Abstract
Activating transcription factor 3 (ATF-3), a cyclic AMP-dependent transcription factor, has been shown to play a regulatory role in melanoma, although its function during tumor progression remains unclear. Here, we demonstrate that ATF-3 exhibits tumor suppressive function in melanoma. Specifically, ATF-3 nuclear expression was significantly diminished with melanoma progression from nevi to primary to metastatic patient melanomas, correlating low expression with poor prognosis. Significantly low expression of ATF-3 was also found in cultured human metastatic melanoma cells lines. Importantly, overexpression of ATF-3 in metastatic melanoma cell lines significantly inhibited cell growth, migration, and invasion in vitro; as well as abrogated tumor growth in human melanoma xenograft mouse model in vivo. RNA-seq analysis revealed downregulation of ERK and AKT pathways and upregulation in apoptotic-related genes in ATF-3 overexpressed melanoma cell lines, which was further validated by Western-blot analysis. In summary, this study demonstrated that diminished ATF-3 expression is associated with melanoma virulence and thus provides a potential target for novel therapies and prognostic biomarker applications.
Keywords: ATF-3, melanoma, ERK, AKT, RNA-seq, xenograft
Introduction:
Melanoma is a common and potentially aggressive form of skin cancer1. One of the major risk factors is excessive ultraviolet (UV) exposure2. Recent studies have provided insights into melanoma pathogenesis, and novel therapies that target pathways and check points that confer virulence have been promising, although overall survival remains poor in advanced metastatic disease3,4. Therefore, novel molecular mechanisms that mediate melanoma behavior require more comprehensive understanding in order to inform effective therapeutic strategies.
Activating transcription factor 3 (ATF-3) is a cyclic AMP-dependent transcription factor that is encoded by stress-inducible gene, ATF-3 5. Similar to other transcription factors in the CREB family, the expression of ATF-3 is associated with various pathophysiological responses, such as inflammation and apoptosis, as well as with various diseases6–8. ATF-3 plays a critical role in DNA damage repair and regulates the expression, activation, or repression of cell-cycle regulators. Downregulation of ATF-3 expression may destabilize p53-induced pathways, resulting in the activation of oncogenes9. ATF-3 is selectively induced by inhibiting calcineurin/NFAT signaling pathways in cutaneous squamous cell carcinomas from cyclosporine-treated patients, further suggesting ATF-3 as a critical transcriptional repressor of p53 and other senescence-associated genes8. Recently, ATF-3 has been detected in melanocytes and shown to be negatively regulated by transient receptor potential vanilloid 1 (TRPV1)-induced calcineurin activation in melanoma, with its oncogenic effect potentiated by calcineurin inhibitors7. Other studies have also shown that ATF-3 expression either promotes or inhibits tumorigenesis, suggesting the regulatory role of ATF-3 to likely be tumor-specific and signal pathway-dependent10,11. However, the detailed role of ATF-3 in melanogenesis remains incompletely understood and the function of ATF-3 in melanoma progression is not well characterized.
Materials and Methods:
Human melanoma samples
This study was conducted with approval of the Institutional Review Board of Brigham and Women’s Hospital, Harvard Medical School. In total, 72 cases were studied retrospectively: A cohort of 63 cases of melanocytic lesions, which included benign nevi (n=10), primary cutaneous (n=38) and metastatic melanomas (n=24) from a tissue microarray previously constructed from the archives of the Department of Pathology at Brigham and Women’s Hospital was obtained. Approval for the archived cases and the tissue microarray study were granted by the Brigham and Women’s Partners Human Research Committee. Informed consent was not necessary, as all tissue samples were discarded and deidentified. The hematoxylin and eosin (H&E) stained sections, pathological diagnoses, and reported prognostic attributes were independently reviewed and confirmed by two dermatopathologists (G.F.M., C.G.L.).
Routine Histology
All melanoma specimens were formalin fixed, paraffin-embedded, sectioned, and stained with H&E for histopathological evaluation.
Immunohistochemistry (IHC) and Immunofluorescence (IF) studies
Immunohistochemistry and immunofluorescence staining were performed according to a standard protocol. Sections were treated with heat-induced epitope retrieval using target retrieval solution (Dako, Cat: S1699, Carpenteria, CA, USA) and heated in a Pascal-pressurized heating chamber (Dako, 125 °C for 30 min, 90 °C for 10 min). After incubation with primary antibodies at 4 °C overnight, sections were incubated with horse radish peroxidase (HRP)-conjugated secondary antibodies for 30 min at room temperature, and signals were visualized with NovaRED HRP substrate (Vector Laboratories, Cat: SK-4800, Burlingame, CA, USA) with a hematoxylin counter stain. Alternatively, cells plated on chamber slides were fixed in 4% paraformaldehyde, penetrated with 1% Tween-20, incubated with primary antibodies at 4 °C overnight, followed by incubation with fluorophore-conjugated secondary antibodies for 30 min at room temperature. Isotype-matched irrelevant antibodies were used in place of primary antibodies as controls. The following primary and secondary antibodies were used for immunofluorescence studies on melanoma cell lines and human tissue specimens: Rabbit anti-ATF-3 antibody (1:300, Abcam, Cat: ab216569, Cambridge, MA), Mouse anti-MART-1 antibody (1:1, BioLegend, Cat: 917902, San Diego, CA), rabbit anti-Ki-67 antibody (1:500, Abcam, Cat: ab15580, Cambridge, MA), DyLight 594 goat anti-rabbit IgG(H+L) (1:500, Multiscience, Cat: GAR5942, Hangzhou, China) and DyLight 488 goat anti-mouse IgG(H+L) (1:500, Multiscience, Cat: GAM4882, Hangzhou, China). Immunofluorescence studies were evaluated by two independent investigators (G.F.M., C.G.L.).
Human melanoma cell culture and ATF3 gene transfection
Melanoma cell lines derived from human primary melanoma (Meljuso, WM115, M14) and metastatic melanoma (A375, SK-MEL-28, SK-MEL-30, UACC62, MeWo) were provided by Prof. David E. Fisher (Cutaneous Biology Research Center, Massachusetts General Hospital, Boston, USA). A2058 and C8161 melanoma cell lines were provided by Dr. Tobias Schatton (Dermatology Department, Boston, MA). C8161 and A2058 exhibited lower ATF-3 expression in comparison to other melanoma cell lines, and therefore, selected for further studies. C8161 and A2058 melanoma cell lines were cultured and transfected with the prepared retrovirus, as previously described8,12.
RNA extraction, Quantitative Real-Time Polymerase Chain Reaction (RT-PCR)
Total RNA extracted by RNAeasy kit (QIAGEN, Cat: 74104, Germantown, Maryland) and the cDNAs were synthesized by SuperScript III first strand kit (Bio-Rad, Cat: 1708891, Hercules, CA). The human melanoma cDNA arrays were obtained from Origen (Cat: MERT301). Relative gene expression was normalized to HPRT. The gene-specific primers and antibodies used for immunoblotting are provided in Supplementary Table 1. Conditions for RNA extraction and quantitative RT-PCR analysis were implemented as previously documented8,13–15.
