In this review, Klein et al. discuss the p53-independent roles of MDM2 and MDMX. First, they review the structural and functional features of MDM2 and MDMX proteins separately and together that could be relevant to their p53-independent activities. Following this, they summarize how these two proteins are regulated and how they can function in cells that lack p53.
Keywords: cancer, MDM2, MDM2 and MDMX inhibitors, MDMX, pathologies, roles, upstream regulation, p53
Abstract
Most well studied as proteins that restrain the p53 tumor suppressor protein, MDM2 and MDMX have rich lives outside of their relationship to p53. There is much to learn about how these two proteins are regulated and how they can function in cells that lack p53. Regulation of MDM2 and MDMX, which takes place at the level of transcription, post-transcription, and protein modification, can be very intricate and is context-dependent. Equally complex are the myriad roles that these two proteins play in cells that lack wild-type p53; while many of these independent outcomes are consistent with oncogenic transformation, in some settings their functions could also be tumor suppressive. Since numerous small molecules that affect MDM2 and MDMX have been developed for therapeutic outcomes, most if not all designed to prevent their restraint of p53, it will be essential to understand how these diverse molecules might affect the p53-independent activities of MDM2 and MDMX.
Mouse double minute 2 (MDM2/HDM2) was first reported in a study identifying potential oncogenes present in a derivative of the NIH3T3 cell line that had acquired numerous double minutes (Fakharzadeh et al. 1991). MDM2 soon rose to some prominence when it was discovered to bind to p53, which prior to that had been revealed to function as a major tumor suppressor when present in its wild-type form. What follows is a very abbreviated description of the highlights of the profound relationship between p53 and MDM2. These findings have been brought into extraordinary detail and focus by tens of thousands of studies so it would not be possible to cite all of the key findings here. Fortunately, they have been summarized and discussed in numerous reviews (Iwakuma and Lozano 2003; Moll and Petrenko 2003; Manfredi 2010; Wade et al. 2013; Karni-Schmidt et al. 2016; Tackmann and Zhang 2017). Perhaps most compelling have been studies in mice, where it was discovered that loss of MDM2 causes lethality in embryos at a very early stage unless the mice lack expression of p53, in which case mice develop normally and display a similar tumor spectrum as mice with MDM2. Several other milestones have been reached that inform our present view of MDM2 as the prime regulator of p53. The discoveries that MDM2 negatively regulates p53 by preventing it from activating transcription, by targeting it for proteasomal degradation, as well as by evicting it from the nucleus were illuminating. They went hand in hand with findings that releasing p53 to do its work when needed requires disruption of MDM2 from p53, which occurs through myriad signaling pathways extending from DNA damage, eliciting modifications that prevent their interactions, to oncogene activation via ARF, a protein that interacts with MDM2 and thereby stabilizes p53. Adding complexity and depth to these revelations was the discovery that there is a homolog of MDM2, namely, MDMX (also known as HDMX/MDM4/HDM4) that works in concert with MDM2 to degrade p53. Indeed, loss of MDMX is also a lethal event in mouse embryos, thereby highlighting its comparable importance in p53 biology with MDM2. Finally, given the potency of p53 as a tumor suppressor and the potential of harnessing it for therapeutic purposes, a key motivation stemming from these discoveries has been to identify molecules that can disrupt the interactions between wild-type p53 and MDM2, thereby releasing p53 to arrest or kill tumor cells.
Despite their profound relationship to p53, it is now becoming clear that MDM2 and MDMX may have quite rich and complex lives outside of p53, and that is the focus of this review. First, we review structural and functional features of MDM2 and MDMX proteins (separately and together) that could be relevant to their p53-independent activities. We then describe how they are each regulated at multiple levels in cells. Following this, we summarize the many roles they can play in cells in the absence of wild-type p53. Finally, we discuss molecules that interact with them and regulate their activities with the main goal of directing attention toward how they might be useful in cells that lack wild-type p53. Since this is certainly not the first article to focus on the topic of p53-independent regulation and functions of MDM2 and MDMX, we focus where possible on findings that have been made over the past 10 yr, although we reach back earlier to cite key findings that inform the more recent discoveries. In particular, in addition to MDM2, we focus on the p53-independent functions of MDMX and the MDM2–MDMX E3 ligase complex, which are relatively less studied in comparison with those of MDM2. We complete this review by posing some questions that might point the way toward future research on this fascinating subject.
MDM2 and MDMX separately and together: the basics
The MDMs—MDM2 and MDMX—are structural homologs, sharing the following key motifs: a p53 binding domain in the N terminus, a relatively unstructured acidic domain, a zinc finger motif, and a RING domain within the C-terminal portion of the protein (Fig. 1). Both proteins also possess short basic regions that can function as cryptic nucleolar signal sequences within their RING domains, as well as Walker A/P loop motifs. MDM2 alone has a nuclear localization signal and a nuclear export signal in the region between the p53 binding domain and the acidic domain. Therefore, in nonstressed situations, MDM2 is often localized in the nucleus, while MDMX, lacking these motifs, is usually found in the cytoplasm (Shadfan et al. 2012). Most studies dealing with these two proteins tend to emphasize one or the other, with the vast majority centering on MDM2 rather than MDMX. Very few deal with their function together as a complex, although one excellent review does focus on this subject (Leslie and Zhang 2016). Note as well that at least some findings about the structure and function of MDM2 and MDMX have been derived from cells that express wild-type p53. Nevertheless, such information is useful in considering whether and how such observations could be relevant in p53-independent settings.
Figure 1.
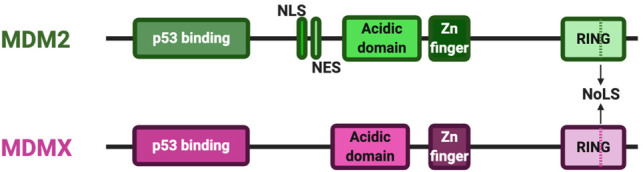
Structural landmarks of MDM2 and MDMX proteins. Schematic representation of the key domains found in MDM2 and MDMX proteins from N terminus (left) to C terminus (right). Shown are their conserved p53 binding domains, acidic domains, and zinc finger regions, as well as their RING domains, which contain an NoLS; MDM2 alone has NLS and NES signal sequences. (NLS) Nuclear localization signal, (NES) nuclear export signal, (NoLS) nucleolar localization signal.
It is well known that MDM2 functions as an E3 ligase that can polyubiquitinate and monoubiquitinate its targets. It can also add ubiquitin-like moieties such as SUMO and NEDD8 to certain substrates. Mouse models harboring a MDM2 mutant deficient in E3 ligase function are similarly embryonic lethal as a fully MDM2-deficient mouse, thereby effectively demonstrating the necessity of this function for MDM2 to effectively inhibit one of its main targets, p53 (Tackmann and Zhang 2017). Interestingly, in some settings p53 is monoubiquitinated by MDM2 and is then polyubiquitinated by other E3 ligases such as Cul4/DDB1 (Banks et al. 2006), UBE4B (Wu et al. 2011), or the histone acetyl transferases (HATs) p300 and CBP (Shi et al. 2009), in which CBP polyubiquitination of p53 requires DBC1 (Akande et al. 2019). Whether MDM2 cooperates with these or different factors to achieve polyubiquitination of targets other than p53 remains to be seen. The RING domain of MDM2 is essential for this E3 ligase activity through chelation of zinc, which is required for MDM2 to transfer ubiquitin from the E2 enzyme onto its target protein (Fang et al. 2000). The MDMX RING domain has no E3 ligase activity of its own, although through mutational swapping of key residues from the MDM2 RING domain, the MDMX RING can acquire E3 ligase activity (Iyappan et al. 2010). The MDM2 acidic and zinc finger domains are also important for its E3 ligase activity, as well as for multiple protein–protein interactions outside of p53 (Bohlman and Manfredi 2014).
Residues within the MDM2 and MDMX RING domains are crucial for their respective abilities to form homodimers and heterodimers (Kawai et al. 2007; Okamoto et al. 2009). There are also five residues at the extreme C termini of both proteins (outside of their RINGs) that are essential for these complexes to be stably formed (Poyurovsky et al. 2007; Uldrijan et al. 2007; Huang et al. 2011). The structures of the MDM2 and MDMX RING domains have been solved by NMR (Kostic et al. 2006) and X-ray crystallography (Linke et al. 2008). The crystal structure revealed the role of their respective extreme C-terminal sequences in stabilizing the formation of heterodimers. Mutations in one of these two regions in either protein can block the formation of the heterocomplex. While in MDM2 alone these mutations can also block its E3 ligase activity, the extreme C terminus of MDMX can rescue the E3 ligase ability of such C-terminal mutant forms of MDM2 (Poyurovsky et al. 2007; Singh et al. 2007; Uldrijan et al. 2007). Relatedly, a patient with segmental progeria syndrome was reported to harbor an antitermination mutation in MDM2, extending the protein by five amino acids (Lessel et al. 2017). This extension likely mimics mutations within the above-mentioned short C-terminal region, as it lacks full E3 ligase activity but can be at least partially rescued by the presence of MDMX (Dolezelova et al. 2012).
The roles of MDMX outside of its regulation of p53 have not been well studied, although by forming a heterocomplex with MDM2, MDMX has the potential to regulate the stability of many proteins via the E3 ligase activity of this complex. It is, however, unclear which of the known E3 ligase targets of MDM2 may also require the presence of MDMX for effective degradation. One hypothesis is that MDMX blocks the ability of MDM2 to ubiquitinate itself and hence stabilizes it (Stad et al. 2001). Instead, MDMX might then focus the E3 ligase activity of MDM2 against its other targets, which has been supported experimentally (Linares et al. 2003; Kawai et al. 2007; Linke et al. 2008; Okamoto et al. 2009), thereby making the MDM2–MDMX heterocomplex a better E3 ligase. In support of this theory, there are several small molecule inhibitors of MDM2 that have been shown to enhance its autoubiquitination ability, and these are described in detail later in this review. There is some controversy, however, as to whether and when MDM2 serves an E3 ligase for itself. In a mouse knock-in model, a RING domain mutant of MDM2 (C462A) that lacks the E3 ligase activity does not have increased stability when compared with wild-type MDM2 (Itahana et al. 2007; He et al. 2014). However, the human counterpart of this mutation (C464A), when ectopically expressed, is more stable than the wild-type human MDM2 both basally (Gu et al. 2003; Inuzuka et al. 2010; Leslie et al. 2015; Xu et al. 2015; Giono et al. 2017; Zhao et al. 2018) and in response to stress (Stommel and Wahl 2004). However, even in the context of stress, there is not a clear consensus among various studies regarding the ability of MDM2 to self-ubiquitinate, and this could be based on the differing methods used (Li and Kurokawa 2015). Taken together, these reports suggest that the extent of difference in stability of the E3 ligase mutant of MDM2 and its wild-type counterpart depends on the context—physiological versus stress-induced conditions and perhaps mouse versus human cells. Therefore, in some settings, the autoubiquitination activity may not be crucial to the stability of MDM2. Furthermore, MDMX may be able to protect MDM2 from ubiquitination by other E3 ligases as well (Li and Kurokawa 2015). A detailed and systematic evaluation of the stability of the E3 ligase-deficient MDM2 under various stresses in different cellular contexts would shed more light on this aspect of regulation. As documented in the next section, several E3 ligases have been reported to be able to target MDM2 for degradation.
MDM2 and MDMX upstream: regulation occurs at every level
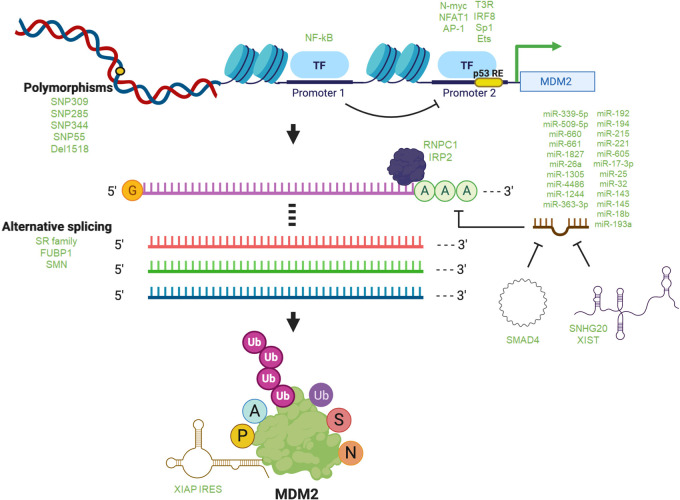
Although best studied as a transcriptional target of p53, abundant information has also accrued as to how MDM2 expression and activity are modulated at virtually all stages that have been documented for other proteins. To a much lesser extent, there are also examples of varied forms of regulation of MDMX. In addition to genomic alterations such as copy number variations, mutations, and polymorphisms, regulation of MDM2 (Fig. 2) and MDMX (Fig. 3) has been documented at multiple levels, extending from transcription all the way to innumerable post-translational modifications.
Figure 2.
Regulation of MDM2. Different factors are involved in the regulation of MDM2 at the DNA, RNA, and protein levels in eukaryotic cells. Listed are the most important or most recently described processes and biomolecules regulating MDM2. (Top) Polymorphisms (SNPs) in the MDM2 gene that have been characterized, as well as transcription factors reported to regulate transcription from the two well-studied MDM2 promoters 1 and 2 and regulation of P2 by P1. (Middle) Factors, including numerous listed microRNAs, that interact with and regulate MDM2 mRNA levels, splicing, and translation. (Bottom) MDM2 protein is extensively modified by ubiquitination (Ub), acetylation (A), phosphorylation (P), SUMOylation (S), and NEDDylation (N).
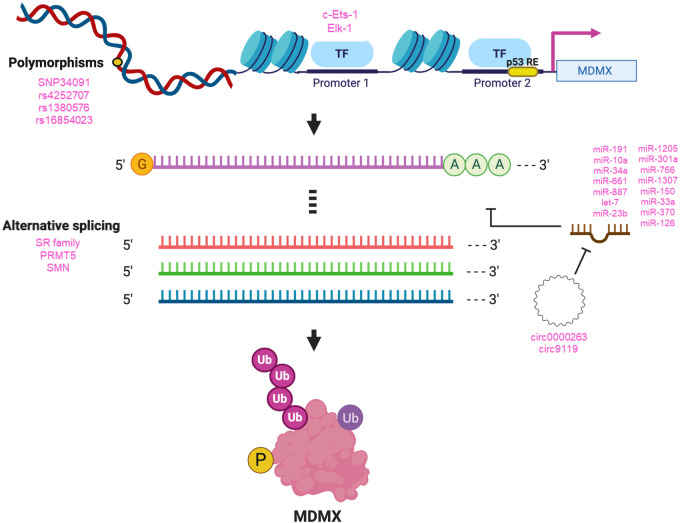
Figure 3.
Regulation of MDMX. Different factors are involved in the regulation of MDMX at the DNA, RNA, and protein levels in eukaryotic cells. Listed are the most important or most recently described processes and biomolecules regulating MDMX. (Top) Polymorphisms in the MDMX gene that have been characterized, as well as factors reported to regulate transcription from the MDMX promoters 1 and 2. (Middle) Factors, including numerous listed microRNAs, that interact with and regulate MDMX mRNA levels, splicing, and translation. (Bottom) MDM2 protein is modified by ubiquitination (Ub) and phosphorylation (P).
Clinical studies have revealed that MDM2 gene amplification occurs in several tumor types (see https://www.cbioportal.org/results/cancerTypesSummary?case_set_id=all&gene_list=MDM2&cancer_study_list=5c8a7d55e4b046111fee2296). MDM2 amplification tends to correlate with the presence of wild-type p53, suggesting that this mode of MDM2 overexpression is relevant primarily due to its function in restraining p53 (Oliner et al. 2016). Nevertheless, there are multiple modes other than gene amplification by which MDM2 and MDMX and their products are regulated; the challenge we face here is that oftentimes studies dealing with this topic have been carried out in cells that express wild-type p53. While the studies we mention have not described an active role for p53, we cannot rule out a possible effect of its presence in these cells. In some cases, however, similar results have been documented in cell lines that either lack p53 or express cancer-related mutant forms of p53, which can mitigate such concerns. Regardless of the context, studies on upstream regulation of MDM2 and MDMX are intriguing and may direct future questions and experimental examination.
Regulation at the DNA level: a dance of promoters and transcription factors
Promoters
Transcription of MDM2 is controlled primarily via two promoters, termed P1 and P2; the former is linked to basal, constitutive MDM2 expression, while expression from the latter is induced by various transcription factors in response to stimuli, most notably p53 (Zhao et al. 2014). A third MDM2 promoter, P3, which has not been widely studied, is also activated independently of p53 and is actually repressed by p53 binding (Liang and Lunec 2005). Transcripts generated from the P1 promoter (located upstream of exon 1) lack exon 2, while transcripts from P2 (located within exon 1) include exon 2, but not exon 1. While the transcript regulated by P1 can be processed to create various isoforms including MDM2A (Alt2), MDM2B (Alt1), and MDM2C (Alt3), as well as full-length MDM2 (Rosso et al. 2014), activation at P2 mostly produces full-length MDM2 (Cheng and Cohen 2007).
The P1 promoter does not possess a p53 response element (RE) and is considered to be virtually p53-independent (Juven et al. 1993). P1 is inhibited by PTEN working through its lipid phosphatase domain (Chang et al. 2004); relatedly, the P1 promoter transcript is modulated by rapamycin (Kao et al. 2009). NF-κB had been shown previously to induce MDM2 expression at the P1 promoter, but it was unclear whether such induction was caused directly or indirectly (Busuttil et al. 2010). However, more recently, a TGF-β family ligand, activin, was reported to induce MDM2 expression and enhance cell migration in colorectal cancer cells by activating the PI3K pathway and promoting the binding of the NF-κB component p65 to the MDM2 promoter (Jana et al. 2017).
