Abstract
Mitochondrial genome content and structure vary widely across the eukaryotic tree of life, with protists displaying extreme examples. Apicomplexan and dinoflagellate protists have evolved highly reduced mitochondrial genome sequences, mtDNA, consisting of only three cytochrome genes and fragmented rRNA genes. Here, we report the independent evolution of fragmented cytochrome genes in Toxoplasma and related tissue coccidia and evolution of a novel genome architecture consisting minimally of 21 sequence blocks (SBs) totaling 5.9 kb that exist as nonrandom concatemers. Single-molecule Nanopore reads consisting entirely of SBs ranging from 0.1 to 23.6 kb reveal both whole and fragmented cytochrome genes. Full-length cytochrome transcripts including a divergent coxIII are detected. The topology of the mitochondrial genome remains an enigma. Analysis of a cob point mutation reveals that homoplasmy of SBs is maintained. Tissue coccidia are important pathogens of man and animals, and the mitochondrion represents an important therapeutic target. The mtDNA sequence has been elucidated, but a definitive genome architecture remains elusive.
Mitochondria are notable for their incredible diversity in genome content and genome topology (Burger et al. 2003; Flegontov and Lukes 2012; Kolesnikov and Gerasimov 2012). The endosymbiont genome sequence that gave rise to the mitochondrial genome sequence, mtDNA, was likely circular, characteristic of its prokaryotic origins (Lang et al. 1997). However, increasing bodies of literature indicate that this structure is not universal and that considerable evolution of both genome structure and gene content has occurred in eukaryotes (Burger et al. 2003; Gissi et al. 2008). At the time of its discovery, the mtDNA of the apicomplexan Plasmodium falciparum was the smallest mtDNA known, at 5967 bp, and among the most unusual with only three protein-encoding genes (cytochrome oxidase subunit I, coxI: cytochrome oxidase subunit III, coxIII; and cytochrome b, cob) and fragmented rRNA genes with many rRNA portions missing (Vaidya and Arasu 1987; Feagin 1992; Feagin et al. 1997, 2012; Wilson and Williamson 1997). At the other end of the size spectrum, the mtDNA of angiosperms ranges from 200 kb to 11 Mb in size and shows a huge variation in structure, content, gene/DNA transfers, and processes such as RNA editing (Gualberto and Newton 2017). Mitochondrial genomes consisting of multiple divergent circular molecules have been noted in the fungus Spizellomyces (Burger and Lang 2003), Columbicola feather lice (Sweet et al. 2020), and certain cnidarian parasites (Yahalomi et al. 2017). Among the protists, the kinetoplastid mtDNA (called kDNA) consists of multiple gene-encoding maxicircles and guide RNA encoding minicircles that form a tight physical network, the kinetoplast (Morris et al. 2001). The mtDNA of the symbiotic protist Amoebidium parasiticum is comprised of hundreds of distinct linear molecules with a common pattern of terminal repeats (Burger et al. 2003).
The alveolates have their own noncanonical mtDNA sequences and structures (Flegontov and Lukes 2012). The ciliates contain a 47-kb linear mtDNA flanked by telomere-like repeats with a number of ciliate-specific open reading frames (ORFs) of unknown function (Slamovits et al. 2007; Nash et al. 2008; Waller and Jackson 2009; Flegontov and Lukes 2012). The mtDNA of dinoflagellates (sister group to the Apicomplexa) share the reduced gene content and fragmented rRNA genes with the Apicomplexa, but their protein-encoding genes are highly fragmented, highly repetitive, contain multiple nonidentical mtDNA molecules, utilize noncanonical start codons, and have evolved trans-splicing and RNA editing (Slamovits et al. 2007; Nash et al. 2008; Waller and Jackson 2009; Flegontov and Lukes 2012).
Whereas all sequenced apicomplexan mtDNAs contain only three protein-coding genes (coxI, coxIII, and cob) and show a high level of rRNA gene fragmentation like the dinoflagellates, the orientation and arrangement of these genes vary across the phylum, as does the genome architecture (Creasey et al. 1993; Hikosaka et al. 2010, 2011, 2012, 2013). The Theileria mtDNA is a 7.1-kb linear monomer with telomere-like termini (Kairo et al. 1994). Members of the genus Babesia have linear mtDNA monomers with a dual flip-flop inversion system that results in four different sequences that are present in equal ratios in the mitochondrion (Hikosaka et al. 2011). The coccidian Eimeria, the species closest to Toxoplasma gondii with a published mtDNA sequence and structure, is detected as a linear concatemer of a repeating 6.2-kb sequence similar to that of Plasmodium, albeit with the genes in a different order.
The mitochondrial genome of the coccidian protist pathogen Toxoplasma gondii has been enigmatic and elusive. Toxoplasma gondii is an incredibly successful zoonotic generalist pathogen which infects 30%–50% of the human population (Flegr et al. 2014) but primarily causes illness in the immunocompromised, or fetuses as a result of transplacental infections (Montoya and Liesenfeld 2004; Dubey 2010). Attempts to assemble the mitochondrial genome from the various organismal genome projects failed, and efforts to isolate the single mitochondrion organelle and identify the complete mtDNA sequence and structure have faced numerous challenges. Attempts to elucidate the mtDNA sequence using sequenced-based approaches, hybridization probes, PCR amplification, or assembly from Sanger or second-generation short-read genome sequence projects were unsuccessful (Minot et al. 2012; Lau et al. 2016; Lorenzi et al. 2016), presumably due to the large number of nuclear-encoded sequence fragments of mitochondrial origin (NUMTs) present in the nuclear genome. NUMTs are insertions of fragments of mtDNA at the location of a nuclear double-strand DNA break (Minot et al. 2012; Lau et al. 2016; Lorenzi et al. 2016). NUMT sequences (which display a range of degeneration relative to organellar mtDNA because they decay with evolutionary time following insertion) can cross-hybridize and interfere with signals from molecular-based mtDNA isolation and amplification methods. NUMTs also interfere with assembly algorithms for short-read sequences (Sanger and Illumina) that were used to generate the numerous existing Toxoplasma genome sequences (Minot et al. 2012; Lau et al. 2016; Lorenzi et al. 2016). Physical attempts to isolate the mitochondrion have also failed thus far (Nishi et al. 2008), but mitochondrial-enriched fractions are obtainable (Miranda et al. 2010).
Despite the community's inability to isolate and determine a mitochondrial genome sequence for this important pathogen, multiple lines of evidence suggest that the T. gondii mitochondrion is functional, including cDNAs for cob and coxI. Mitochondrial membrane-potential markers indicate that the mitochondrion is active (Tanabe 1985; Melo et al. 2000; Weiss and Kim 2007); a complete set of tRNAs are imported into the T. gondii mitochondrion (Esseiva et al. 2004); the mitochondrial protein cob has been proven as an excellent drug target (Vaidya et al. 1993); mutations in cob were found to be associated with atovaquone (McFadden et al. 2000) and endochin-like quinolone (ELQ-316) (Alday et al. 2017) drug resistance; and NUMTs identified in the current T. gondii ME49 nuclear genome sequence, even if combined together, cannot functionally encode any of the cytochrome genes in their entirety—thus, they must be encoded in the mitochondrion (Namasivayam 2015). Here, we employ Sanger, Illumina, and single-molecule Oxford Nanopore sequencing technology (ONT) of DNA and RNA together with molecular and bioinformatics strategies to identify the sequence and structure of the T. gondii mtDNA.
Results
21 discrete sequence blocks constitute the minimal Toxoplasma gondii mitochondrial genome
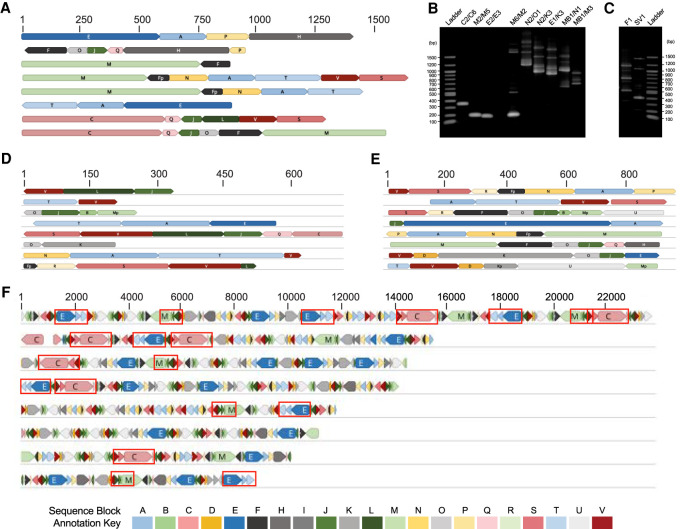
The T. gondii ME49 genome sequence project yielded numerous contigs containing portions of, or in some cases, complete cytochrome genes and mtDNA rRNA gene fragments (Gajria et al. 2008; Lorenzi et al. 2016). These contigs did not assemble into a single mtDNA sequence. However, the sequence information contained in these contigs as well as individual unassembled T. gondii genomic and EST reads (Sanger chemistry) that matched mtDNA from related species, served as a template for PCR primer design that carefully avoided NUMTs (Supplemental Table S1). Mitochondria-enriched cell fractions were used as template. The PCR results were puzzling. No amplicon was larger than ∼3.5 kb. Primer pairs often generated multiple amplicons that, when sequenced and annotated, revealed a high level of sequence identity to each other in some regions (sequence blocks, SBs) but differed in others (Fig. 1A,B, lanes 6–11; Supplemental Fig. S1; Supplemental Data S1). PCR reactions with only a single primer also generated multiple amplicons (Fig. 1C). With difficulty, we did identify primers that avoided NUMTs and would amplify small regions (100–350 bp) within coxI, coxIII, and cob (Fig. 1B, lanes 2–4; Hikosaka et al. 2013).
