Abstract
Molecular genetic and structural studies have revealed the mechanisms of fundamental components of key auxin regulatory pathways consisting of auxin biosynthesis, transport, and signaling. Chemical biology methods applied in auxin research have been greatly expanded through the understanding of auxin regulatory pathways. Many small-molecule modulators of auxin metabolism, transport, and signaling have been generated on the basis of the outcomes of genetic and structural studies on auxin regulatory pathways. These chemical modulators are now widely used as essential tools for dissecting auxin biology in diverse plants. This review covers the structures, primary targets, modes of action, and applications of chemical tools in auxin biosynthesis, transport, and signaling.
The plant hormone auxin plays a crucial role in almost every aspect of plant development and is implicated in cell division, elongation, and differentiation. At the whole-plant level, auxin regulates embryogenesis, apical dominance, lateral root formation, tropic responses to light and gravity, and vascular tissue differentiation (Woodward and Bartel 2005; Enders and Strader 2015). The term auxin was initially derived from the Greek word “auxein,” meaning “to grow,” and auxin was defined as a growth-promoting substance that promotes cell elongation in shoots before the identification of indole-3-acetic acid (IAA) as a genuine natural plant hormone (Abel and Theologis 2010). IAA is a simple small molecule that displays profound and pleiotropic effects on plant growth. During the 1940–1970s, many synthetic compounds were extensively screened for auxin-like activity using physiological bioassays, such as hypocotyl elongation and rooting promotion assays, leading to the identification of chemicals referred to as synthetic auxins (Abel and Theologis 2010; Quareshy et al. 2018b). Synthetic auxins, strictly defined as auxin receptor agonists, are now widely used as agrochemicals, especially as herbicides (Busi et al. 2018).
The diverse auxin responses can be modulated by three major pathways: auxin metabolism, polar auxin transport, and signal transduction. IAA is mainly biosynthesized from tryptophan via indole-3-pyruvic acid (IPA) through the IPA pathway (Zhao 2014; Kasahara 2016). IAA is then transported by the AUXIN1/LIKE-AUX1 (AUX1/LAX) auxin influx transporters, PIN-FORMED (PIN) efflux facilitators, and ATP-BINDING CASSETTE B (ABCB) transporters to form auxin concentration gradients in plant tissues (Petrasek and Friml 2009; Peer et al. 2011). IAA is perceived by a nuclear-localized auxin coreceptor complex comprising TIR1/AFB F-box proteins and Aux/IAA transcriptional repressors, leading to the transactivation of auxin-responsive genes (Hayashi 2012; Salehin et al. 2015; Leyser 2018). IAA is metabolized by several inactivation pathways. GRETCHEN HAGEN3 (GH3) proteins synthesize IAA-amino acid conjugates, such as IAA-Asp and IAA-Glu, as inactive conjugates (Ludwig-Müller 2011). IAA is also oxidized to 2-oxo-indole-3-acetic acid (oxIAA) by DIOXYGENASE FOR AUXIN OXIDATION 1 (DAO1) (Porco et al. 2016; Zhang and Peer 2017). These pathways all contribute to the spatiotemporal dynamics of IAA distribution and allow the broad but subtle range of auxin responses to environmental and developmental cues.
In chemical biology, small-molecule modulators play a central role in dissecting biological functions in living systems (Dejonghe and Russinova 2017; Kinoshita et al. 2018). Small-molecule modulators used in auxin biology are frequently referred to as auxin chemical probes, and they can modulate all the redundant functions of cognate gene families in auxin biosynthesis, transport, and signaling, thus enabling us to dissect the physiological and developmental roles of redundant genes (De Rybel et al. 2009; Ma and Robert 2014). Additionally, fundamental components of auxin pathways have been conserved as pivotal genes over evolutionary history across diverse land plants such as liverworts, mosses, ferns, and vascular plants (Eklund et al. 2015; Flores-Sandoval et al. 2015; Kato et al. 2018; Tao and Estelle 2018). Mutations in these pivotal genes cause severe pleiotropic defects in plant development. Auxin chemical probes can be applied as essential tools for the elucidation of auxin biology in diverse plants, as molecular genetic and genomic approaches are often not available for nonmodel plants.
This review introduces the recent list of representative synthetic auxins and chemical probes that target auxin signaling, biosynthesis, and transport. Additionally, I will summarize the mechanisms of action of some key chemical probes in auxin pathways (Table 1).
Table 1.
List of key modulators of auxin regulatory pathways
| Compound name | Target protein(s)/pathway | CAS registry numbera | Molecular weight | Commercial sourceb | References |
|---|---|---|---|---|---|
| 4-Cl-iMT | TIR1 | 2244499-31-6 | 233.6 | N | Quareshy et al. 2018a |
| RN1 | TIR1/AFBs | 428446-82-6 | 370.3 | B | Vain et al. 2019 |
| RN2 | TIR1/AFBs | 2349373-16-4 | 426.7 | B | Vain et al. 2019 |
| RN3 | TIR1/AFBs | 425609-03-6 | 444.7 | B | Vain et al. 2019 |
| RN4 | TIR1/AFBs | 433950-87-9 | 364.2 | B | Vain et al. 2019 |
| Fipexide | Unknown | 34161-24-5 | 388.8 | B | Nakano et al. 2018 |
| AAL1 | Unknown | 868215-48-9 | 320.4 | B | Li et al. 2018 |
| Auxinole | TIR1/AFBs | 86445-22-9 | 321.4 | A | Hayashi et al. 2008 |
| PEO-IAA | TIR1/AFBs | 6266-66-6 | 293.3 | A | Hayashi et al. 2008 |
| cvxIAA | ccvTIR1 | 2127037-35-6 | 281.3 | A | Uchida et al. 2018 |
| 5-Adamantyl-IAA | ccvTIR1 | 2244426-40-0 | 309.4 | A | Uchida et al. 2018 |
| L-Kynurenine | TAA1/TARs | 2922-83-0 | 208.2 | A | He et al. 2011 |
| KOK1169/AONP | TAA1/TARs | 1394950-09-4 | 231.3 | N | Narukawa-Nara et al. 2016 |
| Pyruvamine 2031 | TAA1/TARs | 1394950-16-3 | 273.3 | N | Kakei et al. 2017 |
| BBo | YUCCA | 5122-94-1 | 198.0 | A | Kakei et al. 2015 |
| PPBo | YUCCA | 51067-38-0 | 214.0 | A | Kakei et al. 2015 |
| Yucasin | YUCCA | 26028-65-9 | 211.7 | A | Nishimura et al. 2014 |
| Yucasin DF | YUCCA | 1094690-87-5 | 213.2 | N | Tsugafune et al. 2017 |
| BUM | ABCB transporters | 5809-23-4 | 313.4 | A | Kim et al. 2010 |
| Gravacin | ABCB19/PGP19 | 188438-05-3 | 283.1 | A | Rojas-Pierce et al. 2007 |
| Bz-IAA | AUX1, PIN, ABCB | 4382-53-0 | 281.3 | A | Tsuda et al. 2011 |
| Bz-NAA | PIN, ABCB | 1270002-60-2 | 292.3 | N | Tsuda et al. 2011 |
| CHPAA | AUX1/LAX | 33697-81-3 | 186.6 | A | Parry et al. 2001 |
| 1-NOA | AUX1/LAX | 2976-75-2 | 202.2 | A | Parry et al. 2001 |
aThe CAS registry number is a unique numerical identifier assigned by the Chemical Abstracts Service to every chemical substance. The compounds in the online catalog of multiple chemical suppliers can be searched by the CAS registry number. The compound name and its abbreviation are not listed in some catalogs.
b(A) commercially available from multiple suppliers, (B) commercially available as a rare chemical from a few suppliers, (N) commercially unavailable, contact author.
