In this study, Ma et al. set out to study the mechanism of centromeric cohesion protection by Moa1/meikin during meiosis I. Using genetic and fluorescence microscopy approaches in fission yeast, the authors show that Moa1-Plo1 phosphorylates Rec8, which is required for Sgo1-PP2A to protect centromeric cohesion by antagonizing CK1-mediated phosphorylation of the cohesion subunit Rec8.
Keywords: meikin, shugoshin, cohesin Rec8, meiosis, fission yeast
Abstract
The conserved meiosis-specific kinetochore regulator, meikin (Moa1 in fission yeast) plays a central role in establishing meiosis-specific kinetochore function. However, the underlying molecular mechanisms remain elusive. Here, we show how Moa1 regulates centromeric cohesion protection, a function that has been previously attributed to shugoshin (Sgo1). Moa1 is known to associate with Plo1 kinase. We explore Plo1-dependent Rec8 phosphorylation and identify a key phosphorylation site required for cohesion protection. The phosphorylation of Rec8 by Moa1-Plo1 potentiates the activity of PP2A associated with Sgo1. This leads to dephosphorylation of Rec8 at another site, which thereby prevents cleavage of Rec8 by separase.
The cohesin complex that mediates sister chromatid cohesion and DNA looping plays an essential role in mitotic and meiotic chromosome segregation (Nasmyth 2001; Peters et al. 2008; Watanabe 2012; Yatskevich et al. 2019). In meiosis, the cohesin subunit Rad21 is replaced by a meiosis-specific version Rec8, and premeiotic DNA replication is followed by reductional, rather than equational, division (Watanabe and Nurse 1999; Watanabe et al. 2001). During the meiosis I division, sister kinetochores are captured by microtubules from the same pole (mono-orientation) and cohesin Rec8 is cleaved along the chromosome arms but not at the centromere (cohesion protection) (Buonomo et al. 2000; Kitajima et al. 2003a). Shugoshin (Sgo1) and PP2A collaboratively antagonize casein kinase 1 (CK1)-dependent Rec8 phosphorylation, a prerequisite for cleavage by separase, thus protecting cohesion at the centromeres (Kitajima et al. 2004, 2006; Ishiguro et al. 2010; Katis et al. 2010). Accumulating evidence suggests that the meiosis-specific kinetochore regulator meikin (Moa1, Spo13 in budding yeast, and MEIKIN in mouse) is also required for cohesion protection in addition to mono-orientation (Shonn et al. 2002; Katis et al. 2004; Lee et al. 2004; Yokobayashi and Watanabe 2005; Kim et al. 2015; Miyazaki et al. 2017).
Sgo1 accumulates at centromeres dependent upon phosphorylation of histone H2A by Bub1 kinase, which localizes at kinetochores only when the kinetochore protein Spc7 (KNL1 homolog) is phosphorylated by Mph1 (MPS1 homolog) (Kawashima et al. 2010; Yamagishi et al. 2012). Moreover, in meiosis I, not only Mph1 but also polo-like kinase 1 (Plo1) associated with Moa1 phosphorylates Spc7 so that Bub1 localizes at kinetochores efficiently, contributing to the robust localization of Sgo1 at centromeres (Miyazaki et al. 2017). However, because a small amount of Bub1 still sustains Sgo1-dependent protection, the reduced Bub1 (and Sgo1) localization cannot explain the defects of cohesion protection in moa1Δ cells (Miyazaki et al. 2017). Therefore, the major mechanism of cohesion protection mediated by Moa1-Plo1 remains elusive.
Results and Discussion
Kinetochore-tethered Plo1 substitutes for Moa1 functions
Mono-orientation is established by cohesion of the core centromere, whereas cohesion protection needs to act at the pericentromeric region (Kitajima et al. 2003b, 2004; Yokobayashi and Watanabe 2005; Sakuno et al. 2009). Our previous studies suggest that Moa1 might use Plo1 to establish both these functions at meiotic kinetochores (Kim et al. 2015; Miyazaki et al. 2017). To delineate Plo1 function at kinetochore/centromeres in meiosis, we engineered a Cnp3-Plo1 fusion protein, which targets Plo1 to the kinetochore assembled on the core centromere through the C-terminal sequence of Cnp3 (CENP-C homolog) (Miyazaki et al. 2017). Similarly, Plo1 was fused with two copies of the chromodomain (CD), which binds to H3K9me (histone H3 methylated on Lys 9), a histone modification predominantly found at the pericentromere (Yamagishi et al. 2008). We sought to examine whether such differentially localized Plo1 can compensate for a loss of Moa1 from kinetochores (Fig. 1A). To express these Plo1 fusion proteins in meiosis I, we used the mei4+ promoter, which is activated during late prophase I (Mata et al. 2002), the period when Moa1 is functional (Fig 1A; Supplemental Fig. S1; Kagami et al. 2011). We thereby expressed Cnp3-Plo1 and CD-Plo1 in moa1Δ cells, which we arrested in meiotic prophase II (by mes1-B44) (Izawa et al. 2005) when centromeric cohesion is normally preserved. As negative controls, we expressed the kinase dead versions (Tanaka et al. 2001), Cnp3-Plo1-K69R and CD-Plo1-K69R, respectively. We monitored GFP fluorescence associated with a lacO array integrated at the centromere of one homolog of chromosome I (imr1-GFP) (Fig. 1B; Sakuno et al. 2009). In moa1Δ cells, although a minority population of cells (17%) underwent equational segregation at meiosis I (because of defects in mono-orientation), the remaining majority (83%) underwent reductional segregation due to the presence of chiasmata and tension exerted across homologs (Fig. 1C, top; Shonn et al. 2002; Yokobayashi and Watanabe 2005). However, because moa1Δ cells are partially defective in cohesion protection (Miyazaki et al. 2017), 18% of the “reductional” moa1Δ cells showed a separation of imr1-GFP in prophase II, compared with 8% in wild-type cells (Fig. 1C, bottom). Remarkably, expression of CD-Plo1 suppressed cohesion defects but hardly suppressed mono-orientation defects of moa1Δ cells (Fig. 1C), suggesting that Plo1 has an ability to promote cohesion protection at pericentromeres. In contrast, expression of Cnp3-Plo1 suppressed defects in both mono-orientation and cohesion protection (Fig. 1C), implying that the kinetochore-tethered Plo1 can substitute for both Moa1 functions.
