Highlights
-
•
Plant metabolic networks are highly complex.
-
•
Engineering the phytochemical pathways fully in heterologous hosts is challenging.
-
•
Single plant cells with amplified multiple fission enable homogeneity.
-
•
Homogeneity and high cell division rate can facilitate stable product scale-up.
Abbreviations: HEx, heterologous expression; PTM, post-translational modification; TEAMP, Transient Expression in Arabidopsis Mesophyll Protoplast; TOR, Target of Rapamycin
Keywords: Heterologous hosts, Metabolic engineering, Single plant cells, Synthetic biology, Transgenesis
Abstract
Plant-based biopreparations are reasonably priced and are devoid of viral, prion and endotoxin contaminants. However, synthesizing these natural plant products by chemical methods is quite expensive. The structural complexity of plant-derived natural products poses a challenge for chemical synthesis at a commercial scale. Failure of commercial-scale synthesis is the chief reason why metabolic reconstructions in heterologous hosts are inevitable. This review discusses plant metabolite pathway reconstructions experimented in various heterologous hosts, and the inherent challenges involved. Plants as native hosts possess enhanced post-translational modification ability, along with rigorous gene edits, unlike microbes. To achieve a high yield of metabolites in plants, increased cell division rate is one of the requisites. This improved cell division rate will promote cellular homogeneity. Incorporation and maintenance of plant cell synchrony, in turn, can program stable product scale-up.
1. Introduction
Plants have been a treasure trove of metabolites. A study has documented that, plants can synthesize more than 1 × 106 metabolites [1]. However, merely 50,000–60,000 metabolites have been recorded in databases such as KNApSAcK, PlantCyc and METLIN, emphasizing that many more compounds await to be mined out [2]. Production of natural plant products has always been challenging because of their stereochemical complexities [3]. About 25 % of drugs that are used by humans are plant-derived and still higher than 350,000 species are yet to be explored in the context of the therapeutics they contain [4]. Metabolomics is one of the principal approaches adopted by phytochemists to explore plant medicinal properties. Multicellularity offers complexity to plants, which limits our understanding towards variable cellular potencies and their symplastic networks. A method that is often used to negate this complexity is, ‘single plant cell assays’ [5]. Generally, in multicellular systems, there is an extracellular matrix, outward of the plasma membrane of every cell. The plant cell wall is a type of extracellular matrix with the fibrillar structure that facilitates expansion, and it chemically modulates coordination between the neighboring cells [6]. The organization and dependency between the cells implicate heterogeneity in plant metabolome.
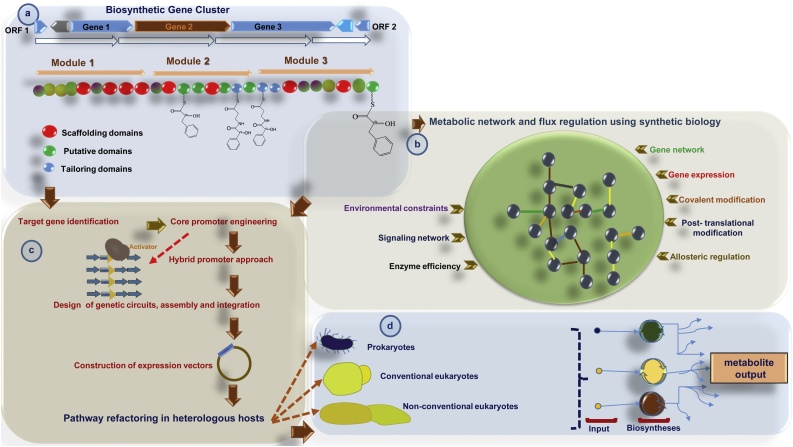
Metabolic engineering is a technique that channelizes upstream precursors from primary metabolic pathways to routes of interest. It is not just the introduction of several genes into the cell, but it carefully balances the pathways, so that the product synthesis is well-regulated, without any precursor deprivation [7]. A fundamental hurdle during metabolic reconstruction is the persistent regulation of gene cluster and metabolic flux balance (Fig. 1c) in heterologous hosts [8,9]. Through meticulous reconstructions, it is possible to produce pharmaceutically relevant compounds in biologically amenable heterologous hosts such as S. cerevisiae, E. coli and Streptomyces coelicolor in large quantities (Fig. 1d). Metabolic engineering encompasses: (1) pathway design and optimization, (2) components of combinatorics and graph theory, (3) thermodynamics of target pathways, (4) routing the pathway fluxes, (5) pathway validation by isotopic tracers, (6) developing genome-scale variants, (7) ascertaining gene expression fingerprints, (8) recognizing pathway kinetics via Metabolic Control Analysis (MCA), and (9) eradicating kinetic blockades [[10], [11], [12], [13], [14]]. Whereas, ‘Inverse metabolic engineering’ involves rational and combinatorial pathway optimizations along with regulation of synthetic gene circuits [15]. Flux detection is the vital theme of metabolic engineering that involves time-reliant kinetics, tracer enrichment assessments and reasonable confidence interval detections [16]. Estimates of optimal flux fingerprints help researchers to enhance pathway performance. In some instances, notably in cell cultures, growth and biosynthesis are often coupled, so that growth optimization can result in maximum product yield [17]. Metabolic engineering operates by (1) utilizing natural native pathways and (2) using non-natural channels, which are constructed by heterologous gene expression through alteration of amino acid scaffolds, that function as upstream precursors [18]. One such example of metabolic engineering is ‘Artemisinin’, a sesquiterpene lactone produced by an Asteraceae member called, Artemisia annua. This plant-derived natural product is effective against the multidrug-resistant malaria parasite Plasmodium spp. Because of its less yield and challenging chemical synthesis, Artemisinin is costly. Therefore, it is recommended to execute semi-synthesis of the compound [19,20] or induce the production of it from its immediate precursor, the artemisinic acid in eukaryotic microbial heterologous hosts, notably yeast [8,21]. This approach is relatively cost-effective and eco-friendly. Metabolic reconstruction of the enzyme, amorpha-4,11-diene synthase (found in Artemisia annua) in S. cerevisiae along with a novel cytochrome P450 mono-oxygenase (CYP71AVI), enabled tri-step oxidation, converting amorpha-4, 11-diene to artemisinic acid. Conversion of farnesyl pyrophosphate [FPP] to sterol in yeast was downregulated by a repressible promoter of methionine [PMET3] [22]. This downregulation step facilitated a two-fold increase in the production of amorphadiene. The engineered yeast could transport extracellular product, artemisinic acid truncating the purification process. The highest artemisinic acid production recorded was 100 mg L−1, which was around thousand-fold higher than the transitory yield reported in Artemisia spp. This successful pathway reconstruction with extensive gene integration in the non-native host is sometimes incompatible with its essential genes and enzymes. The crux of practical metabolic engineering is to produce the desired products in milligram quantities, yet making the process lucrative with sufficient Titer, Rate and Yield, abbreviated as ‘TRY’ [23,24].
Fig. 1.
Metabolic refactoring in heterologous hosts a. Identification of biosynthetic gene clusters b. Consideration of the metabolic network and flux regulations in native hosts and non-native hosts using a synthetic biology approach c. Determination of the gene responsible for the compound of interest and causes for flux perturbations and upgrading the metabolic network using gene editing and promoter engineering followed by pathway refactoring in suitable heterologous hosts d. Post refactoring analysis in non-native hosts along with precursor input and metabolite output evaluations.
Intricate genetic systems within cells have gates and switches that can regulate metabolic pathways more competently. ‘BioBricks’ are such multiple gene links, which aid in building metabolic pathways, cell compartmentalization or even synthetic cell as a whole [25]. These BioBricks promote gene expression assemblage that constitutes recurrent pathway optimization. Instead of including all the complicated networks, it is always strategic to include only relevant pathway initially and check whether the results are promising [[26], [27], [28]]. However, the physiological space available for reviewing the critical outcome is narrow. Hence, we need proof of concept before large-scale program launch. We need to identify the contextual priorities and temporal fitness of BioBricks. After the generation of ‘synthetic DNA’, this is not far from reach. However, recommending the translational aspect (in terms of product yield) of synthetic cells is imprecise at the moment.