SYNTHETIC AUXINS AND AUXIN PRODRUGS
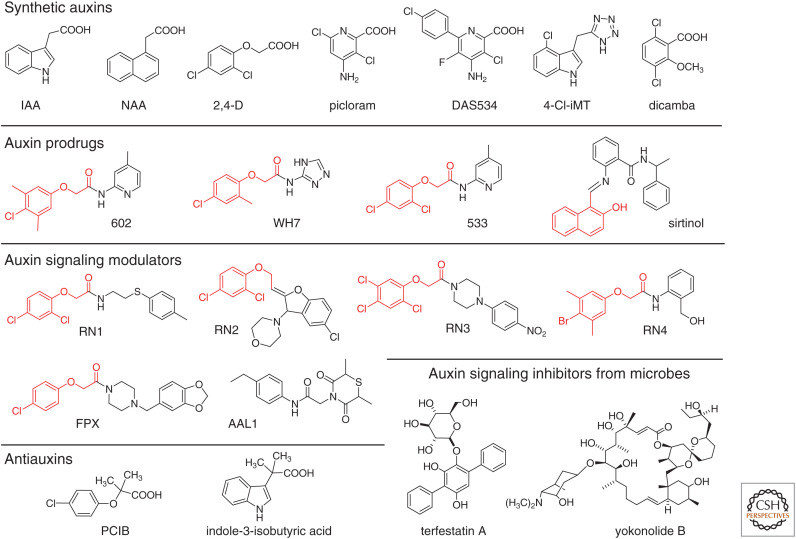
During the 1940–1970s, the potent pleiotropic hormonal activities of IAA led to a rush to explore synthetic chemicals exhibiting IAA-like activity. The synthetic chemical approach was initially applied to the chemical modification of the natural auxins phenylacetic acid (PAA) and IAA. Over time, many structurally divergent chemicals have been screened using phenotype-based bioassays based on auxin-like responses (Quareshy et al. 2018b). Synthetic chemicals with IAA-like activities are designated as synthetic auxins, many of which are widely used as agrochemicals, especially as herbicides to control weeds in arable crops. The synthetic auxins have been categorized into five groups (Ferro et al. 2010): (1) naphthalene-1-acetic acids (NAA); (2) phenoxyacetic acids (e.g., 2,4-dichlorophenoxyacetic acid) (2,4-D); (3) picolinate-type auxins (e.g., picloram); (4) benzoic acids (e.g., dicamba); and (5) indoles (e.g., haloganated IAA) (Fig. 1). The common structural features of synthetic auxins are an aromatic ring and the anion moiety attached to the aromatic ring, which are essential requirements for auxinic activity. These structurally divergent synthetic auxins are expected to bind to the TIR1/AFB-Aux/IAA coreceptor complex and consequently activate auxin signaling, but each synthetic auxin is likely to show different biological behaviors because they present distinct profiles of metabolic stability, polar transport in tissues, and affinity toward specific sets of auxin receptor complexes. Diverse synthetic auxins have been evaluated in the auxin responses of roots and hypocotyls, auxin-responsive gene expression, and auxin transport profiles (Simon et al. 2013), and this comprehensive analysis under identical assay conditions confirms that these synthetic auxins show distinct transport profiles and induce strikingly diverse phenotypes.
Figure 1.
Structure of auxins and modulators of auxin signaling.
Two major synthetic auxins, NAA and 2,4-D, are widely used for plant research because both are chemically stable and show profound hormonal activities similar to IAA. Membrane-permeable NAA can passively move into cells and is actively exported by efflux transport proteins. NAA can rescue the impaired root phenotypes of aux1 auxin influx mutants (Marchant et al. 1999). The AUX1 symporter takes up IAA and 2,4-D into the cell (Yang et al. 2006; Singh et al. 2018). PIN and ABCB efflux transporters export NAA as actively as IAA, but 2,4-D is a poor substrate for efflux transport proteins (Yang and Murphy 2009; Simon et al. 2013). Thus, NAA and 2,4-D can be useful diagnostic tools for dissecting the auxin polar transport machinery. For example, the yuc quintuple mutant (yuc 3, 5, 7, 8, 9) shows severe defects in root phenotypes (Chen et al. 2014). IAA and NAA can fully restore defects in both root growth and gravitropic responses. However, 2,4-D only restores the defects in root growth, not in root gravitropism, suggesting that root gravitropic responses require an asymmetric auxin concentration gradient formed by polar efflux transport (Sugawara et al. 2015). On the other hand, AUX1 does not mediate the uptake of other synthetic auxins, such as picloram and dicamba (Hoyerova et al. 2018).
IAA homeostasis is regulated by two major pathways involving GH3, IAA-amino acid conjugate synthases, and DAO, 2-oxoglutarate-dependent IAA oxidases. 2,4-D and dicamba have been shown to be very poor substrates of GH3 enzymes (Staswick et al. 2005; Chiu et al. 2018). Similarly, non-indole-type synthetic auxins are not oxidized by DAO1 oxidases. Thus, synthetic auxins induce a selective auxin response based on distinct transport and metabolic profiles (Busi et al. 2018; Hoyerova et al. 2018).
Precursors of a synthetic auxin (auxin prodrugs) release active auxin molecules via metabolic activation, for example, through hydrolysis by cellular enzymes. By employing auxin-related phenotype-based screening, auxinic compounds that selectively induce auxin responses were identified from a chemical library. The active compounds were found to be auxin prodrugs (WH-7, compounds 533 and 602 in Fig. 1) that release phenoxyacetic acid-type auxins after metabolic activation by plant amidohydrolases (Christian et al. 2008; Savaldi-Goldstein et al. 2008). These auxin prodrugs preferentially elicit auxin responses in the shoots, although 2,4-D shows auxin activity in both the roots and shoots. It is possible that these auxin prodrugs might be selectively activated in shoots where the hydrolases are expressed. Sirtinol was initially identified as a synthetic activator of auxin signaling because of its distinct structure from known synthetic auxins, but sirtinol was later found to be an auxin prodrug (Zhao et al. 2003). Sirtinol is metabolized to 2-hydroxy-1-naphthoic acid, which is an active auxin (Dai et al. 2005). Auxin prodrugs have important applications in agriculture. For example, auxin prodrugs IBA, 2,4-dichlorophenoxybutyric acid (2,4-BD), and naphthalene-1-acetamide (1-NAA amide) are widely used as agrochemicals.
SELECTIVE MODULATORS OF SCFTIR1/AFB SIGNALING
In canonical SCFTIR1 auxin signaling, IAA is perceived by TIR1 auxin receptors. The SCFTIR1 E3 ubiquitin ligase complex consisting of TIR1, Skp1 (ASK1), and Cullin (CUL1) catalyzes the ubiquitination of Aux/IAA repressor proteins. Auxin promotes the ubiquitination of Aux/IAAs by enhancing the interaction between Aux/IAA and TIR1 receptors (Leyser 2018). Consequently, auxin induces the degradation of Aux/IAA repressors via the 26S proteasome pathway to activate ARF transcriptional activity regulating auxin-responsive gene expression. The crystal structure of the TIR1-auxin-Aux/IAA ternary complex shows that auxin acts as molecular glue through which the two proteins are tightly bound together. In the crystal structure, the domain II motif (WPPV) from the degron peptide of Aux/IAAs covers the auxin-bound cavity in TIR1. The tryptophan and second proline residues of the degron are involved in the hydrophobic interaction with IAA. In this scenario, the auxin-agonist activities of a small molecule are determined by the binding affinity to a hydrophobic cavity formed within the TIR1-Aux/IAA coreceptor.
This perception model suggests that auxinic compounds can be relatively simple small molecules such as IAA and PAA that fit within a small binding cavity. In Arabidopsis, TIR1 and five homologous proteins (AFB1–5) redundantly function as nuclear auxin receptors (Prigge et al. 2020). Interestingly, the afb4 and afb5 loss-of-function mutants are resistant to picolinate-type auxins, picloram and DAS534, but not to 2,4-D or IAA, suggesting that AFB4 and AFB5 are the major targets of picolinic acid-based herbicides (Walsh et al. 2006; Prigge et al. 2016). The binding affinity of synthetic auxins to TIR1/AFB-Aux/IAA coreceptors has been comprehensively investigated through surface-plasmon resonance analysis (Calderón Villalobos et al. 2012; Lee et al. 2014). NAA and 2,4-D show a lower affinity for both TIR1 and AFB5 receptors than IAA, and 2,4-D obviously prefers TIR1 to AFB5. Consistent with genetic evidence, picolinates display a higher affinity for AFB5 than the TIR1 receptor. This indicates that the receptor selectivity of synthetic auxins could be important for the design of auxinic herbicides. The tetrazole group functions as a bioisostere of carboxylic acid. The bioisostere analog of IAA, indole-3-methylene tetrazole was found to be a TIR1-selective agonist that is not recognized by AFB5 (Quareshy et al. 2018a). Through the optimization of indole-3-methylene tetrazole, 4-chloroindole-3-methylene tetrazole (4-Cl-iMT) was identified as a potent TIR1-selective agonist. Interestingly, the tetrazole group of 4-Cl-iMT is not conjugated with amino acids and glucose by the GH3 enzyme and UGT84B1 (IAA-glycoside synthase), suggesting that 4-Cl-iMT is more metabolically stable than IAA.
