Here, Fursova et al. investigated the role of BAP1, an H2AK119ub1 deubiquitylating enzyme, in regulating gene expression in mouse ESCs. Using inducible genetic perturbations and quantitative genomics, they show that BAP1 plays a widespread role in promoting gene expression and that this relies on BAP1 counteracting pervasive H2AK119ub1. In the absence of BAP1, accumulation of H2AK119ub1 throughout the genome leads to widespread reductions in transcription.
Keywords: chromatin, epigenetics, histone modification, gene expression, transcription, BAP1, deubiquitylase, histone monoubiquitylation, H2AK119ub1, Polycomb
Abstract
Histone-modifying systems play fundamental roles in gene regulation and the development of multicellular organisms. Histone modifications that are enriched at gene regulatory elements have been heavily studied, but the function of modifications found more broadly throughout the genome remains poorly understood. This is exemplified by histone H2A monoubiquitylation (H2AK119ub1), which is enriched at Polycomb-repressed gene promoters but also covers the genome at lower levels. Here, using inducible genetic perturbations and quantitative genomics, we found that the BAP1 deubiquitylase plays an essential role in constraining H2AK119ub1 throughout the genome. Removal of BAP1 leads to pervasive genome-wide accumulation of H2AK119ub1, which causes widespread reductions in gene expression. We show that elevated H2AK119ub1 preferentially counteracts Ser5 phosphorylation on the C-terminal domain of RNA polymerase II at gene regulatory elements and causes reductions in transcription and transcription-associated histone modifications. Furthermore, failure to constrain pervasive H2AK119ub1 compromises Polycomb complex occupancy at a subset of Polycomb target genes, which leads to their derepression, providing a potential molecular rationale for why the BAP1 ortholog in Drosophila has been characterized as a Polycomb group gene. Together, these observations reveal that the transcriptional potential of the genome can be modulated by regulating the levels of a pervasive histone modification.
In eukaryotes, DNA is wrapped around histones to form nucleosomes and chromatin, which packages the genome inside the nucleus. In addition to their structural role, histones are subject to a variety of posttranslational modifications (PTMs), which have been proposed to play important roles in regulation of gene expression and other chromosomal processes (Groth et al. 2007; Kouzarides 2007; Bannister and Kouzarides 2011; Zhao and Garcia 2015; Hauer and Gasser 2017). If chromatin-modifying systems are perturbed, this can lead to profound alterations in gene expression, resulting in severe developmental disorders and cancer (Audia and Campbell 2016; Atlasi and Stunnenberg 2017; Bracken et al. 2019; Zhao and Shilatifard 2019). However, for many histone modifications, the mechanisms that control their levels throughout the genome and ultimately how this influences gene expression remain poorly understood.
Genome-wide profiling has revealed that some histone modifications are specifically enriched at gene promoters and distal regulatory elements (Barski et al. 2007; Ernst and Kellis 2010; Kharchenko et al. 2011; Zhou et al. 2011; Ho et al. 2014), where they have been proposed to regulate chromatin accessibility and work with the transcriptional machinery to control gene expression (Lee et al. 1993; Vettese-Dadey et al. 1996; Pray-Grant et al. 2005; Wysocka et al. 2006; Vermeulen et al. 2007; Lauberth et al. 2013; Zhang et al. 2017a). However, it has also emerged that there are other histone modifications that are extremely abundant and cover broad regions of the genome, extending far beyond genes and gene regulatory elements (Kharchenko et al. 2011; Ferrari et al. 2014; Lee et al. 2015; Kahn et al. 2016; Zheng et al. 2016; Carelli et al. 2017; Fursova et al. 2019). Much less effort has been placed on studying these more pervasive histone modifications, raising the possibility that they could also have important and previously underappreciated roles in gene regulation.
The Polycomb repressive complex 1 (PRC1) is an E3 ubiquitin ligase that catalyzes monoubiquitylation of histone H2A (H2AK119ub1) (de Napoles et al. 2004; Wang et al. 2004; Buchwald et al. 2006). PRC1 is targeted to CpG island-associated gene promoters where it can deposit high levels of H2AK119ub1 (Ku et al. 2008; Farcas et al. 2012; He et al. 2013; Wu et al. 2013; Bauer et al. 2016), and this is central to Polycomb-mediated gene repression (Endoh et al. 2012; Blackledge et al. 2014, 2020; Tsuboi et al. 2018; Fursova et al. 2019; Tamburri et al. 2020). A second Polycomb repressive complex, PRC2, is recruited to the same sites (Boyer et al. 2006; Bracken 2006; Li et al. 2017a; Perino et al. 2018), where it deposits histone H3 lysine 27 methylation (H3K27me3) (Cao et al. 2002; Czermin et al. 2002; Kuzmichev 2002; Müller et al. 2002), leading to the formation of transcriptionally repressive Polycomb chromatin domains that have high levels of PRC1, PRC2, and their respective histone modifications (Mikkelsen et al. 2007; Ku et al. 2008). In addition to this punctate high-level enrichment of H2AK119ub1 at Polycomb target gene promoters, we and others have recently demonstrated that H2AK119ub1 is also found broadly throughout the genome, albeit at much lower levels (Lee et al. 2015; Kahn et al. 2016; Fursova et al. 2019). However, whether this genome-wide pool of H2AK119ub1 influences gene expression has remained enigmatic.
Interestingly, H2AK119ub1 is highly dynamic (Seale 1981), and a number of deubiquitylating enzymes (DUBs) have been proposed to regulate its levels (for review, see Belle and Nijnik 2014; Aquila and Atanassov 2020). The most extensively characterized and evolutionarily conserved of these DUBs is BAP1, which interacts with ASXL proteins to form the Polycomb repressive deubiquitinase complex (PR-DUB) (Scheuermann et al. 2010; Wu et al. 2015; Hauri et al. 2016; Kloet et al. 2016; Sahtoe et al. 2016; Campagne et al. 2019). Previous attempts to understand how BAP1 regulates gene expression and whether this relies on its H2AK119ub1 deubiquitylase activity have primarily focused on examining how the PR-DUB complex is targeted to gene promoters and distal regulatory elements and how this regulates binding and/or activity of chromatin-modifying transcriptional coactivators (Li et al. 2017b; Wang et al. 2018; Campagne et al. 2019; Kuznetsov et al. 2019; Kolovos et al. 2020; Szczepanski et al. 2020). While this has revealed that BAP1 can remove H2AK119ub1 at specific loci, its primary site of action in the genome and the mechanisms by which it controls gene expression have appeared to be context-dependent and in some cases difficult to reconcile with the known roles of H2AK119ub1 in gene regulation. Therefore, how H2AK119ub1 levels in the genome are modulated by BAP1 and how this influences transcription remains poorly defined. Addressing these questions is particularly important in the light of the essential role that BAP1 plays as a tumor suppressor (Ventii et al. 2008; Dey et al. 2012; Carbone et al. 2013; Murali et al. 2013; Daou et al. 2015) and could provide important new insight into how BAP1 dysfunction causes cellular transformation.
To dissect how BAP1 controls H2AK119ub1 levels and gene expression, here, we integrate genome editing, inducible genetic perturbations, and quantitative genomics. We found that BAP1 functions to constrain pervasive H2AK119ub1 throughout the genome, with no preference for gene promoters or distal regulatory elements. We demonstrate that by counteracting pervasive H2AK119ub1, BAP1 plays a fundamental role in facilitating gene expression. In the absence of BAP1, elevated H2AK119ub1 broadly inhibits Serine 5 phosphorylation (Ser5P) on the C-terminal domain (CTD) of RNA polymerase II at gene regulatory elements and causes widespread reductions in transcription and transcription-associated histone modifications, without limiting chromatin accessibility. Finally, we discover that a subset of Polycomb target genes rely on BAP1 for their silencing and provide a mechanistic rationale for how BAP1 can indirectly support Polycomb-mediated gene repression. Together, these observations demonstrate how the levels of a pervasive histone modification must be appropriately controlled to enable the transcriptional potential of the genome.
Results
BAP1 functions pervasively throughout the genome to constrain H2AK119ub1
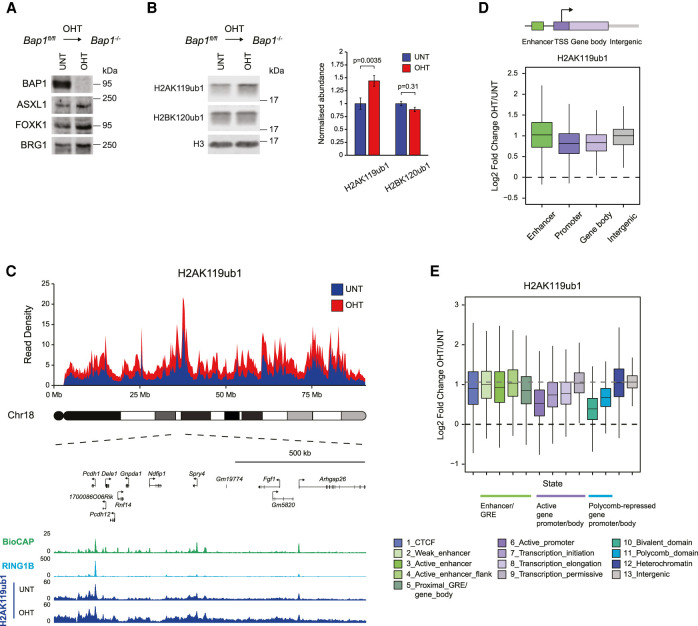
Given our recent discovery that H2AK119ub1 is deposited more broadly throughout the genome than previously appreciated (Fursova et al. 2019), we set out to determine where in the genome BAP1 functions to control the levels of H2AK119ub1 and how this influences gene expression. To address these important questions, we developed a BAP1 conditional knockout mouse embryonic stem cell (ESC) line (Bap1fl/fl) in which addition of tamoxifen (OHT) enables inducible removal of BAP1, allowing us to capture the primary effects that BAP1 loss has on H2AK119ub1 and gene expression. Importantly, tamoxifen treatment of Bap1fl/fl cells resulted in a complete loss of BAP1 protein, while the levels of BAP1-interacting partners were largely unchanged (Fig. 1A). In line with previous observations in BAP1 knockout mouse ESCs and human cancer cell lines (Wang et al. 2018; Campagne et al. 2019; He et al. 2019; Kolovos et al. 2020), Western blot analysis showed that H2AK119ub1 levels were markedly increased following BAP1 removal, whereas H2BK120ub1 was unaffected (Fig. 1B).
Figure 1.
BAP1 functions pervasively throughout the genome to constrain H2AK119ub1. (A) Western blot analysis for BAP1 and other subunits of the PR-DUB complex (ASXL1 and FOXK1) in untreated (UNT) and OHT-treated (OHT) Bap1fl/fl ESCs. BRG1 is shown as a loading control. (B) Western blot analysis (left panel) and quantification (right panel) of H2AK119ub1 and H2BK120ub1 levels relative to histone H3 in untreated and OHT-treated Bap1fl/fl ESCs. Error bars represent SEM (n = 3). P-values denote the result of a paired two-tailed Student's t-test. (C) A chromosome density plot showing H2AK119ub1 cChIP-seq signal across chromosome 18 in Bap1fl/fl ESCs (untreated and OHT-treated) with an expanded snapshot of a region on chromosome 18 shown below. BioCAP-seq and RING1B cChIP-seq in wild-type ESCs are also shown to indicate the location of CGIs that are occupied by PRC1. (D) Box plots comparing log2 fold changes in H2AK119ub1 cChIP-seq signal at gene regulatory elements (enhancers and promoters), gene bodies, and intergenic regions in Bap1fl/fl ESCs following OHT treatment. (E) Box plots comparing log2 fold changes in H2AK119ub1 cChIP-seq signal following OHT treatment in Bap1fl/fl ESCs across different chromatin states derived from unsupervised genome segmentation using ChromHMM. Chromatin states are grouped based on the underlying gene regulatory elements (GREs) and transcriptional activity. The dashed gray line indicates the overall change in H2AK119ub1 levels in the genome as determined by its median value in intergenic regions.
Having shown that conditional knockout of BAP1 leads to an increase in H2AK119ub1 (Fig. 1B), we set out to define where in the genome H2AK119ub1 was elevated using an unbiased quantitative genomic approach. To achieve this, we carried out calibrated ChIP-seq (cChIP-seq) for H2AK119ub1 before and after removal of BAP1. Remarkably, this revealed a major and widespread accumulation of H2AK119ub1, which was evident when we visualized changes in H2AK119ub1 across an entire chromosome and also when we focused on individual regions of chromosomes (Fig. 1C; Supplemental Figs. S1A, S2D). Importantly, the magnitude of H2AK119ub1 accumulation appeared to be largely uniform throughout the genome (Fig. 1C), showing no preference for gene regulatory elements, including promoters and enhancers (Fig. 1D; Supplemental Fig. S2A), where BAP1 has been previously proposed to function (Wang et al. 2018; Campagne et al. 2019; Kuznetsov et al. 2019). To characterize the effect of BAP1 removal on H2AK119ub1 in more detail, we employed an unsupervised ChromHMM classification approach (Ernst and Kellis 2012) to segment the genome into 13 chromatin states encompassing all major functional genomic annotations (Supplemental Fig. S2B) and examined changes in H2AK119ub1 across these distinct states. This revealed that all chromatin states were significantly affected and displayed similar increases in H2AK119ub1 (Fig. 1E; Supplemental Fig. S2C,D). Interestingly, these effects appeared to be less pronounced at Polycomb-enriched chromatin states (Fig. 1E). However, a closer examination revealed that these regions gained similar amounts of H2AK119ub1 to the rest of the genome (Supplemental Fig. S2C), with higher initial levels of H2AK119ub1 likely accounting for the smaller apparent fold changes following BAP1 removal (Fig. 1E). Furthermore, states encompassing actively transcribed genes also showed a less pronounced increase in H2AK119ub1 (Fig. 1E; Supplemental Fig. S2C), possibly due to the previously proposed antagonism between the Polycomb system and transcription (Klose et al. 2013; Riising et al. 2014; Beltran et al. 2016). Together, these observations demonstrate that BAP1 functions pervasively and indiscriminately throughout the genome to constrain H2AK119ub1.