Another application of synthetic biology is, monitoring gene expression by the usage of Green Fluorescent Protein (GFP) at the single-cell level in culture. In synthetic biology, it is crucial to have a clear idea about the gene expression dynamics and population synergy. There is a deep contrast between metabolic engineering and synthetic biology [29,30]. In metabolic engineering, cells function as metabolic reactors, where they convert substrates into products retaining the homeostasis by replenishing the chief enzymes. This method lacks physical separation between the units, reactant-product segregation and unreacted compound recycling. Enzymatic controls might prove to be inadequate if the distal network interactions are not well understood. Synthetic biology fixes this problem through various mechanisms regulated by rigid electric circuits. The chemical reactions that underlie any cellular and metabolic network are distinctly analogous to their kinetics. Synthetic biologists believe that the digital framework of the study may not be sufficient enough to understand the network. Pathway flux distribution is regulated by critical enzymes at various metabolic intersections and is not solely a switch on/off process [31]. The digital framework helps to understand cellular functions via gene expression, as it can enable an association with global transcription data (Fig. 1b). However, the global transcription data may have the following drawbacks: (1) it can only be significant for a particular phenomenon, (2) the data may be direct or indirect, and (3) it may lack profound mechanistic insights of gene expression [30]. In short, metabolic engineering needs synthetic biology to incorporate non-native biosynthetic pathways for the commercialization of known and unknown economically valuable products, viz., fuels, therapeutics and chemicals from agricultural wastes [32]. Synthetic biology furnishes vital information regarding various biological phenomena, whereas metabolic engineering applies this information towards the biosynthesis of a compound of interest [33].
Despite tremendous development in the field of metabolic engineering, there exist many challenges for heterologous protein production. Limited productivity is a significant drawback in plant-based systems. Moreover, due to complex genome architecture and transformation barricades, many alternative platforms are used for the enhanced yield of proteins and bioactives. The main advantage of using microbial systems is their ease of manipulation along with well-characterized genomes.
2. Microbes as heterologous hosts
Microbial bioreactor systems are inevitable for the commercial production of many essential compounds. Engineering Escherichia coli and Saccharomyces cerevisiae demand biosynthetic pathway regulation [34]. For the production of Naringenin and p-Coumaric acid (necessary intermediates of flavonoid biosynthetic pathway), the flavonoid pathway is refactored in E. coli and S. cerevisiae [35].
2.1. Bacterial expression platforms
E. coli has a rapid growth rate with specific expression vectors. There is extensive information about its genome and regulatory networks. Other bacteria that serve as heterologous hosts include Bacillus subtilis [36] and S. coelicolor [37]. E. coli has been extensively used for the synthesis of natural plant products, ranging from simple terpenoids to complex polyketides [38]. For instance, genes encoding enzymes such as 1. phenylalanine ammonia-lyase (PAL) from the yeast [Rhodotorula rubra] [39], 2. 4-coumarate- CoA ligase (4CL) from soil bacteria [S. coelicolor] [40], 3. chalcone synthase (CHS) from the licorice plant [Glycyrrhizae chinata] [41], 4. stilbene synthase (STS) from groundnut [Arachis hypogaea] [42], and 5. chalcone isomerase (CHI) from Kudzu [Pueraria lobata] [43] have been inserted into a single bacterial plasmid (combinatorial synthesis) [44]. Similarly, the carotenoids such as lycopene and β-carotene are synthesized from E. coli platforms by combining the native Methylerythritol 4-Phosphate (MEP) pathway and the heterologous Mevalonate (MVA) pathway [terpenoid biosynthesis pathway native to S. cerevisiae], by fed-batch fermentation [45]. Taxol biosynthetic pathway has been expressed in E. coli for the production of Taxadiene (precursor for Taxol, a potent antineoplastic drug). Bacteria, in general, do not possess the machinery for the production of Benzylisoquinoline alkaloids (BIAs), and hence a heterologous pathway was constructed in E. coli, which produced (S)-reticuline, a precursor for many essential alkaloids [46]. Additionally, the above process involved the expression of critical enzymes (especially methyltransferases) in the Nuclear Localization Signal (NLS) pathway of alkaloid synthesis.
Benefits of E. coli expression platform include: (1) high productivity of target molecule in an eco-friendly manner avoiding organic solvents, heavy metals and strong acids, (2) eased extraction of protein achieved by transporting the proteins to periplasmic space (by fusion of signal peptides), as they are less exposed to the protease activity, (3) known genomic information and regulatory networks (operons) predisposing them for manipulation and metabolic refactoring, (4) availability of multiple expression vectors for transformation (e.g., pET series of plasmids), (5) growth requirements and transformant selection simplicity, and (6) stable platform to rapidly test a considerable number of different coding sequences and optimize the expression levels before the purification step [16]. Shortcomings of E. coli expression platform include (1) absence of PTM (N/O-linked glycosylation) machinery, due to which eukaryotic proteins cannot be synthesized in a functionally active state, (2) secretion of target proteins as inclusion bodies, wherein they are misfolded, demanding correction steps with the help of chaperones, (3) discharge of target proteins in ectopic subcellular sites, hindering disulfide bond leading to misfolding, (4) lysis of secreted target proteins by intracellular proteases, (5) maintenance of fermentation parameters making the production cost expensive, (6) methionine residue retention at the protein amino-terminal ends, compromising the stability, promoting immunogenicity, (7) codon preference, compelling to overlook unusual amino acids, and (8) insolubility of foreign proteins [47].
2.2. Yeast expression platforms
S. cerevisiae, a unicellular eukaryotic organism is one of the most widely used heterologous hosts. Among the microbial eukaryotic hosts, yeasts harbor the properties of prokaryotic organisms, i.e., ease of genetic manipulation and growth and also the ability to perform PTMs being a eukaryote [48]. Owing to their biosafety regulations for human applications and economic efficiency, they are used for the production of vaccines for Hepatitis B. Both the strains of yeasts - S. Cerevisiae and Schizosaccharomyces pombe are employed as expression systems, where the former is extensively studied. The plasmids can be maintained either in the episomal form (yeast episomal vectors) or can be integrated into the genome at a specific locus (yeast integrating vectors) [49].
There are two primary components, which is common to all the yeast vectors - the backbone which is derived from pBR322 (E. coli plasmid), containing the bacterial replication origin and the selectable markers Ampicillin (bla) and tetracycline (tet). The other component includes the selectable yeast marker, allowing the assortment of putative transformants. Most of the yeast-specific selectable markers work by complementing a specific auxotrophy. The most commonly used markers are LEU2 (encoding, β-isopropyl malate dehydrogenase), URA3 (encoding, Orotidine 5′-phosphate decarboxylase), HIS3 (encoding, Imidazole glycerol-phosphate dehydratase). The URA3 marker is used often since it endows both positive and negative selections. The positive selection involves the complementation of the auxotrophy of specific compounds, and the adverse selection involves the ability to survive on a medium containing a compound which inhibits the growth of cells of wild-type origin. For URA3, adverse selection involves growing the cells in 5-fluoroacetic acid (5-FOA). This vector architecture and the mode of selection of the transformants is another reason why yeast platform is preferred for the heterologous gene expression studies [50].
Carotenoid production was achieved by expression of GGPS (geranylgeranyl pyrophosphate synthase) from Capsicum annuum in S. Pombe [51]. S. pombe does not produce carotenoids but synthesizes Ergosterol, which is a precursor for the synthesis of many carotenoids. Genistein biosynthesis involved the concept of combinatorial biosynthesis, wherein both E. coli and S. cerevisiae platforms have been employed. The biosynthesis was divided into two parts: (1) precursor synthesis (naringenin) in E. coli engineered with phenylpropanoid pathway, and (2) conversion of naringenin to genistein in S. cerevisiae expressing isoflavone synthase (IFS). Both the systems were co-incubated in potassium phosphate buffer, and this was termed “One-pot synthesis” [52]. Benefits of yeast expression platform include (1) ability to perform PTMs facilitating the production of functional eukaryotic proteins, (2) occurrence of the enriched endomembrane system, conditioning release of protein from intracellular compartments, thereby simplifying the purification process, (3) well-known functional yeast genome sequencing for heterologous protein production and development of specialized vector systems for transformation, (4) the presence of active promoters which can either be constitutive (PGK1) or inducible (GAL1) is an added gain, where yeast system employs the auxotrophy mediated selection, unlike prokaryotes. Auxotrophic-mediated selection is advantageous over antibiotic-mediated selection as the latter affects ribosomal activity, (5) Absence of endotoxins and oncogenes entitling the host as “Generally Regarded As Safe” [GRAS], (6) high cell density & growth rate providing attractive scale-up opportunities, and (7) curator of biochemical pathways comprising amino acids such as tyrosine and tryptophan, which serve as precursors for flavonoid biosynthesis [53]
2.3. Comparison of conventional yeast strains - S. cerevisiae vs S. pombe
The S. pombe (fission yeast) strain is less explored in terms of commercial protein production, yet it shows tremendous potential and advantages over S. cerevisiae (budding yeast). Both have distinct properties owing to their taxonomical and evolutionary differences [54]. Many molecular, biochemical and genetic features of S. pombe, are comparable to higher eukaryotes. The molecular architecture involving mRNA splicing and PTMs are quite similar in higher eukaryotes and S. pombe. The study of the mammalian introns getting spliced appropriately, when expressed in S. pombe system, justifies the same. Also, at the gene level, the mammalian promoters and poly (A) signals are found to be functional in S. pombe [55].