Figure 1.
Sequence evidence of fragmented mtDNA and full-length protein-encoding genes. (A, D–F) Each row of colored blocks represents individual, annotated T. gondii (A) mtDNA-specific PCR amplicons; (D) EST reads; (E) Sanger genomic reads; or (F) Nanopore reads. Sections of reads containing identical sequence to other mtDNA reads are called “sequence blocks” (SBs) and are colored and labeled with a unique color and letter corresponding to the key, Table 1 and Figure 2. Shades of blue, red, and green represent different SBs found in cob, coxI, and coxIII, respectively. Orientation of a block is indicated by the point on each block. Red boxes in F indicate complete protein-encoding genes. SBs located on the ends of reads may be incomplete. (B) Genomic DNA from mitochondrial-enriched fractions assayed with different primer pairs (Supplemental Table S1) as indicated. Lanes 2–4 represent a small fragment of each cytochrome gene. (C) Single primers produce multiple amplicons. Scale (in bp) is as indicated in each panel. Additional reads are in Supplemental Figures S1–S4.
As PCR is capable of amplifying rare artifacts or a NUMT capable of annealing to the primers, we abandoned approaches that relied on amplification or sequence assembly and examined individual genomic and EST Sanger sequence reads in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/; Benson et al. 2018) to identify putative mtDNA or mitochondrial cDNA. Like the PCR amplicons, identified mtDNA reads and transcripts could be annotated with discrete, reproducible, sequence blocks with similarity to various regions of known apicomplexan mtDNA (Fig. 1D–E; Supplemental Figs. S2, S3). In total, 21 mtDNA SBs that reproducibly share 100% sequence identity were identified. The 21 SBs were named from A to V (Fig. 2; Table 1; Supplemental Data S2). Three SBs (F, K, and M) were observed to also exist in precise, reproducible, truncated forms, indicated with a “p” for partial, for example, Fp (Table 1). Fifteen of the 21 SBs share significant similarity to regions of Eimeria tenella and/or Plasmodium falciparum mtDNA sequences (Supplemental Table S2). The SBs range from 40 to 1050 bp, and none encodes a cytochrome or rRNA gene in its entirety (Fig. 2). Following SB identification, annotation of the short-read data sets (PCR, Sanger genomic, EST) revealed that reads contained one or more SBs, one directly after the other, with no intervening sequence, in distinct, nonrandom permutations (Fig. 1). No full-length cytochrome genes were observed, but many fragments of cytochrome genes were annotated in a variety of contexts and orientations. Some SBs were always observed in specific combinations with other blocks. For example, we found block E to always occur between blocks J and A. However, these blocks could not be assembled into a larger (JEA) SB because J and A each occur in other contexts, such as LJQ and NAP.
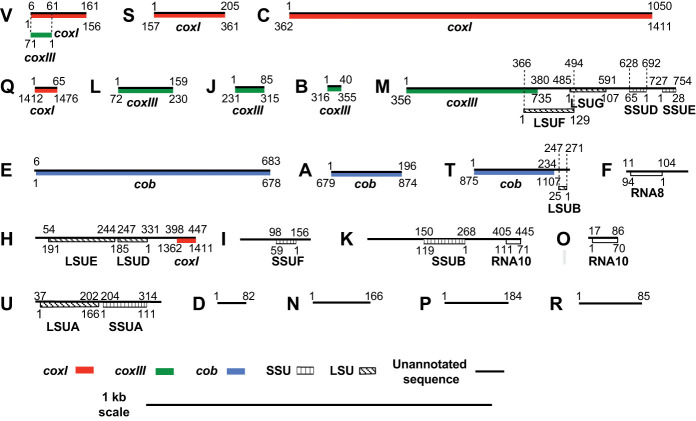
Figure 2.
The 21 minimal T. gondii mtDNA sequence blocks. The DNA sequence is represented by a black line, drawn to scale and named with 21 alphabet characters, A to V (there is no “G”). The coordinates of an SB that encodes a cytochrome or rRNA gene fragment are indicated above the black line and the corresponding coordinates of the assembled gene or rRNA fragment are indicated below the gene fragment; the fragments are colored as defined in the key. Portions of sequence block V contribute to both coxI and coxIII but in different orientations.
Table 1.
Characterization and conservation of the 21 mtDNA sequence blocks in T. gondii, N. caninum, and Hammondia

All SBs including the partial forms of F, K, and M when not located at the beginning or end of a read have the same length and sequence irrespective of the blocks they are flanked by, whether they appear in a PCR product, genomic, or EST sequence read. A full-length block C was not found in the Sanger genomic or EST reads nor in the PCR products, perhaps due to its large size, but it was observed in ONT data described below (Fig. 1F). Exhaustive searches of Sanger and second-generation sequence read data repeatedly identified only the 21 SBs. Occasionally, examination of a read revealed the presence of nuclear sequence, indicating the detected SB(s) was the result of a recent intracellular mtDNA transfer, that is, a NUMT (Ossorio et al. 1991; Lopez et al. 1994; Namasivayam 2015). A NUMT, unlike a SB, can be derived from any mtDNA region and the boundaries of a NUMT usually do not match the boundaries of the identified SBs. NUMTs are usually degenerate relative to mtDNA as a result of evolutionary decay. NUMT-containing reads were not analyzed further in this study.
Finally, we generated ONT long-read single-molecule sequences for T. gondii ME49 to take advantage of the long read lengths and to analyze single DNA molecules without the addition of any amplification steps (Supplemental Table S3) and mined these unassembled and uncorrected data for reads that could be annotated with mtDNA SBs or cytochrome genes. We searched uncorrected reads to avoid the possibility of correction confounding our detection and disambiguation from NUMTs. We identified 269 ONT reads ranging in length from 320 to 23,619 bp that, when annotated, consisted exclusively of nonrandom arrangements of the 21 SBs, and many had the ability to encode one or more full-length cytochrome genes; for example, SBs (EAT) encode cob (Fig. 1F). However, single ONT reads consisting of only one copy of each of the 21 SBs are not observed. Following identification of the ONT mtDNA reads, the reads were corrected with T. gondii ME49 SB-specific Illumina data. It is the corrected ONT mtDNA reads that are analyzed and presented in this work (Supplemental Fig. S4; Supplemental Data S3). An extensive analysis of these ONT mtDNA reads and two additional ONT data sets (see below; Supplemental Table S3) did not reveal any new SBs. Thus, the T. gondii mitochondrial genome consists minimally of 21 SBs that total 5909 bp.
The 21 sequence blocks encode expected mtDNA features
The SBs were subjected to extensive analyses to detect and annotate additional coding, rRNA and ncRNAs. Eleven SBs contribute in whole, or in part, to three identified protein-coding sequences: coxI (VSCQ); coxIII (the first 71 bp of V [opposite strand], LJB); and the first half of M and cob (EAT) (Fig. 2). The small portion of coxI replicated at the end of sequence block H does not appear to contribute to a functional cytochrome. Three blocks encode portions of a cytochrome and fragments of rRNA genes (H, M, and T) (Fig. 2). Six blocks (F, H, I, K, O, and U) encode 14 evolutionarily conserved LSU and SSU rRNA gene fragments (Fig. 2; Supplemental Fig. S5; Feagin et al. 2012). However, the rRNA gene fragment order is not conserved with E. tenella and P. falciparum (Supplemental Table S2). The well-conserved apicomplexan RNAs (RNA8 and RNA10) are detectable, but RNA10 is split onto SBs (O and K), and the short 22-nt LSUC fragment observed in the apicomplexan Plasmodium falciparum could not be detected. Currently, four SBs have no discernable features (D, N, P, and R) (Fig. 2). Several RNA genes observed in other apicomplexans (Feagin et al. 2012) have not been identified (Fig. 2). Overall, 3333 bp of the SBs are annotated as protein-encoding and 1290 bp are annotated as rDNA. Searches for additional RNA or protein features on each SB and the longest T. gondii ME49 ONT read did not reveal any new features.
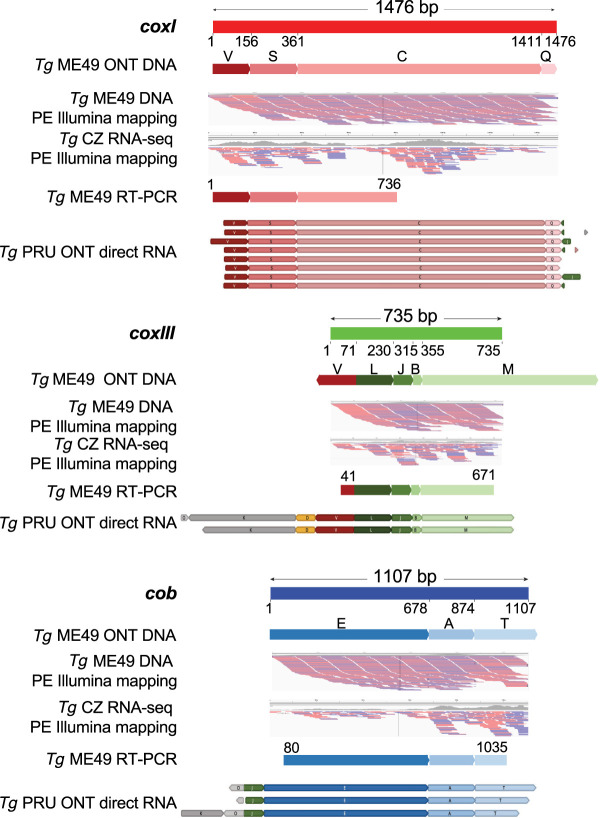
Full-length cytochrome genes are detected in the ONT reads from both T. gondii ME49 and RH (Figs. 1F, 6; Supplemental Fig. S6), and each gene is well supported by Illumina read data (Fig. 3). The encoded T. gondii mtDNA cytochrome proteins are comparable in length and sequence to other apicomplexan cytochrome genes, but there is some doubt about the exact beginning of the cob open reading frame, as two potential starts are possible, one 9 aa longer than the other. COXI and COB are well conserved, but COXIII is less well conserved. As was observed in the dinoflagellates, coxI does not have an ATG start codon. All genes have stop codons. Each sequence begins with a methionine (using mitochondrial translation tables) and ends with a canonical stop codon (Supplemental Fig. S7; Supplemental Data S4). There are no spliceosomal introns located in the full-length CDSs and no indication that RNA editing is required to produce the CDSs.
