Abstract
Extracellular vesicles (EVs) offer a vehicle for diagnostic and therapeutic utility. EVs carry bioactive cargo and an accrued interest in their characterization has emerged. Efforts at identifying EV-enriched protein or RNA led to a surprising realization that EVs are excessively heterogeneous in nature. This diversity was originally attributed to vesicle sizes but it is becoming evident that different classes of EVs vehiculate distinct molecular cargos. Therefore, one of the current challenges in EV research is their selective isolation in quantities sufficient for efficient downstream analyses. Many protocols have been developed, however reproducibility between research groups can be difficult to reach and inter-studies analyses of data from different isolation protocols are unmanageable. Therefore, there is an unmet need to optimize and standardize methods and protocols for the isolation and purification of EVs. This review focuses on the diverse techniques and protocols used over the years to isolate and purify EVs with a special emphasis on their adequacy for proteomics applications. By combining recent advances in specific isolation methods that yield superior quality of EV preparations and mass spectrometry techniques, the field is now prepared for transformative advancements in establishing distinct categorization and cargo identification of subpopulations based on EV surface markers.
Keywords: extracellular vesicles, exosomes, mass spectrometry, proteomics
1. Introduction
Extracellular vesicle (EV) is an umbrella term encompassing various subtypes of cell-released, non-replicating lipid bilayer structures that encloses and protects a highly diverse molecular cargo that is generally composed of bioactive molecules such as lipids, nucleic acid (DNA and RNA), proteins, and metabolites. Since the discoveries of microvesicles and exosomes over 25 years ago [1, 2], advances in imaging, proteomics and physical separation techniques have helped identify several more types of vesicles, of various sizes and cargo composition.
However, the EV research community has yet to reach a consensus on specific markers of EV subtypes [3]. It therefore remains exceptionally contentious to assign an EV to a particular biogenesis mechanism. As such, the International Society for Extracellular Vesicles (ISEV) has released position statements that propose rigorous guidelines for EV experimentation and nomenclature [3, 4]. The main position is for authors to consider using working terms for EV subtypes if and when specific markers of subcellular origin cannot be ascribed in lieu of terms such as exosomes or microvesicles given that the latters have historically been misuse. For example, authors could include working terms such as 1) size characteristics of EVs, with range-defined parameters such as small (<100 or <200 nm), medium (between 200 nm to 1 μm) and large (>1 μm) EVs. Alternatively, EV subtypes can be described by 2) their density in gradient centrifugation systems (low, middle, or high), with each range defined. EV subtypes can also be pinpointed by 3) their biochemical composition such as surface proteins (CD63+- or CD81+-EVs) or lipid composition. In addition, EVs can be defined through a description of source conditions or cell of origin (eg. EVs from cells under hypoxic conditions or melanoma EVs).
With this in mind, there are now well-accepted and reproducibly demonstrated concepts that are foundational in the field [5, 6]. For example, to be recognized as exosomes, EVs need to meet certain molecular and physical criteria. Exosomes are relatively small EVs (with sizes ranging between 30–150 nm diameter) that usually originate within late endosomal compartments. Exosomes have a cup-shaped physical appearance as determined by transmission electron microscopy owing to the dehydration step during the preparation of the sample for electron microscopy [7]. Their biogenesis and release mechanisms involves tetraspanins membrane proteins, Rab proteins, elements of the endosomal sorting complexes required for export (ESCRT), syntenin and other regulators [5, 6], therefore exosomes display expression of specific marker molecules that are associated with endosomal biogenesis and release. Finally, exosomes have an ability to sediment at high centrifugation forces and/or floatation at specific densities of sucrose or iodixanol gradients [3, 5, 6].
On the other hand, ectosomes, which comprise microvesicles, microparticles, oncosomes and migrasomes are much larger structures (>150 nm to several μm in diameter) formed through cellular surface budding of plasma membrane [6] and contain different cargo than exosomes. Large vesicles sediment differently than exosome on gradient centrifugation and are typically devoided of endosomal markers. In addition to membranous particles, cells release solid proteinaceous-nucleic acid amalgam particles called ribonucleoparticles (RNPs), as well as the recently documented structures known as exomeres [8–10]. In general, ectosomes, RNPs and exomeres are much less well-defined biochemically than exosomes currently are, and there is a pressing need to better define these entities biochemically and physically.
There has recently been an accrued interest in characterizing EVs lipid composition and cargo to identify clues to their biogenesis, targeting and cellular effects. One of the major challenges of protein identification from EV samples as opposed to RNA identification, is the absence of amplification techniques for protein cargo making detection of minor proteins dependent on the analytical sensitivity of the methods used. This has forced studies to be designed towards high protein yields, which are associated with higher EV heterogeneity rather than aiming for homogeneity of EV subtype samples but with much lower protein yields. The purpose of this review is to focus on experimental methods currently used to separate EV entities and approaches to study their proteomes. This review does not focus on lipidomics of EV as this topic has recently been covered eloquently in other reviews [11–13].
At the time of writing this review, there are 392 publications that utilized proteomic techniques to analyze EVs (Figure 1A) (Pubmed, search criteria, “proteomic” and “vesicle” or “exosome”). 93 manuscripts were chosen to be curated [14–106] and were included in this review. They were chosen mostly based on availability of their proteomics data through the vesiclepedia deposition database (microvesicles.org). Publication of those 93 manuscripts spans the years (Figure 1B,C) so as to provide an unbiased sampling of source of materials, species, methods of collection, processing and analysis. The manuscripts were curated on the basis of whether the source material (bodily fluids, tissues or cells) came from normal or diseased individuals, the species, the isolation methodology, post-isolation clean up, pre-mass spectrometry processing, mass spectrometry techniques and post-mass spectrometry analysis, and finally, the number of proteins identified (Figure 2 and Supplemental Table 1). These analyses reveal interesting aspects of EV isolation and processing that are covered in the following sections.
Figure 1.
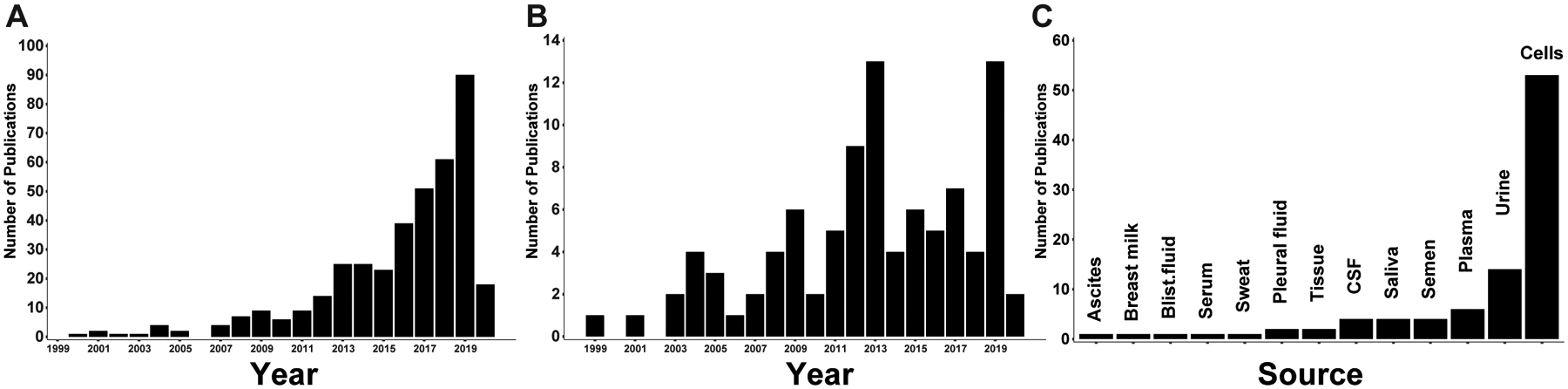
Distribution of publications on proteomics studies of EVs. A) Results of a Pubmed search (search criteria, “proteomic” and “vesicle” or “exosome”) as of 02-01-2020 showing the number of manuscripts per year. B) Distribution of the publication years of the 93 curated manuscripts that sourced Figure 2 and Supplemental Table 1. C) Frequency of the different sources of EVs curated from the 93 studies reviewed herein.
Figure 2.
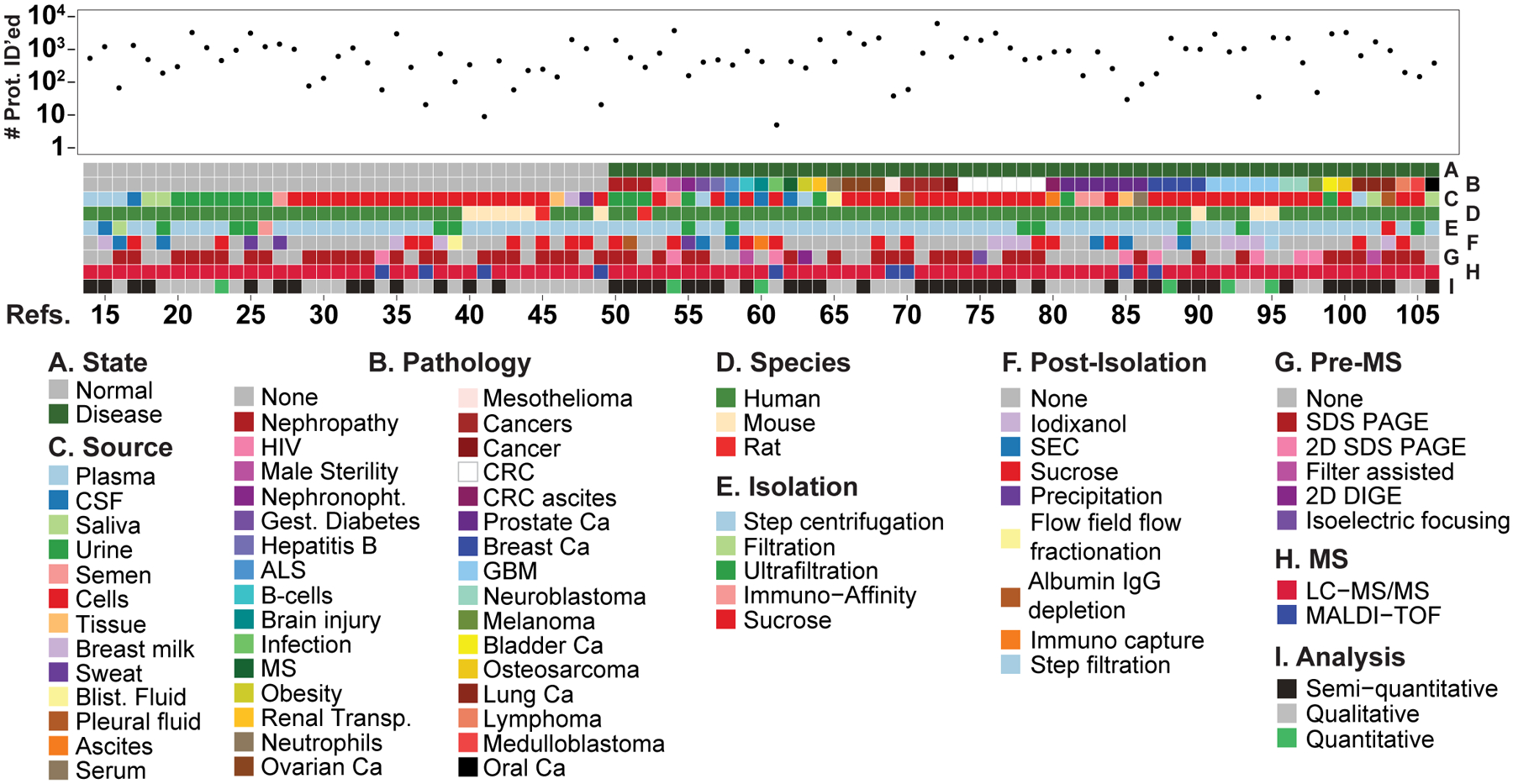
Tile plot depicting the various categories used to analyze the 93 proteomics manuscripts with their number of proteins identified for each. The Refs. numbers [14–106] correspond to the manuscripts listed in the references section. Under MS Analysis, semi-quantitative is defined as MS spectral data converted to absolute data, and quantitative data was obtained from the quantitative methods iTRAQ (Isobaric Tags for Relative and Absolute Quantitation), TMT (Tandem Mass Tag), or SILAC (Stable Isotope Labeling with Amino acids in Cell culture). Abbreviations: CSF; cerebrospinal fluid, Blist.; blister, Nephronopht. Nephronophthisis, Gest. Gestational, ALS; amyotrophic lateral sclerosis, MS; multiple sclerosis, Transp.; transplantation, Ca; cancer, CRC; colorectal cancer, GBM; Glioblastoma multiforme, SEC, size exclusion chromatography, SDS-PAGE; sodium dodecylsulfate polyacrylamide gel electrophoresis, 2D; two dimentional, DIGE; difference gel electrophoresis, LC-MS/MS; liquid chromatography tandem mass spectrometry, MALDI-TOF; Matrix-assisted laser desorption/ionization- time of flight.
