Summary
This protocol for the separation of nuclear and cytoplasmic fractions of cells of Xenopus laevis embryos was developed to study changes in the intracellular localization of the Zyxin and Ybx1 proteins, which are capable of changing localization in response to certain stimuli. Western blot analysis allows the quantification of changes in the distribution of these proteins between the cytoplasm and nucleus, whereas the posttranslational modifications specific to each compartment can be identified by changes in electrophoretic mobility.
For complete details on the use and execution of this protocol, please refer to Parshina et al. (2020).
Subject areas: Cell Biology, Cell separation/fractionation, Model Organisms, Molecular Biology, Protein Biochemistry
Graphical abstract

Highlights
-
•
A simple way to obtain tagged proteins in Xenopus embryos
-
•
Rapid separation of Xenopus embryonic cells into nuclear, cytoplasmic fractions
-
•
Quantifying the distribution of shuttle proteins between the cytoplasm and nucleus
-
•
Protein posttranslational modifications specific to the cytoplasm and nucleus can be studied
This protocol for the separation of nuclear and cytoplasmic fractions of cells of Xenopus laevis embryos was developed to study changes in the intracellular localization of the Zyxin and Ybx1 proteins, which are capable of changing localization in response to certain stimuli. Western blot analysis allows the quantification of changes in the distribution of these proteins between the cytoplasm and nucleus, while the posttranslational modifications specific to each compartment can be identified by changes in electrophoretic mobility.
Before you begin
In this protocol, proteins of interest are translated on synthetic mRNA templates encoding these proteins and injected into Xenopus laevis embryos. Thus, all interactions, modifications, and changes in the localization of these proteins occur in vivo in the cells of the developing Xenopus laevis embryo. Synthetic RNAs are injected at the two-blastomere stage, followed by incubation until the mid-gastrula stage. Fractionation of the crude lysate into nuclear and cytoplasmic extracts and analysis by immunochemical methods (Western, ELISA) makes it possible to quantify changes in the distribution of the studied proteins between the cytoplasm and nucleus. In addition, the use of Xenopus laevis embryos developing in the external environment allows the use of various stimuli: temperature fluctuations, chemical agents, and UV radiation applied to whole embryos or explants obtained from different parts of the embryo.
Using this protocol to study the intracellular distribution of exogenous proteins, cytoskeletal protein Zyxin and Myc-tagged transcriptional regulator Ybx1, a Y-box factor known to be able to protect mRNA from degradation due to its chaperone activity, it was shown that Zyxin can retain Ybx1 in the cytoplasm, preventing its translocation into the nucleus.
In addition, the method allows experimentation with a small amount of material, for example, embryonic explants. This benefit enables comparisons of the intracellular distribution of shuttle proteins that are expressed ubiquitously in different regions of the developing embryo.
An important advantage of this protocol is in its adaptation for simple and reliable separation of nuclear and cytoplasmic fractions, which allows one to study the distribution of endogenous and exogenous shuttle proteins in the Xenopus embryo cells in response to various experimental stimuli.
Obtaining the embryos and microinjecting synthetic RNAs are completed following the protocols in N.M., unpublished data.
Preparation of stock solutions for separation of nuclear and cytoplasmic fractions
Timing: 1–2 h, on the day of the experiment
For the buffers to be used for lysis, washing, and nuclear extraction, we usually use buffers from stock solutions that can be prepared in advance and store them for several months. The solutions must be mixed on the day of the experiment, and protease inhibitors should be added to the buffers just before use in an experiment.