Additionally, the S. pombe mutants were functionally complemented by the mammalian counterparts, which are beneficial for the production of eukaryotic proteins (as they result in very similar or marginal differences in PTMs ensuring proper protein conformation). However, there are variations in the pattern of glycosylations in both the systems, when compared to humans. S. cerevisiae comprises mannose residues, while S. pombe has galactose residues (galactomannan outer chain). Therefore, S. pombe qualifies as an attractive model system for studying the gene expression and regulatory networks. It also aids in analyzing various gene counterparts from evolutionarily distant organisms [56].
Shortcomings of Saccharomyces include (1) excessive mannosylation in the proteins which alters the functional and immunological activities (e.g., EBV gp350 produced from S. cerevisiae with decreased reactivity towards the antibodies), (2) protein folding and disulfide bond formation are entirely different from humans prompting protein misfolding and their confinement in the Golgi apparatus, (3) restraining more abundant proteins within the cell wall, (4) aberrant proteolytic cleavage, producing various non-essential proteins with additional amino acid sequences, (5) fusion of the heterologous protein to the yeast signal sequence (acid phosphatase, invertase and α-factor), resulting in amino acid alteration from C-terminal to the signal cleavage site, which affects proteolytic processing and protein functionality, (6) rare vacuolar recombinant protein degradation mediated by cytosolic proteases reducing their half-life, and (7) fed-batch fermentation to attain high cell densities leading to ethanol-induced cell death in yeasts [57].
2.4. Strategies adopted to improvise the heterologous efficiency of yeasts
The concept of strain improvement comprises multiple gene manipulations in the native yeast genome, resulting in the production of an efficient, and stable superlative strain. The strain improvement techniques are (1) engineering endogenous regulatory mechanisms for enhancing the gene expression, (2) eventuating the protein folding and the ER machinery associated, (3) scheming the intracellular trafficking pathways of the proteins to their desired destinations and enhancing their secretion, (4) avoiding protein exposure to the intracellular proteases, and (5) altering PTMs (notably, glycosylations) [58]. The initial trial adopted was the expression of the target gene using the desired promoters. Galactose regulated gene promoters (GAL1, GAL7 and GAL10) are one of the most tightly-regulated promoters of S. cerevisiae. The drawback of these promoters is the limited availability of GAL4 protein (one or two per cell) and the presence of repressor protein (GAL80) in excess. Hence, the strategies adopted to elevate transcription rates include: (1) improving the expression of trans-activators (cloned into multicopy expression vectors), (2) mutating the galactose regulatory network through the glucose-repression system, and (3) constructing galactose-regulated chimeric promoters. A strain with resistance to glucose repression (reg1) mutation was analyzed, wherein efficient induction occurred even when the galactose was present in trace amounts (Galactose/Glucose: 1/100). The chimeric promoters were created by inserting the GAL upstream activator sequences (UAS) between the UAS and TATA box of Glyceraldehyde-3-phosphate dehydrogenase (GAP) to test the expression of IFN-γ. The fusion promoter showed higher levels of expression when compared to the native GAP promoter [59].
2.5. Enhancing protein folding and associated ER machinery
The major chaperone involved in yeast protein folding is ‘Binding immunoglobulin protein’ (BiP). It stimulates the degradation of misfolded proteins and regulates the unfolded protein responses. Hence, overexpression of BiP along with other co-chaperone Hsp40 proteins such as Sis1p/Sil1p and Lhs1p (Hsp70 homologue), protein disulfide isomerases (PDI) and Hac1p (basic leucine zipper transcription factor), serves enhanced protein folding and secretion. Isolation of “super-secreting” yeast strains can achieve improved secretion of target metabolites [60]. Strain analyses are carried out by random mutagenesis followed by the screening for the superlative mutant. Two superlative secreting strains from chymotrypsin were identified and named as ssc1 and ssc2 [61]. These strains are similar to the fmr1 mutants (deficient in ATPase pump) and are reported to enhance the protein secretion. The effects were found to be additive in double mutants (ssc1-ssc2).
Regarding the modification of the protein trafficking machinery, little alterations have been reported, due to lack of knowledge related to the rate-limiting steps and the critical membrane proteins. Some studies revealed the accumulation of the folded proteins in the vacuoles of the yeast system. This Golgi-vacuole missorting is mediated by a specific receptor called ‘Vps10p’ or the ‘CPY’ [62]. It has been reported that the deletion of this receptor in S. cerevisiae reduces the accumulation of proteins in the vacuole. However, this problem is mainly resolved by fusing the protein sequence with an appropriate signal sequence, α-mating factor signal (MFAα1) at the N-terminal end. Minimization of the post-secretory proteolytic degradation of the target proteins is achieved by deleting the vital intracellular proteases involved in the cytosolic and non-cytosolic (vacuolar-mediated degradation) pathways [63]. In S. cerevisiae, many vacuolar proteases such as PrA (PEP4 gene) and PrB (PRB1 gene) are deleted, and the double mutant showed decreased intracellular protease activity [64]. The mitochondrial metalloendoprotease gene (CYM1) knock-down in S. cerevisiae not only decreased proteolysis but also increased protein secretion [46,65].
2.6. Engineering the post-translational mechanism for improved yeast strains
The generation of mutants avoids the limitation of excessive mannosylation (N-linked glycosylation) in yeast [66]. Production of human proteins through glycoengineering in yeast and filamentous fungi is a developing area of Genetics [67]. Many mutant strains possess deletions in MNN1, MNN4 & MNN9 and are used to produce proteins with lesser mannose residues in the outer chain, without influencing the immunogenic α-1, 3 mannose linkages. Further deletion of immunogenic residues can produce non-immunogenic and non-hyperglycosylated proteins (e.g., MNN1 MNN9 double mutant strain). Another example of mannosylation deficient strain includes the PMR1 strain [68,69]. This strain is refactored with a defective calcium ATPase ion pump, which makes the secreted protein by-pass the Golgi complex resulting in the production of proteins, which are glycosylated only at their core and not in the outer chain. Also, it was reported that OCH1, encoding the vital enzyme α-1,6-mannosyltransferase, when deleted along with MNN1 and MNN4, led to protein production with N-glycan intermediate, structurally analogous to the anthropoid counterpart [70]. Minimal reports have been published about O-linked glycosylation as the deletions of O-mannosyltransferases are found to be lethal [71]. These modifications thus overcome the limitations of the yeast platform. Many other limitations are yet to be addressed and are still in the process of improvement.
2.7. High throughput DNA assembly - a big challenge in heterologous expression
Refactoring BGCs in simple eukaryotic fungal systems (budding yeast) could not succeed despite many advantages, indicating that plant cells would display more diverse challenges. To overcome the hurdles, multiple splice variant testing for cryptic genes along with transcript and protein expression levels is a requisite. Moreover, phosphopantetheinylation of all Acyl Carrier Protein (ACP) domains might clarify the BGC refactoring inadequacies in yeast. The type of the protein, its physical and chemical attributes and many other factors dictate the choice of the system to be employed. However, an ideal heterologous system should be able to fulfil the following elemental and essential conditions: (1) higher product yield, (2) producing the product with right conformation and additional modifications (e.g., PTMs) for it to be functionally active, (3) ease of genetic manipulation, (4) safe and economical, and (5) ease of purification and extraction [downstream processing] [72]. Therefore, there is a requirement of a universal competent system that can serve as an efficient heterologous host for the production of plant-based natural product.
Multiple heterologous expression systems have been developed from simple prokaryotes such as E. coli [73] to higher eukaryotes, including plant [74] and mammalian cells [75]. Among the various eukaryotic heterologous hosts, single plant cells appear to be more promising with regard to error-free PTMs for the synthesis of secondary plant bioactives (Table 1). Proteins and therapeutic compounds expressed in plant systems have reached various levels of clinical trials. For instance, many vaccines, Hepatitis B (tobacco), Cholera (potato), Influenza (tobacco) have been successfully produced and are currently in various levels of clinical trials [[76], [77], [78]]. The most exciting aspect in the field of vaccine development is the emergence of edible vaccines such as the one for Rabies, expressed in tomato plants [79]. Proteins are expressed in the comestible parts of the plant and can be consumed as edible vaccines, thereby eliminating the need of downstream purification of the desired product. Over the past few years, molecular pharming in plants has shown tremendous potential and success [80].
Table 1.