2. EVs Come in Different Shapes and Sizes.
Recent reviews detailing our current knowledge of the mechanisms of biogenesis of EVs [5, 6, 107] show that there exists a remarkable amount of heterogeneity within EV subtypes. Nevertheless, the field now recognizes that EVs can be loosely subdivided into subtypes based on size. The work of many laboratories has been reviewed elsewhere and this section serves as a brief overview of the various accepted subtypes of EVs.
Exosomes are considered the smallest EV subpopulation, ranging from 40–120 nm in diameter. Exosomes are generally understood to originate from the classical endosome and multivesicular bodies (MVBs) axis and are secreted into the extracellular milieu upon fusion of the latter to the plasma membrane of cells. As an example of the ever-evolving EV field, a recent report uncovered distinct vesicle types within an exosomal population. Using asymmetric flow field-flow fractionation, the Lyden group reported the existence of small exosomes of 60–80 nm in diameter and larger exosomes 40–120 nm within the same exosomal preparation and each with distinct proteomic contents [10]. The ability to isolate further refined subpopulations of exosomes through more and more sophisticated methodologies and based on size or other physical properties will not only enhance our understanding of EV biogenesis but also function.
Microvesicles and microparticles are mid-sized EVs of 100–1000 nm in diameter, which are considered to be released from cells through budding and fission of the plasma membrane (reviewed in [5, 6]). As such, they contain different protein cargos than exosomes but with some degree of overlap [35, 76, 95]. A better understanding of the biogenesis of the medium-sized EV population will require the incorporation of additional methods of size separation or other physical attributes with powerful discriminating capacities.
Regardless of the triggering induction mechanisms (extrinsic or intrinsic), the active process of apoptosis occurs through several stages. First, cells undergo nuclear chromatin condensation, nuclear splitting and the frequent formation of micronuclei, then membrane blebbing, followed by the final stage of partition of the cellular contents into distinct membrane-bound vesicles called apoptotic bodies (Abs) or apoptosomes. These irregularly shaped vesicles are between 500–2000 nm in diameter and can be more abundant than other EVs. They are highly variables in size, structure and composition, as they contain a cellular components, including micronuclei, chromatin remnants, cytosolic contents, degraded proteins and DNA fragments and in some cases intact organelles [108]. Upon release into the extracellular space, ABs are phagocytosed by parenchymal cells, macrophages or neoplastic cells and are swiftly degraded within the phagolysosome. ABs have been reported to vehiculate the horizontal transfer of oncogenes in tumors [109], however clearance of ABs is a very rapid process and therefore the presence of interstitial ABs in vivo is often unobserved.
Oncosomes also referred to as Large Oncosomes (LO) are an outsized EV subpopulation (1000–10,000 nm in diameter) that appears to be exclusively shed by cancer cells [110, 111] and originally described in prostate cancer [112]. They are released from large protrusions of the plasma membrane of cancer cells, their composition is unique when compared to large vesicles from normal cells [112], and they have been shown to mediate the functional transfer of miRNAs and proteins to cancer-associated fibroblasts to facilitate the metastatic program of prostate cancer [113–115]. Note that although several tumors release large EVs [116–122], the biology of LO remains understudied outside prostate cancer.
Finally, migrasomes are recently discovered vesicles of various sizes ranging up to 3000 nm in diameter that are released from the tip of retraction fibers from migrating cells and that contain numerous smaller vesicles (50–100 nm) [123]. Proteomics analysis of migrasomes [123, 124] fueled marker discoveries and subsequent emerging mechanistic studies on migrasomes formation and function during cell migration in development and cancer [125–128].
In conclusion, EVs are a remarkably diverse group of structures and careful attention needs to be paid to the details of the isolation procedures and sources of materials when comparing inter-studies results.
3. EV Isolation Methods
As the field of EV biology evolves, more and more separation methodologies are being employed and currently, a typical protocol includes several separation steps before EVs are isolated (Figure 3). Every laboratory has applied slight variations on the basic protocols described below. Some of the techniques are non-specific, meaning that highly heterogeneous populations of EVs will be co-isolated and therefore robust pre- and post-purification steps are important.
Figure 3.
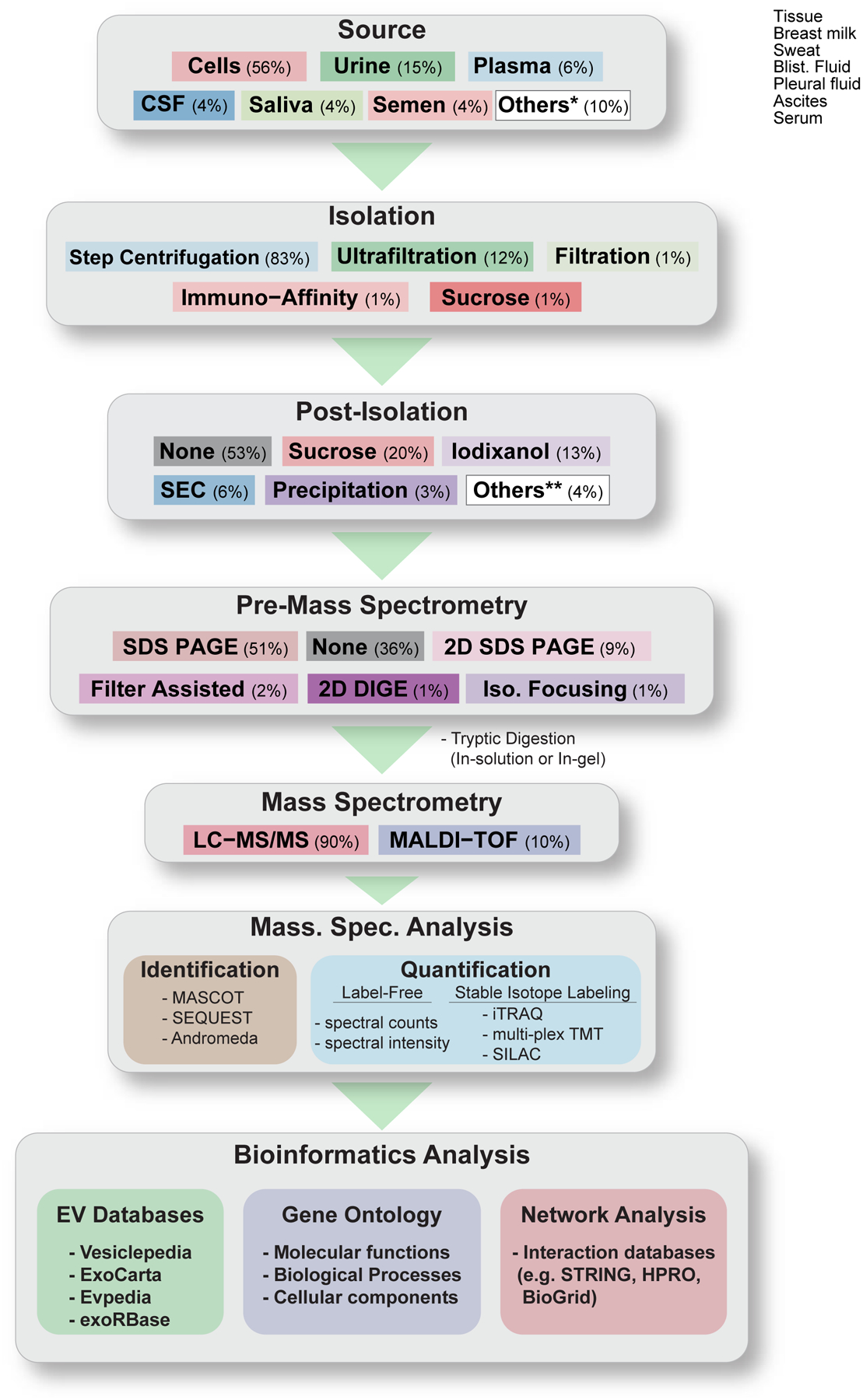
Flowchart depicting the various sources of EVs, and approaches to high-throughput mass spectrometry-based proteomic analysis of EVs from the manuscripts reviewed herein. Percentage (%) indicates the number of studies (out of the 93 studies reviewed) that uses the indicated techniques/methods. Others* comprise: Tissue, Sweat, Serum, Pleural Fluid, Blister Fluid, Breast Milk, and Ascites. Others** comprise: Step Filtration, Immuno Capture, Flow Field Flow Fractionation, Albumin or IgG Depletion.
3.1. Ultracentrifugation
Ultracentrifugation has been and remains a conventional and commonly used method for the isolation of EVs. Initial separation of large contaminants (e.g. dead cells or large debris) is achieved by differential centrifugation of the source material, which in many cases consists of a cell culture supernatant or a bodily fluid, first at low g forces (300–2000 g). In many cases, this is followed by a higher g force centrifugation (10,000g), which will pellet larger vesicles. An often used alternative is subjecting the supernatant to filtration through a 0.22 nm filter to remove particles larger than 200 nm. In cases where large volumes are involved, the pre-cleared supernatant can be concentrated using membrane-based ultrafiltration (see section 3.3). Finally, the cleaned supernatant is ultracentrifuged at high speed (100,000 g) for 1 to 2 hours. The pellet, which comprises small EVs is washed with PBS and ultracentrifuged again. Historically, the EVs present within this pellet have been, and are still referred to as exosomes. According to ISEV however, additional characterization is necessary to label these as exosomes, lacking otherwise, they should be referred to as small EVs. Although highly implemented in many laboratories, this basic method contains key properties that should not be overlooked. For example, the combinations of rotor type, g force and time of centrifugation should be considered carefully to achieve proper sedimentation. In addition, the viscosity of the starting materials is especially important, for sedimentation efficiency differences exist between plasma, serum and conditioned culture media [129].
A key point to remember from ultracentrifugation methods is that even though samples are pre-processed to reduce contamination of the final pellet from larger EVs, dead cells and debris, ultracentrifugation remains non-specific to generate a pure population of exosomes. Some undesirable components such as protein aggregates or aggregated smaller vesicles will co-sediment with the pellet. From the perspective of protein cargo characterization using proteomic techniques, ultracentrifugation has disadvantages and additional purification steps are necessary, otherwise the final results will be affected by these non-exosomal proteins.
3.2. Precipitation
Polymer-based precipitation principles have been adapted to the EV field and popularized in commercial EV isolation kits (e.g. Exosome Isolation kit (Life Technologies), Exoquick (System Bioscience) and Exo-spin (Cell Guidance System)). The isolation is based on the creation of a polymer network that captures all components (incl. EVs) present in the solution under specific salt and temperature conditions, which essentially reduces solubility and causes precipitation of these components. Following low-speed centrifugation, precipitates containing EVs are recovered and washed in PBS before downstream analysis. Although less laborious than ultracentrifugation, this procedure has serious limitations with regards to subsequent proteomics analysis because the samples now contain polymer molecules that are often incompatible with mass spectrometry applications.
3.3. Membrane-Based Ultrafiltration
The main principle of this approach is separation based on size of vesicles. Ultrafiltration units that are commercially available (usually with a molecular weight cut off of 100 kDa but it can vary) function like sieves that keep EVs on their membranous surface and allow flow through of most of the sample buffer resulting in a significant reduction in volume. This approach is especially convenient and efficient in reducing large volume samples such as urine or cell culture media. Low protein binding rated membranes are recommended as they reduce the attachment of EV proteins and therefore facilitate their recovery. In addition, procedures based on centrifugation appear to yield superior outcomes than pressure-driven devices [130, 131]. Given the membrane MWCO values, there are reports [130, 131] of noticeable reduction in high-abundance proteins from biofluids using ultrafiltration, making this approach very attractive for downstream mass spectrometry applications.
3.4. Size Exclusion Chromatography
In addition to ultrafiltration, gel filtration is another method that separates EVs based on size. Briefly, size exclusion chromatography’s basic principle is based on a matrix of polymeric beads, typically crosslinked, beaded form of agarose of diverse diameter ranges, packed in columns. Isolation of target materials is achieved by loading samples onto a packed matrix and gravity flow through the heteroporous resin. Typically, there is no interaction between the matrix and the components of the samples. Materials from samples are separated by differential accessibility to the pores. Larger molecules enter fewer pores than smaller molecules and are therefore exiting the matrix before smaller molecules. Commercially available resins of various pore sizes allow for separation based on diameter of particles. The advantages of this approach to separate EVs based on size are a good reproducibility and the low pressure involved, which leads to a retention of the shape of the vesicles. Vesicles are devoid of protein contaminants e.g. high abundance protein aggregates, and are usable in functional studies [132, 133]. On the other hand, the sample volume typically cannot exceed 10 percent of the resin volume, thus limiting studies where larger volumes are involved. Another disadvantage is that EVs of a given size tend to elute in several fractions and can be rather quite diluted, requiring downstream concentration steps. Isolation of EVs using stand-alone gel filtration isolation from serum for mass spectrometry analysis has been achieved in the past [134, 135], however most of the successful SEC usage for EV analysis has been in combination with ultracentrifugation and/or ultrafiltration.