Pervasive accumulation of H2AK119ub1 in the absence of BAP1 causes widespread reductions in gene expression
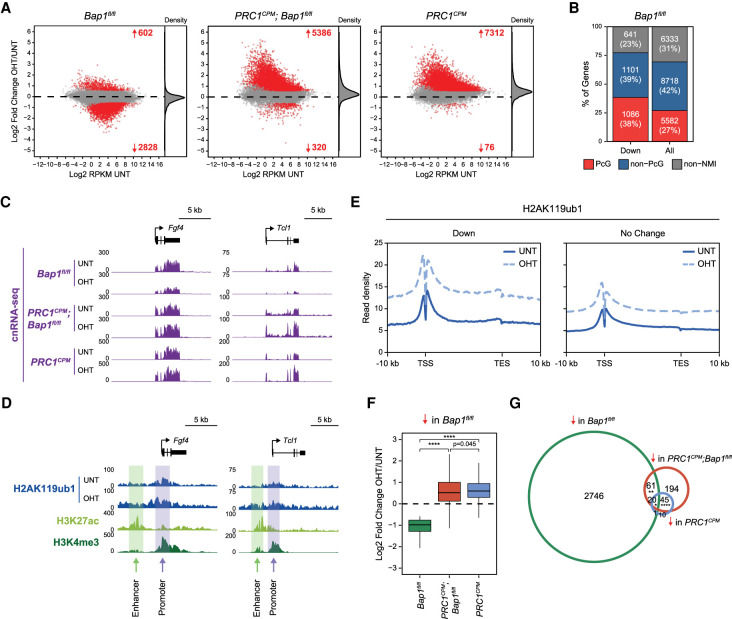
Given that H2AK119ub1 plays a central role in PRC1-mediated gene repression (Endoh et al. 2012; Blackledge et al. 2020; Tamburri et al. 2020), we were curious to determine what effect BAP1 removal and the resulting accumulation of H2AK119ub1 throughout the genome would have on gene expression. Therefore, we carried out calibrated nuclear RNA sequencing (cnRNA-seq) in our conditional BAP1 knockout cells. This revealed that removal of BAP1 caused widespread changes in gene expression, with the majority of genes exhibiting reduced expression (Fig. 2A). Remarkably, we found that 6440 genes (∼44% of all expressed genes) (Supplemental Fig. S3A) showed at least a 20% reduction in expression. Of these, 2828 genes (∼20% of all expressed genes) were significantly reduced by at least 1.5-fold, indicating that BAP1 plays a broad role in promoting gene expression.
Figure 2.
Pervasive accumulation of H2AK119ub1 in the absence of BAP1 causes widespread reductions in gene expression. (A) MA plots showing log2 fold changes in gene expression (cnRNA-seq) in Bap1fl/fl, PRC1CPM;Bap1fl/fl, and PRC1CPM ESCs following OHT treatment. Significant gene expression changes (P-adj < 0.05 and >1.5-fold) for a custom nonredundant set of refGene genes (n = 20,633) are shown in red. The density of gene expression changes is shown at the right. (B) A bar plot illustrating the distribution of different gene classes among genes showing significantly reduced expression following OHT treatment in Bap1fl/fl ESCs based on cnRNA-seq analysis (P-adj < 0.05 and >1.5-fold). (PcG) Polycomb-occupied genes, (Non-PcG) non-Polycomb-occupied genes, (Non-NMI) genes lacking a nonmethylated CGI (NMI) at their promoter. (C) Snapshots of genes whose expression is significantly reduced (P-adj < 0.05 and >1.5-fold) following removal of BAP1, showing gene expression (cnRNA-seq) in Bap1fl/fl, PRC1CPM;Bap1fl/fl, and PRC1CPM ESCs (untreated and OHT-treated). (D) Snapshots of genes whose expression is significantly reduced (P-adj < 0.05 and >1.5-fold) following removal of BAP1, showing H2AK119ub1 cChIP-seq in Bap1fl/fl ESCs (untreated and OHT-treated). Also shown is cChIP-seq for H3K27ac and H3K4me3 in untreated Bap1fl/fl ESCs to highlight the position of promoters (H3K27ac-high, H3K4me3-high) and nearest putative enhancers (H3K27ac-high, H3K4me3-low) for these genes. (E) Metaplots of H2AK119ub1 cChIP-seq signal in Bap1fl/fl ESCs (untreated and OHT-treated) across genes that show a significant reduction (Down, n = 2828) or no change (No Change, n = 17,203) in expression following BAP1 removal based on cnRNA-seq analysis (P-adj < 0.05 and >1.5-fold). (F) Box plots comparing log2 fold changes in expression (cnRNA-seq) following OHT treatment in Bap1fl/fl (green), PRC1CPM;Bap1fl/fl (red), and PRC1CPM (blue) ESCs for genes whose expression is significantly reduced (P-adj < 0.05 and >1.5-fold) in the absence of BAP1. P-values denote the result of a two-tailed Student's t-test. (****) P < 10−100. (G) A Venn diagram of the overlap between genes that show a significant reduction in expression based on cnRNA-seq analysis (P-adj < 0.05 and >1.5-fold) following OHT treatment in Bap1fl/fl (green), PRC1CPM;Bap1fl/fl (red), and PRC1CPM (blue) ESCs. P-values denote the result of a Fisher's exact test for the pairwise overlaps between genes showing reduced expression in PRC1CPM;Bap1fl/fl and PRC1CPM, Bap1fl/fl and PRC1CPM;Bap1fl/fl, as well as Bap1fl/fl and PRC1CPM ESCs. (****) P < 10−100 for PRC1CPM;Bap1fl/fl and PRC1CPM, (**) P < 10−5 for Bap1fl/fl and PRC1CPM;Bap1fl/fl, (*) P < 0.05 for Bap1fl/fl and PRC1CPM.
Although reductions in gene expression following BAP1 removal were widespread, expression of some genes was more severely affected than others. Importantly, the majority of genes showing significantly reduced expression were not classical Polycomb target genes (Fig. 2B; Supplemental Fig. S3B). However, interestingly, these genes were often found in regions of the genome that had higher levels of H2AK119ub1 in wild-type cells and, in the absence of BAP1, also acquired higher levels of H2AK119ub1 than genes that were not significantly affected (Fig. 2C–E; Supplemental Fig. S3C–F). Importantly, the increase in H2AK119ub1 was not specific to the promoters or enhancers of these genes but was evident across the entire gene and flanking regions (Fig. 2D,E; Supplemental Fig. S3C–F). Together, these observations suggest that widespread reductions in gene expression following BAP1 removal likely result from pervasive accumulation of H2AK119ub1, with some genes being more susceptible to these effects than others.
To directly test whether elevated H2AK119ub1 was responsible for gene repression in the absence of BAP1, we developed an inducible mouse ES cell line (PRC1CPM;Bap1fl/fl) in which we could simultaneously disrupt BAP1 and inactivate PRC1 catalysis to remove H2AK119ub1 (Supplemental Fig. S3G,H; Blackledge et al. 2020). We then carried out cnRNA-seq and compared the effects on gene expression caused by concurrent removal of BAP1 and H2AK119ub1 with the effects caused by removing BAP1 or H2AK119ub1 individually (Fig. 2A; Supplemental Fig. S3I). Strikingly, in the absence of H2AK119ub1, removal of BAP1 no longer caused widespread reductions in gene expression (Fig. 2A,C,F,G; Supplemental Fig. S3J), indicating that H2AK119ub1 was required for these effects. In contrast, Polycomb target genes were derepressed following catalytic inactivation of PRC1 regardless of whether BAP1 was disrupted (Fig. 2A; Supplemental Fig. S3K). Therefore, we conclude that BAP1 counteracts accumulation of H2AK119ub1 throughout the genome, and in its absence elevated H2AK119ub1 causes widespread inhibition of gene expression.
BAP1 counteracts pervasive H2AK119ub1 to promote Ser5 phosphorylation on the CTD of RNA polymerase II at gene regulatory elements
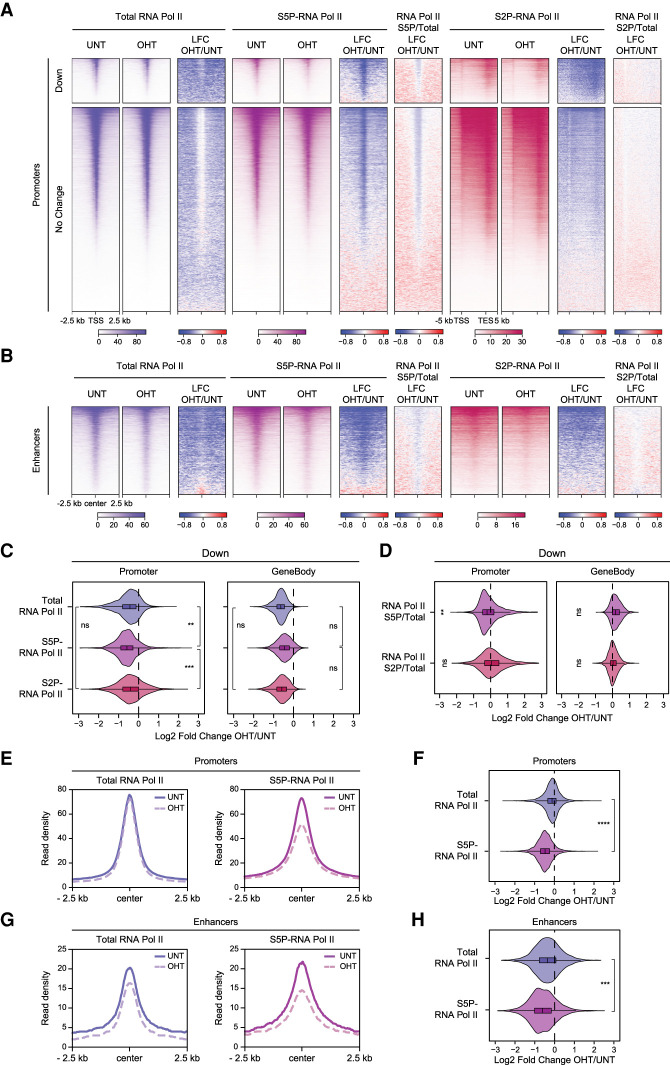
To understand how accumulation of H2AK119ub1 counteracts gene expression, we examined how RNA polymerase II (Pol II) was affected after BAP1 removal. To achieve this, we carried out cChIP-seq to quantitate total Pol II levels and also examined its phosphorylation states, which are associated with transcription initiation (Ser5P) and elongation (Ser2P) (Buratowski 2009; Harlen and Churchman 2017). When we inspected genes whose expression was significantly reduced following BAP1 removal, we found that levels of Pol II and its phosphorylated forms were decreased at promoters and over gene bodies (Fig. 3A,C; Supplemental Fig. S4A–C). The reduction in Ser2P in gene bodies was similar in magnitude to the decrease in total Pol II levels (Fig. 3A,C,D), indicating that elongation-associated phosphorylation was not specifically disrupted, despite reduced transcription. In contrast, the reduction in Ser5P at the promoters of these genes was larger in magnitude than the decrease in Pol II occupancy (Fig. 3A,C,D; Supplemental Fig. S4A–C), suggesting that elevated H2AK119ub1 limits Ser5 phosphorylation on the CTD of Pol II, and this may lead to reduced transcription and gene expression.
Figure 3.
BAP1 counteracts pervasive H2AK119ub1 to promote Ser5 phosphorylation on the CTD of RNA Pol II at gene regulatory elements. (A) Heat maps illustrating cChIP-seq signal for total Pol II occupancy and its Ser5 phosphorylation (S5P) at gene promoters, as well as Pol II Ser2 phosphorylation (S2P) over gene bodies in Bap1fl/fl ESCs (untreated and OHT-treated). Also shown are the log2 fold changes in cChIP-seq signal after BAP1 removal (LFC OHT/UNT) and the log2 fold changes in the abundance of Ser5P and Ser2P relative to total Pol II levels (S5P/Total and S2P/Total). Genes were segregated into those that show a significant reduction (Down, n = 2828) or no change (No Change, n = 17,203) in expression following BAP1 removal based on cnRNA-seq analysis (P-adj < 0.05 and >1.5-fold). Intervals were sorted by total Pol II cChIP-seq signal in untreated Bap1fl/fl ESCs. (B) Heat maps illustrating cChIP-seq signal for total Pol II occupancy, as well as its Ser5P and Ser2P forms, at active enhancers in Bap1fl/fl ESCs (untreated and OHT-treated). As in A, the log2 fold changes in cChIP-seq signal after BAP1 removal (LFC OHT/UNT) are shown together with the log2 fold changes in the abundance of Ser5P and Ser2P relative to total Pol II levels (S5P/Total and S2P/Total). Intervals were sorted by total Pol II cChIP-seq signal in untreated Bap1fl/fl ESCs. (C) Violin plots comparing log2 fold changes in cChIP-seq signal for total Pol II, as well as its Ser5P and Ser2P forms, following OHT treatment in Bap1fl/fl ESCs at the promoters and over the bodies of genes that show a significant reduction (Down, n = 2828) in expression after BAP1 removal based on cnRNA-seq analysis (P-adj < 0.05 and >1.5-fold). P-values denote the result of a one-tailed Student's t-test. (***) P < 10−10, (**) P < 10−5, (ns) P > 0.05. For comparisons of Ser5P/Ser2P with total Pol II, the alternative hypothesis was that the log2 fold change in Ser5P/Ser2P was smaller. For the comparison of Ser5P with Ser2P, the alternative hypothesis was that the log2 fold change in Ser5P was smaller. (D) Violin plots comparing log2 fold changes in the abundance of Ser5P and Ser2P relative to total Pol II levels (S5P/Total and S2P/Total) following OHT treatment in Bap1fl/fl ESCs at the promoters and over the bodies of genes defined in C. P-values denote the result of a one-sample one-tailed Student's t-test to determine whether the log2 fold changes were significantly smaller than 0. (**) P < 10−5, (ns) P > 0.05. (E) Metaplots of total and Ser5P Pol II cChIP-seq signal at active gene promoters in Bap1fl/fl ESCs (untreated and OHT-treated). (F) Violin plots comparing log2 fold changes in total and Ser5P Pol II cChIP-seq signal at active gene promoters in Bap1fl/fl ESCs following OHT treatment. P-value denotes the result of a one-tailed Student's t-test with the alternative hypothesis that the log2 fold change in Ser5P was smaller. (****) P < 10−100. (G) As in E but for active enhancers. (H) As in F but for active enhancers. P-value denotes the result of a one-tailed Student's t-test with the alternative hypothesis that the log2 fold change in Ser5P was smaller. (***) P < 10−10.
