Abstract
Leishmaniases are caused by protozoan parasites of the genus Leishmania transmitted by females blood-feeding phlebotomine insects (Diptera: Psychodidae). In Tunisia, cutaneous and visceral leishmaniases are of public health concern. In Tunisia, 17 species of phlebotomine sand flies are described. Here we investigate natural infection in Tunisian mixed foci regions of leishmaniases. We trap female sandflies during the Leishmania transmission season in the country's central-eastern and northern parts. We investigate Leishmania infection using PCR-RFLP targeting the ITS1 ribosomal DNA, followed by enzymatic digestion with HaeIII; then, we identify sand flies using molecular methodologies. We confirm the presence of Phlebotomus papatasi and Phlebotomus perniciosus infected by L. major and L. infantum parasites in Tunisia.
Keywords: Leishmaniasis, Sand flies, Phylogeny, Cytochrome b, Phlebotomus, ITS1, Leishmania, L. major, L. infantum, Tunisia
1. Introduction
Leishmaniases, caused by protozoan parasites of the Leishmania genus (Kinetoplastida, Trypanosomatidae), represent a significant health problem. They are neglected tropical diseases with a strong link with poverty (Alvar et al., 2006). Leishmaniases present clinical forms ranging from self-curing cutaneous leishmaniasis (CL) to fatal visceral leishmaniasis (VL) (Desjeux, 2001). They are transmitted by phlebotomine sand flies (Killick-Kendrick, 1990).
Three clinico-epidemiological CL forms and the visceral form of leishmaniasis occur in Tunisia. In Tunisia, 17 phlebotomine sand fly species are identified; they belong to the Phlebotomus and Sergentomyia genera (Croset et al., 1978; Depaquit et al., 1998). Among them, 6 species are proven or potential vectors of Leishmania. The zoonotic CL (ZCL) due to L. major is of public health concern in the Center and the South of the country but has recently spread to the northern territories (Aoun et al., 2012) (Haouas et al., 2012). The proven vector of L. major is Phlebotomus papatasi, which is present all over Tunisia (Chelbi et al., 2009) (Ben Ismail et al., 1987) (Ben-Ismail et al., 1987; Ghrab et al., 2006). The anthroponotic cutaneous Leishmaniasis, or chronic CL (CCL), is caused in Tunisia by L. tropica, first described in 2005 in a microfocus of Tataouine in the South-eastern part of the country (Rioux et al., 1986). It is transmitted by P. sergenti, mainly found in the central and southern regions (Tabbabi et al., 2011). The sporadic CL (SCL) is caused by dermotropic variants of L. infantum (zymodeme MON-24) that prevails in northern Tunisia, and sand flies belonging to the Larroussius subgenus are incriminated as a vector (Haouas et al., 2012). The visceral form is caused by L. infantum, with P. perniciosus as the proven vector (Ghrab et al., 2006).
Climate change would affect Leishmaniases' incidence and phlebotomine dispersal (Kholoud et al., 2018). Likewise, ZCL caused by L. major has recently spread over its traditional foci to central and southern parts of the country. Whereas CCL caused by L. tropica is no longer restricted to South-eastern Tunisia (Haouas et al., 2012). Such extension over historical foci has also been recorded in Morocco (Kholoud et al., 2018) (Kholoud et al., 2020) (Guernaoui et al., 2020). -Such expansion in territories has favored the emergence of CL mixed foci of L. tropica and L. major. Besides, P. perniciosus, a vector of visceral L. infantum MON-1, is also present in the North and the South of the country (Ghrab et al., 2006; Guerbouj et al., 2007). To gain insight into the activity of the epidemiological cycle of Leishmania. spp in endemic leishmaniasis foci, we have ascertained the Leishmania infection rate of the collected sand flies and re-investigated the phlebotomine fauna diversity in these foci.
2. Materials and methods
2.1. Study area and sample collection
Male and female sand fly samples were collected from previous work (Ayari et al., 2016). A set of 188 female sand flies collected in various habitats from six governorates (Beja, Zaghouan, Kairouan, Monastir, Sousse, and Mahdia) in the central, eastern, and northern parts of Tunisia, belonging to the sub-humid, semi-arid and arid bioclimatic stages were selected for the study. They were trapped with CDC miniature light traps (John W. Hock, USA) and sticky traps during the summer season of 2010 and 2012. The traps were placed in animal shelters, outside and inside houses. All specimens have been preserved in 95% ethanol or/and in nitrogen liquid.