Western Blot
Western Blot was performed as we previously reported8,14. Briefly, cells lysate was collected in 100 μl lysis buffer (100mM Tris PH 6.8, 2% SDS and 12% glycerinum). A Pierce BCA Protein Assay kit (Thermo Scientific, Cat: 23227, Waltham, MA) was used to collect supernatants and detect protein concentration. Cell lysates were diluted in loading buffer and heated at 100 °C for 5 min, then centrifuged for protein isolation. The proteins were electrophoresed in running buffer on Mini-protean® TGXTM Precast Gel (Bio-Rad, Cat: 456–1094, Hercules, CA) and transferred to Nitrocellulose membranes using transfer buffer. Primary antibodies—ATF-3, ERK, and AKT (Table 1) were immunoblotted at 4 °C overnight on a shaker. After washing, the membranes were then incubated with either goat anti-mouse or goat anti-rabbit IgGHRP antibodies (1:2000, 7074V, Cat: 7076V, Cell Signaling, Danvers, MA) at room temperature for 1 hr. Protein detection and densitometry measurement were completed by ChemiDOCTM XRS+ with Image Lab system (Bio-Rad, Hercules, CA).
Table 1:
Antibodies for Western blot
| Antibody | Species | Company | Category No. | DF |
|---|---|---|---|---|
| ATF-3 | Rabbit | Abcam (Cambridge, MA) | ab207434 | 1:2000 |
| ERK | Rabbit | Cell Signaling (Danvers, MA) | 4695 | 1:1000 |
| p-ERK | Rabbit | Cell Signaling (Danvers, MA) | 4370 | 1:2000 |
| AKT | Rabbit | Cell Signaling (Danvers, MA) | 4691 | 1:1000 |
| p-AKT | Rabbit | Cell Signaling (Danvers, MA) | 4060 | 1:2000 |
| GAPDH | Mouse | BioLegend (San Diego, CA) | 649203 | 1:10000 |
Cell proliferation, cell viability, colony formation and cell migration assays
Melanoma cell lines were analyzed for cell proliferation and cell viability by using EdU kit (RiboBio, Cat: C10310–2, Guangzhou, China) and Cell Counting Kit-8 (Dojindo, Cat: CK04, Japan), respectively, as previously described12,16. EdU-labelled cells were detected by an immunofluorescence microscope (Olympus BX53-DP80, Tokyo, Japan) and viable melanoma cells were quantified using SPECTROstar Nano microplate reader (BMG Labtech, Offenburg, Germany). For colony formation assays, virally-transfected melanoma cells were seeded in triplicates in 6-well plates at a density of 1500 cells/well and cultured as described16. An Olympus CKX41 inverted microscope (Olympus, Tokyo, Japan) was used to count the number of colonies. Cell migration assays were also performed with virally-transfected melanoma cells which were seeded in 6 well plates in triplicates and grown to 90% confluency as previously described12. An inverted microscope (Olympus CKX41, Tokyo, Japan) was used to photograph the cell monolayer at specific time points (0, 16h, 24h) and the migration distance was measured with ImageJ software. All experiments were repeated at least three times.
Matrigel Invasion Assay
Invasion chambers (Falcon, Cat: 353097, Austin, TX) were coated with Corning Matrigel Basement Membrane Matrix (Corning, Cat: 356234, Bedford, MA) per manufacturer’s instructions and 60,0000 ATF-3 over-expressing (OE) and Neomycin control melanoma cells were loaded on the surface and placed entirely onto a 24-well plate (in triplicates). DMEM containing 10% FBS was placed on the outer walls of the Matrigel embedded with either A2058 and C8161 melanoma cells and incubated with 5% CO2 at 37℃ for 16 hr and 24 hr, respectively. To visualize the migrated cells on the lower surface of the insert membranes, the lower aspect of the Matrigel was fixed with 4% paraformaldehyde solution for 30 minutes and stained with 0.1% crystal violet (Solarbio Science and Technology, Cat: G1063, Beijing). Images of the stained cells were captured and counted. The experiments were repeated at least three times.
Xenograft Assay
Tumor xenograft assay was performed as previously described8. 6-week-old female nude/nude mice (Charles River Laboratory, MA, USA) were assigned to two groups: C8161 ATF-3 overexpressing (ATF-3 OE) and C8161 neomycin (NEO) control melanoma cells. Each group comprised of six mice and was subcutaneously injected with either 1.0 × 106 C8161 ATF3 OE melanoma cells or 1.0 × 106 C8161neomycin control (NEO) cells in the left and right anterior and posterior flanks. After 3 weeks, the mice were euthanized, and the melanoma tumor volumes and weights were measured. Tumors were then fixed in formalin and embedded in paraffin for immunofluorescence analysis. The in vivo experiments were repeated 3 times. The same experiments were also performed using A2058 ATF-3 OE and A2058 NEO melanoma cells. All animal studies were approved by the Ethics Committee of Stomatology Hospital, Shandong University (Protocol No. GR201720, Date 02-27-2017) and carried out according to National Institute of Health Guideline for the Care and Use of Laboratory Animals.
RNA-sequencing
RNA-sequencing of the melanoma cell lines A2058 and C8161 OE ATF-3 or NEO was performed at the Beijing Genomics Institute (Shenzhen, China). Three technical replicates of C8161 were sequenced. Bowtie2 was used to align the reads and RSEM17 to quantify gene expression. Gene expression values for the three C8161 technical replicates were averaged. RPKM values ≤ 1 were assigned a value of 1 before log2 transformation. Genes expressed at < 5 RPKM in all samples (row sums) were excluded from further analysis. 10,993 genes remained. To determine genes differentially expressed between A2058 and C8161 ATF-3 OE and NEO cell lines, log2(0.2) fold change was used as a cut-off. Genes meeting this cut-off were used as input for Gene Ontology (GO) Analysis.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA). Student’s unpaired t test was used to compare two groups while one-way ANOVA was used for multiple pairwise comparisons. The data were summarized as mean ± standard error (SEM) for at least three different experiments and a p-value <0.05 was considered statistically significant.
Data Availability
The RNA-sequencing data have been deposited to the NCBI Gene Expression Omnibus under accession number GSE152460.
Results:
Expression of ATF-3 is significantly decreased in metastatic melanoma of human melanoma
We initially evaluated ATF-3 expression (protein immunoreactivity) in human melanocytic lesions, including normal melanocytes of skin, benign nevi and primary melanoma by IF. Strong nuclear positivity was present in normal melanocytes and benign nevi, while primary and metastatic melanoma showed partial and complete loss, respectively (Fig. 1A). Next, we validated the findings with a tissue microarray of 72 patient cases of melanocytic lesions, which included benign nevi (n=10), primary cutaneous melanomas (n=38) and metastatic melanomas (n=24) by IF (Fig. 1B). Multiplex IF revealed that ATF-3 expression decreased significantly with progression from nevi to melanoma (p<0.01), with an apparent qualitative diminution in nuclear staining seen in metastatic melanoma when compared to primary melanomas (p<0.05) (Fig. 1C). Multiplexing IF studies show a significant decrease of nuclear positivity of ATF-3 in the melanoma component in comparison to adjacent dermal nevus in the specimens of melanoma arising in dermal nevus (Supplementary Fig. S1).