Figure 3.
Full-length protein-encoding genes and their expression in the T. gondii mt genome are supported. Segments of an ONT read that can encode a full-length cytochrome gene are annotated with their SB name and shown below the schematic of each gene. Numbers below each gene schematic represent nucleotide start/stop positions of a SB on the gene. All blocks are in the forward orientation except block V in coxIII. Only a portion of SBs V, M, and T is included in the CDS. MtDNA-specific PE-150 Illumina DNA (SRR9200762) and RNA-seq (SRR6493545) reads were independently mapped to each of the cytochrome gene sequences. Red and blue lines below each gene indicate mapped Illumina paired-ends. Both ends were required to map. RT-PCR products and ONT RNA reads with full-length CDSs are shown.
Cytochrome b is well studied in T. gondii (Hikosaka et al. 2011). A partial coxI transcript has been reported, but a convincing coxIII has not. Using RT-PCR, we were able to generate a nearly full-length cob and coxIII and a partial coxI (Fig. 3; Supplemental Data S5). Full-length sequences for all three cytochrome genes can be assembled from Sanger ESTs as well as Illumina RNA-seq data (Fig. 3; Supplemental Table S4). The RNA-seq evidence is less strong, but we note that poly(A)+ RNA was purified for these libraries. T. gondii PRU RNA sequence data from ONT (kindly provided by Stuart Ralph), once corrected, revealed a few reads capable of encoding full-length cytochrome transcripts for all three genes (some missing a few bp at read ends) (Fig. 3). The mtDNA transcripts are short ∼2.8 kb. Of the 30 longest error-corrected RNA ONT molecules that consisted of only mtDNA, three reads were capable of encoding cob, seven coxI, and two coxIII (Supplemental Table S5; Supplemental Data S6).
Based on only seven full-length coxI ONT transcripts, we observe that all begin at the start of the gene and appear to only encode coxI (VSCQ) or coxI plus SB J (VSCQJ), whereas the other transcripts encode numerous SBs in addition to the cytochrome CDS. The coxI transcripts that are not followed by a nearly full block J are polyadenylated, but the pattern is unusual. An additional 12 nucleotides of sequence, specifically the last 12 nucleotides of sequence block J in the reverse orientation (5′-CACAATAGAACT), are present following the coxI TAA stop codon. These 12 nucleotides are followed by a poly(A) stretch of variable length, suggesting that sequences present in J may serve as a degenerate poly(A) signal because sequence block J contains sequence similar to the canonical AAUAAA poly(A) signal. The remainder of sequence block J is capable of forming a prominent but imperfect stem–loop (Supplemental Fig. S8), but the rest of the SBs do not appear to contain long runs of obvious hairpin structures, as evidenced by the dot plots (Fig. 4). There is no evidence of polyadenylation of any other transcripts based on the few available sequences (Supplemental Table S5).
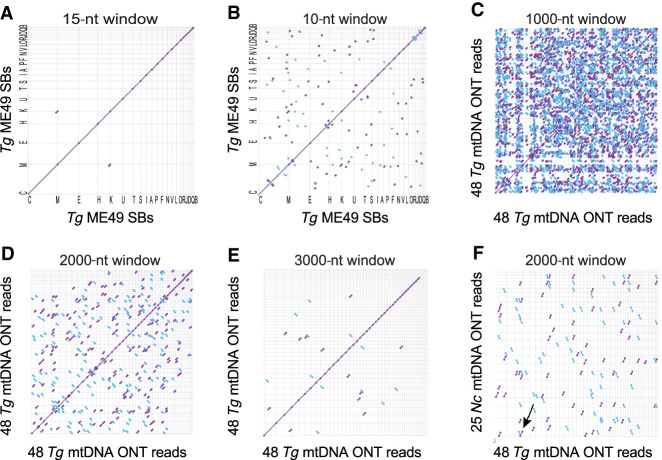
Figure 4.
Dot plot comparisons of long T. gondii ME49 mtDNA SBs and ONT reads against themselves and N. caninum Nc-1. (A,B) T. gondii SBs (5909 nt) compared to themselves at differing window sizes; (C–E) 48 mtDNA-specific T. gondii ONT reads (313,102 nt) (Supplemental Methods; Supplemental Fig. S4; Supplemental Data S3) compared to themselves at differing window sizes; (F) 25 N. caninum ONT mtDNA reads (141,161 nt) (Supplemental Methods; Supplemental Fig. S10; Supplemental Data S8) compared to the same 48 mtDNA-specific T. gondii ONT reads and visualized as a dot plot (see also Supplemental Fig. S11). Matches in the forward and reverse orientation are indicated as purple and blue lines, respectively. Sequences were sorted, from longest to shortest. Window size is indicated above each panel. The arrow in F points to the only match observed to be longer than 3000 bp. The names of the sequences located on each axis are located in the Supplemental Methods.
Homoplasmy of sequence is maintained
Comparative analyses of the multiple copies of the 21 SBs revealed that all internally located SBs, that is, blocks not at the end of a sequence read (where incomplete blocks are often present), are identical in sequence, barring a few SBs that contain small aberrations characteristic of rearrangement artifacts (Supplemental Tables S5, S6). To assess homoplasmy in the context of this unusual fragmented and redundant genome architecture, a T. gondii RH N-ethyl-N-nitrosourea (ENU) cob mutant named ELQ-316 (Alday et al. 2017) was sequenced with Oxford Nanopore technology (Supplemental Table S3). This strain has a point mutation in SB “E” which encodes a Thr222→Pro substitution. SB “E” is only found flanked by SBs “J” and “A” arranged as (JEA) (Table 2). If homoplasmy exists, we expect this mutation to be present in all copies of SB “E.” To avoid bias, only uncorrected Nanopore reads from an early passage of this mutant were analyzed. Seven hundred seventy-nine mtDNA reads were identified (Supplemental Table S3). Of the 138 SB “E” identified in these reads that contained the mutated region, 132 contained the point mutation (Supplemental Fig. S9), indicating homoplasmy is maintained. The six blocks that did not show the mutation are well within the ∼5% error rate of ONT.
Table 2.
Lexicon of triplet block orders in T. gondii and N. caninum mtDNA
mtDNA sequence blocks are evolutionarily conserved in the Toxoplasmatinae but not Eimeria
We analyzed available genomic and transcriptomic data for the closely related Neospora caninum. All 21 mtDNA SBs are highly conserved between the two species in sequence and length (Table 1; Supplemental Table S2; Supplemental Data S7). However, N. caninum does contain an additional partial SB, block Sp, relative to T. gondii. Analysis of N. caninum Sanger genomic and EST reads using the 21 N. caninum SBs revealed the presence of nonrandom arrangements similar to those identified in T. gondii (Supplemental Figs. S2, S3). We generated low-coverage ONT genome data for N. caninum strain Nc-1. mtDNA reads of 0.374–15.6 kb, some of which are capable of encoding cytochromes, were detected (Supplemental Fig. S10; Supplemental Table S3; Supplemental Data S8). N. caninum ESTs encoding mtDNA SBs, which if assembled could encode each of the cytochromes, were also detected (Supplemental Table S4; Supplemental Data S9).
Examination of available sequences from another closely related Toxoplasmatinae, Hammondia (Walzer et al. 2013), also revealed the presence of all 21 mtDNA SBs. The ends of a few blocks could not be detected likely due to the sparsity of data available for this species (Table 1; Supplemental Table S2). However, we were not able to conclusively determine the nature of the mtDNA in the more distantly related Sarcocystis neurona, a member of the Toxoplasmatinae separated by ∼250 million years (Su et al. 2003). We identified hits to a number of the 21 mtDNA SBs, but the block boundaries were rarely conserved (Supplemental Table S2) and we were unable to detect similar permutation patterns. An examination of the more distant coccidian E. tenella (Su et al. 2003) (∼500 million years of divergence), for which a published mtDNA genome sequence exists (Hikosaka et al. 2011), did not reveal fragmented blocks as are observed in T. gondii (Supplemental Table S2). Examination of the S. neurona sequences using the assembled mtDNA and annotated genes of E. tenella does not strongly support an E. tenella-like mtDNA either. Together, the findings suggest that the unusual nature of the mtDNA is a derived state shared among the closest relatives of T. gondii.