Sex identification was able using morphological tools and based on external and internal characters of the head and genitalia (according to the keys of Lewis (Lewis, 1974; Lewis and Hitchocock Jr., 1968) and Artemiev (Artemiev, 1991)).
2.2. DNA extraction, PCR amplification, and Leishmania detection
Genomic DNA from the female specimens was extracted from the thorax and the anterior abdomen, as described by Cornel and Collins (1996) (Cornel and Collins, 1996).
2.2.1. Real-time PCR
The detection and quantification of Leishmania spp. DNA was realized by the real-time PCR (qPCR) technique. It consists of amplifying the conserved regions of the kinetoplast minicircle DNA (kDNA) gene. It is considered one of the most sensitive PCR approaches (Martin-Ezquerra et al., 2009). Each amplification of 5 μL of genomic extracted DNA was performed in duplicate. Eightfold dilution series of DNA from promastigotes (MHOM/ES/04/BCN-61, L. infantum MON-1) was used as calibrators (serial dilution from 105 parasites/ml to 0.01 parasite/ml), allowing plotting of a standard curve. Samples with Ct lower than 36 were considered potentially positives and retained for further analyses.
2.2.2. PCR-RFLP
The ribosomal internal transcribed spacer 1 (ITS1) amplification was performed using LITSR and L5.8S primers (Esseghir et al., 1997). Amplification was performed in a final volume of 50 μL containing 5 μL of DNA added to 5 μL of 10 × BioLabs Buffer, 4 mM MgCl2, 10 mM dNTP, 5 U/μL of TaqBio Labs, and 10 μM of each primer. The cycling was: 95 °C for 120 s, then 30 cycles of 95 °C for 20 s, 54 °C for 30 s, and 72 °C for 60 s. As a control, DNA extracted from reference strains of Leishmania was used as positive controls. Following amplification, PCR products (17 μL) were digested with 1 μL HaeIII enzyme without prior purification and visualized after electrophoresis on a 1% agarose ethidium bromide-stained gel.
2.3. PCR amplification for sand fly identification
We choose to identify sand flies with molecular methods due to the difficulties in identification via morphological criteria. Sandflies share several morphological characters, and their identification can be mistaken. Polymerase chain reactions (PCR) were carried out to amplify into the mitochondrial DNA 336 bp of 3′ end of cytochrome b gene along with 67 bp of complete tRNA for serine, 22 bp interval sequences (including stop codons), and 124 bp of 3′ end of NADH1 gene. Using 50 ng of genomic DNA in 25 μL PCR reaction containing 10 pmol of each primers CB3-PDR(5′GGTA(C/T)(A/T)TTGCCTCGA(T/A)TTCG(T/A)TATGA- 3′) and N1NPDR (5′-CA(T/C)ATTCAACC(A/T)GAATGATA- 3′) (Esseghir et al., 1997), 200 μmol dNTPs, 1 U Taq DNA polymerase (product number: D1806, SIGMA-ALDRICH, USA), 2.5 μL 10× buffer. The amplification cycle was: an initial denaturation step at 94 °C for 5 min, followed by 30 cycles of (denaturation at 94 °C for 60 s, annealing at 46 °C for 60 s and extension at 72 °C for 90 s), and a final extension at 72 °C for 10 min. The PCR products were analyzed after electrophoresis on 1% agarose gel stained with ethidium bromide.
2.4. Sequence analyses and sand fly phylogenetic tree construction
According to the supplier's instructions, amplicons were purified using the QIAquick PCR purification kit (QIAGEN). Sequencing was performed using a Big Dye Terminator cycle sequencing ready reaction kit (Perkin Elmer, Applied Biosystems, Foster City, CA) and an ABI373 Automated DNA Sequencer. Sequences were edited using Clustal-X version 1.81 software (Larkin et al., 2007). Sequences were matched against NCBI using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Genetic diversity and identification of sand fly samples were performed using the MEGA6 software (Tamura et al., 2007) against reference sequences. The tree was constructed following the UPGMA (Sneath and Sokal, 1962) distance analyses using the Jukes-Cantor model. All bootstrap support values are based on 1000 replicates.