Given that removal of BAP1 caused pervasive accumulation of H2AK119ub1 throughout the genome (Fig. 1), we wondered whether the repressive effects of this histone modification on transcription may in fact extend beyond the subset of genes that showed significant reductions in gene expression. When we examined genes whose expression did not change significantly after BAP1 removal, we found that the occupancy of Pol II at their promoters was only marginally affected, but there were widespread reductions in the levels of Pol II and Ser2P over gene bodies (Fig. 3A; Supplemental Fig. S4A,B,D), which were similar in magnitude (Fig. 3A; Supplemental Fig. S4E). Importantly, these effects on Pol II in the gene body correlated well with changes in transcript levels (Supplemental Fig. S4F), indicating that elevated H2AK119ub1 causes widespread reductions in transcription and gene expression (Supplemental Fig. S4G), despite only a subset of genes being captured as having significantly reduced expression in cnRNA-seq analysis. Importantly, in contrast to Pol II occupancy, which was only modestly affected, Ser5P was markedly reduced at the promoters of all genes, including those whose expression did not change significantly after BAP1 removal (Fig. 3A,E,F; Supplemental Fig. S4A–E,H). This suggests that the widespread reductions in transcription following BAP1 removal are likely linked to the observed reductions in Ser5P. Since the accumulation of H2AK119ub1 in the absence of BAP1 is not restricted to genes or their promoters (Fig. 1D), we wondered whether the observed effects on Pol II may in fact extend to other gene regulatory elements, like enhancers, which have been also reported to bind Pol II and initiate transcription (Li et al. 2016; Andersson and Sandelin 2020; Sartorelli and Lauberth 2020). This revealed that there was also a pronounced decrease in total Pol II occupancy and an even larger reduction in Ser5P at enhancers as well (Fig. 3B,G,H; Supplemental Fig. S4A,H). Together, these observations demonstrate that BAP1 functions broadly throughout the genome to support transcription from gene regulatory elements by constraining pervasive H2AK119ub1.
Aberrant accumulation of H2AK119ub1 compromises transcription-associated histone modifications but does not limit chromatin accessibility
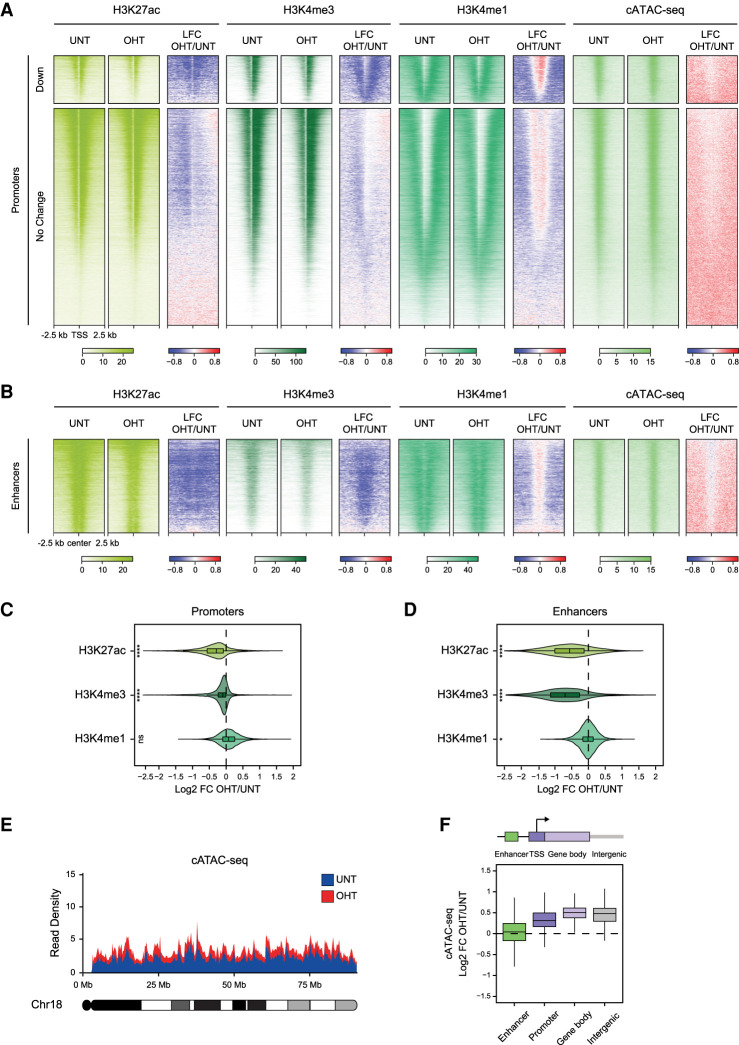
Having established that elevated H2AK119ub1 in the absence of BAP1 broadly inhibits transcription from promoters and enhancers (Fig. 3), we wanted to investigate whether chromatin features associated with transcription were also affected. To address this question, we carried out cChIP-seq for histone modifications that are typically enriched at active promoters (H3K27ac and H3K4me3) or enhancers (H3K27ac and H3K4me1) (Calo and Wysocka 2013; Andersson and Sandelin 2020). Interestingly, we observed a widespread decrease in H3K27ac at both types of gene regulatory elements in the absence of BAP1, with enhancers showing more pronounced reductions (Fig. 4A–D; Supplemental Fig. S5A–C,E,F). Removal of BAP1 also compromised H3K4me3 at gene promoters, but this effect was on average much more modest and mostly limited to genes that showed significant reductions in expression (Fig. 4A,C; Supplemental Fig. S5A–C,E). In contrast, H3K4me3 at enhancers was markedly reduced, despite the starting levels of this modification being considerably lower than at promoters (Fig. 4A,B,D; Supplemental Fig. S5A,E,F). Finally, we also observed a modest but widespread decrease in H3K4me1 around promoters and enhancers, which was accompanied by a slight increase at the center of these regulatory elements (Fig. 4A–D; Supplemental Fig. S5A–C,E,F). Therefore, we found that BAP1 removal leads to moderate and seemingly indiscriminate effects on transcription-associated histone modifications at both promoters and enhancers, which correlate with the effects on gene expression (Supplemental Fig. S5D). This observation differs from previous studies that have implicated BAP1 and other PR-DUB subunits in directly recruiting chromatin-modifying transcriptional coactivators to either promoters or enhancers to specifically affect histone modifications at these sites (Li et al. 2017b; Wang et al. 2018; Szczepanski et al. 2020). Instead, our new findings are more consistent with a model in which, in the absence of BAP1, elevated H2AK119ub1 inhibits early stages of transcription, which then leads to modest but broad effects on transcription-associated histone modifications.
Figure 4.
Aberrant accumulation of H2AK119ub1 compromises transcription-associated histone modifications but not chromatin accessibility at gene regulatory elements. (A) Heat maps illustrating H3K27ac, H3K4me3, and H3K4me1 cChIP-seq signal at gene promoters in Bap1fl/fl ESCs (untreated and OHT-treated). cATAC-seq is shown as a measure of chromatin accessibility. Also shown are the log2 fold changes in cChIP-seq and cATAC-seq signal after BAP1 removal (LFC OHT/UNT). Genes were segregated into those that show a significant reduction (Down, n = 2828) or no change (No Change, n = 17,203) in expression following BAP1 removal based on cnRNA-seq analysis (P-adj < 0.05 and >1.5-fold). Intervals were sorted by total Pol II cChIP-seq signal in untreated Bap1fl/fl ESCs. (B) As in A but for active enhancers. (C) Violin plots comparing log2 fold changes in cChIP-seq signal for H3K27ac, H3K4me3, and H3K4me1 at active gene promoters in Bap1fl/fl ESCs following OHT treatment. P-values denote the result of a one-sample one-tailed Student's t-test to determine whether the log2 fold changes were significantly smaller than 0. (****) P < 10−100, (ns) P > 0.05. (D) As in C but for active enhancers. P-values denote the result of a one-sample one-tailed Student's t-test to determine whether the log2 fold changes were significantly smaller than 0. (****) P < 10−100, (*) P < 0.05. (E) A chromosome density plot showing chromatin accessibility across chromosome 18 as measured by cATAC-seq in Bap1fl/fl ESCs (untreated and OHT-treated). This illustrates a widespread increase in cATAC-seq signal throughout the genome following BAP1 removal. (F) Box plots comparing log2 fold changes in cATAC-seq signal at gene regulatory elements (enhancers and promoters), gene bodies, and intergenic regions in Bap1fl/fl ESCs following OHT treatment.
Given that some chromatin modifications have been proposed to function through making chromatin less accessible to gene regulatory factors (Francis et al. 2001, 2004; Danzer and Wallrath 2004; Soufi et al. 2012; Becker et al. 2016; Fyodorov et al. 2018), we sought to determine whether elevated H2AK119ub1 could elicit its widespread effects on transcription by limiting chromatin accessibility. To test this possibility, we performed calibrated ATAC-seq (cATAC-seq) that measures chromatin accessibility by its susceptibility to tagmentation by Tn5 transposase (Buenrostro et al. 2015). Importantly, we found that accumulation of H2AK119ub1 in the absence of BAP1 did not cause major reductions in chromatin accessibility at gene promoters and enhancers (Fig. 4A,B,F; Supplemental Figs. S5A, S6A–C). Instead, to our surprise, we found that, following BAP1 removal, chromatin accessibility was modestly increased throughout the genome, in a similar manner to the pervasive accumulation of H2AK119ub1 (Fig. 4A,B,E,F; Supplemental Fig. S6D). Importantly, this demonstrates that H2AK119ub1 does not counteract transcription simply by limiting the access of regulatory factors to promoters and enhancers. Instead, pervasive accumulation of H2AK119ub1 in the absence of BAP1 broadly counteracts Ser5 phosphorylation of the Pol II CTD, resulting in widespread reductions in transcription and its associated histone modifications. Together, our findings illustrate how a pervasive histone modification that can inhibit transcription must be appropriately controlled to support the transcriptional potential of the genome.
BAP1 indirectly supports repression of a subset of Polycomb target genes
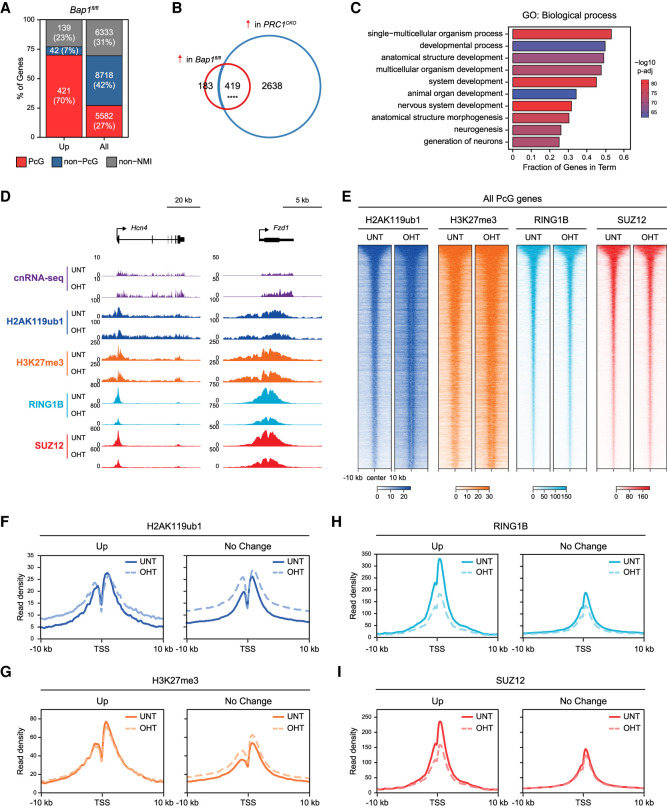
Our finding that BAP1 constrains pervasive H2AK119ub1 to facilitate gene expression is conceptually at odds with genetic characterisation of the Drosophila orthologs of BAP1 (Calypso) and other PR-DUB components as Polycomb group (PcG) transcriptional repressors (Jürgens 1985; Soto et al. 1995; de Ayala Alonso et al. 2007; Scheuermann et al. 2010). Intriguingly, despite the majority of genes showing reduced expression in BAP1-deficient cells, we also identified 602 genes whose expression was significantly increased in the absence of BAP1 (Fig. 2A). Remarkably, when we examined these genes in more detail, we found that the majority were Polycomb target genes enriched in GO categories related to regulation of developmental processes, which are characteristic of PRC1-repressed genes in mouse ESCs (Fig. 5A–D). Therefore, we show that BAP1 is required to repress a subset of Polycomb target genes, consistent with its genetic designation as a PcG gene.
Figure 5.
BAP1 indirectly supports repression of a subset of Polycomb target genes by counteracting pervasive H2AK119ub1 to focus Polycomb complex occupancy at target sites. (A) A bar plot illustrating the distribution of different gene classes among genes that become significantly derepressed (P-adj < 0.05 and >1.5-fold) following OHT treatment in Bap1fl/fl ESCs. (PcG) Polycomb-occupied genes, (Non-PcG) non-Polycomb-occupied genes, (Non-NMI) genes lacking a nonmethylated CGI (NMI) at their promoter. (B) A Venn diagram showing the overlap between genes that become significantly derepressed (P-adj < 0.05 and >1.5-fold) following OHT treatment in Bap1fl/fl (red) or PRC1CKO (blue) ESCs. P-value denotes the result of a Fisher's exact test. (****) P < 10−100. (C) A gene ontology (GO) analysis of biological process term enrichment for genes that become significantly derepressed (P-adj < 0.05 and >1.5-fold) in Bap1fl/fl cells following OHT treatment. (D) Snapshots of Polycomb target genes that become significantly derepressed (P-adj < 0.05 and >1.5-fold) following BAP1 removal, showing gene expression (cnRNA-seq) and cChIP-seq for H2AK119ub1, H3K27me3, RING1B (PRC1), and SUZ12 (PRC2) in Bap1fl/fl ESCs (untreated and OHT-treated). (E) Heat maps of cChIP-seq signal for H2AK119ub1, H3K27me3, RING1B (PRC1), and SUZ12 (PRC2) across Polycomb chromatin domains at the promoters of Polycomb-occupied genes in Bap1fl/fl ESCs (untreated and OHT-treated). Intervals were sorted by RING1B occupancy in untreated Bap1fl/fl ESCs. (F) Metaplots of H2AK119ub1 cChIP-seq signal in Bap1fl/fl ESCs (untreated and OHT-treated) at the promoters of Polycomb-occupied genes that become significantly derepressed (Up, n = 421) or do not change in expression (No Change, n = 4075) following BAP1 removal based on cnRNA-seq analysis (P-adj < 0.05 and >1.5-fold). (G) As in F for H3K27me3 cChIP-seq. (H) As in F for RING1B cChIP-seq. (I) As in F for SUZ12 cChIP-seq.