The order, orientation, and number of the 21 sequence blocks are highly variable but not random
To detect regions of sequence identity within the SBs, dot plot analyses of the reads aligned against each other were performed. In a dot plot analysis, if identical sequence is observed between the sequences on each axis, a diagonal line is drawn with coordinates that span the alignment, using two different colors to indicate the orientation of the match. A dot plot of the SBs against themselves revealed that they are nonredundant except for the last 50 bp of SBs C and H, which are identical (Fig. 4A). Small 10- to 14-bp regions of microhomology are detected between many SBs (Fig. 4B). Larger window sizes that can span multiple SBs were used to compare ONT reads against themselves. A 1000-nt window revealed numerous aligned regions between ONT reads (Fig. 4C). At a window size of 2000 nt, many aligned regions are still observed (Fig. 4D), but at a window size of 3000 nt, few shared sequence regions of at least this length are observed (Fig. 4E). Finally, T. gondii ONT reads were compared to N. caninum ONT reads and fewer alignments are observed than when T. gondii reads are compared to themselves with the same window size (Fig. 4D,F).
To further analyze sequence block order, a lexicon of all triplet block orders was constructed from all ONT mtDNA sequences for both T. gondii and N. caninum (Table 2). Certain SBs are always observed upstream of, or downstream from, specific SBs (e.g., block L), as if they are part of a larger conserved super block of sequence, whereas other blocks (e.g., block J) are observed to be flanked by several different blocks (Table 2), indicating that they occur in multiple distinct contexts. The lexicon also revealed that T. gondii and N. caninum share the majority of their triplets but not all (Table 2). To examine the frequency of each SB junction, we tallied the counts of the element observed directly at the 5′ and 3′ end of each sequence block in the T. gondii and N. caninum ONT mtDNA reads (Supplemental Fig. S12A,B) and T. gondii PRU ONT direct RNA strand sequencing and N. caninum Sanger EST reads to verify the occurrence of the same block relationships in transcripts (Supplemental Fig. S12C,D). The patterns observed in the RNA and DNA are shared for each species and molecule type except for the differences associated with the acquisition of Sp in N. caninum. Thus, the basic units of mtDNA secondary structure are evolutionarily conserved and transcripts are not edited or rearranged.
To examine sequence redundancy, we counted the number of full-length genes in the ONT reads, which yielded 22 and 57 coxI, 26 and 103 coxIII, and 23 and 52 cob in T. gondii ME49 and RH mtDNA reads and three, seven, and two in N. caninum. Of the 1165 ONT mtDNA sequences from two T. gondii and one N. caninum run (Supplemental Table S3), 77% do not encode full-length cytochromes (Supplemental Tables S7, S8). To estimate relative T. gondii ME49 mtDNA gene copy numbers based on read depth, we mapped genomic Illumina PE data from two different T. gondii strains to full-length cytochrome gene sequences and a single-copy nuclear gene, GAPDH, for normalization. coxI, coxIII, and cob were detected at 316, 423, and 297× for ME49 and 369, 523, and 313× for RH88 relative to GAPDH (Supplemental Table S9). For comparison, the E. tenella mtDNA was estimated to exist at ∼50 copies per nuclear genome (Hikosaka et al. 2011).
The topology of the mitochondrial genome is dynamic
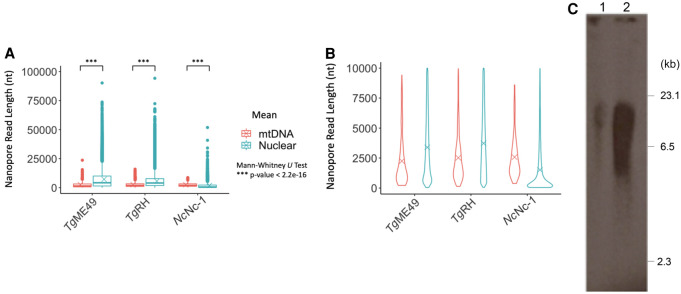
ONT single-molecule sequencing of T. gondii ME49 revealed molecules as long as 23 kb consisting fully of mtDNA SBs, although most ONT reads were shorter (Figs. 1F, 5). To assess whether the ONT reads were informative with respect to naturally occurring mtDNA size (a read cannot be longer than the naturally occurring molecule), we compared mtDNA and nuclear read lengths (Fig. 5A). Toxoplasma mtDNA ONT reads are shorter than nuclear reads, except for N. caninum where the nuclear reads are shorter due to apparent degradation. Yet, we note the N. caninum mtDNA distribution remains similar to T. gondii. The observed T. gondii mtDNA ONT read lengths are similar to the range of sizes observed in a Southern of a CHEF blot probed with a 1021-bp section of the cob gene, but the abundance of ONT reads in the 1–4 kb range (Fig. 5B) do not match the intense smear (∼5–16 kb) observed on the blot (Fig. 5C). A similar Southern smear was observed in P. falciparum (Preiser et al. 1996), where rolling circle replication creates tandem concatemers (Wilson and Williamson 1997). Physically, if the Toxoplasma mtDNA existed as a linear concatemer of tandemly repeating units, the dot plots of ONT reads (Fig. 4) should have revealed this topology. Restriction digest of a tandemly repeating sequence structure with an enzyme that has a single cut site within the unit should result in one major band and a trailing smear of smaller fragments, as is seen in Plasmodium (Preiser et al. 1996). However, when T. gondii genomic DNA was digested with XhoI, which uniquely cuts the cob gene, and probed with a coxI probe, two faint bands of ∼1.35 and 1.65 kb in addition to a large smear were detected (Supplemental Fig. S13). No pattern characteristic of a tandem repeat was observed.
Figure 5.
Size distribution of mtDNA versus nuclear ONT reads. (A) Three ONT runs are plotted and tested for statistical significance. Box plots cover the 25th–75th percentiles. See Supplemental Table S10 for mean and median values. (B) Violin plots of the same ONT reads <10 kb. (C) Southern of a CHEF gel electrophoresis of T. gondii total DNA (lane 1) and mitochondrial-enriched DNA (lane 2) probed with a 1012-bp section of the cob gene. DNA ladder is as indicated.
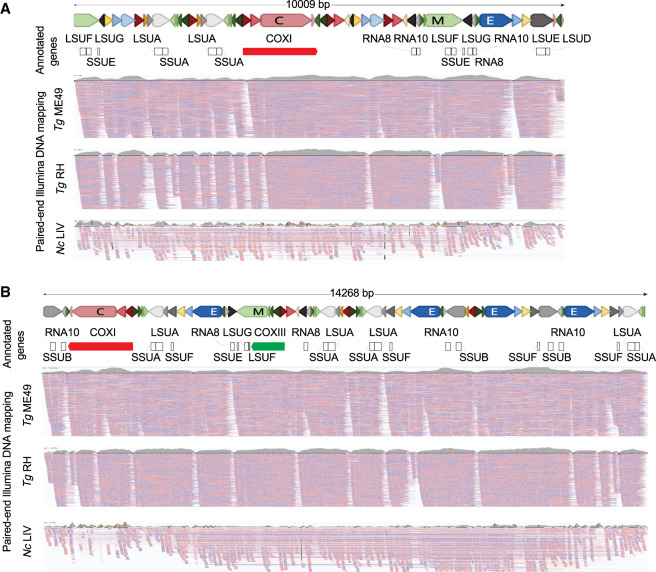
The secondary structure of the mtDNA appears to be dynamic. Several years separated the isolation of T. gondii ME49 DNA for Illumina and ONT sequencing. During this time, the parasites were being passaged. We note that when our Illumina PE reads are mapped to our ONT reads, we observe gaps in the mapping (Fig. 6; Supplemental Fig. S6). The mapping further degenerates when PE Illumina reads from a different T. gondii strain, RH, are mapped, as evidenced by the increase in the number of white gaps in the mapping and dips in the overall coverage plots above the alignments (Fig. 6). The degeneration in mappability is even stronger when N. caninum PE Illumina sequence reads are mapped despite using a lower mapping stringency (Fig. 6). A similar trend was observed when N. caninum and T. gondii Illumina reads were mapped to N. caninum ONT reads (Supplemental Fig. S14). Given the fact that the SBs (Table 1) and lexicon (Table 2) are highly conserved, it seems likely that the long mtDNA molecules evolve rapidly. The mapping experiments also revealed the presence of different populations of PE reads, some that span particular ONT SB orders and others that do not, for example, right-hand edge of Figure 6A or the beginning of the TgRH track in Figure 6B and Supplemental Figure S14.
Figure 6.
ONT and Illumina comparisons reveal sequence block order variability and decay with evolutionary time. (A,B) Two T. gondii ME49 ONT sequence reads were annotated with the gene sequences they contain. Reads and SBs are drawn to scale. The blocks are colored as shown in Table 1. “Annotated genes” track represents the annotation of the cytochrome genes and rRNA gene fragments on the ONT reads. The three “PE Illumina DNA mapping” tracks show PE read mapping of T. gondii ME49, TgRH88, and N. caninum (Nc) LIV mtDNA-specific reads. TgME49 and TgRH reads mapping required 100% nucleotide identity whereas 1% mismatch was allowed for mapping NcLIV reads. Reads were independently mapped to each of the ONT mtDNA reads and visualized using IGV. Red and blue lines below each read indicate the mapped Illumina PE reads. Coverage plots of the mapped reads are shown above each mapping panel in gray to indicate the depth of all reads and not just those shown.