Furthering the complexity, transcripts from the P1 promoter can suppress transcription from the P2 promoter independently of p53 via H3K36 trimethylation; this toggling occurs in different cell lines and also as a natural event during human embryonic stem cell differentiation (Hollerer et al. 2019). MDM2 transcripts that are expressed from the P1 or P2 promoters may be regulated differently. For example, the rapamycin-sensitive eukaryotic translation initiation factor 4E (eIF4E) regulates translation of MDM2 mRNA produced from the P1 but not from the P2 promoter (Kao et al. 2009).
A number of transcription factors can regulate transcription from P2. Besides the classic p53, these include N-myc (Slack et al. 2005), NFAT1 (Zhang et al. 2012), IRF8 (Zhou et al. 2009), T3R (Qi et al. 1999), AP-1 (Pikkarainen et al. 2009), Ets (Truong et al. 2005; Sashida et al. 2009), and Sp1 (Bond et al. 2004). Activity at P2 by these other transcription factors can be independent of wild-type p53. For example, N-myc was found to form a complex with the WRD5 subunit of a histone H3K4 methyltransferase complex and activate MDM2 transcription. Furthermore, expression of WRD5 is necessary for cell proliferation and survival in mutant p53-expressing neuroblastoma cells, although it remains unclear whether this phenotype is due to WRD5 modulation of MDM2 (Sun et al. 2015). These studies provide a tantalizing glimpse of the intricate process through which MDM2 expression is regulated at the promoter level. While transcriptional regulation of MDM2 may likely have an impact on cancer outcomes such as proliferation and migration of malignant cells, the field is still evolving, and these findings raise several questions about MDM2 transcription that merit future study.
As with all other aspects of MDMX, its promoter has not been as well studied as that of MDM2. Interestingly, MDMX also possesses a main promoter (P1) as well as a second promoter (P2) within the first intron that can be regulated by p53 and that generates a novel transcript (HDMX-L) whose product cooperates with MDM2 to efficiently degrade p53 (Phillips et al. 2010). Among the few transcription factors known to regulate the MDMX P1 promoter are c-Ets-1 and Elk-1 (Gilkes et al. 2008).
SNPs
Coupled with discoveries of transcription factors that can regulate the gene have been reports of naturally occurring genetic MDM2 variants in the form of single-nucleotide polymorphisms (SNPs) that regulate its gene expression (for review, see Oliner et al. 2016). The first report of such an MDM2 variant, SNP309, revealed that one variant (GG) creates a strong Sp1 binding site that is correlated with increased predisposition to certain cancers (Bond et al. 2004). The oncogenic outcome of the GG variant was subsequently confirmed in an elegant mouse model (Post et al. 2010). Since then, more recent studies have further examined the role of SNPs in the regulation of MDM2 and MDMX. Although many of these studies used wild-type p53-expressing cells or patient samples that did not stratify p53 status, they nevertheless indicate that sequence variants in MDM2 and MDMX could contribute to p53-independent effects on their expression in cancer. For example, analogous to SNP309, a SNP in the P2 promoter (SNP55C > T; rs2870820) enhances Sp1 binding and increases MDM2 expression (Okamoto et al. 2015). SNP55C > T is associated with an increased risk of colon cancer but not lung, breast, or prostate cancer. Interestingly, neither healthy nor cancerous tissue samples heterozygous for SNP55C > T showed increased MDM2 (Helwa et al. 2016). Another SNP also located in the P2 promoter, SNP344T > A (rs1196333), is similarly capable of altering the affinity of various transcription factors for the MDM2 promoter. The presence of SNP344T > A does not enhance MDM2 expression, nor is it correlated with age of onset, response to therapy, rate of relapse, or overall survival in ovarian, endometrial, breast, or prostate cancers (Knappskog et al. 2012).
The indel del1518 (rs3730485) polymorphism in the MDM2 P1 promoter is correlated with risk of hepatocellular carcinoma (Dong et al. 2012) but not breast or lung cancers (Hu et al. 2006; Ma et al. 2006). This polymorphism was discovered in a small population of Chinese patients and has been predominantly studied in that group. However, a broader study of other populations determined that del1518 is associated with increased risk of colon cancer, but not lung, breast, or prostate cancer, in other ethnicities as well (Gansmo et al. 2016b).
MDMX expression is also affected by sequence variants such as SNP34091C > A (rs4245739) in its 3′ UTR. This SNP decreases the affinity of microRNAs (miRNAs) for the MDMX transcript, which in turn increases MDMX protein expression, and this is correlated with increased cancer risk in ovary, prostate, and lung carcinomas (Gao et al. 2015; Stegeman et al. 2015; Gansmo et al. 2016a) but not in thyroid or endometrial cancers (Gansmo et al. 2016a; Mohammad Khanlou et al. 2017). In breast cancer, the presence of SNP34091 alone does not correlate with cancer risk (Pedram et al. 2016); however, in combination with the miRNA expression profile, it predicts tumor size and lymph node infiltration (Anwar et al. 2017). Other SNPs in MDMX are associated with an increased risk of glioma (rs4252707) (Sun et al. 2018), prostate, and gastric cancer (rs1380576) (Sun et al. 2010; Wang et al. 2017b) and Parkinson's disease (rs16854023) (Cha et al. 2020).
Future studies will hopefully reveal how SNPs in the MDM2 and MDMX genes that correlate with different aspects of tumorigenesis work at the molecular level. The discovery that SNP309 and SNP55 each affect the binding affinity of the Sp1 transcription factor to their respective regions in the MDM2 promoter provides a model for how such mechanistic information could be obtained. Taken together, MDM2 and MDMX SNPs may provide useful markers for detecting increased risk of certain cancers.
Regulation of RNA processing: MDM2 comes in many forms
Human MDM2 can generate at least 72 alternative and aberrant spliced isoforms, although not all result in protein products (for review, see Okoro et al. 2012). A subset of such isoforms creates MDM2 polypeptides that lack the p53 binding domain and thus have p53-independent functions—including the three main isoforms: MDM2A, MDM2B, and MDM2C (Okoro et al. 2012).
The serine arginine (SR) family of proteins regulates both MDM2 and MDMX RNA splicing. SRSF2, a member of the SR family, suppresses expression of the MDM2B (ALT1) splice variant (Comiskey et al. 2020) whereas SRSF1, another member of the SR family, promotes and is necessary for MDM2B (ALT1) splicing (Comiskey et al. 2015). Interestingly, there is cross talk between these family members; mutation of SRSF1 binding sites is rescued by concurrent mutation of SRSF2 binding sites (Comiskey et al. 2020). FUBP1, an RNA binding protein, also suppresses production of the MDM2B (ALT1) spliced form (Jacob et al. 2014). We note that all of these processes were studied with ectopically expressed protein, and in cells harboring wild-type p53. The behavior of the endogenous splicing factors and their impact on generating different RNA isoforms remain to be explored.
The splicing protein survival of motor neuron (SMN) is necessary for assembly of small nuclear ribonucleoproteins (snRNPs) in the spliceosome and, consequently, for proper splicing of exon 3 of MDM2 and exon 7 of MDMX in motor neurons. Defective snRNP biogenesis caused by deficient SMN results in increased alternative splicing of MDM2 and MDMX (Van Alstyne et al. 2018), although the role of SMN in cells lacking wild-type p53 remains to be examined.
MDMX pre-mRNA generates six variant isoforms beside the full-length protein—MDMX-S, MDMX-211, MDMX-G, MDMX-A, MDMX-XAlt1, and MDMX-XAlt2, although regulatory mechanisms for how these isoforms are produced have not yet been well examined (for review, see Mancini et al. 2009). Among these, the best-studied splice variant is MDMX-S, which lacks exon 6. The modulation of MDMX pre-mRNA splicing to MDMX-S is partially controlled by SRSF3 and PRMT5, which are necessary for sustained expression of full-length MDMX, although, again, these have only been studied in cells expressing wild-type p53, leaving open the question of whether alternative splicing of MDMX depends on p53 activity (Bezzi et al. 2013; Dewaele et al. 2016).
Regulation at the mRNA level: stability is the key
Post-transcriptional control of MDM2 and MDMX at the mRNA level has been a topic of considerable interest, in particular the modulation of transcript stability by microRNAs (miRNAs). By 2015, at least 15 miRNAs were known negative regulators of MDM2 and six of MDMX; most miRNAs work by destabilizing MDM2 or MDMX transcripts (for review, see Vijayakumaran et al. 2015). Among these miRNAs, the majority are p53-independent. Subsequently reported miRNAs function the same way to regulate MDM2 expression. For example, miR-194 down-regulates MDM2 expression through its direct interaction with MDM2 transcripts in p53-null cells (Nakamura et al. 2019). Additionally, miR-1827 lowers MDM2 expression through direct interaction with MDM2 mRNA in the presence or absence of p53 (Zhang et al. 2016). Other miRNAs—miR-26a (Zhou et al. 2019), miR-1305 (Cai et al. 2019b), miR-4486 (Liu et al. 2019b), miR-1244 (Yanbin and Jing 2019), and miR-363-3p (Rong et al. 2020)—behave similarly.
More recently reported miRNAs that regulate MDMX through direct binding with its mRNA include miR-23b (Zhao et al. 2019), miR1205 (Yan et al. 2019), miR-301a (Wang et al. 2017a), miR-766 (Wang et al. 2017c; Chen et al. 2019b), miR-1307 (Wang and Zhu 2018), miR-150 (Cai et al. 2019a), miR-33a (Jiang et al. 2019), miR-370 (Shen et al. 2018), and miR-126 (Tian et al. 2020). Of these, miR-1205, miR-33a, and miR-370were tested in cell lines expressing mutant p53, and miR-1205 was tested in p53-null cells. Since other studies used wild-type p53-expressing cell lines, they do not specifically address whether regulation by the respective miRNAs is p53-dependent.
Long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs) can also affect MDM2 through modulation of miRNA activity. The lncRNA XIST targets miR-363-3p, acting as a “sponge” and preventing miR-363-3p activity against MDM2 transcripts, independently of p53 (Rong et al. 2020). The lncRNA SNHG20 also acts as a sponge for miR-4486, preventing it from destabilizing MDM2 transcripts (Liu et al. 2019b). Similarly, circSMAD4A sequesters miR-1244 in cells with wild-type or mutated p53 and in p53-null cells (Yanbin and Jing 2019). circRNAs, such as hsa_circ_0000263 (Cai et al. 2019a) and circ9119 (Tian et al. 2020), can also inhibit miRNAs that destabilize MDMX, although, so far, this has only been tested in cells that express wild-type p53.
RNA binding proteins also regulate MDM2 mRNA stability. For example, RNPC1 binds to the MDM2 3′ UTR and reduces its half-life (Xu et al. 2013). Interestingly, IRP2, an iron-responsive protein that is degraded at high levels of iron, can stabilize MDM2 mRNA by binding at the 3′ UTR, yet this factor can also destabilize MDM2 RNA by binding at the 5′ UTR (Zhang et al. 2020). HBXIP, an oncoprotein from the hepatitis B virus, promotes MDM2 expression by methylation of the promoter of miR-18b, thereby reducing expression of miR-18b and up-regulating MDM2 protein, independently of p53 (Li et al. 2018b).
Regulation at the protein level: small changes can make a big difference
Given their central importance in restraining p53 and the burgeoning evidence that they play myriad other roles in cells, it is not surprising that the MDM2 and MDMX proteins are themselves heavily regulated. This is reflected both in their various post-translational modifications and the plethora of partners with which they interact. Among others, one excellent review on this subject was published a few years ago (Fåhraeus and Olivares-Illana 2014). Here we focus largely on more recent discoveries related to the many kinds and numbers of modifications of the MDMs along with a smaller number of more recently discovered protein partners.
Ubiquitination
The MDM2 E3 ligase ubiquitinates several targets, including itself and its partner MDMX (for review, see Leslie and Zhang 2016). Previously, a number of other E3 ligases—such as PCAF, SCFβ-TrCP complex, APC/C, and NEDD4-1—were shown to target MDM2 for ubiquitination (for review, see Li and Kurokawa 2015). More recently NAT10, typically an acetyltransferase of the GNAT family, was found to have p53-independent E3 ligase activity against MDM2 (Liu et al. 2016), while RNF12 was shown to interact with and ubiquitinated MDM2 in both wild-type p53-expressing and p53-null cell lines (Gao et al. 2016). Interestingly, although polyubiquitination of MDM2 typically results in proteasomal degradation, its polyubiquitination via the lys63 linkage stabilizes the protein, in contrast to the more well studied lys48 linkage, which targets MDM2 for proteasomal degradation. E3 ligases that have been shown to catalyze lys63 ubiquitination of MDM2 are NEDD4-1 (Xu et al. 2015) and MARCH7 (Zhao et al. 2018), although both studies were performed in a wild-type p53 background. Deubiquitinating enzymes (DUBs)—such as USP7 (or HAUSP), USP2a, and USP15—cleave ubiquitin from MDM2 and MDMX (for review, see Li and Kurokawa 2015), and USP26, a testis-specific DUB, was shown to target MDM2 in wild-type p53-expressing cells (Lahav-Baratz et al. 2017).
Ubiquitination of MDMX has not been studied as extensively; nevertheless, in addition to the established MDM2-mediated ubiquitination of MDMX at K442 (Linke et al. 2008), Peli1, another E3 ligase, binds and polyubiquitinates MDMX, resulting in translocation of MDMX to the cytoplasm in a p53-independent manner (Li et al. 2018a).
SUMOylation
SUMOylation, the addition of small ubiquitin-like modifier (SUMO) to lysine residues, is also performed by E3 ligases through interaction with E2 ligases carrying SUMO and the target protein. SUMOylation of MDM2 has been previously reviewed (Li and Kurokawa 2015). In brief, MDM2 (which itself can SUMOylate other substrates) can be SUMOylated at K446; this modification inhibits ubiquitination of MDM2, including autoubiquitination, and stabilizes the protein (Chen and Chen 2003; Stindt et al. 2011; Li and Kurokawa 2015). MDMX is also SUMOylated at K254 and K379, although these modifications do not regulate stability, ubiquitination, or subcellular localization of the protein (Ghosh et al. 2005; Pan and Chen 2005).
NEDDylation
NEDDylation is the addition of the small ubiquitin-like peptide NEDD8 to lysine residues, which is performed by E3 ligases. As with ubiquitination and SUMOylation, MDM2 is both NEDDylated and NEDDylates other proteins, such as PPARγ and HBx (Watson et al. 2010; Park et al. 2016; Liu et al. 2017a). NEDDylation stabilizes MDM2 protein and decreases MDMX protein levels, while deNEDDylation by NEDP1 destabilizes MDM2 (Watson et al. 2010; Xu et al. 2015).
Interestingly, phosphorylation of MDM2 at Y281 and Y302 by c-Src enhances MDM2 stability independently of p53 and enhances MDM2 NEDDylating activity by promoting binding to the NEDD8 E2-ligase Ubc12 (Batuello et al. 2015). MDM2 also NEDDylates MDMX, which promotes MDMX stability; correspondingly, the loss of c-Src leads to increased degradation of MDMX (Hauck et al. 2017). This study, which also found that MDM2 NEDDylation is enhanced by the presence of MDMX, specifically examined enhanced NEDDylation of p53, and NEDDylation of other targets remains to be explored.
Phosphorylation
Of all protein modifications, phosphorylation of MDM2 and MDMX residues has been studied the most extensively, with MDM2 possessing >20 known phosphorylation sites and MDMX having six such sites. Because of the importance of phosphorylation to MDM2 functions, especially during DNA damage, this subject has been comprehensively reviewed (Meek and Knippschild 2003; Meulmeester et al. 2005; Meek and Hupp 2010; Wade et al. 2010; Markey 2011; Li and Kurokawa 2015; Carr and Jones 2016). Phosphorylation of MDM2 regulates its enzymatic activity, subcellular localization, protein stability, and both protein–protein and protein–mRNA interactions.
Within the last few years, several new phosphorylation sites have been discovered. Mps1 kinase was found to phosphorylate MDM2 at T4, T306, and S307 during mitosis in response to oxidative stress; these result in its increased ability to ubiquitinate histones (Yu et al. 2016). As previously described, c-Src also phosphorylates MDM2 at Y281 and Y302, leading to increased stability of MDM2 (Batuello et al. 2015). Bruton's tyrosine kinase (Btk) was found to interact with and to phosphorylate MDM2, leading to inhibition of its autoubiquitination (Rada et al. 2017). At one of the best-known MDM2 phosphorylation sites, S166, several new pathways and stimuli have been described, including elevated glucose and modulation of intracellular glycosylation (Barzalobre-Gerónimo et al. 2015; de Queiroz et al. 2016), the proinflammatory cytokine MIF (Costa et al. 2016), nicotine (Chen and Wang 2019), and CD44 through activation of EGFR (Dhar et al. 2018). The Robo2-Baiap cascade, involved in axon guidance and neuron migration, is required to maintain MDM2-S166 phosphorylation in kidney cells (Li et al. 2019). DNA damage induces phosphorylation at S429 by ATM, thereby enhancing the E3 ligase activity of the MDM2–MDM2 homodimer, but interestingly not the MDM2–MDMX heterodimer (Magnussen et al. 2020). As the MDM2 homodimer preferentially autoubiquitinates (Linke et al. 2008), it is unsurprising that phosphorylation at S429 also promotes MDM2 degradation, but the work by Magnussen et al. (2020) is especially notable for examining the role of the MDM2–MDMX heterodimer, which is generally overlooked in MDM2 studies. Additionally, phosphorylation sites and activity in the absence of wild-type p53 are mostly unexplored since all of the above-mentioned studies except those of Batuello et al. (2015) and de Queiroz et al. (2016) were performed in wild-type p53-expressing cells.