Competence of various hosts (native & non-native) in metabolite refactoring and scale-up.
| Non-Native Host for metabolic refactoring | Plant Natural products | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Bacterial systems |
|
|
|
[47,21,73] |
| Yeast systems |
|
|
|
[57,72] |
| Unicellular algae |
|
|
|
[101] |
| Filamentous fungi |
|
|
|
[67] |
| Native Host for metabolic refactoring | Plant Natural products | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Single isolated cells of higher plants in culture |
|
|
|
[113,114,110] |
| Intact plants/tissues in culture |
|
|
|
[76,77,78] |
2.8. ‘HEx’ platform - an innovative strategy for enriching fungal species
Fungi are abundant producers of therapeutics such as penicillin, cyclosporine and lovastatin, obtained from single cultured isolates. Genome sequencing has revealed the existence of more than 5 million fungal species across the globe, each encrypting ∼80 native biosyntheses. Even though DNA sequencing has become very easy, fungal culture possesses a few bottlenecks. Only a trace of known fungi could be cultured in laboratories. In a majority of those cultured fungi, some of the biosynthetic gene clusters are either transcriptionally silent or underexpressed. The identification and expression of these clusters in any apposite prototype open up new avenues for novel natural product syntheses [81]. Synthesis of any drug (unknown/known) in a heterologous host involves several gene insertions into its biosynthetic cascade. Very often it is done within the native host either through (1) promoter/transcription factor manipulation or, by (2) Clustered Regularly Interspaced Short Palindromic Repeats-CRISPR associated Protein 9 (CRISPR-Cas9) editing or by (3) epigenetic activation. These gene modifications are beneficial only in cultivable and genetically docile native hosts. BGC refactoring is a strategy that enables a cryptic gateway from any potent organism [82]. Lately ‘HEx’, an improved scalable bioinformatics approach, has been developed for expressing cryptic BGC in S. cerevisiae [83]. The gene family chosen for ‘HEx’ platform consists of sequence divergent yeast promoters. The HEx promoters and homologous recombination-based DNA assembly must be congruent. Here the host is equipped with improved growth and expression phenotype. Strains with upgraded BGC are analyzed using LC–MS and NMR [84]. The novelty of ‘HEx’ was the incorporation of 41 gene clusters picked from diverse fungal species in yeast (that included membrane-bound Ubiquitin A like terpene cyclases [UTCs] and Polyketide synthase [PKS]), empowering it to produce non-native metabolites [83].
Alcohol dehydrogenase 2 promoter (pADH2) is one such auto-inducible promoter functional in media supplemented with glucose. Here activation of promoter occurs only after the diauxic shift. Promoters from other species of yeasts were also compared to identify their efficiencies in harnessing expression patterns of BGCs. These shortlisted novel promoters were called ‘HEx promoters’ [83]. The BGC engineering (Fig. 1a) involved inserting different plasmids with 6–7 gene inserts and HEx promoters into yeast species, from which the fold increase in expression was computed [85]. Comparison of constitutive expression patterns in all the engineered yeasts was inspected to check the competence of each promoter. HEx promoters possess obligate activation under aerobic respiration in improved yeast strains with functional mitochondria. The choice of improved host strains is essential because there exist hosts with mitochondrial genome instability and vestigial defects such as sporulation inefficiency. The engineering of hosts is mainly focused on repairing such insufficiencies [85]. Apart from these modifications, specific genes in host strains were obliterated, which enhanced heterologous protein production. Because of mitochondrial stability during expression, there was an improved respiration-linked proliferation. This improved strain of yeast (DHY) with enhanced genetic compliance, growth and expression characteristics, gets habituated as a conventional host for the ‘HEx’ platform. Thus, heterologous hosts can be made competent for improved recombinant protein production in eukaryotic microbes. However, significant challenges await plant-based heterologous expressions. It would be fascinating to monitor the performance of ‘HEx’ promoters in cultured single plant cells at regular intervals, to segregate the elite population of plant cell-lines for expression studies en route product yield.
3. Transgenic plants: advantages and disadvantages
Transgenesis is used for heterologous expression of metabolites, wherein specific foreign genes are introduced or, integrated into the genome. Various methods, such as Agrobacterium and viral-mediated transformations, are adopted for this purpose. The advantage of transgenic plants for molecular pharming is their inexpensive large-scale manufacturing [86]. Another important aspect is the processing and purification of the product. Plant system has the upper hand over other systems in this regard, as several proteins are used in unprocessed or partially processed forms (e.g., recombinant vaccines in the form of edible fruits and vegetables). The transgenic plants can also be developed by introducing a set of genes or gene cluster as in the case of metabolic engineering, to increase the yield of desirable products [87]. The first report of flavonoid metabolic reconstruction in the plant was, introduction of the gene encoding dihydroquercetin 4-reductase of Maize origin into Petunia. This transgene stimulated the production of the flavonoid ‘Pelargonidin’ [88]. Similarly, anthocyanin production was enhanced by regulating the expression of flavonol synthase [89]. The transformation efficiency is dependent on various factors such as the promoters, driving the transgene expression and sequences (e.g., Kozak), regulating the translational initiation [90]. In some cases, the codon usage is modified in the transgene in order to avoid cryptic introns and other instability issues. Plant tissue culture techniques have eased recombinant protein production even in the absence of genetic modifications via the process of elicitation in suspension cultures. For example, the trans-resveratrol yield is augmented in Vitis vinifera suspension cultures by the addition of methyl jasmonate [91] and other stress inducers such as cyclodextrins and chitosan [92]. Moreover, this is a controlled and reliable approach for eliminating the interference of external factors such as climatic conditions, pests and pathogens. Transgenic plant development is achieved by two transformation approaches - ‘stable’ and ‘transient’. While the former is for the large-scale production, the latter is for the rapid production of metabolites at a smaller scale [93]. The transient approach examines the expression constructs in a tissue-specific manner and the product quality, before proceeding with stable gene integration into the genome. Transient expression analysis also inspects the gene functions and protein localization (GFP-fusion), in vivo gene editing (gene silencing and marker gene excision). Nuclear transformation involves the insertion of the gene into the host genome, thereby ensuring stable gene expression, which ultimately results in the regeneration of the whole plant (transgenic), producing seeds or vegetative tissues maintained in controlled conditions. The primary advantages include ensuring: (1) post-translational modification (PTM) of expressed proteins, and (2) proper storage and secretion of proteins to the several subcellular compartments, thereby confirming protein folding, stability and bioactivity. However, there are disadvantages such as, (1) low levels of expression, making the approach expensive and time-consuming, (2) requirement of complex vector architecture for enabling high levels of expression, (3) limited scope for commercialization due to extended production cycles and the ability to cross with native plant species, and (4) gene silencing and the corresponding positional and pleiotropic effects. Plastid transformation involves the transfer of genes into the chloroplast DNA by the process of homologous recombination [94]. The transgene vector designs must comprise: (1) flanking homologous plastid sequences, (2) spacer regions, and (3) regulatory sequences to integrate, stably express without disturbing the integrity and functions of the plastid genome. The plants developed by this method are called ‘transplastomic’, achieved in tobacco for the first time [95]. Plants stably express the transgene from the chloroplast genome, conferring many agronomic traits such as herbicide and disease resistance. Chloroplast-derived pharmaceutical proteins include interferon-gamma (IFN-γ), insulin, somatotropin and vaccines for Cholera, Tetanus and Anthrax [96]. The dominant mode of gene transfer into the plastids is by bioballistic methods, which involve benefits such as, (1) high expression levels owing to multiple copies of plastids present in the photosynthetic plants, (2) proper protein folding due to the formation of disulfide bonds, (3) transgene containment achieved by maternal inheritance, (4) tissue-specific gene expression, (5) multigene engineering or transgene stacking, (6) prevention of gene silencing and other positional effects, (7) avoidance of vector sequences in transgene, and (8) no codon preference [97]. However, the challenges include: (1) extending this technology to other plant species, (2) absence of data on chloroplast genome sequences in species of interest, (3) inadequate tissue culture techniques and regeneration of transplastomic plants via somatic embryogenesis and organogenesis, (4) choice of appropriate regulatory sequences and selectable markers, and (5) intermittent occurrences of paternal inheritance and pollen transfer (tobacco species have been reported to have 0.5–1 % of pollen transfer of plastid traits) [98].
Despite tremendous development in the field of plant transgenics, there exist many challenges for heterologous protein or, metabolite production. Truncated bioactive productivity is the major drawback in the plant-based system [99]. Due to complex genome architecture and transformation barricades, many alternative platforms are used for the enhanced yield of proteins and bioactives. The main advantage of using microbial systems is their ease of manipulation along with well-characterized genomes [100].
4. Equipping plant-based apposite hosts - unicellular algal forms vs single plant cells
The impediment in plant cells is that they form a heterogeneous multicellular cluster and exhibit slow division rate. Higher plant tissues rely on multicellularity for their coordinated functions. A model cell system demands unicellular homogeneity and exponentially steady division rate for metabolite refactoring. The cell cycle regulation in plant cells has to be understood well before metabolic engineering. It is essential to investigate binary and multiple fission regulations in algal systems as they serve in: (1) obtaining synchronous cells by natural means just by altering light/dark regime, (2) studying the cell cycle progression in the absence of environmental cues, (3) demanding the availability of unlimited growth supplements for acquiring high cell density, and (4) simplifying the reverse genetics process, as the cell cycle controllers are often present in single copies [101]. Unicellular algae confer flexibility as it is effortless to alter their cell cycle by the application of 5-fluorodeoxyuridine, a nuclear synthesis inhibitor [102].