3.5. Affinity-Based Purification
The main principle of affinity-based purification methods is to capture EVs by targeting their surface markers. These mostly include, but are not restricted to, members of the tetraspanin family (e.g. CD9, CD63, or CD81), EpCAM or annexin. Specific antibodies against these surface markers are conjugated or immobilized onto various solid carriers that can easily be separated from the sample mixtures being analyzed. Commercial kits that make use of immuno-beads, ELISA plates, chromatographic columns and microfluidic devices as carriers are available. Each of the platforms has intrinsic advantages and disadvantages. In terms of mass spectrometry proteomics analysis, they mostly all fall short of generating quantities of starting materials sufficient for deep and comprehensive MS/MS studies without being prohibitively expansive. Another aspect to bear in mind is the presence of copious amount of Ig protein present in the affinity purified samples post elution and the buffer composition necessary for separating the EVs from the solid carrier are not always compatible with mass spectrometry techniques. The major drawbacks of this approach are the unknown EV types that are not captured by these antibodies. Affinity-based purification methods are not well-suited for unbiased discovery approaches. On the other hand, affinity-based isolations are fulfill the requirements for diagnostic-oriented biomarker studies of disease-specific EVs isolated from biofluids. Finally, it is worth mentioning that affinity-based isolation methods require prior purification and/or concentration of EVs from samples. As such, they are typically viewed as complementary to ultra-centrifugation and precipitation methods.
In addition to antibody-directed isolation methods, there exist a few examples of alternative approaches that are much less selective than antibodies but nevertheless based on affinity purification. The addition of commercially available (ME-kit, New England Peptide) synthetic peptides called venceremin (Vn), which bind heat shock proteins present on the surface of EVs, to cell culture or biological fluids triggers an affinity precipitation of EVs that can be recovered by centrifugation at 16,000 xg and is suitable for high-throughput proteomics. [136, 137]. Another approach is based on the affinity of heparin for EV. Based on previous observations that heparin blocks EV uptake in cells [138], Balaj and colleagues demonstrated that Heparin-conjugated agarose beads can affinity purify EVs from cell culture media and plasma [139]. Finally, some lectins have high affinity for EV surface glycoproteins, and have been used in lectin-induced agglutination methods for the isolation of urinary EVs [140–142]. These methods represent alternatives to traditional approaches and are bound to expand the tool kit used by EV biologists.
3.6. Density-Based Centrifugation
Density gradient centrifugation is an established technique designed to separate particles based on their size and density. Its common use for the isolation of subcellular components has been adapted to isolate EVs. EVs have different sizes and diverse lipid, protein and nucleic acid compositions owing to their varied subcellular origins. This translates into EVs displaying different densities. EVs of identical diameter may have different densities owing to their cargo and lipid composition, and alternatively, EVs of different sizes but of identical cargo and lipid compositions can have the same densities. Therefore, one needs to be cognizant that density-based approaches can enrich and purify vesicles away from protein aggregates and other unwanted contaminants but perhaps are not as stringent to achieve specific vesicle purification based on size alone. Nevertheless, there are two main approaches to density-based centrifugation. Samples are centrifuged through 1) a continuous density gradient or 2) a discontinuous gradient. In the former, the density of the medium seamlessly increases from the top of the tube (less dense) to bottom (more dense). Gradients can be generated using gradient maker devices prior to centrifugation or gradients are established during centrifugation. In discontinuous density gradients, the densities of the medium increase in discrete steps from the top to the bottom of the tube.
Sucrose medium has traditionally been popular for exosomal studies and the EV community recognizes that for example, exosomes are defined as vesicles that sediment at densities between 1.10 and 1.19 g/mL [3]. This characteristic has been extensively employed to purify exosomes from more crude EV preparations. The approach is used in conjunction with ultracentrifugation where high-speed (~100,000 xg) pelleted crude EV preparations are separated from non-vesicular protein aggregates and other vesicles through centrifugation on density gradients of sucrose or sucrose cushions. Crude EV preparations are either top- or bottom-loaded and during centrifugation, vesicles float to their equilibrium density with the sucrose, thus separating vesicles of specific densities away from each other and particles. Vesicles that equilibrate at densities of 1.10–1.19 g/mL are referred to as exosomes. Fractions are collected, diluted and pelleted to yield purified preparations of vesicles.Note that sucrose gradients are hyperosmotic and the harvested EVs from them will not retain their integrity.
Iodixanol is an alternative medium to sucrose with similar parameters, including density range. Iodixanol gradients are iso-osmotic and self-generating, thus maintaining the integrity of EVs and affording high reproducibility. OptiPrep™ is a sterile 60% (w/v) solution of iodixanol with a density of 1.32 g/ml, which has been used to isolate EVs from several laboratories. A modified version of the more traditional iodixanol density gradient was established where EVs are centrifuged through a 17% Optiprep cushion with a superior exosome yield [143]. It is worth noting that current protocols describing density gradient centrifugation call for relatively short (4–16 hrs) ~ 100,000 xg ultracentrifugation preparations while other studies describe much longer (up to 90 hrs) and higher speed (190,000 xg) centrifugations to reach equilibrium density [144, 145]. It thus remains possible that traditional density gradient protocols might not adequately separate exosomes from other EVs and introduce artifacts.
3.7. Sequential Filtration
An approach relying on serial filtration through decreasing pore-sized filters has recently been described [95, 146]. In this method, cell culture conditioned media was serially filtered through 0.8 μm, 0.22 μm and 0.02 μm filters to collect vesicles ≥ 800 nm, ≥220 nm and ≥ 20 nm respectively. Proteins and total RNA were directly harvested on filters for downstream proteomic and RNA-seq analysis. This method is time consuming but the purity of vesicles as gauged by transmission electron microscopy and in terms of size distribution appears to be robust [95, 146].
3.8. Comparative Analysis of EV Isolation Methods
The recent explosion in EV research continues to fuel the diversity of the isolation protocols available to researchers. Despite appeals to more robust standardization protocols [3, 4, 147], unanimity amongst researchers remain unattainable. Perhaps a pressing approach to remedy this discordance is the need for comparative studies of isolation methods. Over the years, several groups have set out to perform side-by-side comparisons of isolation procedures to establish advantages and disadvantages of each technique in terms of purity and yield outputs [131, 143, 148–154]. When differential ultracentrifugation is compared to one-step sucrose cushion ultracentrifugation using cell culture supernatant, it was found that sucrose cushion ultracentrifugation yielded two to three times the amount of exosomes [149]. Similarly, ultracentrifugation through an OptiprepTM cushion yielded twice as much materials as traditional ultracentrifugation [143]. Another study compared exosome purification using ultracentrifugation alone, ultracentrifugation with an iodixanol cushion and ultracentrifugation on an iodixanol density gradient and concluded that the recovery of the gradient method was superior to the other two approaches [154]. Other studies showed that no significant differences in size distribution and morphology were achieved by different methods [131, 153]. Precipitation methods tend to produce higher yield of EVs than other methods [131, 153] but are also carrying protein complexes contaminants.
Finally, if sample purity is defined as mg of proteins per particle number, ultracentrifugation with Optiprep gradient is a superior approach [131, 153] as it separates high abundance contaminating proteins from EVs [150]. Likewise, ultrafiltration concentration coupled to size exclusion chromatography yields similar results [131].
3.9. A Cautionary Note on Purity of EVs
Studies isolating vesicles from cell culture conditioned media have to take into consideration the potential for vesicle contamination from other species, e.g. mainly bovine from the bovine serum supplements typically added to culture media. A simple strategy is to use EV-depleted bovine serum, and that has been shown to curb the cross species problem [4]. Regardless of the methods used to isolate EVs, it is highly recommended to authenticate their purity through several means such as electron microscopy (EM) visualization, which assess size, morphology, integrity and provides a measure of size heterogeneity. Combining EM results to nanoparticle tracking analysis [155] or other technologies capable of determining particle sizes (dynamic light scattering [156] or resistive pulse sensing [157]) and western blotting for the presence of specific markers (e.g. tetraspanning proteins, TSG101, annexins or synthenins) is viewed as the most robust characterization of EV isolation procedures.
Preservation of EV functionality is not necessary for downstream mass spectrometry proteomic analyses. Instead, reduction in contaminating protein complexes and control over buffer compositions are more important features of any isolation protocols. In fact, a few studies have shown that the choice of isolation procedures influence significantly the analysis and interpretation of mass spectrometry data [150, 152, 153]. It is recommended that low quality samples be discarded in order to avoid artifacts and false identification of EV cargo proteins. It is therefore highly recommended to verify, when possible, the biological and chemical purity of samples.
4. Source of Material for EV Isolation
EVs have been isolated from various sources. In its simplest form, EVs are isolated from cultured cells’ conditioned media using the methods described above. Careful considerations need to be given to protein contaminants commonly found in cell culture media supplements, notably fetal bovine/calf serum (FBS) or other animal biofluid derived supplement (e.g. B27). Bovine EVs are present within FBS and they need to be removed prior to EV isolation procedures to avoid contamination. commercially available EV-depleted FBS can be purchased, however these tend to be expensive. A more popular alternative is to deplete EVs from FBS by ultracentrifugation at 100,000 xg for 16–24 hours followed by sterilizing filtration or variations on this approach [158, 159].
For proteomics studies, bovine serum albumin protein contamination is also a major problem in the context of simple ultracentrifugation approaches. Harvesting EVs in serum-free media is often non-optimal as cells can develop stress responses to serum withdrawal, which can translates into unwanted changes in EV production and cargo. Even washing cells with phosphate buffered saline prior to collection incubation in low or no serum media does not completely remove albumin. To completely get rid of albumin prior to proteomics studies from EVs, a size exclusion or density gradient centrifugation based isolation of EVs is warranted.
The isolation of EVs from bodily fluids poses much different challenges than those observed in cell culture systems. First, EVs entering bodily fluids are produced by an immense number of different types of cells in tissue and in varying proportions, increasing the heterogeneity to unimaginable levels. Second, because fluid EVs are circulating they can be modified (chemically or biochemically) during the course of their journey through bodily fluids, which can cause them to aggregate or fuse. Finally, the most important aspect of isolating EVs from bodily fluids is the prominent presence of high amounts of contaminant materials (proteins, lipids, debris etc.), which can impede an accurate and efficient isolation and analysis.
Plasma (or serum) is one of the most commonly used bodily fluid sources to isolate EVs. It is relatively easy to collect and process, however special attention needs to be given during and post drawing for optimal storage and to avoid damaging EVs [147, 160]. Albumin, lipoproteins and immunoglobulins are highly abundant constituent proteins that can co-isolate with EVs and overwhelm the less abundant EV cargo proteins. For discovery-based proteomics studied, it is highly recommended to include additional steps (affinity chromatography, size exclusion chromatography or density gradient centrifugation) in the isolation pipeline to separate these contaminants away from EVs. The ease in collection of plasma/serum is advantageous for biomarker monitoring of cancers located in hard to access anatomies such as lung or brain cancers.
Saliva is made up of fluids from multiple secretory glands within the oral cavity. Saliva contains much less proteins than plasma/serum, thus making it a good source for interrogating low abundance proteins from EVs. It does contain however copious amount of bacteria and high levels of amylase, a viscous enzymatic protein necessary for the initiation of the digestion of carbohydrates (mainly starch), which hampers EV isolation. Clever approaches to circumvent these drawbacks have been established [161] and saliva from oral squamous cell carcinoma and non-small cell lung cancer patients has been used to study EV proteomics [102, 106, 162].
Urine is another easily accessible, non-invasive biofluid that can be collected in large volume. Although it is less biologically complex than plasma/serum, there is a wide range of urinary proteins with high-abundance proteins like the glycoprotein Tamm-Horsfall protein (a.k.a. uromodulin) that require removal prior to EV isolation and proteomics analysis.
Seminal fluid or semen is composed of fluid and constituents from several tissues including the prostate, which makes it an ideal source of material for studies of prostate- and prostate cancer-derived biomarker EVs. In normalcy, seminal EVs or “prostasomes” are important during the sperm maturation process [163]. However, in prostate cancer, seminal EVs have recently been suggested to represent a promising source of prostate cancer biomarkers [27].
