Abstract
Major biotechnological advances have facilitated a tremendous boost to the collection of (gen-/transcript-/prote-/methyl-/metabol-)omics data in very large sample sizes worldwide. Coordinated efforts have yielded a deluge of studies associating diseases with genetic markers (genome-wide association studies) or with molecular phenotypes. Whereas omics–disease associations have led to biologically meaningful and coherent mechanisms, the identified (non-germline) disease biomarkers may simply be correlates or consequences of the explored diseases. To move beyond this realm, Mendelian randomization provides a principled framework to integrate information on omics- and disease-associated genetic variants to pinpoint molecular traits causally driving disease development. In this review, we show the latest advances in this field, flag up key challenges for the future, and propose potential solutions.
Most common diseases have 30%–80% heritability (Ge et al. 2017) and the remaining causes are comprised of modifiable environmental, lifestyle, and molecular factors. The identification of all genetic and nongenetic factors remains elusive for multiple reasons.
The major hurdle in deciphering the genetic basis of complex traits is the necessity of very large sample sizes to identify contributors of a genetic architecture very much resembling an infinitesimal model (where a very large number of genetic variants have increasingly small effects). Nongenetic factors, on the other hand, tend to have larger correlations with diseases. However, accurate measurement of all relevant (molecular and high-level) phenotypes in human populations in a noninvasive fashion is difficult; hence, we often rely on noisy proxies, and dissecting true causes from mere disease correlates has proved to be particularly challenging.
The aim of this review is to provide an overview of how principled causal inference methods can be adapted to combine molecular phenotypes (i.e., omics data) and complex diseases to reveal robust causal biomarkers. First, we introduce genome-wide association studies (GWASs) of complex and molecular traits. We then explain how results generated from these two types of studies can be combined together, with strong emphasis on various forms of Mendelian randomization (MR). Next, we delve into the key findings of transcriptome-, proteome-, and methylome-wide MR studies. We also review key limitations of current approaches (heritable confounding, tissue-specificity, reconciling different omics). Finally, we point out future challenges: linking disease causes and consequences, teasing out sex-specific effects, deriving causal regulatory networks, and leveraging findings for drug repurposing. Because of the extremely advanced state of transcriptomics research and hence the dominating abundance of (publicly available) gene expression data sets (>1.8 million human samples in the Gene Expression Omnibus), our review is slanted toward this type of omics data. However, most concepts, difficulties, and challenges presented for RNA-based biomarkers are, in principle, transferrable to other omics data types.
GENOME-WIDE ASSOCIATION STUDIES
Whereas whole genome sequencing is not yet a viable option in millions of samples (current costs are ∼$1000/sample), measuring common genetic variations has become affordable (cost <$50/sample for genome-wide genotyping arrays, probing >700,000 variants). There is growing evidence that these measured common variants capture the majority of the heritability for several model traits (Yang et al. 2015). GWASs have been designed to identify single-nucleotide polymorphisms (SNPs) associated with traits/diseases (Visscher et al. 2017). Vast amounts of financial and human resources have been invested in the past decade into data collection for large population cohorts, including whole-genome-scale genotyping/sequencing and extensive characterization for dozens of clinically relevant phenotypes. Indeed, these efforts have led to thousands of association studies on hundreds of phenotypes and diseases. These traits include anthropometric traits (e.g., height and body mass index [BMI]; Yengo et al. 2018), blood chemistry variables (e.g., lipid levels; Global Lipids Genetics Consortium et al. 2013), cardiovascular traits (e.g., blood pressure; Evangelou et al. 2018), complex diseases (e.g., type 2 diabetes [T2D]; Mahajan et al. 2018), and cognitive traits (e.g., educational attainment; Lee et al. 2018).
These studies have shown that each identified variant alone has a minute impact (0.01%–0.5% explained variance) on the respective phenotype. However, as study sizes continue to grow larger, the cumulative effect of the increasing number of discovered DNA variants is steadily approaching their respective heritability (Maier et al. 2018) and the remaining gap could possibly be filled by additive effects of rare variants (Wainschtein et al. 2019). GWASs have also shed light on biologically meaningful pathways (Pers et al. 2015) and key tissues (Ongen et al. 2017; Finucane et al. 2018; Richardson et al. 2020). They have also enabled accurate estimation of narrow sense heritability due to common variants (Yang et al. 2010; Bulik-Sullivan et al. 2015b) and genetic correlation (Bulik-Sullivan et al. 2015a).
MOLECULAR TRAITS UNDERLYING DISEASES
Whereas GWASs have revealed many biological insights in the past decade, the underlying mechanisms as to how the effect of SNPs leads to disease are still poorly understood. Even if effect sizes are unbiasedly estimated, the fine-mapping of the association signals is very difficult and largely insufficient for pinpointing the implicated gene or understanding the underlying molecular mechanisms. One opportunity to fill the gap from genetic variants to complex traits is combining GWAS findings with other “omics” (e.g., transcriptomics, methylomics, metabolomics, proteomics) data, to functionally characterize the statistical associations (Passador-Gurgel et al. 2007; Petretto et al. 2008). SNPs influencing gene/protein expression level are referred to as gene/protein expression quantitative trait loci (eQTLs/pQTLs) and genes amenable to genomic regulation are called eGenes (GTEx Consortium 2015). Encouragingly, trait-associated SNPs are three times more likely to be associated with gene expression (Nica et al. 2010; Nicolae et al. 2010; Fehrmann et al. 2011; Hernandez et al. 2012), suggesting strongly shared genetic mechanisms between molecular and complex traits.
CAUSAL INFERENCE
The presence of a causal effect of an exposure on an outcome implies that a modification of the exposure (and the exposure only) would lead to a change in the outcome. Elucidating the existence and the strength of a causal effect is inherently hard because it requires an exposure-specific intervention. Estimating causal effect is primarily hampered by confounding and reverse causation.
In the special case where the exposure is a DNA variant, reverse causation can be excluded (because of temporal considerations), but confounding can arise because of linkage disequilibrium (being only correlated to the causal variant), population stratification (Kang et al. 2008), indirect effects (e.g., parental [Kong et al. 2018] and assortative mating [Morris et al. 2019] effects), and selection or participation bias (Taylor et al. 2018; Hughes et al. 2019). Despite careful efforts to control for population stratification (by using ancestry principal component correction and mixed effect models), large meta-analyses of GWASs have still been influenced by confounding, as suggested by an emerging body of evidence for human height and education level (Abdellaoui et al. 2019; Haworth et al. 2019; Sohail et al. 2019). Family-based designs can be a solution to eliminate most of the confounding effects, which are often shared among siblings (Belsky et al. 2018; Davies et al. 2019).
When the exposure is not a genetic variant, limitations and biases (including residual confounding and reverse causation) in causal effect inference are far more substantial. This means that many of the associations found in classical epidemiological studies are mere correlates of disease risk, rather than causal factors directly involved in disease development (Fewell et al. 2007; Pingault et al. 2018). As nongenetic molecular traits are also vulnerable to the effects of confounding and reverse causation, challenges of causal inference also extend to the investigation of molecular traits as causal risk factors underlying disease.
For example, transcriptome-wide association studies (TWASs) have aimed at identifying genes whose (genetically predicted) expression is strongly associated to complex traits (Nica et al. 2010; Gusev et al. 2016). Later developments were based on a Bayesian sparse linear mixed model (Mancuso et al. 2018), genetic best linear unbiased predictor (GBLUP) (Mancuso et al. 2017), or elastic net (Barbeira et al. 2018). However, these studies were not designed to estimate the strength of the causal effect nor to distinguish causation from horizontal pleiotropy (i.e., when a genetic variant influences multiple phenotypes independently). Nonetheless, when the same genetic variant is associated with both the expression level of a gene and a phenotype, there is some evidence that the implicated gene is causally involved for that trait. To detect such overlapping associations, several methods have been proposed. To quantify the colocalization signal of gene expression and phenotype associations one can use Bayesian model comparison methods, such as various versions of coloc (Wallace et al. 2012; Giambartolomei et al. 2014; Fortune et al. 2015; Guo et al. 2015) and eCAVIAR (Hormozdiari et al. 2016). The disadvantage of such methods is that they are blind to the directionality of effect and sensitive to the presence of heritable confounding factors. A more principled approach based on model comparison aiming to disentangle various causal models has been long proposed (Millstein et al. 2009), but it is based on a single genetic marker as instrument.