Figure 1. ATF3 expression decreased in melanoma.
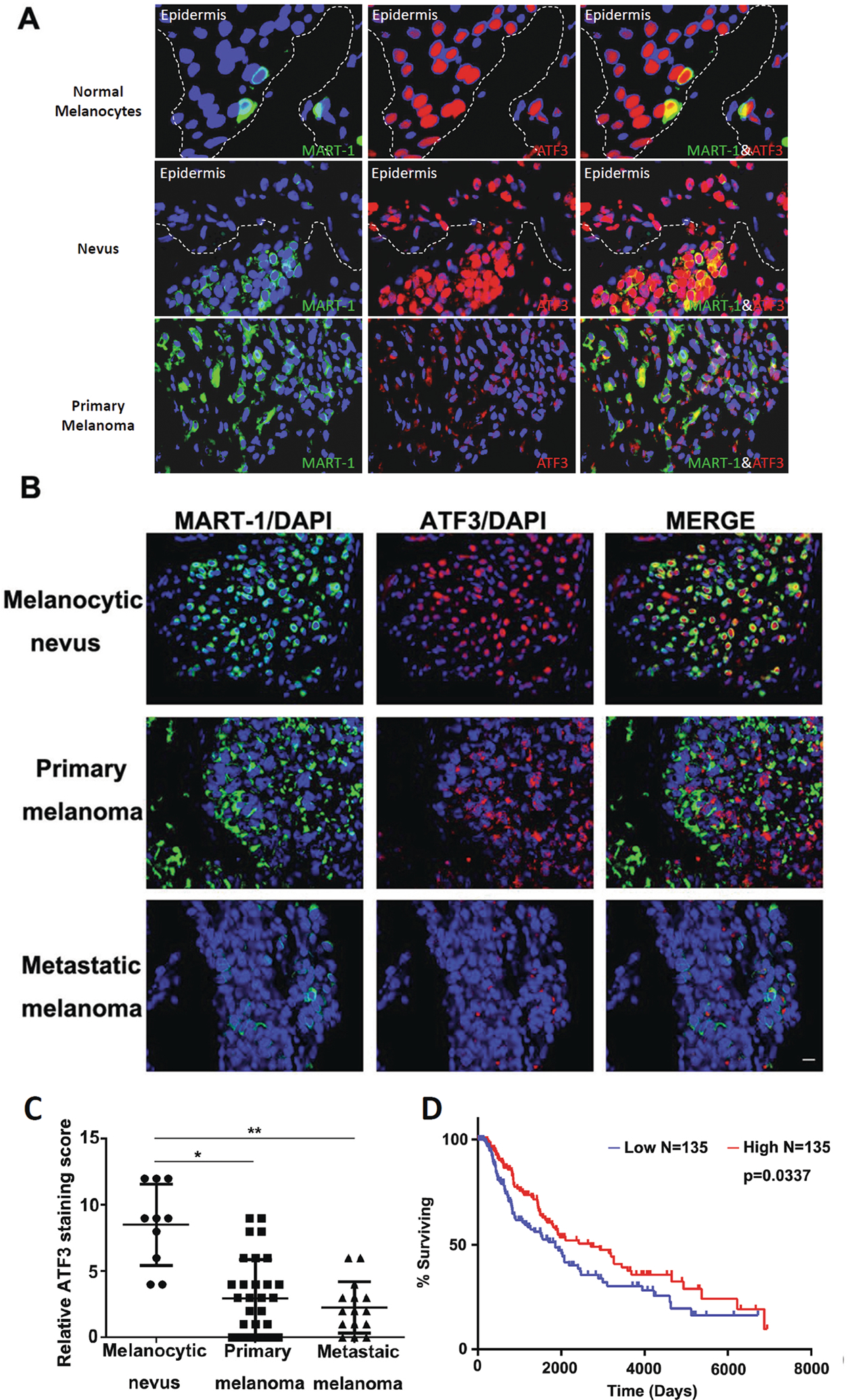
(A and B) Representative IF images of double staining of ATF-3 (red) and MART-1 (green) in human melanocytic specimens and tissue microarray, respectively. (A). Strong nuclear positivity of ATF-3 (red) present in normal melanocytes (40x) and benign nevi (20x), while melanoma showed marked decrease (20x) in the representative images. (B). Representative IF images of tissue microarray of 72 patient cases of melanocytic lesions, which included benign nevi (n=10), primary cutaneous melanomas (n=38) and metastatic melanomas (n=24) by IF are shown (10x) and revealed significant decrease of ATF-3 nuclear positivity (red) with progression from nevi to primary melanoma (p<0.01), with an apparent qualitative diminution in nuclear staining seen in metastatic melanoma when compared to primary melanomas (p<0.05) (C). Kaplan-Meier survival curves on the ATF3 level in the TCGA skin cutaneous melanoma database (D). Data are presented as the mean ± standard deviation, *P < 0.05; **P < 0.01.
Moreover, using the TCGA skin cutaneous melanoma database, a Kaplan-Meier analysis was performed on patients with ATF-3 mRNA level partitioned from the lowest 30% (n=135) and highest 30% (n=135) from a total pool of 458 patients. We found that lower ATF-3 mRNA level group exhibited a worse survival over time than those in the higher ATF-3 mRNA level group (p=0.0337) (Fig. 1D).
We then compared ATF-3 expression by Western Blot in a panel of melanoma cell lines. While some melanoma cell lines (mel-juso and wm115) expressed high levels of ATF-3, the protein levels of ATF-3 declined from primary to metastatic melanoma, with markedly lower expression of ATF-3 in melanoma cell lines derived from human metastatic melanomas (Supplementary Fig. S2A). These results were further confirmed by immunofluorescence studies of melanoma cell lines (Supplementary Fig. S2B).
Over-expression of ATF-3 inhibits melanoma cell proliferation, migration and invasion
Lower expression of ATF-3 in metastatic melanoma may indicate that ATF-3 plays a tumor suppressing function in melanoma. Studies have suggested that ATF-3 has an inverse regulatory relationship with p53, affecting cellular apoptosis and tumor progression7,8. Therefore, we investigated the effect of induced expression of ATF-3 in melanoma cells. We first overexpressed ATF-3 in melanoma cell lines derived from human metastatic melanomas, A2058 and C8161. Increased expression of ATF-3 was confirmed by immunofluorescence, Western Blotting and qRT-PCR analysis (Supplementary Fig. S3A–D). Immunofluorescence analysis revealed that the number of EdU incorporating cells was significantly reduced in melanoma cell lines that overexpressed ATF-3, consistent with suppression of proliferation (Fig.2A–D). Cell Counting Kit-8 and colonogenic assays indicated that the proliferation and survival, respectively, of melanoma cells were significantly decreased in ATF-3 OE melanoma cell lines (Fig. 2E–F) Moreover, overexpression of ATF-3 in melanoma cell lines also attenuated cell migration (Fig 2G–H) and invasion (Fig. 2I–J). Taken together, induced ATF-3 expression inhibited human metastatic melanoma cell growth, migration and invasion in vitro.
