The bacterial flagellum is a unique organelle that functions as a rotary motor. The interaction between the stator and rotor is indispensable for stator assembly into the motor and the generation of motor torque.
KEYWORDS: Escherichia coli, Vibrio, flagella, rotor, stator
ABSTRACT
The bacterial flagellum is the motility organelle powered by a rotary motor. The rotor and stator elements of the motor are located in the cytoplasmic membrane and cytoplasm. The stator units assemble around the rotor, and an ion flux (typically H+ or Na+) conducted through a channel of the stator induces conformational changes that generate rotor torque. Electrostatic interactions between the stator protein PomA in Vibrio (MotA in Escherichia coli) and the rotor protein FliG have been shown by genetic analyses but have not been demonstrated biochemically. Here, we used site-directed photo-cross-linking and disulfide cross-linking to provide direct evidence for the interaction. We introduced a UV-reactive amino acid, p-benzoyl-l-phenylalanine (pBPA), into the cytoplasmic region of PomA or the C-terminal region of FliG in intact cells. After UV irradiation, pBPA inserted at a number of positions in PomA and formed a cross-link with FliG. PomA residue K89 gave the highest yield of cross-links, suggesting that it is the PomA residue nearest to FliG. UV-induced cross-linking stopped motor rotation, and the isolated hook-basal body contained the cross-linked products. pBPA inserted to replace residue R281 or D288 in FliG formed cross-links with the Escherichia coli stator protein, MotA. A cysteine residue introduced in place of PomA K89 formed disulfide cross-links with cysteine inserted in place of FliG residues R281 and D288 and some other flanking positions. These results provide the first demonstration of direct physical interaction between specific residues in FliG and PomA/MotA.
IMPORTANCE The bacterial flagellum is a unique organelle that functions as a rotary motor. The interaction between the stator and rotor is indispensable for stator assembly into the motor and the generation of motor torque. However, the interface of the stator-rotor interaction has only been defined by mutational analysis. Here, we detected the stator-rotor interaction using site-directed photo-cross-linking and disulfide cross-linking approaches. We identified several residues in the PomA stator, especially K89, that are in close proximity to the rotor. Moreover, we identified several pairs of stator and rotor residues that interact. This study directly demonstrates the nature of the stator-rotor interaction and suggests how stator units assemble around the rotor and generate torque in the bacterial flagellar motor.
INTRODUCTION
F-type ATP synthase, V/A-type ATPase, and the bacterial flagellum are well-known examples of ion-driven molecular rotary motors (1, 2). The flagella of bacteria other than spirochetes each have a helical filament that extends from the cell surface and functions as a rotary screw to propel swimming. The rotary motor of the bacterial flagellum consists of a rotor surrounded by a varied numbers of stator units, with both the rotor and stator embedded in the cytoplasmic membrane (3–6). The rotor contains a transmembrane MS-ring and an attached cytoplasmic C-ring below the MS-ring (7, 8). In many bacteria, the C-ring contains the three proteins FliG, FliM, and FliN. Mutations in the genes encoding these proteins can confer fla, mot, and che phenotypes, corresponding to deficiencies in flagellar formation, motor rotation, or the switching between counterclockwise (CCW) and clockwise (CW) rotation (5, 9). FliG is thought to interact with the stator units to generate torque (10, 11).
MotA and MotB in Escherichia coli and PomA and PomB in Vibrio species are the membrane proteins that comprise the stator complex (12–16). The A subunit has four transmembrane segments (TMs) and a large cytoplasmic region between TM2 and TM3. The B subunit has one TM in its N-terminal region and a peptidoglycan-binding (PGB) domain in its C-terminal region (17–19). In E. coli, at least 11 stator units can assemble around, and interact with, the rotor. They are anchored at the proper position by the PGB domain, and once incorporated, the stator unit is activated for ion conduction and motor rotation (20–25). The coupling ion is a proton in the E. coli motor and a sodium ion in the Vibrio motor; it is conducted to the cytoplasm through an ion-transporting pathway in the stator complex (26, 27). A conserved aspartate residue in the TM of the B subunit receives the coupling ion from outside the cytoplasm, and the ion then dissociates into the cytoplasm (28–30). The ion binding and release cycle induces conformational changes in the stator complex that change the interactions between the A subunit of the stator and FliG of the rotor (31, 32).
Earlier biochemical studies demonstrated the stator-rotor interaction using a His tag pulldown assay (33). Stator interactions with the rotor protein have been examined in detail in E. coli using genetic analysis (34–36). MotA and FliG in E. coli have well-conserved charged residues, R90 and E98 in MotA and R281, D288, and D289 in the C-terminal domain of FliG (see Fig. S1A and B in the supplemental material). Charge neutralization or inversion of these residues leads to defects in motility, and the proper combinations of charge reversals between MotA and FliG synergistically rescue motility. The conserved charged residues in the A subunit are important for torque generation and assembly of the stator units into the motor (24, 25, 37). In contrast, charge neutralization or inversion of the corresponding residues in Vibrio alginolyticus did not abolish motility, suggesting that the charged residues are important, but not essential, for flagellar rotation in V. alginolyticus (Fig. S1A and B) (38, 39). This result implies that additional residues contribute to motor rotation. Since the stator-rotor interaction in the Vibrio flagellar motor is likely to be more extensive than that in E. coli (25, 40), Vibrio PomA is better suited for the examination of interactions between the stator A subunit and FliG of the rotor.
Structural information is indispensable for understanding the mechanism that produces rotation of the bacterial flagellar motor. Until recently, we had only low-resolution density maps of the stator unit obtained through single-particle analysis using electron microscopy (41, 42). However, atomic resolution structures of MotA/MotB from Campylobacter jejuni, Clostridium sporogenes, Bacillus subtilis, and other species have been reported recently (43, 44). The structures resemble the structure of ExbB/ExbD (45–47). MotA and PomA share a weak sequence homology with ExbB of the Ton bacterial transport system, which transports relatively large molecules, such as siderophores and vitamin B12, into the cell (43, 44). The MotA/MotB and PomA/PomB complexes exist as a 5:2 heteroheptamer, although the stoichiometry was previously proposed as a 4:2 heterohexamer (14, 16, 42). The atomic resolution structures of the stator provide insight into its organization and its contribution to flagellar rotation. Dynamic interactions between the stator and rotor generate torque that rotates the flagellum. However, the molecular details of the stator-rotor interaction remain obscure.