Epidemiologists have sought to develop statistical methods to tease out causal effects based on observational data, which may lend themselves to evaluating causality of molecular phenotypes. A large family of causal inference methods is instrumental variable (IV) analysis (Lawlor et al. 2008). It requires that beyond the exposure (X) and the outcome (Y) we also measure an IV (G) in a population, which fulfils the following three assumptions: (1) Relevance—G is robustly associated with the exposure. (2) Exchangeability—G is not associated with any confounder of the exposure–outcome relationship. (3) Exclusion restriction—G is independent of the outcome conditional on the exposure (i.e., there is no path between the instrument and the outcome independent of the exposure). A special case of IV analysis is MR, where the instrument is a genetic variant.
In MR with risk factors that are not directly linked to the DNA sequence (e.g., not related to chromatin properties, gene expression, protein, or methylation levels), multiple genetic variants across the genome can be used as instruments to look for a homogenous effect across all instruments (Palmer et al. 2012). However, if the exposure of interest is the expression level of a gene or a protein, then the optimal instrument(s) would be genetic variants lying within or near the gene coding for the protein itself (i.e., in cis). In this way, the presumed effect of the instrument on the outcome can only possibly be through the exposure.
As listed above, many genetic studies now combine genetic information of diseases with one or multiple molecular traits such as gene expression (Zhu et al. 2016; Porcu et al. 2019), metabolomics (Bell et al. 2020), protein data (Al Awam et al. 2015), and DNA methylation (Wahl et al. 2017) to gain a better understanding of complex disease etiology. It has been shown that a large fraction of trait-associated SNPs are also associated with molecular traits (i.e., molecular quantitative trait loci [molQTLs]) (Nica et al. 2010; Hannon et al. 2017; Sun et al. 2018), suggesting their potential involvement in the molecular mechanisms. However, understanding how such variants also influence complex traits is challenging. The most efficient way to address this question is to combine summary-level data from molQTL and GWAS studies in a two-sample MR framework to evaluate whether a molecular trait has a causal influence on a complex trait (Richardson et al. 2018). Given the complexity of human phenotypes, no single molecular feature is expected to fully capture all key biological mechanisms. Furthermore, molecular markers form highly complex networks and disentangling their individual roles in isolation is challenging due to largely shared instruments leading to contamination of horizontal pleiotropic effects.
MR STUDIES WITH OMICS EXPOSURES
Transcriptome-Wide MR
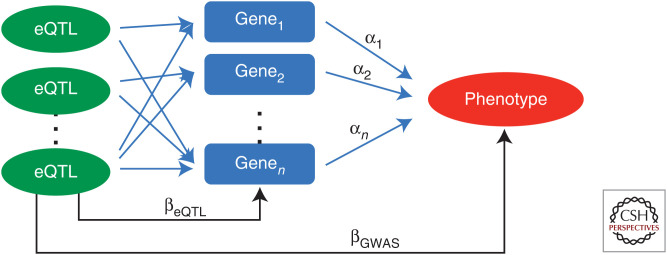
Recently, we adapted a multivariable MR method (Sanderson et al. 2019) tailored to gene expression exposure, termed TWMR (transcriptome-wide Mendelian randomization) (Porcu et al. 2019). Key improved features include approximate conditional analysis to select potentially correlated instruments and iteratively including genes with shared instruments for joint causal analysis. TWMR integrates summary-level data from GWAS and eQTL studies in an MR framework to estimate the multivariate causal effect of gene expression on complex human traits (Fig. 1). The robustness, validity, and interpretation of such a multivariable approach is described in Sanderson (2020). Because proximal genes often have correlated expression levels, multivariate causal effects are key to disentangle direct, mediated, and shared effects.
Figure 1.
Multivariable Mendelian randomization (MR) applied to expression traits. (eQTL) expression quantitative trait loci.
Applying TWMR to complex traits revealed thousands of putative genes with a causal effect on at least one phenotype. Notably, about one-third of these gene–trait associations were not prioritized by previous GWASs (i.e., no SNP reached genome-wide significance level within the gene ±500 kb). For example, while educational attainment GWAS entirely missed the BSCL2 locus, TWMR highlighted this gene as potentially causal (PTWMR = 1.89 × 10−6). BSCL2 has been previously shown to be involved in type 2 congenital generalized lipodystrophy (OMIM [Online Mendelian Inheritance in Man]:#615924) (Guillén-Navarro et al. 2013), which has been associated with some degree of intellectual impairment. It is generally accepted by now that the gene in closest proximity to the lead GWAS SNP may not necessarily be the key player. For instance, in the height-associated 2p21 region (Wood et al. 2014), TWMR results suggested that SOCS5 is not the causal gene, despite being closest to the lead SNP. Instead, TWMR revealed that high expression of CRIPT is causally linked to high stature. Loss-of-function mutations in CRIPT are known to be associated with short stature (OMIM:#615789) (Shaheen et al. 2014), making it a plausible candidate.
Interestingly, TWMR-implicated genes show global trends for functional relevance, defined as being linked to a more severe version of the same trait in the OMIM database. For example, height-, total cholesterol-, and educational attainment–associated TWMR genes are 1.3/3.7/2.6-fold enriched for being linked by OMIM to abnormal skeletal growth syndrome, hypercholesterolemia, and cognitive impairment, respectively. These results provide a hint that both mild (e.g., modified expression level) and strong (e.g., protein truncating mutation) perturbation of the same genes may impact the same trait, but to a different extent.
Metabolome-Wide MR
As emerging technologies have made feasible the assessment of hundreds of metabolites, many studies explored the role of metabolites in several diseases (Sabatine et al. 2005; Shah et al. 2010; Wang et al. 2011). Additionally, many GWASs on metabolite concentrations (metabolite GWAS [mGWAS]) have been performed to investigate how genetic variants affect changes in metabolite levels in human urine (Raffler et al. 2015) and blood (Shin et al. 2014). Such large studies enable two-sample MR approaches to estimate the causal effect of metabolites on complex traits using metabolite QTLs (mQTLs) as genetic instruments.
MR studies have been particularly important in providing insights into the role of low-density lipoprotein cholesterol (LDL-C) on coronary artery disease (CAD) development. For example, many independent trials have shown statins to reduce LDL-C levels and risk of CAD, proportional to the dose of the statin, owing to the causal link between LDL-C and CAD (Cannon et al. 2004). Using all known genetic variants associated with LDL-C as instruments, the causal role of LDL-C has been substantiated by MR (Ference et al. 2012). Furthermore, MR models have been successful in predicting the effect of specific LDL-C-lowering drugs by restricting the analysis to the gene target (e.g., NPC1L1) of the drug (e.g., ezetimibe) in question (Holmes et al. 2013; Myocardial Infarction Genetics Consortium Investigators et al. 2014; Ference et al. 2015; Würtz et al. 2016). Moreover, MR studies have encouraged the development of novel drugs, such as PCSK9 inhibitors, which have been recently shown to reduce cardiovascular events in phase III trials (Giugliano and Sabatine 2015; Sabatine et al. 2017). Further details on these links and broader discussions on the role of MR in drug repositioning can be found in the section “Omics-Based MR to Boost Drug Development and Repositioning” below.