Phosphorylation of MDMX has also not been widely studied in p53-null settings; however, in cell lines with wild-type or mutant p53, the tyrosine kinase receptor AXL stimulates phosphorylation of MDMX by CDK4/6 and p38 at S314, leading to MDMX nuclear localization and increased affinity between MDMX and MDM2 (de Polo et al. 2017).
Acetylation
Compared with other modifications, acetylation of MDM2 has not been widely explored. Over 15 yr ago, p300 and CBP were shown to acetylate MDM2 at K466/467 and K469/470 and thereby inhibit MDM2 E3 ligase activity (Wang et al. 2004). More recently, it was reported that p300 can also acetylate MDM2 at K182 and K185, leading to its stabilization by both inhibiting its autoubiquitination and enhancing its interaction with USP7 (Nihira et al. 2017). Additionally, SIRT1 deacetylates K182 and K185 (Nihira et al. 2017) while HDAC1 and HDAC2 deacetylate MDM2 at K469 and K470, restoring MDM2 binding affinity for the MCL-1 ubiquitin ligase E3 (MULE), resulting in its degradation (Patel et al. 2019). The SIRT7 NAD+-dependent deacetylase removes the acetyl group from PCAF (K720), thereby stimulating PCAF binding to MDM2 and leading to MDM2 degradation (Lu et al. 2020). Note that under certain conditions, HATs such as Tip60, p300, and NAT10 can interact with but do not acetylate MDM2 (Zeng et al. 2003; Dohmesen et al. 2008; Liu et al. 2016). Thus, the conditions under which MDM2 is acetylated and the functional relevance of these modifications—as well as whether and when MDMX is acetylated under any physiologically relevant conditions—pose several interesting questions for future studies.
Regulation at the protein level: partners in crime
While protein–protein interactions that regulate MDM2 have been extensively reviewed (Fåhraeus and Olivares-Illana 2014), several binding partners have been recently described. It has been established for some time that MDM2 binds to an extraordinary number of ribosomal proteins (RPs) (Zhang and Lu 2009; Deisenroth et al. 2016). More recently, ribosomal protein RPL4 was shown to interact with MDM2, leading to decreased ubiquitination of MDM2 targets, and also to promote binding to MDM2 of two well-studied RPs, namely, RPL5 and RPL11, in p53-null cells (He et al. 2016). PHLDB2 directly interacts with MDM2 to inhibit E-cadherin degradation and the epithelial–mesenchymal transition (EMT) in a p53-independent fashion (Chen et al. 2019a). FKBP12, a cytoplasmic binding protein, binds the MDM2 RING domain and stabilizes MDM2, but does not interact with MDMX, in the setting of both wild-type p53 and mutant p53 (Liu et al. 2017b). In wild-type p53-expressing cells, XBP-1 (a transcription factor involved in endoplasmic reticulum stress response) binds MDM2 and impairs its autoubiquitination but does not interact with MDMX; XBP-1 silencing also inhibits MDM2 transcription at the P2 promoter in cells lacking wild-type p53 (Huang et al. 2017). Similarly, XIAP IRES, an mRNA molecule from the XIAP gene, binds to MDM2 and prevents its homodimerization and subsequent autoubiquitination in wild-type p53-expressing cells (Liu et al. 2015).
A number of reports have described p53-independent regulation of MDM2 in the context of different viral strategies for overcoming host resistance. The hepatitis B virus X protein (HBx) regulates MDM2 gene expression and inhibits MDM2 autoubiquitination (Wang et al. 2017d). The HIV trans-activator of transcription regulatory protein (Tat) also blocks MDM2 autoubiquitination and increases MDM2 protein levels (Raja et al. 2017). Last, infection by influenza A can also modulate MDM2 protein levels independently of p53; in the initial stages of infection, MDM2 protein decreases, and in the later stages, MDM2 protein is highly stabilized (Pizzorno et al. 2018).
That MDM2 and its less well studied partner MDMX are so extensively regulated at virtually every known level attests to their relevance not only as inhibitors of p53 but of myriad other cellular activities and outcomes on their own that are described in the next section.
Getting out of p53's shadow: p53-independent roles of MDM2 and MDMX
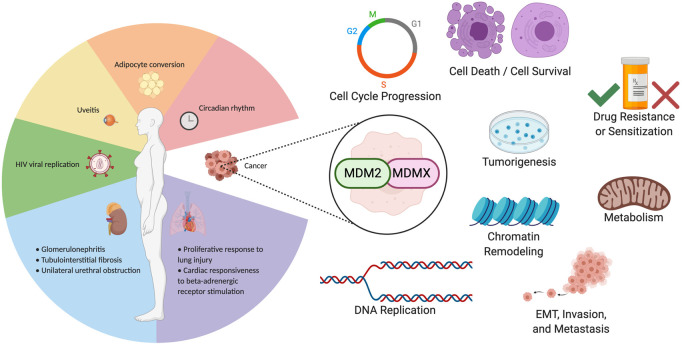
Since the MDMs were discovered, aside from their extensively probed relationship to wild-type p53, numerous studies have examined how these two proteins function independently of that famous tumor suppressor. Indeed, an impressively large number of cellular outcomes are affected by these two proteins on their own. At the present time, most reports in this regard deal largely with MDM2 (the majority) or MDMX (far fewer), rather than considering them together. Studying these two proteins as p53-independent entities is not an empty exercise: p53 is mutated or lost in ∼50% of all tumors (albeit with wide variation across tumor types), which makes understanding their functions separately or together highly relevant. We focus here on their roles in cancer as well as in other pathologies. Figure 4 outlines the many regulatory pathways, functions, and drugs that are relevant to MDM2 and MDMX activity that can be considered to function independently of p53.
Figure 4.
Clinically relevant functions of MDM2 and MDMX. MDM2 and MDMX affect several clinical pathologies in a p53-independent fashion, including nonmalignant disease and cancer-related functions. p53-independent activities of MDM2 and MDMX that have been studied in a clinical context are summarized in this figure. Nonmalignant physiological processes and pathologies are separated by organ system at the left. Cancer-related activities mediated by MDM2 and MDMX are represented graphically at the right.
MDM2 and MDMX play myriad roles in cancer
MDM2 and MDMX as mediators of cell life and death
MDM2 has been known for some time to be able to target several well-known regulators of the cell cycle (for review, see Karni-Schmidt et al. 2016). This has been confirmed and extended in more recent years; ablation or inhibition of MDM2 reduces mitosis (Kundu et al. 2017) and causes G2 arrest (Feeley et al. 2017) in p53-negative or mutant p53-expressing mouse lymphoma and sarcoma cell lines, and human breast cancer cell lines. However, the mechanisms by which MDM2 promotes cell cycle progression have not yet been fully elucidated. In the above-mentioned studies, Kundu et al. (2017) place MDM2 activity within an estrogen-MDM2-Rb-E2F1 pathway, while Feeley et al. (2017) report that MDM2 acts through inhibition of p73. Separately, MDM2 was also shown to be necessary for proliferation of mutant p53-expressing retinoblastoma cells through promotion of MYCN expression (Qi and Cobrinik 2017). Knockdown of MDM2 also reduces primary tumor volumes, although specifically with estrogen receptor α-positive, luminal A subtype breast cancer, but there is no effect on proliferation in triple-negative breast cancer (Gao et al. 2019).
Besides promoting cell growth, MDM2 may also help cells avoid death. The roles of MDM2 in protection from cell cycle arrest and cell death, however, are not straightforward. MDM2 may also inhibit cell cycle progression or promote cell death pathways under certain circumstances. For example, MDM2 promotes proteasome-mediated degradation of Cdc25C in a p53- and ubiquitin-independent manner, and Cdc25C degradation mediated by overexpression of MDM2 delays cell cycle progression through G2/M, although the latter finding has primarily been shown in p53-expressing U2OS cells (Giono et al. 2017). Following inhibition of MEK/ERK signaling in mammary tumor cancer stem cells (CSCs), MDM2 also triggers oncogene-induced senescence and depletion of CSC populations independently of p53 (McGrail et al. 2018). Interestingly, while one study reported that MDM2 promotes cell cycle progression by inhibiting p73 (Feeley et al. 2017), another found that Bruton's tyrosine kinase (BTK) up-regulates MDM2 and induces apoptosis in the absence of p53 and that BTK-induced apoptosis is mediated through p73 activity (Rada et al. 2018).
While MDMX, like MDM2, is typically considered an oncogene and is amplified in many cancers, basal expression of MDMX has numerous antitumor effects in thymus and breast cancers lacking p53: MDMX suppresses proliferation in p53-null thymic tumors (Matijasevic et al. 2008). The central zinc finger domain of MDMX specifically inhibits mitosis, prevents chromosome loss in hyperploid, p53-null tumors, and suppresses growth of mutant p53-expressing breast tumors, while the RING domain of MDMX inhibits proliferation in p53-null cancer cells (Matijasevic et al. 2016). MDMX also blocks proliferation through inhibition of mTORC1, thereby reducing phosphorylation of the mTORC1 target p70S6K1 (Mancini et al. 2017).
Finally, in considering the roles of MDM2 and MDMX in this complex regulatory network of cellular life and death, comparatively little work has focused on the role of the MDM2–MDMX heterocomplex as a unique entity rather than simply a combination of two homologs. Recently, our group demonstrated that ferroptotic cell death is facilitated by MDM2 and MDMX working at least in part as a heterocomplex in select cancer cell lines; they do so through PPARα and lipid peroxidation pathways (Venkatesh et al. 2020). Studied under this new light, the MDM2–MDMX heterocomplex may become an intriguing new target for further research.
Activity at DNA and chromatin
The participation of MDM2 and MDMX in chromatin modification and gene expression has been an intriguing topic of study for some time now (Biderman et al. 2012; Wienken et al. 2017). At the histone level, MDM2 activity recently has been linked to promoting both DNA compaction and DNA relaxation. MDM2 promotes DNA compaction through stabilization of histone deacetylase (Choi et al. 2019) and through association with polycomb repressive complex 2 (PRC), resulting in both histone trimethylation and monoubiquitination (Wienken et al. 2016). However, MDM2 can increase DNA accessibility via degradation of the major methyltransferase suppressor of variegation 3-9 homolog 1 (SUV39H1), which is opposed by USP7 deubiquitination of SUV39H1 (Mungamuri et al. 2016). MDM2 via its RING domain promotes genomic stability by limiting R loop formation (Klusmann et al. 2018). Interestingly, MDMX also interacts with members of the PRC complex and thereby supports histone ubiquitination (Wohlberedt et al. 2020).
MDM2 and MDMX also modulate the cellular response to DNA damage (for reviews, see Lehman and Mayo 2012; Eischen 2017); in that regard, it seems that they engender greater genome instability. MDM2 ubiquitination of the HBP1 transcription factor targets it for degradation, thereby delaying DNA damage repair and enhancing tumorigenesis (Cao et al. 2019). Additionally, MDMX both plays a role in genome instability via association with Nbs1 (Carrillo et al. 2015) and also potentially plays a crucial role in DNA replication; its loss delays replication fork progression and sensitizes tumor cells to gemcitabine, suggesting that it may play a role in malignancy (Wohlberedt et al. 2020).
Invasion and metastasis
MDM2 has been linked to the EMT, a crucial step in metastasis, and in a p53-independent fashion and as mentioned previously, MDM2 regulates E-cadherin, one of the core markers of the EMT (Yang et al. 2006; for review, see Sun and Tang 2016). Further work supports the role of MDM2 in additional steps in the complex process of metastasis and investigates an emerging role for MDMX. Knockdown of MDM2 or MDMX reduces circulating tumor cells in triple-negative breast cancer without affecting proliferation; however, only MDMX is necessary for maintaining levels of metastatic factor CXCR4 (Gao et al. 2019). MDM2 also drives the EMT through the TGF-β–Smad pathway in ovarian cancer by promoting Snail/Slug expression and activating Smad2/3 (Chen et al. 2017). MA242, a small molecule that degrades MDM2 and is discussed in detail in the next section, reduces metastasis of hepatocellular carcinoma cells in vitro and in vivo (Wang et al. 2019).
Roles of MDM2 and MDMX in tumorigenesis
Given the amplification of MDM2 and MDMX in cancers spanning the spectrum of p53 expression, their role in tumorigenesis has been an understandable topic of interest. In the context of p53-independent tumorigenesis, MDM2 induction, along with NRF2-signaling, is linked to transformation of acinar cells and promotes conversion of premalignant pancreatic intraepithelial neoplasia lesions into pancreatic ductal adenocarcinoma (Todoric et al. 2017). In breast cancer cell lines, MDM2 was also shown to be necessary for colony formation in soft agar (Kundu et al. 2017).
MDMX activity is also linked to tumorigenesis: Overexpression of the MDMX gene in p53-null mice decreases survival, increases the number of tumors, and alters the spectrum of tumors in male mice (Xiong et al. 2017). Additionally, previous work identified an MDMX splice variant (MDMX-S) as a possible target of interest in tumorigenesis, due to notable overexpression and concomitant poor prognosis in several cancers (Bartel et al. 2005; Lenos et al. 2012; Liu et al. 2012; Grawenda et al. 2015; Dewaele et al. 2016). However, while MDMX-S is also overexpressed in B-cell chronic lymphocytic leukemia (B-CLL), it does not cause tumor formation and does not contribute to tumor aggressiveness; instead, MDMX-S overexpression in B-CLL is a result of tumorigenesis (Pant et al. 2017). Nevertheless, MDMX may still have potential as a prognostic biomarker.
MDM2 and chemotherapeutic responses
MDM2 confers resistance to the HER2 inhibitor, lapatinib, in HER2+ breast cancer cell lines through ubiquitin-mediated degradation of HUWE1 (Kurokawa et al. 2013). This finding has been expanded subsequently to demonstrate an inverse relationship between MDM2 and HUWE1 protein, but not mRNA, levels in vivo, which may help further establish a mechanism through which HER2+ breast cancers develop drug resistance (Canfield et al. 2016). Additionally, various cancer cell lines with overexpressed MDM2 also have shown resistance to topoisomerase II inhibitors, but not other DNA damaging agents, and this resistance requires intact MDM2 ubiquitin ligase function (Senturk et al. 2017).
MDM2 has been identified as a potentially druggable target in other cancers. In hepatocellular carcinomas, MA242, the inhibitor of MDM2 that can also inhibit NFAT1 as discussed in the next section, inhibits growth and metastasis (Wang et al. 2019). Cotreatment with interferon-α (IFNα) and nutlin (more commonly used to block MDM2-mediated inhibition of p53 but exhibits some inhibitory effects on MDM2 even in the absence of p53 as described in the next section) in p53-null non-small-cell lung carcinoma cells synergistically inhibits proliferation (Shuvalov et al. 2018).
MDM2 and MDMX have clinical roles outside of cancer
While MDM2 and MDMX have been most broadly studied for their cancer-related functions, they have also been linked to a range of nonmalignant diseases, including inflammatory and autoimmune disease, neurodegeneration, kidney disease, diabetes, and cardiovascular disease (Thomasova et al. 2012; Wang et al. 2020). MDM2 in particular has risen as a topic of interest in various other organ systems, including adipocyte conversion mediated through STAT activation (Hallenborg et al. 2016), cardiac responsiveness to β-adrenergic receptor stimulation (Jean-Charles et al. 2017), enhanced HIV-1 Tat protein-mediated viral replication (Raja et al. 2017), control of circadian period length through degradation of PER2 (Liu et al. 2018a), cellular senescence in the premature aging condition, Werner syndrome (Liu et al. 2019a), and the pathogenesis of idiopathic pulmonary arterial hypertension through destabilization of angiotensin-converting enzyme 2 (ACE2) (Shen et al. 2020)
We discuss below additional discoveries of p53-independent activities of MDM2 and MDMX outside of cancer.
Metabolism
While p53 is a well-known regulator of cellular metabolism, MDM2 and MDMX can also independently modulate metabolic pathways. MDM2 was shown to be recruited to chromatin in response to starvation and oxidative stress in a post-transcriptional manner, and chromatin-bound MDM2 cooperates with transcription factors ATF3 and ATF4 to control genes involved in serine metabolism (Riscal et al. 2016). This transcriptional role is independent of MDM2's E3 ligase activity but is negatively regulated by its central acidic domain. Such regulation serves to restore cellular oxidative homeostasis. MDM2 is also able to regulate mitochondrial dynamics to alter the energy homeostasis of cells (Rubio-Patiño et al. 2019). In two recent studies, MDM2 was shown to inhibit the activity of mitochondrial complex 1 to promote oxidative stress (Arena et al. 2018; Elkholi et al. 2019). MDM2 prevents mitochondrial localization of NDUFS1 to cause the destabilization of complex 1 (Elkholi et al. 2019). On the other hand, in response to oxidative stress, MDM2 was shown to be recruited to the mitochondria to down-regulate NADH dehydrogenase 6 (MT-ND6) in order to reduce the activity of complex 1 (Arena et al. 2018).
