lockwood CM, Thompson M, Lutz B, Jackson M, Hughes JP, Boeckh M, Shendure J, Bedford T, Seattle Flu Study Investigators. 2020. Early detection of Covid-19 through a citywide pandemic surveillance platform. N Engl J Med 383:185–187. 10.1056/NEJMc2008646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim AE, Brandstetter E, Wilcox N, Heimonen J, Graham C, Han PD, Starita LM, McCulloch DJ, Casto AM, Nickerson DA, Van de Loo MM, Mooney J, Ilcisin M, Fay KA, Lee J, Sibley TR, Lyon V, Geyer RE, Thompson M, Lutz BR, Rieder MJ, Bedford T, Boeckh M, Englund JA, Chu HY, on behalf of the Seattle Flu Study Investigators . 2021. Evaluating specimen quality and results from a community-wide, home-based respiratory surveillance study. J Clin Microbiol 59:e02934-20. 10.1128/JCM.02934-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tu Y-P, Jennings R, Hart B, Cangelosi G, Wood R, Wehber K, Verma P, Vojta D, Berke EM. 2020. Patient-collected tongue, nasal, and mid-turbinate swabs for SARS-CoV-2 yield equivalent sensitivity to health care worker collected nasopharyngeal swabs. medRxiv 10.1101/2020.04.01.20050005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.