MR analysis also pointed to a potential causal effect of serum leucine concentration on T2D, which has been long known to be an important biomarker (Melnik 2012). High triglyceride levels have also been confirmed by MR studies to be a risk factor for T2D, while another longitudinally implicated biomarker (Vangipurapu et al. 2019), alanine, seems rather to be a consequence of diabetes (Liu et al. 2017). To our knowledge, systematic comparison between MR and longitudinal observational studies is scarce and whereas a rare attempt showed good concordance (Würtz et al. 2014), it still lacked directionality by comparing exposure change to outcome change.
Proteomics MR
Blood-based biomarker measurement is a mainstay for patient management and fundamental for diagnosis, prognosis, and treatment. Biomarkers that are known to be causally linked to a disease and can subsequently be modified are key to lowering a patient's risk. Because of the limitations of most alternative approaches, MR has become an increasingly common technique to identify and shed light on causal relations between protein biomarkers and disease.
We have applied MR to identify novel, causal mediators of CAD in the ORIGIN Trial by examining a comprehensive panel of 237 biomarkers (Sjaarda et al. 2018a). Using genetic variants residing at or near the gene for each biomarker under study, MR analysis revealed six biomarkers, in which there was evidence for a causal effect on CAD, including two novel markers: CSF1 and CXCL12. Both biomarkers had been previously linked to inflammatory processes characteristic of atherosclerosis in both animal models and human studies, consistent with MR results showing a causal link with CAD. These findings were also consistent with results from the CANTOS study (Ridker et al. 2017), which showed that an intervention aimed at decreasing inflammation through interleukin-1β inhibition can lead to lower rates of recurrent cardiovascular events. Together, these studies shed light on the role of inflammation in CAD and highlight the utility of MR in revealing specific protein markers directly involved in disease progression that could be targeted via pharmacological inhibition.
Another MR study revealed new mediators of chronic kidney disease (CKD) whereby human epidermal growth factor receptor 2 (HER2) and uromodulin (UMOD) were both identified as causal mediators of CKD (Sjaarda et al. 2018b). Further MR exploration of the HER2 pathway also revealed ACE as a regulator of HER2 levels. These results implicate HER2 not only as a mediator of ACE inhibitors’ protective effect on CKD, but also as a marker to identify patients who would benefit from ACE/RAAS inhibition. Furthermore, these findings suggest HER2 inhibitors (e.g., gefitinib) as a potential novel treatment for CKD.
Recent proteome-wide work (Zheng et al. 2019) has probed the causal effect of over 1000 proteins on 225 phenotypes and identified 105 putatively causal effects of 64 proteins on 51 phenotypes. Importantly, they have demonstrated that protein–disease links supported by MR and colocalization are far more likely to indicate potentially successful therapeutic targets.
Methylome-Wide MR
As epigenetic processes are putative intermediate mechanisms between socioenvironmental exposures and health outcomes, epidemiologic studies of epigenetic marks may shed light on the biological pathways embodying exposures or provide biomarkers of exposures alternative to self-reported questionnaires (Relton and Davey Smith 2015; Sharp and Relton 2017). Applications of MR in epigenetics have been limited to DNA methylation as, so far, it is the most common and simplest epigenetic process measurable in epidemiologic studies. To perform MR, cis genetic variations related to DNA methylation levels (i.e., mQTLs) are typically used as instruments. Traditional bidirectional MR has been applied to interrogate exposure–methylation associations, such as maternal hyperglycemia (Allard et al. 2015), adiposity (Richmond et al. 2016), or methylation–complex traits associations (Richardson et al. 2018). Other studies have applied a two-step MR framework (Relton and Davey Smith 2012) to elucidate the mediating role of methylation between the exposure–outcome association of interest, such as obesity and cardiometabolic diseases (Mendelson et al. 2017). A recent systematic review of MR studies suggests that DNA methylation may be mediating the causal effect of pre- and postnatal exposures to tobacco smoke on birth weight and inflammation markers, respectively, and prenatal exposure to vitamin B12 on cognitive outcomes (Grau-Perez et al. 2019).
MR has mostly been performed only on a selected set of methylation loci and a limited number of exposures and health outcomes. The reason for this is that currently available platforms (e.g., Illumina Infinium HumanMethylation450 or MethylationEPIC BeadChips) can tag only a small subset of CpGs (∼3% genome-wide). In addition, samples with genome-wide methylation and genomic data are still smaller than those with gene expression measurements, but the gap is closing rapidly (Huan et al. 2019), greatly catalyzing methylation MR studies.
CURRENT LIMITATIONS
The Curse of Unmeasured Heritable Confounders
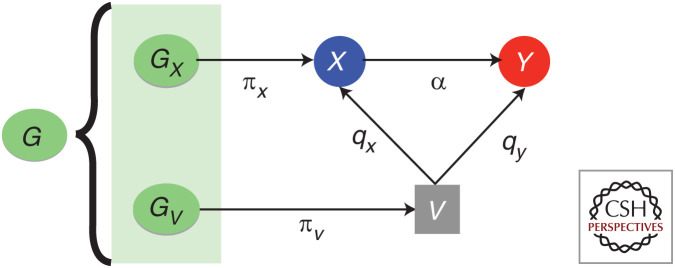
In the case of most MR methods (using multiple instruments), the exclusion restriction assumption can be replaced by the weaker InSIDE assumption, which requires that instrument strength is uncorrelated to the direct (horizontal pleiotropic) effect on outcome. Despite pleiotropy being pervasive, the InSIDE assumption is reasonable when the (alternative) pleiotropic pathway from the instrument to the outcome does not involve the exposure. However, in the presence of a confounder (V) of the exposure–outcome relationship (Fig. 2), all SNPs associated with V would serve as instruments for the exposure (X) and their effect on X will be proportional (qy/qx) to their effects on the outcome (Y). In classical MR, all instruments G for X would be included in the analysis, both direct (Gx) and indirect (Gv). As the genetic basis of the confounder becomes more prominent (i.e., the confounder is heritable), increasing numbers of SNPs will yield such biased estimates. Therefore, the estimated causal effect, which would be a weighted combination of the true causal effect (α) and (qy/qx), will become more biased. To evaluate this bias, more sophisticated methods (Darrous et al. 2020; Morrison et al. 2020) or functional validation is necessary (Lepik et al. 2017).
Figure 2.
A key violation of the InSIDE assumption of Mendelian randomization. An unmeasured heritable confounding factor V and separate direct (πx) and indirect (qxπv) genetic effect for X for a set of single-nucleotide polymorphisms (SNPs) Gx and Gv. The causal effects of V on X, V on Y, and X on Y are denoted by qx, qy, and α, respectively.
Difficulty of Gene Prioritization
Shifting from the classical single-gene view of complex traits biology to a pathway/network perspective can help us to better understand the etiology of the variation in human phenotypes. Pathway enrichment analyses can pinpoint biological mechanisms based on gene lists usually created based on physical proximity to GWAS hits (Pers et al. 2015; Lamparter et al. 2016). Combining GWAS-implicated genes with causal genes could improve the prioritization and subsequently boost the power to detect enrichment in relevant pathways and regulatory networks. However, to do so, several hurdles need to be overcome.
First, TWAS/TWMR approaches are currently limited to only the 17 K established eGenes (Võsa et al. 2018), which substantially decreases power to detect enrichment of the prioritized gene set in relevant pathways and regulatory networks. We expect that larger eQTLs studies will allow for the identification of additional eGenes resulting in stronger enrichment when using causally associated gene sets, rather than selecting genes based on physical proximity to GWAS hits. Despite these promising developments, many genes lead to disease not through change in their gene expression, but via other mechanisms (e.g., modification of the RNA or protein sequence [Marouli et al. 2017]) to which TWAS/TWMR are blind. This problem can be mitigated by the simultaneous application of multiple omics MR. Whereas omics data provide quantifiable readouts for the impact of SNPs, it becomes less straightforward to quantify the impact of coding variants. The suitability of different measures of coding variant severity (based on conservation, pathogenicity, protein folding property, etc.) could be tested in the MR framework.