In this study, we probed residues of PomA for the ability to cross-link with FliG using a site-directed in vivo photo-cross-linking technique. This technique allows p-benzoyl-l-phenylalanine (pBPA), a phenylalanine derivative containing a UV-reactive benzophenone group, to be charged to an amber suppressor tRNA in vivo by a mutated tyrosyl-tRNA synthase from Methanococcus jannaschii. pBPA can be incorporated into any protein of interest by introducing an amber codon into the target position (48). The pBPA incorporated into the protein forms a covalent bond with a close C-H bond upon UV irradiation. Another approach utilizes disulfide bond formation between cysteine residues inserted at desired positions in two interacting proteins. Here, we report that photo-cross-linked and disulfide cross-linked products are formed between targeted residues of PomA and FliG. This work provides the first direct evidence to show which residues in the stator and rotor are in close juxtaposition and suggests interactions that are responsible for stator assembly and torque generation in the flagellar motor.
RESULTS
Effect of PomA with pBPA on E. coli cell motility.
Because we were unable to adapt the technique for introducing pBPA into V. alginolyticus, we performed a photo-cross-linking experiment in E. coli using a chimeric PomA/PotB stator unit that functions in E. coli. PotB is a chimeric protein in which the N-terminal region of V. alginolyticus PomB is fused to the C-terminal region of E. coli MotB (49). We introduced pBPA at each position in PomA from E74 to F104. This region contains the important charged residues, R88 and E96, which are proposed to interact with FliG (see Fig. S1A and C in the supplemental material) (24, 25, 34–36). First, we examined whether PomA with the pBPA insertions confers motility to an E. coli ΔmotAB null strain. The pBPA substitutions at L76, I77, I80, A84, G91, L95, N102, and F104 caused loss of motility (see Fig. S2). None of these proteins other than the one with a substitution at L95 was detected in the cells (see Fig. S3). PomA was detected in the pBPA substitution mutants that retained motility (Fig. S3). PomA with substitutions at A87, R88, and E96 supported much less motility than wild-type PomA (Fig. S2). These results suggest that residues A87, R88, L95, and E96 are at, or are very close to, the sites that are important for motor function.
Detection of photo-cross-linked products of PomA and FliG.
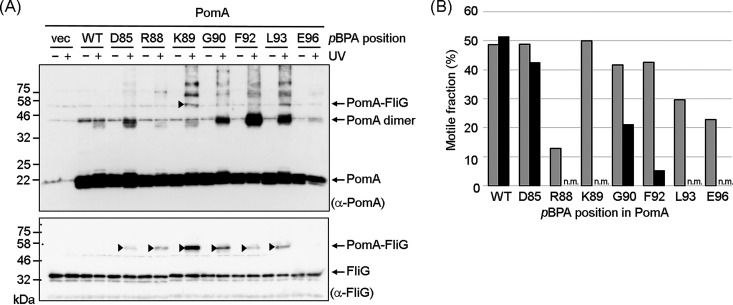
We UV-irradiated cells expressing PomA in which pBPA was introduced at each position from E74 to F104 and then probed by immunoblotting with anti-FliG antibody for cross-linked products. A higher-molecular-weight band was observed when pBPA replaced the residues D85, R88, K89, G90, F92, L93, and E96, whereas it was not observed in the vector control or with wild-type PomA (Fig. 1A, bottom; for the complete set of mutants, see Fig. S3). This result indicates that PomA with pBPA formed photo-cross-links with FliG. The only cross-linked product identified by probing with anti-PomA antibody was observed when pBPA replaced K89, presumably because of the low titer of the anti-PomA antibody (Fig. 1A, top). Consistent with this result, the signal intensity of cross-linked products detected by the anti-FliG antibody was strongest when pBPA replaced K89 (Fig. 1A).
FIG 1.
Photo-cross-linking between plasmid-encoded V. alginolyticus PomA and endogenous E. coli FliG. (A) V. alginolyticus PomA and chimeric PotB were expressed from plasmid pYS3, and the amber suppressor tRNA and the mutated tyrosyl-tRNA synthase were expressed from plasmid pEVOL-pBpF in the E. coli ΔmotAB strain, RP6894. After photo-cross-linking, whole-cell lysates were prepared and analyzed by immunoblotting. The top and bottom images show immunoblots with anti-PomA and anti-FliG antibodies, respectively. The cross-linked products are indicated by arrowheads. We show an image of the photo-cross-linked product of PomA E96-pBPA in Fig. S3C in the supplemental material, because we did not detect it in this immunoblot. Bands with a higher molecular weight were derived from nonspecific cross-linking between stator units or nonspecific cross-linking of PomA with other proteins. (B) Motile fractions of E. coli RP6894 cells expressing PomA/PotB before and after UV irradiation in free-swimming assays. The gray boxes show the motile fractions before UV irradiation. The black boxes show the motile fractions after UV irradiation. A movie of more than 10 s in length was captured for each mutant before and after UV irradiation. At least 30 freely suspended cells were analyzed for each mutant with dark-field microscopy. Immobile cells, such as those stuck to the glass, were excluded. n.m., nonmotile.
We determined the fraction of motile cells before and after UV irradiation, because we expected that the PomA-FliG cross-link would block motor function. With wild-type PomA, almost the same fractions of cells swam before as after UV irradiation. Irradiation of the cells containing pBPA replacing R88, K89, L93, and E96 did not swim after UV irradiation, and cells with pBPA replacing G90 and F92 mutants had a lower motile fraction (Fig. 1B). These results suggest that the residues R88, K89, G90, F92, L93, and E96 of PomA are close enough to the rotor C-ring to form cross-links with FliG.
pBPA-labeled PomA binds to the C-ring after photo-cross-linking.