First eukaryotes rely on binary fission for cell proliferation. However, some algae mother cells divide into multiple daughter cells, which are called multiple fission. Multiple fission exists merely as ‘split’ to ‘consecutive’ and ‘clustered’ types in different plant population [103]. Rapidly flourishing algae can double their cell volume per photoperiod. However, once the critical size is attained, photosynthetic cessation might occur in the dark, which in turn delays the subsequent growth processes. Rescheduling plant cell division until the dark period would prove to be an evolutionary advantage. The evolution of multiple fission in the algal cell cycle evidence that algae are condensed versions of higher plants [104].
Here, nuclear and cellular divisions are naturally disconnected. Light period promotes the algal cell cycle progression. During scotophase, DNA replication, nuclear and cell divisions occur [105]. Thus, multiple fission provides a significant cellular advantage, as the algal forms can dedicate the entire growth phase to light period, postponing the DNA replication and division sequence in the dark. Multiple fission neither wastes time nor impedes cell growth rates. However, in typical plant cell divisions, UV-the natural non-ionizing radiation induces DNA mutations severely affecting the cell growth. In unicellular plants, multiple fission is an extreme case of growth optimization, when exposed to dark/light cycles of variant durations. Mathematical analysis of binary and multiple fission in cell populations and cell size distributions were conducted [106]. The population size distribution by multiple fission does not alter even with high cell growth rates. The mother cells undergoing multiple fission produce daughter cells without any size-reduction, unlike binary fission, where there is cell size reduction. The division rate in binary fission would attain a plateau phase, and even further supplementation of growth nutrients would not increase the population size [107]. Whereas, in multiple fission, the population size continues to rise in consonance with growth nutrient supplementations. Before the metabolic pathway refactoring, multiple fission incorporation in isolated plants cells probably could emerge as a good strategy for promising recombinant protein yields [108]. The regulation of binary and multiple fissions in plant cell systems can be studied in vitro.
Metabolic pathway reconstructions of native natural products in single plant cells are considered to be less complicated when compared to heterologous microbial hosts. Being efficient a eukaryotic system, plants encompass suitable post-translational modifications. However, slow cell division rate and heterogeneous nature often cause an impediment to consistent product retrieval from plant cells. Plant cell synchrony can be attained in cultures developed in vitro.
4.1. Perquisites of single plant cells
A diploid cell has only two potential target site alleles. Mutagenesis efficiency can be determined rapidly by single-cell DNA analysis [109]. There are several reports on single-cell isolation methods. Single-cell analysis has been subjected to both transcriptome and metabolome studies [110]. However, these studies demand technically robust protocols and facilities such as fluorescence-activated cell sorting (FACS) and microinjections. A credible yet conducive single-cell isolation protocol is a perk for plant gene editing experiments. One such single-cell isolations performed in higher plants is protoplast generation. Protoplasts are generally obtained from the suspension cell line of callus generated from mesocotyls, immature embryos, anthers and young leaf bases. It is also one of the ways by which stable nuclear transformation is achieved. Enzymatic digestion of the cell wall and mechanical methods release protoplasts [111]. The gene of interest is directly introduced into the protoplasts with the help of polyethylene glycol (PEG). The PEG treatment alters the osmotic pressure inside the plant cells, thereby facilitating insertion of the transgene. The gene transfer into the protoplasts is mediated by many methods including Agrobacterium, PEG-calcium fusion, electroporation and microinjection. This mode of transformation is employed for both transient and stable gene expression analyses [112]. The former is employed explicitly in functional genomics wherein, the metabolic/signal transduction pathways and the transcriptional/translational machinery can be ephemerally altered or manipulated to study the cell-specific responses.
Since Arabidopsis thaliana serves as the model organism for the plant species, its mesophyll protoplast is often used as a standard tool for studying cell-based functional genomics. The tool thus developed is termed as ‘Transient Expression in Arabidopsis Mesophyll Protoplast’ (TEAMP) [113,114]. Luciferase and β-glucuronidase (GUS) are the reporter assays used in TEAMP. The TEAMP tool has elucidated many vital regulators and the mechanisms of signal transduction systems. For instance, the functional regulation and responses of AUX/IAA proteins in the auxin signalling pathway is proved by this method [115]. Protein-protein interactions, such as the heterodimerization map of transcription factors and the single-cell imaging of fluorescence marker proteins, are some of the additional benefits of this method. Some of the advantages of this technique include: (1) ability to transfer large DNA fragments with simple vector architecture, (2) ectopic protein expression, where the proteins tend to occur in a tissue-specific manner, and (3) provides detailed information on functional and physiological plant aspects, thereby notifying the plant responses to external cues [6]. Since protoplasts are independent units with the partial/fully formed cell walls along with autotrophic ability, they are highly congruent even though not identical to unicellular algae. The similarity could be owing to equivalent conserved developmental modules, notably the total size checkpoint mechanism. In both budding and fission yeast, the critical size for the division is fixed based on nutrient obtainability via the conserved Target Of Rapamycin (TOR) signaling pathway, which has to be evaluated in plant protoplast cultures [116]. The TOR signaling pathway could provide one among many mechanisms of connecting the metabolic status and growth in the size of both unicellular algae and protoplast isolated from land plants. TOR proteins have high sequence similarity between unicellular algae and land plants [117]. Nutrient availability activates TOR, whereas a few growth factors were found to repress TOR1 isoform. The TOR signaling pathway of protoplast is anticipated to be identical to its source plant. However, its regulation in osmoticum at specific time points would deliver many mechanistic insights on division rates, cell wall regeneration, cell reprogramming, the viability of protoplasts and protoplast-derived plant cells. In whole plants, cell division is limited chiefly to meristematic regions. The shoot apical meristem (SAM) is an elaborate dome-shaped arrangement that stocks the stem cells and produces the vegetative and reproductive organs (leaves and flowers) of shoot systems [118]. Development of SAM is tightly regulated with condensed cells at the central zone and bigger cells in the developing organs [116]. Unfortunately, no cell size control mechanisms have been proposed or, identified for plant cells or, tissues during morphogenesis. Furthermore, it is unfamiliar, when and how the plant cell size and control signals are functionally coupled and decoupled. However, the protoplast needs perfect osmoticum, and they are under stress with a short life span.
Overall, if isolated plant protoplasts are made to divide similar to microbes and unicellular eukaryotes, they can potentially enhance the unimpaired yield of target bioactives. Evidences from yeast experiments suggest that‘critical cell size’ and division rates are acute aspects for enhancement machinery, which primarily depend on culture conditions and nutrient obtainability. The cell size control mechanisms in Arabidopsis shoot apical meristem is analogous to yeast notably, fission yeast. If protoplasts isolated from plants are subjected to cell sizer studies and cell cycle progression in culture, it is possible to develop an answer to the underlying molecular mechanisms such as, unicellularity to multicellularity transition states, longevity, senescence, ‘cell-size resetting’ during organogenesis and adaptations to external cues. Again the transformation study efficiency is limited to only a few species. There is a long way to go before we engineer plant protoplasts with an appreciable metabolic output.
5. Conclusions and perspectives
Microbial heterologous hosts, both prokaryotes and eukaryotes, form excellent systems for plant metabolite pathway reconstructions. But, there are plenty of limitations such as the absence of PTM machinery, ectopic secretion of target proteins, misfolding of pathway-related enzymes, excessive mannosylation, altered immunological functions, the formation of non-essential amino acid sequences altering the enzymes that function as pathway regulators. Rare vacuolar recombinant protein degradation mediated by cytosolic proteases reducing their half-life is also quite frequent. These demerits emphasize the importance of native host cells for pathway refactoring. High cell division rate is connected to cell size regulation. Cell size regulators and their functional mechanism in multicellular plant tissues are yet to be deciphered. In multicellular systems, there are cell constraints by tissue structure and cell size changes connected with development and morphogenesis. There are extracellular signals that coordinate with mechanisms operating at the tissue level. Plant cells might rely on its familial affiliation between growth and division not only to achieve its cell size homeostasis within the multiplying cells or, tissues, but also to reset cell size throughout organogenesis. However, plant cell multiplication rates, heterogeneity, cell size controllers and synchronization cues must be studied before biosynthetic pathway engineering in native isolated single plant cells.