Additionally, in a mouse model of lipodystrophy, MDM2 was shown to control certain aspects of adipocyte differentiation independently of p53. The absence of this control led to various metabolic disorders, many of which are related to dysfunctional lipid metabolism (Liu et al. 2018b). MDM2 has also been shown to regulate certain members of the PPAR family that are well-known master regulators of lipid homeostasis (Kersten 2008; Gopinathan et al. 2009). By controlling the transcriptional activity of PPARα through ubiquitination (Gopinathan et al. 2009) and the stability of PPARγ through NEDDylation (Park et al. 2016), MDM2 can have a diverse influence on the global lipid metabolism of cells. In fact, during ferroptosis, the MDM2–MDMX heterocomplex can modulate the lipid profile of cells under stress through mediating the transcriptional activity of PPARα (Venkatesh et al. 2020). MDMX has also been reported to promote the excessive accumulation of fat in mice (Kon et al. 2018). These reports therefore suggest that MDM2 and MDMX might have the potential of being targeted to treat metabolic disorders.
Inflammation
MDM2 has a complex relationship with the well-studied inflammatory factor NF-κB, serving to either induce or suppress NF-κB signaling (Thomasova et al. 2012). p53-independent MDM2 regulation of NF-κB signaling has been demonstrated to attenuate ocular inflammation, although since some of the pathological uveitis findings were performed on p53-expressing mice, the extent to which MDM2 may influence uveitis independently of p53 remains to be seen (Fan et al. 2018). MDM2 also promotes progression of inflammatory kidney disease in a two-pronged attack: It stimulates glomerular inflammation through NF-κB-mediated cytokine induction in a p53-independent fashion while also promoting proliferation in parietal epithelial cells and crescent formation where, in this case, p53 is now required (Mulay et al. 2016).
MDM2 and Notch
The Notch signaling pathway, most well studied as a regulator of cell fate in development and as being dysregulated in cancer, has been connected to MDM2 in several interesting ways. Notch1 stimulates MDM2 activity, and through MDM2 binding at the Notch intracellular domain (NICD), Notch1 is also ubiquitinated and activated by MDM2 (Wade et al. 2010; Pettersson et al. 2013). Numb, a negative regulator of Notch1, also binds and inhibits MDM2 (Juven-Gershon et al. 1998; Wade et al. 2010).
Clinically, MDM2 is further implicated in kidney disease beyond the previously described inflammatory pathways. In particular, MDM2 is up-regulated in tubulointerstitial fibrosis and unilateral urethral obstruction and is necessary for activation of collagen-producing fibroblasts by ubiquitinating Notch1, leading to proteasome degradation (Ye et al. 2017). In contrast to previous findings that MDM2 activates Notch via ubiquitination in cancer cells, Ye et al. (2017) suggest that MDM2-mediated ubiquitination of Notch1 specifically in fibroblasts leads to proteasome-mediated degradation. However, other work indicates that MDM2 is necessary for Notch1 activation in glomerular mesangial cells in the setting of hyperglycemia and diabetic kidney disease (Lei et al. 2017).
Outside of the kidney, MDM2 activates Notch1 signaling in lung club and alveolar cells and induces DNA replication and proliferation in lung progenitor cells in response to chemical- or radiation-induced injury (Singh et al. 2019). These varied findings suggest that MDM2 may have a wide range of effects on Notch1 signaling and physiological activity, depending on disease, organ, and cell type.
In summary, the p53-independent roles of MDM2 and MDMX are myriad and complex. This suggests that compounds that can affect their activities separately or together may be harnessed to combat their deleterious roles in cancer or other pathologies. Indeed, to date a plethora of small molecules have been shown to affect the MDMs that are described in the next part of our review.
Inhibitors of MDM2 and MDMX: strength in numbers
Given the complex nature of MDM2 and MDMX roles in many diseases, it follows that isolation or synthesis of drugs that might mitigate these roles is worthy of significant research and development. Fortunately, efforts to produce MDM2 inhibitors (and to a lesser extent agents that inhibit MDMX) have been underway for many years, albeit in the context of blocking the abilities of these two proteins to inhibit p53, and by now, numerous pharmacological antagonists have been designed to reactivate p53 for cancer treatment (Qin et al. 2012; Tisato et al. 2017; Fang et al. 2020).
Small molecules, often referred to as MDM2 or MDMX inhibitors, are generally also shown to be either MDM2-p53 or MDMX-p53 inhibitors. The first such agent to be identified, nutlin, has been invaluable for the p53 research community and has the added distinction of being one of the first small molecules that can efficiently disrupt the interaction between two full-length proteins (Vassilev et al. 2004). While nutlin and the many subsequent drugs that were developed to specifically separate MDM2 (and more rarely MDMX) from p53 have not generally been considered useful outside the context of p53, even nutlin can disrupt MDM2 from binding the p53 homolog p73 (Lau et al. 2008), as well as E2F (Ambrosini et al. 2007). Note that another quite commonly used inhibitor, RITA, separates p53 and MDM2 but does so by binding to p53 (Issaeva et al. 2004) and as such would not be useful for studies on p53-independent regulation of MDM2 and MDMX.
Several MDM2 or MDMX inhibitors that do not disrupt their interactions with p53 have also been discovered and these are summarized in Table 1 along with those that separate p53 from MDM2 but can also disrupt the interactions of MDM2 with other proteins. We have categorized these into three main classes of MDM2 or MDMX antagonists that affect (1) the transcriptional regulation of MDM2 or MDMX, (2) the protein stability or post-translational regulation of MDM2 or MDMX, and (3) the E3 ligase function of either the MDM2 homodimer or the MDM2–MDMX heterodimer. The varying mechanisms of action of these inhibitors provide different ways of targeting MDM2 and MDMX, depending on the needs of the biological question at hand.
Table 1.
Inhibitors of MDM2 and MDMX
There are some inhibitors that have been primarily categorized into any one of the above classes but are able to inhibit multiple aspects of MDM2 or MDMX as well. While some of these inhibitors truly have multiple independent functions (such as SQ, MA242, CP1-7C, and serdemetan), there are others where the functions could be interdependent. For example, adriamycin is reported to lower the levels of MDM2 mRNA and to also cause a proteasome-independent decrease in MDM2 protein (Ma et al. 2000). Similarly, tanespimycin causes the destabilization of MDMX, down-regulation of MDM2 protein, and disruption of the MDM2–MDMX heterocomplex (Vaseva et al. 2011). In these cases, the latter effect(s) could simply be due to the ability of MDMX to regulate the stability of MDM2 and the necessity of both binding partners to be present in order to have a functional heterocomplex. As another example, the MMRi compounds also cause the degradation of both MDM2 and MDMX apart from blocking their RING interactions (Wu et al. 2015). It is quite possible that this is the main reason for prevention of complex formation, but the exact sequence of events and reasons behind the degradation are yet unknown. This also suggests that the MMRi compounds may additionally affect functions of MDM2 and MDMX that are independent of each other. The challenge in these situations lies in effectively decoupling the multiple effects of these small molecules.
It is important to evaluate the effect of these compounds in a truly p53-null setting in order to determine the uniquely p53-independent roles of MDM2 or MDMX. While CRISPR-Cas9 genetic ablation or RNA interference-based depletion of protein levels are great tools to experimentally create such contexts in cancer cell lines, a series of available mouse embryonic fibroblast lines with loss of MDM2 and p53, MDMX and p53, as well as loss of MDM2, MDMX, and p53 are also prominent resources (Barboza et al. 2008). Usage of such tools has proven to be very useful in the discovery of another class of inhibitors that comprises compounds mainly known for their ability to disrupt the interactions of MDM2 or MDMX with p53 but can also inhibit some p53-independent interactions of MDM2 or MDMX (such as nutlin and LQFM030). This sparks the need to evaluate whether other well-known and successful p53–MDM2/MDMX inhibitors can also have such multipronged effects on the functions of MDM2 or MDMX. For example, azadiractin is a natural product obtained from the neem tree that competitively binds to the N-terminal p53 binding site of MDM2 and has additionally been shown to induce p53-independent apoptosis through disrupted NF-κB signaling (Gupta et al. 2018). Given the known roles of MDM2 in regulating the NF-κB pathway (Thomasova et al. 2012), it would be interesting to evaluate the potential of azadiractin to impede this p53-independent role of MDM2, akin to the effect of nutlin (Mulay et al. 2012, 2016; Fan et al. 2018). DS-5272 is another inhibitor of p53–MDM2 interaction, which has also been shown to inhibit the regulation of NF-κB by MDM2, likely in a p53-independent manner (Fujikura et al. 2018). However, DS-5272 also needs to be evaluated in the complete absence of p53 as done in comparable studies with nutlin (Mulay et al. 2012, 2016) in order to confirm that this is indeed a p53-independent effect on the function of MDM2. In line with this, it is important to note that a few of the small molecules listed in Table 1 have not yet been tested in cells devoid of p53, and instead, cells having mutant p53 were used to determine their p53-independent roles, thus necessitating further evaluation of these inhibitors as well.
Apart from the biological question at hand, the choice of these tools should also be based on their limitations. Since the various antagonists can also have other effects based on the system of use, they must be carefully assessed before being used to infer the functions of MDM2 and MDMX. As listed in Table 1, while there are some inhibitors whose nonspecific off-target effects have come to light (such as serdemetan, HLI, and NSC 207895), there are others that are known to specifically target other proteins apart from MDM2 or MDMX (such as adriamycin, tanespimycin, SP141, and SQ). There are also some inhibitors whose exact mechanism of action is not yet fully elucidated, and they could potentially have multiple effects beyond what is reported, even if they only affect other interactions of MDM2 or MDMX. Since the p53-independent roles of MDMX and the MDM2–MDMX complex are yet largely unexplored, it would also be interesting to test the effect of inhibitors that are only reported to affect the functions of MDM2 alone on various aspects of MDMX and the MDM2–MDMX heterocomplex.
These limitations highlight the need to use multiple approaches that include numerous small molecules with different mechanisms complemented with genetic techniques in order to make robust conclusions. Even though each method has its own drawbacks, if multiple methods concur on the core observations, there would be higher confidence in the conclusions.
Apart from the inhibitors listed here, we refer the reader to two comprehensive reviews of various natural products that are robust inhibitors of MDM2 and MDMX, both in the context of p53 and otherwise (Qin et al. 2012, 2018a). For example, JapA, gambogic acid, InuA, and berberine can cause a down-regulation of the MDM2 transcript as well as promote the degradation of MDM2 protein, thus also effectively blocking the formation of the MDM2–MDMX complex (Qin et al. 2018a). On the other hand, sempervirine is another natural product that only inhibits the E3 ligase activity of MDM2 (Sasiela et al. 2008), and its effect on the activity of the MDM2–MDMX heterocomplex is yet unknown.
Currently, there is no definitive proof of the clinical utility of any of the MDM2-X inhibitors listed in Table 1, as most of the MDM2/X inhibitors tested in the clinic are those that primarily target the interaction of MDM2/X with p53 (Tisato et al. 2017; Jiang and Zawacka-Pankau 2020). In support of a therapeutic advantage of targeting the p53-independent roles of MDM2/X, some of these inhibitors have also shown an effect in certain patients’ tumors harboring a mutation in p53 (Burgess et al. 2016). For example, the clinical activity of RG7112, a molecule that exert its effects via competitive binding to the p53 pocket of MDM2 (Vu et al. 2013), in AML patients correlated with the expression levels of MDM2 but not the status of p53 (Andreeff et al. 2016). In this study, two patients harboring different mutations of p53 did respond positively to the drug. While it is possible that the MDM2 inhibitor was somehow able to restore wild-type p53 activity in these patients, it is also suggestive of the involvement of p53-independent roles of MDM2 in the malignancy of these cases. Additionally, the MDM2 E3 ligase inhibitor serdemetan has been tested in phase 1 clinical trials (ClinicalTrials.gov no. NCT00676810). Although these trials also revealed issues of toxicity and potential off-target effects (Karni-Schmidt et al. 2016), it is possible that these off-target effects were simply dependent on MDM2 but not p53, as serdemtan is also reported to target the p53-independent functions of MDM2 (these reports are listed in Table 1). Taken together with the study where serdemetan elicited an anticancerous response in multiple patient-derived xenografts harboring either mutant or wild-type p53 (Chargari et al. 2011), there is a need for additional research into its clinical effectiveness. Given the myriad p53-independent functions of MDM2 and MDMX both in tumorigenesis and in physiological maladies, as highlighted in this review, we believe that targeting MDM2/X has significant clinical potential outside of the context of p53. That said, a lot more research is required before strong conclusions can be drawn in this regard.
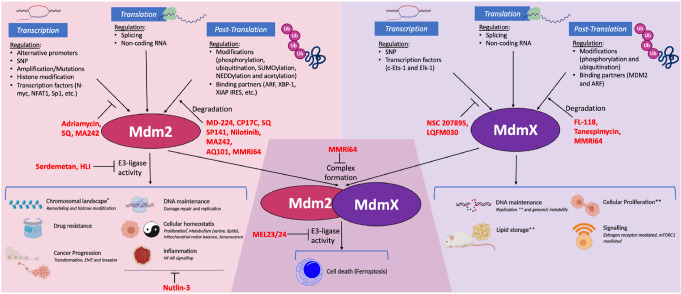
We propose that future research on the p53-independent activities and interactions of MDM2 and MDMX will benefit from wider use of the rather impressive tools that have been described in this section. Figure 5 depicts both the varied inhibitors and different points of attack that the compounds tabulated in this section use to interfere with MDM2 and MDMX and, in turn, how each might affect the many distinct roles that these two proteins can play in cells.
Figure 5.
A look at MDM2 and MDMX with p53-blind glasses. Several proteins other than p53 regulate MDM2 and MDMX at the transcriptional, translational, and post-translational levels through distinct mechanisms. These mechanisms are summarized at the top. The bottom depicts some of the most prominent p53-independent functions of MDM2 and MDMX, both individually and together as a complex. In red are the different inhibitors of these two proteins that are capable of modifying the ability of MDM2 and MDMX to undertake these p53-independent roles. The figure also briefly indicates the mechanism of action of these drugs; while some of them impair the regulation of MDM2 and/or MDMX (e.g., SQ, SP141, NSC 207895, and FL-118), others can directly affect their functionality (e.g., HLI, MEL23, and Nutlin-3).
Epilog: what the future of research on MDM2 and MDMX may hold
The vast majority of studies published on the MDMs have focused on their respective abilities to restrain wild-type p53. However, as outlined in this review, MDM2 and MDMX have a significant impact on cells that lack wild-type p53. Here, in summary, we pose questions that might inform future research.
When does regulation of MDM2 differ in cells that contain or lack p53?
One of the first discoveries concerning MDM2 was that it is a transcriptional target of p53, thereby establishing the negative feedback circuit that must be broken in order for p53 to be unleashed. As we outline earlier in this review, since then, there have been a multitude of studies documenting the complex ways that these proteins are controlled that do not directly involve p53, extending from gene expression at several levels to protein modification. We still do not understand the extent to which p53 regulates these many modes of regulation. Furthermore, when MDM2 is released from p53, its own levels rise dramatically due to activation of p53, and depending on the stimulus, it can become modified in myriad ways. Reciprocally, we still do not understand how basal MDM2 gene expression is regulated, nor do we understand how MDMX is expressed either in basal conditions or after different stimuli. At the protein level, it will be imperative to understand the function (or lack thereof) of all MDM2 and MDMX isoforms found in cells. Most relevant to this review, to what extent do altered levels and modifications of MDM2 matter when cells lack p53? Is one of the functions of activated p53 to alter the cellular response to having more or differently modified versions of MDM2? Experimentally, the use of either knockout mice or the generation of human cell lines via gene editing can begin to address this and is highly relevant to the next question we pose.
When do p53-independent activities of MDM2 or MDMX come into play?
Aside from experimentally (and therefore artificially) engineered cells and animals that lack wild-type p53, likely the only naturally occurring situations where cells have lost wild-type p53 are those that occur as a result of oncogenic transformation. Since solid tumors lacking wild-type p53 range from very high frequency (e.g., ovarian cancer) to very low (e.g., melanoma, testicular cancer, sarcoma, and cervical cancer) (Olivier et al. 2010), the question is highly context dependent. In many but not all such cases, tumors express high levels of mutant p53, and there is mounting evidence that in some settings, such mutant p53 proteins are eliciting protumorigenic activities sometimes referred to as “gain of function” (Brosh and Rotter 2009; Freed-Pastor and Prives 2012; Muller and Vousden 2014). Do mutant p53 proteins cooperate with the MDMs to elicit their gain-of-function activities? If so, do different mutant p53 alleles differ in their ability to do so? Do MDM2 or MDMX work similarly in cells that lack any p53 protein compared with those that express mutant proteins with documented oncogenic activities? Conversely, do different MDM2 spliced isoforms play critical roles in mutant p53 pro-oncogenic activities as suggested in a previous study (Zheng et al. 2013)?
Furthermore, it has been speculated that wild-type p53-mediated survival functions (such as promoting DNA repair) may provide a survival advantage to those tumors that harbor wild-type p53. In either setting, do the p53-independent activities of MDM2 and MDMX contribute to the evolution of such tumors that harbor wild-type p53? Questions such as these will require a combination of experimental and patients’ data sets, prior to reaching any firm conclusions.
In a larger sense, we pose the question as to how these p53-independent functions drive tumorigenesis. How do the roles and regulation of MDM2 and MDMX drive their functions separately and together in clinical pathology? It will also be critical to link mechanisms that control expression and functions of MDM2 and MDMX to their regulation of pathophysiological processes. While numerous upstream regulators and downstream effects of MDM2 and MDMX have been identified as described in this review, it is not yet known which of such myriad processes have more significant effects on disease
When do MDM2 and MDMX work separately or together in the absence of wild-type p53?