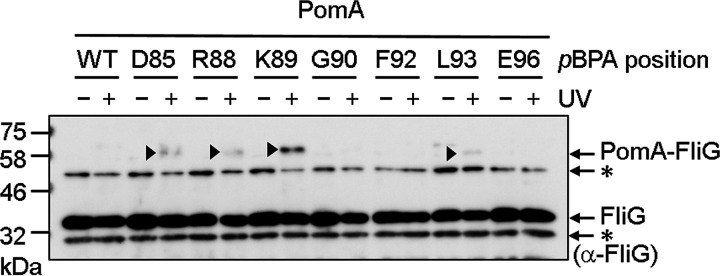
Most of the photo-cross-linked products described above could have arisen through cross-linking between PomA freely diffusing in the cytoplasmic membrane and FliG freely diffusing in the cytoplasm (please see Fig. S4, as described below). Therefore, we examined whether photo-cross-linked PomA was associated with the isolated hook-basal body (HBB) fraction. After UV irradiation of cells expressing PomA with pBPA replacing D85, R88, K89, G90, F92, L93, or E96, the cells were solubilized using Triton X-100, and then HBBs were isolated using ultracentrifugation. In immunoblots developed using the anti-FliG antibody, the HBB fractions contained a small fraction of photo-cross-linked products when pBPA replaced D85, R88, K89, and L93 (Fig. 2). The cross-linked products were most evident when pBPA replaced PomA K89. Unfortunately, we could not see PomA/PotB bound to the C-ring in electron micrographs. The immunoblot analysis of the isolated HBB fractions confirms that the PomA/PotB complex associates with FliG assembled into the rotor and supports the suggestion that that K89 is the closest in proximity to FliG.
FIG 2.
Photo-cross-linking between plasmid-borne V. alginolyticus PomA and endogenous E. coli FliG in the isolated hook-basal body. V. alginolyticus PomA and chimeric PotB were expressed from plasmid pYS3, and the amber suppressor tRNA and the mutated tyrosyl-tRNA synthase were expressed from plasmid pEVOL-pBpF in the E. coli ΔmotAB strain RP6894. The panel shows immunoblot images visualized with an anti-FliG antibody. The cross-linked products are indicated by arrowheads. Asterisks indicate nonspecific signals.
pBPA-labeled PomA binds to FliG diffusing freely in the cytoplasm.
We next examined whether PomA/PotB interacts with freely diffusing FliG. We expressed PomA with pBPA replacing D85, R88, K89, G90, F92, L93, or E96, PotB, and E. coli FliG from plasmid pTSK170 in E. coli ΔflhDC cells, which lack all flagellar proteins. After UV irradiation of these cells, we probed the photo-cross-linked products by immunoblotting with anti-PomA and anti-FliG antibodies. PomA with pBPA replacing K89 produced a large amount of cross-linked product, whereas the other proteins showed fewer cross-linked products (Fig. S4). We speculate that photo-cross-linked products could be detected in this experiment using the anti-PomA antibody because FliG was produced in great excess.
FliG residues that interact with MotA.
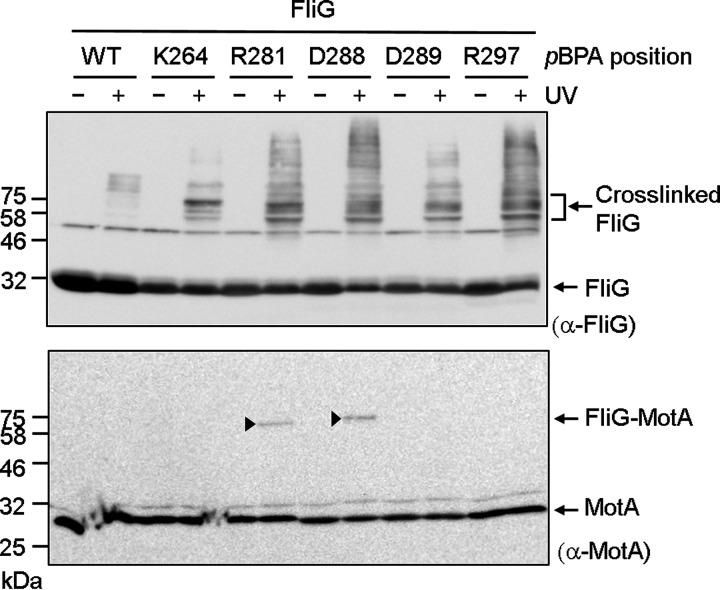
Residues R281 and D288 of E. coli FliG have been implicated in interacting with MotA. First, we investigated whether FliG with pBPA introduced at these two positions formed cross-linked products with endogenous E. coli MotA (Fig. 3; Fig. S1B). Indeed, when these proteins were expressed as the sole FliG, we detected cross-linked FliG-MotA using anti-MotA antibody, whereas the vector control and wild-type FliG did not form the cross-linked product. Furthermore, FliG with pBPA replacing K264, D289, and R297, which are additional conserved charged residues, did not support motility (K264amber was only barely motile) and did not form the cross-linked products. This result suggests that R281 and D288 are in close proximity to MotA.
FIG 3.
Photo-cross-linking between plasmid-borne E. coli FliG and endogenous E. coli MotA. E. coli FliG was expressed from plasmid pTY801, and the amber suppressor tRNA and the mutated tyrosyl-tRNA synthase were expressed from plasmid pEVOL-pBpF in the E. coli ΔfliG strain DFB225. The top and bottom images show immunoblots with anti-MotA and anti-E. coli FliG antibodies, respectively. The cross-linked products are indicated by arrowheads. Bands with a higher molecular weight in the immunoblot with anti-FliG antibody were derived from nonspecific interactions of FliG with other proteins.
Interaction pair between PomA and FliG residues.