Pleural effusion is the accumulation of fluid in the pleural cavity that is often associated with malignancy or other diseases, and can be a useful tool for diagnosis. Few studies have utilized pleural fluid-derived EVs for proteomic profiling [70, 103], demonstrating the potential for this fluid in biomarker studies. Like most other bodily fluids, highly abundant proteins (e.g. immunoglobulin and complement proteins) dominate the samples and additional separation steps are necessary to improve EV purification.
Ascites refers to the abnormal accumulation of fluid in the abdominal cavity. This condition is associated with various diseases including advanced-stage cancers. Cancer ascites is rich in tumor cells and is a potential source of cancer-derived EVs. To date, only one study has demonstrated its use in EV proteomic profiling for cancer EV marker discovery [80]. Another study demonstrated that ovarian cancer ascites contain higher numbers of EVs than non-cancer ascites [164], however EV cargo profiling was not performed.
Cerebrospinal fluid (CSF) is another biofluid source of EVs. Although not as accessible as other biofluids, CSF has been used to isolate EVs for proteome analysis with the aim of biomarker identification for multiple sclerosis [62, 165, 166], and chronic traumatic head injuries [60, 167].
Finally, sweat [48] and tears [166, 168] are two additional biofluid sources that have been utilized for EV isolation and proteomic analysis. Overall, EVs can be isolated from nearly all biofluids in sufficient numbers to allow for proteomic analyses of biomarker discovery and diagnostication. Based on the analysis of ~25% of all EV proteomic publication (93/392), by far the most common source of EVs is conditioned media from cell culture systems and the most common sources of biofluids are urine and plasma (Figure 1C).
5. Processing of Samples for MS.
Proper preparation of samples prior to mass spectrometry analysis is crucial. Remnant chemical contaminants from methods for pre-processing and isolation of EVs can be devastating and obtaining a high quality sample remains a challenge. An important first step towards mass spectrometry analysis of EV cargo is the enzymatic (trypsin) digestion of protein samples. There are 3 methods that are consistently used in the field.
In gel digestion is a very popular approach in which protein samples are separated by polyacrylamide gel electrophoresis, followed by staining (e.g. coomassie) and bands or gel slices excised, and processed (dehydration, reduction, alkylation and rehydration) for tryptic digest. Extracted peptides (acetonitrile) are then loaded onto LC-MS/MS or processed for MALDI-TOF and sequencing. This 1 dimension gel electrophoresis removes abundant contaminants. An alternative still in use is 2-dimensional gel electrophoresis, where proteins are first separated based on their isoelectric point, followed by a second dimension SDS-PAGE. Proteins are visualized by Coomassie blue or silver staining and individual spots (proteins) are identified using 2D-PAGE databases and/or excised and identified by sequencing.
In addition to in-gel digestion, in-solution and filter-aided digestion are two methods of sample preparation for mass spectrometry. With in-solution tryptic digestion approach, proteins can be purified from detergents, salts or lipid components by precipitation (trichloroacetic acid, acetone or chloroform/methanol), and the downstream procedures (reduction, alkylation and digestion) can be performed in single tube format. Note that in-solution digestion result in a high complexity sample, unlike in-gel where proteins are pre-fractionated. Finally, the filter-aided digestion strategy [169, 170] utilizes denatured protein in urea-based buffer passed through a high molecular weight cut-off filter to discard detergents and other impurities in the effluent. Trapped proteins are washed and treated with trypsin for digestion on filter and peptides are recovered by centrifugation.
Overall, the above three strategies are highly suitable for preparing EV protein samples with high quality for downstream mass spectrometry analysis. Experimenters should focus their choice of method based on the complexity and size of the samples to be analyzed as well as the type of protein, salts, and detergents contaminants present.
6. Functional Properties of EVs in Normalcy and Cancer.
There exists numerous studies that demonstrate a role for EVs in intercellular communication, which have been reviewed elsewhere [107, 171]. However, there have been limited advances in our understanding of the various mechanisms of how horizontal transfer of EVs is capable of changing the physiology of the microenvironment. The field is still in a descriptive phase and several high profile studies have not been reproducibly validated by alternate groups. More rigorous genetic approaches to validate the purported intercellular communication roles of specific RNAs or proteins are warranted for the field to advance beyond observational phenomenology.
Nevertheless, it has been shown that EVs, RNPs and exomeres have a unique ability to vehiculate bioactive cargo between cell populations. As such, EVs and RNPs are suggested to be implicated in homeostatic and pathological processes. In cancer, they have been shown to impact cellular growth [172, 173]} and to influence the immune response to tumor cells (reviewed in [174–176]). EVs can mediate directional migration [177], cell invasion [92, 178, 179], angiogenesis [180–182], and metastasis [183–186]. Mechanistic insights into these studies remain to be brought forward.
An undeniable usage of EVs is perhaps their capacity to serve as circulating biomarkers of diseases. In the context of cancer, EVs serve as bio-carriers of tumor-specific mutations in the form of mutated nucleic acids (genomic DNA and/or RNA) and proteins that can readily be detected using highly sensitive and readily available technologies. In fact, the use of EV to monitor cancer progression and response to therapy is an active area of research, which has recently been reviewed [187], and will not be covered in details. However, rigorous experimentation is an important facet of cancer EV-based biomarker discovery and usage. For example, the use of a Glypican-1 as a single marker of pancreatic cancer detection and progression using liquid biopsy exosomes [188], was not duplicated from another group and in fact, the latter study showed the robustness and superiority of a combination of exosomal cancer markers to diagnose pancreatic cancer [189].
As mentioned, the majority of studies that aim at demonstrating EVs as vehicle of horizontal transfer of information are mostly descriptive in nature. This is also true for most EV proteomics studies reported so far. For instances, the studies highlighted in the following sections 7, 8 and 9, which are taken from the list of 93 studies from Figure 2, demonstrate this fact. For organizational purposes, studies have been grouped according to sources, healthy donor or normal cell lines in section 7, and diseases from patient biofluids or cell lines in section 8. A few studies reported on comparing the proteomes of EVs isolated from disease sources and control normal sources. In addition, a few comparative studies that analyzed EV proteomics of biofluids from several patients or a given disease or multiple cell lines from the same disease were highlighted in section 9.
7. EV Protein Cargos Sourced from Healthy or Non-Disease Models.
All studies that used bodily fluids (e.g. plasma, urine, semen, saliva) from asymptomatic normal individuals or normal cells to isolate EVs and identify their protein cargo by mass spectrometry reported the results without enrichment parameters or simply characterized their results by comparing to existing vesicle databases or other bioinformatics resources (e.g. DAVID [190, 191], Gene Ontology GO [192, 193], Ingenuity Pathway Analysis (QIAGEN Inc., https://www.qiagenbio-informatics.com/products/ingenuity-pathway-analysis), The Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis [194]). The following studies simply list proteins identified in EV preparations isolated from normal, healthy cell line sources; from a B-cell culture [37], microglia cell culture [43], hepatocytes in culture [45], human neural stem cells [39], trophoblast cells [36], and human Mesenchymal stem cells [38].
These following studies utilized various biofluids from normal healthy individuals to perform EV proteomic studies. Looze et al, showed that peroxisome proliferator-activated receptor-gamma (PPARγ is expressed in EVs isolated from normal human plasma, perhaps representing an exosomal-associated marker [16]. Utleg and colleagues studied EVs sourced from normal semen [82] and Thimon et al. used EVs isolated from epididymal interstitial fluid obtained during reversal vasectomies [46]. A study by Chiasserini and co-workers reports on EV proteins from normal cerebrospinal fluid [17]. Similarly, the following studies simply report on proteins identified in EVs isolated from various biofluid sources. Pisitkun and colleagues and Gonzales et al. identified protein cargo of urine EVs from healthy donors [20, 22] and reported that many proteins identified in urinary vesicles are associated with kidney diseases or hypertension, perhaps leading to the use of urinary EVs for early detection of renal diseases. Prunotto and co-workers studied the proteome of podocyte-derived EVs from urine [26]. Ogawa reported the protein cargo of EVs from saliva [19] and van Herwijnen and colleagues reported on the proteome of breast milk EVs [47]. Wu and colleagues reported on the proteome of EVs isolated from the sweat of healthy individuals after physical exercise [48].
8. EV Cargo Composition of Pathological Sources.
Many studies simply report protein compositions of EVs from disease sources, sampling from cancer cell lines, or bodily fluids (e.g. plasma, urine, pleural efflusion etc.) from patients with cancers or other afflictions.
Mears et al., listed a few melanoma cell line-derived EV proteins [98], Miguet et al. listed several proteins expressed in EVs isolated from a T cell leukemia cell line [33] and a proteomic analysis of EVs isolated from lymphoma cells (Raji cells) was performed by Yao and colleagues [104]. Sinha et al., reported the proteome of EVs isolated from 4 ovarian cancer cell lines [66] and Choi et al. listed EV proteins from a colorectal cancer (CRC) cell line [79]. Graner and colleagues listed proteins isolated from a glioblastoma cell line [94], Epple and co-workers reported on a list of EV proteins from medulloblastoma cell lines [105], Liang and colleagues profiled EVs isolated from two ovarian cancer cell lines [68], and Marimpietri et al. studied the proteome of EVs isolated from a neuroblastoma cell line [97].
Choi et al. reported on the isolation and characterization of the proteomes of CRC ascites EVs from patients [80] where they demonstrated a significant number of overlapping EV proteins between patients. Bard and colleagues isolated EVs from pleural effusions from lung cancer, breast cancer and mesothelioma patients and showed little overlap between the three cancers [70]. The proteome of pleural effusion EVs isolated from a mesothelioma patient does not overlap with EVs isolated from mesothelioma cell lines [69]. In the latter studies, the number of identified proteins from EVs were relatively modest and the lack of mesothelioma-specific EV markers perhaps reflect the difficulties associated with EV heterogeneity and inter-laboratory consistency.
In all of these studies, the authors simply reported the proteomics data “as is” without comparisons to cellular proteomes or other vesicle types and performed rudimentary bioinformatics analyses. Some of the studies are more informative in the sense that they provided additional proteins to the various vesicle databases, however these studies did not reveal important insights in terms of new EV-based markers.
9. Comparative EV Proteomics Studies.
Finally, the more advanced EV proteomics research aims at leveraging the power of MS proteomics to quantitatively compare different vesicle sources or types to one another. This approach yields insightful information on the biological questions originally intended to be addressed in those studies. The followings are examples of comparative proteomics studies.
Palazzolo et al. reported on the proteomes of cells and EVs of the breast cancer cell line MDA-MB-231 and showed a distinctive profile of proteins in EVs compared to cells [87]. Liem and colleagues studied the effects of activation of PI3K/Akt signaling by insulin in a colorectal cancer cell line on the proteome of EVs secreted [74]. The authors demonstrated that the protein cargo of EVs is modified upon activation of the insulin-PI3K-Akt signaling pathway, with enrichment in proteins implicated in cell proliferation. These results led the authors to hypothesize that EVs can amplify signals provided by growth factors in the tumor microenvironment.
Giri and colleagues demonstrated that M. tuberculosis-infected macrophages in culture secrete EVs that contain highly immunogenic mycobacterial proteins, an observation that led the authors to suggest that EVs containing M. tuberculosis antigens may serve as an alternative approach to tuberculosis vaccine development [195]. Peterson and colleagues compared the EV cargos of endothelial cells stimulated with plasminogen activator inhibitor type 1 (PAI-1) or tumor necrosis factor-alpha (TNF-α). They identified common EV proteins and unique to control and treated endothelial cells [28]. A more in-depth comparison with cellular proteins would have revealed mechanistic insights into the biological effects of PAI-1 and TNF-α on cargo composition. A study by Mathivanan et al. compared the protein profiles of EVs isolated from a colorectal cancer cell line to a murine mast cell and human urine-derived EVs and reported that a small subset of proteins were common to all EVs [78], including proteins involved in endosomal sorting complex required for transport (ESCRT) function, tetraspanins, signaling, trafficking and cytoskeletal proteins.
A study by Meckes et al. report on significant changes in EV proteomes upon infection with two human gamma herpesviruses [59]. EVs isolated from Kaposi sarcoma-associated virus contained a selective enrichment in proteins that would affect cellular metabolism. On the other hand, Epstein-Barr virus infected cells produce EVs that carry a proteome that would activate cellular signaling mediated through integrins, actin, IFN, and NFκB. The differences in EV contents upon infection by these two viruses suggest mechanism by which they modify their tumor microenvironment. Forterre and co-workers have defined the changes in EV proteins during the process of C2C12 myoblast cells proliferation and differentiation into myotubes, revealing stage-preferred enrichment of proteins in EVs from each state [42].