To better understand the interplay between BAP1 and the Polycomb system, we investigated the effect that BAP1 removal has on Polycomb chromatin domains by examining the binding of PRC1 (RING1B), PRC2 (SUZ12), and levels of their respective histone modifications (H2AK119ub1 and H3K27me3) by cChIP-seq. This showed that, in the absence of BAP1, H2AK119ub1 increased across Polycomb chromatin domains at target gene promoters (Fig. 5E), although the magnitude of this effect was slightly smaller than at other regions of the genome, in agreement with ChromHMM analysis (Fig. 1E). Furthermore, H3K27me3 was also modestly elevated (Fig. 5E), consistent with an essential role for H2AK119ub1 in shaping H3K27me3 at Polycomb target gene promoters (Blackledge et al. 2014, 2020; Cooper et al. 2014; Kalb et al. 2014; Illingworth et al. 2015; Tamburri et al. 2020). In contrast, at the subset of Polycomb target genes that become derepressed in the absence of BAP1, the levels of H2AK119ub1 and H3K27me3 at their promoters remained largely unchanged (Fig. 5D,F,G; Supplemental Fig. S7A,D), suggesting that reactivation of these genes following BAP1 removal is not due to reductions in these histone modifications. We then examined PRC1 and PRC2 occupancy at Polycomb target gene promoters and found that it was modestly reduced, despite the observed increases in H2AK119ub1 and H3K27me3 (Fig. 5E), and this was not due to reductions in RING1B and SUZ12 protein levels (Supplemental Fig. S7B). However, strikingly, when we focused on the promoters of Polycomb target genes that were derepressed in the absence of BAP1, we found that they were on average occupied by much higher levels of PRC1 and PRC2 in untreated cells and showed much more dramatic reductions in their occupancy after BAP1 removal (Fig. 5D,H,I; Supplemental Fig. S7A,C,D). Based on these observations, we conclude that this subset of Polycomb target genes are particularly reliant on high-level occupancy of PRC1 and PRC2 for their silencing and that the major decrease in Polycomb complex binding at their promoters caused by removal of BAP1 leads to their derepression. Given that both Polycomb repressive complexes can directly bind to H2AK119ub1 (Arrigoni et al. 2006; Kalb et al. 2014; Cooper et al. 2016; Zhao et al. 2020; Kasinath et al. 2021), we envisage that the reductions in PRC1 and PRC2 occupancy at this subset of genes are likely caused by elevated H2AK119ub1 elsewhere in the genome competing for their binding. Together, these findings provide a potential molecular rationale for the counterintuitive observation that disruption of the BAP1 ortholog in Drosophila (Calypso) gives rise to PcG phenotypes in genetic assays, despite its role in counteracting H2AK119ub1. Furthermore, it reveals that limiting pervasive H2AK119ub1 throughout the genome is important for focusing Polycomb repressive complexes at target gene promoters, while enabling transcription elsewhere in the genome.
Discussion
Chromatin-modifying enzymes can function at defined gene regulatory elements to support cell type-specific gene expression patterns (Atlasi and Stunnenberg 2017; Yadav et al. 2018). Their recruitment to these sites often relies on DNA- and chromatin-binding activities (Smith and Shilatifard 2010), and these mechanisms underpin how PRC1 creates high-level enrichment of H2AK119ub1 at Polycomb target gene promoters to enable repression (Endoh et al. 2012; Blackledge et al. 2015, 2020; Fursova et al. 2019; Scelfo et al. 2019; Cohen et al. 2020; Tamburri et al. 2020). In addition to this punctate pool of H2AK119ub1, we and others have recently discovered that PRC1 also places low levels of H2AK119ub1 broadly throughout the genome (Lee et al. 2015; Kahn et al. 2016; Fursova et al. 2019). However, whether pervasive H2AK119ub1 contributes to gene regulation or other chromosomal processes has remained unclear. Here, we found that BAP1 plays a central role in counteracting pervasive H2AK119ub1, and in its absence, accumulation of H2AK119ub1 throughout the genome leads to widespread reductions in transcription that are likely related to defects in Ser5 phosphorylation on the CTD of Pol II at gene regulatory elements. This reveals an important and previously underappreciated mechanism for chromatin-based gene regulation, whereby a pervasive histone modification can broadly control the function of gene regulatory elements without the need for elaborate site-specific targeting mechanisms.
We envisage that this generic mode of gene regulation could be particularly relevant during cellular differentiation and development when the transcriptional activity of the genome or large genomic regions needs to be coordinately modulated to support acquisition and maintenance of cell type-specific transcriptional states. In fact, support for this concept has recently emerged from studies of X inactivation where H2AK119ub1 was shown to accumulate across the entire silenced X chromosome to drive transcriptional repression and enable dosage compensation (Bousard et al. 2019; Fursova et al. 2019; Nesterova et al. 2019; Żylicz et al. 2019). Furthermore, our discoveries also indicate that there exists an important balance between the enzymes that place and remove pervasive H2AK119ub1, with the levels of this histone modification regulating the capacity of the genome to be transcribed. Given that the composition and expression of PRC1 and BAP1 complexes changes extensively during development (Fisher et al. 2006; Morey et al. 2012, 2015; O'Loghlen et al. 2012; Kloet et al. 2016), in future work, it will be interesting to investigate how the balance between these two opposing activities is regulated at different developmental stages and how cell type-specific H2AK119ub1 levels influence the transcriptional potential of the genome. Given that BAP1 and other PR-DUB subunits are frequently mutated in a variety of cancers with diverse origins (Wiesner et al. 2011; Dey et al. 2012; Carbone et al. 2013; Katoh 2013; Murali et al. 2013; Masoomian et al. 2018; Zhang et al. 2020), our findings also suggest that maintaining the cell type-specific balance between the activities that control H2AK119ub1 levels could play an important role in protecting cells from transformation.
BAP1 has previously been proposed to regulate gene expression through diverse mechanisms, some of which are thought to function independently of H2AK119ub1 (Yu et al. 2010; Dey et al. 2012; Li et al. 2017b; Wang et al. 2018; Campagne et al. 2019; Kuznetsov et al. 2019). We now discover that BAP1 plays a widespread role in supporting gene expression and show that this relies on BAP1 counteracting H2AK119ub1, as catalytic inactivation of PRC1 reverts the effects of BAP1 removal on gene expression, consistent with previous reports of epistasis between BAP1 and PRC1 (Campagne et al. 2019; He et al. 2019). This raises the important question of how pervasive H2AK119ub1 can function at such a broad scale to constrain gene expression. Many histone modifications are thought to regulate gene expression through reader proteins that bind to modified nucleosomes and directly affect transcription (Musselman et al. 2012; Patel and Wang 2013). This is particularly relevant for histone modifications that are of low abundance yet highly enriched at gene regulatory elements. However, we estimate that ∼5.9 × 106 H2AK119ub1 molecules decorate the genome of ES cells (Huseyin and Klose 2021), and this number increases ∼1.5-fold to twofold after conditional removal of BAP1 (Fig. 1), which is comparable with the at least twofold increase in H2AK119ub1 levels reported previously in constitutive BAP1 knockout cells (Campagne et al. 2019; Kolovos et al. 2020). If a reader protein was required for the widespread repressive effects of H2AK119ub1, we envisage that it would also need to be immensely abundant, a requirement that none of the proposed H2AK119ub1-binding proteins fulfill (Arrigoni et al. 2006; Richly et al. 2010; Beck et al. 2011; Schwanhäusser et al. 2011; Kalb et al. 2014; Wiśniewski et al. 2014; Qin et al. 2015; Cooper et al. 2016; Zhang et al. 2017b). Alternatively, addition of a bulky ubiquitin moiety to histone H2A could simply restrict access of the transcriptional machinery to gene regulatory elements. However, in agreement with previous work (Hodges et al. 2018; King et al. 2018), we found that accumulation of H2AK119ub1 does not limit chromatin accessibility, and if anything, the genome becomes slightly more accessible when H2AK119ub1 levels are increased. Based on these observations, we favor the possibility that H2AK119ub1 controls gene expression by counteracting the process of transcription more directly, which is in agreement with previous work showing that PRC1 and H2AK119ub1 can inhibit various aspects of transcription, including initiation, pause release, and elongation (Dellino et al. 2004; Stock et al. 2007; Nakagawa et al. 2008; Zhou et al. 2008; Lehmann et al. 2012; Aihara et al. 2016).
To investigate how H2AK119ub1 might regulate the process of transcription, we examined Pol II and its phosphorylated forms by cChIP-seq. This revealed that elevated H2AK119ub1 preferentially compromised Ser5 phosphorylation on the CTD of Pol II at gene regulatory elements, which is primarily carried out by the CDK7 subunit of the general transcription factor TFIIH (Phatnani and Greenleaf 2006; Buratowski 2009; Harlen and Churchman 2017). It is thought that during transcription initiation, CDK7 catalyzes Ser5 phosphorylation of the Pol II CTD to promote the transition of Pol II into early elongation (Larochelle et al. 2012). Therefore, given the disproportionate effect of elevated H2AK119ub1 on Ser5 phosphorylation, we speculate that pervasive H2AK119ub1 may regulate the function of gene regulatory elements by limiting productive transcription initiation or the transition from initiation to early elongation. Consistent with this, we have recently shown that rapid depletion of H2AK119ub1 leads to increased Polycomb target gene expression, which results from a higher rate of transcription initiation, as suggested by elevated burst frequency (Dobrinić et al. 2020). These findings are also in agreement with observations from in vitro studies where installation of H2AK119ub1 into chromatin templates was sufficient to impede transcription initiation (Nakagawa et al. 2008). In future work, examining how BAP1 removal affects transcriptionally engaged Pol II using high-resolution techniques (Wissink et al. 2019) will help to pinpoint the step of transcription that is disrupted by elevated H2AK119ub1.
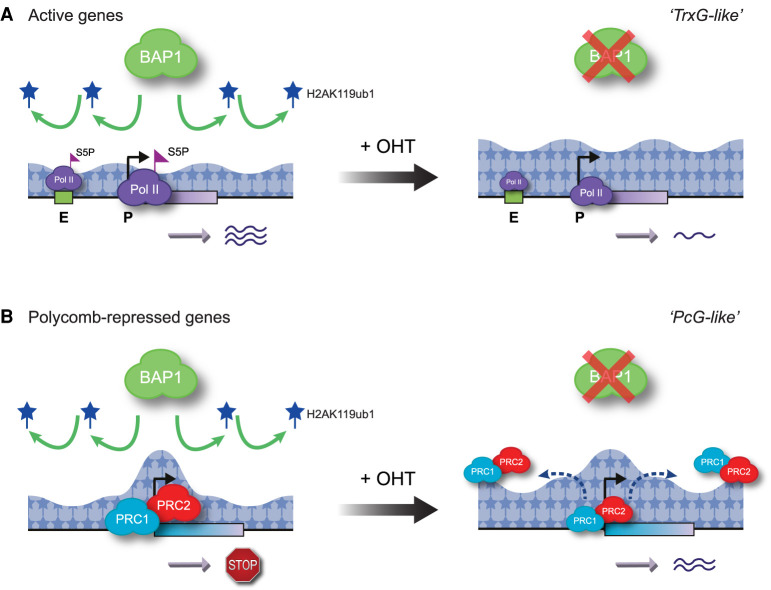
Our molecular understanding of how BAP1 and its H2AK119ub1 deubiquitylase activity contribute to gene regulation has been complicated by the initial characterization of the Drosophila ortholog of BAP1 as a PcG gene in genetic experiments (de Ayala Alonso et al. 2007; Scheuermann et al. 2010). Given that PcG genes are known to maintain Polycomb target gene repression during development, it was puzzling why BAP1, which removes H2AK119ub1, would be required for this process. Initially, it was proposed that BAP1 functioned at Polycomb target gene promoters to appropriately balance H2AK119ub1 levels by enabling its dynamic turnover and that this would somehow facilitate repression of these genes (Scheuermann et al. 2010; Schuettengruber and Cavalli 2010). While alternative and less direct mechanisms have also been previously considered (Schuettengruber and Cavalli 2010; Gutiérrez et al. 2012; Scheuermann et al. 2012), here, we demonstrate that BAP1 indirectly supports repression of a subset of Polycomb target genes by limiting pervasive H2AK119ub1 elsewhere in the genome to promote high-level occupancy of Polycomb complexes at target gene promoters. This mechanism has conceptual similarities to regulation of telomeric gene silencing in budding yeast where the Dot1 histone methyltransferase has been proposed to deposit H3K79me pervasively throughout the genome to counteract promiscuous binding of SIR proteins and focus their repressive activity at telomeric regions (van Leeuwen and Gottschling 2002; van Leeuwen et al. 2002; Verzijlbergen et al. 2009). Interestingly, mutations in the BAP1 and ASXL components of the PR-DUB complex can also lead to phenotypes that are reminiscent of mutations in Trithorax group (TrxG) genes, which are known to oppose PcG gene activity and facilitate gene expression (Sinclair et al. 1992; Milne et al. 1999; Gildea et al. 2000; Baskind et al. 2009; Fisher et al. 2010; Scheuermann et al. 2010). In line with these observations, we demonstrate at the molecular level that, by counteracting pervasive H2AK119ub1, BAP1 directly promotes gene expression, akin to a TrxG gene (Fig. 6A), while also indirectly supporting repression of a subset of Polycomb target genes, akin to a PcG gene (Fig. 6B). Together, these new discoveries provide a potential mechanistic rationale for the dual role that BAP1 has in gene regulation based on genetic assays and reveal that the balance between activities that place and remove pervasive H2AK119ub1 is essential for supporting the expression of some genes, while maintaining the repression of others.
Figure 6.
A model illustrating how BAP1 can regulate gene expression by constraining pervasive H2AK119ub1. (A) BAP1 facilitates gene expression by constraining the pervasive sea of H2AK119ub1 that covers the genome. Inducible removal of BAP1 (+OHT) results in a broad accumulation of H2AK119ub1 throughout the genome. Elevated H2AK119ub1 indiscriminately counteracts Ser5 phosphorylation (S5P) on the CTD of Pol II at gene regulatory elements ([P] promoters, [E] enhancers), and causes widespread reductions in transcription and gene expression. This explains why disruption of BAP1 and other PR-DUB subunits can lead to Trithorax group (TrxG)-like phenotypes. (B) BAP1 also indirectly supports repression of a subset of Polycomb target genes by counteracting pervasive H2AK119ub1 and focusing high levels of Polycomb complexes at target gene promoters. In the absence of BAP1, PRC1/PRC2 occupancy at Polycomb target sites is reduced, presumably due to the increased binding of these complexes to elevated H2AK119ub1 elsewhere in the genome. This leads to derepression of a subset of Polycomb target genes that appear to rely on high-level Polycomb complex occupancy for their silencing, providing a potential molecular rationale for why the BAP1 ortholog in Drosophila has been originally characterized as a Polycomb group (PcG) gene.
Materials and methods
Cell culture conditions and treatments
E14TG2a mouse embryonic stem cells (ESCs) were grown on gelatin-coated plates at 37°C and 5% CO2 in Dulbecco's modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (Labtech), 2 mM L-glutamine (Life Technologies), 1× penicillin/streptomycin (Life Technologies), 1× nonessential amino acids (Life Technologies), 0.5 mM β-mercaptoethanol (Life Technologies), and 10 ng/mL leukemia inhibitory factor (in-house). To induce conditional removal of BAP1, on its own or in combination with PRC1 catalytic activity, Bap1fl/fl and PRC1CPM;Bap1fl/fl cells were treated with 800 nM 4-hydroxytamoxifen (OHT) for 96 h. To induce conditional removal of PRC1 catalytic activity on its own, PRC1CPM cells were treated with 800 nM OHT for 72 h. Cells were regularly tested for the presence of mycoplasma.