Figure 1.
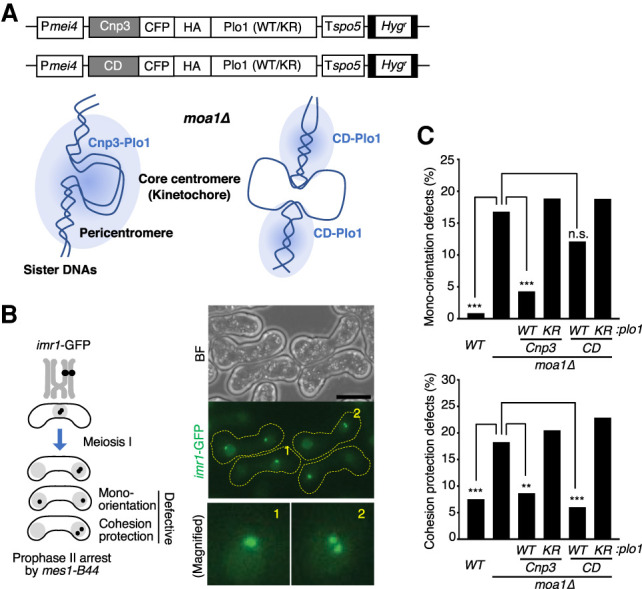
Plo1 plays distinct roles at the core centromere and pericentromeres. (A) Schematic of constructs used for expression of plo1+ and plo1-KR (kinase dead mutation) fusion proteins. (Pmei4) mei4+ promoter, (Cnp3) Cnp3 C-terminal domain, (CD) two copies of chromodomain, (CFP) cyan fluorescent protein, (HA) three copies of HA tag, (Tspo5) terminator of the spo5+ gene, (Hygr) hygromycin resistant marker. Schematic depiction of the centromeric domain of moa1Δ cells, highlighting position of tethered Plo1. Blue clouds represent the distribution of Plo1 kinase activity. (B, left) Schematic of chromosome segregation assay. The segregation pattern of imr1-GFP marked on one homolog was monitored in prophase II by arresting cells with the mes1-B44 mutation. (Right) Representative bright-field (BF; top) and fluorescent images (optionally maximum z-dimension projections; middle) of the assay. (Bottom right) Numbered nuclei are magnified representing normal reductional segregation with an intact (1) or defective (2) cohesion protection. Scale bar, 5 μm. (C) Defects in mono-orientation (top) and cohesion protection (bottom) were monitored in the indicated cells. Cell numbers used for assay: WT = 231, moa1Δ = 375, CD-plo1-WT = 322, CD-plo1-KR = 468, Cnp3-plo-WT = 374, Cnp3-plo1-KR = 355. (n.s.) Not significant, (**) P < 0.001, (***) P < 0.0001, Fisher's exact test for moa1Δ.
Plo1 kinase phosphorylates Rec8
Because cohesin Rec8 is directly involved in the regulation of both mono-orientation and cohesion protection (Watanabe et al. 2001; Kitajima et al. 2003b), we thought it possible that Rec8 might be a functional target of Plo1 at centromeres. To investigate this possibility, we examined the in vitro phosphorylation of Rec8 by Plo1. Plo1, purified from bacteria, was shown to phosphorylate the full length of recombinant Rec8 protein (Supplemental Fig. S2A). We introduced alanine substitutions in the putative polo-kinase consensus E/D-X-S/T or related sequences of Rec8N, Rec8M, and Rec8C fragments (Supplemental Fig. S2B). Although all fragments were phosphorylated by Plo1 in vitro, their mutant versions, Rec8N-11A (alanine substitutions at 11 sites), Rec8M-13A (alanine substitutions at 13 sites), and Rec8C-S450A (single alanine substitution at S450), exhibited reduced levels of phosphorylation (Supplemental Fig. S2C). To evaluate the biological significance of phosphorylation of Rec8 by Plo1, we introduced the nonphosphorylatable mutations into the endogenous rec8+ gene and examined chromosome segregation during meiosis I. Although rec8-11A and rec8-13A cells showed apparently no defects in meiosis I, rec8-S450A cells showed defects in cohesion protection but not in mono-orientation (Fig. 2A). This was unexpected because all rec8 or cohesin-related mutants so far showed defects in both functions or a stronger defect in mono-orientation (Yokobayashi and Watanabe 2005). By raising phospho-specific antibodies, we confirmed that Rec8-S450 is indeed phosphorylated in meiotic cells (Fig. 2B), and nearly half of the cellular phosphorylation depends on Moa1 (Fig. 2C).