In a rat model of diabetes, Raimondo studied the proteomes of urine EVs isolated from diabetic rats at different ages and normal control animals to study the evolution of diabetes [52]. Although no specific proteins demonstrated longitudinal changes associated with diabetes progression, urine-isolated EVs from advanced diabetic rats can serve as a source of diabetes biomarkers. Gonzalez-Begne and colleagues have compared EV proteome of parotid glands to that of saliva and found that half of the EV proteins from the former are observed in saliva EV proteome [18]. The diagnosis of mature B-cell malignancies can be difficult and Miguet and colleagues performed proteomic analyses on EVs isolated from patients’ cells with chronic lymphocytic leukemia (CLL), small cell lymphoma (SLL) and mantle cell lymphoma (MCL) to identify diagnostic markers within EVs that are potentially relevant and useful. Comparison of the EV proteomes of CLL, MCL, SLL and normal controls identified CD148 as a discriminating MCL biomarker candidate [73].
A report by Moon and colleagues reported the proteomes of urine EVs from patients associated with early IgA nephropathy (IgAN), thin basement membrane nephropathy (TBMN) and control healthy individuals [50]. Among the differentially expressed proteins, vasorin precursor, alpha-1-antitrypsin, aminopeptidase N and ceruloplasmin were identified as biomarkers that differentiate early IgAN from TBMN. Chen and co-workers reported on urine EVs from bladder cancer patients and healthy individuals for the purpose of biomarker discovery. The authors discovered 24 signature proteins that changed significantly between bladder cancer and normal individuals [99]. Raj et al. showed that certain proteins from EVs isolated from urine of older healthy individuals differ from those of younger people [23] and Wang and colleagues demonstrated the ability to identify ~1000 proteins common in EVs from multiple urine samples from many individuals [21]. Dalli and colleagues studied the proteomes of microvesicles from neutrophils grown under adherent or suspension conditions and found that a substantial number of proteins are preferentially expressed in EVs isolated from one or the other conditions [65]. The authors found that alpha-2-macroglobulin and ceruloplasmin were enriched in the microvesicles isolated from adherent neutrophils, while heat shock 70kDa protein 1 was elevated in EVs isolated from suspension grown neutrophils. Annexin A1, Lactoferrin, and actin were observed at equal levels in the two subsets.
An analysis of the proteome of plasma EVs from lung adenocarcinoma patients and matched healthy subject controls identified proteins with significant differential expression [101]. The authors further demonstrated that a set of four EV-associated proteins (TPM3, HUWE1, SRGN, and THBS1) can distinguish adenocarcinoma cases from controls, supporting the notion that EV derived proteins can be a source of biomarkers for diagnosis and other tumor assessment. Finally, a study of CSF-isolated EVs from amyotrophic lateral sclerosis (ALS) patients and age- and gender-matched control healthy individuals reveal very few proteins that are either up or down regulated in EVs form CSF of ALS patients [58]. The significance of these in the etiology of ALS remains to be determined
A few studies have compared EV cargos of disease versus normal or between cancers. Li and colleagues reported on changes in EV proteomes of lymphocytes infected with HIV-1 versus non-infected controls and found a small number (14/770) of proteins to be significantly differentially expressed in EVs from HIV-1 infected cells, of which modulators of cellular apoptosis and proliferation are included [53]. Duijvesz et al. reported on differences in EV proteomes isolated from immortalized prostatic epithelia and prostate cancer cell lines [84]. Although the cancer cell lines were not derivatives of the immortalized prostatic epithelia cells, the authors nevertheless identified 9 proteins that are more abundant in prostate cancer cell lines than epithelia including PDCD6IP, FASN, XPO1 and ENO1, prompting the authors to suggest these as new candidate biomarkers for prostate cancer.
A study by Ji and colleagues report on proteome differences between EVs isolated from metastatic and non-metastatic isogenic colorectal cancer cell lines [77]. Not surprisingly, the authors observed a selective enrichment of metastatic factors and signaling pathway components in EVs from the metastatic cell line, which contributes to our understanding of the horizontal communication between tumor and normal stroma within the tumor microenvironment. Winck et al. studied the proteomes of salivary EVs isolated from normal healthy individuals and oral cavity squamous cell carcinoma patients and found a handful of proteins (e.g. prolactin-inducible protein, alpha-2-macroglobulin, lalectin-3-binding protein, mucin-5B) to be differentially expressed [106].
A significant study from David Meckes’ group identified the proteomes of EVs isolated from the NCI-60 cell panel, and uncovered common cargo proteins and cancer type-specific EV proteins [72]. The authors established EV markers such as vesicular trafficking proteins (Rab GTPases) and tetraspanins, and importantly, compared EV proteomes to whole-cell proteomes, which revealed high similarities but also several EV- and cancer-specific enrichments. Mallawaaratchy et al., studied the proteomes of EVs isolated from low grade glioma and high grade glioma tissues and revealed that a high level of invasion-related proteins were enriched in EVs of high grade tumors when compared to low grade gliomas [92]. Chandran and co-workers have defined the proteomic changes occurring in EVs isolated from a glioblastoma cell line grown in normoxic and hypoxic conditions [91]. In this study, the authors identified nine proteins that strongly denoted the hypoxic condition. The biological significance of these proteins during the hypoxic process needs to be further studied.
In a proteomic study comparing EVs isolated from semen of asthenozoospermic (a common cause of male infertility) and normozoospermic men, Lin and colleagues identified eleven proteins that were significantly upregulated and eighty that were downregulated [54]. The functions of these proteins were mainly associated with transport, metabolism and signaling pathways. This study provided more insights into the involvement of seminal EVs in asthenozoospermia and male infertility. Panigrahi et al. performed proteomic studies on serum-isolated EVs from African American and Caucasian men with prostate cancer and age- and ethnicity-matched healthy controls. The authors identified Isoform 2 of Filamin A protein strongly enriched in serum EVs of African American prostate cancer men whilst excluded from serum EVs of Caucasian prostate cancer men [86]. Further studies are required to validate this protein as a potential marker of ethnocentric prostate cancer. A study by Cheng and co-workers identified proteins that differ in their expression levels in EVs isolated from an ovarian cancer cell line when compared to EVs isolated from ovarian surface epithelial cells [67]. Although the ill-defined relationship between these two cell lines makes interpretation of these results in the context of ovarian cancer development difficult, the study nevertheless provided ovarian cancer EV markers.
In addition to these studies, there are a number of reports that compared EV proteomes of parental cell lines and their isogenic variants that carry specific oncogenic mutations. For examples, the proteomes of a parental glioblastoma cell line and its isogenically variant expressing EGFRvIII were compared and contrasted [93], revealing specific EV cargo changes that purport a more malignant phenotype. Similarly, isogenic breast cancer cell lines for HER2 overexpression alters the EV proteome [196] when compared to parental.
There are a few studies that showed the dramatic changes in EV composition when cells undergo the cancer hallmark of epithelial to mesenchymal transition (EMT). EVs isolated from MDCK cells following oncogenic H-Ras-induced EMT [197] demonstrate drastic differences in cargo proteins that are in line with phenotypic EMT. In another EMT study using the squamous cell carcinoma cell line A431, EV proteomic analysis of parental A431 and A431 cells having undergone EMT by treatment with the EGFR ligand TGFα and an E-cadherin neutralizing antibody, showed significant changes in EV cargo that are indicative of EMT [71]. Finally, comparative proteomics of EVs isolated from patental MDCK cells, and its isogenic MDCK cells intermediate for EMT, stably expressing the YBX1 gene, and Ras-transformed MDCK cells (complete EMT) showed that the latter EVs contained VEGF-associated proteins, while YBX1 expressing cells’ EVs were enriched with activated Rac1 and PAK2. [198]
Proteomic analysis of EVs from the colorectal cancer DLD-1 cell line that contain both wild-type and G13D mutant KRAS alleles and their isogenically matched derivative cell lines, DKO-1 (mutant KRAS allele only) and DKs-8 (wild-type KRAS allele only) has been performed [75]. The results demonstrate that mutant KRAS status of cells dramatically affects the composition of the EV proteome. [75]. Conditional ectopic expression of the TGFβ receptor TGFBR2 in CRC cell line HCT116 also changes the proteomic composition of EVs [199]. Moreover, isogenically mutating (R173H) or deleting TP53 in HCT116 significantly reduced EV sizes and changed EV protein cargo considerably, of which hepatocyte growth factor-regulated tyrosine kinase substrate was down-regulated in the EVs from the mutated and deleted TP53 isogenic cells.[200].
Finally, several studies have reported important changes in proteomics of EV isolated from pairs of non-metastatic and metastatic cell derivatives of colon, breast, pancreatic and bladder cancers [90, 183, 201–204]. Validation studies of biological function for given EV markers within the metastatic program remain to be perform for all of these studies and is necessary to solidify credence in EV biology.
Gangoda and colleagues have generated proteomics data on EVs isolated from mouse breast cancer cell lines with different metastatic potentials (nonmetastatic, weakly metastatic, or highly metastatic) [90]. The authors found that EVs derived from metastatic cells contain proteins that are implicated in promoting migration, proliferation, invasion and angiogenesis. In contrast, nonmetastatic cell-derived EVs comprise proteins involved in cell-cell and cell-matrix adhesion and maintenance of cell polarity.
In most if not all of these studies however, the degree of differences between two populations of EVs did not take into account the degree of differences between the respective cell lines. In other words, are the up- and down-regulated proteins observed in the different EVs similarly up- and down-regulated in the cells? This latter analysis is crucial to our understanding of the mechanisms that direct EV cargo production and should be seriously considered in future studies.
10. Comparative Studies of Proteomes of Diverse Vesicles.
The following reports compare the proteomic cargos of different sized EVs from a single source. These manuscripts are very informative because they helped us realize how heterogeneous EVs are in term of size and composition. In some instances, these studies identified EV size-centric protein markers. Turiak et al reported on proteomes of microvesicles (100–800 nm) and apoptotic bodies (>800 nm) from mouse thymus tissue and found that the majority of proteins detected were common between the two preparations or vesicles with fewer unique to each vesicle types [44]. This is a surprising observation given the divergent biogenesis of each vesicle types. Ogawa et al. showed different protein contents in two exosomal EV populations from normal saliva isolated by gel filtration. Both types of EVs contained the canonical EV markers Alix, Tsg101 and Hsp70 but also contained type-specific proteins, demonstrating the heterogeneous nature of EVs.
Clark and co-workers compared the proteomes of large EVs to those of two small EVs preparations from the breast cancer cell line SKBR3B [88] revealing a preferential expression of canonical exosomal markers in the small EV fractions. A study of EV proteomes isolated from non-mineralizing and mineralizing human osteoblasts showed a comparable (97 percent) EV proteome between the two types [32] with the remaining 3 percent of proteins that are preferentially expressed in the different type of vesicles requiring further studies to determine their role(s) in the mineralization of osteoblast process.
Kowal and colleagues have compared the dendritic cells proteomes of large and small EVs and of gradient centrifugation fractions and quantitatively demonstrated that several classically used exosome markers (e.g. frotillin, hsp70, MHCII) are present in all EVs [35]. The authors also identified proteins specifically enriched in small EVs and they have described a set of five protein categories that show different relative abundance in individual EV populations. Defining exosomes as vesicles expressing CD63, or CD81 or CD9, this study also unveil the presence of non-exosomal subpopulations within small EVs. This seminal study lays the foundation for future in-depth characterization of the heterogeneity of EVs. Finally, Sun et al. demonstrated that large and small EV proteomes from saliva of lung cancer patients differ in their contents and that some salivary EV proteins (CRNN, BPIFA1, IQGAP, and MUC5B) are of lung tissue origins [102].
11. Conclusions and the Future of Proteomics of Small Samples
Over the past few years, numerous studies have been published on different aspects of EV proteomics. In reviewing the literature, it was found that the number of proteins identified from mass spectrometry output is independent of sample provenance or preparation method. For the most part, the studies are highly descriptive and qualitative in nature and the field is now poised for the discovery of more biologically meaningful proteomic markers of EVs. In fact, a hot area of research is comparative proteomics of EV heterogeneity and markers of tissue provenance of EVs within biofluids. As mass spectrometry methods continue to advance and be perfected, enhanced protein identification with less input sample materials will increase. This will allow the community to envisage new isolation approaches that are directly addressing the heterogeneity problems that currently dampens EV research.
Supplementary Material
Supplementary Table 1. Tabulated version of Figure 2. Parameters curated from literature.
Acknowledgements
This work was supported by NIH grants to AC (P01 CA069246 and UG3 CA241685). The author is indebted to Dr. Xiaohu Zhang and Ms. Arianna Salazar Miranda of the Senseable City Lab at Massachusetts Institute of Technology for their coding contribution to Figure 2. Apologies are presented to our colleagues whose pertinent work was not included due to electronic search results oversight or space constraints.