Second, as opposed to standard GWAS where magnitude of SNP-trait correlation and evidence (P-value) have a monotonic relationship, MR P-values and causal effect estimates across genes are not directly comparable. The reason for this is that the evidence for causal effect depends on the number and strength of instruments and the magnitude of the effect. Therefore, it is not clear whether genes should be prioritized based on estimated causal effect size or P-value. Moreover, the threshold above which a causal effect size is deemed of clinical interest may vary from gene to gene.
Third, a further limitation of TWAS/TWMR is the presence of high correlation between coregulated genes, which in turn results in shared eQTLs. Many of these shared eQTLs may represent direct effects on a common transcription factor and indirectly relate to the gene of interest. Larger eQTL studies may be able to reveal unique instruments for such genes and hence disentangle their multivariate causal effects.
Tissue Specificity
Typically, the methods integrating data from GWASs and omics data are focused on eQTLs/mQTLs/pQTLs in whole blood, because it is the easiest tissue to collect and summary statistics from large studies are available (e.g., Westra et al. 2013; Võsa et al. 2018). However, because gene regulation is tissue-specific and many diseases manifest themselves only in certain tissues, the possibility to interrogate more tissues could unravel causative genes whose whole blood expression is not disease-relevant.
For example, the causal effect of SORT1 on LDL levels is only detectable when eQTLs derived from liver are used (Porcu et al. 2019; Richardson et al. 2020). Furthermore, FBN2 expression is driving systolic blood pressure in heart tissues, but is associated to forced vital capacity when using lung eQTLs (Richardson et al. 2020). Finally, for CAD, TWMR pointed to MRAS and PHACTR1 as causal genes exclusively in arterial tissues (Porcu et al. 2019).
The gene expression data collected in 54 tissues by the GTEx Consortium (Aguet et al. 2019), ranging in sample size up to 706 (muscle) genotyped individuals, provide an extremely valuable resource for tissue-specific analyses. With the increased sample size, increased cis allelic heterogeneity has been observed (providing more independent instruments) and more robust allele-specific expression QTLs have been identified. The new data have also revealed fundamental differences in the genetic architecture of gene expression and splicing. Importantly, this work identified cell-type composition as a key driver for tissue-specific eQTLs. Unfortunately, the sample size for any given tissue in GTEx is still >30 times lower than meta-analyzed data from whole blood by the eQTLGen Consortium (Võsa et al. 2018). This represents a considerable limitation to identifying tissue-specific causal genes given the limited number of eGenes shared between the tissues. However, whereas blood may not necessarily be the causal tissue, gene expression in blood may be a sufficient proxy for expression levels in other tissues. For example, cis eQTL effects are highly correlated (r = 0.7) between brain and whole blood for genes expressed in both tissues (Qi et al. 2018). Until larger tissue-specific samples become available, maximizing the available resources in whole blood studies can thus be a viable strategy.
Cis versus Trans QTLs as Instruments
It is estimated that ∼70% of gene expression heritability is via trans effects, which is probably induced by the modulation of upstream genes (Liu et al. 2019). This observation is compatible with an underlying model whereby only a relatively small number of (partly correlated) core genes have direct effects on gene expression, while the bulk of eQTLs exert their effect through regulating these core genes. Thus, using cis eQTLs would be the most appropriate variants to estimate the impact of gene expression, because trans eQTLs are more likely to represent indirect effects and hence are subject to pleiotropy, thus violating MR assumptions. Still, a potential weakness of using cis eQTLs as instruments is that the signals they represent are all derived from the same region. While large eQTL data sets provide evidence that most genes have several statistically independent eQTLs, they may still be influenced by shared haplotype effects (i.e., a common confounder). In addition, cis eQTLs are sometimes shared among neighboring genes, hindering the distinction between causal effects of coregulated biomarkers. Identifying independent and not shared secondary associations in a multivariable MR setting can circumvent such issues, but this requires larger sample sizes (Porcu et al. 2019).
FUTURE DIRECTIONS
Sex-Specific Analysis
The extant literature shows that sex could modify the effect of causal variants (Ober et al. 2008; Randall et al. 2013; Winkler et al. 2015). Such sex-specific associations could arise as a result of sexual dimorphism in gene/protein expression. To explore this hypothesis and to better understand the genetic basis of sexual dimorphism, applying a sex-specific omics-MR, combining sex-specific summary statistics for both omics-QTLs and complex human traits would be necessary. Such analysis would reveal whether sexual dimorphism observed for complex traits (such as waist-to-hip ratio) is already present at the transcriptome/methylome/proteome level or appears only downstream.
Diseases Modify Gene Expression
Most TWMR efforts have focused on using cis eQTLs as instruments to tease out the causal effect of gene expression on a complex trait. The (causal) impact of diseases on the transcriptome program has only very recently been investigated in a large eQTL study (Võsa et al. 2018). It was found that disease-associated genetic variants affect expression levels more often in trans than in cis. Interestingly, only 4% of trans eQTL effects could be explained by mediation of a cis eQTL effect, indicating that in addition to gene–gene regulation, many other nontranscriptional stimuli play a role in gene expression modification. These facts imply that diseases may have more pronounced impact on gene expression than the reverse, thus comparing gene expression levels of diseased and healthy subjects may reveal the transcriptomic fingerprint of a disease. In fact, the identification of such differentially expressed genes (DEGs) has long been the prevailing approach to tease out disease biology (e.g., Rodriguez-Esteban and Jiang 2017). However, DEGs may be causes, consequences, or mere correlates of the disease under scrutiny.
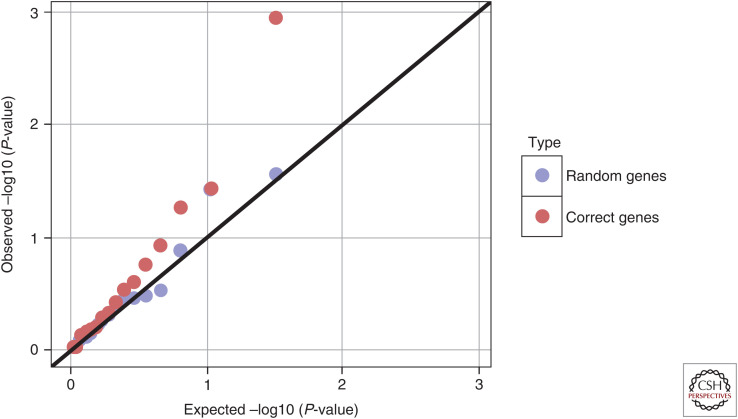
As a proof-of-concept, we asked how highly TWMR-identified causal genes would rank in a DEG analysis. To address this question for LDL cholesterol, we computed the correlation between the expression levels of causally implicated genes for LDL (by TWMR) and the actual LDL level in an Estonian population (N = 490), part of the EGCUT biobank. We found that TWMR-implicated genes have only marginally stronger correlation with LDL than a random gene set of the same size (Fig. 3).
Figure 3.
QQ plot of the correlation P-values. Observational correlations were calculated between low-density lipoprotein (LDL) levels and whole blood expression levels of causal genes for LDL (by transcriptome-wide Mendelian randomization [TWMR]) in the EGCUT study (in red). Correlations were also calculated between LDL levels and a set of random genes (in blue).
Although the gene expression study was very small and many more traits need to be studied, the results indicate that analyzing DEGs might reveal disease-induced changes in the transcriptome rather than disease-causing genes.