3. Results
3.1. Leishmania infection identification
Forty-three samples with Ct lower than 36 were retained for Leishmania species identification. Amplification of ITS1 detected Leishmania's DNA presence in 17 out of the 43 sand fly specimens analyzed (Table S1). The Enzymatic digestion of the ITS1-PCR product with the restriction enzyme HaeIII allowed identifying two Leishmania species, L. infantum (10) and L. major (7). Most of the positive phlebotomes for Leishmania DNA are from Zaghouan and Kairouan. Infected specimens are P. perniciosus, P. perfiliewi carrying L. infantum DNA, and P. papatasi carrying L. major. Surprisingly, L. infantum DNA was detected in one P. papatasi specimen. Finally, P. longicuspis was infected with L. major.
3.2. Molecular sandflies identification
The selected female specimens display the expected amplicon of 560 bp in size. The specimens were captured in the 14 stations of the North, East, and Center of the country. Five samples could not be sequenced. The obtained sequences present homology higher than 98% for six sand fly species that belong to the Phlebotomus (P. papatasi, P. perniciosus, P. perfiliewi, P. longicuspis, and P. chabaudi) and Sergentomyia (S. minuta) genera. All the sequences were deposited into the Gene Bank database (http://www.ncbi.nlm.nih.gov/) under the accession number MW305396 to MW305433.
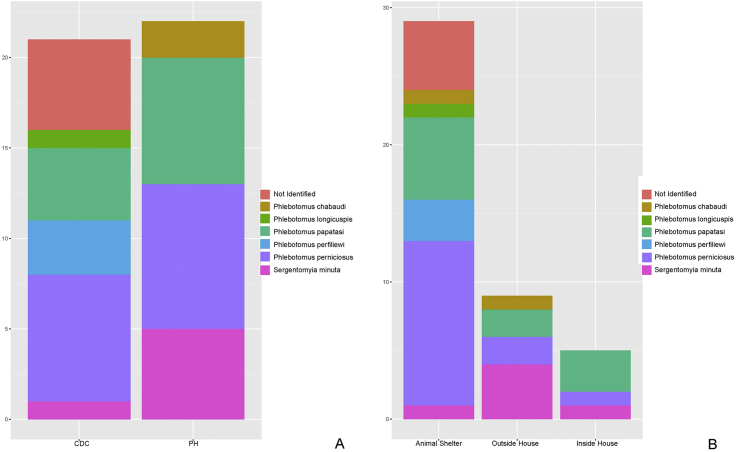
An equal number of phlebotomes was collected using sticky (22) or CDC traps (21). In our survey, Phlebotomus chabaudi was identified only in sticky traps, P. longicuspis and P. perfiliewi were identified solely in CDC light traps (Fig. 1A). Two-thirds (29) were caught in animal shelters, the others from outside and inside houses (Fig. 1A).
Fig. 1.
Sandflies Phylogenetic tree based on UPGMA distance analyses using the Jukes–Cantor Model. Values on branches indicate bootstrap values.
A cladistic analysis was performed to identify sand fly specimens against reference sequences collected from NCBI. The tree's topology discloses two clades with high bootstrap values (Fig. 2). From our samples, Phlebotomus and Sergentomyia genera were separated. Six specimens from North and East of Tunisia gathered together within the Sergentomyia clade. The elven specimens belong to the Phlebotomus subgenus represented by a single species in Tunisia P. papatasi. These specimens were collected in various areas distributed over four bioclimatic stages: semi-arid medium, arid upper, and semi-arid lower. For the subgenus Paraphlebotomus represented in Tunisia by four species, only two P. chabaudi specimens were captured. In our tree, species belonging to the Larroussius subgenus are individualized with high Bootstrap values (98% to 100%). Thirteen specimens cluster with P. perniciosus reference sequences, 3 with P. perfiliewi, and 1 with P. longicuspis (Fig. 2).
Fig. 2.
Sand flies sampling according to the trap type (A) and location (B).