Footnotes
Conflict of interest
The author declares that he has no conflicts of interest.
References
- 1.Johnstone RM, Adam M, Hammond JR, Orr L, and Turbide C, J Biol Chem 1987, 262, 9412. [PubMed] [Google Scholar]
- 2.Wolf P, Br J Haematol 1967, 13, 269. [DOI] [PubMed] [Google Scholar]
- 3.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG Jr., Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ Jr., Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D and Zuba-Surma EK, J Extracell Vesicles 2018, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, and Thery C, J Extracell Vesicles 2014, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo M, Raposo G, and Thery C, Annu Rev Cell Dev Biol 2014, 30, 255. [DOI] [PubMed] [Google Scholar]
- 6.Cocucci E and Meldolesi J, Trends Cell Biol 2015, 25, 364. [DOI] [PubMed] [Google Scholar]
- 7.Chuo ST, Chien JC, and Lai CP, J Biomed Sci 2018, 25, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Higginbotham JN, Jeppesen DK, Yang YP, Li W, McKinley ET, Graves-Deal R, Ping J, Britain CM, Dorsett KA, Hartman CL, Ford DA, Allen RM, Vickers KC, Liu Q, Franklin JL, Bellis SL, and Coffey RJ, Cell Rep 2019, 27, 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H and Lyden D, Nat Protoc 2019, 14, 1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, Mark MT, Molina H, Martin AB, Bojmar L, Fang J, Rampersaud S, Hoshino A, Matei I, Kenific CM, Nakajima M, Mutvei AP, Sansone P, Buehring W, Wang H, Jimenez JP, Cohen-Gould L, Paknejad N, Brendel M, Manova-Todorova K, Magalhaes A, Ferreira JA, Osorio H, Silva AM, Massey A, Cubillos-Ruiz JR, Galletti G, Giannakakou P, Cuervo AM, Blenis J, Schwartz R, Brady MS, Peinado H, Bromberg J, Matsui H, Reis CA, and Lyden D, Nat Cell Biol 2018, 20, 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi DS, Kim DK, Kim YK, and Gho YS, Proteomics 2013, 13, 1554. [DOI] [PubMed] [Google Scholar]
- 12.Kreimer S, Belov AM, Ghiran I, Murthy SK, Frank DA, and Ivanov AR, J Proteome Res 2015, 14, 2367. [DOI] [PubMed] [Google Scholar]
- 13.Skotland T, Hessvik NP, Sandvig K, and Llorente A, J Lipid Res 2019, 60, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostergaard O, Nielsen CT, Iversen LV, Jacobsen S, Tanassi JT, and Heegaard NH, J Proteome Res 2012, 11, 2154. [DOI] [PubMed] [Google Scholar]
- 15.Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, Lotvall J, and Lasser C, Cell Mol Life Sci 2018, 75, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Looze C, Yui D, Leung L, Ingham M, Kaler M, Yao X, Wu WW, Shen RF, Daniels MP, and Levine SJ, Biochem Biophys Res Commun 2009, 378, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiasserini D, van Weering JR, Piersma SR, Pham TV, Malekzadeh A, Teunissen CE, de Wit H, and Jimenez CR, J Proteomics 2014, 106, 191. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Begne M, Lu B, Han X, Hagen FK, Hand AR, Melvin JE, and Yates JR, J Proteome Res 2009, 8, 1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogawa Y, Miura Y, Harazono A, Kanai-Azuma M, Akimoto Y, Kawakami H, Yamaguchi T, Toda T, Endo T, Tsubuki M, and Yanoshita R, Biol Pharm Bull 2011, 34, 13. [DOI] [PubMed] [Google Scholar]
- 20.Pisitkun T, Shen RF, and Knepper MA, Proc Natl Acad Sci U S A 2004, 101, 13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Hill S, Luther JM, Hachey DL, and Schey KL, Proteomics 2012, 12, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, and Knepper MA, J Am Soc Nephrol 2009, 20, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raj DA, Fiume I, Capasso G, and Pocsfalvi G, Kidney Int 2012, 81, 1263. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Chinello C, Musante L, Cazzaniga M, Tataruch D, Calzaferri G, James Smith A, De Sio G, Magni F, Zou H, and Holthofer H, Proteomics Clin Appl 2015, 9, 568. [DOI] [PubMed] [Google Scholar]
- 25.Bijnsdorp IV, Maxouri O, Kardar A, Schelfhorst T, Piersma SR, Pham TV, Vis A, van Moorselaar RJ, and Jimenez CR, J Extracell Vesicles 2017, 6, 1313091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prunotto M, Farina A, Lane L, Pernin A, Schifferli J, Hochstrasser DF, Lescuyer P, and Moll S, J Proteomics 2013, 82, 193. [DOI] [PubMed] [Google Scholar]
- 27.Yang C, Guo WB, Zhang WS, Bian J, Yang JK, Zhou QZ, Chen MK, Peng W, Qi T, Wang CY, and Liu CD, Andrology 2017, 5, 1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson DB, Sander T, Kaul S, Wakim BT, Halligan B, Twigger S, Pritchard KA Jr., Oldham KT, and Ou JS, Proteomics 2008, 8, 2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banfi C, Brioschi M, Wait R, Begum S, Gianazza E, Pirillo A, Mussoni L, and Tremoli E, Proteomics 2005, 5, 4443. [DOI] [PubMed] [Google Scholar]
- 30.Xiao Z, Camalier CE, Nagashima K, Chan KC, Lucas DA, de la Cruz MJ, Gignac M, Lockett S, Issaq HJ, Veenstra TD, Conrads TP, and Beck GR Jr., J Cell Physiol 2007, 210, 325. [DOI] [PubMed] [Google Scholar]
- 31.Prokopi M, Pula G, Mayr U, Devue C, Gallagher J, Xiao Q, Boulanger CM, Westwood N, Urbich C, Willeit J, Steiner M, Breuss J, Xu Q, Kiechl S, and Mayr M, Blood 2009, 114, 723. [DOI] [PubMed] [Google Scholar]
- 32.Morhayim J, van de Peppel J, Demmers JA, Kocer G, Nigg AL, van Driel M, Chiba H, and van Leeuwen JP, FASEB J 2015, 29, 274. [DOI] [PubMed] [Google Scholar]
- 33.Miguet L, Pacaud K, Felden C, Hugel B, Martinez MC, Freyssinet JM, Herbrecht R, Potier N, van Dorsselaer A, and Mauvieux L, Proteomics 2006, 6, 153. [DOI] [PubMed] [Google Scholar]
- 34.Sander TL, Ou JS, Densmore JC, Kaul S, Matus I, Twigger S, Halligan B, Greene AS, Pritchard KA Jr., and Oldham KT, Shock 2008, 29, 504. [DOI] [PubMed] [Google Scholar]
- 35.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, and Thery C, Proc Natl Acad Sci U S A 2016, 113, E968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atay S, Gercel-Taylor C, Kesimer M, and Taylor DD, Exp Cell Res 2011, 317, 1192. [DOI] [PubMed] [Google Scholar]
- 37.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, and Stoorvogel W, J Biol Chem 2003, 278, 10963. [DOI] [PubMed] [Google Scholar]
- 38.Kim HS, Choi DY, Yun SJ, Choi SM, Kang JW, Jung JW, Hwang D, Kim KP, and Kim DW, J Proteome Res 2012, 11, 839. [DOI] [PubMed] [Google Scholar]
- 39.Kang D, Oh S, Ahn SM, Lee BH, and Moon MH, J Proteome Res 2008, 7, 3475. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Chen R, Kemper S, and Brigstock DR, Cells 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, and Amigorena S, J Cell Biol 1999, 147, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forterre A, Jalabert A, Berger E, Baudet M, Chikh K, Errazuriz E, De Larichaudy J, Chanon S, Weiss-Gayet M, Hesse AM, Record M, Geloen A, Lefai E, Vidal H, Coute Y, and Rome S, PLoS One 2014, 9, e84153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potolicchio I, Carven GJ, Xu X, Stipp C, Riese RJ, Stern LJ, and Santambrogio L, J Immunol 2005, 175, 2237. [DOI] [PubMed] [Google Scholar]
- 44.Turiak L, Misjak P, Szabo TG, Aradi B, Paloczi K, Ozohanics O, Drahos L, Kittel A, Falus A, Buzas EI, and Vekey K, J Proteomics 2011, 74, 2025. [DOI] [PubMed] [Google Scholar]
- 45.Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M, Elortza F, Lu SC, Mato JM, and Falcon-Perez JM, J Proteome Res 2008, 7, 5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thimon V, Frenette G, Saez F, Thabet M, and Sullivan R, Hum Reprod 2008, 23, 1698. [DOI] [PubMed] [Google Scholar]
- 47.van Herwijnen MJ, Zonneveld MI, Goerdayal S, Nolte-’t Hoen EN, Garssen J, Stahl B, Maarten Altelaar AF, Redegeld FA, and Wauben MH, Mol Cell Proteomics 2016, 15, 3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu CX and Liu ZF, J Invest Dermatol 2018, 138, 89. [DOI] [PubMed] [Google Scholar]
- 49.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, and Amigorena S, J Immunol 2001, 166, 7309. [DOI] [PubMed] [Google Scholar]
- 50.Moon PG, Lee JE, You S, Kim TK, Cho JH, Kim IS, Kwon TH, Kim CD, Park SH, Hwang D, Kim YL, and Baek MC, Proteomics 2011, 11, 2459. [DOI] [PubMed] [Google Scholar]
- 51.Zubiri I, Posada-Ayala M, Sanz-Maroto A, Calvo E, Martin-Lorenzo M, Gonzalez-Calero L, de la Cuesta F, Lopez JA, Fernandez-Fernandez B, Ortiz A, Vivanco F, and Alvarez-Llamas G, J Proteomics 2014, 96, 92. [DOI] [PubMed] [Google Scholar]
- 52.Raimondo F, Corbetta S, Morosi L, Chinello C, Gianazza E, Castoldi G, Di Gioia C, Bombardi C, Stella A, Battaglia C, Bianchi C, Magni F, and Pitto M, Mol Biosyst 2013, 9, 1139. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Aliotta JM, Asara JM, Tucker L, Quesenberry P, Lally M, and Ramratnam B, Proteomics 2012, 12, 2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Y, Liang A, He Y, Li Z, Li Z, Wang G, and Sun F, Mol Reprod Dev 2019, 86, 1094. [DOI] [PubMed] [Google Scholar]
- 55.Stokman MF, Bijnsdorp IV, Schelfhorst T, Pham TV, Piersma SR, Knol JC, Giles RH, Bongers E, Knoers N, Lilien MR, Jimenez CR, and Renkema KY, J Proteomics 2019, 192, 27. [DOI] [PubMed] [Google Scholar]
- 56.Jayabalan N, Lai A, Nair S, Guanzon D, Scholz-Romero K, Palma C, McIntyre HD, Lappas M, and Salomon C, Proteomics 2019, 19, e1800164. [DOI] [PubMed] [Google Scholar]
- 57.Zhao X, Wu Y, Duan J, Ma Y, Shen Z, Wei L, Cui X, Zhang J, Xie Y, and Liu J, J Proteome Res 2014, 13, 5391. [DOI] [PubMed] [Google Scholar]
- 58.Hayashi N, Doi H, Kurata Y, Kagawa H, Atobe Y, Funakoshi K, Tada M, Katsumoto A, Tanaka K, Kunii M, Nakamura H, Takahashi K, Takeuchi H, Koyano S, Kimura Y, Hirano H, and Tanaka F, Neurosci Res 2019. [DOI] [PubMed] [Google Scholar]
- 59.Meckes DG Jr., Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, and Raab-Traub N, Proc Natl Acad Sci U S A 2013, 110, E2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muraoka S, Jedrychowski MP, Tatebe H, DeLeo AM, Ikezu S, Tokuda T, Gygi SP, Stern RA, and Ikezu T, Front Neurosci 2019, 13, 1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JJ, Chen C, Xie PF, Pan Y, Tan YH, and Tang LJ, Microbes Infect 2014, 16, 283. [DOI] [PubMed] [Google Scholar]
- 62.Lee J, McKinney KQ, Pavlopoulos AJ, Han MH, Kim SH, Kim HJ, and Hwang S, Clin Chim Acta 2016, 462, 118. [DOI] [PubMed] [Google Scholar]
- 63.Barrachina MN, Sueiro AM, Casas V, Izquierdo I, Hermida-Nogueira L, Guitian E, Casanueva FF, Abian J, Carrascal M, Pardo M, and Garcia A, Proteomics 2019, 19, e1800248. [DOI] [PubMed] [Google Scholar]
- 64.Pisitkun T, Gandolfo MT, Das S, Knepper MA, and Bagnasco SM, Proteomics Clin Appl 2012, 6, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dalli J, Montero-Melendez T, Norling LV, Yin X, Hinds C, Haskard D, Mayr M, and Perretti M, Mol Cell Proteomics 2013, 12, 2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinha A, Ignatchenko V, Ignatchenko A, Mejia-Guerrero S, and Kislinger T, Biochem Biophys Res Commun 2014, 445, 694. [DOI] [PubMed] [Google Scholar]