Credit authorship contribution statement
Ipsita Pujari: Performed the literature search, Drafted the article. Abitha Thomas: Performed the literature search, Drafted the article. Vidhu Sankar Babu: Performed the literature search, Drafted the article, Ideated the manuscript. Ipsita Pujari, Abitha Thomas, Vidhu Sankar Babu critically revised the review. Ipsita Pujari, Abitha Thomas, Vidhu Sankar Babu contributed to finalizing the manuscript.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
In this study, we did not deal with the human participants, and we did not need for informed consent.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The authors thank the Science and Engineering Research Board - Extra Mural Research (SERB-EMR) (presently called Core Research Grant [CRG]), Government of India, File No. EMR/2015/001816 for funding the research project. I.P. and A.T. thank Manipal Academy of Higher Education, Manipal, India, for providing the prestigious Dr T. M. A. Pai PhD Scholarship. I.P. acknowledges SERB for granting the Junior Research Fellowship from May 2017 to December 2019.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2021.e00619.
Contributor Information
Ipsita Pujari, Email: ipsitapujari@gmail.com.
Abitha Thomas, Email: thabi.tom@gmail.com.
Vidhu Sankar Babu, Email: vidhu.sankar@manipal.edu.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Tan D.-X., Reiter R.J. An evolutionary view of melatonin synthesis and metabolism related to its biological functions in plants. J. Exp. Bot. 2020;71:4677–4689. doi: 10.1093/jxb/eraa235. [DOI] [PubMed] [Google Scholar]
- 2.Arkorful E., Hu S., Zou Z. Metabolomic analyses provide new insights into signaling mechanisms for nutrient uptake by lateral roots of pruned tea plant (Camellia sinensis) J. Agric. Food Chem. 2020;68:7890–7903. doi: 10.1021/acs.jafc.0c02053. [DOI] [PubMed] [Google Scholar]
- 3.Cravens A., Payne J., Smolke C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 2019;10:2142. doi: 10.1038/s41467-019-09848-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isah T. Stress and defense responses in plant secondary metabolites production. Bio. Res. 2019;52:39. doi: 10.1186/s40659-019-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C.S., Hsu C.T., Yang L.H. Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single‐cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018;16:1295–1310. doi: 10.1111/pbi.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marx V. Plants: a tool box of cell-based assays. Nat. Methods. 2016;13:551–554. doi: 10.1038/nmeth.3900. [DOI] [PubMed] [Google Scholar]
- 7.Li M., Hou F., Wu T. Recent advances of metabolic engineering strategies in natural isoprenoid production using cell factories. Nat. Prod. Rep. 2020;37:80–99. doi: 10.1039/C9NP00016J. [DOI] [PubMed] [Google Scholar]
- 8.Krivoruchko A., Nielsen J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 2015;35:7–15. doi: 10.1016/j.copbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Machado D., Andrejev S., Tramontano M., Patil K.R. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46:7542–7553. doi: 10.1093/nar/gky537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angelani C.R., Carabias P., Cruz K.M. A metabolic control analysis approach to introduce the study of systems in biochemistry: the glycolytic pathway in the red blood cell. Biochem. Mol. Biol. Educ. 2018;46:502–515. doi: 10.2307/623917. [DOI] [PubMed] [Google Scholar]
- 11.Gopalakrishnan S., Pakrasi H.B., Maranas C.D. Elucidation of photoautotrophic carbon flux topology in Synechocystis PCC 6803 using genome-scale carbon mapping models. Metab. Eng. 2018;47:190–199. doi: 10.1016/j.ymben.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Halper S.M., Cetnar D.P., Salis H.M. Methods in Molecular Biology. Humana Press; New York, NY: 2018. An automated pipeline for engineering many-enzyme pathways: computational sequence design, pathway expression-flux mapping, and scalable pathway optimization; pp. 39–61. [DOI] [PubMed] [Google Scholar]
- 13.Nagai H., Masuda A., Toya Y. Metabolic engineering of mevalonate-producing Escherichia coli strains based on thermodynamic analysis. Metab. Eng. 2018;47:1–9. doi: 10.1016/j.ymben.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Skraly F.A., Ambavaram M.M.R., Peoples O., Snell K.D. Metabolic engineering to increase crop yield: from concept to execution. Plant Sci. 2018;273:23–32. doi: 10.1016/j.plantsci.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Lian J., Mishra S., Zhao H. Recent advances in metabolic engineering of Saccharomyces cerevisiae: new tools and their applications. Metab. Eng. 2018;50:85–108. doi: 10.1016/j.ymben.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Yang D., Park S.Y., Park Y.S. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020;38:745–765. doi: 10.1016/j.tibtech.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Rutledge P.J., Challis G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 18.Wu G., Yan Q., Jones J.A. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 2016;34:652–664. doi: 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Paddon C.J., Keasling J.D. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 2014;12:355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- 20.O’Connor S.E. Engineering of secondary metabolism. Annu. Rev. Genet. 2015;49:71–94. doi: 10.1146/annurev-genet-120213-092053. [DOI] [PubMed] [Google Scholar]
- 21.Jensen M.K., Keasling J.D. Recent applications of synthetic biology tools for yeast metabolic engineering. FEMS Yeast Res. 2015;15:1–10. doi: 10.1111/1567-1364.12185. [DOI] [PubMed] [Google Scholar]
- 22.Shiba Y., Kirby J., Paradise E.M., Keasling J.D. 2009. Genetically modified host cells and use of same for producing isoprenoid compounds. US 2009/0053797. [Google Scholar]
- 23.Jarboe L.R. Improving the success and impact of the metabolic engineering design, build, test, learn cycle by addressing proteins of unknown function. Curr. Opin. Biotechnol. 2018;53:93–98. doi: 10.1016/j.copbio.2017.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Klamt S., Müller S., Regensburger G., Zanghellini J. A mathematical framework for yield (vs. rate) optimization in constraint-based modeling and applications in metabolic engineering. Metab. Eng. 2018;47:153–169. doi: 10.1016/j.ymben.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng M., Chen S., Yin X., Zhang G. Systems and Synthetic Metabolic Engineering. 2020. System metabolic engineering strategies for cell factories construction; pp. 125–151. [Google Scholar]
- 26.Ellinger J., Schmidt-Dannert C. Construction of a BioBrickTM compatible vector system for Rhodococcus. Plasmid. 2017;90:1–4. doi: 10.1016/j.plasmid.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Jia H., Heymann M., Bernhard F. Cell-free protein synthesis in micro compartments: building a minimal cell from biobricks. N. Biotechnol. 2017;39:199–205. doi: 10.1016/j.nbt.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 28.Popp P.F., Dotzler M., Radeck J. The Bacillus BioBrick Box 2.0: expanding the genetic toolbox for the standardized work with Bacillus subtilis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-15107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymanski E., Calvert J. Designing with living systems in the synthetic yeast project. Nat. Commun. 2018;9:2950. doi: 10.1038/s41467-018-05332-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calero P., Nikel P.I. Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms. Microb. Biotechnol. 2019;12:98–124. doi: 10.1111/1751-7915.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capeness M., Horsfall L. Synthetic biology approaches towards the recycling of metals from the environment. Biochem. Soc. Trans. 2020;48:1367–1378. doi: 10.1042/BST20190837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson A.D., Jez J.M. Synthetic biology meets plant metabolism. Plant Sci. 2018;273:1–2. doi: 10.1016/j.plantsci.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 33.García-Granados R., Lerma-Escalera J.A., Morones-Ramírez J.R. Metabolic engineering and synthetic biology: synergies, future, and challenges, Front Bioeng. Biotechnol. 2019;7:36. doi: 10.3389/fbioe.2019.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotopka B.J., Li Y., Smolke C.D. Synthetic biology strategies toward heterologous phytochemical production. Nat. Prod. Rep. 2018;35:902–920. doi: 10.1039/c8np00028j. [DOI] [PubMed] [Google Scholar]