The single-molecule ONT reads are riddled with inverted SBs throughout, but they do not appear to be located at the ends of the molecules where they could serve as telomeres (Smith and Keeling 2013). A few ONT reads have portions of the same sequence block on their ends, a feature characteristic of rolling-circle replication (Supplemental Fig. S15A), but most do not (Supplemental Fig. S15C). Attempts to circularize the ONT reads using the same program that was able to circularize the P. falciparum mtDNA revealed many potential circular chromosome candidates. The candidates range in size from 1.6 to 5.6 kb, some of which are shown in Supplemental Figures S15 and S16, but the putative circular chromosomes (Supplemental Figs. S15, S16) do not contain any measurable degree of support from other ONT reads. They do have support from PE-Illumina reads, but it is not always uniform (Supplemental Fig. S15A,C, top and bottom panels, respectively). Notably, full-length cytochrome genes are detected on the putative circular chromosomes. Finally, to address the possibility of a population of very small circular chromosomes, we identified all short ONT mtDNA reads 0.6–2.0 kb from the 779 mtDNA reads identified in the T. gondii RH ELQ-316 mutant strain and asked if they could be clustered into significantly overlapping populations. They could not, suggesting the reads were not derived from one or more identical short templates.
Discussion
We show here how long-read single-molecule ONT sequences elucidated novel mtDNA sequence arrangements for T. gondii and its closest relatives that consist of redundant, highly rearranged strings of sequence blocks that do not appear to be the direct result of rolling-circle replication. The SBs are evolutionarily conserved with a few minor variations in Hammondia hammondi and the more divergent N. caninum but not Sarcocystis. We also show that this lineage has independently evolved fragmentation of the coxI, coxIII, and cob genes, a feature previously detected only in dinoflagellates (Flegontov et al. 2015; Obornik and Lukes 2015; Gagat et al. 2017), perhaps as a mechanism to generate novelty in these streamlined mtDNA genomes.
The evidence indicates that the mtDNA became shattered and redundantly rearranged by some unknown event during the evolution of the tissue coccidia. Single-molecule mtDNA ONT reads of 0.10–23.6 kb comprised exclusively of SBs, in nonrandom arrangements, are observed across libraries. The fragmented nature of the T. gondii mtDNA is supported by four different lines of evidence: (1) individual unassembled Sanger genomic and EST reads; (2) complex PCR patterns from mtDNA enriched cell fractions; (3) single-molecule ONT long-read sequences of DNA and RNA; and (4) evolutionary conservation of nearly identical sequence blocks in the related tissue coccidia, Hammondia and Neospora. Despite the novel genome sequence(s) which includes many redundant fragments and full-length copies of coxI, coxIII, and cob genes and rRNA genes, additional RNA and protein features were not detected. The CDSs of coxI and cob are well conserved with other apicomplexans, but coxIII, which is reported here for the first time, is divergent. Low conservation was also reported for the coxIII of a free-living ancestor of the Apicomplexa, Chromera (Obornik and Lukes 2015). T. gondii, like several species in the alveolata, has evolved overlapping CDSs in its mtDNA. Sequence block V is required to form the full open reading frame (using opposite DNA strands) for both coxI and coxIII, reminiscent of the fused coxI and coxIII in chromerids (Jackson et al. 2007), albeit a different fusion.
In addition to full-length transcripts, evidence for the expression of every single sequence block was seen in the ONT direct RNA strand sequences including SBs for which we have no annotation (blocks D, N, P, and R). As was observed in the dinoflagellates, partial cytochrome genes are expressed. There is no evidence of trans-splicing or RNA-editing. There is no evidence of self-splicing introns, a feature often observed in mtDNA (Lang et al. 2007). Limited ONT direct RNA transcripts revealed poly(A) tails on the subset of coxI transcripts flanked by block J which may encode a cryptic poly(A) signal. In the dinoflagellates, the cytochrome transcripts are polyadenylated, but this occurs upstream of the stop codon (Jackson et al. 2007). Dinoflagellates display strong stem–loop and multiple hairpin features (Jackson et al. 2007) in noncoding regions; however, we did not find strong evidence for long, nearly perfect hairpins other than a structure in sequence block J. Regulation of mtDNA gene expression in this highly polymorphic mtDNA context is unknown. All SBs are expressed.
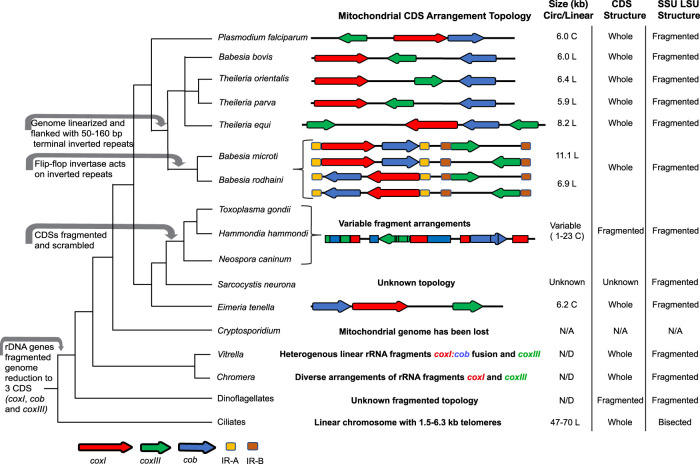
Fragmented mtDNA sequences have been observed in other organisms, most notably the sister phylum to the Apicomplexa, the dinoflagellates, which also share fragmented rRNA genes and a reduced mtDNA gene content of only coxI, coxIII, and cob. What is fascinating from an evolutionary perspective is the fact that many dinoflagellates, including a basal dinoflagellate, Hematodinium, have also redundantly fragmented their cytochrome genes and contain both full-length and fragmented copies (Ling and Shibata 2004; Bedoya et al. 2009). Examination of Hematodinium cytochrome gene fragments reveals that their sequences have been shattered in different locations relative to T. gondii and that there are a greater number of different, often overlapping fragments in the dinoflagellates. The capacity to both generate and survive structural rearrangements within such a small genome is a prominent feature of the Apicomplexa and dinoflagellates (Fig. 7).
Figure 7.
Alveolate mtDNA evolution and characteristics. (Left) Cladogram of alveolate relationships with major mitochondrial genome events indicated with gray arrows. (Center) Schematic of mtDNA CDS (red, green, and blue arrows) or inverted repeats (gold and brown) indicated. Spacing is approximate with CDS lengths exaggerated for ease of viewing. Ribosomal RNA and other RNA genes or fragments thereof are not represented. (Right) mtDNA size and topology. (L) Linear, (C) concatemer, presumably from circular progenitors. Status of CDS sequences and status of Short (SSU) and Long Subunit (LSU) rRNA are indicated.
The data do not reveal the exact topology of the mitochondrial genome or how it replicates. Our inability to assemble a single full-length mitochondrial genome sequence and the presence of redundant sequence block order and orientation variants suggests that the T. gondii mtDNA could exist as a number of distinct chromosomes as is seen in dinoflagellates and other organisms (Burger et al. 2003; Flegontov and Lukes 2012; Dong et al. 2014; Yahalomi et al. 2017). It is also likely that these chromosomes are recombining with each other to produce the larger molecules and variety of SB patterns that are observed in this study. Theoretically, the mtDNA of Toxoplasma and its relatives could be circular, linear, or some combination of both, or it could be contained on a single chromosome, as is observed in all Apicomplexa to date (Fig. 7). However, the data do not support a single circular topology containing all SBs that replicates via rolling-circle replication, as evidenced by sequencing, the dot plot comparisons, and Southern analysis of restricted DNA. Formally, each of the 21 SBs could exist on its own chromosome and replicates could become concatenated nonrandomly via some undiscovered process that guides particular ligations. We consider this unlikely because inverted repeats are not detected at the ends of individual SBs. We do, however, detect support from our copy number analysis for the hypothesis that, when the mtDNA shattered into smaller pieces, it shattered redundantly into many different units of sequence, each of which is capable of being annotated with more than one of the 21 SBs. Minimally, the mtDNA consists of these 21 nonredundant SBs, none of which individually encodes a full gene, for a total of 5909 bp of sequence in T. gondii and 5908 in N. caninum.
Most apicomplexan mtDNAs are ∼ 6 kb. In P. falciparum, a Southern blot smear ranging from 6 to 23 kb was attributed to rolling-circle replication intermediates (Weissig and Rowe 1999) of the mtDNA. Smears of 6–10 kb or more were observed in dinoflagellates (Sandhu et al. 2007; Chen 2013; Gualberto and Newton 2017). T. gondii showed a smear ranging from 2 to 16 kb with greatest signal above 5 kb. mtDNA ONT reads are statistically shorter than ONT nuclear reads. Thus, the sizes are likely indicative of the biological reality that mtDNA molecules are mostly <10 kb. The fact that N. caninum mtDNA ONT reads do not appear to be degraded in contrast to nuclear reads could indicate circular molecules that were protected from exonucleases.