4. Discussion
This study reported the molecular identification of phlebotomes trapped in the North, East, and Center of Tunisia, targeting a portion of mitochondrial DNA: cytochrome b, PCR-sequencing technique. Mitochondrial DNA (mtDNA) genes are commonly employed in population genetics studies (Mahamdallie et al., 2011) (Ready, 2013) (Ebrahimi et al., 2016). They have been widely used for population genetics analyses of the New World leishmaniasis vector sandfly, Lutzomyia longipalpis (Coutinho-Abreu et al., 2008), to highlight molecular variation within Phlebotomine species from a wide geographical range (Pesson et al., 2004) (Yahia et al., 2004) (Perrotey et al., 2005) (Depaquit, 2014). As already reported (Carta et al., 2020), a predominance of species belonging to the subgenus Larroussius (P. perniciosus, P. perfiliewi, P. longicuspis) is attracted to CDC light traps that might be due to the phototropisms of these species.
The majority of specimens are captured around animal shelters, while there are fewer in and around houses (Fig. 1B), that sign a preference of sand fly for animal baits and the environmental condition, temperature, and humidity, also the presence of food source and blood for coprophagous larvae (Ximenes et al., 1999) (Feliciangeli, 2004).
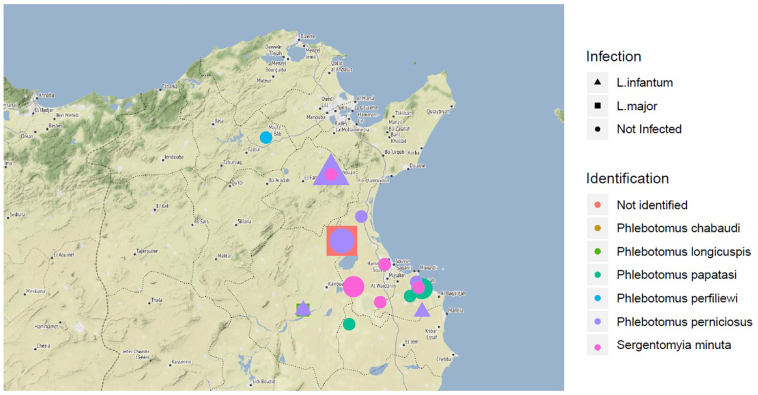
A majority of samples carrying Leishmania DNA originates from Zaghouan and Kairouan governorates (Fig. 3), an endemic focus for Cutaneous and Visceral Leishmaniases. Most of the infected sandflies were captured between August and September, which corresponds to the highest transmission period. Interestingly, most of the positive females were not found engorged by blood, highlighting their sustained infection and points to their role as a vector of leishmaniasis (Ghrab et al., 2006).
Fig. 3.
Geographical distribution and natural Leishmania infection of captured sand flies.
The specimens of P. perniciosus carrying Leishmania DNA were trapped in Kairouan and Zaghouan. Phlebotomus perniciosus is the primary vector of L. infantum MON-24, which causes human visceral leishmaniasis. This species is widespread in the North, extending towards the country's center (Benallal et al., 2017). The molecular analysis revealed molecular variant P. perniciosus specimen (Highlighted in Fig. 2). This molecular variant corresponds to an atypical P. perniciosus (PNA), previously confused with P. longicuspis, mainly present in the semi-arid and arid bioclimatic regions. Therefore, the question arises of the vectorial capacity of the atypical form of P. perniciosus in the southern regions (Benallal et al., 2017). The distribution of the atypical form of P. perniciosus in the south can be attributed to adaptation to these arid conditions. This is currently under investigation (data not published), and more specimens are presently analyzed in Tunisia. Phlebotomus perfiliewi, a known vector of L. infantum responsible for SCL (Izri and Belazzoug, 1993), was sampled in the north's historical foci (Beja and Zaghouan, belonging to the sub-humid bioclimatic stage). In Kairouan (arid bioclimatic stage), we found P. papatasi, P. longicuspis, and unidentified sand fly specimens carrying L. major DNA and P. perniciosus with L. infantum DNA. This region is a hyper-endemic focus of ZCL due to L. major and is also affected by VL due to L. infantum (Pousse et al., 1995) (Aissi et al., 2015). From Monastir (semi-arid bioclimatic), an endemic region for LCZ (Haouas et al., 2012), only one L. infantum infection was reported in P. perniciosus. Surprisingly, In Zaghouan, L. infantum DNA was detected in an engorged P. papatasi specimen. Such observation, although rare, has already been reported in Greece (Aransay et al., 1933-1938), Iran (Yavar et al., 2013), and recently in Italy (Latrofa et al., 2018). However, L. infantum does complete its developmental life cycle in P. papatasi as it is eliminated after blood digestion and defecation (Pimenta et al., 1994).