Depending on the auxin perception module, 6 TIR/AFB receptors and 24 Aux/IAAs harboring a domain II motif (WPPV) could establish specific subsets of TIR/AFB–Aux/IAA coreceptor complexes that could regulate specific ARFs (Parry et al. 2009). However, the physiological roles of each pair of TIR1/AFB-Aux/IAA coreceptor complexes cannot be easily dissected due to the redundant function of each Aux/IAA and TIR1/AFB. Vain et al. performed a chemical biology screen to identify auxin-like small molecules (Vain et al. 2019). A synthetic chemical library was screened based on the root and shoot phenotypes of wild-type (WT) and auxin-resistant 1 (axr1) mutants. Four chemicals, RN1–RN4, were identified as active compounds that showed auxin-like activity specific to roots or shoots. RN compounds are derivatives of phenoxyacetic acids (Fig. 1), and physiological and biochemical analyses showed that RN1, RN3, and RN4 partially function as prodrugs of 2,4-D or 2,4,5-T in planta. Pull-down experiments with TIR1 and Aux/IAA demonstrated that RN3 and RN4 selectively induce the interaction of specific TIR1–Aux/IAA pairs. Therefore, RN3 and RN4 appear to act as selective molecular glues that can assemble specific subsets of TIR1 and Aux/IAA. These RN probes could be useful to dissect the physiological function of specific combinations of TIR1/AFB and Aux/IAAs.
Another phenotype-based systematic screen from a large chemical library identified two synthetic compounds exhibiting auxin-like activity. From screening 24,275 compounds, fipexide (FPX) was shown to inhibit hypocotyl elongation of etiolated Arabidopsis seedlings (Nakano et al. 2018). The inhibition of etiolated hypocotyl elongation is a typical auxin response, suggesting that FPX exhibits auxin-like activity. In general, efficient callus formation requires both auxin and cytokinin. Interestingly, FPX alone promotes potent callus formation and shoot regeneration. FPX contains a 4-chlorophenoxyacetyl amide moiety that can be converted by certain amidases to 4-chlorophenoxyacetic acid, a synthetic auxin. The complete molecular mechanism of FPX action has remained elusive. However, FPX shows structural similarity to RN3, implying that FPX might also be a selective agonist of particular sets of TIR1-Aux/IAA coreceptors. AAL1 was also identified as an inhibitor of etiolated hypocotyl extension from a set of 12,000 structurally diverse compounds (Li et al. 2018). A chemical genetic screen for an AAL1-resistant mutant identified a gain-of-function iaa3/shy2 Aux/IAA mutant. AAL1 induces DR5::GUS expression and promotes the degradation of DII-VENUS auxin-reporter proteins. Interestingly, aux1 mutation and NPA treatment reduce hypocotyl inhibition by AAL1, suggesting that the auxin-like activity of AAL1 depends on polar auxin transport.
RATIONALLY DESIGNED AUXIN AGONISTS AND ANTAGONISTS OF THE TIR1/AFB RECEPTOR
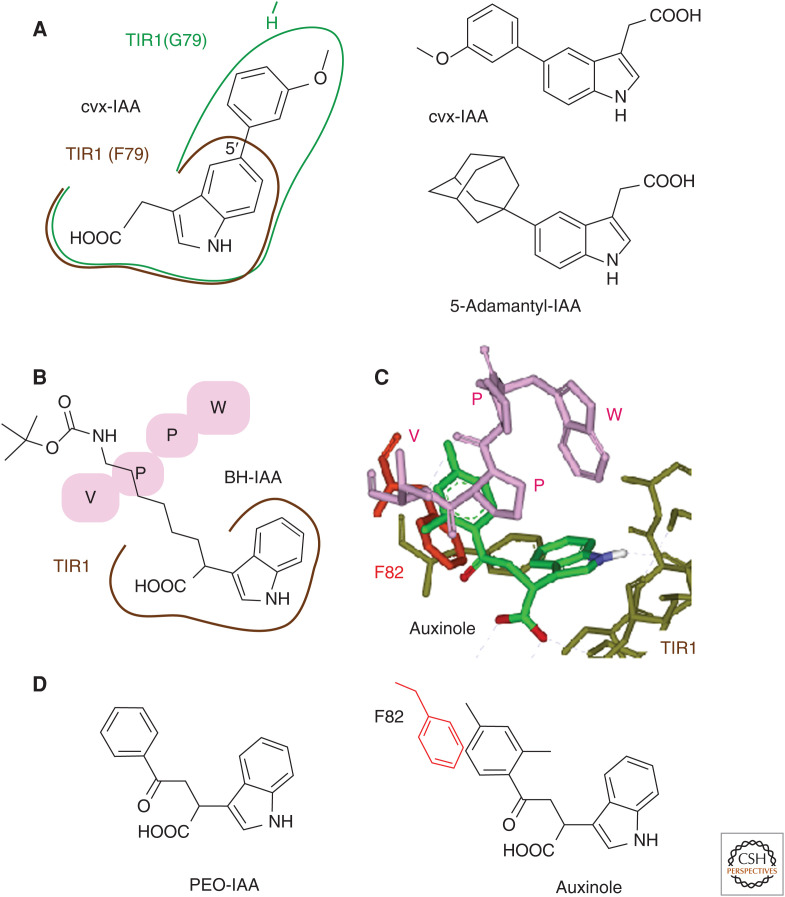
Synthetic chemistry approaches have improved the selectivity, affinity, and stability of synthetic auxins and greatly contributed to auxin biology. Uchida et al. (2018) created a new engineered TIR1-auxin module referred to as “concaveTIR1-convexIAA.” The modified IAA molecule and engineered TIR1 mutant receptor were designed according to a bump-and-hole strategy. The crystal structure of the TIR1-IAA complex suggests that phenylalanine 79 (F79) in the auxin-binding site is essential for the interaction with the aromatic ring of IAA. Indeed, the point mutation (F79G) resulted in a small hole in the TIR1 pocket, thereby disrupting the binding affinity for IAA. This TIR1 (F79G) mutant was designated concaveTIR1 (ccvTIR1), and it was also found to enable the binding of 5-aryl-substituted IAAs (Fig. 2A), including 5-methoxyphenyl IAA (convexIAA, cvxIAA). The large phenyl group (bump) of cvxIAA orients toward glycine 79 (G79) and fills the hole caused by G79 in ccvTIR1, thereby forming the ternary complex of ccvTIR1-cvxIAA-Aux/IAA. Thus, cvxIAA can selectively elicit auxin responses in transgenic plants expressing ccvTIR1 but can be completely inert in WT plants. ccvTIR1-cvxIAA can recover the impaired auxin responses of tir1 afb2 mutants, suggesting that the ccvTIR1 and cvxIAA pair can function as a surrogate auxin signaling system. The highlights of the use of this system include its application for auxin-induced rapid hypocotyl elongation, referred to as acid growth. The ccvTIR1-cvxIAA system elegantly demonstrates that cvxIAA-induced acid growth is fully dependent on ccvTIR1, implying that TIR1 might mediate the rapid elongation response through a noncanonical pathway. Thus, the ccvTIR1-cvxIAA system can be an excellent tool for studying transcription-independent auxin signaling via TIR1 (Fendrych et al. 2016, 2018). The structural optimization of 5-substituted cvxIAA led to the generation of 5-adamantyl-IAA as a super-strong cvxIAA in a yeast two-hybrid system expressing another cvxTIR1(F79A) with activity in the picomolar concentration range (Yamada et al. 2018). 5-Adamantyl-IAA and cvxTIR1 (F79A) are additional promising candidates for bump-and-hole synthetic auxin switches.
Figure 2.