Answering this question is quite challenging. Despite a number of elegant studies documenting how MDM2 and MDMX interact with and influence each other's activities, there are scant reports documenting their functions as a heterocomplex in cells lacking wild-type p53. With the possible exception of MEL23, most compounds that inhibit MDM2 or MDMX activity do so by changing the levels of one or the other. Searching for more compounds that uniquely affect the heterocomplex without changing the levels of either of the two proteins will extend our ability to address this question. In the meantime, there are experimentally derived mutant forms of both proteins that cannot form the heterocomplex; introducing these either as ectopic proteins or perhaps using gene editing technology to allow them to be endogenously expressed (under conditions where the presence of such mutant proteins is compatible with cell viability) would help to address this query.
Other questions as well need to be answered. What is the extent of the E3 ligase activity of the MDM2–MDMX complex, and importantly, what are the key targets in cells that lack p53? Does the complex also add other UBLs, such as ATGs or FAT10? What is the biological relevance of the different post-translational modifications that the complex can mediate? We will need as full a compendium of MDM2/MDMX targets as current state of the art proteomic screens and protocols can provide to approach these questions.
Another complicated aspect concerning the functionality of the MDM2–MDMX complex resides in their ability to both polyubiquitylate and monoubiquitylate their targets. As described earlier in this review, MDM2 is able to regulate different aspects of its E3 ligase targets, including p53, outside of mere degradation depending on whether it performs polyubiquitylation or monoubiquitylation (Li et al. 2003; Marine and Lozano 2010). Since there are only a few physiologically confirmed targets of the MDM2–MDMX complex outside of p53, it is not clear whether the complex can regulate the same proteins as MDM2. A considerable undertaking for the future would be to identify both ubiquitylation targets and binding partners of the MDM2–MDMX heterocomplex.
Can MDM2 and MDMX be tumor suppressive?
The vast majority of studies on MDM2 and MDMX are consistent with their functioning to promote oncogenesis, either by suppressing p53 or, as we have reviewed herein, on their own. However, since the first surprising report that showed that overexpressed MDM2 can inhibit growth (Brown et al. 1998), there have been a handful of reports that are consistent with MDM2 at least playing roles that are actually growth and tumor suppressive, which is discussed in an earlier excellent review (Manfredi 2010). Our discovery that MDM2 and MDMX promote ferroptosis (Venkatesh et al. 2020), a potentially tumor-suppressive process (Stockwell et al. 2017), supports the suggestion that the context-dependent role of MDM2 and MDMX in tumor suppression is worth serious consideration. Indeed, a recent review pointed out that MDM2 or MDMX overexpression is a relatively rare and tumor type-restricted occurrence (Dobbelstein and Levine 2020) when compared with the extremely frequent occurrence of p53 mutations across a wide swath of cancers. The same trend applies to SNPs in MDM2 and MDMX, which are only present in some specific tumor types. Since, as we have outlined in this review, there are a plethora of inhibitors of MDM2 and MDMX, it may become important to define the context in which they may function to prevent tumor formation when considering whether or not to use such agents.
As MDM2 and MDMX continue to emerge from the shadow of p53 in their own right, we hope that future studies will address these interesting questions. With the increasing number of tools in hand that we present in Table 1, it is hoped that harnessing the answers to such questions can lead to better and more effective therapies to alter the roles of these two proteins in disease.
Acknowledgments
This work was supported by grant CA220526 to C.P. The figures in this review were prepared using BioRender software.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.347872.120.
Competing interest statement
The authors declare no competing interests.
References
- Akande OE, Damle PK, Pop M, Sherman NE, Szomju BB, Litovchick LV, Grossman SR. 2019. DBC1 regulates p53 stability via inhibition of CBP-dependent p53 polyubiquitination. Cell Rep 26: 3323–3335.e4. 10.1016/j.celrep.2019.02.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G, Sambol EB, Carvajal D, Vassilev LT, Singer S, Schwartz GK. 2007. Mouse double minute antagonist nutlin-3a enhances chemotherapy-induced apoptosis in cancer cells with mutant p53 by activating E2F1. Oncogene 26: 3473–3481. 10.1038/sj.onc.1210136 [DOI] [PubMed] [Google Scholar]
- Andreeff M, Kelly KR, Yee K, Assouline S, Strair R, Popplewell L, Bowen D, Martinelli G, Drummond MW, Vyas P, et al. 2016. Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin Cancer Res. 10.1158/1078-0432.ccr-15-0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar SL, Wulaningsih W, Watkins J. 2017. Profile of the breast cancer susceptibility marker rs4245739 identifies a role for miRNAs. Cancer Biol Med 14: 387–395. 10.20892/j.issn.2095-3941.2017.0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arena G, Cissé MY, Pyrdziak S, Chatre L, Riscal R, Fuentes M, Arnold JJ, Kastner M, Gayte L, Bertrand-Gaday C, et al. 2018. Mitochondrial MDM2 regulates respiratory complex I activity independently of p53. Mol Cell 69: 594–609.e8. 10.1016/j.molcel.2018.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts J, Page M, Valckx A, Blattner C, Kulikov R, Floren W, van Nuffel L, Janssen L, King P, Masure S, et al. 2008. JNJ-26854165—a novel hdm2 antagonist in clinical development showing broad-spectrum preclinical antitumor activity against solid malignancies. Cancer Res 68: 1592. [Google Scholar]
- Banks D, Wu M, Higa LA, Gavrilova N, Quan J, Ye T, Kobayashi R, Sun H, Zhang H. 2006. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle 5: 1719–1729. 10.4161/cc.5.15.3150 [DOI] [PubMed] [Google Scholar]
- Barboza JA, Iwakuma T, Terzian T, El-Naggar AK, Lozano G. 2008. Mdm2 and Mdm4 loss regulates distinct p53 activities. Mol Cancer Res 6: 947–954. 10.1158/1541-7786.MCR-07-2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel F, Schulz J, Böhnke A, Blümke K, Kappler M, Bache M, Schmidt H, Würl P, Taubert H, Hauptmann S. 2005. Significance of HDMX-S (or MDM4) mRNA splice variant overexpression and HDMX gene amplification on primary soft tissue sarcoma prognosis. Int J Cancer 117: 469–475. 10.1002/ijc.21206 [DOI] [PubMed] [Google Scholar]
- Barzalobre-Gerónimo R, Flores-López LA, Baiza-Gutman LA, Cruz M, García-Macedo R, Ávalos-Rodriguez A, Contreras-Ramos A, Díaz-Flores A, Ortega-Camarillo C. 2015. Hyperglycemia promotes p53-Mdm2 interaction but reduces p53 ubiquitination in RINm5F cells. Mol Cell Biochem 405: 257–264. 10.1007/s11010-015-2416-0 [DOI] [PubMed] [Google Scholar]
- Batuello CN, Hauck PM, Gendron JM, Lehman JA, Mayo LD. 2015. Src phosphorylation converts Mdm2 from a ubiquitinating to a neddylating E3 ligase. Proc Natl Acad Sci 112: 1749–1754. 10.1073/pnas.1416656112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. 2013. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev 27: 1903–1916. 10.1101/gad.219899.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biderman L, Manley JL, Prives C. 2012. Mdm2 and MdmX as regulators of gene expression. Genes Cancer 3: 264–273. 10.1177/1947601912455331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlman S, Manfredi JJ. 2014. p53-independent effects of Mdm2. Subcell Biochem 85: 235–246. 10.1007/978-94-017-9211-0_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119: 591–602. 10.1016/j.cell.2004.11.022 [DOI] [PubMed] [Google Scholar]
- Brosh R, Rotter V. 2009. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer 9: 701–713. 10.1038/nrc2693 [DOI] [PubMed] [Google Scholar]
- Brown DR, Thomas CA, Deb SP. 1998. The human oncoprotein MDM2 arrests the cell cycle: elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J 17: 2513–2525. 10.1093/emboj/17.9.2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. 2016. Clinical overview of MDM2/X-targeted therapies. Front Oncol 6: 7. 10.3389/fonc.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busuttil V, Droin N, McCormick L, Bernassola F, Candi E, Melino G, Green DR. 2010. NF-κB inhibits T-cell activation-induced, p73-dependent cell death by induction of MDM2. Proc Natl Acad Sci 107: 18061–18066. 10.1073/pnas.1006163107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Zhang P, Xu M, Yan L, Liu N, Wu X. 2019a. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol 234: 11391–11400. 10.1002/jcp.27796 [DOI] [PubMed] [Google Scholar]
- Cai Y, Hao Y, Ren H, Dang Z, Xu H, Xue X, Gao Y. 2019b. miR-1305 inhibits the progression of non-small cell lung cancer by regulating MDM2. Cancer Manag Res 11: 9529–9540. 10.2147/CMAR.S220568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield K, Wells W, Geradts J, Kinlaw WB, Cheng C, Kurokawa M. 2016. Inverse association between MDM2 and HUWE1 protein expression levels in human breast cancer and liposarcoma. Int J Clin Exp Pathol 9: 6342–6349. [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Xue J, Cheng Y, Wang J, Liu Y, Li H, Jiang W, Li G, Gui Y, Zhang X. 2019. MDM2 promotes genome instability by ubiquitinating the transcription factor HBP1. Oncogene 38: 4835–4855. 10.1038/s41388-019-0761-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr MI, Jones SN. 2016. Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis. Transl Cancer Res 5: 707–724. 10.21037/tcr.2016.11.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo AM, Bouska A, Arrate MP, Eischen CM. 2015. Mdmx promotes genomic instability independent of p53 and Mdm2. Oncogene 34: 846–856. 10.1038/onc.2014.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha PC, Satake W, Ando-Kanagawa Y, Yamamoto K, Murata M, Toda T. 2020. Genome-wide association study identifies zonisamide responsive gene in Parkinson's disease patients. J Hum Genet 65: 693–704. 10.1038/s10038-020-0760-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Freeman DJ, Wu H. 2004. PTEN regulates Mdm2 expression through the P1 promoter. J Biol Chem 279: 29841–29848. 10.1074/jbc.M401488200 [DOI] [PubMed] [Google Scholar]
- Chargari C, Leteur C, Angevin E, Bashir T, Schoentjes B, Arts J, Janicot M, Bourhis J, Deutsch E. 2011. Preclinical assessment of JNJ-26854165 (serdemetan), a novel tryptamine compound with radiosensitizing activity in vitro and in tumor xenografts. Cancer Lett 312: 209–218. 10.1016/j.canlet.2011.08.011 [DOI] [PubMed] [Google Scholar]
- Chen L, Chen J. 2003. MDM2-ARF complex regulates p53 sumoylation. Oncogene 22: 5348–5357. 10.1038/sj.onc.1206851 [DOI] [PubMed] [Google Scholar]
- Chen L, Wang H. 2019. Nicotine promotes human papillomavirus (HPV)-immortalized cervical epithelial cells (H8) proliferation by activating RPS27a-Mdm2-P53 pathway in vitro. Toxicol Sci 167: 408–418. 10.1093/toxsci/kfy246 [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang DD, Wu YP, Su D, Zhou TY, Gai RH, Fu YY, Zheng L, He QJ, Zhu H, et al. 2017. MDM2 promotes epithelial–mesenchymal transition and metastasis of ovarian cancer SKOV3 cells. Br J Cancer 117: 1192–1201. 10.1038/bjc.2017.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Zhou T, Ma T, Cao T, Yu Z. 2019a. Oncogenic effect of PHLDB2 is associated with epithelial–mesenchymal transition and E-cadherin regulation in colorectal cancer. Cancer Cell Int 19: 184. 10.1186/s12935-019-0903-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Cai G, Liao Z, Lin K, Li G, Li Y. 2019b. miRNA-766 induces apoptosis of human colon cancer cells through the p53/Bax signaling pathway by MDM4. Exp Ther Med 17: 4100–4108. 10.3892/etm.2019.7436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TH, Cohen SN. 2007. Human MDM2 isoforms translated differentially on constitutive versus p53-regulated transcripts have distinct functions in the p53/MDM2 and TSG101/MDM2 feedback control loops. Mol Cell Biol 27: 111–119. 10.1128/MCB.00235-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YM, An S, Bae S, Jung JH. 2019. Mdm2 is required for HDAC3 monoubiquitination and stability. Biochem Biophys Res Commun 517: 353–358. 10.1016/j.bbrc.2019.07.052 [DOI] [PubMed] [Google Scholar]
- Comiskey DF Jr, Jacob AG, Singh RK, Tapia-Santos AS, Chandler DS. 2015. Splicing factor SRSF1 negatively regulates alternative splicing of MDM2 under damage. Nucleic Acids Res 43: 4202–4218. 10.1093/nar/gkv223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comiskey DF Jr, Montes M, Khurshid S, Singh RK, Chandler DS. 2020. SRSF2 regulation of MDM2 reveals splicing as a therapeutic vulnerability of the p53 pathway. Mol Cancer Res 18: 194–203. 10.1158/1541-7786.MCR-19-0541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa AF, Gomes SZ, Lorenzon-Ojea AR, Martucci M, Faria MR, Pinto Ddos S Jr, Oliveira SF, Ietta F, Paulesu L, Bevilacqua E. 2016. Macrophage migration inhibitory factor induces phosphorylation of Mdm2 mediated by phosphatidylinositol 3-kinase/Akt kinase: role of this pathway in decidual cell survival. Placenta 41: 27–38. 10.1016/j.placenta.2016.03.001 [DOI] [PubMed] [Google Scholar]
- da Mota MF, Cortez AP, Benfica PL, Rodrigues BDS, Castro TF, Macedo LM, Castro CH, Lião LM, de Carvalho FS, Romeiro LA, et al. 2016. Induction of apoptosis in Ehrlich ascites tumour cells via p53 activation by a novel small-molecule MDM2 inhibitor—LQFM030. J Pharm Pharmacol 68: 1143–1159. 10.1111/jphp.12573 [DOI] [PubMed] [Google Scholar]
- da Mota MF, de Carvalho FS, de Ávila RI, de Ávila PHM, Cortez AP, Menegatti R, Sabino JR, Dos Santos TRM, Gomes SA, da Cunha LC, et al. 2018. LQFM030 reduced Ehrlich ascites tumor cell proliferation and VEGF levels. Life Sci 201: 1–8. 10.1016/j.lfs.2017.12.029 [DOI] [PubMed] [Google Scholar]
- Deisenroth C, Franklin DA, Zhang Y. 2016. The evolution of the ribosomal protein-MDM2-p53 pathway. Cold Spring Harb Perspect Med 6: a026138. 10.1101/cshperspect.a026138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Ribeiro H, Cortez AP, de Ávila RI, da Silva ACG, de Carvalho FS, Menegatti R, Lião LM, Valadares MC. 2020. Small-molecule MDM2 inhibitor LQFM030-induced apoptosis in p53-null K562 chronic myeloid leukemia cells. Fundam Clin Pharm 34: 444–457. 10.1111/fcp.12540 [DOI] [PubMed] [Google Scholar]
- de Polo A, Luo Z, Gerarduzzi C, Chen X, Little JB, Yuan ZM. 2017. AXL receptor signalling suppresses p53 in melanoma through stabilization of the MDMX–MDM2 complex. J Mol Cell Biol 9: 154–165. 10.1093/jmcb/mjw045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz RM, Madan R, Chien J, Dias WB, Slawson C. 2016. Changes in O-linked N-acetylglucosamine (O-GlcNAc) homeostasis activate the p53 pathway in ovarian cancer cells. J Biol Chem 291: 18897–18914. 10.1074/jbc.M116.734533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewaele M, Tabaglio T, Willekens K, Bezzi M, Teo SX, Low DH, Koh CM, Rambow F, Fiers M, Rogiers A, et al. 2016. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest 126: 68–84. 10.1172/JCI82534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D, Antonucci L, Nakagawa H, Kim JY, Glitzner E, Caruso S, Shalapour S, Yang L, Valasek MA, Lee S, et al. 2018. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell 33: 1061–1077.e6. 10.1016/j.ccell.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelstein M, Levine AJ. 2020. Mdm2: open questions. Cancer Sci 111: 2203–2211. 10.1111/cas.14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmesen C, Koeppel M, Dobbelstein M. 2008. Specific inhibition of Mdm2-mediated neddylation by Tip60. Cell Cycle 7: 222–231. 10.4161/cc.7.2.5185 [DOI] [PubMed] [Google Scholar]
- Dolezelova P, Cetkovska K, Vousden KH, Uldrijan S. 2012. Mutational analysis of Mdm2 C-terminal tail suggests an evolutionarily conserved role of its length in Mdm2 activity toward p53 and indicates structural differences between Mdm2 homodimers and Mdm2/MdmX heterodimers. Cell Cycle 11: 953–962. 10.4161/cc.11.5.19445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Gao X, Zhu Z, Yu Q, Bian S, Gao Y. 2012. A 40-bp insertion/deletion polymorphism in the constitutive promoter of MDM2 confers risk for hepatocellular carcinoma in a Chinese population. Gene 497: 66–70. 10.1016/j.gene.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Eischen CM. 2017. Role of Mdm2 and Mdmx in DNA repair. J Mol Cell Biol 9: 69–73. 10.1093/jmcb/mjw052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkholi R, Abraham-Enachescu I, Trotta AP, Rubio-Patiño C, Mohammed JN, Luna-Vargas MPA, Gelles JD, Kaminetsky JR, Serasinghe MN, Zou C, et al. 2019. MDM2 integrates cellular respiration and apoptotic signaling through NDUFS1 and the mitochondrial network. Mol Cell 74: 452–465.e7. 10.1016/j.molcel.2019.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus R, Olivares-Illana V. 2014. MDM2's social network. Oncogene 33: 4365–4376. 10.1038/onc.2013.410 [DOI] [PubMed] [Google Scholar]