The required materials and equipment are presented in full in the Key Resources Table and in Materials and Equipment, Experimental Model and Subject Details sections.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Zyxin 1:500 |
A.G. Zaraisky Lab | (Martynova et al., 2008) |
| Mouse monoclonal anti-c-Myc, AP-conjugated 1:1000 (for WB) | Sigma-Aldrich | Cat#A5963; RRID:AB_258265 |
| Anti-rabbit IgG, AP-conjugated, produced in goat 1:30000 (for WB) | Sigma-Aldrich | Cat#A3937; RRID:AB_258122 |
| Mouse monoclonal anti-alpha-tubulin 1:1000 (for WB) | Sigma-Aldrich | Cat#T9026; RRID:AB_477593 |
| Anti-mouse IgG, AP-conjugated 1:1000 (for WB) |
Sigma-Aldrich | Cat#A3562; RRID:AB_258091 |
| Chemicals, peptides, and recombinant proteins | ||
| Human chorionic gonadotropin | Sigma-Aldrich | Cat#CG10 |
| Western Blue® stabilized substrate for alkaline phosphatase | Promega | Cat#S3841 |
| Sodium chloride (NaCl) | Helicon | Cat#H-1401 |
| Potassium chloride (KCl) | Helicon | Cat#H-1101 |
| Сalcium chloride dihydrate (CaCl2∗2H2O) | Amresco | Cat#0556 |
| Magnesium chloride hexahydrate (MgCl₂∗6H₂O) | Panreac | Cat#141396 |
| HEPES | Panreac | Cat#A3724 |
| Sucrose | Sigma-Aldrich | Cat#S9378 |
| L-Cysteine | Dia-m | Cat#M52904 |
| Ficoll | Dia-m | Cat#PS400 |
| Sodium hydroxide (NaOH) | Dia-m | Cat#145881 |
| Ethanol | N/A | N/A |
| Chloroform | N/A | N/A |
| Methanol | N/A | N/A |
| Igepal Nonidet P-40 | Sigma-Aldrich | Cat#I-30201 |
| DTT | Fermentas | Cat#RO861 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340 |
| Tris | Helicon | Cat#Am-O497 |
| Acrylamide 2K | Helicon | Cat#H-0104 |
| Bis-Acrylamide | Helicon | Cat#Am-O172 |
| Ammonium peroxodisulfate ((NH4)2S2O6(O2)) | Merck | Cat#1.01201 |
| TEMED (N,N,N′,N′-tetramethylethylenediamine) | Helicon | Cat#L-0847 |
| EDTA | Helicon | Cat#Am-O105B |
| Glycerol | Helicon | Cat#I-800687 |
| Sodium dodecyl sulfate (SDS) powder | Sigma-Aldrich | Cat# L4509 |
| Tween-20 | Helicon | Cat# Am-O777 |
| Fluorescein lysine dextran (FLD) 40 kD | Invitrogen | Cat#D1845 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat#M3148 |
| Experimental models: organisms/strains | ||
| Wild-type Xenopus laevis frogs (males and females aged 1–3 years) | Nasco | Cat#LM00456; RRID:XEP_Xla100 |
| Software and algorithms | ||
| ImageJ | N/A | https://imagej.nih.gov/ij/ |
| Other | ||
| 2 mL Syringe with 23G×1¼” needle | Medical Products | Cat#V160218 |
| Mini-centrifuge-vortex Microspin | BioSan | Cat#FV-2400 |
| Flake ice machine | Porkka | Cat#KF45 |
| Surgical scissors | N/A | N/A |
| Forceps | N/A | N/A |
| Microcentrifuge | Eppendorf | Cat#5415 |
| Thermostat “Gnom” | DNA-Technology | N/A |
| Microcentrifuge MiniSpin | Eppendorf | Cat#5452 000 |
| Implen NanoPhotometer С | Implen | Cat# C40∗ |
| Pipetmans | Gilson | N/A |
| Nutator | Clay Adams | Cat#421105 |
| Electrophoresis power supply | Amersham Pharmacia Biotech | Cat#EPS 301 |
| Hoefer Mighty Small Dual Mini Gel Caster | Hoefer | N/A |
| Hoefer Mighty Small II Mini Vertical Electrophoresis System | Hoefer | N/A |
| Amersham ECL Semi-Dry Blotters | Amersham Biosciences | Cat#TE77PWR |
| Microinjector FemtoJet | Eppendorf | Cat# 5253000017 |
| Petri dish | Greiner | Cat#628102 |
| Terasaki plate | Greiner | Cat#659190 |
| Serum pipette | Greiner | Cat#612301 |
| Microcentrifuge tubes | SSI | Cat#1260-00 |
| Pipette tips | SSI | N/A |
Materials and equipment
| NaOH | Final concentration | Amount |
|---|---|---|
| NaOH | 1 M | 20 g |
| Milli-Q water | To a final volume of 50 mL | |
| Total | 1 M | 50 mL |
Can be stored at room temperature for several months.
| 20× MMR | Final concentration | Amount |
|---|---|---|
| NaCl | 2 M | 58.45 g |
| KCl | 0.04 M | 1.43 g |
| CaCl2∗2H2O | 0.04 M | 2.94 g |
| MgCl₂∗6H₂O | 0.02 M | 2.03 g |
| Milli-Q water | To a final volume of 500 mL | |
| Total | n/a | 500 mL |
Can be stored at +4°C for several months.
| 200× Hepes | Final concentration | Amount |
|---|---|---|
| Hepes | 1 M | 119.15 g |
| Milli-Q water | To a final volume of 500 mL | |
| Total | 1 M | 500 mL |
Adjust pH to 7.4 with NaOH. Can be stored at +4°C for several months.
| 1× MMR | Final concentration | Amount |
|---|---|---|
| 20× MMR | N/A | 5 mL |
| 200× Hepes | 5 mM | 0.5 mL |
| ddH2O | To a final volume of 100mL | |
| Total | n/a | 100 mL |
Can be stored at +4°C for several months.