Examples of the structure-based design of auxin analogs. (A) cvxIAA was designed as a specific agonist of the engineered ccvTIR1 (F79G) receptor. The methoxyphenyl group can fit within a vacant pocket in the TIR1 mutant (F79G). 5-Adamantyl-IAA is a more potent analog of cvxIAA. (B) The IAA moiety in BH-IAA binds to the auxin-binding site of TIR1 in the same manner as IAA. The flexible long alkyl chain is oriented toward the Aux/IAA-binding site and blocks the access of the WPPV motif to the binding pocket. (C,D) Auxinole shows high affinity for the TIR1 auxin-binding site via a strong hydrophobic interaction between the dimethylphenyl ring and F82, which is essential for the interaction with the WPPV degron motif of Aux/IAAs. PEO-IAA is a lead compound of auxinole.
Detailed knowledge of the molecular mechanism of auxin perception has also allowed the structure-based design of TIR1/AFB auxin antagonists. A crystal structure showed that the formation of the ternary complex TIR1-IAA-Aux/IAA did not affect the conformation of TIR1, indicating that the IAA-binding site was a rigid and inflexible structure (Tan et al. 2007). Therefore, ligands specific to TIR1/AFB could be rationally designed according to the rigid structural model of the ternary complex. BH-IAA, tert-butoxycarbonylaminohexyl-IAA, was demonstrated as the first rationally designed TIR1 antagonist (Hayashi et al. 2008). BH-IAA inhibited almost all the typical auxin responses of Arabidopsis plants in a competitive manner with auxin. The crystal structure of the complex of BH-IAA and TIR1 demonstrated that the IAA moiety of BH-IAA sits in the auxin-binding site of TIR1 in the same binding position as IAA, and the long alkyl chain is oriented toward the Aux/IAA-binding cavity. This flexible long alkyl chain occupies the Aux/IAA-binding site to efficiently prevent access of the Aux/IAA WPPV degron motif (Fig. 2B). TIR1-Aux/IAA coreceptors tightly capture auxin within a small cavity in the coreceptor. On the other hand, TIR1 receptors interact solely with the IAA moiety of BH-IAA and not with the flexible alkyl chain. Thus, BH-IAA is not a potent antagonist (Hayashi et al. 2008). To explore potent TIR1 antagonists, an in silico molecular docking screen was performed with an indole-focused chemical library, and α-(phenylethyl-2-oxo)-IAA (PEO-IAA) was identified as a lead compound (Fig. 2; Hayashi et al. 2012). Auxinole, the most potent derivative of PEO-IAA, was generated by the modification of the phenyl group of PEO-IAA. A phenyl ring at the α-position of auxinole interacts with the phenylalanine residue (F82) of the TIR1 auxin-binding site via a strong π-π stacking interaction. This F82 residue is conserved in TIR1/AFB members in diverse land plants and plays a crucial role in the interaction with the WPPV motif in domain II of Aux/IAAs. Thus, auxinole shows a high affinity to TIR1 and efficiently blocks Aux/IAA binding, explaining its strong antagonistic activity in auxin-responsive gene expression. Auxinole spatiotemporally blocks all the auxin responses mediated by redundant TIR1/AFB members in diverse land plants, including monocots, rice, and the moss Physcomitrella patens, and has been widely used as a TR1/AFB blocker in plant biology.
INHIBITORS OF AUXIN SIGNALING PATHWAYS
Several auxin derivatives, such as p-chlorophenoxyisobutylic acid (PCIB) (Oono et al. 2003), indole-3-isobutyric acid (ÅBerg 1951), and α-(5,7-dichloroindole-3-) isobutyric acid (Hatano et al. 1989), have been reported to be antiauxins that are expected to bind to a receptor in competition with auxin and thereby block the action of auxin (Fig. 1). The molecular mechanism of the antiauxin activity of PCIB has been extensively investigated in Arabidopsis plants. PCIB represses the IAA-induced degradation of Aux/IAAs, auxin-responsive gene expression, and typical physiological auxin responses, such as lateral root formation and root gravitropism. However, IAA-induced root phenotypes are not restored by PICB, suggesting that PCIB is not only an antagonist in auxin signaling but might also affect auxin transport and metabolism. Before the structural determination of the TIR1-IAA-Aux/IAA complex, auxin signaling inhibitors were screened from natural products using transgenic Arabidopsis auxin-responsive reporter systems (BA3::GUS and DR5::GUS) (Oono et al. 1998). These reporter assays are highly specific, rapid, and sensitive to auxin, making them very powerful for large library screens. Yokonolide A and B are macrolide-type compounds and were the first auxin signaling inhibitors isolated from the soil bacterium Streptomyces diastatochromogenes (Fig. 2; Hayashi et al. 2001, 2003). Similarly, microbial-origin terfestatin A and toyocamycin were identified by an auxin-responsive reporter screen (Fig. 1; Yamazoe et al. 2005). All these signaling inhibitors block Aux/IAA degradation and auxin-induced gene expression and repress various auxin responses in planta. However, the target proteins of these signaling inhibitors have not yet been determined.
TIR1/AFB are also thought to mediate nontranscriptional auxin signaling. This type of noncanonical auxin signaling is related to the very rapid inhibition of root elongation (Fendrych et al. 2018), vacuolar remodeling (Löfke et al. 2015), and auxin-induced Ca2+ signaling (Dindas et al. 2018; Li et al. 2019). However, little is known about the downstream pathway(s) of noncanonical TIR1/AFB signaling. A chemical screen for inhibitors of auxin-induced cytosolic Ca2+ dynamics was carried out using transgenic tobacco BY-2 cell lines expressing yellow fluorescent protein-apoaequorin as a reporter of Ca2+ signaling (De Vriese et al. 2019). Bepridil, a calcium channel blocker, was found to inhibit auxin-induced cytosolic Ca2+ accumulation in root cells and to affect auxin-induced vacuolar remodeling. Bepridil thus represents a valuable tool for exploring auxin-regulated Ca2+ signaling in plants.
AUXIN BIOSYNTHESIS INHIBITORS
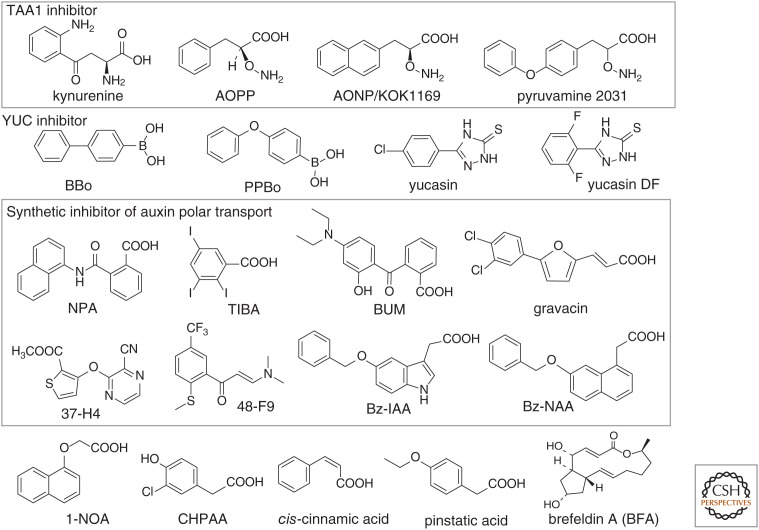
The IPA pathway is the predominant IAA biosynthetic pathway and is composed of two sequential enzymatic steps (Mashiguchi et al. 2011; Won et al. 2011). In the initial step, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/TRYPTOPHAN AMINOTRANSFERASE RELATED (TAA1/TAR) converts Trp to IPA. The YUCCA (YUC) flavin monooxygenases then catalyzes the oxidative decarboxylation of IPA to IAA. YUC genes belong to a large redundant gene family including 11 genes in Arabidopsis and 14 genes in Zea mays and Oryza sativa. Therefore, auxin biosynthesis inhibitors would be powerful tools for studying auxin biology. Both TAA1 and YUC in the IPA pathway can be considered as targets for auxin biosynthesis inhibitors. L-Kynurenine (L-Kyn), which is the major metabolite of Trp in animals, was identified as the first competitive TAA1 inhibitor (He et al. 2011). L-Kyn was initially identified from a screen for an inhibitor of ethylene responses in Arabidopsis roots. Consistent with the structural similarity of L-Kyn and Trp, L-Kyn is regarded as an analog of Trp and shows competitive inhibition (Ki = 11 μM) of TAA1 activity. Intriguingly, other Trp derivatives, such as α-methyltryptophan and 5-methyltryptophan, have never been reported as IAA biosynthesis inhibitors, although TAA1 shows broad substrate specificity in vitro and will accept alternative amino acids such as Phe and Tyr. L-Kyn is commercially available and has been widely used to block IAA synthesis in various land plants, including the liverwort Marchantia polymorpha and the moss Physcomitrella patens (Eklund et al. 2015; Tsugafune et al. 2017). L-2-aminooxy-3-phenylpropionic acid (AOPP) was initially identified as an inhibitor of pyridoxyl-5-phosphate (PLP)-dependent enzymes, such as phenylalanine ammonia-lyase. Subsequently, AOPP was reported to inhibit the PLP-dependent enzymes TAA1/TAR (Soeno et al. 2010) and to reduce both endogenous IAA levels and root gravitropism in Arabidopsis. However, AOPP is not specific to TAA1/TAR enzymes and also represses other PLP-dependent enzymes, such as phenylalanine ammonia-lyase. Aminooxy-naphthylpropionic acid (AONP/KOK1169) was designed as a specific inhibitor of TAA1 (Narukawa-Nara et al. 2016). AONP confers typical auxin-deficient phenotypes and inhibits auxin-responsive gene expression. AONPs competitively inhibit TAA1 (Ki = 76.8 nM). Another aminooxy compound, pyruvamine 2031 (KOK2031), has also been identified as a potent inhibitor of rice OsTAR1 and represses endogenous IAA synthesis, resulting in apparent auxin-deficient phenotypes in rice (Kakei et al. 2017).