Figure 2. ATF3 over-expression inhibited melanoma cell growth, migration and invasion.
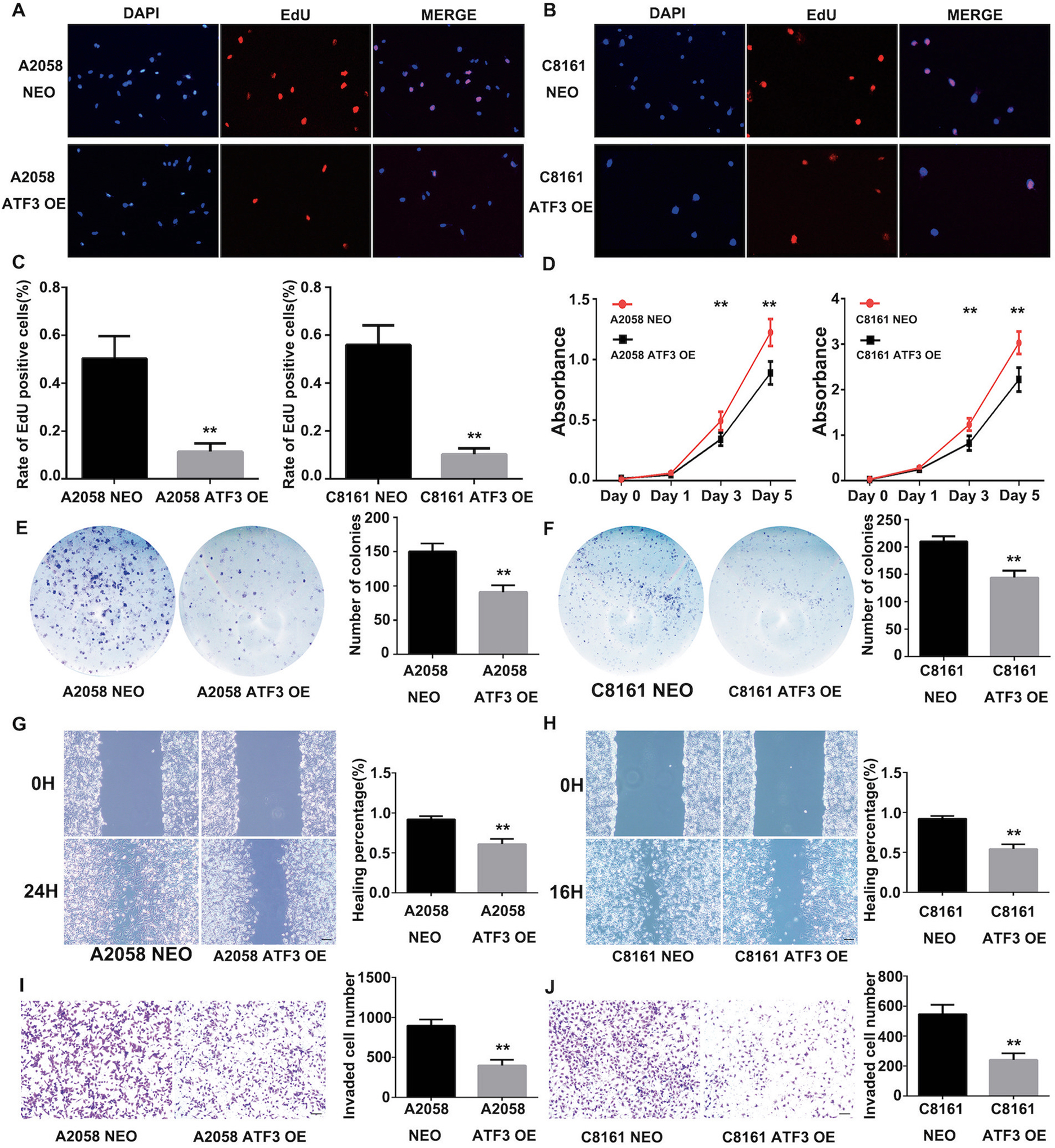
(A), (B) and (C) EdU incorporation assay in A2058 and C8161 ATF3 overexpression cell lines. The nuclei of EdU-positive cells were labeled red and cell nuclei were labeled blue with DAPI. The column summarized the staining scores of EdU-positive cells. Scale bar represent 100μm. (D) CCK-8 assay in A2058 and C8161 ATF3 overexpression cell lines. (E) and (F) Cell colony assay in A2058 and C8161 ATF3 overexpression cell lines. The column summarized the colonies of the cells. (G) and (H) Wound healing assay in A2058 and C8161 ATF3 overexpression cell lines. The column summarized the healing percentages compared with NEO control group. Scale bar represent 100μm. (I) and (J) Transwell assay for A2058 and C8161 ATF3 overexpression cell lines invasion tests. The column summarized the invaded cells. Scale bar represent 100μm. Data are presented as the mean ± standard deviation, **P < 0.01.
Over-expression ATF-3 significantly inhibits melanoma growth and tumor formation in a xenograft mouse model in vivo
In order to confirm our in vitro findings, next we investigated the effect of ATF-3 on melanoma growth in vivo. We deployed a xenograft model by subcutaneously injecting ATF-3 OE or control NEO C8161 melanoma cells into the left and right, anterior and posterior flanks of 6-week-old female nude mice. During 3 weeks of observation, the size, weight and rate of tumor growth of melanomas with ATF-3 overexpression were significantly less than that of the control group (Fig. 3A–C). These tumors were subjected to IF staining for proliferation marker Ki-67, which revealed a lower proliferative index in melanomas with ATF-3 overexpression (Fig. 3G–H). We repeated the same xenograft mouse model using ATF-3 OE or control NEO A2058 melanoma cells. The results were similar to those seen using ATF-3 OE and control C8161 melanoma cells (Fig. 3D–F, I–J). Therefore, the data altogether supported our in vitro findings, suggesting ATF-3 as a tumor suppressor in metastatic melanomas.
Figure 3. ATF3 over-expression inhibit melanoma tumor formation and growth in vivo.