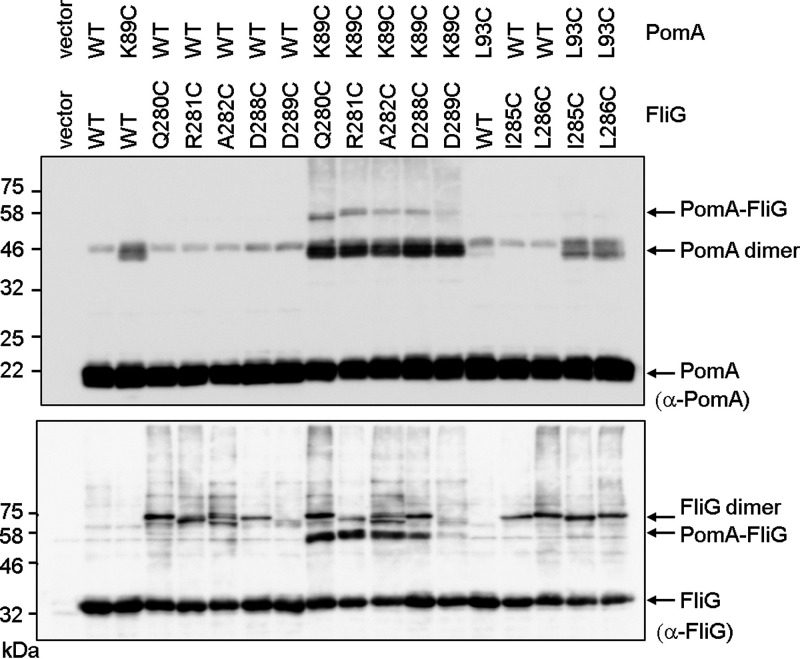
Next, we investigated whether PomA containing cysteine replacements for residues K89 and L93 could form disulfide cross-links with FliG containing cysteine replacements at critical residues. Previous genetic studies (25) suggested that PomA K89 interacts with residues R301, D308, and D309 of V. alginolyticus FliG. Therefore, we coexpressed PomA K89C/PotB together with the Q280C, R281C, A282C, D288C, or D289C variants of E. coli FliG in a ΔmotA ΔfliG E. coli mutant. These FliG residues correspond to K300, R301, A302, D308, and D309 of V. alginolyticus. We also expressed PomA L93C/PotB with I285C and L286C variants of E. coli FliG. I285 and L286 are hydrophobic residues located between R281 and D288 (Fig. S1B). Cells expressing only R281C FliG or L93C PomA lost motility in soft agar (see Fig. S5), suggesting that these residues are critical for motor function. Cells expressing K89C PomA with wild-type FliG and Q280C, A282C, D288C, or D289C FliG with wild-type PomA retained motility (Fig. S5). Cells coexpressing PomA K89C and FliG Q280C, A282C, or D288C had reduced motility (Fig. S5).
We next tried to detect disulfide cross-linked products between PomA and FliG. After oxidation with copper phenanthroline, we detected disulfide cross-linked products of PomA K89C with FliG Q280C, R281C, A282C, and D288C with anti-PomA or anti-FliG antibody (Fig. 4). The cross-linked products disappeared upon treatment with the reducing agent β-mercaptoethanol (see Fig. S6). In contrast, PomA L93C did not form disulfide cross-links with either FliG I285C or L286C (Fig. 4).
FIG 4.
Disulfide cross-linking between plasmid-borne V. alginolyticus PomA and E. coli FliG. V. alginolyticus PomA, chimeric PotB, and E. coli FliG were coexpressed from plasmid pTSK170 in the E. coli ΔmotA ΔfliG strain DFB245. Top and bottom images show immunoblots with anti-PomA and anti-FliG antibodies, respectively.
Effect of the conserved aspartate residue in the B subunit on cross-linking efficiency.
Flagellar rotation is powered by conformational changes of the stator units that are driven by ion conduction. Therefore, we thought that the interaction pattern between PomA and FliG might be different in the presence and absence of Na+. We expressed PomA with various pBPA substitutions and PotB in E. coli ΔmotAB cells and irradiated these cells with UV in the presence of Na+ or K+, followed by immunoblotting. Unfortunately, there were no reproducible differences in cross-linking (see Fig. S7). Next, we examined photo-cross-linking when the pBPA-substituted PomA proteins were coexpressed with PotB D24N, which has no ability to bind Na+ or support ion flow (30). The equivalent D32N substitution in E. coli (D33N in Salmonella) confers a dominant negative effect on motility (28, 31, 50, 51). It has been speculated that aspartate-to-asparagine substitution mimics the protonated or Na+-bound state of the aspartyl residue. The cross-linking patterns with D24N PotB were similar to those seen with wild-type PotB (Fig. 5; see also Fig. S8), but the signal intensities of the cross-linked products with PotB D24N were stronger than those seen with wild-type PotB, implying that the stator with the D24N mutant interacts with FliG more stably (Fig. 5).
FIG 5.
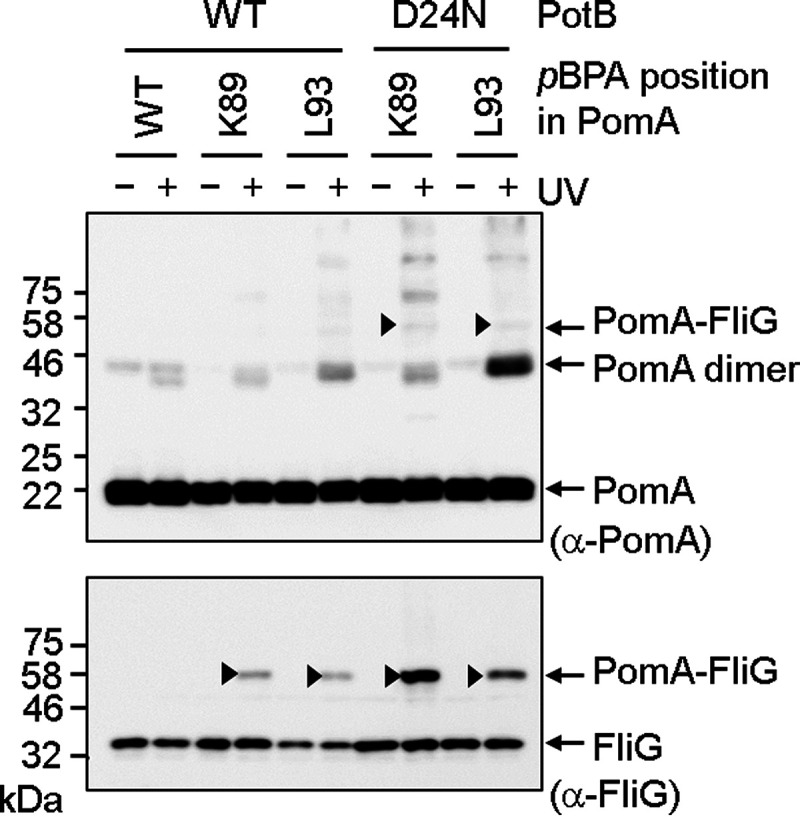
Photo-cross-linking between plasmid-borne V. alginolyticus PomA and endogenous E. coli FliG in the presence of PotB D24N. V. alginolyticus PomA and chimeric PotB were expressed from plasmid pYS3, and the amber suppressor tRNA and the mutated tyrosyl-tRNA synthase were expressed from plasmid pEVOL-pBpF in the E. coli ΔmotAB strain RP6894. Top and bottom images show immunoblots of whole-cell lysates with anti-PomA and anti-FliG antibodies, respectively. The cross-linked products are indicated by arrowheads.