YUCs are flavin-containing monooxygenases that function at final step in the IPA pathway, making YUCs important targets of IAA biosynthesis inhibitors. Yucasin was discovered as a YUC inhibitor through a chemical library screen in maize coleoptiles (Nishimura et al. 2014). Yucasin has a similar structure to methimazole, another inhibitor of flavin-containing monooxygenases (Eswaramoorthy et al. 2006). Although yucasin counters the high-auxin phenotype of YUC overexpression lines, severe auxin-deficient phenotypes are not induced in WT plants by yucasin treatment. To improve the inhibitory activity of yucasin, the structure of yucasin was optimized by chemical modification (Tsugafune et al. 2017). Difluorinated yucasin, yucasin DF (YDF), causes severe auxin-deficient phenotypes in WT plants, but YDF shows slightly less inhibitory activity than yucasin against in vitro YUC1 enzyme. YDF was shown to be a chemically and metabolically stable compound, probably because of the difluorinated phenyl ring, suggesting that cellular YDF levels are sustained long enough to fully inhibit IAA biosynthesis. YDF was found to be effective in the moss Physcomitrella patens and the liverwort Marchantia polymorpha, demonstrating that YDF is a promising tool for modulating auxin biosynthesis in various land plants.
The phenylboronic acids 4-biphenylboronic acid (BBo) and 4-phenoxyphenylboronic acid (PPBo) have been reported to be potent IAA biosynthesis inhibitors targeting YUCs (Kakei et al. 2015). BBo and PPBo cause auxin-deficient phenotypes in Arabidopsis seedlings. These boronic acids competitively inhibit YUC enzyme activity against the substrate IPyA. Additionally, these boronic acids recover the extreme high-auxin phenotype of 35S::YUC1 seedlings and repress IAA biosynthesis and growth in the model monocot plant Brachypodium distachyon.
SYNTHETIC MODULATORS OF AUXIN TRANSPORT
N-1-naphthylphthalamic acid (NPA) and 2,3,5-triiodobenzoic acid (TIBA) are classical phytotropins and auxin polar transport inhibitors that have been widely used to study plant biology (Fig. 3). NPA blocks the auxin export activity of ABCB transporters expressed in both yeast and mammalian cells (Geisler et al. 2005; Blakeslee et al. 2007; Yang and Murphy 2009). Additionally, NPA represses PIN2-mediated IAA efflux in cultured tobacco cells (Barbez et al. 2013). However, the direct binding of NPA to PIN proteins has been unclear because of inconsistent results for NPA in relation to the activity of PINs in heterologous systems. The immunophilin-like protein TWD1 interacts with ABCBs to modulate auxin efflux. NPA binds to TWD1 to perturb the interaction of TWD1 with ABCB1 (Bailly et al. 2008; Wang et al. 2013). Additionally, NPA modulates the membrane localization of PINs and ABCBs by affecting the TWD1-ACTIN7 axis (Zhu et al. 2016). Thus, NPA efficiently inhibits polar auxin transport by modulating multiple targets of the polar auxin transport machinery. Interestingly, TIBA was also shown to disrupt PIN vesicle trafficking by perturbing the stability of actin (Dhonukshe et al. 2008). However, the molecular target of TIBA has remained elusive.
Figure 3.
Structures of auxin biosynthesis inhibitors and auxin polar transport modulators.
Gravacin (3-(5-[3,4-dichlorophenyl]-2-furyl) acrylic acid) and BUM (2-[4-(diethylamino)-2-hydroxybenzoyl] benzoic acid) (Fig. 3) were identified from a phenotype-based screen of synthetic chemicals. Gravacin perturbs root and shoot gravitropism in Arabidopsis. The primary target of gravacin was identified as ABCB19 by a chemical genetic approach (Rojas-Pierce et al. 2007), with gravacin requiring ABCB19 for its activity. BUM selectively targets ABCB transporters and causes pleiotropic defects in auxin-related phenotypes similar to what is observed in NPA-treated plants. BUM was demonstrated to compete at the NPA-binding sites of ABCB1 and ABCB19 (Kim et al. 2010). Two other auxin transport inhibitors, 37-H4 and 48-F9, were discovered by screening a chemical library using an IAA transport assay in maize coleoptiles (Nishimura et al. 2012). These two inhibitors exert NPA-like effects on Arabidopsis seedlings, but 37-H4 and 48-F9 are structurally distinct from the inhibitors NPA and BUM. All of these synthetic inhibitors inhibit polar auxin transport and affect the auxin-regulated response of plants. However, the molecular mechanisms of these inhibitors are still poorly understood due to the complicated regulation of IAA efflux and the interplay of PINs, TWD1, and ABCBs.
The auxin analogs 5-benzyloxy-IAA (Bz-IAA) and 7-benzyloxy-NAA (Bz-NAA) are inactive auxin analogs that cannot bind to the auxin-binding pocket of TIR1/AFB auxin receptors (Tsuda et al. 2011). The benzyloxy group of Bz-IAA and Bz-NAA (Bz-auxins) would likely compete with Phe79 in the binding pocket of TIR1/AFB in a similar way to cvxIAA (Tsuda et al. 2011). Bz-auxins do, however, show similar effects to NPA regarding auxin-regulated phenotypes in Arabidopsis, such as the inhibition of the gravitropic response and primary root growth. Additionally, Bz-auxins inhibited IAA movement in Arabidopsis and maize, but Bz-auxins do not affect the subcellular localization of PIN proteins in Arabidopsis. Bz-auxins inhibit the transport activity of PINs, ABCBs, and AUX1 expressed in yeast cells. Consistent with the transport profiles of the synthetic auxin NAA, Bz-NAA selectively acts on PINs and ABCBs, but not on AUX/LAX proteins. Therefore, Bz-auxins could be reorganized as active auxin analogs by auxin transport proteins and reduce endogenous IAA transport by competing with endogenous IAA movement, indicating that Bz-auxins are a new class of transport inhibitors.
1-Naphthoxyacetic acid (1-NOA) and 3-chloro-4-hydroxyphenylacetic acid (CHPAA) have been found to be selective inhibitors of auxin influx transport by the AUX1 importer (Hoyerova et al. 2018). 1-NOA and CHPAA repress the root gravitropic response without affecting root growth; phenocopying the aux1 mutant (Parry et al. 2001) and 1-NOA can inhibit auxin uptake by AUX1 expressed in Xenopus oocytes (Yang et al. 2006).
Pinstatic acid, 4-ethoxyphenylacetic acid (PISA), was identified as another disruptor of auxin transport from a screen of modulators of PIN localization (Oochi et al. 2019). PISA shows auxin-like activity, such as the promotion of hypocotyl elongation and adventitious root formation. Interestingly, PISA does not directly activate the SCFTIR1 pathway as an active auxin. PISA inhibited gravitropic response and root hair formation in a manner similar to that observed when PIN1 is overexpressed in roots. It appears that PISA enhances the auxin flow by blocking PIN endocytosis and reorientation. PISA dramatically increases lateral root development under cotreatment with IAA, suggesting that PISA enhances the rate of shootward auxin transport, making PISA notably different from other known auxin chemical tools. Therefore, PISA represents a promising tool for studying the complicated regulation of PIN trafficking by auxin.