The observation that short stretches of mtDNA <3 kb are observed to occur multiple times within the ONT reads implies that regions of mtDNA longer than single SBs exist. The ONT reads and resulting lexicon prove that multiple distinct SB arrangements exist. More importantly, evolution of the lexicon in N. caninum to contain Sp, (and VSpD) indicates that these stretches of sequence are encoded together as a larger single unit. The data support a model in which parts of the mitochondrial genome, including at least one full-length copy of each cytochrome gene, are redundantly present on different chromosomes. We speculate that it is much less risky to the organism if it maintains at least one full-length copy of each gene or rRNA gene fragment. The situation is more complex when considering stretches of DNA >3 kb. It is probable and parsimonious to hypothesize that larger sequences are constructed, albeit uniquely, from smaller sequences. A possible mechanism is homologous recombination between circular chromosomes to create a larger single chromosome. If the mtDNA of T. gondii exists as a population of redundant circular chromosomes, each of which contains multiple portions of the ancestral mtDNA redundantly, then homologous recombination between two different circular chromosomes sharing a homologous region (or a stretch of microhomology) will create a novel larger molecule. Recombination among such a population is a plausible explanation for the variety of sequences that are detected by ONT. It would also explain some of the errors observed in the ONT reads where we see damaged SBs. Further, gaps in mapping of Illumina data to ONT reads generated from T. gondii ME49 DNA extracted several years apart is indicative of rapidly evolving mtDNA molecules, providing support for recombination events. Recombination has been observed in P. falciparum during mtDNA replication (Preiser et al. 1996), and it is a widely reported phenomenon within and between mitochondria across eukaryotes (Sandhu et al. 2007; Chen 2013; Gualberto and Newton 2017).
We do not yet know how much variation, if any, exists within a single parasite mitochondrion versus population level variation. The data presented here cannot address that question as DNA was extracted from millions of parasites and as of yet, individual mitochondrial organelle sequencing is not yet possible. However, the ONT reads from the T. gondii RH ENU mutant strain are quite informative. DNA from the T. gondii ELQ-316 mutant was from a recently derived clone presumably originating from a single resistant parasite. The ONT reads from this mutant also yielded a myriad of long, unique sequence block concatemers, suggesting that they either already existed in the parent mitochondrion or are easily regenerated. We also observed that the point mutation was present in nearly all copies of sequence block E on the ONT reads. We do not know if this finding indicates that each mutant sequence block E (which is always flanked by J and A, i.e., JEA) originates from the same source molecule, or if T. gondii has the machinery needed to maintain homoplasmy, perhaps via MSH1, a MutS homolog that targets to the mitochondrion in Toxoplasma (Garbuz and Arrizabalaga 2017).
Although the lack of a single mtDNA sequence raises difficulties for those targeting the T. gondii mitochondrion, the presence of a possible mechanism for maintaining homoplasmy at the level of a sequence block was observed. This finding opens the door to further exploration of the Toxoplasma mitochondrion, as it is an important drug target. The need to be able to purify mitochondria in Toxoplasma remains strong. Elucidation of the T. gondii mtDNA might have happened years ago were it not for NUMTs. We now know that “REP” elements in T. gondii (Reid et al. 2012) are NUMTs that can be annotated with SBs (Supplemental Table S11). Toxoplasma gondii contains >9000 NUMTs in its nuclear genome (Namasivayam 2015), thwarting both molecular and sequence-based mtDNA identification. The NUMTs together with the unique architecture presented here generated a difficult puzzle that required long single-molecule sequencing for a breakthrough. ONT offers the opportunity for re-analysis of dinoflagellate mtDNAs and further exploration of mitochondrial biology and evolution. Perhaps, the independent evolution of redundant fragmented mtDNA in the dinoflagellates and Apicomplexa provides a mechanism for the generation of evolutionary novelty in these tiny, streamlined organellar genome sequences.
Methods
Species and strains
T. gondii RH and ME49 tachyzoites were maintained in human fibroblast reverse transcriptase (hTERT) cells as described in Weiss and Kim (2007). Extracellular tachyzoites, washed twice in PBS, were used for mitochondrial enrichment or DNA extraction. Neospora caninum Nc-1 parasites were cultured on Vero cells using RPMI 1640 supplemented with 5% normal horse serum, purified by centrifugation through 30% isotonic Percoll and washed twice in PBS.
Enriched mitochondrial fractions
T. gondii tachyzoites (∼2–4 × 109) were purified as in Miranda et al. (2010) and Supplemental Methods.
Nucleic acid extraction, PCR, RT-PCR, and sequencing
DNA for PCR was isolated through standard Proteinase K and RNase A digestion followed by phenol/chloroform extraction. Primer pairs are in Supplemental Table S1. PCR conditions are as in Namasivayam (2015) and Supplemental Methods.
T. gondii RH Δku80 RNA extraction used the RNeasy Mini Kit (Qiagen) Quick-Start protocol with the DNase digestion step. RT-PCR used the SuperScript III first-strand synthesis kit (Invitrogen) and cytochrome gene-specific primers (coxI V2, coxIII M9, cob E5) (Supplemental Table S1) to amplify the three cytochrome transcripts which were sequenced with primers C9, M9, and T3 (Supplemental Table S1) to verify identity.
DNA for sequencing was extracted from T. gondii ME49 and N. caninum Nc-1 parasites using the DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer's cultured cells protocol and sequenced with Illumina or an ONT flowcell 9.4.1 according to the manufacturer's instructions (see Supplemental Methods). MinKNOW version 18.03.1 was used to call bases. ONT Direct RNA sequence data generated from poly(A)+ purified RNA were provided by Stuart Ralph. Supplemental Table S3 summarizes all sequencing yields. Corrected mtDNA ONT reads are in Supplemental Data S3, S6, S8, and S10).
Sequence analysis
Sequenced PCR amplicons, public unassembled Sanger mtDNA-like EST, and genome sequence reads were compared to each other using BLASTN (E-value: e−10, nucleotide identity ≥98% in all instances unless noted). Mitochondrial SBs were determined based on their reproducible occurrence, sequence boundaries, and arrangements in the sequences. T. gondii reference sequences were obtained from ToxoDB (https://toxodb.org/) release 11 (Gajria et al. 2008). These contigs and all unassembled T. gondii Sanger genomic and EST reads from NCBI were screened via BLASTN for any additional mtDNA sequences using the determined mtDNA SBs as queries. A read was classified as mitochondrial if no adjoining nuclear sequence was found. All reads classified as mitochondrial were analyzed for length, sequence boundaries, and SB arrangements using BLASTN.
NUMT identification
RepeatMasker version 4.0.5 (Smit et al. 2015) with a repeat library composed of the 21 T. gondii mtDNA SBs and search engine “crossmatch” was used to identify nuclear sequences of mitochondrial origin in the nuclear genome sequences of T. gondii and N. caninum (Namasivayam 2015).
Analysis of ONT reads
Two strategies were used to identify mtDNA reads. First, the ONT reads were screened for the 21 mtDNA SBs using BLASTN (nucleotide identity ≥60% and ≤10% alignment length mismatch). Second, the reads were aligned to the 21 mtDNA SBs using Exonerate v2.4.0 (Slater and Birney 2005). The best 2000 alignments with an alignment ≥60% were classified as putative mtDNA. Reads containing NUMTs were removed and error correction was performed with Illumina reads (Supplemental Data S6; see Supplemental Materials for details).
Mapping of genomic Illumina reads
T. gondii ME49 DNA PE-Illumina reads were mapped requiring 100% identity to the 21 mtDNA SBs using BWA v0.7.17. mtDNA-containing reads were further examined for any new mtDNAs. To assess Illumina support for the ONT reads, T. gondii ME49 (SRR9200762), RH88 (SRR521957), and N. caninum LIV (ERR012900) mtDNA reads were independently mapped to corrected T. gondii ME49 ONT reads using BWA v0.7.17 (local alignment, requiring both pairs to match at 100% identity) and visualized using the Integrative Genomics Viewer (IGV) (Robinson et al. 2011), Any methodological deviation is indicated in figure legends. When estimating copy numbers for mtDNA relative to a single-copy nuclear gene, negative controls included mapping to manually created SB fusions not observed in our lexicon.
Gene prediction and annotation
Annotation was based on similarity to the cytochrome genes and rRNA fragments of P. falciparum and E. tenella (Feagin 1992; Feagin et al. 1997, 2012; Hikosaka et al. 2011). TBLASTN and ORF prediction (NCBI ORF finder, genetic code for protozoan mitochondria) was used to identify and annotate SB protein-coding regions. ORFs >50 aa from the SBs and the longest T. gondii ONT read were searched with InterProScan V5.31-70.0 to identify protein features. PE T. gondii RNA-seq reads obtained from NCBI (SRR6493545) were mapped to mtDNA SBs as described above to identify mtDNA-specific reads. These reads were then mapped to cytochrome CDSs using BWA v0.7.17 (local alignment, requiring both pairs to map at 100% identity). Toxoplasma gondii PRU Δku80 ONT direct RNA strand reads (SRR9200760) (Supplemental Data S6) were processed like the genomic ONT reads and annotated. rRNA genes that did not align well with the rRNA gene fragments of P. falciparum were identified using conserved nucleotides, manually folded, and compared to E. coli rRNA secondary structures and their Plasmodium counterparts. All SBs and the longest ONT read were used to search Rfam v14.2 to identify any additional RNA genes or RNA features like catalytic RNAs or self-splicing introns.
Mitochondrial genome assembly
CAP3 with default parameters (Huang and Madan 1999) was used to assemble PCR amplicons and unassembled mtDNA Sanger reads. SPAdes 3.12.0 (Bankevich et al. 2012) was used to assemble mtDNA PE Illumina reads. Canu v1.9 (Koren et al. 2017), Flye v2.6 (Kolmogorov et al. 2019), CAP3, and Geneious Prime 2019.1.3 (Kearse et al. 2012) were used with corrected mtDNA ONT reads.