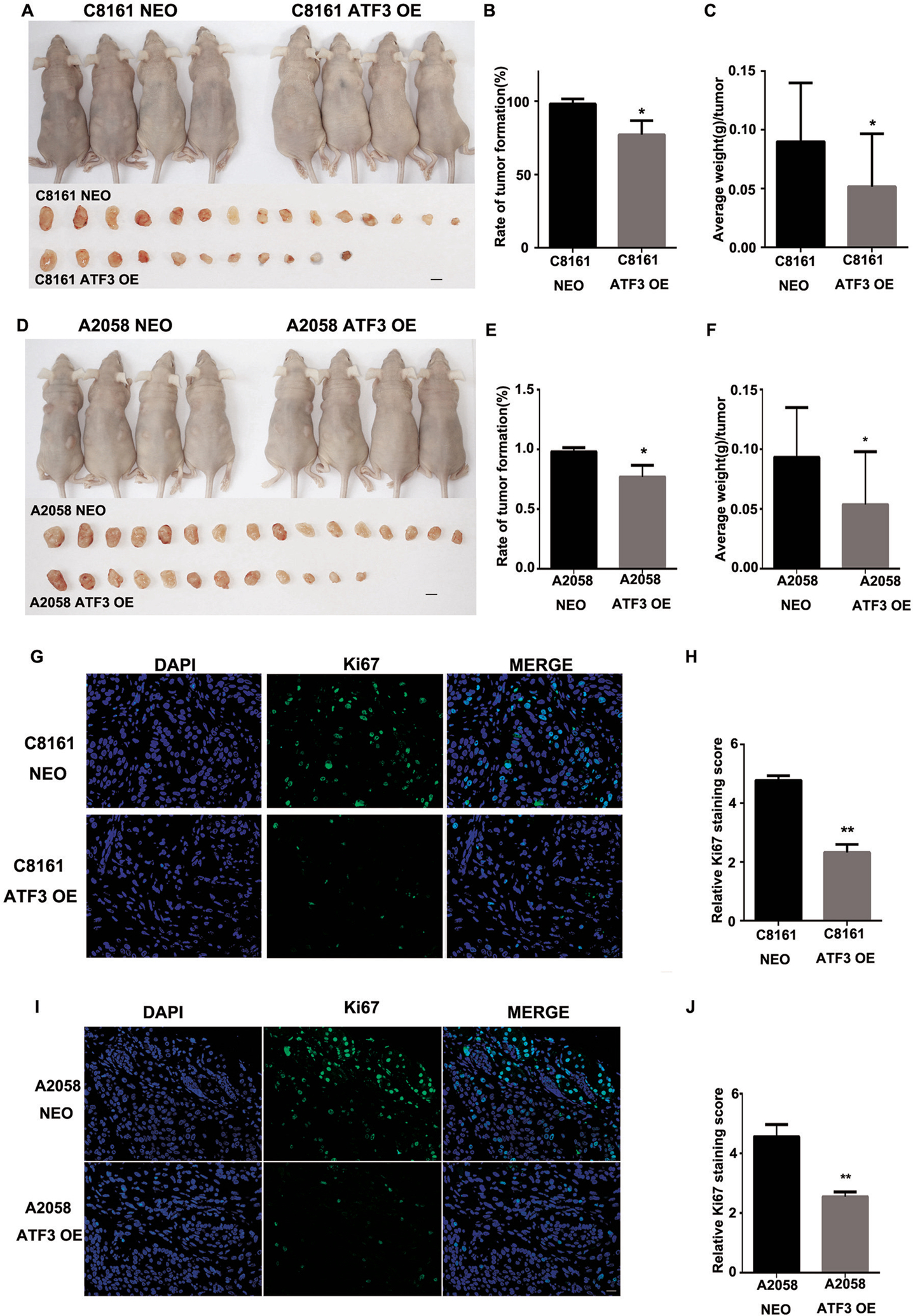
(A) The representative of images of mice at 3 wk after subcutaneously injection of either C8161 NEO and C8161 ATF3 OE cells, and each tumor was collected shown in the lower panel. (B) The percentage of C8161 NEO and C8161ATF3 OE tumor formation and (C) average weight of each tumor was quantified. (D) Representative images of mice at 3 wk after subcutaneously injection of either A2058 NEO and A2058 ATF3 OE cells, and each tumor was collected shown in the lower panel. (E) The percentage of A2058 NEO and A2058 OE tumor formation and (F) average weight of each tumor was quantified. (G) Representative images of Ki67 immunofluorescence staining in C8161 NEO and C8161 ATF3 OE melanoma tumors and (H) summarized staining score of Ki67 expression. (I) Representative images of Ki67 immunofluorescence staining in A2058 NEO and A2058 ATF3 OE melanoma tumors and (J) summarized staining score of Ki67 expression. Data are presented as the mean ± standard deviation, *P<0.05; **P < 0.01.
Enhanced expression of ATF-3 inhibits melanoma tumor growth by increasing apoptosis and downregulating ERK and AKT phosphorylation
The development and progression of melanoma have been linked to genetic alterations in a variety of pathways4,7,18. To investigate potential molecular mechanisms responsible for ATF-3-mediated inhibition of metastatic melanoma cell growth, migration, and invasion in vitro, we performed RNA-sequencing (RNA-seq) of A2058 and C8161 ATF-3 OE and NEO cell lines. Principal component analysis (PCA) performed on all genes detected by RNA-seq showed separation specific to the individual cell lines on PC1 while PC2 and PC3 separated the ATF-3 OE and NEO cells (Figure 4A). Prominent genes associated with apoptosis and ERK and AKT signaling pathways were identified using Homo sapiens Gene Summary from NCBI. In particular, genes found to be upregulated in both A2058 and C8161 ATF3-OE cells include, in addition to ATF-3, were the tumor suppressor ING4, and genes involved in promoting apoptosis and cell death such as CDKN1A and FOS. Gene Ontology (GO) analysis revealed pathways such as GO:0034976 ‘response to endoplasmic reticulum stress’ and GO:0008625 ‘extrinsic apoptotic signaling pathway via death domain receptors’ to be upregulated in C8161 ATF3-OE cells compared to NEO cells (FDR-adjusted p-value = 4.79 × 10−8 and 5.4 × 10−4, respectively) (Figure 4B).
Figure 4. ATF3 over-expression inhibits melanoma cell growth, migration and invasion by down regulating ERK and AKT phosphorylation.