- 35.Lu H., Villada J.C., Lee P.K.H. Modular metabolic engineering for biobased chemical production. Trends Biotechnol. 2018;37:152–166. doi: 10.1016/j.tibtech.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Lim J.H., Hwang H.H., Lee N.J. Enhanced biosynthesis of 2-deoxy-scyllo-inosose in metabolically engineered Bacillus subtilis recombinants. Front. Microbiol. 2018;9:2333. doi: 10.3389/fmicb.2018.02333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C., Yang C., Zhang W. Molecular basis of dimer formation during the biosynthesis of benzofluorene-containing atypical angucyclines. Nat. Commun. 2018;9:2088. doi: 10.1038/s41467-018-04487-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang D., Kim W.J., Yoo S.M. Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. U. S. A. 2018;115:9835–9844. doi: 10.1073/pnas.1808567115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Yang Y., Yan Y. Biotechnology of Natural Products. Springer international publishing; Cham: 2017. Bioproduction of resveratrol; pp. 61–79. [DOI] [Google Scholar]
- 40.Yang J.E., Park S.J., Kim W.J. One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nat. Commun. 2018;9:79. doi: 10.1038/s41467-017-02498-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura Y., Aoki T., Ayabe S.I. Chalcone isomerase isozymes with different substrate specificities towards 6′-hydroxy- and 6′-deoxychalcones in cultured cells of Glycyrrhiza echinata, a leguminous plant producing 5-deoxyflavonoids. Plant Cell Physiol. 2001;42:1169–1173. doi: 10.1093/pcp/pce130. [DOI] [PubMed] [Google Scholar]
- 42.Shomura Y., Torayama I., Suh D.Y. Crystal structure of stilbene synthase from Arachis hypogaea. Proteins Struct. Funct. Genet. 2005;60:803–806. doi: 10.1002/prot.20584. [DOI] [PubMed] [Google Scholar]
- 43.He X.Z., Blount J.W., Ge S. A genomic approach to isoflavone biosynthesis in kudzu (Pueraria lobata) Planta. 2011;233:843–855. doi: 10.1007/s00425-010-1344-1. [DOI] [PubMed] [Google Scholar]
- 44.Thuan N.H., Chaudhary A.K., Van Cuong D., Cuong N.X. Engineering co-culture system for production of apigetrin in Escherichia coli, J Ind. Microbiol. Biotechnol. 2018;45:175–185. doi: 10.1007/s10295-018-2012-x. [DOI] [PubMed] [Google Scholar]
- 45.Yoon S.H., Lee S.H., Das A. Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. J. Biotechnol. 2009;140:218–226. doi: 10.1016/j.jbiotec.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Leonard E., Runguphan W., O’Connor S., Prather K.J. Opportunities in metabolic engineering to facilitate scalable alkaloid production. Nat. Chem. Biol. 2009;5:292–300. doi: 10.1038/nchembio.160. [DOI] [PubMed] [Google Scholar]
- 47.Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parapouli M., Vasileiadis A., Afendra A.S., Hatziloukas E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020;6:1–31. doi: 10.3934/microbiol.2020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Böer E., Steinborn G., Kunze G., Gellissen G. Yeast expression platforms. Appl. Microbiol. Biotechnol. 2007;77:513–523. doi: 10.1007/s00253-007-1209-0. [DOI] [PubMed] [Google Scholar]
- 50.Song J.Y., Park J.S., Kang C.D. Introduction of a bacterial acetyl-CoA synthesis pathway improves lactic acid production in Saccharomyces cerevisiae. Metab. Eng. 2016;35:38–45. doi: 10.1016/j.ymben.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 51.Ye Y., Fujii M., Hirata A. Geranylgeranyl diphosphate synthase in fission yeast is a heteromer of Farnesyl Diphosphate Synthase (FPS), Fps1, and an FPS-like protein, Spo9, essential for sporulation. Mol. Biol. Cell. 2007;18:3568–3581. doi: 10.1091/mbc.E07-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ricca E., Brucher B., Schrittwieser J.H. Multi-enzymatic cascade reactions: overview and perspectives. Adv. Synth. Catal. 2011;353:2239–2262. doi: 10.1002/adsc.201100256. [DOI] [Google Scholar]
- 53.Mikkelsen M.D., Buron L.D., Salomonsen B. Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 2012;14:104–111. doi: 10.1016/j.ymben.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Dujon B., Sherman D., Fischer G. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 55.Yin J., Li G., Ren X., Herrler G. Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 2007;127:335–347. doi: 10.1016/j.jbiotec.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 56.Albertsen L., Chen Y., Bach L.S. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl. Environ. Microbiol. 2011;77:1033–1040. doi: 10.1128/AEM.01361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demain A.L., Vaishnav P. Production of recombinant proteins by microbes and higher organisms. Biotechnol. Adv. 2009;27:297–306. doi: 10.1016/j.biotechadv.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Keasling J.D. Manufacturing molecules through metabolic engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 59.Mattanovich D., Branduardi P., Dato L. Recombinant protein production in yeasts. Methods Mol. Biol. 2012;824:329–358. doi: 10.1007/978-1-61779-433-9_17. [DOI] [PubMed] [Google Scholar]
- 60.Macauley-Patrick S., Fazenda M.L., McNeil B., Harvey L.M. Heterologous protein production using the Pichia pastoris expression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- 61.Becerra M., Díaz Prado S., Cerdán E., González Siso M.I. Heterologous Kluyveromyces lactis β-galactosidase secretion by Saccharomyces cerevisiae super-secreting mutants. Biotechnol. Lett. 2001;23:33–40. doi: 10.1023/A:1026795706520. [DOI] [Google Scholar]
- 62.Kama R., Gabriely G., Kanneganti V., Gerst J.E. Cdc48 and ubiquilins confer selective anterograde protein sorting and entry into the multivesicular body in yeast. Mol. Biol. Cell. 2018;29:948–963. doi: 10.1091/mbc.E17-11-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kroukamp H., den Haan R., van Zyl J.H., van Zyl W.H. Rational strain engineering interventions to enhance cellulase secretion by Saccharomyces cerevisiae, Biofuels. Bioprod. Biorefining. 2018;12:108–124. doi: 10.1002/bbb.1824. [DOI] [Google Scholar]
- 64.Zheng X., Li K., Shi X. Potential characterization of yeasts isolated from Kazak artisanal cheese to produce flavoring compounds. MicrobiologyOpen. 2018;7 doi: 10.1002/mbo3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fidan O., Zhan J. Recent advances in engineering yeast for pharmaceutical protein production. RSC Adv. 2015;5:86665–86674. doi: 10.1039/c5ra13003d. [DOI] [Google Scholar]
- 66.Conde R., Pablo G., Cueva R., Larriba G. Screening for new yeast mutants affected in mannosylphosphorylation of cell wall mannoproteins. Yeast. 2003;20:1189–1211. doi: 10.1016/j.clgc.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 67.Gerngross T.U. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- 68.Mandal D., Woolf T.B., Rao R. Manganese selectivity of Pmr1, the yeast secretory pathway ion pump, is defined by residue Gln783 in transmembrane segment 6: residue Asp778 is essential for cation transport. J. Biol. Chem. 2000;275:23933–23938. doi: 10.1074/jbc.M002619200. [DOI] [PubMed] [Google Scholar]
- 69.Wei Y., Chen J., Rosas G. Phenotypic screening of mutations in Pmr1, the yeast secretory pathway Ca2+/Mn2+-ATPase, reveals residues critical for ion selectivity and transport. J. Biol. Chem. 2000;275:23927–23932. doi: 10.1074/jbc.M002618200. [DOI] [PubMed] [Google Scholar]
- 70.Yoko-o T., Tsukahara K., Watanabe T. Schizosaccharomyces pombe och1+encodes α-1,6-mannosyltransferase that is involved in outer chain elongation of N-linked oligosaccharides. FEBS Lett. 2001;489:75–80. doi: 10.1016/S0014-5793(01)02082-8. [DOI] [PubMed] [Google Scholar]
- 71.Thak E.J., Lee S.-B., Vanpala S.X. Core N-glycan structures are critical for the pathogenicity of cryptococcus neoformans by modulating host cell death. MBio. 2020;11:e00711–e00720. doi: 10.1128/mBio.00711-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gandier J.-A., Master E. Pichia pastoris is a suitable host for the heterologous expression of predicted class I and class II hydrophobins for discovery, study, and application in biotechnology. Microorganisms. 2018;6:3. doi: 10.3390/microorganisms6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mesa-Pereira B., Rea M.C., Cotter P.D. Heterologous expression of biopreservative bacteriocins with a view to low cost production. Front. Microbiol. 2018;9:1654. doi: 10.3389/fmicb.2018.01654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valeeva L.R., Nyamsuren C., Sharipova M.R., Shakirov E.V. Heterologous expression of secreted bacterial BPP and HAP phytases in plants stimulates Arabidopsis thaliana growth on phytate. Front. Plant Sci. 2018;9:186. doi: 10.3389/fpls.2018.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cognato B.B., Handali S., Morassutti A.L. Heterologous expression of three antigenic proteins from Angiostrongylus cantonensis: ES-7, Lec-5, and 14-3-3 in mammalian cells. Mol. Biochem. Parasitol. 2018;221:32–35. doi: 10.1016/j.molbiopara.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 76.Kumar G., Karthik L., Rao K.V.B. Microbial Bioprospecting for Sustainable Development. Springer; Singapore, Singapore: 2018. Plant vaccines: an overview; pp. 249–263. [DOI] [Google Scholar]