DISCUSSION
Previous genetic studies in E. coli and Salmonella showed that electrostatic interactions between the conserved charged residues of the stator A subunit and FliG in the rotor are important both for assembly of stator units into the motor and for torque generation (24, 34–37). Similar studies in Vibrio alginolyticus suggested that interactions in addition to the electrostatic interactions contribute to motor rotation (25, 38–40, 52). In this study, we used two different chemical cross-linking approaches to identify the residues in the V. alginolyticus PomA subunit that are in close proximity to FliG and the residues in E. coli FliG that are in close proximity to PomA. Cross-linked products were found in the isolated hook-basal body, indicating that cross-linking occurred in the intact motor. We also observed motility defects caused by photo-cross-linking, indicating that the cross-linking occurred, at least in part, within the motor.
The three-dimensional structure of the A subunit at an atomic resolution has been revealed by two independent groups (43, 44). The PDB structural data for MotA/MotB in C. jejuni were kindly supplied by N. M. I. Taylor before being available to the general public (Fig. 6). The residues corresponding to L76, I77, I80, A84, L95, N102, and F104, at which substitution with pBPA led to a complete loss of motility, were at positions internal to the MotA structure, suggesting that they are important for proper folding and stability. The residues corresponding to D85, R88, K89, G90, F92, L93, and E96 in PomA, which showed photo-cross-linking with FliG when replaced with pBPA, were arrayed on helices H1 and H2 and the H1-H2 linker. These residues are located on the most external and membrane-distal portion of MotA/PomA bound to the B subunit (see Fig. S1C in the supplemental material). The cross-linking data indicate that this surface interacts closely with FliG and is important for stator assembly and motor rotation.
FIG 6.
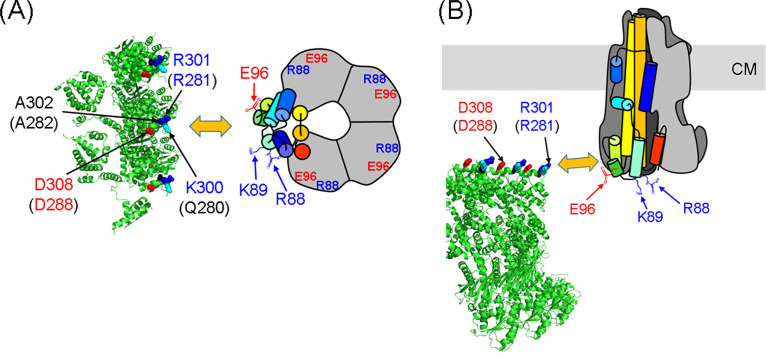
Model for the interaction between the C-ring and PomA from a top view (A) and a side view (B). The model incorporates a schematic of the PomA pentamer based on the cryo-electron microscopy (cryo-EM) structure (43). One PomA subunit is shown in rainbow colors, and the other four PomA subunits are outlined in gray. The C-ring of Vibrio alginolyticus is based on a model previously reported (56). The positions of R88, K89, and E96 are indicated by arrows. FliG residues K300, R301, and A302 in V. alginolyticus, corresponding to residues Q280, R281, and A282 in E. coli (the residue numbers for E. coli FliG are given in brackets), are shown as cyan, blue, and red spheres, respectively. CM, cytoplasmic membrane.
Cross-linking was also observed between FliG and PomA in nonflagellated cells overexpressing soluble FliG, showing that stators freely diffusing in the cytoplasmic membrane interact with cytoplasmic FliG that is not assembled into a C-ring. Once the C-ring forms, FliG molecules in the C-ring do not exchange with cytoplasmic FliG monomers (53). Therefore, it is unlikely that the stator unit binds to the cytoplasmic FliG before the stator-FliG complex assembles into the motor. This result implies that a region around K89 is the first site in PomA to have access to FliG assembled into the rotor. Since PomA with pBPA replacing R88 in the chimeric PomA/PotB stator conferred a severe motility defect, we speculate that the stator complex containing PomA with pBPA replacing R88 assembles poorly into the motor. Previous studies have also suggested that the region around PomA R88 and K89 seems to be important for stator assembly into the motor rather than for torque generation (24, 25).
We found that the conserved motif RXXGΦΦXLE, which spans the region from PomA R88 to PomA E96, is important for motility (Fig. S1A). The motif contains a completely conserved G91 residue followed by two hydrophobic residues (Φ). Leucine (or, less often, isoleucine) at residue 95 is also highly conserved, and when it was replaced with pBPA, motility was abolished, suggesting that an aliphatic residue at this position is important for motor rotation. The hydrophobic residues F92 and L93 as well as the charged residues seem to contribute to the stator-rotor interaction. PomA with pBPA replacing L93 still supported motility, whereas the PomA L93C mutant did not support motility, suggesting that hydrophobicity at residue 93 is important for motor function. We showed the strong disulfide cross-linking between V. alginolyticus PomA K89C and E. coli FliG R281C or D288C. This result is consistent with the idea that electrostatic repulsion and attraction between K89 in PomA and R281 and D288, respectively, contribute to torque generation. However, the residue at the position corresponding to K89 in PomA is Q in E. coli and Salmonella MotA, suggesting that these electrostatic interactions are not essential for motility in all cases. Overall, it seems that both electrostatic and hydrophobic interactions between the stator and rotor contribute to torque generation.
The coupling ion for motility binds to an absolutely conserved aspartate residue in the B subunit. Substitution of this residue with asparagine, the D24N replacement in V. alginolyticus PomA, may mimic the electrically neutral protonated state of MotA/MotB or the Na+-bound state of PomA/PomB. Therefore, the stator with the D24N mutation may not undergo conformational changes accompanied by Na+ binding or Na+ release from the aspartate residue. The chimeric PomA/PotB stator with the D24N variant of PotB formed a more-cross-linked product with FliG than the stator containing the wild-type form of PotB. This result may imply that the D24N stator interaction with FliG is more stable than that with the wild-type stator due to less movement in the region of the A subunit that interacts with FliG. However, this increased cross-linking may also reflect interactions of PomA/PotB D24N freely diffusing in the cytoplasmic membrane with cytoplasmic FliG.