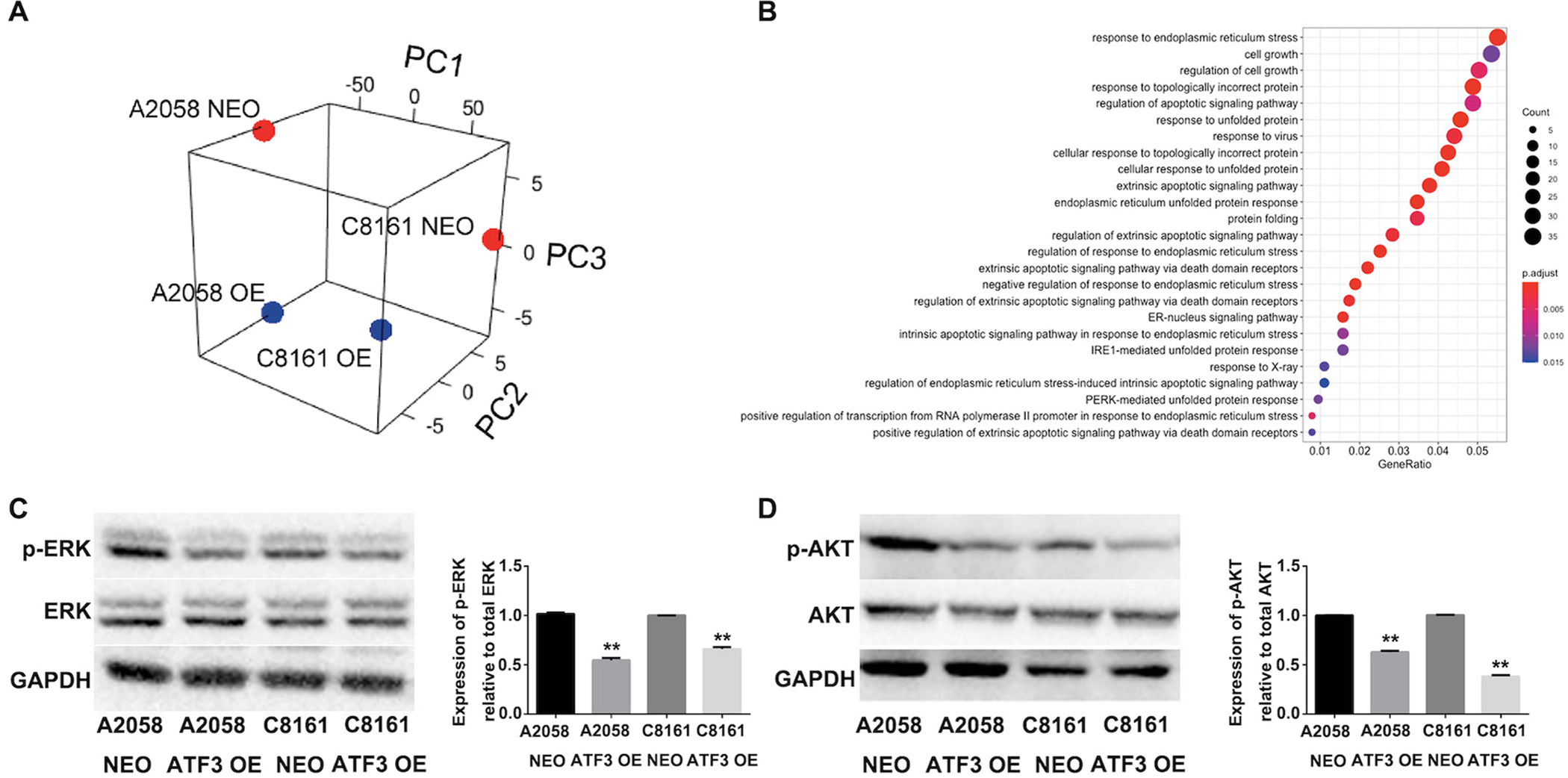
(A) PCA of all genes detected by RNA-seq of A2058 and C8161 ATF-3 overexpressing (OE) and neomycin control (NEO) cell lines. (n=2). (B) Dot plot of enriched GO terms. The 25 GO processes with the largest gene ratios are shown in order of gene ratio. Dot size represents the number of genes with increased expression in C8161 ATF3 OE cells compared to NEO associated with that GO term. Dot color indicates the FDR-adjusted p-value. (C) and (D) Total and phosphorylation level of ERK, and AKT changes in A2058 and C8161 NEO control and ATF3 overexpressed cell lines. The column summarized the protein phosphorylation level in melanoma cells, which normalized to total protein respectively. Data are presented as the mean ± standard deviation, **P < 0.01.
Downregulated genes, such as GDNF and TNFRSF6B involved with proliferation were also identified, suggesting a possible link to ERK and AKT signaling pathways. Western-Blotting analysis confirmed that levels of phosphorylated ERK and AKT were significantly downregulated in melanoma cell lines in which ATF-3 was overexpression (Fig. 4C–D), suggesting that ATF-3 plays a role in negatively regulating ERK and AKT pathways in melanoma cells.
Discussion:
ATF-3 is part of the ATF/CREB family and can act as either a transcriptional activator or repressor21. ATF-3 plays a dichotomous role in melanoma progression by either preventing transcription by stabilizing inhibitory co-factors at the promoter site or activating transcription of target genes via the C-Jun NH2-terminal kinase/stress-activated protein kinase (JNK/SAPK) stress-inducible signaling pathway22,23. In general, the expression of ATF-3 can be induced by anticancer drugs, cellular stress, toxic chemicals, UV irradiation and oncogenic signaling that ultimately triggers cell cycle arrest and apoptosis24–27.
ATF-3 partakes in a number of cellular signal transduction pathways. Previous studies have shown that the activation of various toll-like receptors (TLRs) may elevate the expression level of ATF-3 in bone marrow-derived macrophages28. Similarly, the upregulation of ATF-3 in response to Streptococcus pneumoniae infection via TLR2 and TLR4 in vitro suggests that ATF-3 plays a role in TLR signaling29. Consistent with previous literature, ATF-3 negatively regulates TLR signaling pathways evidenced by elevated IL-12 and IL-6 expression levels in macrophages from ATF-3−/− mice28. ATF-3 has also shown to positively regulate tumor suppressor gene, p53, in response to DNA damage and ATF3−/− mice exhibits genomic instability and spontaneous tumorigenesis30. Recently, ATF-3 has been reported to a be a direct target of Wnt/ β-catenin pathway and differentially regulates MMP and TIMPs in human colon cancer cells31.
In the past decade, the dysregulation of ATF-3 has been shown to play different roles in various tumors. Studies have shown that ATF-3 expression is downregulated in hepatocellular carcinoma, prostate cancer, and colon cancer32–34. Similarly, ATF-3 inhibits tumorigenesis and progression of esophageal squamous cell carcinoma by downregulating inhibitor of DNA-binding 1 (ID-1)35. In epithelial cells, ATF-3 appears to bind to ID-1 promoter and mitigates its expression level in response to cellular stress signals36. Analogous to these studies, ATF-3 inhibits ID-1 transcription and protein expression in metastatic melanoma and is partly mediated by melanoma cell adhesion molecules (MCAM/MUC18)37. However, the role of ATF-3 in signaling pathways for cancer progression is quite complex. The migration and proliferation rate of bladder cancer cells is diminished in metastatic bladder cancer cells with ATF-3 overexpression, suggesting that ATF-3 suppresses metastasis of bladder cancer cells through the upregulation of actin filament severing protein gelsolin (GSN)38. Similar to this data, we found ATF-3 expression to be significantly decreased with melanoma progression (Fig 1A–B). It is worth noting, though, that several other reports have found elevated ATF-3 expression in melanoma and lung cancer7,10. Since ATF-3 induction has been associated with oncogenic stress26, early stages of tumorigenesis may induce high levels of ATF-3 expression, which we also observed in this study (Supplementary Fig. S2A). Nevertheless, the contradictory role of ATF-3 in various malignancies, and even among various melanoma cell lines, as observed in our study, may relate to intrinsic heterogeneity among various melanoma primary sources. However, the ability to stratify melanoma patients based on ATF-3 expression based on survival outcomes is consistent with an important role for the ATF-3 pathway in generalized mediation of tumor virulence (Fig. 1D).