The specificity of the transmission cycle in these regions relies on the compatibility and the frequency of encounters between the reservoirs carrying the parasite and the sand fly vectors favored by the overlapping ecological niches (Esseghir et al., 1997). Moreover, the environmental modifications due to climate change and human intervention have probably induced an increase in the reservoir and vector population densities and a change in their geographic location (Moo-Llanes et al., 2013). This study provides an updated distribution map of phlebotomine species and validates cyt b for Tunisian sand fly molecular identification. Although molecular analysis is very effective, issues have been raised. In our study, from the initial sampling, a reduced number of individuals were analyzed; this is firstly due to the selection performed based on the Real-time PCR results and then to the molecular steps, which can also reduce the number of successfully analyzed samples. For this, special attention should be paid to the sampling and pre-sequencing steps to optimize molecular identification.
Author contributions
Conceptualization, Melek Chaouch and Souha Ben Abderrazak; Data curation, Melek Chaouch and Amal Chaabane; Formal analysis, Chiraz Ayari; Funding acquisition, Souha Ben Abderrazak; Investigation, Melek Chaouch, Chiraz Ayari, Souad Ben Othman, Jomaa Chemkhi and Souha Ben Abderrazak; Methodology, Melek Chaouch, Amal Chaabane, Chiraz Ayari, Jomaa Chemkhi and Souha Ben Abderrazak; Project administration, Souha Ben Abderrazak; Supervision, Souha Ben Abderrazak; Validation, Melek Chaouch and Souha Ben Abderrazak; Visualization, Melek Chaouch; Writing – original draft, Melek Chaouch and Souha Ben Abderrazak; Writing – review & editing, Melek Chaouch, Denis Sereno and Souha Ben Abderrazak.
Appendix A. Supplementary data
The following is the supplementary data related to this article.
List of captured sand flies.
Declaration of Competing Interest
The authors have declared that no competing interests exist.
Acknowledgments
The study was supported by the Ministry of higher education and scientific research (Tunisia) in the frame of the research lab LR 11-IPT-06 with a special thanks to the H3Africa Bioinformatics Network (H3ABioNet).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.parepi.2021.e00212.
References
- Aissi W., Ben Hellel K., Habboul Z., Ben Sghaier I., Harrat Z., Bouratbine A., Aoun K. Epidemiological, clinical and biological features of infantile visceral leishmaniasis at Kairouan hospital (Tunisia): about 240 cases. Bull. Soc. Pathol. Exot. 2015;108:265–271. doi: 10.1007/s13149-015-0438-110.1007/s13149-015-0438-1. (pii) [DOI] [PubMed] [Google Scholar]
- Alvar J., Yactayo S., Bern C. Leishmaniasis and poverty. Trends Parasitol. 2006;22:552–557. doi: 10.1016/j.pt.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Aoun K., Ben Abda I., Bousslimi N., Bettaieb J., Siala E., Ben Abdallah R., Benmously R., Bouratbine A. Comparative characterization of skin lesions observed in the three endemic varieties of cutaneous leishmaniasis in Tunisia. Ann. Dermatol. Venereol. 2012;139:452–458. doi: 10.1016/j.annder.2012.04.154. [DOI] [PubMed] [Google Scholar]
- Aransay A.M., Scoulica E., Tselentis Y. Detection and identification of Leishmania DNA within naturally infected sand flies by seminested PCR on minicircle kinetoplastic DNA. Appl. Environ. Microbiol. 1933-1938;2000:66. doi: 10.1128/aem.66.5.1933-1938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemiev M.M. A classification of the subfamily Phlebotominae. Parassitologia. 1991;33:69–77. [PubMed] [Google Scholar]