Because we detected the interaction between E. coli FliG and V. alginolyticus PomA within E. coli cells using the chimeric stator system, we cannot rule out the unlikely possibility that the residues in PomA and FliG that we have identified are not in close proximity in V. alginolyticus cells. The multiple alignment of C-terminal regions of FliG from E. coli and V. alginolyticus showed that they are quite similar; therefore, we assumed V. alginolyticus FliG interacts with PomA in a similar manner (Fig. S1B). To further investigate this, we would need to do comprehensive disulfide cross-linking experiments with cysteine-substituted PomA and FliG in V. alginolyticus cells.
Recently, a rotational gear model for stator function has been proposed by several groups (43, 44, 54). A similar rotational model has been reported for ExbB/ExbD in the Ton molecular motor. ExbB/ExbD shares a weak homology with MotA/MotB and PomA/PomB (43–47, 55). In the rotational gear model, the A subunit pentamer rotates around the B subunit dimer, and this rotation transmits torque to the rotor via interaction with the FliG ring. Based on our results, we can suggest which residues at the PomA (MotA) interface with FliG are involved in the interaction. We predicted that PomA K89 interacts with both R281 and D288 in E. coli FliG. We speculate that PomA residues R88 and K89 first interact with FliG D288 (D308 in V. alginolyticus FliG) during assembly of a stator unit into the motor. This interaction activates ion conduction through the stator (24) and brings PomA E96 close to FliG R281 (R301 in V. alginolyticus). Next, the A subunit begins to rotate in response to ion influx. Finally, PomA residues R88 and K89 repel the positive charge of FliG R281, and PomA E96 repels the negative charge of FliG D288 (D308 in V. alginolyticus). As the stator rotation proceeds, the next PomA subunit interacts with the next FliG subunit on the C-ring (Fig. 6; see also Fig. S9). PomA residue L93 may also contribute to torque generation through hydrophobic interactions with FliG.
In summary, we provide the first biochemical evidence for the close proximity of specific residues in the PomA/MotA stator with specific residues of FliG in the C-ring rotor. The presence of both charged and hydrophobic residues at these positions suggests that both electrostatic and hydrophobic interactions contribute to stator assembly and the generation of rotation. These results provide insight into the fundamental molecular mechanisms of stator assembly around the rotor and torque generation within the flagellar motor.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids are listed in Table S1 in the supplemental material. E. coli was cultured in LB broth (1% [wt/vol] Bacto tryptone, 0.5% [wt/vol] yeast extract, 0.5% [wt/vol] NaCl), and TG broth (1% [wt/vol] Bacto tryptone, 0.5% [wt/vol] NaCl, 0.5% [wt/vol] glycerol). Chloramphenicol was added to a final concentration of 25 μg/ml for E. coli. Ampicillin was added to a final concentration of 100 μg/ml for E. coli.
Swimming assay in semisoft agar.
E. coli RP6894 cells harboring both pYS3 and pEVOL-pBpF, or E. coli DFB245 cells harboring pTSK170, were plated on LB agar with antibiotics. A single colony from the LB plate was inoculated onto TG agar plates (TG containing 0.3% [wt/vol] Bacto agar and 0.02% [wt/vol] arabinose) and incubated at 30°C for 24 h. For the dominant negative experiment with PotB D24N, E. coli RP437 cells expressing PomA/PotB D24N were precultured in LB broth with 0.2% (wt/vol) arabinose overnight at 30°C, and then 1 μl of the overnight cell culture was inoculated in TG semisoft agar (TG containing 0.3% [wt/vol] Bacto agar and 0.2% [wt/vol] arabinose) at 30°C for 7 h.
Photo-cross-linking experiment.
E. coli cells harboring two different plasmids, pEVOL-pBpF and a pBAD24-based plasmid, were cultured in TG broth containing 1 mM p-benzoyl-l-phenylalanine (pBPA) (Bachem AG, Switzerland) at 30°C for 2 h from an initial optical density at 600 nm (OD600) of 0.1. Arabinose was added to a final concentration of 0.02% (wt/vol) to express mutated tyrosyl-tRNA synthase, amber suppressor tRNA, and PomA/PotB and/or FliG and further cultivated for 4 h. The cells were collected by centrifugation (3,400 × g, 5 min), resuspended in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4), recollected by centrifugation, and resuspended in PBS. In the experiment shown in Fig. S7, we used sodium buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl) or potassium buffer (20 mM Tris-HCl [pH 8.0], 150 mM KCl) instead of PBS. UV irradiation was performed with a B-100AP UV lamp (Analytik Jena US, Upland, CA, USA) for 5 min. The cells were collected by centrifugation (3,400 × g, 5 min) and resuspended in sodium dodecyl sulfate (SDS) loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% [wt/vol] SDS, 10% [wt/vol] glycerol, 0.01% [wt/vol] bromophenol blue) containing 5% (vol/vol) β-mercaptoethanol. The samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes. The proteins were detected using rabbit anti-PomA antibody and rabbit anti-Salmonella FliG antibody (a gift from T. Minamino at Osaka University). For the experiment shown in Fig. 3, rabbit anti-E. coli FliG antibody (a gift from D. F. Blair at University of Utah) was used. The rabbit anti-Salmonella FliG antibody cross-reacts with E. coli FliG and can detect E. coli FliG at chromosomal levels.
Purification of the hook-basal body complex.
The hook-basal body complex was isolated as described previously (8), with several modifications. After UV irradiation for photo-cross-linking, the cells were suspended in 100 μl sucrose solution (0.5 M sucrose, 50 mM Tris-HCl [pH 8.0]). EDTA, lysozyme, and DNase were added to final concentrations of 10 mM, 1 mg/ml, and 1 mg/ml, respectively. The suspension was left on ice for 30 min, and then spheroplasts were lysed by adding Triton X-100 and MgSO4 to final concentrations of 1% (wt/vol) and 15 mM, respectively. The lysate was then incubated on ice for 1 h. After removal of the cell debris by centrifugation at 15,000 × g for 10 min, hook-basal bodies were precipitated by centrifugation at 60,000 × g for 60 min. The precipitate was resuspended in the SDS loading buffer.