Extension to (Gene Regulatory) Network of Causal Effects
Despite the fact that the basic network MR concept has been introduced (Burgess et al. 2015), its application to real biological/clinical data remains scarce. MR could be extended to estimate bidirectional causal effects for every pair of eGenes and then iteratively identify and eliminate indirect edges through a generalized version of summary statistics-based mediation analysis (Burgess et al. 2017). The resulting gene–gene regulatory causal network should be comparable to tissue-specific regulatory circuits built based on promoter-enhancer activity (Marbach et al. 2016). Networks should be drawn for each layer of omics data and their relative causal relationship to complex traits needs to be examined via mediation analysis, revealing direct and indirect effects.
Causal networks would allow the dissection of direct and indirect effects in GWAS by adjusting the observed SNP-trait associations by the sum of the total SNP effects acting through all the incoming causal edges to that focal trait. In a previous work (McDaid et al. 2017), we used incoming causal edges to create prior effect estimates, but these priors could as well be subtracted from the observed effects to classify SNPs into core and peripheral genes—such a distinction has been promoted by the omnigenic model (Boyle et al. 2017).
More recent work on the omnigenic model proposed trans eQTLs as an explanation for the observed complex trait architecture (Liu et al. 2019). More research is needed toward this direction as their proposed model ignores reverse causality from trait to gene expression and assumes that most trans eQTLs are downstream effects of cis eQTLs, which has not been convincingly evidenced (Võsa et al. 2018). Linking eQTLs and traitQTL in a causal model setting has gained attention. An elegant extension of the LD score regression allowed the estimation of the fraction of trait heritability propagated through cis gene expression regulation (Yao et al. 2020). They estimated that ∼11% of trait heritability could be explained by cis eQTL regulation on average across 42 traits, with up to 30% for CAD.
Omics-Based MR to Boost Drug Development and Repositioning
It has been tempting to exploit disease-associated loci identified by GWAS to aid drug discovery (Yin et al. 2018). For this, not only disease onset, but disease progression genetics need to be exploited (Paternoster et al. 2017). Under ideal settings, the effect of an SNP might share mechanisms with the impact of a drug. Therefore, if an allele of a disease-associated SNP is predisposing to disease through increased gene/protein expression, a drug suppressing the level of the same gene/protein may be beneficial for that disease. Several examples exist where GWASs identified disease-relevant genes that are targets of efficient drugs. These include the statin-targeted HMGCR gene, which is associated with LDL levels (Swerdlow et al. 2015), psoriasis, and inflammatory bowel disease–associated IL12, T2D-associated ATM, and the LDL-associated PCSK9 gene (Robinson et al. 2018). In addition, phenome-wide association studies (pheWASs) may support the elucidation of side effect profiles (e.g., Neuraz et al. 2013).
Recent genome-wide approaches proposed to match a pharmagenic enrichment score of a drug with the polygenic risk score for a disease (Reay et al. 2020). Others (So et al. 2017) used MetaXcan (Barbeira et al. 2018) to impute genetically determined expression in disease cases versus controls. The differential gene expression is then contrasted to the impact of a drug on the transcriptome profile (obtained from the Connectivity Map [Subramanian et al. 2017]).
Notably, many of these approaches can lend themselves to predict off-target effects of drugs. For example, a recent MR study (Richardson et al. 2020) has shown that high HMGCR expression also lowers T2D risk, explaining why HMGCR-lowering drugs (e.g., statins) present diabetes-related side effects. The same study also demonstrated that ACHE—a target for several Alzheimer's medications—expression is positively correlated with blood pressure.
CONCLUDING REMARKS
With the increasing sample size of GWAS and sequenced reference panels, fine-mapping frequently yields credible sets containing only a handful of causal genetic variants. Still the functional characterization and the molecular consequences of those genetic variants mostly remain unclear. Although mapping leads SNPs to genes based on physical distance has already revealed meaningful insights (especially for coding variants), it is an oversimplification of the underlying biology. The emergence of large omics data sets (transcriptomics, proteomics, methylomics, metabolomics, chromatin states, etc.) with genomic information have led to the discovery of tens of thousands of omics QTLs (eQTL, splicing QTLs [Aguet et al. 2019], single-cell eQTL [Van der Wijst et al. 2019], pQTL [Suhre et al. 2017], mQTL [Bonder et al. 2017], meQTL [Shin et al. 2014], and cQTL [Delaneau et al. 2019]). The central challenge is to combine these QTLs with GWAS results of complex traits in a tissue-specific manner (Aguet et al. 2019). Clearly, competing risk factors need to be simultaneously included in a multivariable MR setting (Sanderson et al. 2019; Sanderson 2020). The MR framework lends itself to extensions integrating various sources of information and generate hypotheses on a massive scale. The novel insights gained from such an approach has tremendous potential to reveal the basis of the underlying genetic network, which in turn can be leveraged to boost drug repositioning and discovery.
ACKNOWLEDGMENTS
Z.K. was funded by the Swiss National Science Foundation (31003A-143914, 310030-189147). E.P. received support from the Swiss National Science Foundation (32003B-173092).
Footnotes
Editors: George Davey Smith, Rebecca Richmond, and Jean-Baptiste Pingault
Additional Perspectives on Combining Human Genetics and Causal Inference to Understand Human Disease and Development available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Abdellaoui A, Hugh-Jones D, Yengo L, Kemper KE, Nivard MG, Veul L, Holtz Y, Zietsch BP, Frayling TM, Wray NR, et al. 2019. Genetic correlates of social stratification in Great Britain. Nat Hum Behav 3: 1332–1342. 10.1038/s41562-019-0757-5 [DOI] [PubMed] [Google Scholar]
- Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B, Kasela S, Kim-Hellmuth S, Liang Y, Oliva M, et al. 2019. The GTEx Consortium atlas of genetic regulatory effects across human tissues. bioRxiv 787903. 10.1101/787903 [DOI] [Google Scholar]
- Al Awam K, Haußleiter IS, Dudley E, Donev R, Brüne M, Juckel G, Thome J. 2015. Multiplatform metabolome and proteome profiling identifies serum metabolite and protein signatures as prospective biomarkers for schizophrenia. J Neural Transm (Vienna) 122: 111–122. 10.1007/s00702-014-1224-0 [DOI] [PubMed] [Google Scholar]
- Allard C, Desgagné V, Patenaude J, Lacroix M, Guillemette L, Battista MC, Doyon M, Ménard J, Ardilouze JL, Perron P, et al. 2015. Mendelian randomization supports causality between maternal hyperglycemia and epigenetic regulation of leptin gene in newborns. Epigenetics 10: 342–351. 10.1080/15592294.2015.1029700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards TL, et al. 2018. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 9: 1825. 10.1038/s41467-018-03621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JA, Bull CJ, Gunter MJ, Carslake D, Mahajan A, Davey Smith G, Timpson NJ, Vincent EE. 2020. Early metabolic features of genetic liability to type 2 diabetes: cohort study with repeated metabolomics across early life. Diabetes Care 43: 1537–1545. 10.2337/dc19-2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Domingue BW, Wedow R, Arseneault L, Boardman JD, Caspi A, Conley D, Fletcher JM, Freese J, Herd P, et al. 2018. Genetic analysis of social-class mobility in five longitudinal studies. Proc Natl Acad Sci 115: E7275–E7284. 10.1073/pnas.1801238115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, van Iterson M, van Dijk F, van Galen M, Bot J, et al. 2017. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet 49: 131–138. 10.1038/ng.3721 [DOI] [PubMed] [Google Scholar]