Identification of mtDNA sequences in N. caninum, Hammondia, and S. neurona
N. caninum genomic sequences were obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/Neospora/caninum/NEOS.contigs.072303. A single contig containing mtDNA sequences was identified using the 21 T. gondii mtDNA SBs via BLASTN (E-value: e−10). This contig was not present in later genome assemblies. N. caninum mtDNA SBs were annotated based on this contig, and the SBs were used to identify additional mtDNA sequence contigs. N. caninum ONT data were queried as above for T. gondii, and these reads were error-corrected using N. caninum PE Illumina genomic reads (ERR012900) and annotated using N. caninum SBs (Supplemental Data S8). Similarly, the genome sequence of Hammondia hammondi H.H.34 (Walzer et al. 2013) (ToxoDB version 2014-06-03) as well as sequences of other Hammondia species available in NCBI were mined for mtDNA sequences via BLASTN. Sarcocystis neurona SN3 and SN1 genome sequences (versions 2015-04-13 and 2015-07-23, respectively) obtained from ToxoDB and sequence data in NCBI were mined for mtDNA using T.gondii mtDNA SBs and the mitochondrial genome sequence of E. tenella (AB564272.1) using BLASTN and TBLASTX.
Other bioinformatic analyses and data visualization
The arrangements and permutations of SBs in the data sets were obtained via BLASTN and manually inspected. A GFF file containing the annotation of genes and SBs for each read and PCR amplicon was generated using BLASTN and a Perl script (bp_search2gff.pl) available from APPRIS (Rodriguez et al. 2018) and visualized using Geneious Prime v.2019 (Kearse et al. 2012). DotPlots were performed with NUCmer (MUMmer 4.0.0beta2-foss-2019b using the -maxmatch option) and plotted using mummerplot (Kurtz et al. 2004) and gnuplot 5.2.8. Alignments of Illumina reads to SBs, ONT, and cytochrome sequences were visualized using IGV and coverage was determined using BEDTools 2.21.0 genomeCoverageBed (Quinlan and Hall 2010).
Contour-clamped homogeneous electric field (CHEF) electrophoresis
T. gondii RH tachyzoite parasites were isolated and suspended in 1.0% low-melting agarose plugs and digested with 0.5 M EDTA, 1% lauroylsarcosine, 2 mg/mL Proteinase K, and 0.2 mg/mL RNase A at pH 8.0, 50°C for 48 h. Electrophoresis was performed using a CHEF Mapper (Bio-Rad Laboratories) in a 1% agarose gel (Sigma-Aldrich) with the following conditions: 6V/cm, linear 11s–22s, 15 h, and 4°C with a switching angle of 120°. A radioactive 1012-bp cob probe was synthesized through PCR, gel-extracted, and cleaned using mini Quick spin columns (Roche). Southern analysis was performed as described in Supplemental Methods.
Data access
The Illumina, ONT, and nucleotide data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA546244, PRJNA436351, and PRJNA560064.
Supplementary Material
Acknowledgments
We thank Silvia Moreno for providing mitochondrial-enriched fractions of T. gondii; Stuart Ralph for sharing his T. gondii PRU Δku80 direct RNA strand sequence data; David Sibley for providing the T. gondii ME49 strain used in the genome project; Tobias Lilja and Faruk Dube for assistance with ONT sequencing; and Yiran Li for assistance with plots. This work benefited from discussions with Kissinger Research Group members both past and present. This work was supported in part through National Institutes of Health funding (NIH R01 AI068908) and resources from the Georgia Advanced Computing Resource Center. J.S.D. received support from BX002440 from the U.S. Department of Veterans Affairs Biomedical Laboratory Research.
Author contributions: S.N. and J.C.K. designed research; S.N., J.C.K., W.X., and E.M.H. performed research; K.T. and J.S.D. contributed new reagents or analytic tools; S.N., R.P.B., and J.C.K. analyzed data; S.N., R.P.B., K.T., and J.C.K. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.266403.120.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Alday PH, Bruzual I, Nilsen A, Pou S, Winter R, Ben Mamoun C, Riscoe MK, Doggett JS. 2017. Genetic evidence for cytochrome b Qi site inhibition by 4(1H)-quinolone-3-diarylethers and antimycin in Toxoplasma gondii. Antimicrob Agents Chemother 61: e01866-16. 10.1128/AAC.01866-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedoya F, Medveczky MM, Lund TC, Perl A, Horvath J, Jett SD, Medveczky PG. 2009. Identification of mitochondrial genome concatemers in AIDS-associated lymphomas and lymphoid cell lines. Leuk Res 33: 1499–1504. 10.1016/j.leukres.2009.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Ostell J, Pruitt KD, Sayers EW. 2018. Genbank. Nucleic Acids Res 46: D41–D47. 10.1093/nar/gkx1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Lang BF. 2003. Parallels in genome evolution in mitochondria and bacterial symbionts. IUBMB Life 55: 205–212. 10.1080/1521654031000137380 [DOI] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF. 2003. Mitochondrial genomes: anything goes. Trends Genet 19: 709–716. 10.1016/j.tig.2003.10.012 [DOI] [PubMed] [Google Scholar]
- Chen XJ. 2013. Mechanism of homologous recombination and implications for aging-related deletions in mitochondrial DNA. Microbiol Mol Biol Rev 77: 476–496. 10.1128/MMBR.00007-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey AM, Ranford-Cartwright LC, Moore DJ, Williamson DH, Wilson RJ, Walliker D, Carter R. 1993. Uniparental inheritance of the mitochondrial gene cytochrome b in Plasmodium falciparum. Curr Genet 23: 360–364. 10.1007/BF00310900 [DOI] [PubMed] [Google Scholar]
- Dong WG, Song S, Jin DC, Guo XG, Shao R. 2014. Fragmented mitochondrial genomes of the rat lice, Polyplax asiatica and Polyplax spinulosa: intra-genus variation in fragmentation pattern and a possible link between the extent of fragmentation and the length of life cycle. BMC Genomics 15: 44. 10.1186/1471-2164-15-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP. 2010. Toxoplasmosis of animals and humans. Taylor & Francis Group, Boca Raton, FL. [Google Scholar]
- Esseiva AC, Naguleswaran A, Hemphill A, Schneider A. 2004. Mitochondrial tRNA import in Toxoplasma gondii. J Biol Chem 279: 42363–42368. 10.1074/jbc.M404519200 [DOI] [PubMed] [Google Scholar]
- Feagin JE. 1992. The 6-kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol Biochem Parasitol 52: 145–148. 10.1016/0166-6851(92)90046-M [DOI] [PubMed] [Google Scholar]
- Feagin JE, Mericle BL, Werner E, Morris M. 1997. Identification of additional rRNA fragments encoded by the Plasmodium falciparum 6 kb element. Nucleic Acids Res 25: 438–446. 10.1093/nar/25.2.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, Cannone JJ, Tami G, Schnare MN, Gutell RR. 2012. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS One 7: e38320. 10.1371/journal.pone.0038320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegontov P, Lukes J. 2012. Mitochondrial genomes of photosynthetic euglenids and alveolates. In Advances in botanical research (ed. Maréchal-Drouard L), Vol. 63, pp. 127–153. Academic Press, Cambridge, MA. [Google Scholar]
- Flegontov P, Michálek J, Janouškovec J, Lai DH, Jirků M, Hajdušková E, Tomčala A, Otto TD, Keeling PJ, Pain A, et al. 2015. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol Biol Evol 32: 1115–1131. 10.1093/molbev/msv021 [DOI] [PubMed] [Google Scholar]
- Flegr J, Prandota J, Sovičková M, Israili ZH. 2014. Toxoplasmosis – a global threat. Correlation of latent toxoplasmosis with specific disease burden in a set of 88 countries. PLoS One 9: e90203. 10.1371/journal.pone.0090203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagat P, Mackiewicz D, Mackiewicz P. 2017. Peculiarities within peculiarities – dinoflagellates and their mitochondrial genomes. Mitochondrial DNA Part B 2: 191–195. 10.1080/23802359.2017.1307699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, et al. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res 36: D553–D556. 10.1093/nar/gkm981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuz T, Arrizabalaga G. 2017. Lack of mitochondrial MutS homolog 1 in Toxoplasma gondii disrupts maintenance and fidelity of mitochondrial DNA and reveals metabolic plasticity. PLoS One 12: e0188040. 10.1371/journal.pone.0188040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G. 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity (Edinb) 101: 301–320. 10.1038/hdy.2008.62 [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Newton KJ. 2017. Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annu Rev Plant Biol 68: 225–252. 10.1146/annurev-arplant-043015-112232 [DOI] [PubMed] [Google Scholar]
- Hikosaka K, Watanabe Y, Tsuji N, Kita K, Kishine H, Arisue N, Palacpac NM, Kawazu S, Sawai H, Horii T, et al. 2010. Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol Biol Evol 27: 1107–1116. 10.1093/molbev/msp320 [DOI] [PubMed] [Google Scholar]