- Fakharzadeh SS, Trusko SP, George DL. 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 10: 1565–1569. 10.1002/j.1460-2075.1991.tb07676.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Zhang W, Ni A, Mahato B, Chavala SH. 2018. Inhibition of noncanonical murine double minute 2 homolog abrogates ocular inflammation through NF-κB suppression. Am J Pathol 188: 2087–2096. 10.1016/j.ajpath.2018.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. 2000. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. Journal of Biological Chemistry 275: 8945–8951. 10.1074/jbc.275.12.8945 [DOI] [PubMed] [Google Scholar]
- Fang Y, Liao G, Yu B. 2020. Small-molecule MDM2/X inhibitors and PROTAC degraders for cancer therapy: advances and perspectives. Acta Pharmaceutica Sinica B. 10: 1253–1278. 10.1016/j.apsb.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley KP, Adams CM, Mitra R, Eischen CM. 2017. Mdm2 is required for survival and growth of p53-deficient cancer cells. Cancer Res 77: 3823–3833. 10.1158/0008-5472.CAN-17-0809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C. 2012. Mutant p53: one name, many proteins. Genes Dev 26: 1268–1286. 10.1101/gad.190678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura T, Yasuda H, Iwakura T, Tsuji T, Anders H-J. 2018. MDM2 inhibitor ameliorates cisplatin-induced nephropathy via NFκΒ signal inhibition. Pharmacol Res Perspect 7: e00450. 10.1002/prp2.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansmo LB, Bjørnslett M, Halle MK, Salvesen HB, Dørum A, Birkeland E, Hveem K, Romundstad P, Vatten L, Lønning PE, et al. 2016a. The MDM4 SNP34091 (rs4245739) C-allele is associated with increased risk of ovarian—but not endometrial cancer. Tumour Biol 37: 10697–10702. 10.1007/s13277-016-4940-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansmo LB, Vatten L, Romundstad P, Hveem K, Ryan BM, Harris CC, Knappskog S, Lønning PE. 2016b. Associations between the MDM2 promoter P1 polymorphism del1518 (rs3730485) and incidence of cancer of the breast, lung, colon and prostate. Oncotarget 7: 28637–28646. 10.18632/oncotarget.8705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Xiong X, Pan W, Yang X, Zhou C, Yuan Q, Zhou L, Yang M. 2015. A regulatory MDM4 genetic variant locating in the binding sequence of multiple microRNAs contributes to susceptibility of small cell lung cancer. PLoS One 10: e0135647. 10.1371/journal.pone.0135647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K, Wang C, Jin X, Xiao J, Zhang E, Yang X, Wang D, Huang H, Yu L, Zhang P. 2016. RNF12 promotes p53-dependent cell growth suppression and apoptosis by targeting MDM2 for destruction. Cancer Lett 375: 133–141. 10.1016/j.canlet.2016.02.013 [DOI] [PubMed] [Google Scholar]
- Gao C, Xiao G, Piersigilli A, Gou J, Ogunwobi O, Bargonetti J. 2019. Context-dependent roles of MDMX (MDM4) and MDM2 in breast cancer proliferation and circulating tumor cells. Breast Cancer Res 21: 5. 10.1186/s13058-018-1094-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Weghorst K, Berberich SJ. 2005. Mdmx inhibits ARF mediated Mdm2 sumoylation. Cell Cycle 4: 604–608. [PubMed] [Google Scholar]
- Gilkes DM, Pan Y, Coppola D, Yeatman T, Reuther GW, Chen J. 2008. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol 28: 1999–2010. 10.1128/MCB.01633-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giono LE, Resnick-Silverman L, Carvajal LA, St Clair S, Manfredi JJ. 2017. Mdm2 promotes Cdc25C protein degradation and delays cell cycle progression through the G2/M phase. Oncogene 36: 6762–6773. 10.1038/onc.2017.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinathan L, Hannon DB, Peters JM, Vanden Heuvel JP. 2009. Regulation of peroxisome proliferator-activated receptor-α by MDM2. Toxicol Sci 108: 48–58. 10.1093/toxsci/kfn260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawenda AM, Møller EK, Lam S, Repapi E, Teunisse AF, Alnæs GI, Børresen-Dale AL, Kristensen VN, Goding CR, Jochemsen AG, et al. 2015. Interaction between p53 mutation and a somatic HDMX biomarker better defines metastatic potential in breast cancer. Cancer Res 75: 698–708. 10.1158/0008-5472.CAN-14-2637 [DOI] [PubMed] [Google Scholar]
- Gu L, Ying H, Zheng H, Murray SA, Xiao Z-XJ. 2003. The MDM2 RING finger is required for cell cycle-dependent regulation of its protein expression. FEBS Lett 544: 218–222. 10.1016/S0014-5793(03)00502-7 [DOI] [PubMed] [Google Scholar]
- Gu L, Zhang H, Liu T, Draganov A, Yi S, Wang B, Zhou M. 2018. Inhibition of MDM2 by a rhein-derived compound AQ-101 suppresses cancer development in SCID mice. Mol Cancer Ther 17: 497–507. 10.1158/1535-7163.MCT-17-0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Zaidi AH, Manna SK. 2018. Suppression of IKK, but not activation of p53 is responsible for cell death mediated by naturally occurring oxidized tetranortriterpenoid. J Cell Biochem 119: 6828–6841. 10.1002/jcb.26879 [DOI] [PubMed] [Google Scholar]
- Hallenborg P, Siersbæk M, Barrio-Hernandez I, Nielsen R, Kristiansen K, Mandrup S, Grøntved L, Blagoev B. 2016. MDM2 facilitates adipocyte differentiation through CRTC-mediated activation of STAT3. Cell Death Dis 7: e2289. 10.1038/cddis.2016.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck PM, Wolf ER, Olivos DJ 3rd, McAtarsney CP, Mayo LD. 2017. The fate of murine double minute X (MdmX) is dictated by distinct signaling pathways through murine double minute 2 (Mdm2). Oncotarget 8: 104455–104466. 10.18632/oncotarget.22320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes JJ, Nerva JD, Reilly KM. 2008. Novel dual-reporter preclinical screen for antiastrocytoma agents identifies cytostatic and cytotoxic compounds. J Biomol Screen 13: 795–803. 10.1177/1087057108321085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Tollini L, Kim T-H, Itahana Y, Zhang Y. 2014. The anaphase-promoting complex/cyclosome is an E3 ubiquitin ligase for Mdm2. Cell Cycle 13: 2101–2109. 10.4161/cc.29106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Li Y, Dai MS, Sun XX. 2016. Ribosomal protein L4 is a novel regulator of the MDM2-p53 loop. Oncotarget 7: 16217–16226. 10.18632/oncotarget.7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwa R, Gansmo LB, Romundstad P, Hveem K, Vatten L, Ryan BM, Harris CC, Lønning PE, Knappskog S. 2016. MDM2 promoter SNP55 (rs2870820) affects risk of colon cancer but not breast-, lung-, or prostate cancer. Sci Rep 6: 33153. 10.1038/srep33153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman AG, Hayano M, Poyurovsky MV, Shimada K, Skouta R, Prives C, Stockwell BR. 2011. Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discov 1: 312–325. 10.1158/2159-8290.CD-11-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollerer I, Barker JC, Jorgensen V, Tresenrider A, Dugast-Darzacq C, Chan LY, Darzacq X, Tjian R, Unal E, Brar GA. 2019. Evidence for an integrated gene repression mechanism based on mRNA isoform toggling in human cells. G3 9: 1045–1053. 10.1534/g3.118.200802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Ma H, Lu D, Qian J, Zhou J, Chen Y, Xu L, Wang X, Wei Q, Shen H. 2006. Genetic variants in the MDM2 promoter and lung cancer risk in a Chinese population. Int J Cancer 118: 1275–1278. 10.1002/ijc.21463 [DOI] [PubMed] [Google Scholar]
- Huang L, Yan Z, Liao X, Li Y, Yang J, Wang Z-G, Zuo Y, Kawai H, Shadfan M, Ganapathy S, et al. 2011. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci 108: 12001–12006. 10.1073/pnas.1102309108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wu S, Ji H, Yan X, Xie Y, Murai S, Zhao H, Miyagishi M, Kasim V. 2017. Identification of XBP1-u as a novel regulator of the MDM2/p53 axis using an shRNA library. Sci Adv 3: e1701383. 10.1126/sciadv.1701383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, et al. 2010. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCFβ-TRCP ubiquitin ligase. Cancer Cell 18: 147–159. 10.1016/j.ccr.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. 2004. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat Med 10: 1321–1328. 10.1038/nm1146 [DOI] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindström MS, Bhat Krishna P, Godfrey VL, Evan GI, Zhang Y. 2007. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12: 355–366. 10.1016/j.ccr.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Iwakuma T, Lozano G. 2003. MDM2, an introduction. Mol Cancer Res 1: 993. [PubMed] [Google Scholar]
- Iyappan S, Wollscheid H-P, Rojas-Fernandez A, Marquardt A, Tang H-C, Singh RK, Scheffner M. 2010. Turning the RING domain protein MdmX into an active ubiquitin-protein ligase. Journal of Biological Chemistry 285: 33065–33072. 10.1074/jbc.M110.115113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AG, Singh RK, Mohammad F, Bebee TW, Chandler DS. 2014. The splicing factor FUBP1 is required for the efficient splicing of oncogene MDM2 pre-mRNA. J Biol Chem 289: 17350–17364. 10.1074/jbc.M114.554717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana A, Krett NL, Guzman G, Khalid A, Ozden O, Staudacher JJ, Bauer J, Baik SH, Carroll T, Yazici C, et al. 2017. NFkb is essential for activin-induced colorectal cancer migration via upregulation of PI3K-MDM2 pathway. Oncotarget 8: 37377–37393. 10.18632/oncotarget.16343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Charles PY, Yu SM, Abraham D, Kommaddi RP, Mao L, Strachan RT, Zhang ZS, Bowles DE, Brian L, Stiber JA, et al. 2017. Mdm2 regulates cardiac contractility by inhibiting GRK2-mediated desensitization of β-adrenergic receptor signaling. JCI Insight 2: e95998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zawacka-Pankau J. 2020. The p53/MDM2/MDMX-targeted therapies—a clinical synopsis. Cell Death Dis 11: 237. 10.1038/s41419-020-2445-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Sun F, Zhu J, Luo G, Ban Y, Zhang P. 2019. miR-33a inhibits cell growth in renal cancer by downregulation of MDM4 expression. Mol Genet Genomic Med 7: e833. 10.1002/mgg3.833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RJ, Gu D, Bjorklund CC, Kuiatse I, Remaley AT, Bashir T, Vreys V, Orlowski RZ. 2013. The novel anticancer agent JNJ-26854165 induces cell death through inhibition of cholesterol transport and degradation of ABCA1. J Pharmacol Exp Ther 346: 381–392. 10.1124/jpet.113.204958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven T, Barak Y, Zauberman A, George DL, Oren M. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8: 3411–3416. [PubMed] [Google Scholar]
- Juven-Gershon T, Shifman O, Unger T, Elkeles A, Haupt Y, Oren M. 1998. The Mdm2 oncoprotein interacts with the cell fate regulator numb. Mol Cell Biol 18: 3974–3982. 10.1128/MCB.18.7.3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CL, Hsu HS, Chen HW, Cheng TH. 2009. Rapamycin increases the p53/MDM2 protein ratio and p53-dependent apoptosis by translational inhibition of mdm2 in cancer cells. Cancer Lett 286: 250–259. 10.1016/j.canlet.2009.05.031 [DOI] [PubMed] [Google Scholar]
- Karni-Schmidt O, Lokshin M, Prives C. 2016. The roles of MDM2 and MDMX in cancer. Annu Rev Pathol 11: 617–644. 10.1146/annurev-pathol-012414-040349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. 2007. RING domain-mediated interaction is a requirement for MDM2's E3 ligase activity. Cancer Res 67: 6026–6030. 10.1158/0008-5472.CAN-07-1313 [DOI] [PubMed] [Google Scholar]
- Kersten S. 2008. Peroxisome proliferator activated receptors and lipoprotein metabolism. PPAR Res 2008: 132960. 10.1155/2008/132960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagaki J, Agama KK, Pommier Y, Yang Y, Weissman AM. 2008. Targeting tumor cells expressing p53 with a water-soluble inhibitor of Hdm2. Mol Cancer Ther 7: 2445–2454. 10.1158/1535-7163.MCT-08-0063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusmann I, Wohlberedt K, Magerhans A, Teloni F, Korbel JO, Altmeyer M, Dobbelstein M. 2018. Chromatin modifiers Mdm2 and RNF2 prevent RNA:DNA hybrids that impair DNA replication. Proc Natl Acad Sci 115: E11311–E11320. 10.1073/pnas.1809592115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappskog S, Gansmo LB, Romundstad P, Bjørnslett M, Trovik J, Sommerfelt-Pettersen J, Løkkevik E, Norwegian Breast Cancer Group trial NBCG VI, Tollenaar RA, Seynaeve C, et al. 2012. MDM2 promoter SNP344T > A (rs1196333) status does not affect cancer risk. PLoS One 7: e36263. 10.1371/journal.pone.0036263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Wang D, Li T, Jiang L, Qiang L, Gu W. 2018. Inhibition of Mdmx (Mdm4) in vivo induces anti-obesity effects. Oncotarget 9: 7282–7297. 10.18632/oncotarget.23837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic M, Matt T, Martinez-Yamout MA, Dyson HJ, Wright PE. 2006. Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53. J Mol Biol 363: 433–450. 10.1016/j.jmb.2006.08.027 [DOI] [PubMed] [Google Scholar]
- Kundu N, Brekman A, Kim JY, Xiao G, Gao C, Bargonetti J. 2017. Estrogen-activated MDM2 disrupts mammary tissue architecture through a p53-independent pathway. Oncotarget 8: 47916–47930. 10.18632/oncotarget.18147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M, Kim J, Geradts J, Matsuura K, Liu L, Ran X, Xia W, Ribar TJ, Henao R, Dewhirst MW, et al. 2013. A network of substrates of the E3 ubiquitin ligases MDM2 and HUWE1 control apoptosis independently of p53. Sci Signal 6: ra32. 10.1126/scisignal.2003741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav-Baratz S, Kravtsova-Ivantsiv Y, Golan S, Ciechanover A. 2017. The testis-specific USP26 is a deubiquitinating enzyme of the ubiquitin ligase Mdm2. Biochem Biophys Res Commun 482: 106–111. 10.1016/j.bbrc.2016.10.135 [DOI] [PubMed] [Google Scholar]
- Lau LM, Nugent JK, Zhao X, Irwin MS. 2008. HDM2 antagonist nutlin-3 disrupts p73-HDM2 binding and enhances p73 function. Oncogene 27: 997–1003. 10.1038/sj.onc.1210707 [DOI] [PubMed] [Google Scholar]
- Lehman JA, Mayo LD. 2012. Integration of DNA damage and repair with murine double-minute 2 (Mdm2) in tumorigenesis. Int J Mol Sci 13: 16373–16386. 10.3390/ijms131216373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei CT, Tang H, Ye C, You CQ, Zhang J, Zhang CY, Xiong W, Su H, Zhang C. 2017. MDM2 contributes to high glucose-induced glomerular mesangial cell proliferation and extracellular matrix accumulation via Notch1. Sci Rep 7: 10393. 10.1038/s41598-017-10927-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenos K, Grawenda AM, Lodder K, Kuijjer ML, Teunisse AF, Repapi E, Grochola LF, Bartel F, Hogendoorn PC, Wuerl P, et al. 2012. Alternate splicing of the p53 inhibitor HDMX offers a superior prognostic biomarker than p53 mutation in human cancer. Cancer Res 72: 4074–4084. 10.1158/0008-5472.CAN-12-0215 [DOI] [PubMed] [Google Scholar]
- Leslie PL, Zhang Y. 2016. MDM2 oligomers: antagonizers of the guardian of the genome. Oncogene 35: 6157–6165. 10.1038/onc.2016.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie PL, Ke H, Zhang Y. 2015. The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2–MDMX protein heterodimerization. J Biol Chem 290: 12941–12950. 10.1074/jbc.M115.644435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D, Wu D, Trujillo C, Ramezani T, Lessel I, Alwasiyah MK, Saha B, Hisama FM, Rading K, Goebel I, et al. 2017. Dysfunction of the MDM2/p53 axis is linked to premature aging. J Clin Invest 127: 3598–3608. 10.1172/JCI92171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kurokawa M. 2015. Regulation of MDM2 stability after DNA damage. J Cell Physiol 230: 2318–2327. 10.1002/jcp.24994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. 2003. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302: 1972–1975. 10.1126/science.1091362 [DOI] [PubMed] [Google Scholar]
- Li D, Tavana O, Sun SC, Gu W. 2018a. Peli1 modulates the subcellular localization and activity of Mdmx. Cancer Res 78: 2897–2910. 10.1158/0008-5472.CAN-17-3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang Z, Jiang M, Fang RP, Shi H, Shen Y, Cai XL, Liu Q, Ye K, Fan SJ, et al. 2018b. The oncoprotein HBXIP promotes human breast cancer growth through down-regulating p53 via miR-18b/MDM2 and pAKT/MDM2 pathways. Acta Pharmacol Sin 39: 1787–1796. 10.1038/s41401-018-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Cui S, Ma Q, Liu Y, Yu H, Geng G, Agborbesong E, Ren C, Wei K, Zhang Y, et al. 2019. Disruption of Robo2-Baiap2 integrated signaling drives cystic disease. JCI Insight 4: e127602. 10.1172/jci.insight.127602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Lunec J. 2005. Characterisation of a novel p53 down-regulated promoter in intron 3 of the human MDM2 oncogene. Gene 361: 112–118. 10.1016/j.gene.2005.07.018 [DOI] [PubMed] [Google Scholar]
- Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. 2003. Hdmx stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci 100: 12009–12014. 10.1073/pnas.2030930100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X, Xu C, Fan C, Zhong K, Li F, Wang X. 2014. FL118 induces p53-dependent senescence in colorectal cancer cells by promoting degradation of MdmX. Cancer Res 74: 7487–7497. 10.1158/0008-5472.CAN-14-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. 2008. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ 15: 841–848. 10.1038/sj.cdd.4402309 [DOI] [PubMed] [Google Scholar]
- Liu L, Fan L, Fang C, Zou ZJ, Yang S, Zhang LN, Li JY, Xu W. 2012. S-MDM4 mRNA overexpression indicates a poor prognosis and marks a potential therapeutic target in chronic lymphocytic leukemia. Cancer Sci 103: 2056–2063. 10.1111/cas.12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhang H, Xiong J, Yi S, Gu L, Zhou M. 2015. Inhibition of MDM2 homodimerization by XIAP IRES stabilizes MDM2, influencing cancer cell survival. Mol Cancer 14: 65. 10.1186/s12943-015-0334-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tan Y, Zhang C, Zhang Y, Zhang L, Ren P, Deng H, Luo J, Ke Y, Du X. 2016. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep 17: 349–366. 10.15252/embr.201540505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhang J, Yang X, Jiao T, Zhao X, Li W, Zhu J, Yang P, Jin J, Peng J, et al. 2017a. HDM2 promotes NEDDylation of hepatitis B virus HBx to enhance its stability and function. J Virol 91: e00340-17. 10.1128/JVI.00340-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Xiong J, Yi S, Zhang H, Zhou S, Gu L, Zhou M. 2017b. FKBP12 enhances sensitivity to chemotherapy-induced cancer cell apoptosis by inhibiting MDM2. Oncogene 36: 1678–1686. 10.1038/onc.2016.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zou X, Gotoh T, Brown AM, Jiang L, Wisdom EL, Kim JK, Finkielstein CV. 2018a. Distinct control of PERIOD2 degradation and circadian rhythms by the oncoprotein and ubiquitin ligase MDM2. Sci Signal 11: eaau0715. 10.1126/scisignal.aau0715 [DOI] [PubMed] [Google Scholar]
- Liu Z, Jin L, Yang J-K, Wang B, Wu KKL, Hallenborg P, Xu A, Cheng KKY. 2018b. The dysfunctional MDM2–p53 axis in adipocytes contributes to aging-related metabolic complications by induction of lipodystrophy. Diabetes 67: 2397–2409. 10.2337/db18-0684 [DOI] [PubMed] [Google Scholar]
- Liu B, Yi J, Yang X, Liu L, Lou X, Zhang Z, Qi H, Wang Z, Zou J, Zhu WG, et al. 2019a. MDM2-mediated degradation of WRN promotes cellular senescence in a p53-independent manner. Oncogene 38: 2501–2515. 10.1038/s41388-018-0605-5 [DOI] [PubMed] [Google Scholar]
- Liu J, Cheng LG, Li HG. 2019b. LncRNA SNHG20 promoted the proliferation of glioma cells via sponging miR-4486 to regulate the MDM2-p53 pathway. Eur Rev Med Pharmacol Sci 23: 5323–5331. 10.26355/eurrev_201906_18199 [DOI] [PubMed] [Google Scholar]
- Lu YF, Xu XP, Lu XP, Zhu Q, Liu G, Bao YT, Wen H, Li YL, Gu W, Zhu WG. 2020. SIRT7 activates p53 by enhancing PCAF-mediated MDM2 degradation to arrest the cell cycle. Oncogene 39: 4650–4665. 10.1038/s41388-020-1305-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yuan R, Meng Q, Goldberg ID, Rosen EM, Fan S. 2000. P53-independent down-regulation of Mdm2 in human cancer cells treated with Adriamycin. Mol Cell Biol Res Commun 3: 122–128. 10.1006/mcbr.2000.0201 [DOI] [PubMed] [Google Scholar]
- Ma H, Hu Z, Zhai X, Wang S, Wang X, Qin J, Jin G, Liu J, Wang X, Wei Q, et al. 2006. Polymorphisms in the MDM2 promoter and risk of breast cancer: a case-control analysis in a Chinese population. Cancer Lett 240: 261–267. 10.1016/j.canlet.2005.09.019 [DOI] [PubMed] [Google Scholar]
- Magnussen HM, Ahmed SF, Sibbet GJ, Hristova VA, Nomura K, Hock AK, Archibald LJ, Jamieson AG, Fushman D, Vousden KH, et al. 2020. Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain. Nat Commun 11: 2094. 10.1038/s41467-020-15783-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F, Di Conza G, Moretti F. 2009. MDM4 (MDMX) and its transcript variants. Curr Genomics 10: 42–50. 10.2174/138920209787581280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F, Teveroni E, Di Conza G, Monteleone V, Arisi I, Pellegrino M, Buttarelli M, Pieroni L, D'Onofrio M, Urbani A, et al. 2017. MDM4 actively restrains cytoplasmic mTORC1 by sensing nutrient availability. Mol Cancer 16: 55. 10.1186/s12943-017-0626-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi JJ. 2010. The Mdm2–p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev 24: 1580–1589. 10.1101/gad.1941710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Lozano G. 2010. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Diff 17: 93–102. 10.1038/cdd.2009.68 [DOI] [PubMed] [Google Scholar]
- Markey MP. 2011. Regulation of MDM4. Front Biosci (Landmark Ed) 16: 1144–1156. 10.2741/3780 [DOI] [PubMed] [Google Scholar]
- Matijasevic Z, Steinman HA, Hoover K, Jones SN. 2008. Mdmx promotes bipolar mitosis to suppress transformation and tumorigenesis in p53-deficient cells and mice. Mol Cell Biol 28: 1265–1273. 10.1128/MCB.01108-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matijasevic Z, Krzywicka-Racka A, Sluder G, Gallant J, Jones SN. 2016. The Zn-finger domain of MdmX suppresses cancer progression by promoting genome stability in p53-mutant cells. Oncogenesis 5: e262. 10.1038/oncsis.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail DJ, Lin CC, Dai H, Mo W, Li Y, Stephan C, Davies P, Lu Z, Mills GB, Lee JS, et al. 2018. Defective replication stress response is inherently linked to the cancer stem cell phenotype. Cell Rep 23: 2095–2106. 10.1016/j.celrep.2018.04.068 [DOI] [PubMed] [Google Scholar]
- Meek DW, Hupp TR. 2010. The regulation of MDM2 by multisite phosphorylation—opportunities for molecular-based intervention to target tumours? Semin Cancer Biol 20: 19–28. 10.1016/j.semcancer.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Meek DW, Knippschild U. 2003. Posttranslational modification of MDM2. Mol Cancer Res 1: 1017–1026. [PubMed] [Google Scholar]
- Meulmeester E, Pereg Y, Shiloh Y, Jochemsen AG. 2005. ATM-mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle 4: 1166–1170. 10.4161/cc.4.9.1981 [DOI] [PubMed] [Google Scholar]
- Mohammad Khanlou Z, Pouladi N, Hosseinpour Feizi M, Pedram N. 2017. Lack of associations of the MDM4 rs4245739 polymorphism with risk of thyroid cancer among Iranian-Azeri patients: a case-control study. Asian Pac J Cancer Prev 18: 1133–1138. 10.22034/APJCP.2017.18.4.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Petrenko O. 2003. The MDM2–p53 interaction. Mol Cancer Res 1: 1001–1008. [PubMed] [Google Scholar]
- Mulay SR, Thomasova D, Ryu M, Anders H-J. 2012. MDM2 (murine double minute-2) links inflammation and tubular cell healing during acute kidney injury in mice. Kidney Int 81: 1199–1211. 10.1038/ki.2011.482 [DOI] [PubMed] [Google Scholar]
- Mulay SR, Romoli S, Desai J, Honarpisheh MM, Kumar SV, Anders HJ, Thomasova D. 2016. Murine double minute-2 inhibition ameliorates established crescentic glomerulonephritis. Am J Pathol 186: 1442–1453. 10.1016/j.ajpath.2016.01.017 [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. 2014. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25: 304–317. 10.1016/j.ccr.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungamuri SK, Qiao RF, Yao S, Manfredi JJ, Gu W, Aaronson SA. 2016. USP7 enforces heterochromatinization of p53 target promoters by protecting SUV39H1 from MDM2-mediated degradation. Cell Rep 14: 2528–2537. 10.1016/j.celrep.2016.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Sawada K, Miyamoto M, Kinose Y, Yoshimura A, Ishida K, Kobayashi M, Shimizu A, Nakatsuka E, Hashimoto K, et al. 2019. Downregulation of miR-194-5p induces paclitaxel resistance in ovarian cancer cells by altering MDM2 expression. Oncotarget 10: 673–683. 10.18632/oncotarget.26586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihira NT, Ogura K, Shimizu K, North BJ, Zhang J, Gao D, Inuzuka H, Wei W. 2017. Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function. Sci Signal 10: eaai8026. 10.1126/scisignal.aai8026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Taya Y, Nakagama H. 2009. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett 583: 2710–2714. 10.1016/j.febslet.2009.07.021 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Tsunematsu R, Tahira T, Sonoda K, Asanoma K, Yagi H, Yoneda T, Hayashi K, Wake N, Kato K. 2015. SNP55, a new functional polymorphism of MDM2-P2 promoter, contributes to allele-specific expression of MDM2 in endometrial cancers. BMC Med Genet 16: 67. 10.1186/s12881-015-0216-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro DR, Rosso M, Bargonetti J. 2012. Splicing up mdm2 for cancer proteome diversity. Genes Cancer 3: 311–319. 10.1177/1947601912455323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Saiki AY, Caenepeel S. 2016. The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb Perspect Med 6: a026336. 10.1101/cshperspect.a026336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. 2010. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2: a0010088. 10.1101/cshperspect.a001008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Chen J. 2005. Modification of MDMX by sumoylation. Biochem Biophys Res Commun 332: 702–709. 10.1016/j.bbrc.2005.05.012 [DOI] [PubMed] [Google Scholar]
- Pant V, Larsson CA, Aryal N, Xiong S, You MJ, Quintas-Cardama A, Lozano G. 2017. Tumorigenesis promotes Mdm4-S overexpression. Oncotarget 8: 25837–25847. 10.18632/oncotarget.15552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Ju UI, Park JW, Song JY, Shin DH, Lee KH, Jeong LS, Yu J, Lee HW, Cho JY, et al. 2016. PPARγ neddylation essential for adipogenesis is a potential target for treating obesity. Cell Death Differ 23: 1296–1311. 10.1038/cdd.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N, Wang J, Shiozawa K, Jones KB, Zhang Y, Prokop JW, Davenport GG, Nihira NT, Hao Z, Wong D, et al. 2019. HDAC2 regulates site-specific acetylation of MDM2 and its ubiquitination signaling in tumor suppression. iScience 13: 43–54. 10.1016/j.isci.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedram N, Pouladi N, Feizi MA, Montazeri V, Sakhinia E, Estiar MA. 2016. Analysis of the association between MDM4 rs4245739 single nucleotide polymorphism and breast cancer susceptibility. Clin Lab 62: 1303–1308. 10.7754/Clin.Lab.2016.151128 [DOI] [PubMed] [Google Scholar]
- Pettersson S, Sczaniecka M, McLaren L, Russell F, Gladstone K, Hupp T, Wallace M. 2013. Non-degradative ubiquitination of the Notch1 receptor by the E3 ligase MDM2 activates the Notch signalling pathway. Biochem J 450: 523–536. 10.1042/BJ20121249 [DOI] [PubMed] [Google Scholar]
- Phillips A, Teunisse A, Lam S, Lodder K, Darley M, Emaduddin M, Wolf A, Richter J, de Lange J, Verlaan-de Vries M, et al. 2010. HDMX-L is expressed from a functional p53-responsive promoter in the first intron of the HDMX gene and participates in an autoregulatory feedback loop to control p53 activity. J Biol Chem 285: 29111–29127. 10.1074/jbc.M110.129726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikkarainen S, Kennedy RA, Marshall AK, Tham el L, Lay K, Kriz TA, Handa BS, Clerk A, Sugden PH. 2009. Regulation of expression of the rat orthologue of mouse double minute 2 (MDM2) by H2O2-induced oxidative stress in neonatal rat cardiac myocytes. J Biol Chem 284: 27195–27210. 10.1074/jbc.M109.037887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishas KI, Adwal A, Neuhaus SJ, Clayer MT, Farshid G, Staudacher AH, Callen DF. 2015. XI-006 induces potent p53-independent apoptosis in Ewing sarcoma. Sci Rep 5: 11465. 10.1038/srep11465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzorno A, Dubois J, Machado D, Cartet G, Traversier A, Julien T, Lina B, Bourdon JC, Rosa-Calatrava M, Terrier O. 2018. Influenza A viruses alter the stability and antiviral contribution of host E3-ubiquitin ligase Mdm2 during the time-course of infection. Sci Rep 8: 3746. 10.1038/s41598-018-22139-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post SM, Quintás-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, Maccio DR, Bond GL, Johnson DG, Levine AJ, et al. 2010. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell 18: 220–230. 10.1016/j.ccr.2010.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. 2007. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J 26: 90–101. 10.1038/sj.emboj.7601465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi DL, Cobrinik D. 2017. MDM2 but not MDM4 promotes retinoblastoma cell proliferation through p53-independent regulation of MYCN translation. Oncogene 36: 1760–1769. 10.1038/onc.2016.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi JS, Yuan Y, Desai-Yajnik V, Samuels HH. 1999. Regulation of the mdm2 oncogene by thyroid hormone receptor. Mol Cell Biol 19: 864–872. 10.1128/MCB.19.1.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JJ, Nag S, Voruganti S, Wang W, Zhang R. 2012. Natural product MDM2 inhibitors: anticancer activity and mechanisms of action. Curr Med Chem 19: 5705–5725. 10.2174/092986712803988910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JJ, Li X, Hunt C, Wang W, Wang H, Zhang R. 2018a. Natural products targeting the p53-MDM2 pathway and mutant p53: recent advances and implications in cancer medicine. Genes Dis 5: 204–219. 10.1016/j.gendis.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JJ, Wang W, Li X, Deokar H, Buolamwini JK, Zhang R. 2018b. Inhibiting β-catenin by β-carboline-type MDM2 inhibitor for pancreatic cancer therapy. Front Pharmacol 9: 5. 10.3389/fphar.2018.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada M, Althubiti M, Ekpenyong-Akiba AE, Lee KG, Lam KP, Fedorova O, Barlev NA, Macip S. 2017. BTK blocks the inhibitory effects of MDM2 on p53 activity. Oncotarget 8: 106639–106647. 10.18632/oncotarget.22543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada M, Barlev N, Macip S. 2018. BTK modulates p73 activity to induce apoptosis independently of p53. Cell Death Discov 4: 30. 10.1038/s41420-018-0097-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja R, Ronsard L, Lata S, Trivedi S, Banerjea AC. 2017. HIV-1 Tat potently stabilises Mdm2 and enhances viral replication. Biochem J 474: 2449–2464. 10.1042/BCJ20160825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riscal R, Schrepfer E, Arena G, Cissé MY, Bellvert F, Heuillet M, Rambow F, Bonneil E, Sabourdy F, Vincent C, et al. 2016. Chromatin-bound MDM2 regulates serine metabolism and redox homeostasis independently of p53. Mol Cell 62: 890–902. 10.1016/j.molcel.2016.04.033 [DOI] [PubMed] [Google Scholar]
- Rong H, Chen B, Wei X, Peng J, Ma K, Duan S, He J. 2020. Long non-coding RNA XIST expedites lung adenocarcinoma progression through upregulating MDM2 expression via binding to miR-363-3p. Thorac Cancer 11: 659–671. 10.1111/1759-7714.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M, Okoro DE, Bargonetti J. 2014. Splice variants of MDM2 in oncogenesis. Subcell Biochem 85: 247–261. 10.1007/978-94-017-9211-0_14 [DOI] [PubMed] [Google Scholar]
- Rubio-Patiño C, Trotta AP, Chipuk JE. 2019. MDM2 and mitochondrial function: one complex intersection. Biochem Pharmacol 162: 14–20. 10.1016/j.bcp.2018.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G, Liu Y, Elf S, Miyata Y, Ohyashiki K, Izumi M, Menendez S, Nimer SD. 2009. ELF4/MEF activates MDM2 expression and blocks oncogene-induced p16 activation to promote transformation. Mol Cell Biol 29: 3687–3699. 10.1128/MCB.01551-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasiela CA, Stewart DH, Kitagaki J, Safiran YJ, Yang Y, Weissman AM, Oberoi P, Davydov IV, Goncharova E, Beutler JA, et al. 2008. Identification of inhibitors for MDM2 ubiquitin ligase activity from natural product extracts by a novel high-throughput electrochemiluminescent screen. J Biomol Screen 13: 229–237. 10.1177/1087057108315038 [DOI] [PubMed] [Google Scholar]
- Senturk JC, Bohlman S, Manfredi JJ. 2017. Mdm2 selectively suppresses DNA damage arising from inhibition of topoisomerase II independent of p53. Oncogene 36: 6085–6096. 10.1038/onc.2017.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadfan M, Lopez-Pajares V, Yuan Z-M. 2012. MDM2 and MDMX: alone and together in regulation of p53. Transl Cancer Res 1: 88–89. [PMC free article] [PubMed] [Google Scholar]