- Boyle EA, Li YI, Pritchard JK. 2017. An expanded view of complex traits: from polygenic to omnigenic. Cell 169: 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium; Duncan L, et al. 2015a. An atlas of genetic correlations across human diseases and traits. Nat Genet 47: 1236–1241. 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J; Working Group of the Psychiatric Genomics Consortium; Patterson N, Daly MJ, Price AL, Neale BM. 2015b. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47: 291–295. 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Daniel RM, Butterworth AS, Thompson SG; EPIC-InterAct Consortium. 2015. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol 44: 484–495. 10.1093/ije/dyu176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S, Thompson DJ, Rees JMB, Day FR, Perry JR, Ong KK. 2017. Dissecting causal pathways using Mendelian randomization with summarized genetic data: application to age at menarche and risk of breast cancer. Genetics 207: 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon CP, Braunwald E, McCabe CH, Rader DJ, Rouleau JL, Belder R, Joyal SV, Hill KA, Pfeffer MA, Skene AM, et al. 2004. Intensive versus moderate lipid lowering with statins after acute coronary syndromes. N Engl J Med 350: 1495–1504. 10.1056/NEJMoa040583 [DOI] [PubMed] [Google Scholar]
- Darrous L, Mounier N, Kutalik Z. 2020. Simultaneous estimation of bi-directional causal effects and heritable confounding from GWAS summary statistics. medRxiv 10.1101/2020.01.27.20018929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies NM, Howe LJ, Brumpton B, Havdahl A, Evans DM, Davey Smith G. 2019. Within family Mendelian randomization studies. Hum Mol Genet 28: R170–R179. 10.1093/hmg/ddz204 [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zazhytska M, Borel C, Giannuzzi G, Rey G, Howald C, Kumar S, Ongen H, Popadin K, Marbach D, et al. 2019. Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science 364: eaat8266. 10.1126/science.aat8266 [DOI] [PubMed] [Google Scholar]
- Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, Ntritsos G, Dimou N, Cabrera CP, Karaman I, et al. 2018. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet 50: 1412–1425. 10.1038/s41588-018-0205-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, et al. 2011. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 7: e1002197. 10.1371/journal.pgen.1002197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference BA, Yoo W, Alesh I, Mahajan N, Mirowska KK, Mewada A, Kahn J, Afonso L, Williams KA Sr, Flack JM. 2012. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. J Am Coll Cardiol 60: 2631–2639. 10.1016/j.jacc.2012.09.017 [DOI] [PubMed] [Google Scholar]
- Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. 2015. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 × 2 factorial Mendelian randomization study. J Am Coll Cardiol 65: 1552–1561. 10.1016/j.jacc.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell Z, Davey Smith G, Sterne JA. 2007. The impact of residual and unmeasured confounding in epidemiologic studies: a simulation study. Am J Epidemiol 166: 646–655. 10.1093/aje/kwm165 [DOI] [PubMed] [Google Scholar]
- Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh PR, Lareau C, Shoresh N, et al. 2018. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet 50: 621–629. 10.1038/s41588-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune MD, Guo H, Burren O, Schofield E, Walker NM, Ban M, Sawcer SJ, Bowes J, Worthington J, Barton A, et al. 2015. Statistical colocalization of genetic risk variants for related autoimmune diseases in the context of common controls. Nat Genet 47: 839–846. 10.1038/ng.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Chen CY, Neale BM, Sabuncu MR, Smoller JW. 2017. Phenome-wide heritability analysis of the UK Biobank. PLoS Genet 13: e1006711. 10.1371/journal.pgen.1006711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. 2014. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10: e1004383. 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giugliano RP, Sabatine MS. 2015. Are PCSK9 inhibitors the next breakthrough in the cardiovascular field? J Am Coll Cardiol 65: 2638–2651. 10.1016/j.jacc.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Global Lipids Genetics Consortium; Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat Genet 45: 1274–1283. 10.1038/ng.2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Perez M, Agha G, Pang Y, Bermudez JD, Tellez-Plaza M. 2019. Mendelian randomization and the environmental epigenetics of health: a systematic review. Curr Environ Health Rep 6: 38–51. 10.1007/s40572-019-0226-3 [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. 2015. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Navarro E, Sánchez-Iglesias S, Domingo-Jiménez R, Victoria B, Ruiz-Riquelme A, Rábano A, Loidi L, Beiras A, González-Méndez B, Ramos A, et al. 2013. A new seipin-associated neurodegenerative syndrome. J Med Genet 50: 401–409. 10.1136/jmedgenet-2013-101525 [DOI] [PubMed] [Google Scholar]
- Guo H, Fortune MD, Burren OS, Schofield E, Todd JA, Wallace C. 2015. Integration of disease association and eQTL data using a Bayesian colocalisation approach highlights six candidate causal genes in immune-mediated diseases. Hum Mol Genet 24: 3305–3313. 10.1093/hmg/ddv077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ, Boomsma DI, Wright FA, et al. 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48: 245–252. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E, Weedon M, Bray N, O'Donovan M, Mill J. 2017. Pleiotropic effects of trait-associated genetic variation on DNA methylation: utility for refining GWAS loci. Am J Hum Genet 100: 954–959. 10.1016/j.ajhg.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth S, Mitchell R, Corbin L, Wade KH, Dudding T, Budu-Aggrey A, Carslake D, Hemani G, Paternoster L, Smith GD, et al. 2019. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat Commun 10: 333. 10.1038/s41467-018-08219-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Moore M, Chong S, Dillman A, Trabzuni D, Gibbs JR, Ryten M, Arepalli S, Weale ME, et al. 2012. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol Dis 47: 20–28. 10.1016/j.nbd.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MV, Simon T, Exeter HJ, Folkersen L, Asselbergs FW, Guardiola M, Cooper JA, Palmen J, Hubacek JA, Carruthers KF, et al. 2013. Secretory phospholipase A2-IIA and cardiovascular disease: a Mendelian randomization study. J Am Coll Cardiol 62: 1966–1976. 10.1016/j.jacc.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, van de Bunt M, Segrè AV, Li X, Joo JWJ, Bilow M, Sul JH, Sankararaman S, Pasaniuc B, Eskin E. 2016. Colocalization of GWAS and eQTL signals detects target genes. Am J Hum Genet 99: 1245–1260. 10.1016/j.ajhg.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, Yao C, Liu C, Ma J, Richard M, et al. 2019. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun 10: 4267. 10.1038/s41467-019-12228-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes RA, Davies NM, Davey Smith G, Tilling K. 2019. Selection bias when estimating average treatment effects using one-sample instrumental variable analysis. Epidemiology 30: 350–357. 10.1097/EDE.0000000000000972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. 10.1534/genetics.107.080101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, Benonisdottir S, Oddsson A, Halldorsson BV, Masson G, et al. 2018. The nature of nurture: effects of parental genotypes. Science 359: 424–428. 10.1126/science.aan6877 [DOI] [PubMed] [Google Scholar]
- Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. 2016. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol 12: e1004714. 10.1371/journal.pcbi.1004714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. 2008. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 27: 1133–1163. 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linner R, et al. 2018. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 50: 1112–1121. 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepik K, Annilo T, Kukuškina V; eQTLGen Consortium; Kisand K, Kutalik Z, Peterson P, Peterson H. 2017. C-reactive protein upregulates the whole blood expression of CD59—an integrative analysis. PLoS Comput Biol 13: e1005766. 10.1371/journal.pcbi.1005766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, van Klinken JB, Semiz S, van Dijk KW, Verhoeven A, Hankemeier T, Harms AC, Sijbrands E, Sheehan NA, van Duijn CM, et al. 2017. A Mendelian randomization study of metabolite profiles, fasting glucose, and type 2 diabetes. Diabetes 66: 2915–2926. 10.2337/db17-0199 [DOI] [PubMed] [Google Scholar]