- Hikosaka K, Nakai Y, Watanabe Y, Tachibana S, Arisue N, Palacpac NM, Toyama T, Honma H, Horii T, Kita K, et al. 2011. Concatenated mitochondrial DNA of the coccidian parasite Eimeria tenella. Mitochondrion 11: 273–278. 10.1016/j.mito.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Hikosaka K, Tsuji N, Watanabe Y, Kishine H, Horii T, Igarashi I, Kita K, Tanabe K. 2012. Novel type of linear mitochondrial genomes with dual flip-flop inversion system in apicomplexan parasites, Babesia microti and Babesia rodhaini. BMC Genomics 13: 622. 10.1186/1471-2164-13-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka K, Kita K, Tanabe K. 2013. Diversity of mitochondrial genome structure in the phylum Apicomplexa. Mol Biochem Parasitol 188: 26–33. 10.1016/j.molbiopara.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res 9: 868–877. 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Norman JE, Schnare MN, Gray MW, Keeling PJ, Waller RF. 2007. Broad genomic and transcriptional analysis reveals a highly derived genome in dinoflagellate mitochondria. BMC Biol 5: 41. 10.1186/1741-7007-5-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kairo A, Fairlamb AH, Gobright E, Nene V. 1994. A 7.1 kb linear DNA molecule of Theileria parva has scrambled rDNA sequences and open reading frames for mitochondrially encoded proteins. EMBO J 13: 898–905. 10.1002/j.1460-2075.1994.tb06333.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov AA, Gerasimov ES. 2012. Diversity of mitochondrial genome organization. Biochemistry (Mosc) 77: 1424–1435. 10.1134/S0006297912130020 [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37: 540–546. 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5: R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang BF, Burger G, O'Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. 1997. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 387: 493–497. 10.1038/387493a0 [DOI] [PubMed] [Google Scholar]
- Lang BF, Laforest MJ, Burger G. 2007. Mitochondrial introns: a critical view. Trends Genet 23: 119–125. 10.1016/j.tig.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Lau YL, Lee WC, Gudimella R, Zhang G, Ching XT, Razali R, Aziz F, Anwar A, Fong MY. 2016. Deciphering the draft genome of Toxoplasma gondii RH strain. PLoS One 11: e0157901. 10.1371/journal.pone.0157901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling F, Shibata T. 2004. Mhr1p-dependent concatemeric mitochondrial DNA formation for generating yeast mitochondrial homoplasmic cells. Mol Biol Cell 15: 310–322. 10.1091/mbc.e03-07-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez JV, Yuhki N, Masuda R, Modi W, O'Brien SJ. 1994. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol 39: 174–190. 10.1007/BF00163806 [DOI] [PubMed] [Google Scholar]
- Lorenzi H, Khan A, Behnke MS, Namasivayam S, Swapna LS, Hadjithomas M, Karamycheva S, Pinney D, Brunk BP, Ajioka JW, et al. 2016. Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat Commun 7: 10147. 10.1038/ncomms10147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DC, Tomavo S, Berry EA, Boothroyd JC. 2000. Characterization of cytochrome b from Toxoplasma gondii and Qo domain mutations as a mechanism of atovaquone-resistance. Mol Biochem Parasitol 108: 1–12. 10.1016/S0166-6851(00)00184-5 [DOI] [PubMed] [Google Scholar]
- Melo EJ, Attias M, De Souza W. 2000. The single mitochondrion of tachyzoites of Toxoplasma gondii. J Struct Biol 130: 27–33. 10.1006/jsbi.2000.4228 [DOI] [PubMed] [Google Scholar]
- Minot S, Melo MB, Li F, Lu D, Niedelman W, Levine SS, Saeij JP. 2012. Admixture and recombination among Toxoplasma gondii lineages explain global genome diversity. Proc Natl Acad Sci 109: 13458–13463. 10.1073/pnas.1117047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda K, Pace DA, Cintron R, Rodrigues JC, Fang J, Smith A, Rohloff P, Coelho E, de Haas F, de Souza W, et al. 2010. Characterization of a novel organelle in Toxoplasma gondii with similar composition and function to the plant vacuole. Mol Microbiol 76: 1358–1375. 10.1111/j.1365-2958.2010.07165.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya JG, Liesenfeld O. 2004. Toxoplasmosis. The Lancet 363: 1965–1976. 10.1016/S0140-6736(04)16412-X [DOI] [PubMed] [Google Scholar]
- Morris JC, Drew ME, Klingbeil MM, Motyka SA, Saxowsky TT, Wang Z, Englund PT. 2001. Replication of kinetoplast DNA: an update for the new millennium. Int J Parasitol 31: 453–458. 10.1016/S0020-7519(01)00156-4 [DOI] [PubMed] [Google Scholar]
- Namasivayam S. 2015. “Repetitive DNA and nuclear integrants of organellar DNA shape the evolution of coccidian genomes.” PhD thesis, University of Georgia, Athens, GA. https://getd.libs.uga.edu/pdfs/namasivayam_sivaranjani_201508_phd.pdf [Google Scholar]
- Nash EA, Nisbet RE, Barbrook AC, Howe CJ. 2008. Dinoflagellates: a mitochondrial genome all at sea. Trends Genet 24: 328–335. 10.1016/j.tig.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Nishi M, Hu K, Murray JM, Roos DS. 2008. Organellar dynamics during the cell cycle of Toxoplasma gondii. J Cell Sci 121: 1559–1568. 10.1242/jcs.021089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oborník M, Lukeš J. 2015. The organellar genomes of Chromera and Vitrella, the phototrophic relatives of Apicomplexan parasites. Annu Rev Microbiol 69: 129–144. 10.1146/annurev-micro-091014-104449 [DOI] [PubMed] [Google Scholar]
- Ossorio PN, Sibley LD, Boothroyd JC. 1991. Mitochondrial-like DNA sequences flanked by direct and inverted repeats in the nuclear genome of Toxoplasma gondii. J Mol Biol 222: 525–536. 10.1016/0022-2836(91)90494-Q [DOI] [PubMed] [Google Scholar]
- Preiser PR, Wilson RJ, Moore PW, McCready S, Hajibagheri MA, Blight KJ, Strath M, Williamson DH. 1996. Recombination associated with replication of malarial mitochondrial DNA. EMBO J 15: 684–693. 10.1002/j.1460-2075.1996.tb00401.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AJ, Vermont SJ, Cotton JA, Harris D, Hill-Cawthorne GA, Könen-Waisman S, Latham SM, Mourier T, Norton R, Quail MA, et al. 2012. Comparative genomics of the Apicomplexan parasites Toxoplasma gondii and Neospora caninum: Coccidia differing in host range and transmission strategy. PLoS Pathog 8: e1002567. 10.1371/journal.ppat.1002567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JM, Rodriguez-Rivas J, Di Domenico T, Vázquez J, Valencia A, Tress ML. 2018. APPRIS 2017: principal isoforms for multiple gene sets. Nucleic Acids Res 46: D213–D217. 10.1093/nar/gkx997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu AP, Abdelnoor RV, Mackenzie SA. 2007. Transgenic induction of mitochondrial rearrangements for cytoplasmic male sterility in crop plants. Proc Natl Acad Sci 104: 1766–1770. 10.1073/pnas.0609344104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. 2007. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol 372: 356–368. 10.1016/j.jmb.2007.06.085 [DOI] [PubMed] [Google Scholar]
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2015. RepeatMasker Open-4.0. 2013–2015. https://www.repeatmasker.org. [Google Scholar]
- Smith DR, Keeling PJ. 2013. Gene conversion shapes linear mitochondrial genome architecture. Genome Biol Evol 5: 905–912. 10.1093/gbe/evt059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Evans D, Cole RH, Kissinger JC, Ajioka JW, Sibley LD. 2003. Recent expansion of Toxoplasma through enhanced oral transmission. Science 299: 414–416. 10.1126/science.1078035 [DOI] [PubMed] [Google Scholar]
- Sweet AD, Johnson KP, Cameron SL. 2020. Mitochondrial genomes of Columbicola feather lice are highly fragmented, indicating repeated evolution of minicircle-type genomes in parasitic lice. PeerJ 8: e8759. 10.7717/peerj.8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K. 1985. Visualization of the mitochondria of Toxoplasma gondii-infected mouse fibroblasts by the cationic permeant fluorescent dye rhodamine 123. Experientia 41: 101–102. 10.1007/BF02005897 [DOI] [PubMed] [Google Scholar]
- Vaidya AB, Arasu P. 1987. Tandemly arranged gene clusters of malarial parasites that are highly conserved and transcribed. Mol Biochem Parasitol 22: 249–257. 10.1016/0166-6851(87)90056-9 [DOI] [PubMed] [Google Scholar]
- Vaidya AB, Lashgari MS, Pologe LG, Morrisey J. 1993. Structural features of Plasmodium cytochrome b that may underlie susceptibility to 8-aminoquinolines and hydroxynaphthoquinones. Mol Biochem Parasitol 58: 33–42. 10.1016/0166-6851(93)90088-F [DOI] [PubMed] [Google Scholar]
- Waller RF, Jackson CJ. 2009. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. Bioessays 31: 237–245. 10.1002/bies.200800164 [DOI] [PubMed] [Google Scholar]
- Walzer KA, Adomako-Ankomah Y, Dam RA, Herrmann DC, Schares G, Dubey JP, Boyle JP. 2013. Hammondia hammondi, an avirulent relative of Toxoplasma gondii, has functional orthologs of known T. gondii virulence genes. Proc Natl Acad Sci 110: 7446–7451. 10.1073/pnas.1304322110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LM, Kim K. 2007. Toxoplasma gondii: the model apicomplexan. Perspectives and methods, 1st ed. (ed. Weiss LM, Kim K). Elsevier Academic Press, London. [Google Scholar]
- Weissig V, Rowe TC. 1999. Analysis of Plasmodium falciparum mitochondrial six-kilobase DNA by pulse-field electrophoresis. J Parasitol 85: 386–389. 10.2307/3285656 [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Williamson DH. 1997. Extrachromosomal DNA in the Apicomplexa. Microbiol Mol Biol Rev 61: 1–16. 10.1128/.61.1.1-16.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahalomi D, Haddas-Sasson M, Rubinstein ND, Feldstein T, Diamant A, Huchon D. 2017. The multipartite mitochondrial genome of Enteromyxum leei (Myxozoa): eight fast-evolving megacircles. Mol Biol Evol 34: 1551–1556. 10.1093/molbev/msx072 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.