Human HEK293T cells used for spike-in calibration of cChIP-seq were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (Labtech), 2 mM L-glutamine (Life Technologies), 1× penicillin/streptomycin (Life Technologies), and 0.5 mM β-mercaptoethanol (Life Technologies). Drosophila S2 (SG4) cells used for spike-in calibration of native cChIP-seq, cnRNA-seq, and cATAC-seq were grown adhesively at 25°C in Schneider's Drosophila medium (Life Technologies), supplemented with 1× penicillin/streptomycin (Life Technologies) and 10% heat-inactivated fetal bovine serum (Labtech).
Cell line generation
Bap1fl/fl cells were derived from E14TG2a ESCs by a two-step process. First, parallel loxP sites flanking exon 4 of the Bap1 gene were inserted using a targeting construct with homology arms of ∼1 kb and three different Cas9 guides specific for the Bap1 locus (sgRNA target sequences: TCAAATGGATCGAAGAGCGC, CAAGGTAGGGACACAATAAA, TAAAACACCACCAACTACAG). Second, CreERT2 was inserted into the Rosa26 locus using a Rosa26-specific Cas9 guide (sgRNA target sequence: CGCCCATCTTCTAGAAAGAC). The same BAP1-specific targeting construct and Cas9 guides were used to generate PRC1CPM;Bap1fl/fl cells from a PRC1CPM parental cell line. Loss of BAP1 in response to OHT treatment in Bap1fl/fl and PRC1CPM;Bap1fl/fl ESCs was confirmed using RT-qPCR and Western blot analysis.
PRC1CPM cells were generated and characterized in a previous study (Blackledge et al. 2020). Briefly, a targeting construct for this cell line comprised exon 3 of Ring1b in forward orientation (flanked by 100 bp of Ring1b intron 2/intron 3), followed by a mutant copy of exon 3 (encoding I53A and D56K mutations) in reverse orientation (flanked by splice donor and acceptor sites from mouse IgE gene). Both the wild-type and mutant versions of exon 3 were codon-optimized at wobble positions to minimize sequence similarity. The wild-type/mutant exon 3 pair was flanked by doubly inverted LoxP/Lox2272 sites and ∼1-kb homology arms. The targeting construct was transfected into E14TG2a ESCs in combination with three different Cas9 guides specific for the Ring1b gene. Correctly targeted homozygous clones were identified by PCR screening, followed by RT-PCR and sequencing to check for splicing defects. Using a similar approach, the I50A/D53K mutation was constitutively knocked-in into both copies of the endogenous Ring1a gene. Finally, CreERT2 was inserted into the Rosa26 locus using a Rosa26-specific Cas9 guide.
Genome engineering by CRISPR/homology-directed repair (HDR)
The pSptCas9(BB)-2A-Puro(PX459)-V2.0 vector was obtained from Addgene (62988), and sgRNAs were designed using the CRISPOR online tool (http://crispor.tefor.net/crispor.py). Targeting constructs with appropriate homology arms were generated by Gibson assembly using the Gibson Assembly Master Mix kit (NEB). Targeting constructs were designed such that Cas9 recognition sites were disrupted by the presence of the LoxP sites. ESCs (one well of a six-well plate) were transfected with 0.5 μg of each Cas9 guide and 2 μg of targeting construct using Lipofectamine 3000 (ThermoFisher) according to the manufacturer's guidelines. The day after transfection, cells were passaged at a range of densities and subjected to selection with 1 μg/mL puromycin for 48 h to eliminate nontransfected cells. Approximately 1 wk later, individual clones were isolated, expanded and PCR-screened for the desired genomic modifications.
Preparation of nuclear and histone extracts for immunoblotting analysis
For nuclear extraction, ESCs were washed with 1× PBS and resuspended in 10 volumes of buffer A (10 mM HEPES at pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF, 1× protease inhibitor cocktail [PIC] [Roche]). After 10 min incubation on ice, cells were recovered by centrifugation at 1500g for 5 min and resuspended in 3 vol of buffer A supplemented with 0.1% NP-40. The released nuclei were pelleted by centrifugation at 1500g for 5 min, followed by resuspension in 1 vol of buffer C (5 mM HEPES at pH 7.9, 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1× PIC [Roche], and 0.5 mM DTT) supplemented with 400 mM NaCl. The extraction was allowed to proceed on ice for 1 h with occasional agitation, then the nuclei were pelleted by centrifugation at 16,000g for 20 min at 4°C. The supernatant was taken as the nuclear extract. The Bradford protein assay (Bio-Rad) was used to compare protein concentrations across samples.
For histone extraction, ESCs were washed in RSB (10 mM Tris HCl at pH 8, 10 mM NaCl, 3 mM MgCl2) supplemented with 20 mM N-ethylmaleimide (NEM), incubated for 10 min on ice in RSB with 0.5% NP-40 and 20 mM NEM, pelleted by centrifugation at 800g for 5 min, and incubated in 2.5 mM MgCl2, 0.4 M HCl, and 20 mM NEM for 30 min on ice. After that, cells were pelleted by centrifugation at 16,000g for 20 min at 4°C, the supernatant recovered and precipitated on ice with 25% TCA for 30 min, followed by centrifugation at 16,000g for 15 min at 4°C to recover histones. Following two acetone washes, the histones were resuspended in 1× SDS loading buffer (2% SDS, 100 mM Tris at pH 6.8, 100 mM DTT, 10% glycerol, 0.1% bromophenol blue) and boiled for 5 min at 95°C. Finally, any insoluble precipitate was pelleted by centrifugation at 16,000g for 10 min and the soluble fraction retained as the histone extract. Histone concentrations across samples were compared using SDS-PAGE followed by Coomassie blue staining. Western blot analysis of nuclear and histone extracts was performed using LI-COR IRDye secondary antibodies, and imaging was done using the LI-COR Odyssey Fc system. The list of antibodies used in this study for Western blot and cChIP-seq analysis is in Supplemental Table S1.
Calibrated ChIP sequencing (cChIP-seq)
For RING1B and SUZ12, cChIP-seq was performed as described previously (Fursova et al. 2019; Blackledge et al. 2020). Briefly, 5 × 107 mouse ESCs (untreated and OHT-treated) were cross-linked in 10 mL of 1× PBS with 2 mM DSG (Thermo Scientific) for 45 min at 25°C, and then with 1% formaldehyde (methanol-free; Thermo Scientific) for a further 15 min. Cross-linking was stopped by quenching with 125 mM glycine. Cross-linked ESCs were mixed with 2 × 106 human HEK293T cells, which have been similarly double-cross-linked, and incubated in lysis buffer (50 mM HEPES at pH 7.9, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100, 1× PIC [Roche]) for 10 min at 4°C. Released nuclei were washed (10 mM Tris-HCl at pH 8, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× PIC [Roche]) for 5 min at 4°C. Chromatin was then resuspended in 1 mL of sonication buffer (10 mM Tris-HCl at pH 8, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na deoxycholate, 0.5% N-lauroylsarcosine, 1× PIC [Roche]) and sonicated for 30 min using the BioRuptor Pico (Diagenode), shearing genomic DNA to an average size of 0.5 kb. Following sonication, Triton X-100 was added to a final concentration of 1%, followed by centrifugation at 20,000g for 10 min at 4°C to collect the clear supernatant fraction.
For Pol II and its phosphorylated forms, cChIP-seq was done as described previously (Turberfield et al. 2019). Briefly, 5 × 107 ESCs (untreated and OHT-treated) were cross-linked in 10 mL of 1× PBS with 1% formaldehyde (methanol-free; Thermo Scientific) for 10 min at 25°C and then quenched by addition of 125 mM glycine. Cross-linked ESCs were mixed with 2 × 106 human HEK293T cells, which have been similarly single-cross-linked, and then incubated in FA-lysis buffer (50 mM HEPES at pH 7.9, 150 mM NaCl, 2 mM EDTA, 0.5 mM EGTA, 0.5% NP40, 0.1% sodium deoxycholate, 0.1% SDS, 10 mM NaF, 1 mM AEBSF, 1× PIC) for 10 min at 4°C. Chromatin was sonicated for 30 min using the BioRuptor Pico (Diagenode), followed by centrifugation at 20,000g for 10 min at 4°C to collect the clear supernatant fraction.
For RING1B and SUZ12 ChIP, sonicated chromatin was diluted 10-fold with ChIP dilution buffer (1% Triton X-100, 1 mM EDTA, 20 mM Tris-HCl at pH 8, 150 mM NaCl, 1× PIC). For Pol II ChIP, 300 ug of chromatin per one IP was diluted in FA-lysis buffer up to a final volume of 1 mL. Diluted chromatin was precleared for 1 h using Protein A agarose beads (Repligen) that were preblocked with 1 mg/mL BSA and 1 mg/mL yeast tRNA. For each ChIP reaction, 1 mL of diluted and precleared chromatin was incubated overnight with the appropriate antibody, anti-RING1B (3 μL; CST D22F2), anti-SUZ12 (3 μL; CST D39F6), anti-Rpb1-NTD (15 μL; CST D8L4Y) as a measure of total Pol II occupancy, anti-Rpb1-CTD-Ser5P (12.5 μL; CST D9N5I), and anti-Rpb1-CTD-Ser2P (12.5 μL; CST E1Z3G) as a measure of Pol II phosphorylation levels. To capture antibody-bound chromatin, ChIP reactions were incubated with preblocked protein A agarose beads (Repligen) for 2 h (RING1B and SUZ12) or 3 h (Pol II) at 4°C. For RING1B and SUZ12, ChIP washes were performed as described previously (Farcas et al. 2012). For Pol II, washes were performed with FA-lysis buffer, FA-lysis buffer containing 500 mM NaCl, DOC buffer (250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 2 mM EDTA, 10 mM Tris-HCl at pH 8), followed by two washes with TE buffer (pH 8). ChIP DNA was eluted in elution buffer (1% SDS, 0.1 M NaHCO3) and cross-linking was reversed overnight at 65oC with 200 mM NaCl and 2 μL of RNase A (Sigma). Matched input samples (10% of original ChIP reaction) were treated identically. The following day, ChIP samples and inputs were incubated with Proteinase K (Sigma) for at least 1.5 h at 56oC and purified using a ChIP DNA Clean and Concentrator kit (Zymo Research).
cChIP-seq libraries for both ChIP and Input samples were prepared using a NEBNext Ultra II DNA library preparation kit for Illumina, following the manufacturer's guidelines. Samples were indexed using NEBNext Multiplex oligos. The average size and concentration of all libraries were analysed using the 2100 Bioanalyzer high-sensitivity DNA kit (Agilent) followed by qPCR quantification using SensiMix SYBR (Bioline) and KAPA Illumina DNA standards (Roche). Libraries were sequenced as 40-bp paired-end reads in biological triplicate or quadruplicate on the Illumina NextSeq 500 platform.
Native cChIP sequencing
Native cChIP-seq for H2AK119ub1, H3K27me3, H3K27ac, H3K4me3, and H3K4me1 was performed as described previously (Fursova et al. 2019). Briefly, 5 × 107 mouse ESCs (untreated and OHT-treated) were mixed with 2 × 107 Drosophila SG4 cells in 1× PBS. Mixed cells were pelleted and nuclei were released by resuspending in ice-cold lysis buffer (10 mM Tris-HCl at pH 8, 10 mM NaCl, 3 mM MgCl2, 0.1% NP40, 5 mM sodium butyrate, 5 mM N-ethylmaleimide). Nuclei were then washed and resuspended in 1 mL of MNase digestion buffer (10 mM Tris-HCl at pH 8.0, 10 mM NaCl, 3 mM MgCl2, 0.1% NP40, 0.25 M sucrose, 3 mM CaCl2, 10 mM sodium butyrate, 10 mM N-ethylmaleimide, 1× PIC [Roche]). Each sample was incubated with 200 U of MNase (Fermentas) for 5 min at 37°C, followed by the addition of 4 mM EDTA to halt MNase digestion. Following centrifugation at 1500g for 5 min at 4°C, the supernatant (S1) was retained. The remaining pellet was incubated with 300 µL of nucleosome release buffer (10 mM Tris-HCl at pH 7.5, 10 mM NaCl, 0.2 mM EDTA, 10 mM sodium butyrate, 10 mM N-ethylmaleimide, 1× PIC [Roche]) for 1 h at 4°C, passed five times through a 27G needle using a 1-mL syringe, and spun at 1500g for 5 min at 4°C. The second supernatant (S2) was collected and combined with the corresponding S1 sample from above. Digestion to mostly mononucleosomes was confirmed by agarose gel electrophoresis of purified S1/S2 DNA.
For ChIP, S1/S2 nucleosomes were diluted 10-fold in native ChIP incubation buffer (70 mM NaCl, 10 mM Tris at pH 7.5, 2 mM MgCl2, 2 mM EDTA, 0.1% Triton X-100, 10 mM sodium butyrate [for H3K27ac and H3K4me3 ChIPs], 10 mM N-ethylmaleimide, 1× PIC [Roche]). For each ChIP reaction, 1 mL of diluted nucleosomes was incubated overnight at 4oC with the appropriate antibody, anti-H2AK119ub1 (5 μL; CST D27C4), anti-H3K27me3 (5 μL; in-house), anti-H3K27ac (3 μL CST D5E4), anti-H3K4me3 (4 μL; in-house), or anti-H3K4me1 (5 μL; CST D1A9). Antibody-bound nucleosomes were captured for 1 h at 4°C using Protein A agarose (Repligen) beads, preblocked in native ChIP incubation buffer supplemented with 1 mg/mL BSA and 1 mg/mL yeast tRNA, and collected by centrifugation. Immunoprecipitated material was washed four times with native ChIP wash buffer (20 mM Tris at pH 7.5, 2 mM EDTA, 125 mM NaCl, 0.1% Triton X-100) and once with TE buffer (pH 8). ChIP DNA was eluted using 100 μL of elution buffer (1% SDS, 0.1 M NaHCO3) for 30 min at room temperature, and then purified using a ChIP DNA Clean and Concentrator kit (Zymo Research). For each ChIP sample, DNA from a matched input control (corresponding to 10% of original ChIP reaction) was purified in the same way. Native cChIP-seq library preparation and sequencing was performed as described above for cChIP-seq.