- Ayari C., Ben Othman S., Chemkhi J., Tabbabi A., Fisa R., Ben Salah A., BenAbderrazak S. First detection of Leishmania major DNA in Sergentomyia (Sintonius) clydei (Sinton, 1928, Psychodidae: Phlebotominae), from an outbreak area of cutaneous leishmaniasis in Tunisia. Infect. Genet. Evol. 2016;39:241–248. doi: 10.1016/j.meegid.2015.10.030. [DOI] [PubMed] [Google Scholar]
- Ben Ismail R., Gramiccia M., Gradoni L., Helal H., Ben Rachid M.S. Isolation of Leishmania major from Phlebotomus papatasi in Tunisia. Trans. R. Soc. Trop. Med. Hyg. 1987;81:749. doi: 10.1016/0035-9203(87)90018-6. [DOI] [PubMed] [Google Scholar]
- Benallal K.E., Benikhlef R., Garni R., Gassen B., Dedet J.P., Harrat Z. Presence of Phlebotomus perniciosus atypical form in Algeria. J. Arthropod. Borne Dis. 2017;11:139–146. [PMC free article] [PubMed] [Google Scholar]
- Ben-Ismail R., Helal H., Bach-Hamba D., Ben Rachid M.S. Natural infestation of Phlebotomus papatasi in a focus of zoonotic cutaneous leishmaniasis in Tunisia. Bull. Soc. Pathol. Exot. Filiales. 1987;80:613–614. [PubMed] [Google Scholar]
- Carta S., Sanna D., Scarpa F., Varcasia A., Cavallo L., Meloni M.P., Tamponi C., Cabras P.A., Dessi G., Casu M. Species diversity and molecular insights into phlebotomine sand flies in Sardinia (Italy)-an endemic region for leishmaniasis. Parasitol. Res. 2020;119:63–73. doi: 10.1007/s00436-019-06528-y. [DOI] [PubMed] [Google Scholar]
- Chelbi I., Kaabi B., Bejaoui M., Derbali M., Zhioua E. Spatial correlation between Phlebotomus papatasi Scopoli (Diptera: Psychodidae) and incidence of zoonotic cutaneous leishmaniasis in Tunisia. J. Med. Entomol. 2009;46:400–402. doi: 10.1603/033.046.0229. [DOI] [PubMed] [Google Scholar]
- Cornel A.J., Collins F.H. PCR of the ribosomal DNA intergenic spacer regions as a method for identifying mosquitoes in the Anopheles gambiae complex. Methods Mol. Biol. 1996;50:321–332. doi: 10.1385/0-89603-323-6:321. [DOI] [PubMed] [Google Scholar]
- Coutinho-Abreu I.V., Sonoda I.V., Fonseca J.A., Melo M.A., Balbino V.Q., Ramalho-Ortigao M. Lutzomyia longipalpis s.l. in Brazil and the impact of the Sao Francisco River in the speciation of this sand fly vector. Parasit Vectors. 2008;1:37. doi: 10.1186/1756-3305-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset H., Rioux J.A., Maistre M., Bayar N. The phlebotomines of Tunisia (Diptera-Phlebotominae). A revision of the systematics, distribution and behaviour (author’s transl) Ann. Parasitol. Hum. Comp. 1978;53:711–749. [PubMed] [Google Scholar]
- Depaquit J. Molecular systematics applied to Phlebotomine sand fies: review and perspectives. Infect. Genet. Evol. 2014;28:744–756. doi: 10.1016/j.meegid.2014.10.027. [DOI] [PubMed] [Google Scholar]
- Depaquit J., Leger N., Killick-Kendrick R. Description of Phlebotomus (Paraphlebotomus) riouxi n. sp. (Diptera:Psychodidae) of northern Africa. Parasite. 1998;5:151–158. doi: 10.1051/parasite/1998052151. [DOI] [PubMed] [Google Scholar]
- Desjeux P. The increase in risk factors for leishmaniasis worldwide. Trans. R. Soc. Trop. Med. Hyg. 2001;95:239–243. doi: 10.1016/s0035-9203(01)90223-8. [DOI] [PubMed] [Google Scholar]
- Ebrahimi S., Bordbar A., Parvizi P. Genetic dynamics in the sand fly (Diptera: Psychodidae) nuclear and mitochondrial genotypes: evidence for vector adaptation at the border of Iran with Iraq. Parasit. Vectors. 2016;9:319. doi: 10.1186/s13071-016-1603-510.1186/s13071-016-1603-5. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseghir S., Ready P.D., Killick-Kendrick R., Ben-Ismail R. Mitochondrial haplotypes and phylogeography of Phlebotomus vectors of Leishmania major. Insect Mol. Biol. 1997;6:211–225. doi: 10.1046/j.1365-2583.1997.00175.x. [DOI] [PubMed] [Google Scholar]