| 0,1× MMR | Final concentration | Amount |
|---|---|---|
| 20× MMR | N/A | 5 mL |
| 200× Hepes | 5 mM | 5 mL |
| ddH2O | To a final volume of 1000 mL | |
| Total | n/a | 1000 mL |
Can be stored at room temperature for several months.
| L-Cysteine | Final concentration | Amount |
|---|---|---|
| L-Cysteine | 2% (w/v) | 2 g |
| 0,1× MMR | To a final volume of 100 mL | |
| Total | n/a | 100 mL |
Adjust pH to 7.8 with NaOH. Prepare on the day of microinjection.
| Ficoll | Final concentration | Amount |
|---|---|---|
| Ficoll | 4% (w/v) | 4 g |
| 0,1× MMR | To a final volume of 100 mL | |
| Total | n/a | 100 mL |
Can be stored at +4°C for a month.
| 50× FLD | Final concentration | Amount |
|---|---|---|
| Fluorescein Lysine Dextran | 50 μg/μL | 25 mg |
| Milli-Q water | n/a | 500 μL |
| Total | 50 μg/μL | 500 μL |
Can be stored at −20°C for several years.
| KCl | Final concentration | Amount |
|---|---|---|
| KCl | 1 M | 3.72 g |
| Milli-Q water | To a final volume of 50 mL | |
| Total | 1 M | 50 mL |
Can be stored at room temperature for several years.
| MgCl2 | Final concentration | Amount |
|---|---|---|
| MgCl₂∗6H₂O | 2 M | 20.33 g |
| Milli-Q water | To a final volume of 50 mL | |
| Total | 2 M | 50 mL |
Can be stored at +4°C for several years.
| NaCl | Final concentration | Amount |
|---|---|---|
| NaCl | 5 M | 14.61 g |
| Milli-Q water | To a final volume of 50 mL | |
| Total | 5 M | 50 mL |
Can be stored at +4°C for several years.
| DTT | Final concentration | Amount |
|---|---|---|
| DTT | 1 M | 1.54 g |
| Milli-Q water | To a final volume of 10 mL | |
| Total | 1 M | 10 mL |
Can be stored 1 mL aliquots at −20°C for several years.
| Tris-HCl | Final concentration | Amount |
|---|---|---|
| Tris | 1 M | 6.05 g |
| Milli-Q water | To a final volume of 50 mL | |
| Total | 1 M | 50 mL |
Adjust pH to 6.5 with HCl. Can be stored at +4°C for several years.
| Buffer N | Final concentration | Amount |
|---|---|---|
| 200× Hepes | 20 mM | 20 μL |
| Sucrose | 2% (w/v) | 0.02 g |
| KCl (1 M) | 10 mM | 10 μL |
| MgCl2 (2 M) | 1.5 mM | 3 μL |
| EDTA (0,5 M) | 0.2 mM | 0.4 μL |
| DTT (1 M) | 0.5 mM | 0.5 μL |
| Protease Inhibitor Cocktail | 2% (v/v) | 20 μL |
| Milli-Q water | To a final volume of 1 mL | |
| Total | n/a | 1 mL |
Prepare on the day of experiment.
| Buffer E | Final concentration | Amount |
|---|---|---|
| 200× Hepes | 20 mM | 20 μL |
| Sucrose | 2% (w/v) | 0.02 g |
| KCl (1 M) | 150 mM | 150 μL |
| MgCl2 (2 M) | 1.5 mM | 3 μL |
| EDTA (0,5 M) | 0.2 mM | 0.4 μL |
| DTT (1 M) | 0.5 mM | 0.5 μL |
| Protease Inhibitor Cocktail | 2% (v/v) | 20 μL |
| NP-40 | 0.5% (v/v) | 5 μL |
| Milli-Q water | To a final volume of 1 mL | |
| Total | n/a | 1 mL |
Prepare on the day of experiment.
| EDTA | Final concentration | Amount |
|---|---|---|
| EDTA | 500 mM | 9.31 g |
| Milli-Q water | To a final volume of 50 mL | |
| Total | 500 mM | 50 mL |
Adjust pH to 8.0 with NaOH. Can be stored at +4°C for several years.
| 4× Laemmli buffer | Final concentration | Amount |
|---|---|---|
| Glycerol | 10% (v/v) | 1 mL |
| Tris-HCl (1M, pH 6.5) | 125 mM | 1.25 mL |
| SDS | 8% (w/v) | 0.8 g |
| 2-Mercaptoethanol | 5% (v/v) | 500 μL |
| Milli-Q water | To a final volume of 10 mL | |
| Total | n/a | 10 mL |
Can be stored 1 mL aliquots at −20°C for several years.