Figure 2.
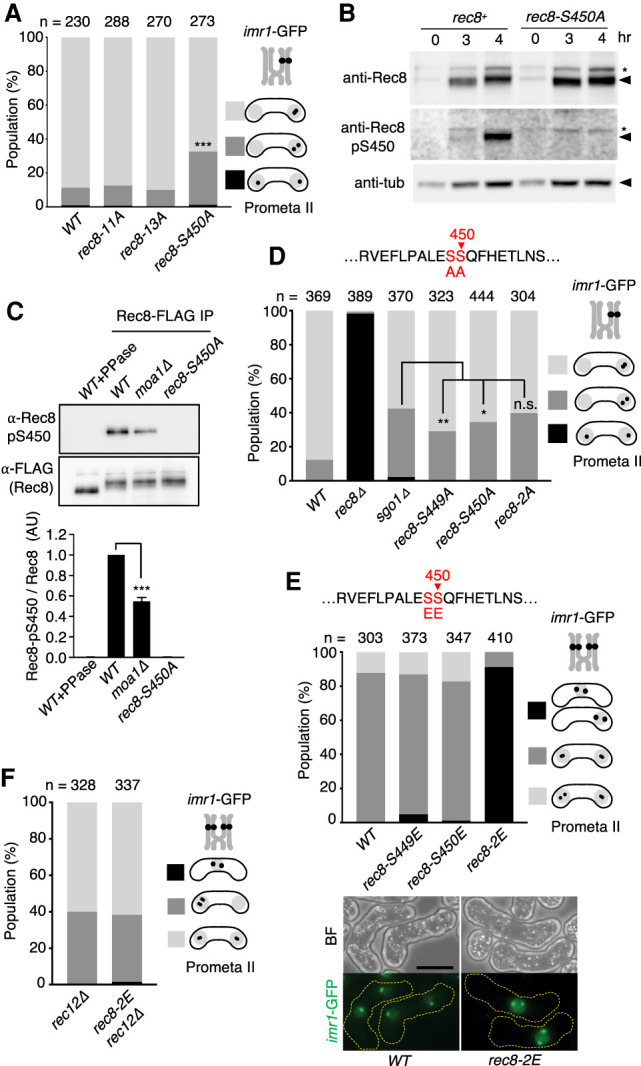
Rec8 is phosphorylated by Plo1 in vitro and in vivo. (A) The segregation pattern of imr1-GFP marked on one homolog was monitored in prophase II-arrested cells. n = cell number used for assay. (***) P < 0.0001, Fisher's exact test for WT. (B) Haploid rec8+ (left) and rec8-450A (right) cells were induced to undergo synchronous meiosis by inactivating pat1-114. (Middle panels) Immunoblot analysis was performed with anti-Rec8, anti-Rec8-pS450, and anti-tubulin antibodies by using lysates prepared from meiotic cells at each time point. Asterisks indicate a weak cross-reacting band that is not reflecting S450 phosphorylation. (C) The indicated haploid rec8-FLAG cells were induced to undergo synchronous meiosis by inactivating pat1-114. Extracts were immunoprecipitated with anti-FLAG M2 beads. Immunoprecipitates (IP) were subjected to SDS-PAGE and analyzed by immunoblot. Band intensity of anti-Rec8-pS450 was normalized to anti-FLAG blotting, and the values were normalized to WT ratio in each experiment. Error bars, SD from three independent experiments. (***) P < 0.0001, one-way ANOVA with Dunnett's multiple comparisons test relative to WT. (D) The segregation pattern of imr1-GFP marked on one homolog was monitored in prophase II-arrested cells. n = cell number used for assay. (n.s.) Not significant, (*) P < 0.05, (**) P < 0.001, Fisher's exact test. (E, top) The segregation pattern of imr1-GFP marked on one homolog was monitored in prophase II-arrested cells. (n) Cell number used for assay. (Bottom) Representative BF and maximum projection fluorescent images of WT and rec8-2E cells are shown. (F) The indicated cells were arrested at prophase II and analyzed for the segregation pattern of imr1-GFP present on both homologs. (n) Cell number used for the assay. Note that nondisjunction of rec8-2E cells was suppressed by rec12Δ. Scale bar, 5μm.