- 67.Cheng L, Zhang K, Qing Y, Li D, Cui M, Jin P, and Xu T, J Ovarian Res 2020, 13, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, Yang J, Li H, Gui T, Li X, and Shen K, J Proteomics 2013, 80, 171. [DOI] [PubMed] [Google Scholar]
- 69.Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, Zitvogel L, Burgers SA, Hoogsteden HC, and Lambrecht BN, Am J Pathol 2004, 164, 1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA, Hoogsteden HC, and Lambrecht BN, Am J Respir Cell Mol Biol 2004, 31, 114. [DOI] [PubMed] [Google Scholar]
- 71.Garnier D, Magnus N, Meehan B, Kislinger T, and Rak J, Exp Cell Res 2013, 319, 2747. [DOI] [PubMed] [Google Scholar]
- 72.Hurwitz SN, Rider MA, Bundy JL, Liu X, Singh RK, and Meckes DG Jr., Oncotarget 2016, 7, 86999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miguet L, Bechade G, Fornecker L, Zink E, Felden C, Gervais C, Herbrecht R, Van Dorsselaer A, Mauvieux L, and Sanglier-Cianferani S, J Proteome Res 2009, 8, 3346. [DOI] [PubMed] [Google Scholar]
- 74.Liem M, Ang CS, and Mathivanan S, Proteomics 2017, 17. [DOI] [PubMed] [Google Scholar]
- 75.Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, and Coffey RJ, Mol Cell Proteomics 2013, 12, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, and Coffey RJ, Cell 2019, 177, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, Xu R, Adda C, Mathivanan S, Zhao W, Xue Y, Xu T, Zhu HJ, and Simpson RJ, Proteomics 2013, 13, 1672. [DOI] [PubMed] [Google Scholar]
- 78.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, and Simpson RJ, Mol Cell Proteomics 2010, 9, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi DS, Lee JM, Park GW, Lim HW, Bang JY, Kim YK, Kwon KH, Kwon HJ, Kim KP, and Gho YS, J Proteome Res 2007, 6, 4646. [DOI] [PubMed] [Google Scholar]
- 80.Choi DS, Park JO, Jang SC, Yoon YJ, Jung JW, Choi DY, Kim JW, Kang JS, Park J, Hwang D, Lee KH, Park SH, Kim YK, Desiderio DM, Kim KP, and Gho YS, Proteomics 2011, 11, 2745. [DOI] [PubMed] [Google Scholar]
- 81.Principe S, Jones EE, Kim Y, Sinha A, Nyalwidhe JO, Brooks J, Semmes OJ, Troyer DA, Lance RS, Kislinger T, and Drake RR, Proteomics 2013, 13, 1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Utleg AG, Yi EC, Xie T, Shannon P, White JT, Goodlett DR, Hood L, and Lin B, Prostate 2003, 56, 150. [DOI] [PubMed] [Google Scholar]
- 83.Dubois L, Ronquist KK, Ek B, Ronquist G, and Larsson A, Mol Cell Proteomics 2015, 14, 3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider T, Pasa-Tolic L, and Jenster G, PLoS One 2013, 8, e82589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ronquist KG, Ronquist G, Larsson A, and Carlsson L, Anticancer Res 2010, 30, 285. [PubMed] [Google Scholar]
- 86.Panigrahi GK, Praharaj PP, Kittaka H, Mridha AR, Black OM, Singh R, Mercer R, van Bokhoven A, Torkko KC, Agarwal C, Agarwal R, Abd Elmageed ZY, Yadav H, Mishra SK, and Deep G, Cancer Med 2019, 8, 1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palazzolo G, Albanese NN, G DIC, Gygax D, Vittorelli ML, and Pucci-Minafra I, Anticancer Res 2012, 32, 847. [PubMed] [Google Scholar]
- 88.Clark DJ, Fondrie WE, Liao Z, Hanson PI, Fulton A, Mao L, and Yang AJ, Anal Chem 2015, 87, 10462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lane RE, Korbie D, Trau M, and Hill MM, Proteomics 2019, 19, e1800156. [DOI] [PubMed] [Google Scholar]
- 90.Gangoda L, Liem M, Ang CS, Keerthikumar S, Adda CG, Parker BS, and Mathivanan S, Proteomics 2017, 17. [DOI] [PubMed] [Google Scholar]
- 91.Indira Chandran V, Welinder C, Goncalves de Oliveira K, Cerezo-Magana M, Mansson AS, Johansson MC, Marko-Varga G, and Belting M, J Neurooncol 2019, 144, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mallawaaratchy DM, Hallal S, Russell B, Ly L, Ebrahimkhani S, Wei H, Christopherson RI, Buckland ME, and Kaufman KL, J Neurooncol 2017, 131, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choi D, Montermini L, Kim DK, Meehan B, Roth FP, and Rak J, Mol Cell Proteomics 2018, 17, 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, and Bigner DD, FASEB J 2009, 23, 1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gyuris A, Navarrete-Perea J, Jo A, Cristea S, Zhou S, Fraser K, Wei Z, Krichevsky AM, Weissleder R, Lee H, Gygi SP, and Charest A, Cell Rep 2019, 27, 3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nasiri Kenari A, Kastaniegaard K, Greening DW, Shambrook M, Stensballe A, Cheng L, and Hill AF, Proteomics 2019, 19, e1800161. [DOI] [PubMed] [Google Scholar]
- 97.Marimpietri D, Petretto A, Raffaghello L, Pezzolo A, Gagliani C, Tacchetti C, Mauri P, Melioli G, and Pistoia V, PLoS One 2013, 8, e75054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, Patel P, Selby PJ, and Banks RE, Proteomics 2004, 4, 4019. [DOI] [PubMed] [Google Scholar]
- 99.Chen CL, Lai YF, Tang P, Chien KY, Yu JS, Tsai CH, Chen HW, Wu CC, Chung T, Hsu CW, Chen CD, Chang YS, Chang PL, and Chen YT, J Proteome Res 2012, 11, 5611. [DOI] [PubMed] [Google Scholar]
- 100.Jerez S, Araya H, Thaler R, Charlesworth MC, Lopez-Solis R, Kalergis AM, Cespedes PF, Dudakovic A, Stein GS, van Wijnen AJ, and Galindo M, J Cell Biochem 2017, 118, 351. [DOI] [PubMed] [Google Scholar]
- 101.Vykoukal J, Sun N, Aguilar-Bonavides C, Katayama H, Tanaka I, Fahrmann JF, Capello M, Fujimoto J, Aguilar M, Wistuba II, Taguchi A, Ostrin EJ, and Hanash SM, Oncotarget 2017, 8, 95466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun Y, Huo C, Qiao Z, Shang Z, Uzzaman A, Liu S, Jiang X, Fan LY, Ji L, Guan X, Cao CX, and Xiao H, J Proteome Res 2018, 17, 1101. [DOI] [PubMed] [Google Scholar]
- 103.Park JO, Choi DY, Choi DS, Kim HJ, Kang JW, Jung JH, Lee JH, Kim J, Freeman MR, Lee KY, Gho YS, and Kim KP, Proteomics 2013, 13, 2125. [DOI] [PubMed] [Google Scholar]
- 104.Yao Y, Wei W, Sun J, Chen L, Deng X, Ma L, and Hao S, Eur J Med Res 2015, 20, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Epple LM, Griffiths SG, Dechkovskaia AM, Dusto NL, White J, Ouellette RJ, Anchordoquy TJ, Bemis LT, and Graner MW, PLoS One 2012, 7, e42064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Winck FV, Prado Ribeiro AC, Ramos Domingues R, Ling LY, Riano-Pachon DM, Rivera C, Brandao TB, Gouvea AF, Santos-Silva AR, Coletta RD, and Paes Leme AF, Sci Rep 2015, 5, 16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tkach M and Thery C, Cell 2016, 164, 1226. [DOI] [PubMed] [Google Scholar]
- 108.Hauser P, Wang S, and Didenko VV, Methods Mol Biol 2017, 1554, 193. [DOI] [PubMed] [Google Scholar]
- 109.Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, and Holmgren L, Proc Natl Acad Sci U S A 2001, 98, 6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meehan B, Rak J, and Di Vizio D, J Extracell Vesicles 2016, 5, 33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ciardiello C, Migliorino R, Leone A, and Budillon A, Cytokine Growth Factor Rev 2020, 51, 69. [DOI] [PubMed] [Google Scholar]
- 112.Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, True LD, Rubin MA, Adam RM, Beroukhim R, Demichelis F, and Freeman MR, Cancer Res 2009, 69, 5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Di Vizio D, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, Mulholland D, Rotinen M, Hager MH, Insabato L, Moses MA, Demichelis F, Lisanti MP, Wu H, Klagsbrun M, Bhowmick NA, Rubin MA, D’Souza-Schorey C, and Freeman MR, Am J Pathol 2012, 181, 1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morello M, Minciacchi VR, de Candia P, Yang J, Posadas E, Kim H, Griffiths D, Bhowmick N, Chung LW, Gandellini P, Freeman MR, Demichelis F, and Di Vizio D, Cell Cycle 2013, 12, 3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Minciacchi VR, Spinelli C, Reis-Sobreiro M, Cavallini L, You S, Zandian M, Li X, Mishra R, Chiarugi P, Adam RM, Posadas EM, Viglietto G, Freeman MR, Cocucci E, Bhowmick NA, and Di Vizio D, Cancer Res 2017, 77, 2306. [DOI] [PubMed] [Google Scholar]
- 116.Ciardiello C, Leone A, Lanuti P, Roca MS, Moccia T, Minciacchi VR, Minopoli M, Gigantino V, De Cecio R, Rippa M, Petti L, Capone F, Vitagliano C, Milone MR, Pucci B, Lombardi R, Iannelli F, Di Gennaro E, Bruzzese F, Marchisio M, Carriero MV, Di Vizio D, and Budillon A, J Exp Clin Cancer Res 2019, 38, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wright PK, Jones SB, Ardern N, Ward R, Clarke RB, Sotgia F, Lisanti MP, Landberg G, and Lamb R, Oncotarget 2014, 5, 3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fraser K, Jo A, Giedt J, Vinegoni C, Yang KS, Peruzzi P, Chiocca EA, Breakefield XO, Lee H, and Weissleder R, Neuro Oncol 2019, 21, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yekula A, Minciacchi VR, Morello M, Shao H, Park Y, Zhang X, Muralidharan K, Freeman MR, Weissleder R, Lee H, Carter B, Breakefield XO, Di Vizio D, and Balaj L, J Extracell Vesicles 2020, 9, 1689784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Patton MC, Zubair H, Khan MA, Singh S, and Singh AP, J Cell Biochem 2020, 121, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kang M, Kim S, and Ko J, FASEB J 2019, 33, 4248. [DOI] [PubMed] [Google Scholar]
- 122.Surman M, Hoja-Lukowicz D, Szwed S, Drozdz A, Stepien E, and Przybylo M, Life Sci 2018, 207, 395. [DOI] [PubMed] [Google Scholar]
- 123.Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y, Chen L, Yan X, Du Y, and Yu L, Cell Res 2015, 25, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao X, Lei Y, Zheng J, Peng J, Li Y, Yu L, and Chen Y, Cell Discov 2019, 5, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang Y, Zucker B, Zhang S, Elias S, Zhu Y, Chen H, Ding T, Li Y, Sun Y, Lou J, Kozlov MM, and Yu L, Nat Cell Biol 2019, 21, 991. [DOI] [PubMed] [Google Scholar]
- 126.Jiang D, Jiang Z, Lu D, Wang X, Liang H, Zhang J, Meng Y, Li Y, Wu D, Huang Y, Chen Y, Deng H, Wu Q, Xiong J, Meng A, and Yu L, Nat Cell Biol 2019, 21, 966. [DOI] [PubMed] [Google Scholar]
- 127.Chen Y, Li Y, Ma L, and Yu L, Methods Mol Biol 2018, 1749, 43. [DOI] [PubMed] [Google Scholar]