ATF-3 plays a dynamic role in DNA damage repair and its regulation in activating or repressing cell-cycle regulators in various types of tissues. Previous studies have also shown that elevated ATF-3 expression either promotes or inhibits tumorigenesis10,11. In melanoma, one study shows that TRPV1-induced calcineurin activation regulates p53 via ATF-3, where elevated ATF-3 expression in melanoma cell lines transcriptionally repress apoptotic-related genes7. However, the effect of ATF-3 on apoptosis in melanoma cells for this particular study are treated with TRPV1, capsaicin, and a calcineurin inhibitor, creating a completely different microenvironment from our study. It is well known that tumor microenvironments in melanoma are heterogenous and may be associated with distinctive distributions of immune cells which can potentially affect specific biological processes, e.g. apoptotic signaling pathways39,40. Moreover, another study further demonstrates that ATF-3 suppresses human melanoma growth by inducing the expression of ATF-3 in human dermal fibroblasts41.
An intriguing finding from this study is the interrelationship among ATF-3, ERK and AKT signaling pathways and p53-dependent apoptosis. Our experiments involving melanoma cell lines with retrovirally-induced ATF-3 overexpression revealed that ATF-3 suppressed the proliferation, migration, and invasion of melanoma cells (Fig. 2A–J) presumably by upregulating apoptotic-related genes in part by downregulating ERK and AKT pathways (Fig. 4A–D). It is well known that the risk of melanoma is increased with excessive UV light exposure. Studies have shown that UV-related DNA damage leads to a prompt response of ATF-3-induction which triggers p53-dependent apoptosis24,27. In response to UV radiation, ATF-3 can either protect p53 wild-type cells by enhancing DNA repair or promote programmed cell death of p53-defective cells by binding to histone acetyltransferase,Tip6027. Our RNA seq analysis of C8161 and A2058 ATF-3 OE melanoma cells exhibited an upregulation of genes, such as ING4 and CDKN1A, which are involved in regulating p53-mediated apoptosis (Fig. 4A–B). ING4 is a member of the inhibitor of growth (ING) family and acts as a tumor suppressor protein by increasing p53 acetylation and promoting apoptosis42. An elevated level of p53 expression, in turn, leads to an increased expression in cyclin-dependent kinase inhibitor p21 (CDKN1A), which negatively regulates p53 transcription43. Our study shows that ATF-3 plays inhibits melanoma tumor progression by enhancing apoptosis.
Interestingly, MAPK signaling pathway is also involved with ATF-3 activation in response to genotoxic stress24. Mitogen-activated protein kinase (MAPK)/ERK and AKT pathways have been extensively studied and heavily targeted by drugs to treat metastatic melanoma. The Raf/MEK/ERK signaling pathway mediates cell proliferation, cell-cycle arrest, terminal differentiation and apoptosis44. Activating mutations within the ERK or AKT signaling pathway can lead to progression of melanoma and affect its cell proliferation, growth, and survival45. A recent study has further shown that deficiency in ATF-3 expression leads to a dramatic increase in phosphorylated AKT expression levels, indicating that ATF-3 may act as a tumor suppressor within the AKT signaling pathway in prostate cancer34. Similarly, our study demonstrates that the tumor suppressive role of ATF-3 is rescued retrovirally by deactivating AKT and ERK pathways (Fig 4C–D). However, a study involving nerve damage has shown that the overexpression of ATF-3 does not affect ERK activity while the combination of ATF-3 overexpression and activation of C-Jun N-Terminal Kinase (JNK) upregulates expression levels of the heat shock protein 27 (Hsp27), which plays a direct or indirect role in activating AKT pathways46. These studies reflect the multifaceted role of ATF-3 in various signaling pathways, suggesting that more studies need to be implemented to truly understand the precise mechanistic role of ATF-3 in ERK and AKT pathways. The ability to validate our in vitro findings in a xenograft mouse model further suggests that ATF-3 acts as a tumor suppressor in melanoma in vivo and pave the way for translational approaches focused on manipulation of ATF-3/ERK/AKT signaling pathways as a novel therapeutic means of controlling human melanomagenesis.
In the past decade, the emergence of targeted therapy and immune check point inhibitors to treat metastatic melanoma have been widely studied. However, melanoma is known to exhibit intratumor and intertumor heterogeneity, which further complicates its management. The heterogenous microenvironments found within melanoma are also associated with a diverse spatial distribution of immune cells40. Similarly, ATF-3 plays a regulatory role in immune responses28,29,47. A recent study shows that ATF-3 screening may be used to determine the efficacy of PD-1 therapy in melanoma48. Thus, future studies to explore the potential role of ATF-3 in tumor response to immune checkpoint inhibitors is necessary.
Supplementary Material
Acknowledgments
This work was supported to X.W. by the National Key Research and Development Program of China (2017YFA0104604), the General Program of National Natural Science Foundation of China (81772093), the Key Program of Shandong Province Natural Science Foundation (ZR2019ZD36) and the Key Research and Development Program of Shandong Province (2019GSF108107). C.A.A.L is currently funded by T32 EB016652-06.
Footnotes
Conflict of interest:
The authors declare no conflicts of interest.
References:
- 1.Lomas A, Leonardi-Bee J, Bath-Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol 2012;166:1069–1080. [DOI] [PubMed] [Google Scholar]
- 2.Koh HK, Geller AC, Miller DR, Grossbart TA, Lew RA. Prevention and early detection strategies for melanoma and skin cancer. Current status. Arch Dermatol 1996;132:436–443. [PubMed] [Google Scholar]
- 3.Bandarchi B, Jabbari CA, Vedadi A, Navab R. Molecular biology of normal melanocytes and melanoma cells. J Clin Pathol 2013;66:644–648. [DOI] [PubMed] [Google Scholar]
- 4.Millet A, Martin AR, Ronco C, Rocchi S, Benhida R. Metastatic Melanoma: Insights Into the Evolution of the Treatments and Future Challenges. Med Res Rev 2017;37:98–148. [DOI] [PubMed] [Google Scholar]
- 5.Hai T, Wolford CC, Chang YS. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr 2010;15:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson MR, Xu D, Williams BR. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med (Berl) 2009;87:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Guo W, Ma J, Xu P, Zhang W, Guo S, et al. Downregulated TRPV1 Expression Contributes to Melanoma Growth via the Calcineurin-ATF3-p53 Pathway. J Invest Dermatol 2018;138:2205–2215. [DOI] [PubMed] [Google Scholar]
- 8.Wu X, Nguyen BC, Dziunycz P, Chang S, Brooks Y, Lefort K, et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature 2010;465:368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan C, Boyd DD. ATF3 regulates the stability of p53: a link to cancer. Cell Cycle 2006;5:926–929. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Zhou X, Li Y, Zu L, Pan H, Liu B, et al. Activating transcription factor 3 promotes malignance of lung cancer cells in vitro. Thorac Cancer 2017;8:181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janz M, Hummel M, Truss M, Wollert-Wulf B, Mathas S, Johrens K, et al. Classical Hodgkin lymphoma is characterized by high constitutive expression of activating transcription factor 3 (ATF3), which promotes viability of Hodgkin/Reed-Sternberg cells. Blood 2006;107:2536–2539. [DOI] [PubMed] [Google Scholar]