Disulfide cross-linking experiment.
E. coli DFB245 strain harboring the pTSK170 plasmid was cultured in TG broth containing arabinose at a final concentration of 0.02% (wt/vol) at 30°C for 5 h from an initial OD600 of 0.05. The cells were collected by centrifugation (3,400 × g, 5 min), resuspended in PBS, collected by centrifugation, and resuspended in PBS. To form the disulfide cross-link, 1 mM copper phenanthroline was added to the cell suspension, which was then incubated for 5 min. To stop the cross-linking, 3 mM N-methylmaleimide was added to the cell suspension and further incubated for 5 min. The cells were then collected by centrifugation (3,400 × g, 5 min) and suspended in the SDS loading buffer without β-mercaptoethanol. The procedures for SDS-PAGE and immunoblotting were the same as those used in the photo-cross-linking experiment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter G. Schultz for the kind gift of pEVOL-pBpF for photo-cross-linking, Tohru Minamino for the kind gift of rabbit anti-Salmonella FliG antibody, David F. Blair for the kind gift of rabbit anti-E. coli FliG antibody, T. Yorimitsu for the construction of plasmid pTY801, and Nicholas M. I. Taylor for the kind gift of PDB data for MotA/MotB (6YKM, 6YKP, and 6YKR) before it was available to the public. We also thank Mike Manson for critical reading of the manuscript and K. Maki, Y. Kawase, and A. Abe for technical support.
This work was supported in part by JSPS KAKENHI grant numbers 18K07108 (to H.T.), 16H04774 and 18K19293 (to S.K.), and 20H03220 (to M.H.).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Stewart AG, Laming EM, Sobti M, Stock D. 2014. Rotary ATPases—dynamic molecular machines. Curr Opin Struct Biol 25:40–48. 10.1016/j.sbi.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Nakanishi A, Kishikawa JI, Mitsuoka K, Yokoyama K. 2019. Cryo-EM studies of the rotary H+-ATPase/synthase from Thermus thermophilus. Biophys Physicobiol 16:140–146. 10.2142/biophysico.16.0_140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 4.Terashima H, Kojima S, Homma M. 2008. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol 270:39–85. 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 5.Morimoto YV, Minamino T. 2014. Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 4:217–234. 10.3390/biom4010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takekawa N, Imada K, Homma M. 2020. Structure and energy-conversion mechanism of bacterial Na+-driven flagellar motor. Trends Microbiol 28:719–731. 10.1016/j.tim.2020.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Ueno T, Oosawa K, Aizawa SI. 1992. M-ring, S-ring and proximal rod of the flagellar basal body of Salmonella-typhimurium are composed of subunits of a single protein. J Mol Biol 227:672–677. 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 8.Francis NR, Sosinsky GE, Thomas D, Derosier DJ. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol 235:1261–1270. 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi S, Aizawa S, Kihara M, Isomura M, Jones CJ, Macnab RM. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol 168:1172–1179. 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd SA, Tang H, Wang X, Billings S, Blair DF. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol 178:223–231. 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas DR, Francis NR, Xu C, DeRosier DJ. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol 188:7039–7048. 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean GD, Macnab RM, Stader J, Matsumura P, Burks C. 1984. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol 159:991–999. 10.1128/JB.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stader J, Matsumura P, Vacante D, Dean GE, Macnab RM. 1986. Nucleotide sequence of the Escherichia coli MotB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol 166:244–252. 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojima S, Blair DF. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34. 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 15.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. 1997. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J Bacteriol 179:5104–5110. 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato K, Homma M. 2000. Multimeric structure of PomA, the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem 275:20223–20228. 10.1074/jbc.M002236200. [DOI] [PubMed] [Google Scholar]
- 17.De Mot R, Vanderleyden J. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol 12:333–334. 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 18.Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, Homma M, Namba K. 2009. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol Microbiol 73:710–718. 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhu S, Takao M, Li N, Sakuma M, Nishino Y, Homma M, Kojima S, Imada K. 2014. Conformational change in the periplasmic region of the flagellar stator coupled with the assembly around the rotor. Proc Natl Acad Sci U S A 111:13523–13528. 10.1073/pnas.1324201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leake MC, Chandler JH, Wadhams GH, Bai F, Berry RM, Armitage JP. 2006. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443:355–358. 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 21.Reid SW, Leake MC, Chandler JH, Lo C-J, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci U S A 103:8066–8071. 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima S, Takao M, Almira G, Kawahara I, Sakuma M, Homma M, Kojima C, Imada K. 2018. The helix rearrangement in the periplasmic domain of the flagellar stator B subunit activates peptidoglycan binding and ion influx. Structure 26:590–598. 10.1016/j.str.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Zhu S, Nishikino T, Takekawa N, Terashima H, Kojima S, Imada K, Homma M, Liu J. 2019. In situ structure of the Vibrio polar flagellum reveals a distinct outer membrane complex and its specific interaction with the stator. J Bacteriol 202:e00592-19. 10.1128/JB.00592-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto YV, Nakamura S, Hiraoka KD, Namba K, Minamino T. 2013. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J Bacteriol 195:474–481. 10.1128/JB.01971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takekawa N, Kojima S, Homma M. 2014. Contribution of many charged residues at the stator-rotor interface of the Na+-driven flagellar motor to torque generation in Vibrio alginolyticus. J Bacteriol 196:1377–1385. 10.1128/JB.01392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blair DF, Berg HC. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439–449. 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 27.Sato K, Homma M. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J Biol Chem 275:5718–5722. 10.1074/jbc.275.8.5718. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol 180:2729–2735. 10.1128/JB.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudo Y, Kitade Y, Furutani Y, Kojima M, Kojima S, Homma M, Kandori H. 2009. Interaction between Na+ ion and carboxylates of the PomA-PomB stator unit studied by ATR-FTIR spectroscopy. Biochemistry 48:11699–11705. 10.1021/bi901517n. [DOI] [PubMed] [Google Scholar]