Experimental model and subject details
Mature Xenopus laevis frogs (males and females aged 1–3 years) were obtained from NASCO (Fort Atkinson, WI) and were maintained in a recirculating tank system with regularly monitored temperature and water quality at 18°C. All experimental protocols involving frogs were performed in accordance with guidelines approved by the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry (Moscow, Russia) Animal Committee and handled in accordance with the Animals (Scientific Procedures) Act 1986 and Helsinki Declaration.
Step-by-step method details
Preparation of crude extract from embryonic cells
Timing: 1–2 h for embryo injection, and 16–20 h for incubation, followed by 30 min of crude extract preparation
-
1.Xenopus embryos at the 2–4 cell stage are injected with synthetic mRNAs (300–400 pg per blastomere). Usually, we inject 30 embryos with each injection mixture. Synthetic RNAs are microinjected according to the protocol of N.M., unpublished data. The main stages of microinjection are shown in Figure 1.
-
a.Embryos are incubated up to the stage needed for the research. In our work, we incubate embryos up to stage 12 gastrula. Photos of embryos at the stage of the onset of cleavage (two blastomeres) and the gastrula stage are shown in Figures 1E–1GNote: Working with X. laevis embryos is convenient because their development can be controlled by temperature. Using the development table (Nieuwkoop and Faber, 1994), the incubation time can be adjusted by changing the temperature. Usually, at room temperature (18°C–20°C), embryos injected at the 2-blastomere stage reach the gastrula stage by the morning of the next day.
-
b.After incubation to the gastrula stage, the embryos are pretreated by incubation with 0.1× MMR containing 150 mg/mL cycloheximide for 1 h at 23°C in a petri dish (Lemaitre et al., 1995).
-
a.
-
2.
Live embryos are carefully collected in a 1.5-mL Eppendorf tube, and all the liquid is carefully collected using a pipette with a fine tip (see Figure 2A).
CRITICAL: Dead embryos appear as white balls with no clearly visible cells (see Figure 1H). In the case of microinjections, FLD should also be selected because it is a vital dye observable with a binocular fluorescence microscope. (The use of FLD is described in more detail in the protocol N.M., unpublished data ). Usually, approximately 10% of the embryos are lost at this stage, depending on the quality of the eggs and the properties of the proteins that are encoded by the injected RNA. As noted, incubation in cycloheximide for one hour does not lead to increased embryonic death. In the case where few embryos are viable, this stage can be omitted, but when included, this step provides a clearer picture of protein distribution.
Figure 1.
The main steps of embryo preparation
(A) Fertilized eggs before cleavage begins.
(B) Embryos in Terasaki plates before manipulation.
(C)Capillary preparation for microinjection.
(D) Microinjection in embryos: general view of the workspace.
(E) Two-cell stage embryo.
(F) Embryo at the gastrula stage (stage 12), top (animal) view.
(G) Embryo at the gastrula stage (stage 12), bottom (vegetal) view.
(H) Alive (four on the left) and dead (two on the right, indicated by a red dashed line) embryos.
Scale bars are 500 μm.
Figure 2.
The main steps of extracts preparation
(A) 60 embryos collected in Eppendorf tube to obtain crude extract (step 2)
(B) Crude extract after centrifugation of embryos (step 3)
(C) Crude extract, diluted 10 times with buffer N, before centrifugation
(D) Separated fractions: left -cytoplasmic fraction, right - the pellet containing the cell nuclei (step 4)
(E) Separated fractions before washing: on the left, the cytoplasmic fraction in buffer E (150 mM KCl), on the right, the nuclei in buffer with 0.8 M sucrose (step 5).
(F) Fractions after extraction in buffer E and centrifugation: on the left is a cytoplasmic extract without any pellet, on the right is a nuclear extract containing a pellet of pigment and insoluble material.
 Pause point: The resulting embryos can be frozen and stored at −70°C for several weeks.
Pause point: The resulting embryos can be frozen and stored at −70°C for several weeks.
-
3.
The crude extract is obtained by centrifuging the Eppendorf tube containing embryos for 10 min at 14000 g and 4°C. (see Figure 2B).
The resulting extract (30 μL from 30 embryos) is diluted 10-fold in N-buffer (300 μL) with the addition of a cocktail of protease inhibitors (Sigma-Aldrich, Critical Reagent) at a dilution of 1:50. (see Figure 2C).
-
4.
The nuclei are pelleted by 8-min centrifugation at 5000 g and 4°C, nuclei are in the pellet, and the cytoplasmic fraction is in the supernatant. (see Figure 2D).
Nuclear extract preparation
Timing: 90 min
-
5.