- Liu X, Li YI, Pritchard JK. 2019. Trans effects on gene expression can drive omnigenic inheritance. Cell 177: 1022–1034.e6. 10.1016/j.cell.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, et al. 2018. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 50: 1505–1513. 10.1038/s41588-018-0241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier RM, Zhu Z, Lee SH, Trzaskowski M, Ruderfer DM, Stahl EA, Ripke S, Wray NR, Yang J, Visscher PM, et al. 2018. Improving genetic prediction by leveraging genetic correlations among human diseases and traits. Nat Commun 9: 989. 10.1038/s41467-017-02769-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B. 2017. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am J Hum Genet 100: 473–487. 10.1016/j.ajhg.2017.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N, Gayther S, Gusev A, Zheng W, Penney KL, Kote-Jarai Z, Eeles R, Freedman M, Haiman C, Pasaniuc B, et al. 2018. Large-scale transcriptome-wide association study identifies new prostate cancer risk regions. Nat Commun 9: 4079. 10.1038/s41467-018-06302-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Lamparter D, Quon G, Kellis M, Kutalik Z, Bergmann S. 2016. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat Methods 13: 366–370. 10.1038/nmeth.3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marouli E, Graff M, Medina-Gomez C, Lo KS, Wood AR, Kjaer TR, Fine RS, Lu Y, Schurmann C, Highland HM, et al. 2017. Rare and low-frequency coding variants alter human adult height. Nature 542: 186–190. 10.1038/nature21039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaid AF, Joshi PK, Porcu E, Komljenovic A, Li H, Sorrentino V, Litovchenko M, Bevers RPJ, Rüeger S, Reymond A, et al. 2017. Bayesian association scan reveals loci associated with human lifespan and linked biomarkers. Nat Commun 8: 15842. 10.1038/ncomms15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik BC. 2012. Leucine signaling in the pathogenesis of type 2 diabetes and obesity. World J Diabetes 3: 38–53. 10.4239/wjd.v3.i3.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelson MM, Marioni RE, Joehanes R, Liu C, Hedman AK, Aslibekyan S, Demerath EW, Guan W, Zhi D, Yao C, et al. 2017. Association of body mass index with DNA methylation and gene expression in blood cells and relations to cardiometabolic disease: a Mendelian randomization approach. PLoS Med 14: e1002215. 10.1371/journal.pmed.1002215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein J, Zhang B, Zhu J, Schadt EE. 2009. Disentangling molecular relationships with a causal inference test. BMC Genet 10: 23. 10.1186/1471-2156-10-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TT, Davies NM, Hemani G, Smith GD. 2019. Why are education, socioeconomic position and intelligence genetically correlated? bioRxiv 630426. 10.1101/630426 [DOI] [Google Scholar]
- Morrison J, Knoblauch N, Marcus JH, Stephens M, He X. 2020. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat Genet 10.1038/s41588-020-0631-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myocardial Infarction Genetics Consortium Investigators; Stitziel NO, Won HH, Morrison AC, Peloso GM, Do R, Lange LA, Fontanillas P, Gupta N, Duga S, et al. 2014. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med 371: 2072–2082. 10.1056/NEJMoa1405386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuraz A, Chouchana L, Malamut G, Le Beller C, Roche D, Beaune P, Degoulet P, Burgun A, Loriot MA, Avillach P. 2013. Phenome-wide association studies on a quantitative trait: application to TPMT enzyme activity and thiopurine therapy in pharmacogenomics. PLoS Comput Biol 9: e1003405. 10.1371/journal.pcbi.1003405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, Dermitzakis ET. 2010. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet 6: e1000895. 10.1371/journal.pgen.1000895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6: e1000888. 10.1371/journal.pgen.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Loisel DA, Gilad Y. 2008. Sex-specific genetic architecture of human disease. Nat Rev Genet 9: 911–922. 10.1038/nrg2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongen H, Brown AA, Delaneau O, Panousis NI, Nica AC; Consortium GTEx; Dermitzakis ET. 2017. Estimating the causal tissues for complex traits and diseases. Nat Genet 49: 1676–1683. 10.1038/ng.3981 [DOI] [PubMed] [Google Scholar]
- Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, Davey Smith G, Sterne JA. 2012. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res 21: 223–242. 10.1177/0962280210394459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passador-Gurgel G, Hsieh WP, Hunt P, Deighton N, Gibson G. 2007. Quantitative trait transcripts for nicotine resistance in Drosophila melanogaster. Nat Genet 39: 264–268. 10.1038/ng1944 [DOI] [PubMed] [Google Scholar]
- Paternoster L, Tilling K, Davey Smith G. 2017. Genetic epidemiology and Mendelian randomization for informing disease therapeutics: conceptual and methodological challenges. PLoS Genet 13: e1006944. 10.1371/journal.pgen.1006944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T, et al. 2015. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun 6: 5890. 10.1038/ncomms6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petretto E, Sarwar R, Grieve I, Lu H, Kumaran MK, Muckett PJ, Mangion J, Schroen B, Benson M, Punjabi PP, et al. 2008. Integrated genomic approaches implicate osteoglycin (Ogn) in the regulation of left ventricular mass. Nat Genet 40: 546–552. 10.1038/ng.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingault JB, O'Reilly PF, Schoeler T, Ploubidis GB, Rijsdijk F, Dudbridge F. 2018. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet 19: 566–580. 10.1038/s41576-018-0020-3 [DOI] [PubMed] [Google Scholar]
- Porcu E, Rüeger S, Lepik K; eQTLGen Consortium; BIOS Consortium; Santoni FA, Reymond A, Kutalik Z. 2019. Mendelian randomization integrating GWAS and eQTL data reveals genetic determinants of complex and clinical traits. Nat Commun 10: 3300. 10.1038/s41467-019-10936-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L, Zhu Z, Kemper K, Yengo L, Zheng Z, et al. 2018. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun 9: 2282. 10.1038/s41467-018-04558-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffler J, Friedrich N, Arnold M, Kacprowski T, Rueedi R, Altmaier E, Bergmann S, Budde K, Gieger C, Homuth G, et al. 2015. Genome-wide association study with targeted and non-targeted NMR metabolomics identifies 15 novel loci of urinary human metabolic individuality. PLoS Genet 11: e1005487. 10.1371/journal.pgen.1005487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall JC, Winkler TW, Kutalik Z, Berndt SI, Jackson AU, Monda KL, Kilpeläinen TO, Esko T, Mägi R, Li S, et al. 2013. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet 9: e1003500. 10.1371/journal.pgen.1003500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay WR, Atkins JR, Carr VJ, Green MJ, Cairns MJ. 2020. Pharmacological enrichment of polygenic risk for precision medicine in complex disorders. Sci Rep 10: 879. 10.1038/s41598-020-57795-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton CL, Davey Smith G. 2012. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol 41: 161–176. 10.1093/ije/dyr233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relton CL, Davey Smith G. 2015. Mendelian randomization: applications and limitations in epigenetic studies. Epigenomics 7: 1239–1243. 10.2217/epi.15.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond RC, Sharp GC, Ward ME, Fraser A, Lyttleton O, McArdle WL, Ring SM, Gaunt TR, Lawlor DA, Davey Smith G, et al. 2016. DNA methylation and BMI: investigating identified methylation sites at HIF3A in a causal framework. Diabetes 65: 1231–1244. 10.2337/db15-0996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson TG, Haycock PC, Zheng J, Timpson NJ, Gaunt TR, Davey Smith G, Relton CL, Hemani G. 2018. Systematic Mendelian randomization framework elucidates hundreds of CpG sites which may mediate the influence of genetic variants on disease. Hum Mol Genet 27: 3293–3304. 10.1093/hmg/ddy210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson TG, Hemani G, Gaunt TR, Relton CL, Davey Smith G. 2020. A transcriptome-wide Mendelian randomization study to uncover tissue-dependent regulatory mechanisms across the human phenome. Nat Commun 11: 185. 10.1038/s41467-019-13921-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, Fonseca F, Nicolau J, Koenig W, Anker SD, et al. 2017. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 377: 1119–1131. 10.1056/NEJMoa1707914 [DOI] [PubMed] [Google Scholar]