- 128.Wu D, Xu Y, Ding T, Zu Y, Yang C, and Yu L, Cell Res 2017, 27, 1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Momen-Heravi F, Balaj L, Alian S, Trachtenberg AJ, Hochberg FH, Skog J, and Kuo WP, Front Physiol 2012, 3, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS, and Star RA, Am J Physiol Renal Physiol 2007, 292, F1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, and Moller A, J Extracell Vesicles 2015, 4, 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hong CS, Muller L, Whiteside TL, and Boyiadzis M, Front Immunol 2014, 5, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Muller L, Hong CS, Stolz DB, Watkins SC, and Whiteside TL, J Immunol Methods 2014, 411, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A, and Nieuwland R, J Extracell Vesicles 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.de Menezes-Neto A, Saez MJ, Lozano-Ramos I, Segui-Barber J, Martin-Jaular L, Ullate JM, Fernandez-Becerra C, Borras FE, and Del Portillo HA, J Extracell Vesicles 2015, 4, 27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ghosh A, Davey M, Chute IC, Griffiths SG, Lewis S, Chacko S, Barnett D, Crapoulet N, Fournier S, Joy A, Caissie MC, Ferguson AD, Daigle M, Meli MV, Lewis SM, and Ouellette RJ, PLoS One 2014, 9, e110443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Knol JC, de Reus I, Schelfhorst T, Beekhof R, de Wit M, Piersma SR, Pham TV, Smit EF, Verheul HMW, and Jimenez CR, EuPA Open Proteom 2016, 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Atai NA, Balaj L, van Veen H, Breakefield XO, Jarzyna PA, Van Noorden CJ, Skog J, and Maguire CA, J Neurooncol 2013, 115, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Balaj L, Atai NA, Chen W, Mu D, Tannous BA, Breakefield XO, Skog J, and Maguire CA, Sci Rep 2015, 5, 10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Echevarria J, Royo F, Pazos R, Salazar L, Falcon-Perez JM, and Reichardt NC, Chembiochem 2014, 15, 1621. [DOI] [PubMed] [Google Scholar]
- 141.Kosanovic M and Jankovic M, Biotechniques 2014, 57, 143. [DOI] [PubMed] [Google Scholar]
- 142.Samsonov R, Shtam T, Burdakov V, Glotov A, Tsyrlina E, Berstein L, Nosov A, Evtushenko V, Filatov M, and Malek A, Prostate 2016, 76, 68. [DOI] [PubMed] [Google Scholar]
- 143.Xu Y, Ku X, Wu C, Cai C, Tang J, and Yan W, Biochem Biophys Res Commun 2018, 499, 856. [DOI] [PubMed] [Google Scholar]
- 144.Aalberts M, van Dissel-Emiliani FM, van Adrichem NP, van Wijnen M, Wauben MH, Stout TA, and Stoorvogel W, Biol Reprod 2012, 86, 82. [DOI] [PubMed] [Google Scholar]
- 145.Palma J, Yaddanapudi SC, Pigati L, Havens MA, Jeong S, Weiner GA, Weimer KM, Stern B, Hastings ML, and Duelli DM, Nucleic Acids Res 2012, 40, 9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, Rabinovsky R, Balaj L, Chen CC, Hochberg F, Carter B, Breakefield XO, and Krichevsky AM, Nat Commun 2017, 8, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, and Hochberg F, J Extracell Vesicles 2013, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Alvarez ML, Khosroheidari M, Kanchi Ravi R, and DiStefano JK, Kidney Int 2012, 82, 1024. [DOI] [PubMed] [Google Scholar]
- 149.Gupta S, Rawat S, Arora V, Kottarath SK, Dinda AK, Vaishnav PK, Nayak B, and Mohanty S, Stem Cell Res Ther 2018, 9, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, Hulett MD, and Mathivanan S, Proteomics 2013, 13, 3354. [DOI] [PubMed] [Google Scholar]
- 151.Lane RE, Korbie D, Anderson W, Vaidyanathan R, and Trau M, Sci Rep 2015, 5, 7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, and Simpson RJ, Methods 2012, 56, 293. [DOI] [PubMed] [Google Scholar]
- 153.Van Deun J, Mestdagh P, Sormunen R, Cocquyt V, Vermaelen K, Vandesompele J, Bracke M, De Wever O, and Hendrix A, J Extracell Vesicles 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yamashita T, Takahashi Y, Nishikawa M, and Takakura Y, Eur J Pharm Biopharm 2016, 98, 1. [DOI] [PubMed] [Google Scholar]
- 155.Mehdiani A, Maier A, Pinto A, Barth M, Akhyari P, and Lichtenberg A, J Vis Exp 2015, 50974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, Minar L, Weinberger V, and Bryja V, J Extracell Vesicles 2015, 4, 25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Maas SL, De Vrij J, and Broekman ML, J Vis Exp 2014, e51623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shelke GV, Lasser C, Gho YS, and Lotvall J, J Extracell Vesicles 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thery C, Amigorena S, Raposo G, and Clayton A, Curr Protoc Cell Biol 2006, Chapter 3, Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 160.Clayton A, Boilard E, Buzas EI, Cheng L, Falcon-Perez JM, Gardiner C, Gustafson D, Gualerzi A, Hendrix A, Hoffman A, Jones J, Lasser C, Lawson C, Lenassi M, Nazarenko I, O’Driscoll L, Pink R, Siljander PR, Soekmadji C, Wauben M, Welsh JA, Witwer K, Zheng L, and Nieuwland R, J Extracell Vesicles 2019, 8, 1647027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sun Y, Xia Z, Shang Z, Sun K, Niu X, Qian L, Fan LY, Cao CX, and Xiao H, Sci Rep 2016, 6, 24669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sun Y, Liu S, Qiao Z, Shang Z, Xia Z, Niu X, Qian L, Zhang Y, Fan L, Cao CX, and Xiao H, Anal Chim Acta 2017, 982, 84. [DOI] [PubMed] [Google Scholar]
- 163.Ronquist G and Nilsson BO, Prostate Cancer Prostatic Dis 2004, 7, 21. [DOI] [PubMed] [Google Scholar]
- 164.Shender VO, Pavlyukov MS, Ziganshin RH, Arapidi GP, Kovalchuk SI, Anikanov NA, Altukhov IA, Alexeev DG, Butenko IO, Shavarda AL, Khomyakova EB, Evtushenko E, Ashrafyan LA, Antonova IB, Kuznetcov IN, Gorbachev AY, Shakhparonov MI, and Govorun VM, Mol Cell Proteomics 2014, 13, 3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Welton JL, Loveless S, Stone T, von Ruhland C, Robertson NP, and Clayton A, J Extracell Vesicles 2017, 6, 1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pieragostino D, Lanuti P, Cicalini I, Cufaro MC, Ciccocioppo F, Ronci M, Simeone P, Onofrj M, van der Pol E, Fontana A, Marchisio M, and Del Boccio P, J Proteomics 2019, 204, 103403. [DOI] [PubMed] [Google Scholar]
- 167.Manek R, Moghieb A, Yang Z, Kumar D, Kobessiy F, Sarkis GA, Raghavan V, and Wang KKW, Mol Neurobiol 2018, 55, 6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Aqrawi LA, Galtung HK, Guerreiro EM, Ovstebo R, Thiede B, Utheim TP, Chen X, Utheim OA, Palm O, Skarstein K, and Jensen JL, Arthritis Res Ther 2019, 21, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Manza LL, Stamer SL, Ham AJ, Codreanu SG, and Liebler DC, Proteomics 2005, 5, 1742. [DOI] [PubMed] [Google Scholar]
- 170.Wisniewski JR, Zougman A, Nagaraj N, and Mann M, Nat Methods 2009, 6, 359. [DOI] [PubMed] [Google Scholar]
- 171.van Niel G, D’Angelo G, and Raposo G, Nat Rev Mol Cell Biol 2018, 19, 213. [DOI] [PubMed] [Google Scholar]
- 172.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr., Carter BS, Krichevsky AM, and Breakefield XO, Nat Cell Biol 2008, 10, 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, and Rak J, Nat Cell Biol 2008, 10, 619. [DOI] [PubMed] [Google Scholar]
- 174.Bobrie A, Colombo M, Raposo G, and Thery C, Traffic 2011, 12, 1659. [DOI] [PubMed] [Google Scholar]
- 175.Filipazzi P, Burdek M, Villa A, Rivoltini L, and Huber V, Semin Cancer Biol 2012, 22, 342. [DOI] [PubMed] [Google Scholar]
- 176.Zhang HG, Zhuang X, Sun D, Liu Y, Xiang X, and Grizzle WE, Biotech Histochem 2012, 87, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sung BH, Ketova T, Hoshino D, Zijlstra A, and Weaver AM, Nat Commun 2015, 6, 7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ginestra A, Monea S, Seghezzi G, Dolo V, Nagase H, Mignatti P, and Vittorelli ML, J Biol Chem 1997, 272, 17216. [DOI] [PubMed] [Google Scholar]
- 179.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, and D’Souza-Schorey C, Curr Biol 2009, 19, 1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, and Rak J, Proc Natl Acad Sci U S A 2009, 106, 3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, Morgelin M, Bourseau-Guilmain E, Bengzon J, and Belting M, Proc Natl Acad Sci U S A 2013, 110, 7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sheldon H, Heikamp E, Turley H, Dragovic R, Thomas P, Oon CE, Leek R, Edelmann M, Kessler B, Sainson RC, Sargent I, Li JL, and Harris AL, Blood 2010, 116, 2385. [DOI] [PubMed] [Google Scholar]
- 183.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, and Lyden D, Nature 2015, 527, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, and Lyden D, Nat Med 2012, 18, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hood JL, San RS, and Wickline SA, Cancer Res 2011, 71, 3792. [DOI] [PubMed] [Google Scholar]
- 186.Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M, and Wrana JL, Cell 2012, 151, 1542. [DOI] [PubMed] [Google Scholar]
- 187.De Rubis G, Rajeev Krishnan S, and Bebawy M, Trends Pharmacol Sci 2019, 40, 172. [DOI] [PubMed] [Google Scholar]
- 188.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, and Kalluri R, Nature 2015, 523, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang KS, Im H, Hong S, Pergolini I, Del Castillo AF, Wang R, Clardy S, Huang CH, Pille C, Ferrone S, Yang R, Castro CM, Lee H, Del Castillo CF, and Weissleder R, Sci Transl Med 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, and Lempicki RA, Curr Protoc Bioinformatics 2009, Chapter 13, Unit 13 11. [DOI] [PubMed] [Google Scholar]
- 191.Huang da W, Sherman BT, and Lempicki RA, Nat Protoc 2009, 4, 44. [DOI] [PubMed] [Google Scholar]
- 192.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, and Sherlock G, Nat Genet 2000, 25, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.The Gene Ontology C, Nucleic Acids Res 2019, 47, D330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kanehisa M and Goto S, Nucleic Acids Res 2000, 28, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Giri PK, Kruh NA, Dobos KM, and Schorey JS, Proteomics 2010, 10, 3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Amorim M, Fernandes G, Oliveira P, Martins-de-Souza D, Dias-Neto E, and Nunes D, Proteomics 2014, 14, 1472. [DOI] [PubMed] [Google Scholar]
- 197.Tauro BJ, Mathias RA, Greening DW, Gopal SK, Ji H, Kapp EA, Coleman BM, Hill AF, Kusebauch U, Hallows JL, Shteynberg D, Moritz RL, Zhu HJ, and Simpson RJ, Mol Cell Proteomics 2013, 12, 2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gopal SK, Greening DW, Hanssen EG, Zhu HJ, Simpson RJ, and Mathias RA, Oncotarget 2016, 7, 19709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fricke F, Lee J, Michalak M, Warnken U, Hausser I, Suarez-Carmona M, Halama N, Schnolzer M, Kopitz J, and Gebert J, Cell Commun Signal 2017, 15, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sun Y, Zheng W, Guo Z, Ju Q, Zhu L, Gao J, Zhou L, Liu F, Xu Y, Zhan Q, Zhou Z, Sun W, and Zhao X, Sci Rep 2016, 6, 28083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Choi DS, Choi DY, Hong BS, Jang SC, Kim DK, Lee J, Kim YK, Kim KP, and Gho YS, J Extracell Vesicles 2012, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Guo J, Cui Y, Yan Z, Luo Y, Zhang W, Deng S, Tang S, Zhang G, He QY, and Wang T, J Proteome Res 2016, 15, 4060. [DOI] [PubMed] [Google Scholar]
- 203.Jeppesen DK, Nawrocki A, Jensen SG, Thorsen K, Whitehead B, Howard KA, Dyrskjot L, Orntoft TF, Larsen MR, and Ostenfeld MS, Proteomics 2014, 14, 699. [DOI] [PubMed] [Google Scholar]
- 204.Yu Z, Zhao S, Ren L, Wang L, Chen Z, Hoffman RM, and Zhou J, Oncotarget 2017, 8, 63461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Tabulated version of Figure 2. Parameters curated from literature.