The pellet containing the cell nuclei is washed with a solution of 0.8 M sucrose (0.253 g of sucrose is added per 1 mL of buffer N), the volume of the wash is 0.5–1 mL, and it is centrifuged at 14000 g 15 min at 4°C. Washes can be repeated from two to four times.
CRITICAL: The most critical point is the correct concentration of sucrose (Critical Reagent) in the buffer for use in the primary precipitation of nuclei and nuclei washing. Loss of nuclei is possible if the concentration of sucrose in the wash buffer is too high.
-
6.
To obtain the nuclear extract, the washed nuclei are extracted in 300 μL of buffer E with a cocktail of protease inhibitors (Sigma-Aldrich) at a dilution of 1:50. Buffer E is buffer N with KCl increased to 150 mM (150 μL 1 M KCl per 1 mL of buffer N) and the addition of NP-40 0.5% (5 μL per 1 mL of buffer N) placed on a rotator for 45 min at 4°C.
-
7.
The nuclear extract is centrifuged at 14000 g for 10 min at 4°C.
Note: The resulting supernatant is the nuclear extract and can be used for various tests, EMSA, etc. In our study, we concentrate the extract using chloroform-methanol (step 9) precipitation of the proteins and analyze the precipitates by Western blotting according to the standard protocol.
Preparation of cytoplasmic extract
Timing: 1 h
-
8.
The supernatant from step 4 (300 μL) is used to prepare a cytoplasmic extract. To clarify the extract and remove nuclei the most thoroughly, the KCl concentration in the supernatant is increased to 150 mM (150 μL 1 M KCl per 1 mL of buffer N) and centrifuged for 10 min at 14000 g at 4°C.
The resulting supernatant can be used for the preparation of Western samples, as it contains all the cytoplasmic proteins.
Note: For further extraction of cytoplasmic proteins in the case of a functional study and for the removal of insoluble materials, the supernatant is diluted 2 times with buffer E: the KCl concentration in the supernatant is increased to 150 mM (150 μL 1 M KCl per 1 mL of buffer N), and 1% NP-40 is added (10 μL per 1 mL). The diluted extract is incubated for 45 min on a rotator and centrifuged for 10 min at 14000 g at 4°C. The resulting supernatant is the cytoplasmic extract.
Concentration of samples for Western analysis
Timing: 2 h
-
9.To concentrate the samples, the protein fraction from the extracts is precipitated with methanol-chloroform:
-
a.methanol and chloroform are added to the resulting extract in equal volumes. Typically, 30 embryos produce 300 μL of nuclear extract and 300 μL of cytoplasmic extract.( see Figure 3A)Therefore, 300 μL of methanol is added to 1.5-mL Eppendorf tubes containing either extract and mixed by inversion, and then, 300 μL of chloroform is added, and it is shaken vigorously by hand or by vortexing.
-
b.centrifuge the fraction for 5 min at 10000 g. The mixture is divided into two phases; the precipitated protein is between the phases in the form of a film floating on the phase boundary. (Figure 3B)
-
c.the upper methanol-chloroform phase is carefully removed to avoid removing the film.
-
d.a triple volume of methanol (900–1000 μL) is added to the Eppendorf tube and centrifuged for 5 min at 10000 g.
-
e.the proteins collected at the bottom of the tube appear as a white precipitate. (Figure 3C)
-
f.the protein precipitate is air-dried for 1–2 h, depending on the mass of the pellet.
 CRITICAL: The protein precipitate should be dried for no more than 120 min. It is important to not let the precipitate air dry for a long time period, as this could negatively impact protein solubilization.
CRITICAL: The protein precipitate should be dried for no more than 120 min. It is important to not let the precipitate air dry for a long time period, as this could negatively impact protein solubilization. -
g.the dried pellet is dissolved in 100 μL of 2× Laemmli SDS sample buffer. The samples are boiled for 10 min at 80°C and stored at −20°C until use.
-
h.the samples are analyzed by SDS-PAGE followed by Western blotting. Detection is performed with the following antibodies: monoclonal anti-Myc alkaline phosphatase conjugated antibodies (Sigma) for Myc-tagged Ybx1 detection and anti-Zyxin rabbit polyclonal monospecific antibodies for Zyxin detection (Martynova et al., 2008).
-
a.
Note: Samples for Western blotting can be made from extracts without precipitation. To do this, a 50-μL aliquot is taken from both extracts and an equal volume of 4× Laemmli buffer is added to them. The sample volume for Western blot analysis depends on the volume of the well, and we usually load 20–25 μL samples onto a 1.5-mm gel using 10-well gel combs.
Figure 3.
Methanol/chloroform protein precipitation
(A) The resulting nuclear (right) and cytoplasmic (left) extracts.
(B) Methanol/chloroform precipitation. The protein layer appears on the interface between the methanol/water and chloroform phases (step 9, b).