- 77.Mardanova E.S., Ravin N.V. Plant-produced recombinant influenza A vaccines based on the M2e peptide. Curr. Pharm. Des. 2018;24:1317–1324. doi: 10.2174/1381612824666180309125344. [DOI] [PubMed] [Google Scholar]
- 78.Sciutto E., Hernández M., Cervantes-Torres J. Prospects of Plant-Based Vaccines in Veterinary Medicine. Springer international publishing; Cham: 2018. Toward the optimization of a plant-based oral vaccine against cysticercosis; pp. 227–237. [DOI] [Google Scholar]
- 79.Rybicki E. Prospects of Plant-Based Vaccines in Veterinary Medicine. Springer international publishing; Cham: 2018. History and promise of plant-made vaccines for animals; pp. 1–22. [DOI] [Google Scholar]
- 80.Rybicki E.P. Plant molecular farming of virus-like nanoparticles as vaccines and reagents. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020;12:e1587. doi: 10.1002/wnan.1587. [DOI] [PubMed] [Google Scholar]
- 81.Zhao X., Yuan X., Chen S. Role of the tomato TAGL1 gene in regulating fruit metabolites elucidated using RNA sequence and metabolomics analyses. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osbourn A., Papadopoulou K.K., Qi X. Finding and analyzing plant metabolic gene clusters. Methods Enzymol. 2012;517:113–138. doi: 10.1016/B978-0-12-404634-4.00006-1. [DOI] [PubMed] [Google Scholar]
- 83.Harvey C.J.B., Tang M., Schlecht U. HEx: a heterologous expression platform for the discovery of fungal natural products. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aar5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jackson S.A., Crossman L., Almeida E.L. Diverse and abundant secondary metabolism biosynthetic gene clusters in the genomes of marine sponge derived Streptomyces spp. Isolates, Mar. Drugs. 2018;16:67. doi: 10.3390/md16020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiong L., Zeng Y., Tang R.Q. Condition-specific promoter activities in Saccharomyces cerevisiae. Microb. Cell Fact. 2018;17:58. doi: 10.1186/s12934-018-0899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 87.Marsafari M., Samizadeh H., Rabiei B. Biotechnological production of flavonoids: an update on plant metabolic engineering, microbial host selection, and genetically encoded biosensors. Biotechnol. J. 2020;15 doi: 10.1002/biot.201900432. [DOI] [PubMed] [Google Scholar]
- 88.Haselmair-Gosch C., Miosic S., Nitarska D. Great cause—small effect: undeclared genetically engineered prange Petunias harbor an inefficient Dihydroflavonol 4-reductase. Front. Plant Sci. 2018;9:149. doi: 10.3389/fpls.2018.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhatia C., Pandey A., Gaddam S.R. Low temperature enhanced flavonol synthesis requires light-associated regulatory components in Arabidopsis thaliana. Plant Cell Physiol. 2018;59:2099–2112. doi: 10.1093/pcp/pcy132/5053954. [DOI] [PubMed] [Google Scholar]
- 90.Kesidis A., Depping P., Lodé A. Expression of eukaryotic membrane proteins in eukaryotic and prokaryotic hosts. Methods. 2020;180:3–18. doi: 10.1016/j.ymeth.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 91.Chastang T., Pozzobon V., Taidi B. Resveratrol production by grapevine cells in fed-batch bioreactor: experiments and modelling. Biochem. Eng. J. 2018;131:9–16. doi: 10.1016/j.bej.2017.12.009. [DOI] [Google Scholar]
- 92.Chu M.Y., Almagro L., Chen B.H. Recent trends and comprehensive appraisal for the biotechnological production of trans-resveratrol and its derivatives. Phytochem. Rev. 2018;17:491–508. doi: 10.1007/s11101-017-9546-9. [DOI] [Google Scholar]
- 93.Khatodia S. S.M. Paul Khurana, Genetic engineering for plant transgenesis: focus to pharmaceuticals. Omi. Technol. Bio-Eng. 2018;2:71–86. doi: 10.1016/B978-0-12-815870-8.00005-X. [DOI] [Google Scholar]
- 94.Fernandez A., Paoletti C. Unintended effects in genetically modified food/feed safety: a way forward. Trends Biotechnol. 2018;36:872–875. doi: 10.1016/j.tibtech.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 95.Fernández-San Millán A., Aranjuelo I., Douthe C. Physiological performance of transplastomic tobacco plants overexpressing aquaporin AQP1 in chloroplast membranes. J. Exp. Bot. 2018;69:3661–3673. doi: 10.1093/jxb/ery148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Franconi R., Demurtas O.C., Massa S. Plant-derived vaccines and other therapeutics produced in contained systems. Expert Rev. Vaccines. 2010;9:877–892. doi: 10.1586/erv.10.91. [DOI] [PubMed] [Google Scholar]
- 97.Dyall S.D., Brown M.T., Johnson P.J. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 98.Fuentes P., Armarego-Marriott T., Bock R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 2018;49:10–15. doi: 10.1016/j.copbio.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 99.de Bruijn W.J.C., Levisson M., Beekwilder J. Plant aromatic prenyltransferases: tools for microbial cell factories. Trends Biotechnol. 2020;38:917–934. doi: 10.1016/j.tibtech.2020.02.006. [DOI] [PubMed] [Google Scholar]
- 100.Alok A., Tiwari S., Kaur J. Molecular Aspects of Plant Beneficial Microbes in Agriculture. 2020. CRISPR/Cas9 mediated genome engineering in microbes and its application in plant beneficial effects; pp. 351–359. [DOI] [Google Scholar]
- 101.Bišová K., Zachleder V. Cell-cycle regulation in green algae dividing by multiple fission. J. Exp. Bot. 2014;65:2585–2602. doi: 10.1093/jxb/ert466. [DOI] [PubMed] [Google Scholar]
- 102.Kobayashi Y., Kanesaki Y., Tanaka A. Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:803–807. doi: 10.1073/pnas.0804270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cvrčková F. Plant Cell Monographs. Springer; Cham: 2018. A brief history of eukaryotic cell cycle research; pp. 67–93. [DOI] [Google Scholar]
- 104.Millar A.J. The Intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu. Rev. Plant Biol. 2016;67:595–618. doi: 10.1146/annurev-arplant-043014-115619. [DOI] [PubMed] [Google Scholar]
- 105.Vitova M., Bisova K., Kawano S., Zachleder V. Accumulation of energy reserves in algae: from cell cycles to biotechnological applications. Biotechnol. Adv. 2015;33:1204–1218. doi: 10.1016/j.biotechadv.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 106.Concas A., Pisu M., Cao G. A novel mathematical model to simulate the size-structured growth of microalgae strains dividing by multiple fission. Chem. Eng. J. 2016;287:252–268. doi: 10.1016/j.cej.2015.11.021. [DOI] [Google Scholar]
- 107.Bonnett H.T. CRC Handbook of Symbiotic Cyanobacteria. CRC Press; 1990. The Nostoc-Gunnera association; pp. 161–171. [DOI] [Google Scholar]
- 108.Li M., Liu C., Yang J. Common problems associated with the microbial productions of aromatic compounds and corresponding metabolic engineering strategies. Biotechnol. Adv. 2020;41 doi: 10.1016/j.biotechadv.2020.107548. [DOI] [PubMed] [Google Scholar]
- 109.Bock C., Farlik M., Sheffield N.C. Multi-omics of single cells: strategies and applications. Trends Biotechnol. 2016;34:605–608. doi: 10.1016/j.tibtech.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Heath J.R., Ribas A., Mischel P.S. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 2016;15:204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomas A., Pujari I., Shetty V. Dendrobium protoplast co-culture promotes phytochemical assemblage in vitro. Protoplasma. 2017;254:1517–1528. doi: 10.1007/s00709-016-1043-2. [DOI] [PubMed] [Google Scholar]
- 112.Planchais S., Camborde L., Jupin I. Methods in Molecular Biology. Humana Press; New York, NY: 2016. Protocols for studying protein stability in an Arabidopsis protoplast transient expression system; pp. 175–194. [DOI] [PubMed] [Google Scholar]
- 113.Yoo S.D., Cho Y.H., Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 114.Im J.H., Yoo S.D. Transient expression in Arabidopsis leaf mesophyll protoplast system for cell-based functional analysis of MAPK cascades signaling. Methods Mol. Biol. 2014;1171:3–12. doi: 10.1007/978-1-4939-0922-3_1. [DOI] [PubMed] [Google Scholar]
- 115.Guilfoyle T.J. The PB1 Domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell. 2015;27:33–43. doi: 10.1105/tpc.114.132753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Son O., Kim S., Kim D. Involvement of TOR signaling motif in the regulation of plant autophagy. Biochem. Biophys. Res. Commun. 2018;501:643–647. doi: 10.1016/J.BBRC.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 117.John F., Roffler S., Wicker T., Ringli C. Plant TOR signaling components. Plant Signal. Behav. 2011;6:1700–1705. doi: 10.4161/psb.6.11.17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu Q., Xu F., Jackson D. All together now, a magical mystery tour of the maize shoot meristem. Curr. Opin. Plant Biol. 2018;45:26–35. doi: 10.1016/j.pbi.2018.0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.