- 12.Chang F, Zhang Y, Mi J, Zhou Q, Bai F, Xu X, et al. ROCK inhibitor enhances the growth and migration of BRAF-mutant skin melanoma cells. Cancer Sci 2018;109:3428–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Tommasi di Vignano A, Zhou Q, Michel-Dziunycz PJ, Bai F, Mi J, et al. The ARE-binding protein Tristetraprolin (TTP) is a novel target and mediator of calcineurin tumor suppressing function in the skin. PLoS Genet 2018;14:e1007366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CW, Zhan Q, Lezcano C, Frank MH, Huang J, Larson AR, et al. Nestin depletion induces melanoma matrix metalloproteinases and invasion. Lab Invest 2014;94:1382–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012;150:1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji Y, Jia L, Zhang Y, Xing Y, Wu X, Zhao B, et al. Antitumor activity of the plant extract morin in tongue squamous cell carcinoma cells. Oncol Rep 2018;40:3024–3032. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116:855–867. [DOI] [PubMed] [Google Scholar]
- 19.Lehraiki A, Cerezo M, Rouaud F, Abbe P, Allegra M, Kluza J, et al. Increased CD271 expression by the NF-kB pathway promotes melanoma cell survival and drives acquired resistance to BRAF inhibitor vemurafenib. Cell Discov 2015;1:15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao H, Liu R, Sun X. STAT3-induced upregulation of lncRNA SNHG17 predicts a poor prognosis of melanoma and promotes cell proliferation and metastasis through regulating PI3K-AKT pathway. Eur Rev Med Pharmacol Sci 2019;23:8000–8010. [DOI] [PubMed] [Google Scholar]
- 21.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 2001;273:1–11. [DOI] [PubMed] [Google Scholar]
- 22.Chen BP, Liang G, Whelan J, Hai T. ATF3 and ATF3 delta Zip. Transcriptional repression versus activation by alternatively spliced isoforms. J Biol Chem 1994;269:15819–15826. [PubMed] [Google Scholar]
- 23.Liang G, Wolfgang CD, Chen BP, Chen TH, Hai T. ATF3 gene. Genomic organization, promoter, and regulation. J Biol Chem 1996;271:1695–1701. [DOI] [PubMed] [Google Scholar]
- 24.Fan F, Jin S, Amundson SA, Tong T, Fan W, Zhao H, et al. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene 2002;21:7488–7496. [DOI] [PubMed] [Google Scholar]
- 25.Yan J, Zhong N, Liu G, Chen K, Liu X, Su L, et al. Usp9x- and Noxa-mediated Mcl-1 downregulation contributes to pemetrexed-induced apoptosis in human non-small-cell lung cancer cells. Cell Death Dis 2014;5:e1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 27.Cui H, Li X, Han C, Wang QE, Wang H, Ding HF, et al. The Stress-responsive Gene ATF3 Mediates Dichotomous UV Responses by Regulating the Tip60 and p53 Proteins. J Biol Chem 2016;291:10847–10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitmore MM, Iparraguirre A, Kubelka L, Weninger W, Hai T, Williams BR. Negative regulation of TLR-signaling pathways by activating transcription factor-3. J Immunol 2007;179:3622–3630. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen CT, Kim EH, Luong TT, Pyo S, Rhee DK. TLR4 mediates pneumolysin-induced ATF3 expression through the JNK/p38 pathway in Streptococcus pneumoniae-infected RAW 264.7 cells. Mol Cells 2015;38:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, He Y, Deng W, Lang L, Yang H, Jin B, et al. Atf3 deficiency promotes genome instability and spontaneous tumorigenesis in mice. Oncogene 2018;37:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue M, Uchida Y, Edagawa M, Hirata M, Mitamura J, Miyamoto D, et al. The stress response gene ATF3 is a direct target of the Wnt/beta-catenin pathway and inhibits the invasion and migration of HCT116 human colorectal cancer cells. PLoS One 2018;13:e0194160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C, Ge C, Liu Z, Li L, Zhao F, Tian H, et al. ATF3 inhibits the tumorigenesis and progression of hepatocellular carcinoma cells via upregulation of CYR61 expression. J Exp Clin Cancer Res 2018;37:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park GH, Song HM, Jeong JB. Kahweol from Coffee Induces Apoptosis by Upregulating Activating Transcription Factor 3 in Human Colorectal Cancer Cells. Biomol Ther (Seoul) 2017;25:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Yan C. Emerging roles of ATF3 in the suppression of prostate cancer. Mol Cell Oncol 2016;3:e1010948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Yang Z, Chen Z, Bao Y, Zhang H, Fang X, et al. ATF3 suppresses ESCC via downregulation of ID1. Oncol Lett 2016;12:1642–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell 2003;11:915–926. [DOI] [PubMed] [Google Scholar]
- 37.Zigler M, Villares GJ, Dobroff AS, Wang H, Huang L, Braeuer RR, et al. Expression of Id-1 is regulated by MCAM/MUC18: a missing link in melanoma progression. Cancer Res 2011;71:3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan X, Yu L, Li J, Xie G, Rong T, Zhang L, et al. ATF3 suppresses metastasis of bladder cancer by regulating gelsolin-mediated remodeling of the actin cytoskeleton. Cancer Res 2013;73:3625–3637. [DOI] [PubMed] [Google Scholar]
- 39.Grzywa TM, Paskal W, Wlodarski PK. Intratumor and Intertumor Heterogeneity in Melanoma. Transl Oncol 2017;10:956–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan Y, Leontovich AA, Gerdes MJ, Desai K, Dong J, Sood A, et al. Understanding heterogeneous tumor microenvironment in metastatic melanoma. PLoS One 2019;14:e0216485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zu T, Wen J, Xu L, Li H, Mi J, Li H, et al. Up-Regulation of Activating Transcription Factor 3 in Human Fibroblasts Inhibits Melanoma Cell Growth and Migration Through a Paracrine Pathway. Front Oncol 2020;10:624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res 2003;63:2373–2378. [PubMed] [Google Scholar]
- 43.Lohr K, Moritz C, Contente A, Dobbelstein M. p21/CDKN1A mediates negative regulation of transcription by p53. J Biol Chem 2003;278:32507–32516. [DOI] [PubMed] [Google Scholar]
- 44.Peyssonnaux C, Eychene A. The Raf/MEK/ERK pathway: new concepts of activation. Biol Cell 2001;93:53–62. [DOI] [PubMed] [Google Scholar]
- 45.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis 2004;9:667–676. [DOI] [PubMed] [Google Scholar]
- 46.Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H. Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J Neurosci 2003;23:5187–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jadhav K, Zhang Y. Activating transcription factor 3 in immune response and metabolic regulation. Liver Res 2017;1:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H, Kuang X, Zhang Y, Ye Y, Li J, Liang L, et al. ADORA1 Inhibition Promotes Tumor Immune Evasion by Regulating the ATF3-PD-L1 Axis. Cancer Cell 2020;37:324–339 e328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data have been deposited to the NCBI Gene Expression Omnibus under accession number GSE152460.