(C) The protein pellet is collected at the bottom of the tube (step 9,e).
 Pause point: Samples in Laemmli buffer can be stored for several months.
Pause point: Samples in Laemmli buffer can be stored for several months.
Expected outcomes
This protocol makes it easy to analyze the distribution of the studied proteins between the nucleus and cytoplasm during the development of Xenopus laevis embryos. In addition to allowing quantification of the distribution of proteins by measuring the integrated density of the Western blot bands, it also allows analyses of various posttranslational protein modifications specific to the cytoplasm and nucleus by enabling observation of the alterations manifested by the electrophoretic mobility of the proteins in the corresponding fractions. We present some examples of how the described protocol is used in our laboratory.
Earlier, we indicated that coexpression of Zyxin in Xenopus laevis embryonic cells with the Sonic hedgehog (Shh) nuclear effector Gli1 resulted in decreased Shh signaling cascade activity (Martynova et al., 2013). To confirm this result, we determined the nuclear-to-cytoplasmic ratio of the fluorescent signal emitted by Myc-Gli1 or EGFP-Zyxin in Xenopus laevis fibroblasts transfected with either or both of the respective plasmids. This experiment was very difficult since the nuclear-to-cytoplasmic fluorescence density ratio (DFR) must be calculated for many cells based on transfection type in two independent experiments.
Taking this experience into account in subsequent next work (Martynova et al., 2018), we developed and used the presented protocol and showed that Zyxin can stabilize the complex formed by Zic1 with Gli1. The formation of this Zic1-carrying complex leads to the nuclear accumulation of Gli1 and Zyxin; however, an excess of Zyxin leads to the retention of Gli1 in the cytoplasm.
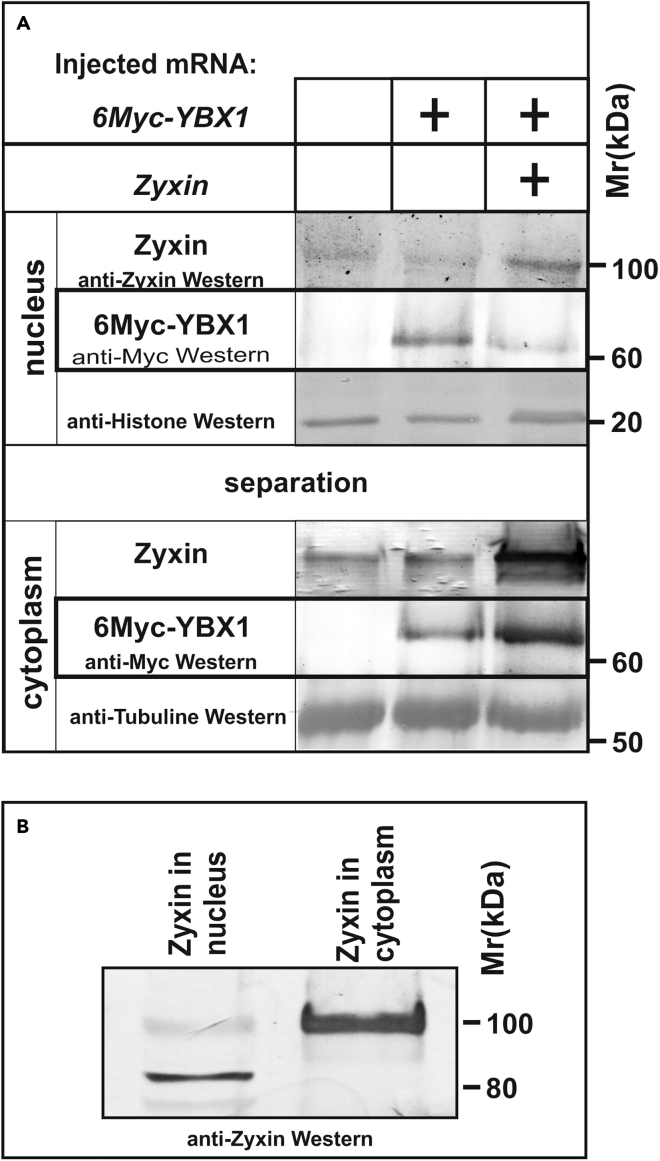
In a recent work, when studying the interaction of Zyxin with Ybx1, we demonstrated that when Zyxin interacts with Myc-tagged Ybx1, it retains Ybx1 in the cytoplasm, thus reducing its concentration in the nucleus (Figure 4A) (Parshina et al., 2020).
Figure 4.