Although S450 (E-S-S-Q) of Rec8 matches the Polo consensus phosphorylation motif and is indeed phosphorylated by Plo1 in vitro, we noticed that the adjacent serine at S449 fits with the DDK (Cdc7/Dbf4 kinase) consensus motif S/T-pS/T-Q once S450 is phosphorylated by Plo1 (Randell et al. 2010). DDK has been shown to be required for phosphorylation of Rec8 and meiotic chromosome segregation in fission and budding yeasts (Matos et al. 2008; Katis et al. 2010; Le et al. 2013), although the precise role of the phosphorylation is unknown. Accordingly, we substituted S449 to alanine and found that rec8-S449A caused defects in cohesion protection. The double mutant rec8-2A (S449A, S450A) showed defects that were comparable with those of sgo1Δ cells, in which cohesion protection is abolished (Fig. 2D). To further explore the possibility that phosphorylation of both S449 and S450 might be relevant to the regulation of cohesion protection, we substituted each serine residue with glutamic acid (i.e., a phospho-mimetic residue). Although rec8-S449E or rec8-S450E cells showed little defects in meiotic chromosome segregation, rec8-2E (S449E, S450E) cells showed extensive nondisjunction at meiosis I (Fig. 2E), indicating that homolog segregation was blocked. The meiotic nondisjunction of rec8-2E cells was suppressed by deleting rec12 (the SPO11 homolog in fission yeast) (Keeney et al. 1997), which abolishes recombination and therefore chiasmata formation (Fig. 2F). This suggests that the prevention of homolog segregation in rec8-2E cells might be caused by persistent cohesion along the chromosome arms in anaphase I, which does not allow chiasmata to resolve. These results further imply that Rec8 might be phosphorylated on both S449 and S450 at centromeres but not at the arm region during normal meiosis I.
Phosphorylation of Rec8 by Plo1 potentiates PP2A
To monitor cellular Rec8, we tagged the endogenous Rec8 with mCherry and observed meiotic cells arrested in prophase II. Because Rec8 is protected at centromeres throughout anaphase I until prophase II, Rec8 (mCherry) signal was detected as two well-separated dots in the zygote (Fig. 3A, rec8+). The centromeric signals of Rec8 decreased in prophase II cells depleted for the cohesin protector Sgo1 (Kitajima et al. 2004). rec8-2A cells also showed diminished Rec8 signals in prophase II but high levels of signal during prophase of meiosis I (Fig. 3A,B; Supplemental Fig. S3), suggesting that Rec8-2A was properly expressed but not protected at centromeres during anaphase I. In sharp contrast to rec8-2A cells, rec8-2E cells showed distinct signals of Rec8 at centromeres in prophase II as well as elevated signals within a single undivided nucleus (Fig. 3B,C). This supports the notion that homologs were not separated due to persistent cohesion by Rec8-2E. Strikingly, the phenotype of rec8-2E was completely suppressed by sgo1Δ (Fig. 3B,C), indicating that Rec8-2E was protected by Sgo1 not only at centromeres but also along the chromosome arm. An explanation for this is that the phosphatase activity of Sgo1-PP2A might be enhanced by Rec8-2E even along the chromosome arms. We speculate that because Plo1 is enriched at kinetochores, dependent on Moa1 in physiological meiosis (Kim et al. 2015), efficient phosphorylation of Rec8 by Plo1 might normally be restricted to centromeres. To further investigate mechanisms of Rec8-2E protection, we expressed rec8-2E ectopically in mitotic cells and induced Par1-CD (the PP2A-B56 subunit fused with CD) by a thiamine-repressible promoter (Pnmt81), which enabled the enrichment of the PP2A complex at the pericentromeres (Kitajima et al. 2006). Previously, we showed that coexpression of Sgo1 and Par1-CD, but not Par1-CD alone, rendered rec8+-expressing mitotic cells inviable because of ectopic protection of Rec8 at centromeres (Kitajima et al. 2006). Our new experiments revealed that induction of Par1-CD alone did render rec8-2E-expressing cells inviable but not rec8+- or rec8-2A-expressing cells. Forced expression of Sgo1 showed the same effect (Fig. 3D). Considering that Sgo1 is normally absent from mitotic cells, these results suggest that Rec8-2E potentiated the action of the PP2A complex at the pericentromeres, thereby protecting Rec8-2E from separase. However, in meiosis, Sgo1 is required to protect Rec8-2E, implying that either Sgo1 or CD tagging can potentiate PP2A activity to protect Rec8-2E. We reason that the mutant Rec8-2E might have gained an increased affinity for the PP2A complex (Supplemental Fig. S4).
Figure 3.