- Shen X, Zuo X, Zhang W, Bai Y, Qin X, Hou N. 2018. MiR-370 promotes apoptosis in colon cancer by directly targeting MDM4. Oncol Lett 15: 1673–1679. 10.3892/ol.2017.7524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhang J, Wang C, Jain PP, Xiong M, Shi X, Lei Y, Chen S, Yin Q, Thistlethwaite PA, et al. 2020. MDM2-Mediated ubiquitination of angiotensin-converting enzyme 2 contributes to the development of pulmonary arterial hypertension. Circulation 142: 1190–1204. 10.1161/CIRCULATIONAHA.120.048191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. 2009. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci 106: 16275–16280. 10.1073/pnas.0904305106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuvalov O, Kizenko A, Shakirova A, Fedorova O, Petukhov A, Aksenov N, Vasileva E, Daks A, Barlev N. 2018. Nutlin sensitizes lung carcinoma cells to interferon-α treatment in MDM2-dependent but p53-independent manner. Biochem Biophys Res Commun 495: 1233–1239. 10.1016/j.bbrc.2017.11.118 [DOI] [PubMed] [Google Scholar]
- Singh RK, Iyappan S, Scheffner M. 2007. Hetero-oligomerization with MdmX rescues the ubiquitin/Nedd8 ligase activity of RING finger mutants of Mdm2. J Biol Chem 282: 10901–10907. 10.1074/jbc.M610879200 [DOI] [PubMed] [Google Scholar]
- Singh AK, Chauhan SS, Singh SK, Verma VV, Singh A, Arya RK, Maheshwari S, Akhtar MS, Sarkar J, Rangnekar VM, et al. 2016. Dual targeting of MDM2 with a novel small-molecule inhibitor overcomes TRAIL resistance in cancer. Carcinogenesis 37: 1027–1040. 10.1093/carcin/bgw088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Vaughan CA, Rabender C, Mikkelsen R, Deb S, Palit Deb S. 2019. DNA replication in progenitor cells and epithelial regeneration after lung injury requires the oncoprotein MDM2. JCI Insight 4: e128194. 10.1172/jci.insight.128194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, Shohet JM. 2005. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci 102: 731–736. 10.1073/pnas.0405495102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stad R, Little NA, Xirodimas DP, Frenk R, van der Eb AJ, Lane DP, Saville MK, Jochemsen AG. 2001. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep 2: 1029–1034. 10.1093/embo-reports/kve227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegeman S, Moya L, Selth LA, Spurdle AB, Clements JA, Batra J. 2015. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocr Relat Cancer 22: 265–276. 10.1530/ERC-15-0013 [DOI] [PubMed] [Google Scholar]
- Stindt MH, Carter S, Vigneron AM, Ryan KM, Vousden KH. 2011. MDM2 promotes SUMO-2/3 modification of p53 to modulate transcriptional activity. Cell Cycle 10: 3176–3188. 10.4161/cc.10.18.17436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, et al. 2017. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171: 273–285. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM. 2004. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 23: 1547–1556. 10.1038/sj.emboj.7600145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Tang L. 2016. MDM2 increases drug resistance in cancer cells by inducing EMT independent of p53. Curr Med Chem 23: 4529–4539. 10.2174/0929867323666160926150820 [DOI] [PubMed] [Google Scholar]
- Sun T, Lee GS, Oh WK, Pomerantz M, Yang M, Xie W, Freedman ML, Kantoff PW. 2010. Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population. Clin Cancer Res 16: 5244–5251. 10.1158/1078-0432.CCR-10-1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Bell JL, Carter D, Gherardi S, Poulos RC, Milazzo G, Wong JW, Al-Awar R, Tee AE, Liu PY, et al. 2015. WDR5 supports an N-Myc transcriptional complex that drives a protumorigenic gene expression signature in neuroblastoma. Cancer Res 75: 5143–5154. 10.1158/0008-5472.CAN-15-0423 [DOI] [PubMed] [Google Scholar]
- Sun P, Yan F, Fang W, Zhao J, Chen H, Ma X, Song J. 2018. MDM4 contributes to the increased risk of glioma susceptibility in Han Chinese population. Sci Rep 8: 11093. 10.1038/s41598-018-29468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackmann NR, Zhang Y. 2017. Mouse modelling of the MDM2/MDMX−p53 signalling axis. J Mol Cell Biol 9: 34–44. 10.1093/jmcb/mjx006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomasova D, Mulay SR, Bruns H, Anders HJ. 2012. p53-independent roles of MDM2 in NF-κB signaling: implications for cancer therapy, wound healing, and autoimmune diseases. Neoplasia 14: 1097–1101. 10.1593/neo.121534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Xu Z, Fu J. 2020. CircularRNA-9119 promotes the proliferation of cervical cancer cells by sponging miR-126/MDM4. Mol Cell Biochem 470: 53–62. 10.1007/s11010-020-03745-3 [DOI] [PubMed] [Google Scholar]
- Tisato V, Voltan R, Gonelli A, Secchiero P, Zauli G. 2017. MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer. J Hematol Oncol 10: 133. 10.1186/s13045-017-0500-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todoric J, Antonucci L, Di Caro G, Li N, Wu X, Lytle NK, Dhar D, Banerjee S, Fagman JB, Browne CD, et al. 2017. Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell 32: 824–839.e8. 10.1016/j.ccell.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AH, Cervi D, Lee J, Ben-David Y. 2005. Direct transcriptional regulation of MDM2 by Fli-1. Oncogene 24: 962–969. 10.1038/sj.onc.1208323 [DOI] [PubMed] [Google Scholar]
- Uldrijan S, Pannekoek W-J, Vousden KH. 2007. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J 26: 102–112. 10.1038/sj.emboj.7601469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Alstyne M, Simon CM, Sardi SP, Shihabuddin LS, Mentis GZ, Pellizzoni L. 2018. Dysregulation of Mdm2 and Mdm4 alternative splicing underlies motor neuron death in spinal muscular atrophy. Genes Dev 32: 1045–1059. 10.1101/gad.316059.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Yallowitz AR, Marchenko ND, Xu S, Moll UM. 2011. Blockade of Hsp90 by 17AAG antagonizes MDMX and synergizes with nutlin to induce p53-mediated apoptosis in solid tumors. Cell Death Dis 2: e156. 10.1038/cddis.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- Venkatesh D, O'Brien NA, Zandkarimi F, Tong DR, Stokes ME, Dunn DE, Kengmana ES, Aron AT, Klein AM, Csuka JM, et al. 2020. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev 34: 526–543. 10.1101/gad.334219.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumaran R, Tan KH, Miranda PJ, Haupt S, Haupt Y. 2015. Regulation of mutant p53 protein expression. Front Oncol 5: 284. 10.3389/fonc.2015.00284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu B, Wovkulich P, Pizzolato G, Lovey A, Ding Q, Jiang N, Liu J-J, Zhao C, Glenn K, Wen Y, et al. 2013. Discovery of RG7112: a small-molecule MDM2 inhibitor in clinical development. ACS Med Chem Lett 4: 466–469. 10.1021/ml4000657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM. 2010. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 20: 299–309. 10.1016/j.tcb.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Li Y-C, Wahl GM. 2013. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13: 83–96. 10.1038/nrc3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yan C. 2011. A small-molecule p53 activator induces apoptosis through inhibiting MDMX expression in breast cancer cells. Neoplasia 13: 611–619. 10.1593/neo.11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu J. 2018. Mir-1307 regulates cisplatin resistance by targeting Mdm4 in breast cancer expressing wild type P53. Thorac Cancer 9: 676–683. 10.1111/1759-7714.12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Taplick J, Geva N, Oren M. 2004. Inhibition of p53 degradation by Mdm2 acetylation. FEBS Lett 561: 195–201. 10.1016/S0014-5793(04)00168-1 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma X, Ren S, Buolamwini JK, Yan C. 2011. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther 10: 69–79. 10.1158/1535-7163.MCT-10-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Qin J-J, Voruganti S, Srivenugopal KS, Nag S, Patil S, Sharma H, Wang M-H, Wang H, Buolamwini JK, et al. 2014a. The pyrido[b]indole MDM2 inhibitor SP-141 exerts potent therapeutic effects in breast cancer models. Nat Commun 5: 5086. 10.1038/ncomms6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WEI, Qin J-J, Voruganti S, Wang M-H, Sharma H, Patil S, Zhou J, Wang HUI, Mukhopadhyay D, Buolamwini JK, et al. 2014b. Identification of a new class of MDM2 inhibitor that inhibits growth of orthotopic pancreatic tumors in mice. Gastroenterology 147: 893–902.e2. 10.1053/j.gastro.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li H, Qiu S, Dong Z, Xiang X, Zhang D. 2017a. MBD2 upregulates miR-301a-5p to induce kidney cell apoptosis during vancomycin-induced AKI. Cell Death Dis 8: e3120. 10.1038/cddis.2017.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MY, Jia M, He J, Zhou F, Qiu LX, Sun MH, Yang YJ, Wang JC, Jin L, Wang YN, et al. 2017b. MDM4 genetic variants and risk of gastric cancer in an Eastern Chinese population. Oncotarget 8: 19547–19555. 10.18632/oncotarget.14666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Selth LA, Callen DF. 2017c. MiR-766 induces p53 accumulation and G2/M arrest by directly targeting MDM4. Oncotarget 8: 29914–29924. 10.18632/oncotarget.15530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang MD, Cheng P, Huang H, Dong W, Zhang WW, Li PP, Lin C, Pan ZY, Wu MC, et al. 2017d. Hepatitis B virus X protein promotes the stem-like properties of OV6+ cancer cells in hepatocellular carcinoma. Cell Death Dis 8: e2560. 10.1038/cddis.2016.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cheng JW, Qin JJ, Hu B, Li X, Nijampatnam B, Velu SE, Fan J, Yang XR, Zhang R. 2019. MDM2-NFAT1 dual inhibitor, MA242: effective against hepatocellular carcinoma, independent of p53. Cancer Lett 459: 156–167. 10.1016/j.canlet.2019.114429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Qin JJ, Rajaei M, Li X, Yu X, Hunt C, Zhang R. 2020. Targeting MDM2 for novel molecular therapy: beyond oncology. Med Res Rev 40: 856–880. 10.1002/med.21637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson IR, Li BK, Roche O, Blanch A, Ohh M, Irwin MS. 2010. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene 29: 297–304. 10.1038/onc.2009.314 [DOI] [PubMed] [Google Scholar]
- Wienken M, Dickmanns A, Nemajerova A, Kramer D, Najafova Z, Weiss M, Karpiuk O, Kassem M, Zhang Y, Lozano G, et al. 2016. MDM2 associates with polycomb repressor complex 2 and enhances stemness-promoting chromatin modifications independent of p53. Mol Cell 61: 68–83. 10.1016/j.molcel.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken M, Moll UM, Dobbelstein M. 2017. Mdm2 as a chromatin modifier. J Mol Cell Biol 9: 74–80. 10.1093/jmcb/mjw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlberedt K, Klusmann I, Derevyanko PK, Henningsen K, Choo J, Manzini V, Magerhans A, Giansanti C, Eischen CM, Jochemsen AG, et al. 2020. Mdm4 supports DNA replication in a p53-independent fashion. Oncogene 39: 4828–4843. 10.1038/s41388-020-1325-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Pomeroy SL, Ferreira M, Teider N, Mariani J, Nakayama KI, Hatakeyama S, Tron VA, Saltibus LF, Spyracopoulos L, et al. 2011. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat Med 17: 347–355. 10.1038/nm.2283 [DOI] [PubMed] [Google Scholar]
- Wu W, Xu C, Ling X, Fan C, Buckley BP, Chernov MV, Ellis L, Li F, Muñoz IG, Wang X. 2015. Targeting RING domains of Mdm2–MdmX E3 complex activates apoptotic arm of the p53 pathway in leukemia/lymphoma cells. Cell Death Dis 6: e2035. 10.1038/cddis.2015.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Pant V, Zhang Y, Aryal NK, You MJ, Kusewitt D, Lozano G. 2017. The p53 inhibitor Mdm4 cooperates with multiple genetic lesions in tumourigenesis. J Pathol 241: 501–510. 10.1002/path.4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E, Zhang J, Chen X. 2013. MDM2 expression is repressed by the RNA-binding protein RNPC1 via mRNA stability. Oncogene 32: 2169–2178. 10.1038/onc.2012.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan CD, Wang X. 2015. Regulation of Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase. Oncogene 34: 281–289. 10.1038/onc.2013.557 [DOI] [PubMed] [Google Scholar]
- Xu J, Han M, Shen J, Guan Q, Bai Z, Lang B, Zhang H, Li Z, Zuo D, Zhang W, et al. 2016. 2-methoxy-5((3,4,5-trimethosyphenyl)seleninyl) phenol inhibits MDM2 and induces apoptosis in breast cancer cells through a p53-independent pathway. Cancer Lett 383: 9–17. 10.1016/j.canlet.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Yan H, Chen X, Li Y, Fan L, Tai Y, Zhou Y, Chen Y, Qi X, Huang R, Ren J. 2019. MiR-1205 functions as a tumor suppressor by disconnecting the synergy between KRAS and MDM4/E2F1 in non-small cell lung cancer. Am J Cancer Res 9: 312–329. [PMC free article] [PubMed] [Google Scholar]
- Yanbin Z, Jing Z. 2019. CircSAMD4A accelerates cell proliferation of osteosarcoma by sponging miR-1244 and regulating MDM2 mRNA expression. Biochem Biophys Res Commun 516: 102–111. 10.1016/j.bbrc.2019.05.182 [DOI] [PubMed] [Google Scholar]
- Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, et al. 2005. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 7: 547–559. 10.1016/j.ccr.2005.04.029 [DOI] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, Broglio K, Berry DA, Hung MC. 2006. MDM2 promotes cell motility and invasiveness by regulating E-cadherin degradation. Mol Cell Biol 26: 7269–7282. 10.1128/MCB.00172-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Tang H, Zhao Z, Lei CT, You CQ, Zhang J, Gao P, He FF, Chen S, Wang YM, et al. 2017. MDM2 mediates fibroblast activation and renal tubulointerstitial fibrosis via a p53-independent pathway. Am J Physiol Renal Physiol 312: F760–F768. 10.1152/ajprenal.00528.2016 [DOI] [PubMed] [Google Scholar]
- Yu ZC, Huang YF, Shieh SY. 2016. Requirement for human Mps1/TTK in oxidative DNA damage repair and cell survival through MDM2 phosphorylation. Nucleic Acids Res 44: 1133–1150. 10.1093/nar/gkv1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng SX, Jin Y, Kuninger DT, Rotwein P, Lu H. 2003. The acetylase activity of p300 is dispensable for MDM2 stabilization. J Biol Chem 278: 7453–7458. 10.1074/jbc.M209030200 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu H. 2009. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16: 369–377. 10.1016/j.ccr.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang Z, Cheng J, Li M, Wang W, Xu W, Wang H, Zhang R. 2012. Transcription factor NFAT1 activates the mdm2 oncogene independent of p53. J Biol Chem 287: 30468–30476. 10.1074/jbc.M112.373738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gu L, Liu T, Chiang KY, Zhou M. 2014. Inhibition of MDM2 by nilotinib contributes to cytotoxicity in both Philadelphia-positive and negative acute lymphoblastic leukemia. PLoS One 9: e100960. 10.1371/journal.pone.0100960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Liu J, Tan C, Yue X, Zhao Y, Peng J, Wang X, Laddha SV, Chan CS, Zheng S, et al. 2016. microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget 7: 8783–8796. 10.18632/oncotarget.7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kong X, Zhang Y, Sun W, Xu E, Chen X. 2020. Mdm2 is a target and mediator of IRP2 in cell growth control. FASEB J 34: 2301–2311. 10.1096/fj.201902278RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu H, Hu W. 2014. The regulation of MDM2 oncogene and its impact on human cancers. Acta Biochim Biophys Sin (Shanghai) 46: 180–189. 10.1093/abbs/gmt147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Yang Y, Zhang G, Wang C, Wang D, Wu M, Mei Y. 2018. Regulation of the Mdm2–p53 pathway by the ubiquitin E3 ligase MARCH7. EMBO Rep 19: 305–319. 10.15252/embr.201744465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Guan JZ, Wu M, Lai GH, Zhu ZL. 2019. Downregulation of microRNA-23b protects against ischemia-reperfusion injury via p53 signaling pathway by upregulating MDM4 in rats. J Cell Biochem 120: 4599–4612. 10.1002/jcb.27748 [DOI] [PubMed] [Google Scholar]
- Zheng T, Wang J, Zhao Y, Zhang C, Lin M, Wang X, Yu H, Liu L, Feng Z, Hu W. 2013. Spliced MDM2 isoforms promote mutant p53 accumulation and gain-of-function in tumorigenesis. Nat Commun 4: 2996. 10.1038/ncomms3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JX, Lee CH, Qi CF, Wang H, Naghashfar Z, Abbasi S, Morse HC 3rd. 2009. IFN regulatory factor 8 regulates MDM2 in germinal center B cells. J Immunol 183: 3188–3194. 10.4049/jimmunol.0803693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li Z, Huang Y, Ju W, Wang D, Zhu X, He X. 2019. MicroRNA-26a targets the mdm2/p53 loop directly in response to liver regeneration. Int J Mol Med 44: 1505–1514. [DOI] [PubMed] [Google Scholar]