MODULATORS OF AUXIN POLAR TRANSPORT FROM NATURAL PRODUCTS
cis-Cinnamic acid has been reported as a natural auxin efflux inhibitor. cis-Cinnamic acid represses the efflux transport of NAA in tobacco BY-2 cells and inhibits root gravitropism in Arabidopsis (Steenackers et al. 2017). Intriguingly, cis-cinnamic acid promotes lateral root formation, in contrast to the effects of NPA and TIBA, probably because cis-cinnamic acid disrupts shootward auxin transport, leading to the accumulation of endogenous IAA in the root meristem. Flavonoid-deficient transparent testa4 mutant shows enhanced basipetal auxin transport, demonstrating that flavonoids such as quercetin and naringenin are endogenous negative regulators of auxin transport (Peer and Murphy 2007). However, the mode of action and the primary targets of these natural inhibitors have remained obscure.
The polar localization of PIN proteins at the plasma membrane is crucial for the regulation of the rate and direction of cellular auxin export, which ultimately determines auxin gradients in the tissue. The polar distribution of PIN is maintained by the recycling of PIN between endosomal compartments and the plasma membrane. Clathrin-mediated endocytosis is involved in the internalization of PIN proteins from the plasma membrane (Adamowski and Friml 2015). Brefeldin A (BFA) is a fungal toxin that has been widely used to dissect auxin carrier trafficking pathways. BFA inhibits ADP ribosylation factor guanine nucleotide exchange factors (ARF-GEFs), which are modulators of vesicle formation. Thus, in tissues with BFA-sensitive ARF-GEFs, BFA represses the exocytosis of PIN by inhibiting ARF-GEFs, leading to the accumulation of PIN1 in artificial intracellular aggregates referred to as “BFA bodies” (Doyle et al. 2015). TIBA inhibits the accumulation of PINs in BFA bodies by disrupting the endocytosis of PINs (Geldner et al. 2001).
CONCLUDING REMARKS
Fundamental molecular mechanisms of auxin regulatory pathways involving metabolism, signaling, and polar transport have been unveiled by extensive genetic and structural biology studies in Arabidopsis. The core modules of all auxin regulatory pathways are well conserved in land plants. Small-molecule modulators of these auxin regulatory pathways have been generated as auxin chemical probes and are widely used in Arabidopsis biology. Given pathway conservation, auxin chemical probes can specifically modulate the auxin-related pathways of diverse land plants from the liverworts to vascular plants, making these probes valuable tools for research over a broad range of species, including in agriculture and horticulture. Furthermore, auxin chemical probes can be efficiently applied for the dissection of cross talk mechanisms between auxin and other hormones and environmental cues. The structural study of TIR1-Aux/IAA has led to the rational design of auxin agonists and antagonists, and structural information related to auxin biosynthesis (TAA1) (Tao et al. 2008) and metabolism (DAO, GH3, and ILL2) (Bitto et al. 2009; Peat et al. 2012; Takehara et al. 2020) could be applied to design more potent inhibitors of these pathways.
How rapid auxin responses are mediated by nontranscriptional auxin signaling remains one of the major questions in auxin biology (Kubeš and Napier 2019). By a chemical biology approach using the ccvTIR1-cvxIAA system, TIR1 was demonstrated to be involved in rapid auxin responses such as hypocotyl elongation and primary root growth inhibition, some of which are likely nontranscriptional (Fendrych et al. 2016, 2018; Uchida et al. 2018). In combination with new tools from synthetic biology, chemical biology continues to offer promising approaches for the elucidation of such new signaling pathways. Novel auxin chemical tools will continue to provide us with novel insights into auxin-regulated developmental processes.
ACKNOWLEDGMENTS
We thank Prof. Yunde Zhao (University of California San Diego), Prof. Richard Napier (University of Warwick), Dr. Kosuke Fukui (Okayama University of Science), and Prof. Hiroyuki Kasahara (Tokyo University of Agriculture and Technology) for helpful comments on the manuscript. This work was supported by a grant from JSPS KAKENHI (Grant No. JP19H03253).
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Abel S, Theologis A. 2010. Odyssey of auxin. Cold Spring Harb Perspect Biol 2: a004572. 10.1101/cshperspect.a004572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ÅBerg B. 1951. The interaction of some auxin antagonists and 2,4-D in root growth. Physiol Plant 4: 627–640. 10.1111/j.1399-3054.1951.tb07698.x [DOI] [Google Scholar]
- Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M. 2008. Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J Biol Chem 283: 21817–21826. 10.1074/jbc.M709655200 [DOI] [PubMed] [Google Scholar]
- Barbez E, Laňková M, Pařezová M, Maizel A, Zažímalová E, Petrášek J, Friml J, Kleine-Vehn J. 2013. Single-cell-based system to monitor carrier driven cellular auxin homeostasis. BMC Plant Biol 13: 20. 10.1186/1471-2229-13-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto E, Bingman CA, Bittova L, Houston NL, Boston RS, Fox BG, Phillips GN Jr. 2009. X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana. Proteins 74: 61–71. 10.1002/prot.22124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. 2007. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147. 10.1105/tpc.106.040782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi R, Goggin DE, Heap IM, Horak MJ, Jugulam M, Masters RA, Napier RM, Riar DS, Satchivi NM, Torra J, et al. 2018. Weed resistance to synthetic auxin herbicides. Pest Manag Sci 74: 2265–2276. 10.1002/ps.4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485. 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Dai X, De-Paoli H, Cheng Y, Takebayashi Y, Kasahara H, Kamiya Y, Zhao Y. 2014. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55: 1072–1079. 10.1093/pcp/pcu039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu LW, Heckert MJ, You Y, Albanese N, Fenwick T, Siehl DL, Castle LA, Tao Y. 2018. Members of the GH3 family of proteins conjugate 2,4-D and dicamba with aspartate and glutamate. Plant Cell Physiol 59: 2366–2380. [DOI] [PubMed] [Google Scholar]
- Christian M, Hannah WB, Lüthen H, Jones AM. 2008. Identification of auxins by a chemical genomics approach. J Exp Bot 59: 2757–2767. 10.1093/jxb/ern133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y. 2005. Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci 102: 3129–3134. 10.1073/pnas.0500185102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejonghe W, Russinova E. 2017. Plant chemical genetics: from phenotype-based screens to synthetic biology. Plant Physiol 174: 5–20. 10.1104/pp.16.01805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Audenaert D, Beeckman T, Kepinski S. 2009. The past, present, and future of chemical biology in auxin research. ACS Chem Biol 4: 987–998. 10.1021/cb9001624 [DOI] [PubMed] [Google Scholar]
- De Vriese K, Himschoot E, Dünser K, Nguyen L, Drozdzecki A, Costa A, Nowack MK, Kleine-Vehn J, Audenaert D, Beeckman T, et al. 2019. Identification of novel inhibitors of auxin-induced Ca2+ signaling via a plant-based chemical screen. Plant Physiol 180: 480–496. 10.1104/pp.18.01393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Grigoriev I, Fischer R, Tominaga M, Robinson DG, Hasek J, Paciorek T, Petrasek J, Seifertova D, Tejos R, et al. 2008. Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc Natl Acad Sci 105: 4489–4494. 10.1073/pnas.0711414105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindas J, Scherzer S, Roelfsema MRG, von Meyer K, Müller HM, Al-Rasheid KAS, Palme K, Dietrich P, Becker D, Bennett MJ, et al. 2018. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat Commun 9: 1174. 10.1038/s41467-018-03582-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Vain T, Robert S. 2015. Small molecules unravel complex interplay between auxin biology and endomembrane trafficking. J Exp Bot 66: 4971–4982. 10.1093/jxb/erv179 [DOI] [PubMed] [Google Scholar]