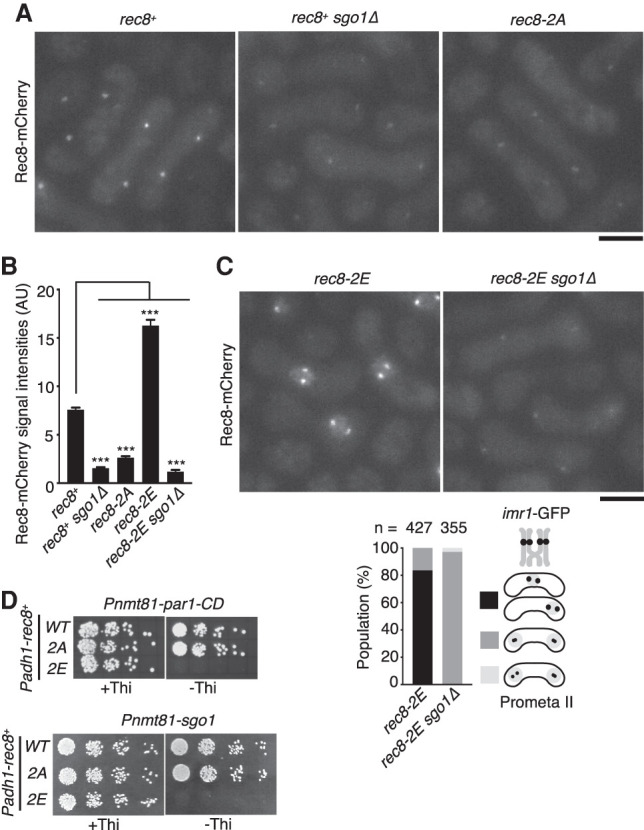
Phosphorylation of Rec8 by Plo1 promotes PP2A-dependent protection. (A) Rec8-mCherry signals were observed in indicated prophase II-arrested cells. Scale bar, 5 μm. (B) Quantification of Rec8-mCherry signals in the centromeric regions. One-hundred to 240 centromeres were measured per sample. Error bars indicate SEM. (***) P < 0.0001, one-way ANOVA with Dunnett's multiple comparisons test relative to rec8+. (C) Rec8-mCherry signals were observed in indicated prophase II-arrested cells. The segregation pattern of imr1-GFP marked on both homologs was monitored in prophase II-arrested cells. (n) Cell number used for assay. Scale bar, 5 μm. (D) Serial fivefold dilutions of the indicated cells were spotted on MM plates including and lacking thiamine (Thi) and grown at 30°C. Note that Par1-CD and Sgo1 are expressed only in the absence of thiamine.
Opposite effect of Plo1 and CK1 on Rec8 protection
We have previously shown that casein kinase 1 (CK1)-dependent phosphorylation on Rec8-S412 and adjacent sites (S400, S402, S404, S410, T414, and S416) plays an essential role in promoting Rec8 cleavage by separase (Ishiguro et al. 2010). Accordingly, the single rec8-412A mutation causes extensive nondisjunction of homologs in meiosis I, exhibiting distinct Rec8 signals at centromeres as well as elevated nuclear signals in prophase II (Fig. 4A), similar to rec8-2E cells (Fig. 3C). Unlike for rec8-2E, the phenotype of rec8-412A was never suppressed by sgo1Δ (Fig. 4A). These results are consistent with a scenario in which Moa1-Plo1-dependent phosphorylation of Rec8-S450 promotes the action of Sgo1-PP2A and thereby antagonizes CK1-dependent phosphorylation of Rec8, a prerequisite for separase-dependent cleavage (Fig. 4B). To further examine this possibility, we reconstituted Rec8 dephosphorylation in vitro using immunoprecipitated Par1-containing PP2A complexes. For this purpose, we prepared a recombinant Rec8 C-terminal fragment (Rec8C1-WT, 390–561 amino acids), which contains both CK1 and Plo1 phosphorylation sites (Fig. 4B). We also prepared the mutant version Rec8C1-2E, in which the Plo1-phosphorylation and adjacent sites (S449 and S450) were substituted with glutamic acids. These two peptides were mixed and phospho-labeled in vitro with recombinant CK1 kinase (Fig. 4C). Subsequently they were treated with the Par1-FLAG immunoprecipitation beads prepared from S. pombe cell extracts, and their dephosphorylation was monitored by SDS-PAGE and autoradiography (Fig. 4D). The results indicate that the half-time of dephosphorylation of phospho-Rec8C-2E was considerably shorter (<5 min) than that of phospho-Rec8C-WT (>60 min) (Fig. 4D). Therefore, consistent with the genetic analyses, this biochemical analysis supports the notion that phosphorylation at Rec8-S450 and the adjacent site plays a role in promoting the PP2A-dependent removal of CK1-dependent phosphorylation of Rec8 (Fig. 4B). Although underlying molecular mechanisms of this selective dephosphorylation remain unclear, we speculate that the interaction of Par1 with the S450p surface of Rec8 may in turn facilitate interaction of the PP2A catalytic domain with the S412p surface.
Figure 4.