Calibrated nuclear RNA sequencing (cnRNA-seq)
For cnRNA-seq, 1 × 107 ESCs (untreated and OHT-treated) were mixed with 4 × 106 Drosophila SG4 cells in 1× PBS. Nuclei were isolated in 1 mL HS lysis buffer (50 mM KCl, 10 mM MgSO4.7H20, 5 mM HEPES, 0.05% NP40 [IGEPAL CA630], 1 mM PMSF, 3 mM DTT, 1× PIC [Roche]) for 1 min at room temperature, and then recovered by centrifugation at 1000g for 5 min at 4°C, followed by a total of three washes with ice-cold RSB buffer (10 mM NaCl, 10 mM Tris at pH 8, 3 mM MgCl2). Nuclei integrity was assessed using 0.4% Trypan blue staining (Thermo Scientific). Pelleted nuclei were resuspended in 1 mL of TRIzol reagent (Thermo Scientific), and RNA was extracted according to the manufacturer's protocol, followed by treatment with the TURBO DNA-free kit (Thermo Scientific) to remove any contaminating DNA. Quality of RNA was assessed using the 2100 Bioanalyzer RNA 6000 Pico kit (Agilent). RNA samples were depleted of rRNA with the NEBNext rRNA depletion kit (NEB) prior to preparing cnRNA-seq libraries using the NEBNext Ultra (for Bap1fl/fl and PRC1CPM ESCs) or Ultra II (for PRC1CPM;Bap1fl/fl ESCs) Directional RNA library preparation kit (NEB). To quantitate the consistency of spike-in cell mixing for each individual sample, a small aliquot of nuclei was saved to isolate genomic DNA using phenol-chloroform extraction. This was followed by sonication of DNA for 15 min using the BioRuptor Pico (Diagenode), shearing genomic DNA to an average size of <1 kb. Libraries from sonicated genomic DNA were constructed as described above for cChIP-seq. Both cnRNA-seq and gDNA-seq libraries were sequenced as 80-bp paired-end reads on the Illumina NextSeq 500 platform in biological triplicate.
Calibrated ATAC-seq (cATAC-seq)
To assay chromatin accessibility, calibrated ATAC-seq was performed as described previously (Turberfield et al. 2019). First, 1 × 107 ESCs (untreated and OHT-treated) were mixed with 4 × 106 Drosophila SG4 cells in 1× PBS and then lysed in 1 mL of HS lysis buffer (50 mM KCl, 10 mM MgSO4.7H20, 5 mM HEPES, 0.05% NP40 [IGEPAL CA630], 1 mM PMSF, 3 mM DTT, 1× PIC [Roche]) for 1 min at room temperature. Nuclei were recovered by centrifugation at 1000g for 5 min at 4°C and washed three times in ice-cold RSB buffer (10 mM NaCl, 10 mM Tris at pH 7.4, 3 mM MgCl2). The concentration and integrity of nuclei were assessed using 0.4% Trypan blue staining (Thermo Scientific). Next, 5 × 105 nuclei were resuspended in Tn5 reaction buffer (10 mM TAPS, 5 mM MgCl2, 10% dimethylformamide) and incubated with Tn5 transposase (25 µM, generated in-house as previously described) (King and Klose 2017) for 30 min at 37°C. Tagmented DNA was purified using MinElute columns (Qiagen) and eluted in 10 μL of elution buffer. To control for the Tn5 transposase sequence bias and to determine the exact spike-in ratio for each individual sample, 50 ng of genomic DNA, isolated from the same nuclei preparation by phenol-chloroform extraction, was tagmented with Tn5 transposase (25 µM) for 30 min at 55°C and purified using MinElute columns (Qiagen).
Libraries for cATAC-seq and gDNA-seq were prepared by PCR amplification using the NEBNext high-fidelity 2× PCR Master Mix and custom-made Illumina barcodes (Buenrostro et al. 2015). Libraries were purified by two rounds of Agencourt AMPure XP bead cleanup (1.5× bead:sample ratio). Library concentration and fragment size distribution were determined as described above for cChIP-seq. Libraries were sequenced using the Illumina NextSeq 500 platform in biological quadruplicate using 80-bp paired-end reads.
Data processing and normalization for massive parallel sequencing
Following massive parallel sequencing, reads were mapped and processed as described previously (Fursova et al. 2019; Turberfield et al. 2019). Briefly, for cChIP-seq, cATAC-seq, and gDNA-seq, Bowtie 2 (Langmead and Salzberg 2012) was used to align paired-end reads to the concatenated mouse and spike-in genome sequences (mm10 + dm6 for native cChIP-seq, cATAC-seq, and gDNA-seq; mm10 + hg19 for cross-linked cChIP-seq) with the “‐‐no-mixed” and “‐‐no-discordant” options. Nonuniquely mapping reads were discarded, and PCR duplicates were removed with Sambamba (Tarasov et al. 2015). For cATAC-seq, reads mapping to the mitochondrial chromosome and other genomic regions with artificially high counts or low mappability, derived from the ENCODE blacklist (Amemiya et al. 2019), were also discarded.
For cnRNA-seq, first, Bowtie 2 was used with “‐‐very-fast,” “‐‐no-mixed,” and “‐‐no-discordant” options to identify and discard reads mapping to the concatenated mm10 + dm6 rDNA genomic sequences (GenBank BK000964.3 and M21017.1). Next, all unmapped reads were aligned against the concatenated mm10 + dm6 genome using STAR (Dobin et al. 2013). To improve mapping of intronic sequences, reads that failed to be mapped by STAR were further aligned with Bowtie 2 (with “‐‐sensitive-local,” “‐‐no-mixed,” and “‐‐no-discordant” options). Uniquely aligned reads from the last two steps were combined, and PCR duplicates were removed using Sambamba (Tarasov et al. 2015). A list of all genomics data sets produced in this study and the number of uniquely aligned reads in each experiment are in Supplemental Table S2.
To visualize the cChIP-seq, cATAC-seq, and cnRNA-seq and quantitatively compare genome enrichment profiles, chromatin accessibility, and gene expression between conditions, the data were internally calibrated using dm6 or hg19 spike-in as described previously (Fursova et al. 2019; Turberfield et al. 2019). Briefly, uniquely aligned mm10 reads were randomly subsampled based on the total number of spike-in (dm6 or hg19) reads in each sample. To account for variations in the spike-in cell mixing, we used the ratio of spike-in/mouse total read counts in the corresponding Input/gDNA-seq samples to correct the subsampling factors. After normalization, read coverages across genomic regions of interest (RING1B peaks for H2AK119ub1, H3K27me3, RING1B and SUZ12 cChIP-seq, TSS ± 2.5 kb for cATAC-seq and H3K27ac, H3K4me3 and H3K4me1 cChIP-seq, or gene bodies for total Pol II, Ser5P-Pol II and Ser2P-Pol II cChIP-seq, and cnRNA-seq) were compared for individual biological replicates using multiBamSummary and plotCorrelation from deepTools (Ramírez et al. 2014). For each experimental condition, biological replicates showed a good correlation (Pearson correlation coefficient > 0.9) (see Supplemental Table S3) and were merged for downstream analysis. Genome coverage tracks were generated using the pileup function from MACS2 (Zhang et al. 2008) for cChIP-seq, bamCoverage from deeptools (Ramírez et al. 2014) for cATAC-seq, and genomeCoverageBed from BEDTools (Quinlan and Hall 2010) for cnRNA-seq, and visualized using the UCSC genome browser (Kent et al. 2002). BigwigCompare from deepTools was used to make differential genome coverage tracks (log2 ratio of two conditions or ratio of Pol II phosphorylated forms to its total levels).
Genome segmentation by ChromHMM
ChromHMM (Ernst and Kellis 2012) was used to perform unsupervised segmentation of the genome into distinct chromatin states, enriched with different combinations of histone modifications and other chromatin features, as described previously, but without extension of the reads (Ernst and Kellis 2017). Briefly, to build the model, cATAC-seq and cChIP-seq for H2AK119ub1, H3K27me3, H3K27ac, H3K4me3, and H3K4me1 from this study (GEO: GSE161996), together with published ChIP-seq data sets for CTCF (GEO: GSE153400) (Huang et al. 2020), OCT4, NANOG, SOX2 (GEO: GSE87822) (King and Klose 2017), H3K36me3 and H3K9me3 (GEO: GSE120376) (Ramisch et al. 2019) in wild-type ESCs were aligned as described above, subsampled to the same number of reads, and binarized using the binarizeBam function in a paired-end mode. These data were used to learn ChromHMM models with 10–15 states using the LearnModel function with default parameters. Finally, a 13-state ChromHMM model was selected for the downstream analysis, as this was the minimal number of states required to accurately segregate an active enhancer state based on the high frequency of OCT4/NANOG/SOX2 binding. This resulted in 13 different chromatin states, which were interpreted as described in Supplemental Figure S2B based on the underlying functional genomic annotations and their transcriptional activity. These included CTCF-bound insulators, distal gene regulatory elements (weak to highly active enhancers), actively transcribed promoters and gene body regions, Polycomb-repressed and bivalent chromatin domains, H3K9me3-enriched heterochromatin, and intergenic regions that are not enriched with any examined chromatin features. The genomic coordinates of chromatin states identified by ChromHMM were deposited in GEO: GSE161993.
Peak calling and annotation of genomic regions
To define active gene regulatory elements, we first performed peak calling for H3K27ac cChIPseq in Bap1fl/fl ESCs (untreated and OHT-treated) for four biological replicates using the dpeak function (-kd 500, -kw 300, -p 1e-30) from DANPOS2 (Chen et al. 2013), discarding peaks that overlapped with a custom set of blacklisted genomic regions. We then intersected ATAC peaks defined previously (King et al. 2018) with our H3K27ac peak set. This resulted in a set of active gene regulatory elements, which were further segregated into active enhancers (non-TSS, n = 12,006) or active promoters (TSS, n = 10,840), based on their overlap with TSS ± 1-kb regions. To obtain the most complete set of TSS positions, we combined TSS annotations from the UCSC refGene (n = 34,852), NCBI RefSeq (n = 106,520), and GENCODE VM24 (n = 67,573) databases. To eliminate the contribution of gene body signal for Pol II cChIP-seq, only intergenic active enhancers (n = 4156), which did not overlap with any of the above gene annotations, were considered for the downstream analysis. To associate genes with putative distal regulatory elements, for each gene promoter we identified the nearest intergenic enhancer located within the 250-kb distance using the closest function from BEDTools. For differential gene expression analysis and quantification of cChIP-seq and cATAC-seq signal at promoters or over the bodies of genes, we used a custom nonredundant set of genes (n = 20,633), derived from mm10 UCSC refGene genes by removing very short genes with poor sequence mappability and highly similar transcripts as described previously (Rose et al. 2016). For the purposes of read quantification, promoters were defined as TSS ± 2.5-kb intervals, with the exception of Pol II cChIP-seq, in which case they were defined as TSS ± 0.5 kb to specifically capture the promoter signal and eliminate the contribution of the gene body signal. A set of intergenic intervals was obtained using the complement function from BEDTools as regions of the genome that do not overlap with any of the genes from a complete UCSC refGene set.
Mouse genes in a custom nonredundant set (n = 20,633) (Rose et al. 2016) were classified into three groups based on the overlap of their gene promoters with nonmethylated CpG islands (NMI), as well as RING1B- and SUZ12-bound sites. NMIs (n = 27,047) were identified using MACS2 peak calling with the matching input control from BioCAP-seq (Long et al. 2013). All genes with promoters (TSS ± 1 kb) not overlapping with NMIs were referred to as non-NMI genes (n = 6333). NMI-overlapping genes were further subdivided into PcG-occupied genes (n = 5582), if their promoters overlapped with both RING1B- and SUZ12-bound sites defined in a previous study (Blackledge et al. 2020), and non-PcG-occupied genes (n = 8718), if they did not. The overlap between NMIs, RING1B- and SUZ12-bound regions with gene promoters (TSS ± 1 kb) was determined using the closest function from BEDTools. In the text, we refer to PcG-occupied genes as Polycomb target genes.
Read count quantitation and analysis
For cChIP-seq and cATAC-seq, computeMatrix and plotProfile/plotHeatmap from deepTools were used to perform metaplot and heat map analysis of read density at regions of interest. Metaplot profiles represented the mean read density over a set of genomic regions, except for Pol II cChIP-seq, for which the median read density was plotted due to an extremely broad range of signal intensities across the intervals of interest. For chromosome-wide density plots, read coverage in 250-kb bins was calculated using a custom R script using GenomicRanges, GenomicAlignments, and Rsamtools Bioconductor packages (Huber et al. 2015) and visualized using ggplot2. For cChIP-seq and cATAC-seq, target regions of interest were annotated with read counts from merged spike-in normalized replicates using multiBamSummary from deepTools (“‐‐outRawCounts”). For differential gene expression analysis, we used a custom Perl script using SAMtools (Li et al. 2009) to obtain read counts from individual biological replicates prior to spike-in normalization for a custom nonredundant mm10 gene set (n = 20,633).
Normalized read counts and log2 fold changes for different genomic intervals were visualized using custom R scripts and ggplot2. For box plot analysis of cChIP-seq and cATAC-seq signal, read counts were normalized to the genomic region size (in kb) and log2 transformed following addition of a pseudocount of 1. For box plots, boxes show the interquartile range (IQR) and whiskers extend by 1.5 × IQR. ggcor function from the GGally R package was used to generate a correlation matrix for the association between log2 fold changes in gene expression (cnRNA-seq) and log2 fold changes in cChIP-seq signal for Pol II and transcription-associated histone modifications at gene promoters or bodies following OHT treatment in Bap1fl/fl ESCs. All correlation analyses used the Pearson correlation coefficient to measure the strength of the association between the variables and were visualized using scatterplots colored by density with stat_density2d. Linear regression was plotted using the stat_poly_eq function from the ggpmisc R package, together with the model's R2adj coefficient of determination. To calculate statistical significance, we carried out Student's t-test using R, with samples considered to be independent and a two-tailed alternative hypothesis, unless specified otherwise.