- Feliciangeli M.D. Natural breeding places of phlebotomine sand fies. Med. Vet. Entomol. 2004;18:71–80. doi: 10.1111/j.0269-283x.2004.0487.x. [DOI] [PubMed] [Google Scholar]
- Ghrab J., Rhim A., Bach-Hamba D., Chahed M.K., Aoun K., Nouira S., Bouratbine A. Phlebotominae (Diptera: Psychodidae) of human leishmaniosis sites in Tunisia. Parasite. 2006;13:23–33. doi: 10.1051/parasite/2006131023. [DOI] [PubMed] [Google Scholar]
- Guerbouj S., Chemkhi J., Kaabi B., Rahali A., Ben Ismail R., Guizani I. Natural infection of Phlebotomus (Larroussius) langeroni (Diptera: Psychodidae) with Leishmania infantum in Tunisia. Trans. R. Soc. Trop. Med. Hyg. 2007;101:372–377. doi: 10.1016/j.trstmh.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Guernaoui S., Hamarsheh O., Garcia D., Fontenille D., Sereno D. Population genetics of Phlebotomus papatasi from endemic and nonendemic areas for zoonotic cutaneous leishmaniasis in Morocco, as revealed by cytochrome oxidase gene subunit I sequencing. Microorganisms. 2020;8 doi: 10.3390/microorganisms8071010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouas N., Chaker E., Chargui N., Gorcii M., Belhadj S., Kallel K., Aoun K., Akrout F.M., Ben Said M., Pratlong F. Geographical distribution updating of Tunisian leishmaniasis foci: about the isoenzymatic analysis of 694 strains. Acta Trop. 2012;124:221–228. doi: 10.1016/j.actatropica.2012.08.012. [DOI] [PubMed] [Google Scholar]
- Izri M.A., Belazzoug S. Phlebotomus (Larroussius) perfiliewi naturally infected with dermotropic Leishmania infantum at Tenes, Algeria. Trans. R. Soc. Trop. Med. Hyg. 1993;87:399. doi: 10.1016/0035-9203(93)90011-e. [DOI] [PubMed] [Google Scholar]
- Kholoud K., Denis S., Lahouari B., El Hidan M.A., Souad B. Management of Leishmaniases in the era of climate change in Morocco. Int. J. Environ. Res. Public Health. 2018;15 doi: 10.3390/ijerph15071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholoud K., Bounoua L., Sereno D., El Hidan M., Messouli M. Emerging and re-emerging leishmaniases in the mediterranean area: what can be learned from a retrospective review analysis of the situation in Morocco during 1990 to 2010? Microorganisms. 2020;8 doi: 10.3390/microorganisms8101511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. Med. Vet. Entomol. 1990;4:1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Latrofa M.S., Iatta R., Dantas-Torres F., Annoscia G., Gabrielli S., Pombi M., Gradoni L., Otranto D. Detection of Leishmania infantum DNA in phlebotomine sand flies from an area where canine leishmaniosis is endemic in southern Italy. Vet. Parasitol. 2018;253:39–42. doi: 10.1016/j.vetpar.2018.02.006. [DOI] [PubMed] [Google Scholar]
- Lewis D.J. The biology of Phlebotomidae in relation to leishmaniasis. Annu. Rev. Entomol. 1974;19:363–384. doi: 10.1146/annurev.en.19.010174.002051. [DOI] [PubMed] [Google Scholar]
- Lewis D.J., Hitchocock J.C., Jr. Phlebotomine sand fies of Chad. Ann. Trop. Med. Parasitol. 1968;62:117–121. doi: 10.1080/00034983.1968.11686537. [DOI] [PubMed] [Google Scholar]