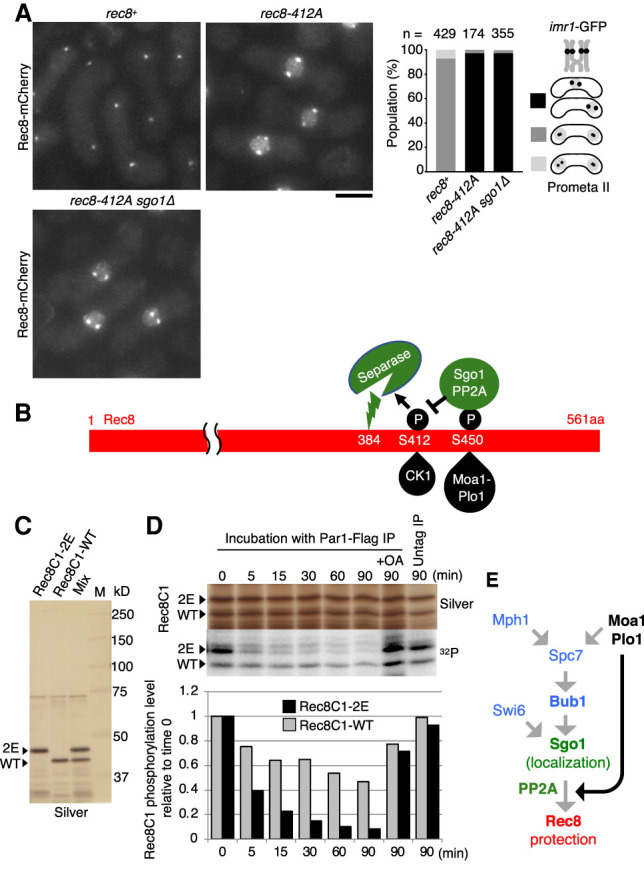
Opposite effect of Plo1 and CK1 on Rec8 protection at centromeres. (A) Rec8-mCherry signals were observed in the indicated prophase II-arrested cells. The segregation pattern of imr1-GFP marked on both homologs was monitored in prophase II-arrested cells. (n) Cell number used for assay. Scale bar, 5 μm. (B) Schematic depiction of the interplay of Plo1 phosphorylation and CK1 phosphorylation of Rec8 through the action of Sgo1-PP2A. (C) GST-Rec8C1-WT (Rec8 residues 391–561 amino acids), GST-Rec8C1-2E, and their mixtures were subjected to SDS-PAGE (silver stain). (D) Purified recombinant proteins (mixtures of GST-Rec8C1-WT and GST-Rec8C1-2E) were phosphorylated by GST-CK1 in the presence of [γ-32P] ATP and dephosphorylated by Par1-FLAG immunoprecipitate (PP2A complex) beads. The radioactive phosphate groups were visualized by autoradiography (32P) and compared with protein levels (silver stain). The autoradiographic signals were quantitated using an image analyzer and the signal strength was normalized to that at time 0. (OA) Okadaic acid. (E) Schematic depiction of hierarchical regulation of centromeric protection of cohesin by meikin and shugoshin during meiosis I. Moa1-Plo1 together with Mph1 phosphorylate Spc7, which thereby recruits Bub1 to kinetochores and enhances Sgo1 localization (Miyazaki et al. 2017). The present study revealed that Moa1-Plo1 phosphorylates Rec8-S450 to potentiate Sgo1-PP2A function (dark arrow).
In budding yeast, PLK (Cdc5) is required to remove some Rec8-including cohesin from arm regions through the “prophase pathway” (Challa et al. 2019), although the requirement of Cdc5 for Rec8 protection in anaphase I is under debate (Katis et al. 2010; Attner et al. 2013; Galander et al. 2019). However, in fission yeast and mammals, PLK (Plo1) plays an important role in both mono-orientation and cohesion protection at centromeres during meiosis I (Kim et al. 2015; Miyazaki et al. 2017). Our tethering experiments here validate this conclusion for fission yeast (Fig. 1). Importantly, we identify the key substrate of Moa1-Plo1, which is required for cohesion protection but not mono-orientation. Therefore, our current results enable us to dissect the Moa1-Plo1 function at kinetochores. Our genetic and biochemical analyses disclose that Moa1-Plo1-dependent phosphorylation of Rec8 contributes to Sgo1-associated PP2A action, which in turn removes CK1-dependent phosphorylation of Rec8, a prerequisite for Rec8 cleavage by separase. In conclusion, our results suggest that, in addition to Bub1-dependent Sgo1 enrichment, Moa1-Plo1-dependent phosphorylation of Rec8 ultimately acts to protect cohesion at the centromere (Fig. 4E). In oocytes, age-related dissociation of Rec8 from chromosomes leads to premature loss of centromeric cohesion, thereby causing aberrant meiotic chromosome segregation (Chiang et al. 2010; Lister et al. 2010; Sakakibara et al. 2015). Because the mechanisms of meiotic chromosome segregation are well conserved (Kim et al. 2015; Ogushi et al. 2020), we anticipate that our findings in fission yeast will contribute to the advancement of understanding of age-related birth defects in humans.
Materials and methods
Schizosaccharomyces pombe strains and media
Unless otherwise stated, all media and growth conditions were as described previously (Moreno et al. 1991). S. pombe strains used in this study are described in Supplemental Table S1. Complete medium (YE), minimal medium (SD and MM), and sporulation media (SPA and SSA) were used. Deletion of sgo1+and moa1+, as well as tagging of rec8+ by mCherry, were carried out according to the PCR-based gene targeting method for S. pombe using the kanMX6 (kanR), hphMX6 (hygR), and natMX6 (natR) genes as selection markers. The plasmids used for Plo1 tethering were linearized and integrated into the genome at the Z locus (Sakuno et al. 2009).