Differential gene expression analysis
To identify significant gene expression changes in cnRNA-seq, we used a custom R script that incorporates spike-in calibration into DESeq2 analysis (Love et al. 2014) as described previously (Fursova et al. 2019; Blackledge et al. 2020). Briefly, dm6 read counts were obtained for unique dm6 refGene genes to calculate DESeq2 size factors for normalization of raw mm10 read counts for a custom nonredundant mm10 gene set. Prior to quantification, dm6 reads were prenormalized using the dm6/mm10 total read ratio in the corresponding gDNA-seq samples in order to account for variations in spike-in cell mixing. For visualization and ranking of the effect sizes, we performed shrinking of log2 fold changes using the original DESeq2 shrinkage estimator with an adaptive normal distribution as prior (Love et al. 2014). For visualization of DESeq2-normalized read counts, they were averaged across the replicates and used to calculate RPKM following addition of a pseudocount of 1. For a change to be called significant, we applied a threshold of P-adj < 0.05 and fold change > 1.5. To distinguish genes that are robustly expressed in ESCs from genes that show no or low expression, we used a threshold of RPKM = 2.838, which was defined based on the density distribution of log2 transformed RPKM gene expression levels from cnRNA-seq in untreated Bap1fl/fl ESCs (Supplemental Fig. S3A). This resulted in a set of 14,779 robustly expressed genes, which was used to estimate the proportion of expressed genes that showed reductions in expression following BAP1 removal. Log2 fold change values were visualized using R and ggplot2 with MA plots, heat maps, and box plots/violin plots. ComplexHeatmap R package (Gu et al. 2016) was used to plot heat maps of log2 fold gene expression changes. For MA plots, the density of the data points across the y-axis was shown to reflect the general direction of gene expression changes. Venn diagrams were plotted using Vennerable R package. The significance of pairwise overlaps for Venn diagrams was calculated by a Fisher's exact test using GeneOverlap R package. HOMER v4.9.1 (Heinz et al. 2010) was used to perform gene ontology (GO) term enrichment analysis for differentially expressed genes, with the custom nonredundant mm10 gene set used as background.
Data and software availability
The high-throughput sequencing data reported in this study have been deposited in GEO under the accession number GSE161996. Published data used in this study include BioCAP-seq, GEO: GSE43512 (Long et al. 2013); cnRNA-seq in PRC1CKO ESCs, GEO: GSE119619 (Fursova et al. 2019); RING1B- and SUZ12-bound regions in ESCs, GEO: GSE132752 (Blackledge et al. 2020); ATAC peaks from E14 ESCs, GEO: GSE98403 (King et al. 2018); CTCF ChIP-seq in wild-type ESCs, GEO: GSE153400 (Huang et al. 2020); ChIP-seq for OCT4, NANOG, and SOX2 in untreated Brg1fl/fl ESCs, GEO: GSE87822 (King and Klose 2017); and H3K9me3 and H3K36me3 ChIP-seq in LIF-grown ESCs, GEO: GSE120376 (Ramisch et al. 2019). Custom R and Perl scripts used for data analysis in this study have been deposited to GitHub and are available at https://github.com/nFursova/Calibrated_ChIPseq_RNAseq.
Supplementary Material
Acknowledgments
We thank Amanda Williams at the Department of Zoology, University of Oxford, for sequencing support on the NextSeq 500. We thank Dr Emilia Dimitrova for critical reading of the manuscript. Work in the Klose laboratory is supported by the Wellcome Trust (209400/Z/17/Z), the European Research Council (681440), and the Lister Institute of Preventive Medicine.
Author contributions: N.A.F., N.P.B., and R.J.K. conceived the study. N.A.F., A.H.T., and N.P.B. developed the methodology. N.A.F., A.H.T., E.L.F., N.P.B., and P.D. performed the investigation. N.A.F. performed formal analysis and curated the software and data. N.A.F., A.L., N.P.B., and M.K.H. obtained the resources. N.A.F. and R.J.K. wrote the original draft of the manuscript. N.A.F. and R.J.K. reviewed and edited the manuscript with input from all of the other authors. R.J.K. acquired the funding. R.J.K. supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.347005.120.
Freely available online through the Genes & Development Open Access Option.
Competing interest statement
The authors declare no competing interests.
References
- Aihara H, Nakagawa T, Mizusaki H, Yoneda M, Kato M, Doiguchi M, Imamura Y, Higashi M, Ikura T, Hayashi T, et al. 2016. Histone H2A T120 phosphorylation promotes oncogenic transformation via upregulation of cyclin D1. Mol Cell 64: 176–188. 10.1016/j.molcel.2016.09.012 [DOI] [PubMed] [Google Scholar]
- Amemiya HM, Kundaje A, Boyle AP. 2019. The ENCODE blacklist: identification of problematic regions of the genome. Sci Rep 9: 9354. 10.1038/s41598-019-45839-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Sandelin A. 2020. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet 21: 71–87. 10.1038/s41576-019-0173-8 [DOI] [PubMed] [Google Scholar]
- Aquila L, Atanassov BS. 2020. Regulation of histone ubiquitination in response to DNA double strand breaks. Cells 9: 1699. 10.3390/cells9071699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N. 2006. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett 580: 6233–6241. 10.1016/j.febslet.2006.10.027 [DOI] [PubMed] [Google Scholar]
- Atlasi Y, Stunnenberg HG. 2017. The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet 18: 643–658. 10.1038/nrg.2017.57 [DOI] [PubMed] [Google Scholar]
- Audia JE, Campbell RM. 2016. Histone modifications and cancer. Cold Spring Harb Perspect Biol 8: a019521. 10.1101/cshperspect.a019521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Baskind HA, Na L, Ma Q, Patel MP, Geenen DL, Wang QT. 2009. Functional conservation of Asxl2, a murine homolog for the Drosophila enhancer of trithorax and polycomb group gene Asx. PLoS One 4: e4750. 10.1371/journal.pone.0004750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Trupke J, Ringrose L. 2016. The quest for mammalian Polycomb response elements: are we there yet? Chromosoma 125: 471–496. 10.1007/s00412-015-0539-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. 2011. The quantitative proteome of a human cell line. Mol Syst Biol 7: 549. 10.1038/msb.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JS, Nicetto D, Zaret KS. 2016. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet TIG 32: 29–41. 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle JI, Nijnik A. 2014. H2A-DUBbing the mammalian epigenome: expanding frontiers for histone H2A deubiquitinating enzymes in cell biology and physiology. Int J Biochem Cell Biol 50: 161–174. 10.1016/j.biocel.2014.03.004 [DOI] [PubMed] [Google Scholar]
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. 2016. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res 26: 896–907. 10.1101/gr.197632.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LLP, Ito S, Cooper S, Kondo K, Koseki Y, et al. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157: 1445–1459. 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, Klose RJ. 2015. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 16: 643–649. 10.1038/nrm4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A, Klose RJ. 2020. PRC1 catalytic activity is central to Polycomb system function. Mol Cell 77: 857–874.e9. 10.1016/j.molcel.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousard A, Raposo AC, Żylicz JJ, Picard C, Pires VB, Qi Y, Gil C, Syx L, Chang HY, Heard E, et al. 2019. The role of Xist -mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep 20: e48019. 10.15252/embr.201948019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Bracken AP. 2006. Genome-wide mapping of polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136. 10.1101/gad.381706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Brien GL, Verrijzer CP. 2019. Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev 33: 936–959. 10.1101/gad.326066.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. 2006. Structure and E3-ligase activity of the Ring–Ring complex of Polycomb proteins Bmi1 and Ring1b. EMBO J 25: 2465–2474. 10.1038/sj.emboj.7601144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J, Wu B, Chang H, Greenleaf W. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546. 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837. 10.1016/j.molcel.2013.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne A, Lee M-K, Zielinski D, Michaud A, Le Corre S, Dingli F, Chen H, Shahidian LZ, Vassilev I, Servant N, et al. 2019. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat Commun 10: 348. 10.1038/s41467-018-08255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. 2013. BAP1 and cancer. Nat Rev Cancer 13: 153–159. 10.1038/nrc3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli FN, Sharma G, Ahringer J. 2017. Broad chromatin domains: an important facet of genome regulation. Bioessays 39: 1700124. 10.1002/bies.201700124 [DOI] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W. 2013. DANPOS: dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res 23: 341–351. 10.1101/gr.142067.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I, Bar C, Ezhkova E. 2020. Activity of PRC1 and histone H2AK119 monoubiquitination: revising popular misconceptions. Bioessays 42: 1900192. 10.1002/bies.201900192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. 2014. Targeting Polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7: 1456–1470. 10.1016/j.celrep.2014.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Grijzenhout A, Underwood E, Ancelin K, Zhang T, Nesterova TB, Anil-Kirmizitas B, Bassett A, Kooistra SM, Agger K, et al. 2016. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun 7: 13661. 10.1038/ncomms13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196. 10.1016/S0092-8674(02)00975-3 [DOI] [PubMed] [Google Scholar]
- Danzer JR, Wallrath LL. 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131: 3571–3580. 10.1242/dev.01223 [DOI] [PubMed] [Google Scholar]
- Daou S, Hammond-Martel I, Mashtalir N, Barbour H, Gagnon J, Iannantuono NVG, Nkwe NS, Motorina A, Pak H, Yu H, et al. 2015. The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. J Biol Chem 290: 28643–28663. 10.1074/jbc.M115.661553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ayala Alonso AG, Gutiérrez L, Fritsch C, Papp B, Beuchle D, Müller J. 2007. A genetic screen identifies novel polycomb group genes in Drosophila. Genetics 176: 2099–2108. 10.1534/genetics.107.075739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SCR, Pirrotta V. 2004. Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893. 10.1016/S1097-2765(04)00128-5 [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676. 10.1016/j.devcel.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, et al. 2012. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337: 1541–1546. 10.1126/science.1221711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrinić P, Szczurek AT, Klose RJ. 2020. PRC1 drives Polycomb-mediated gene repression by controlling transcription initiation and burst frequency. bioRxiv 10.1101/2020.10.09.333294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, et al. 2012. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8: e1002774. 10.1371/journal.pgen.1002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. 10.1038/nbt.1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. 10.1038/nmeth.1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2017. Chromatin-state discovery and genome annotation with ChromHMM. Nat Protoc 12: 2478–2492. 10.1038/nprot.2017.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. 2012. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 1: e00205. 10.7554/eLife.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stützer A, Fischle W, Bonaldi T, Pasini D. 2014. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53: 49–62. 10.1016/j.molcel.2013.10.030 [DOI] [PubMed] [Google Scholar]
- Fisher CL, Randazzo F, Humphries RK, Brock HW. 2006. Characterization of Asxl1, a murine homolog of Additional sex combs, and analysis of the Asx-like gene family. Gene 369: 109–118. 10.1016/j.gene.2005.10.033 [DOI] [PubMed] [Google Scholar]
- Fisher CL, Lee I, Bloyer S, Bozza S, Chevalier J, Dahl A, Bodner C, Helgason CD, Hess JL, Humphries RK, et al. 2010. Additional sex combs-like 1 belongs to the enhancer of trithorax and polycomb group and genetically interacts with Cbx2 in mice. Dev Biol 337: 9–15. 10.1016/j.ydbio.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE. 2001. Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556. 10.1016/S1097-2765(01)00316-1 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. 2004. Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577. 10.1126/science.1100576 [DOI] [PubMed] [Google Scholar]
- Fursova NA, Blackledge NP, Nakayama M, Ito S, Koseki Y, Farcas AM, King HW, Koseki H, Klose RJ. 2019. Synergy between variant PRC1 complexes defines Polycomb-mediated gene repression. Mol Cell 74: 1020–1036.e8. 10.1016/j.molcel.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Zhou B-R, Skoultchi AI, Bai Y. 2018. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol 19: 192–206. 10.1038/nrm.2017.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A. 2000. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156: 645–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G. 2007. Chromatin challenges during DNA replication and repair. Cell 128: 721–733. 10.1016/j.cell.2007.01.030 [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Gutiérrez L, Oktaba K, Scheuermann JC, Gambetta MC, Ly-Hartig N, Müller J. 2012. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Dev Camb Engl 139: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlen KM, Churchman LS. 2017. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol 18: 263–273. 10.1038/nrm.2017.10 [DOI] [PubMed] [Google Scholar]
- Hauer MH, Gasser SM. 2017. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev 31: 2204–2221. 10.1101/gad.307702.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri S, Comoglio F, Seimiya M, Gerstung M, Glatter T, Hansen K, Aebersold R, Paro R, Gstaiger M, Beisel C. 2016. A high-density map for navigating the human polycomb complexome. Cell Rep 17: 583–595. 10.1016/j.celrep.2016.08.096 [DOI] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. 2013. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15: 373–384. 10.1038/ncb2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Chaurushiya MS, Webster JD, Kummerfeld S, Reja R, Chaudhuri S, Chen Y-J, Modrusan Z, Haley B, Dugger DL, et al. 2019. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science 364: 283–285. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JWK, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn K-A, Minoda A, Tolstorukov MY, Appert A, et al. 2014. Comparative analysis of metazoan chromatin organization. Nature 512: 449–452. 10.1038/nature13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges HC, Stanton BZ, Cermakova K, Chang C-Y, Miller EL, Kirkland JG, Ku WL, Veverka V, Zhao K, Crabtree GR. 2018. Dominant-negative SMARCA4 mutants alter the accessibility landscape of tissue-unrestricted enhancers. Nat Struct Mol Biol 25: 61–72. 10.1038/s41594-017-0007-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zhu Q, Jussila A, Han Y, Bintu B, Kern C, Conte M, Zhang Y, Bianco S, Chiariello A, et al. 2020. CTCF mediates dosage and sequence-context-dependent transcriptional insulation through formation of local chromatin domains. bioRxiv 10.1101/2020.07.07.192526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12: 115–121. 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseyin MK, Klose RJ. 2021. Live-cell single particle tracking of PRC1 reveals a highly dynamic system with low target site occupancy. Nat Commun 12: 887. 10.1038/s41467-021-21130-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Moffat M, Mann AR, Read D, Hunter CJ, Pradeepa MM, Adams IR, Bickmore WA. 2015. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 29: 1897–1902. 10.1101/gad.268151.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316: 153–155. 10.1038/316153a0 [DOI] [Google Scholar]
- Kahn TG, Dorafshan E, Schultheis D, Zare A, Stenberg P, Reim I, Pirrotta V, Schwartz YB. 2016. Interdependence of PRC1 and PRC2 for recruitment to Polycomb response elements. Nucleic Acids Res 44: 10132–10149. 10.1093/nar/gkw701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PWTC, Müller CW, Vermeulen M, Müller J. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in polycomb repression. Nat Struct Mol Biol 21: 569–571. 10.1038/nsmb.2833 [DOI] [PubMed] [Google Scholar]