Examples of using the protocol for separating the nuclear and cytoplasmic fractions to study changes in the localization and mobility of the cytoskeletal protein Zyxin and its effect on the localization of the transcription factor Ybx1
(A) While Zyxin reduces the nuclear concentration of 6Myc-Ybx1, it increases the cytoplasmic concentration of 6Myc-Ybx1. Xenopus laevis embryos at the 2–4 cell stage were injected with either 6 myc-ybx1 (200 pg per blastomere) and zyxin (300 pg per blastomere) or only 6 myc-ybx1 mRNA (200 pg per blastomere) and incubated at 18°C until the late gastrula stage. Nuclear and cytoplasmic fractions and precipitated samples were prepared as described in the protocol. The samples were analyzed by SDS-PAGE in a 10% gel according to the method of Laemmli and electroblotted onto PVDF membranes. Detection was performed with the following antibodies: monoclonal anti-Myc alkaline phosphatase conjugated antibodies (Sigma) for Myc-tagged protein Ybx1 and anti-Zyxin rabbit polyclonal monospecific antibodies for Zyxin (Figure reprinted with permission from Parshina et al. 2020).
(B) Changes in the mobility of the band corresponding to Zyxin upon separation into nuclear and cytoplasmic fractions. An equal volume of 4× SDS Laemmli sample buffer was added to 50 μL of the obtained nuclear and cytoplasmic extracts, and then, the samples were boiled for 10 min at 80°C. The samples were analyzed by SDS-PAGE in 7.5% Laemmli gels and electroblotted onto a PVDF membrane. Detection was carried out using rabbit polyclonal monospecific antibodies against the C-terminal Lim domain of Zyxin.
In addition, when analyzing the distribution of endogenous Zyxin between the nucleus and cytoplasm of Xenopus laevis embryonic cells during the latter work, we found that the nuclear band of Zyxin was much shorter than that of its cytoplasmic counterpart (Figure 4B). This result confirms the data reported for Zyxin in cells in porcine wound tissues during healing (Sabino et al., 2020). As the authors of this paper established, this shortening of nuclear Zyxin is the result of its site-specific proteolysis.
It is important that, in this study, we realized that it was possible to work with very small amounts of embryonic implants as the study material.
This example demonstrates how our protocol allows the detection of posttranslational modifications of proteins in Xenopus laevis embryo cells.
Quantification and statistical analysis
For statistical analysis of Zyxin or Ybx1 amounts, integral densities of the bands, corresponding to certain protein and α-Histone or α-Tubulin, were measured using ImageJ software (Schneider et al., 2012). The expression levels of Zyxin were determined using the ImageJ program for a band with a molecular weight of 100 kD. Data obtained in at least three independent experiments were analyzed in Microsoft Excel: the ratios of the integral densities of the protein and a reference proteins bands were analyzed using Student’s t test in Excel. The n’s and p values for all statistical tests can be found in the corresponding figure legends.
Limitations
Due to the simplicity of the protocol, there are practically no restrictions on its use. This protocol can be applied to cell cultures, tissue cultures, whole embryos, and embryonic explants. Even 1-2 Xenopus laevis embryos or 6-10 excised explants are sufficient for reliable preparation of the extract using this protocol. The main limitation is the concentration of sucrose used during the precipitation and washing of the nuclei. This protocol specifies the sedimentation conditions for cell nuclei obtained from Xenopus laevis embryos. It may be necessary to adjust the sucrose concentration and centrifugation conditions when washing the nuclei obtained from other eukaryotic organisms.
Troubleshooting
Problem 1
Problems associated with obtaining embryos (step 1).
Potential solution
Possible problems associated with obtaining embryos are discussed in the protocol: Immunoprecipitation protocol for searching mRNAs associated with mRNA-binding proteins in the Xenopus laevis embryos.
Problem 2
Weak expression of exogenous proteins translated from synthetic mRNAs injected into embryos (step 9h).
Potential solution
As a result of checking the expression of the tagged protein in the lysate of embryonic cells using Western blotting, if there is no band on the blot, or the band does not correspond to the expected molecular weight of the protein, it is necessary:
First, check by sequencing the vector that was used for in vitro transcription for reading frame errors, stop codons, and the presence of a sequence for the synthesis of poly-A, Kozak consensus, etc.
Second, check for the presence of restriction enzymes used to linearize the vector in frame.
Third, check the quality and quantity of synthetic RNA after in vitro transcription.
Forth, use a cap analog for transcription.
Tag size and location (N or C terminus) can have an impact in protein folding and/or function and should be taken into consideration.
Problem 3
Problems with protein detection — there are no or very weak bands with a molecular weight corresponding to the target protein on the membrane upon Western blot detection, but the physiological effects of the protein expression are normal (step 9h).
Potential solution
First, this result may be due to the dilution of the antibodies, which is especially important for specific antibodies used to detect endogenous proteins, provided that the samples are concentrated. Second, it is necessary to compare the quality of the transfer to the PVDF membrane to the molecular weight standards (it is convenient to use colored standards). Third, the quality of the Western blot substrate should be confirmed.