Synchronous cultures of fission yeast meiotic cells
For microscopic observation of imr1-GFP and Rec8-mCherry, logarithmically growing cells were collected, suspended in 20 mg/mL leucine, spotted on sporulation-inducing medium (SPA), and incubated for 12–15 h at 26°C (imr1-GFP and Rec8-mCherry were observed in the mes1-B44 mutant, which arrests after meiosis I). For immunoprecipitation of the Par1 complex, we used mitotic asynchronous cells containing par1-FLAG. We used pat1-114 cells, with shutting off slp1+ by Prad21 (Prad21-slp1+ allele) to induce synchronous meiosis with arrest at metaphase I. Cells were grown in MM liquid medium including NH4Cl (MM + N) to a density of 1 × 107 cells/mL at 25°C, then resuspended in MM medium lacking NH4Cl (MM − N) at a density of 1 × 107 cells/mL for 16 h at 25°C. To induce meiosis, cells were incubated at 34°C, and collected at 0, 3, and 4 h after the induction of meiosis, reaching the period of metaphase I.
Construction of rec8 mutant strains
We made several short fragments from the N-terminal and C-terminal domains and narrowed down the phosphorylation domain into 98–198 amino acids and 441–501 amino acids, respectively (data not shown). Therefore, we introduced mutations mainly in these regions in Rec8N and Rec8C (Fig 2A). To generate alanine substitutions in rec8+, we cloned cDNA of rec8+ into a plasmid and mutagenized using the Kunkel method and PrimeSTAR mutagenesis basal kit (TaKaRa) (Ishiguro et al. 2010). All tagging at the C terminus of the rec8+ gene was accompanied by the 3′ UTR of either the rec8+ or spo5+ gene to prevent the leakage of expression during mitosis (Harigaya et al. 2006). Correct integration was confirmed by PCR and/or DNA sequencing.
Fluorescence microscopy
All live cell fluorescence microscopy was performed using a Nikon Ti12 inverted microscope with Perfect Focus System and Okolab environmental chamber and a Prime sCMOS camera (Photometrics) (Edelstein et al. 2014). The microscope was controlled with Micro-Manager v2.0 software (Open-imaging). Fluorescence excitation was performed using a SpectraX LED light engine (Lumencor) fitted with standard filters. ImageJ software (NIH) was used to measure pixel intensity, adjust brightness and contrast and render maximum projection images (Schindelin et al. 2012). Images were taken using a 100× Plan Apochromat oil-immersion objective (NA 1.45) at 25°C. Nine z sections (spaced by 0.5 μm each) of the fluorescent images were converted into a single two-dimensional image by maximum intensity projection. Fluorescence intensity measurements for Rec8-mChery and CFP-Plo1 were performed on maximum projection images. We measured the average intensity of the centromeric domain (0.34 μm2 size) and subtracted the average background intensity in the adjacent cytoplasm domains. Chromosome segregation assays monitoring imr1-GFP were performed also using maximum projection images.
Supplementary Material
Acknowledgments
We thank Silke Hauf for critically reading the manuscript, and Iain Hagan, Masamitsu Sato, Taro Nakamura, and the Yeast Genetic Resource Center (YGRC) for yeast strains and plasmids. We also thank Tadashi Ishiguro and Seira Miyazaki for assisting with the in vitro dephosphorylation assays. Y.W. especially thanks Paul Nurse for providing a laboratory space in the Francis Crick Institute, Scott Curran for microscope assistance, and all members in P. Nurse's and A.M.C.’s laboratories for their kind support and help. A.M.C. acknowledges Wellcome Trust award 110047/Z/15/Z. This work was supported by the National Key Research and Development Program of China (2017YFC1600403), and the National Natural Science Foundation of China (Key Program, 31830068).