- Eklund DM, Ishizaki K, Flores-Sandoval E, Kikuchi S, Takebayashi Y, Tsukamoto S, Hirakawa Y, Nonomura M, Kato H, Kouno M, et al. 2015. Auxin produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. Plant Cell 27: 1650–1669. 10.1105/tpc.15.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders TA, Strader LC. 2015. Auxin activity: past, present, and future. Am J Bot 102: 180–196. 10.3732/ajb.1400285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S. 2006. Mechanism of action of a flavin-containing monooxygenase. Proc Natl Acad Sci 103: 9832–9837. 10.1073/pnas.0602398103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J. 2016. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048. 10.7554/eLife.19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro N, Bredow T, Jacobsen HJ, Reinard T. 2010. Route to novel auxin: auxin chemical space toward biological correlation carriers. Chem Rev 110: 4690–4708. 10.1021/cr800229s [DOI] [PubMed] [Google Scholar]
- Flores-Sandoval E, Eklund DM, Bowman JL. 2015. A simple auxin transcriptional response system regulates multiple morphogenetic processes in the liverwort Marchantia polymorpha. PLoS Genet 11: e1005207. 10.1371/journal.pgen.1005207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. 2005. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194. 10.1111/j.1365-313X.2005.02519.x [DOI] [PubMed] [Google Scholar]
- Geldner N, Friml J, Stierhof YD, Jürgens G, Palme K. 2001. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. 10.1038/35096571 [DOI] [PubMed] [Google Scholar]
- Hatano T, Kato Y, Katayama M, Marumo S. 1989. A new indolic antiauxin, α-(5,7-dichloroindole-3-)isobutyric acid: its chemical synthesis and biological activity. Experientia 45: 400–402. 10.1007/BF01957494 [DOI] [Google Scholar]
- Hayashi K. 2012. The interaction and integration of auxin signaling components. Plant Cell Physiol 53: 965–975. 10.1093/pcp/pcs035 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ogino K, Oono Y, Uchmiya H, Nozaki H. 2001. Yokonolide A, a new inhibitor of auxin signal transduction, from Streptomyces diastatochromogenes B59. J Antibiot (Tokyo) 54: 573–581. 10.7164/antibiotics.54.573 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Jones AM, Ogino K, Yamazoe A, Oono Y, Inoguchi M, Kondo H, Nozaki H. 2003. Yokonolide B, a novel inhibitor of auxin action, blocks degradation of AUX/IAA factors. J Biol Chem 278: 23797–23806. 10.1074/jbc.M300299200 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Tan X, Zheng N, Hatate T, Kimura Y, Kepinski S, Nozaki H. 2008. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc Natl Acad Sci 105: 5632–5637. 10.1073/pnas.0711146105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H. 2012. Rational design of an auxin antagonist of the SCFTIR1 auxin receptor complex. ACS Chem Biol 7: 590–598. 10.1021/cb200404c [DOI] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. 2011. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960. 10.1105/tpc.111.089029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyerova K, Hosek P, Quareshy M, Li J, Klima P, Kubes M, Yemm AA, Neve P, Tripathi A, Bennett MJ, et al. 2018. Auxin molecular field maps define AUX1 selectivity: many auxin herbicides are not substrates. New Phytol 217: 1625–1639. 10.1111/nph.14950 [DOI] [PubMed] [Google Scholar]
- Kakei Y, Yamazaki C, Suzuki M, Nakamura A, Sato A, Ishida Y, Kikuchi R, Higashi S, Kokudo Y, Ishii T, et al. 2015. Small-molecule auxin inhibitors that target YUCCA are powerful tools for studying auxin function. Plant J 84: 827–837. 10.1111/tpj.13032 [DOI] [PubMed] [Google Scholar]
- Kakei Y, Nakamura A, Yamamoto M, Ishida Y, Yamazaki C, Sato A, Narukawa-Nara M, Soeno K, Shimada Y. 2017. Biochemical and chemical biology study of rice OsTAR1 revealed that tryptophan aminotransferase is involved in auxin biosynthesis: identification of a potent OsTAR1 inhibitor, Pyruvamine2031. Plant Cell Physiol 58: 598–606. [DOI] [PubMed] [Google Scholar]
- Kasahara H. 2016. Current aspects of auxin biosynthesis in plants. Biosci Biotechnol Biochem 80: 34–42. 10.1080/09168451.2015.1086259 [DOI] [PubMed] [Google Scholar]
- Kato H, Nishihama R, Weijers D, Kohchi T. 2018. Evolution of nuclear auxin signaling: lessons from genetic studies with basal land plants. J Exp Bot 69: 291–301. 10.1093/jxb/erx267 [DOI] [PubMed] [Google Scholar]
- Kim JY, Henrichs S, Bailly A, Vincenzetti V, Sovero V, Mancuso S, Pollmann S, Kim D, Geisler M, Nam HG. 2010. Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. J Biol Chem 285: 23309–23317. 10.1074/jbc.M110.105981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, McCourt P, Asami T, Torii KU. 2018. Plant chemical biology. Plant Cell Physiol 59: 1483–1486. 10.1093/pcp/pcy142 [DOI] [PubMed] [Google Scholar]
- Kubeš M, Napier R. 2019. Non-canonical auxin signalling: fast and curious. J Exp Bot 70: 2609–2614. 10.1093/jxb/erz111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sundaram S, Armitage L, Evans JP, Hawkes T, Kepinski S, Ferro N, Napier RM. 2014. Defining binding efficiency and specificity of auxins for SCFTIR1/AFB-Aux/IAA co-receptor complex formation. ACS Chem Biol 9: 673–682. 10.1021/cb400618m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. 2018. Auxin signaling. Plant Physiol 176: 465–479. 10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li H, Xu P, Xie Z, Ye Y, Li L, Li D, Zhang Y, Li L, Zhao Y. 2018. Identification of Auxin Activity Like 1, a chemical with weak functions in auxin signaling pathway. Plant Mol Biol 98: 275–287. 10.1007/s11103-018-0779-9 [DOI] [PubMed] [Google Scholar]
- Li T, Yan A, Bhatia N, Altinok A, Afik E, Durand-Smet P, Tarr PT, Schroeder JI, Heisler MG, Meyerowitz EM. 2019. Calcium signals are necessary to establish auxin transporter polarity in a plant stem cell niche. Nat Commun 10: 726. 10.1038/s41467-019-08575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Dünser K, Scheuring D, Kleine-Vehn J. 2015. Auxin regulates SNARE-dependent vacuolar morphology restricting cell size. eLife 4: e05868. 10.7554/eLife.05868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J. 2011. Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot 62: 1757–1773. 10.1093/jxb/erq412 [DOI] [PubMed] [Google Scholar]
- Ma Q, Robert S. 2014. Auxin biology revealed by small molecules. Physiol Plant 151: 25–42. 10.1111/ppl.12128 [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. 1999. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073. 10.1093/emboj/18.8.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, et al. 2011. The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci 108: 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Tanaka S, Ohtani M, Yamagami A, Takeno S, Hara N, Mori A, Nakano A, Hirose S, Himuro Y, et al. 2018. FPX is a novel chemical inducer that promotes callus formation and shoot regeneration in plants. Plant Cell Physiol 59: 1555–1567. 10.1093/pcp/pcy139 [DOI] [PubMed] [Google Scholar]
- Narukawa-Nara M, Nakamura A, Kikuzato K, Kakei Y, Sato A, Mitani Y, Yamasaki-Kokudo Y, Ishii T, Hayashi K, Asami T, et al. 2016. Aminooxy-naphthylpropionic acid and its derivatives are inhibitors of auxin biosynthesis targeting l-tryptophan aminotransferase: structure-activity relationships. Plant J 87: 245–257. 10.1111/tpj.13197 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Matano N, Morishima T, Kakinuma C, Hayashi K, Komano T, Kubo M, Hasebe M, Kasahara H, Kamiya Y, et al. 2012. Identification of IAA transport inhibitors including compounds affecting cellular PIN trafficking by two chemical screening approaches using maize coleoptile systems. Plant Cell Physiol 53: 1671–1682. 10.1093/pcp/pcs112 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, et al. 2014. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J 77: 352–366. 10.1111/tpj.12399 [DOI] [PubMed] [Google Scholar]