- 30.Onoue Y, Iwaki M, Shinobu A, Nishihara Y, Iwatsuki H, Terashima H, Kitao A, Kandori H, Homma M. 2019. Essential ion binding residues for Na+ flow in stator complex of the Vibrio flagellar motor. Sci Rep 9:11216. 10.1038/s41598-019-46038-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima S, Blair DF. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041–13050. 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 32.Mino T, Nishikino T, Iwatsuki H, Kojima S, Homma M. 2019. Effect of sodium ions on conformations of the cytoplasmic loop of the PomA stator protein of Vibrio alginolyticus. J Biochem 166:331–341. 10.1093/jb/mvz040. [DOI] [PubMed] [Google Scholar]
- 33.Tang H, Braun TF, Blair DF. 1996. Motility protein complexes in the bacterial flagellar motor. J Mol Biol 261:209–221. 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 34.Zhou JD, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci U S A 95:6436–6441. 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd SA, Blair DF. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol 266:733–744. 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 36.Zhou JD, Blair DF. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol 273:428–439. 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 37.Morimoto YV, Nakamura S, Kami-Ike N, Namba K, Minamino T. 2010. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol 78:1117–1129. 10.1111/j.1365-2958.2010.07391.x. [DOI] [PubMed] [Google Scholar]
- 38.Yorimitsu T, Sowa Y, Ishijima A, Yakushi T, Homma M. 2002. The systematic substitutions around the conserved charged residues of the cytoplasmic loop of Na+-driven flagellar motor component PomA. J Mol Biol 320:403–413. 10.1016/S0022-2836(02)00426-6. [DOI] [PubMed] [Google Scholar]
- 39.Yorimitsu T, Mimaki A, Yakushi T, Homma M. 2003. The conserved charged residues of the C-terminal region of FliG, a rotor component of Na+-driven flagellar motor. J Mol Biol 334:567–583. 10.1016/j.jmb.2003.09.052. [DOI] [PubMed] [Google Scholar]
- 40.Yakushi T, Yang J, Fukuoka H, Homma M, Blair DF. 2006. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J Bacteriol 188:1466–1472. 10.1128/JB.188.4.1466-1472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yonekura K, Maki-Yonekura S, Homma M. 2011. The structure of the flagellar motor protein complex PomAB: implications for the torque-generating conformation. J Bacteriol 193:3863–3870. 10.1128/JB.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takekawa N, Terahara N, Kato T, Gohara M, Mayanagi K, Hijikata A, Onoue Y, Kojima S, Shirai T, Namba K, Homma M. 2016. The tetrameric MotA complex as the core of the flagellar motor stator from hyperthermophilic bacterium. Sci Rep 6:31526. 10.1038/srep31526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santiveri M, Roa-Eguiara A, Kühne C, Wadhwa N, Hu H, Berg HC, Erhardt M, Taylor NMI. 2020. Structure and function of stator units of the bacterial flagellar motor. Cell 183:244–257. 10.1016/j.cell.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 44.Deme JC, Johnson S, Vickery O, Aron A, Monkhouse H, Griffiths T, Hennell James R, Berks BC, Coulton JW, Stansfeld PJ, Lea SM. 2020. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat Microbiol 5:1553–1564. 10.1038/s41564-020-0788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Celia H, Noinaj N, Zakharov SD, Bordignon E, Botos I, Santamaria M, Barnard TJ, Cramer WA, Lloubes R, Buchanan SK. 2016. Structural insight into the role of the Ton complex in energy transduction. Nature 538:60–65. 10.1038/nature19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maki-Yonekura S, Matsuoka R, Yamashita Y, Shimizu H, Tanaka M, Iwabuki F, Yonekura K. 2018. Hexameric and pentameric complexes of the ExbBD energizer in the Ton system. Elife 7:e35419. 10.7554/eLife.35419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Celia H, Botos I, Ni X, Fox T, De Val N, Lloubes R, Jiang J, Buchanan SK. 2019. Cryo-EM structure of the bacterial Ton motor subcomplex ExbB-ExbD provides information on structure and stoichiometry. Commun Biol 2:358. 10.1038/s42003-019-0604-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chin JW, Martin AB, King DS, Wang L, Schultz PG. 2002. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A 99:11020–11024. 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asai Y, Yakushi T, Kawagishi I, Homma M. 2003. Ion-coupling determinants of Na+-driven and H+-driven flagellar motors. J Mol Biol 327:453–463. 10.1016/S0022-2836(03)00096-2. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura S, Kami-Ike N, Yokota JP, Minamino T, Namba K. 2010. Evidence for symmetry in the elementary process of bidirectional torque generation by the bacterial flagellar motor. Proc Natl Acad Sci U S A 107:17616–17620. 10.1073/pnas.1007448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakamura S, Minamino T, Kami-Ike N, Kudo S, Namba K. 2014. Effect of the MotB(D33N) mutation on stator assembly and rotation of the proton-driven bacterial flagellar motor. Biophysics (Nagoya-Shi) 10:35–41. 10.2142/biophysics.10.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukuoka H, Yakushi T, Homma M. 2004. Concerted effects of amino acid substitutions in conserved charged residues and other residues in the cytoplasmic domain of PomA, a stator component of Na+-driven flagella. J Bacteriol 186:6749–6758. 10.1128/JB.186.20.6749-6758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukuoka H, Yakushi T, Kusumoto A, Homma M. 2005. Assembly of motor proteins, PomA and PomB, in the Na+-driven stator of the flagellar motor. J Mol Biol 351:707–717. 10.1016/j.jmb.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 54.Chang Y, Zhang K, Carroll BL, Zhao X, Charon NW, Norris SJ, Motaleb MA, Li C, Liu J. 2020. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat Struct Mol Biol 27:1041–1047. 10.1038/s41594-020-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celia H, Noinaj N, Buchanan SK. 2020. Structure and stoichiometry of the ton molecular motor. Int J Mol Sci 21:375. 10.3390/ijms21020375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishikino T, Hijikata A, Miyanoiri Y, Onoue Y, Kojima S, Shirai T, Homma M. 2018. Rotational direction of flagellar motor from the conformation of FliG middle domain in marine Vibrio. Sci Rep 8:17793. 10.1038/s41598-018-35902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.