- Robinson JR, Denny JC, Roden DM, Van Driest SL. 2018. Genome-wide and phenome-wide approaches to understand variable drug actions in electronic health records. Clin Transl Sci 11: 112–122. 10.1111/cts.12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Esteban R, Jiang X. 2017. Differential gene expression in disease: a comparison between high-throughput studies and the literature. BMC Med Genomics 10: 59. 10.1186/s12920-017-0293-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatine MS, Liu E, Morrow DA, Heller E, McCarroll R, Wiegand R, Berriz GF, Roth FP, Gerszten RE. 2005. Metabolomic identification of novel biomarkers of myocardial ischemia. Circulation 112: 3868–3875. 10.1161/CIRCULATIONAHA.105.569137 [DOI] [PubMed] [Google Scholar]
- Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, et al. 2017. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 376: 1713–1722. 10.1056/NEJMoa1615664 [DOI] [PubMed] [Google Scholar]
- *.Sanderson E. 2020. Multivariable Mendelian randomization and mediation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a038984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson E, Davey Smith G, Windmeijer F, Bowden J. 2019. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol 48: 713–727. 10.1093/ije/dyy262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, Dungan J, Newby LK, Hauser ER, Ginsburg GS, et al. 2010. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet 3: 207–214. 10.1161/CIRCGENETICS.109.852814 [DOI] [PubMed] [Google Scholar]
- Shaheen R, Faqeih E, Ansari S, Abdel-Salam G, Al-Hassnan ZN, Al-Shidi T, Alomar R, Sogaty S, Alkuraya FS. 2014. Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res 24: 291–299. 10.1101/gr.160572.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp GC, Relton CL. 2017. Epigenetics and noncommunicable diseases. Epigenomics 9: 789–791. 10.2217/epi-2017-0045 [DOI] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, et al. 2014. An atlas of genetic influences on human blood metabolites. Nat Genet 46: 543–550. 10.1038/ng.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjaarda J, Gerstein H, Chong M, Yusuf S, Meyre D, Anand SS, Hess S, Paré G. 2018a. Blood CSF1 and CXCL12 as causal mediators of coronary artery disease. J Am Coll Cardiol 72: 300–310. 10.1016/j.jacc.2018.04.067 [DOI] [PubMed] [Google Scholar]
- Sjaarda J, Gerstein HC, Yusuf S, Treleaven D, Walsh M, Mann JFE, Hess S, Paré G. 2018b. Blood HER2 and uromodulin as causal mediators of CKD. J Am Soc Nephrol 29: 1326–1335. 10.1681/ASN.2017070812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Chau CK, Chiu WT, Ho KS, Lo CP, Yim SH, Sham PC. 2017. Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nat Neurosci 20: 1342–1349. 10.1038/nn.4618 [DOI] [PubMed] [Google Scholar]
- Sohail M, Maier RM, Ganna A, Bloemendal A, Martin AR, Turchin MC, Chiang CWK, Hirschhorn JN, Daly MJ, Patterson N, et al. 2019. Polygenic adaptation on height is overestimated due to uncorrected population stratification in genome-wide association studies. eLife 8: e39702. 10.7554/eLife.39702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. 2017. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171: 1437–1452.e17. 10.1016/j.cell.2017.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, et al. 2017. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 8: 14357. 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, et al. 2018. Genomic atlas of the human plasma proteome. Nature 558: 73–79. 10.1038/s41586-018-0175-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow DI, Preiss D, Kuchenbaecker KB, Holmes MV, Engmann JE, Shah T, Sofat R, Stender S, Johnson PC, Scott RA, et al. 2015. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet 385: 351–361. 10.1016/S0140-6736(14)61183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AE, Jones HJ, Sallis H, Euesden J, Stergiakouli E, Davies NM, Zammit S, Lawlor DA, Munafò MR, Davey Smith G, et al. 2018. Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 47: 1207–1216. 10.1093/ije/dyy060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wijst M, de Vries DH, Groot HE, Trynka G, Hon CC, Bonder MJ, Stegle O, Nawijn MC, Idaghdour Y, van der Harst P, et al. 2020. The single-cell eQTLGen consortium. eLife 9: e52155. 10.7554/eLife.52155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangipurapu J, Stancáková A, Smith U, Kuusisto J, Laakso M. 2019. Nine amino acids are associated with decreased insulin secretion and elevated glucose levels in a 7.4-year follow-up study of 5,181 Finnish men. Diabetes 68: 1353–1358. 10.2337/db18-1076 [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 2017. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Võsa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, Kirsten H, Saha A, Kreuzhuber R, Kasela S, et al. 2018. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv 10.1101/447367 [DOI] [Google Scholar]
- Wahl S, Drong A, Lehne B, Loh M, Scott WR, Kunze S, Tsai PC, Ried JS, Zhang W, Yang Y, et al. 2017. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 541: 81–86. 10.1038/nature20784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainschtein P, Jain DP, Yengo L, Zheng Z, Cupples LA, Shadyab AH, McKnight B, Shoemaker BM, Mitchell BD, Psaty BM, et al. 2019. Recovery of trait heritability from whole genome sequence data. bioRxiv 588020. 10.1101/588020 [DOI] [Google Scholar]
- Wallace C, Rotival M, Cooper JD, Rice CM, Yang JH, McNeill M, Smyth DJ, Niblett D, Cambien F; The Cardiogenics Consortium, et al. 2012. Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum Mol Genet 21: 2815–2824. 10.1093/hmg/dds098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. 2011. Metabolite profiles and the risk of developing diabetes. Nat Med 17: 448–453. 10.1038/nm.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, Christiansen MW, Fairfax BP, Schramm K, Powell JE, et al. 2013. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45: 1238–1243. 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, Czajkowski J, Esko T, Fall T, Kilpeläinen TO, et al. 2015. The influence of age and sex on genetic associations with adult body size and shape: a large-scale genome-wide interaction study. PLoS Genet 11: e1005378. 10.1371/journal.pgen.1005378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Chu AY, Estrada K, Luan J, Kutalik Z, et al. 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 46: 1173–1186. 10.1038/ng.3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtz P, Wang Q, Kangas AJ, Richmond RC, Skarp J, Tiainen M, Tynkkynen T, Soininen P, Havulinna AS, Kaakinen M, et al. 2014. Metabolic signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS Med 11: e1001765. 10.1371/journal.pmed.1001765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtz P, Wang Q, Soininen P, Kangas AJ, Fatemifar G, Tynkkynen T, Tiainen M, Perola M, Tillin T, Hughes AD, et al. 2016. Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J Am Coll Cardiol 67: 1200–1210. 10.1016/j.jacc.2015.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. 2010. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42: 565–569. 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee SH, Robinson MR, Perry JR, Nolte IM, van Vliet-Ostaptchouk JV, et al. 2015. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 47: 1114–1120. 10.1038/ng.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao DW, O'Connor LJ, Price AL, Gusev A. 2020. Quantifying genetic effects on disease mediated by assayed gene expression levels. Nat Genet 52: 626–633. doi:10.1038/s41588-020-0625-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM, et al. 2018. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum Mol Genet 27: 3641–3649. 10.1093/hmg/ddy271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Gao C, Xu Y, Li B, Ruderfer DM, Chen Y. 2018. Learning opportunities for drug repositioning via GWAS and PheWAS findings. AMIA Jt Summits Transl Sci Proc 2017: 237–246. [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Haberland V, Baird D, Walker V, Haycock P, Gutteridge A, Richardson TG, Staley J, Elsworth B, Burgess S, et al. 2019. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. bioRxiv 627398. 10.1101/627398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, Montgomery GW, Goddard ME, Wray NR, Visscher PM, et al. 2016. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 48: 481–487. 10.1038/ng.3538 [DOI] [PubMed] [Google Scholar]