- Mahamdallie S.S., Pesson B., Ready P.D. Multiple genetic divergences and population expansions of a Mediterranean sand fly, Phlebotomus ariasi, in Europe during the Pleistocene glacial cycles. Heredity (Edinb) 2011;106:714–726. doi: 10.1038/hdy.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moo-Llanes D., Ibarra-Cerdena C.N., Rebollar-Tellez E.A., Ibanez-Bernal S., Gonzalez C., Ramsey J.M. Current and future niche of North and Central American sand flies (Diptera: psychodidae) in climate change scenarios. PLoS Negl. Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002421PNTD-D-13-00174. (pii) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotey S., Mahamdallie S.S., Pesson B., Richardson K.J., Gallego M., Ready P.D. Postglacial dispersal of Phlebotomus perniciosus into France. Parasite. 2005;12:283–291. doi: 10.1051/parasite/2005124283. [DOI] [PubMed] [Google Scholar]
- Pesson B., Ready J.S., Benabdennbi I., Martin-Sanchez J., Esseghir S., Cadi-Soussi M., Morillas-Marquez F., Ready P.D. Sand fies of the Phlebotomus perniciosus complex: mitochondrial introgression and a new sibling species of P. longicuspis in the Moroccan Rif. Med. Vet. Entomol. 2004;18:25–37. doi: 10.1111/j.0269-283x.2004.0471.x. [DOI] [PubMed] [Google Scholar]
- Pimenta P.F.P., Saraiva E.M.B., Rowton E., Modi G.B., Garraway L.A., Beverley S.M., Turco S.J., Sacks D.L. Evidence that the vectorial competence of phlebotomine sand flies for different species of Leishmania is controlled by structural polymorphisms in the surface lipophosphoglycan. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9155–9159. doi: 10.1073/pnas.91.19.9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pousse H., Besbes A., Ben Said M., Ghenimi L., Kharrat H. Epidemiology of human visceral leishmaniasis in Tunisia. J. Trop. Pediatr. 1995;41:191–192. doi: 10.1093/tropej/41.3.191. [DOI] [PubMed] [Google Scholar]
- Ready P.D. Biology of phlebotomine sand flies as vectors of disease agents. Annu. Rev. Entomol. 2013;58:227–250. doi: 10.1146/annurev-ento-120811-153557. [DOI] [PubMed] [Google Scholar]
- Rioux J.A., Moreno G., Lanotte G., Pratlong F., Dereure J., Rispail P. Two episodes of cutaneous leishmaniasis in man caused by different zymodemes of Leishmania infantum s.l. Trans. R. Soc. Trop. Med. Hyg. 1986;80:1004–1005. doi: 10.1016/0035-9203(86)90300-7. [DOI] [PubMed] [Google Scholar]
- Sneath P.H., Sokal R.R. Numerical taxonomy. Nature. 1962;193:855–860. doi: 10.1038/193855a0. [DOI] [PubMed] [Google Scholar]
- Tabbabi A., Bousslimi N., Rhim A., Aoun K., Bouratbine A. First report on natural infection of Phlebotomus sergenti with Leishmania promastigotes in the cutaneous leishmaniasis focus in southeastern Tunisia. Am. J. Trop. Med. Hyg. 2011;85:646–647. doi: 10.4269/ajtmh.2011.10-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Ximenes M.F., Souza M.F., Castellon E.G. Density of sand flies (Diptera: psychodidae) in domestic and wild animal shelters in an area of visceral Leishmaniasis in the state of Rio Grande do Norte, Brazil. Mem. Inst. Oswaldo Cruz. 1999;94:427–432. doi: 10.1590/s0074-02761999000400001. [DOI] [PubMed] [Google Scholar]
- Yahia H., Ready P.D., Hamdani A., Testa J.M., Guessous-Idrissi N. Regional genetic differentiation of Phlebotomus sergenti in three Moroccan foci of cutaneous leishmaniasis caused by Leishmania tropica. Parasite. 2004;11:189–199. doi: 10.1051/parasite/2004112189. [DOI] [PubMed] [Google Scholar]
- Yavar R., Hadi K., Reza A.M., Hasan B., Ali O.M., Sina R., Habib B.H., Abodolrahim H., Manuchehr G. First detection of Leishmania infantum DNA in wild caught Phlebotomus papatasi in endemic focus of cutaneous leishmaniasis, South of Iran. Asian Pac. J. Trop. Biomed. 2013;3:825–829. doi: 10.1016/S2221-1691(13)60163-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of captured sand flies.