- Oochi A, Hajny J, Fukui K, Nakao Y, Gallei M, Quareshy M, Takahashi K, Kinoshita T, Harborough SR, Kepinski, et al. 2019. Pinstatic acid promotes auxin transport by inhibiting PIN internalization. Plant Physiol 180: 1152–1165. 10.1104/pp.19.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Chen QG, Overvoorde PJ, Köhler C, Theologis A. 1998. age Mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10: 1649–1662. 10.1105/tpc.10.10.1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi K, Tanaka A, Uchimiya H. 2003. p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133: 1135–1147. 10.1104/pp.103.027847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann C, Bennett MJ. 2001. Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J 25: 399–406. 10.1046/j.1365-313x.2001.00970.x [DOI] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. 2009. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci 106: 22540–22545. 10.1073/pnas.0911967106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat TS, Böttcher C, Newman J, Lucent D, Cowieson N, Davies C. 2012. Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell 24: 4525–4538. 10.1105/tpc.112.102921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. 2007. Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12: 556–563. 10.1016/j.tplants.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Peer WA, Blakeslee JJ, Yang H, Murphy AS. 2011. Seven things we think we know about auxin transport. Mol Plant 4: 487–504. 10.1093/mp/ssr034 [DOI] [PubMed] [Google Scholar]
- Petrasek J, Friml J. 2009. Auxin transport routes in plant development. Development 136: 2675–2688. 10.1242/dev.030353 [DOI] [PubMed] [Google Scholar]
- Porco S, Pěnčík A, Rashed A, Voß U, Casanova-Sáez R, Bishopp A, Golebiowska A, Bhosale R, Swarup R, Swarup K, et al. 2016. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci 113: 11016–11021. 10.1073/pnas.1604375113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Greenham K, Zhang Y, Santner A, Castillejo C, Mutka AM, O'Malley RC, Ecker JR, Kunkel BN, Estelle M. 2016. The Arabidopsis auxin receptor F-Box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda) 6: 1383–1390. 10.1534/g3.115.025585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, et al. 2020. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9: e54740. 10.7554/eLife.54740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quareshy M, Prusinska J, Kieffer M, Fukui K, Pardal AJ, Lehmann S, Schafer P, Del Genio CI, Kepinski S, Hayashi K, et al. 2018a. The tetrazole analogue of the auxin indole-3-acetic acid binds preferentially to TIR1 and not AFB5. ACS Chem Biol 13: 2585–2594. 10.1021/acschembio.8b00527 [DOI] [PubMed] [Google Scholar]
- Quareshy M, Prusinska J, Li J, Napier R. 2018b. A cheminformatics review of auxins as herbicides. J Exp Bot 69: 265–275. 10.1093/jxb/erx258 [DOI] [PubMed] [Google Scholar]
- Rojas-Pierce M, Titapiwatanakun B, Sohn EJ, Fang F, Larive CK, Blakeslee J, Cheng Y, Cutler SR, Peer WA, Murphy AS, et al. 2007. Arabidopsis P-glycoprotein19 participates in the inhibition of gravitropism by gravacin. Chem Biol 14: 1366–1376. 10.1016/j.chembiol.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M. 2015. SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19. 10.1105/tpc.114.133744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Baiga TJ, Pojer F, Dabi T, Butterfield C, Parry G, Santner A, Dharmasiri N, Tao Y, Estelle M, et al. 2008. New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc Natl Acad Sci 105: 15190–15195. 10.1073/pnas.0806324105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Kubeš M, Baster P, Robert S, Dobrev PI, Friml J, Petrášek J, Zažímalová E. 2013. Defining the selectivity of processes along the auxin response chain: a study using auxin analogues. New Phytol 200: 1034–1048. 10.1111/nph.12437 [DOI] [PubMed] [Google Scholar]
- Singh G, Retzer K, Vosolsobe S, Napier R. 2018. Advances in understanding the mechanism of action of the auxin permease AUX1. Int J Mol Sci 19: 3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno K, Goda H, Ishii T, Ogura T, Tachikawa T, Sasaki E, Yoshida S, Fujioka S, Asami T, Shimada Y. 2010. Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol 51: 524–536. 10.1093/pcp/pcq032 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627. 10.1105/tpc.104.026690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenackers W, Klíma P, Quareshy M, Cesarino I, Kumpf RP, Corneillie S, Araújo P, Viaene T, Goeminne G, Nowack MK, et al. 2017. cis-Cinnamic acid is a novel, natural auxin efflux inhibitor that promotes lateral root formation. Plant Physiol 173: 552–565. 10.1104/pp.16.00943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara S, Mashiguchi K, Tanaka K, Hishiyama S, Sakai T, Hanada K, Kinoshita-Tsujimura K, Yu H, Dai X, Takebayashi Y, et al. 2015. Distinct characteristics of indole-3-acetic acid and phenylacetic acid, two common auxins in plants. Plant Cell Physiol 56: 1641–1654. 10.1093/pcp/pcv088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehara S, Sakuraba S, Mikami B, Yoshida H, Yoshimura H, Itoh A, Endo M, Watanabe N, Nagae T, Matsuoka M, et al. 2020. A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin. Nat Commun 11: 2143. 10.1038/s41467-020-16068-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. 10.1038/nature05731 [DOI] [PubMed] [Google Scholar]
- Tao S, Estelle M. 2018. Mutational studies of the Aux/IAA proteins in Physcomitrella reveal novel insights into their function. New Phytol 218: 1534–1542. 10.1111/nph.15039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. 10.1016/j.cell.2008.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda E, Yang H, Nishimura T, Uehara Y, Sakai T, Furutani M, Koshiba T, Hirose M, Nozaki H, Murphy AS, et al. 2011. Alkoxy-auxins are selective inhibitors of auxin transport mediated by PIN, ABCB, and AUX1 transporters. J Biol Chem 286: 2354–2364. 10.1074/jbc.M110.171165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugafune S, Mashiguchi K, Fukui K, Takebayashi Y, Nishimura T, Sakai T, Shimada Y, Kasahara H, Koshiba T, Hayashi K-i. 2017. Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci Rep 7: 13992. 10.1038/s41598-017-14332-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, Endo TA, Kimura S, Zhang H, Nomoto M, Tada Y, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat Chem Biol 14: 299–305. 10.1038/nchembio.2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vain T, Raggi S, Ferro N, Barange DK, Kieffer M, Ma Q, Doyle SM, Thelander M, Pařízková B, Novák O, et al. 2019. Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development. Proc Natl Acad Sci 116: 6463–6472. 10.1073/pnas.1809037116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP. 2006. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142: 542–552. 10.1104/pp.106.085969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Bailly A, Zwiewka M, Henrichs S, Azzarello E, Mancuso S, Maeshima M, Friml J, Schulz A, Geisler M. 2013. Arabidopsis TWISTED DWARF1 functionally interacts with auxin exporter ABCB1 on the root plasma membrane. Plant Cell 25: 202–214. 10.1105/tpc.112.105999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. 2011. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci 108: 18518–18523. 10.1073/pnas.1108436108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Ann Bot 95: 707–735. 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Murai K, Uchida N, Takahashi K, Iwasaki R, Tada Y, Kinoshita T, Itami K, Torii KU, Hagihara S. 2018. A super strong engineered auxin-TIR1 pair. Plant Cell Physiol 59: 1538–1544. 10.1093/pcp/pcy127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe A, Hayashi K, Kepinski S, Leyser O, Nozaki H. 2005. Characterization of terfestatin A, a new specific inhibitor for auxin signaling. Plant Physiol 139: 779–789. 10.1104/pp.105.068924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Murphy AS. 2009. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J 59: 179–191. 10.1111/j.1365-313X.2009.03856.x [DOI] [PubMed] [Google Scholar]
- Yang Y, Hammes UZ, Taylor CG, Schachtman DP, Nielsen E. 2006. High-affinity auxin transport by the AUX1 influx carrier protein. Curr Biol 16: 1123–1127. 10.1016/j.cub.2006.04.029 [DOI] [PubMed] [Google Scholar]
- Zhang J, Peer WA. 2017. Auxin homeostasis: the DAO of catabolism. J Exp Bot 68: 3145–3154. 10.1093/jxb/erx221 [DOI] [PubMed] [Google Scholar]
- Zhao Y. 2014. Auxin biosynthesis. Arabidopsis Book 12: e0173. 10.1199/tab.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. 2003. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110. 10.1126/science.1084161 [DOI] [PubMed] [Google Scholar]
- Zhu J, Bailly A, Zwiewka M, Sovero V, Di Donato M, Ge P, Oehri J, Aryal B, Hao P, Linnert M, et al. 2016. TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 28: 930–948. 10.1105/tpc.15.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]