- Kasinath V, Beck C, Sauer P, Poepsel S, Kosmatka J, Faini M, Toso D, Aebersold R, Nogales E. 2021. JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Science 371: eabc3393. 10.1126/science.abc3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. 2013. Functional and cancer genomics of ASXL family members. Br J Cancer 109: 299–306. 10.1038/bjc.2013.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Klose RJ. 2017. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 6: e22631. 10.7554/eLife.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King HW, Fursova NA, Blackledge NP, Klose RJ. 2018. Polycomb repressive complex 1 shapes the nucleosome landscape but not accessibility at target genes. Genome Res 28: 1494–1507. 10.1101/gr.237180.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet SL, Makowski MM, Baymaz HI, van Voorthuijsen L, Karemaker ID, Santanach A, Jansen PWTC, Di Croce L, Vermeulen M. 2016. The dynamic interactome and genomic targets of polycomb complexes during stem-cell differentiation. Nat Struct Mol Biol 23: 682–690. 10.1038/nsmb.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. 2013. Chromatin sampling—an emerging perspective on targeting polycomb repressor proteins. PLoS Genet 9: e1003717. 10.1371/journal.pgen.1003717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolovos P, Nishimura K, Sankar A, Sidoli S, Cloos PA, Helin K, Christensen J. 2020. PR-DUB maintains the expression of critical genes through FOXK1/2- and ASXL1/2/3-dependent recruitment to chromatin and H2AK119ub1 deubiquitination. Genome Res 30: 1119–1130. 10.1101/gr.261016.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev 16: 2893–2905. 10.1101/gad.1035902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov JN, Aguero TH, Owens DA, Kurtenbach S, Field MG, Durante MA, Rodriguez DA, King ML, Harbour JW. 2019. BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci Adv 5: eaax1738. 10.1126/sciadv.aax1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle S, Amat R, Glover-Cutter K, Sansó M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. 2012. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 19: 1108–1115. 10.1038/nsmb.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. 2013. H3k4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152: 1021–1036. 10.1016/j.cell.2013.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72: 73–84. 10.1016/0092-8674(93)90051-Q [DOI] [PubMed] [Google Scholar]
- Lee H-G, Kahn TG, Simcox A, Schwartz YB, Pirrotta V. 2015. Genome-wide activities of polycomb complexes control pervasive transcription. Genome Res 25: 1170–1181. 10.1101/gr.188920.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann L, Ferrari R, Vashisht AA, Wohlschlegel JA, Kurdistani SK, Carey M. 2012. Polycomb repressive complex 1 (PRC1) disassembles RNA polymerase II preinitiation complexes. J Biol Chem 287: 35784–35794. 10.1074/jbc.M112.397430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Rosenfeld MG. 2016. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 17: 207–223. 10.1038/nrg.2016.4 [DOI] [PubMed] [Google Scholar]
- Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P, Zhang W, He Q, Patel DJ, Bulyk ML, et al. 2017a. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549: 287–291. 10.1038/nature23881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Hodgson JW, Petruk S, Mazo A, Brock HW. 2017b. Additional sex combs interacts with enhancer of zeste and trithorax and modulates levels of trimethylation on histone H3K4 and H3K27 during transcription of hsp70. Epigenetics Chromatin 10: 43. 10.1186/s13072-017-0151-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grützner F, Odom DT, Patient R, Ponting CP, et al. 2013. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife 2: e00348. 10.7554/eLife.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoomian B, Shields JA, Shields CL. 2018. Overview of BAP1 cancer predisposition syndrome and the relationship to uveal melanoma. J Curr Ophthalmol 30: 102–109. 10.1016/j.joco.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Sinclair DAR, Brock HW. 1999. The Additional sex combs gene of Drosophila is required for activation and repression of homeotic loci, and interacts specifically with Polycomb and super sex combs. Mol Gen Genet MGG 261: 753–761. 10.1007/s004380050018 [DOI] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. 2012. Nonoverlapping functions of the polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10: 47–62. 10.1016/j.stem.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Morey L, Santanach A, Blanco E, Aloia L, Nora EP, Bruneau BG, Di Croce L. 2015. Polycomb regulates mesoderm cell fate-specification in embryonic stem cells through activation and repression mechanisms. Cell Stem Cell 17: 300–315. 10.1016/j.stem.2015.08.009 [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. 10.1016/S0092-8674(02)00976-5 [DOI] [PubMed] [Google Scholar]
- Murali R, Wiesner T, Scolyer RA. 2013. Tumours associated with BAP1 mutations. Pathology 45: 116–126. [DOI] [PubMed] [Google Scholar]
- Musselman CA, Lalonde M-E, Côté J, Kutateladze TG. 2012. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19: 1218–1227. 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, et al. 2008. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev 22: 37–49. 10.1101/gad.1609708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova TB, Wei G, Coker H, Pintacuda G, Bowness JS, Zhang T, Almeida M, Bloechl B, Moindrot B, Carter EJ, et al. 2019. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat Commun 10: 3129. 10.1038/s41467-019-11171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Loghlen A, Muñoz-Cabello AM, Gaspar-Maia A, Wu H-A, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F, et al. 2012. MicroRNA regulation of Cbx7 mediates a switch of polycomb orthologs during ESC differentiation. Cell Stem Cell 10: 33–46. 10.1016/j.stem.2011.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DJ, Wang Z. 2013. Readout of epigenetic modifications. Annu Rev Biochem 82: 81–118. 10.1146/annurev-biochem-072711-165700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perino M, van Mierlo G, Karemaker ID, van Genesen S, Vermeulen M, Marks H, van Heeringen SJ, Veenstra GJC. 2018. MTF2 recruits Polycomb repressive complex 2 by helical-shape-selective DNA binding. Nat Genet 50: 1002–1010. 10.1038/s41588-018-0134-8 [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 20: 2922–2936. 10.1101/gad.1477006 [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, Grant PA. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433: 434–438. 10.1038/nature03242 [DOI] [PubMed] [Google Scholar]
- Qin W, Wolf P, Liu N, Link S, Smets M, Mastra FL, Forné I, Pichler G, Hörl D, Fellinger K, et al. 2015. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell Res 25: 911–929. 10.1038/cr.2015.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramisch A, Heinrich V, Glaser LV, Fuchs A, Yang X, Benner P, Schöpflin R, Li N, Kinkley S, Römer-Hillmann A, et al. 2019. CRUP: a comprehensive framework to predict condition-specific regulatory units. Genome Biol 20: 227. 10.1186/s13059-019-1860-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L. 2010. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468: 1124–1128. 10.1038/nature09574 [DOI] [PubMed] [Google Scholar]
- Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. 2014. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55: 347–360. 10.1016/j.molcel.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Rose NR, King HW, Blackledge NP, Fursova NA, Ember KJ, Fischer R, Kessler BM, Klose RJ. 2016. RYBP stimulates PRC1 to shape chromatin-based communication between polycomb repressive complexes. Elife 5: e18591. 10.7554/eLife.18591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahtoe DD, van Dijk WJ, Ekkebus R, Ovaa H, Sixma TK. 2016. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat Commun 7: 10292. 10.1038/ncomms10292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Lauberth SM. 2020. Enhancer RNAs are an important regulatory layer of the epigenome. Nat Struct Mol Biol 27: 521–528. 10.1038/s41594-020-0446-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scelfo A, Fernández-Pérez D, Tamburri S, Zanotti M, Lavarone E, Soldi M, Bonaldi T, Ferrari KJ, Pasini D. 2019. Functional landscape of PCGF proteins reveals both RING1A/B-dependent-and RING1A/B-independent-specific activities. Mol Cell 74: 1037–1052.e7. 10.1016/j.molcel.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247. 10.1038/nature08966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, Gutiérrez L, Müller J. 2012. Histone H2A monoubiquitination and Polycomb repression: the missing pieces of the puzzle. Fly 6: 162–168. 10.4161/fly.20986 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. 2010. The DUBle life of Polycomb complexes. Dev Cell 18: 878–880. 10.1016/j.devcel.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Seale RL. 1981. Rapid turnover of the histone-ubiquitin conjugate, protein A24. Nucleic Acids Res 9: 3151–3158. 10.1093/nar/9.13.3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Campbell RB, Nicholls F, Slade E, Brock HW. 1992. Genetic analysis of the additional sex combs locus of Drosophila melanogaster. Genetics 130: 817–825. 10.1093/genetics/130.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. 2010. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40: 689–701. 10.1016/j.molcel.2010.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto MC, Chou TB, Bender W. 1995. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140: 231–243. 10.1093/genetics/140.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. 2012. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 151: 994–1004. 10.1016/j.cell.2012.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435. 10.1038/ncb1663 [DOI] [PubMed] [Google Scholar]
- Szczepanski AP, Zhao Z, Sosnowski T, Goo YA, Bartom ET, Wang L. 2020. ASXL3 bridges BRD4 to BAP1 complex and governs enhancer activity in small cell lung cancer. Genome Med 12: 63. 10.1186/s13073-020-00760-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, Pasini D. 2020. Histone H2AK119 mono-ubiquitination is essential for polycomb-mediated transcriptional repression. Mol Cell 77: 840–856.e5. 10.1016/j.molcel.2019.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, Prins P. 2015. Sambamba: fast processing of NGS alignment formats. Bioinformatics 31: 2032–2034. 10.1093/bioinformatics/btv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M, Kishi Y, Yokozeki W, Koseki H, Hirabayashi Y, Gotoh Y. 2018. Ubiquitination-independent repression of PRC1 targets during neuronal fate restriction in the developing mouse neocortex. Dev Cell 47: 758–772.e5. 10.1016/j.devcel.2018.11.018 [DOI] [PubMed] [Google Scholar]
- Turberfield AH, Kondo T, Nakayama M, Koseki Y, King HW, Koseki H, Klose RJ. 2019. KDM2 proteins constrain transcription from CpG island gene promoters independently of their histone demethylase activity. Nucleic Acids Res 47: 9005–9023. 10.1093/nar/gkz607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gottschling DE. 2002. Genome-wide histone modifications: gaining specificity by preventing promiscuity. Curr Opin Cell Biol 14: 756–762. 10.1016/S0955-0674(02)00393-9 [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756. 10.1016/S0092-8674(02)00759-6 [DOI] [PubMed] [Google Scholar]
- Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, Wilkinson KD. 2008. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res 68: 6953–6962. 10.1158/0008-5472.CAN-08-0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers H. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69. 10.1016/j.cell.2007.08.016 [DOI] [PubMed] [Google Scholar]
- Verzijlbergen KF, Faber AW, Stulemeijer IJ, van Leeuwen F. 2009. Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae. BMC Mol Biol 10: 76. 10.1186/1471-2199-10-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettese-Dadey M, Grant PA, Hebbes TR, Crane-Robinson C, Allis CD, Workman JL. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J 15: 2508–2518. 10.1002/j.1460-2075.1996.tb00608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878. 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao Z, Ozark PA, Fantini D, Marshall SA, Rendleman EJ, Cozzolino KA, Louis N, He X, Morgan MA, et al. 2018. Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat Med 24: 758–769. 10.1038/s41591-018-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, Windpassinger C, Wackernagel W, Loy S, Wolf I, et al. 2011. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 43: 1018–1021. 10.1038/ng.910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Hein MY, Cox J, Mann M. 2014. A ‘proteomic ruler’ for protein copy number and concentration estimation without spike-in standards. Mol Cell Proteomics 13: 3497–3506. 10.1074/mcp.M113.037309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink EM, Vihervaara A, Tippens ND, Lis JT. 2019. Nascent RNA analyses: tracking transcription and its regulation. Nat Rev Genet 20: 705–723. 10.1038/s41576-019-0159-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. 2013. Fbxl10/Kdm2b recruits Polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49: 1134–1146. 10.1016/j.molcel.2013.01.016 [DOI] [PubMed] [Google Scholar]
- Wu X, Bekker-Jensen IH, Christensen J, Rasmussen KD, Sidoli S, Qi Y, Kong Y, Wang X, Cui Y, Xiao Z, et al. 2015. Tumor suppressor ASXL1 is essential for the activation of INK4B expression in response to oncogene activity and anti-proliferative signals. Cell Res 25: 1205–1218. 10.1038/cr.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90. 10.1038/nature04815 [DOI] [PubMed] [Google Scholar]
- Yadav T, Quivy J-P, Almouzni G. 2018. Chromatin plasticity: a versatile landscape that underlies cell fate and identity. Science 361: 1332–1336. 10.1126/science.aat8950 [DOI] [PubMed] [Google Scholar]
- Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJ, Drobetsky E, Milot E, et al. 2010. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol 30: 5071–5085. 10.1128/MCB.00396-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Erler J, Langowski J. 2017a. Histone acetylation regulates chromatin accessibility: role of H4K16 in inter-nucleosome interaction. Biophys J 112: 450–459. 10.1016/j.bpj.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones AE, Wu W, Kim J, Kang Y, Bi X, Gu Y, Popov IK, Renfrow MB, Vassylyeva MN, et al. 2017b. Role of remodeling and spacing factor 1 in histone H2A ubiquitination-mediated gene silencing. Proc Natl Acad Sci 114: E7949–E7958. 10.1073/pnas.1711158114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Xu M, Yang F-C. 2020. The role of ASXL1/2 and their associated proteins in malignant hematopoiesis. Curr Stem Cell Rep 6: 6–15. 10.1007/s40778-020-00168-0 [DOI] [Google Scholar]
- Zhao Y, Garcia BA. 2015. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol 7: a025064. 10.1101/cshperspect.a025064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Shilatifard A. 2019. Epigenetic modifications of histones in cancer. Genome Biol 20: 245. 10.1186/s13059-019-1870-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang M, Chang L, Yu J, Song A, Liu C, Huang W, Zhang T, Wu X, Shen X, et al. 2020. RYBP/YAF2-PRC1 complexes and histone H1-dependent chromatin compaction mediate propagation of H2AK119ub1 during cell division. Nat Cell Biol 22: 439–452. 10.1038/s41556-020-0484-1 [DOI] [PubMed] [Google Scholar]
- Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, Li Y, Wang Q, Ma J, Peng X, et al. 2016. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell 63: 1066–1079. 10.1016/j.molcel.2016.08.032 [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80. 10.1016/j.molcel.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18. 10.1038/nrg2905 [DOI] [PubMed] [Google Scholar]
- Żylicz JJ, Bousard A, Žumer K, Dossin F, Mohammad E, da Rocha ST, Schwalb B, Syx L, Dingli F, Loew D, et al. 2019. The implication of early chromatin changes in X chromosome inactivation. Cell 176: 182–197.e23. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-throughput sequencing data reported in this study have been deposited in GEO under the accession number GSE161996. Published data used in this study include BioCAP-seq, GEO: GSE43512 (Long et al. 2013); cnRNA-seq in PRC1CKO ESCs, GEO: GSE119619 (Fursova et al. 2019); RING1B- and SUZ12-bound regions in ESCs, GEO: GSE132752 (Blackledge et al. 2020); ATAC peaks from E14 ESCs, GEO: GSE98403 (King et al. 2018); CTCF ChIP-seq in wild-type ESCs, GEO: GSE153400 (Huang et al. 2020); ChIP-seq for OCT4, NANOG, and SOX2 in untreated Brg1fl/fl ESCs, GEO: GSE87822 (King and Klose 2017); and H3K9me3 and H3K36me3 ChIP-seq in LIF-grown ESCs, GEO: GSE120376 (Ramisch et al. 2019). Custom R and Perl scripts used for data analysis in this study have been deposited to GitHub and are available at https://github.com/nFursova/Calibrated_ChIPseq_RNAseq.