Problem 4
Poor quality of separation of the crude lysate — cytoplasmic markers are detected in the nuclear fraction, and nuclear markers are detected in the cytoplasmic fraction (steps 5 and 8).
Potential solution
For a good-quality cytoplasmic extract, the centrifugation parameters and sucrose concentration must be carefully checked when the crude extract is first diluted. To improve quality, we recommend additional centrifugation of the samples in a buffer with 150 mM KCl. For the nuclear extract, additional washes in 0.8 M sucrose can be performed.
Problem 5
When concentrating samples for Western blotting using the chloroform-methanol protein precipitation method, no protein film is visible at the interface (step 9b).
Potential solution
This occurs when a small amount of the studied material is used, for example, when using only 2-3 embryos or explants. However, this does not mean that it is impossible to detect proteins. However, it is necessary to discard the upper phase incompletely to avoid touching the invisible protein film. Then, three volumes of methanol (in relation to the volume remaining in the tube) are added, the tube is centrifuged, and the sample is dried and prepared for Western blotting.
Resource availability
Lead contact
Further information and requests for and reagents should be directed to and will be fulfilled by the lead contact, Natalia Martynova (martnat61@gmail.com).
Materials availability
No materials were generated in this study.
Data and code availability
No data or code was generated in this study.
Acknowledgments
This work was supported by RFBR grants: 20-34-90017 (N.Y.M.), 18-04-00674 А (N.Y.M.), and RSCF grant 19-14-00098 (A.G.Z.). We thank Fedor Eroshkin and Eugeny Orlov for the lab photos creation.
Author contributions
Conceptualization, N.Y.M.; methodology, N.Y.M., E.A.P., and A.G.Z.; investigation, E.A.P. and N.Y.M.; writing original draft, N.Y.M. and E.A.P.; writing review and editing, N.Y.M. and A.G.Z.; funding acquisition, A.G.Z. and N.Y.M.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Natalia Y. Martynova, Email: martnat61@gmail.com.
Andrey G. Zaraisky, Email: azaraisky@yahoo.com.
References
- Lemaitre J.M., Bocquet S., Buckle R., Mechali M. Selective and rapid nuclear translocation of a c- Myc-containing complex after fertilization of Xenopus laevis eggs. Mol. Cell. Biol. 1995;15:5054–5062. doi: 10.1128/mcb.15.9.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynova N.Y., Eroshkin F.M., Ermolina L.V., Ermakova G.V., Korotaeva A.L., Smurova K.M., Gyoeva F.K., Zaraisky A.G. The LIM-domain protein Zyxin binds the homeodomain factor Xanf1/Hesx1 and modulates its activity in the anterior neural plate of Xenopus laevis embryo. Dev. Dyn. 2008;237:736–749. doi: 10.1002/dvdy.21471. [DOI] [PubMed] [Google Scholar]
- Martynova N.Y., Ermolina L.V., Ermakova G.V., Eroshkin F.M., Gyoeva F.K., Baturina N.S., Zaraisky A.G. The cytoskeletal protein Zyxin inhibits Shh signaling during the CNS patterning in Xenopus laevis through interaction with the transcription factor Gli1. Dev. Biol. 2013;380:37–48. doi: 10.1016/j.ydbio.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P.D., Faber J. Garland Science; 1994. Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. ISBN 9780815318965. [Google Scholar]
- Parshina E.A., Eroshkin F.M., Оrlov E.E., Gyoeva F.K., Shokhina A.G., Staroverov D.B., Belousov V.V., Zhigalova N.A., Prokhortchouk E.B., Zaraisky A.G., Martynova N.Y. Cytoskeletal protein Zyxin inhibits the activity of genes responsible for embryonic stem cell status. Cell Rep. 2020;33:108396. doi: 10.1016/j.celrep.2020.108396. [DOI] [PubMed] [Google Scholar]
- Sabino F., Madzharova E., Auf dem Keller U. Cell density-dependent proteolysis by HtrA1 induces translocation of zyxin to the nucleus and increased cell survival. Cell Death Dis. 2020;11:674. doi: 10.1038/s41419-020-02883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012 Jul;9 doi: 10.1038/nmeth.2089. 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynova N.Y., Parshina E.A., Ermolina L.V., Zaraisky A.G. The cytoskeletal protein Zyxin interacts with the zinc-finger transcription factor Zic1 and plays the role of a scaffold for Gli1 and Zic1 interactions during early development of Xenopus laevis. Biochem Biophys Res Commun. 2018 Sep 26;504:251–256. doi: 10.1016/j.bbrc.2018.08.164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data or code was generated in this study.