Author contributions: Y.W. and W.M. conceived and performed experiments. J.Z., A.M.C., and J.C. supervised the study. Y.W. wrote the paper, with input from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348052.120.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Attner MA, Miller MP, Ee LS, Elkin SK, Amon A. 2013. Polo kinase Cdc5 is a central regulator of meiosis I. Proc Natl Acad Sci 110: 14278–14283. 10.1073/pnas.1311845110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103: 387–398. 10.1016/S0092-8674(00)00131-8 [DOI] [PubMed] [Google Scholar]
- Challa K, Fajish VG, Shinohara M, Klein F, Gasser SM, Shinohara A. 2019. Meiosis-specific prophase-like pathway controls cleavage-independent release of cohesin by Wapl phosphorylation. PLoS Genet 15: e1007851. 10.1371/journal.pgen.1007851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang T, Duncan FE, Schindler K, Schultz RM, Lampson MA. 2010. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr Biol 20: 1522–1528. 10.1016/j.cub.2010.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. 2014. Advanced methods of microscope control using µManager software. J Biol Methods 1: e10. 10.14440/jbm.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galander S, Barton RE, Borek WE, Spanos C, Kelly DA, Robertson D, Rappsilber J, Marston AL. 2019. Reductional meiosis I chromosome segregation is established by coordination of key meiotic kinases. Dev Cell 49: 526–541.e5. 10.1016/j.devcel.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. 2006. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442: 45–50. 10.1038/nature04881 [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y. 2010. Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol 12: 500–506. 10.1038/ncb2052 [DOI] [PubMed] [Google Scholar]
- Izawa D, Goto M, Yamashita A, Yamano H, Yamamoto M. 2005. Fission yeast Mes1p ensures the onset of meiosis II by blocking degradation of cyclin Cdc13p. Nature 434: 529–533. 10.1038/nature03406 [DOI] [PubMed] [Google Scholar]
- Kagami A, Sakuno T, Yamagishi Y, Ishiguro T, Tsukahara T, Shirahige K, Tanaka K, Watanabe Y. 2011. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep 12: 1189–1195. 10.1038/embor.2011.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Matos J, Mori S, Shirahige K, Zachariae W, Nasmyth K. 2004. Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr Biol 14: 2183–2196. 10.1016/j.cub.2004.12.020 [DOI] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W. 2010. Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell 18: 397–409. 10.1016/j.devcel.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327: 172–177. 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kim J, Ishiguro K, Nambu A, Akiyoshi B, Yokobayashi S, Kagami A, Ishiguro T, Pendas AM, Takeda N, Sakakibara Y, et al. 2015. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature 517: 466–471. 10.1038/nature14097 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y. 2003a. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. Embo J 22: 5643–5653. 10.1093/emboj/cdg527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Yokobayashi S, Yamamoto M, Watanabe Y. 2003b. Distinct cohesin complexes organize meiotic chromosome domains. Science 300: 1152–1155. 10.1126/science.1083634 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510–517. 10.1038/nature02312 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52. 10.1038/nature04663 [DOI] [PubMed] [Google Scholar]
- Le AH, Mastro TL, Forsburg SL. 2013. The C-terminus of S. pombe DDK subunit Dfp1 is required for meiosis-specific transcription and cohesin cleavage. Biol Open 2: 728–738. 10.1242/bio.20135173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Kiburz BM, Amon A. 2004. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr Biol 14: 2168–2182. 10.1016/j.cub.2004.12.033 [DOI] [PubMed] [Google Scholar]
- Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, et al. 2010. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 20: 1511–1521. 10.1016/j.cub.2010.08.023 [DOI] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bähler J. 2002. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147. 10.1038/ng951 [DOI] [PubMed] [Google Scholar]
- Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. 2008. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135: 662–678. 10.1016/j.cell.2008.10.026 [DOI] [PubMed] [Google Scholar]
- Miyazaki S, Kim J, Yamagishi Y, Ishiguro T, Okada Y, Tanno Y, Sakuno T, Watanabe Y. 2017. Meikin-associated polo-like kinase specifies Bub1 distribution in meiosis I. Genes Cells 22: 552–567. 10.1111/gtc.12496 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast schizosaccharomyces pombe. Methods Enzymol 194: 795–823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35: 673–745. 10.1146/annurev.genet.35.102401.091334 [DOI] [PubMed] [Google Scholar]
- Ogushi S, Rattani A, Godwin J, Metson J, Schermelleh L, Nasmyth K. 2020. Loss of sister kinetochore co-orientation and peri-centromeric cohesin protection after meiosis I depends on cleavage of centromeric REC8. bioRxiv 10.1101/2020.02.06.935171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114. 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP. 2010. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell 40: 353–363. 10.1016/j.molcel.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y, Hashimoto S, Nakaoka Y, Kouznetsova A, Höög C, Kitajima TS. 2015. Bivalent separation into univalents precedes age-related meiosis I errors in oocytes. Nat Commun 6: 7550. 10.1038/ncomms8550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T, Tada K, Watanabe Y. 2009. Kinetochore geometry defined by cohesion within the centromere. Nature 458: 852–858. 10.1038/nature07876 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn MA, McCarroll R, Murray AW. 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev 16: 1659–1671. 10.1101/gad.975802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Petersen J, MacIver F, Mulvihill DP, Glover DM, Hagan IM. 2001. The role of Plo1 kinase in mitotic commitment and septation in Schizosaccharomyces pombe. EMBO J 20: 1259–1270. 10.1093/emboj/20.6.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. 2012. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat Rev Mol Cell Biol 13: 370–382. 10.1038/nrm3349 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461–464. 10.1038/22774 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. 2001. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409: 359–363. 10.1038/35053103 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. 2008. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature 455: 251–255. 10.1038/nature07217 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol 14: 746–752. 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Yatskevich S, Rhodes J, Nasmyth K. 2019. Organization of chromosomal DNA by SMC complexes. Annu Rev Genet 53: 445–482. 10.1146/annurev-genet-112618-043633 [DOI] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. 2005. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123: 803–817. 10.1016/j.cell.2005.09.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


