Bacterial cell division has been a successful target for antimicrobial agents for decades. How different pathogens regulate cell division is, however, poorly understood.
KEYWORDS: Streptococcus pneumoniae, EloR, MltG, cell division, regulation, cell wall synthesis
ABSTRACT
The ellipsoid shape of Streptococcus pneumoniae is determined by the synchronized actions of the elongasome and the divisome, which have the task of creating a protective layer of peptidoglycan (PG) enveloping the cell membrane. The elongasome is necessary for expanding PG in the longitudinal direction, whereas the divisome synthesizes the PG that divides one cell into two. Although there is still little knowledge about how these two modes of PG synthesis are coordinated, it was recently discovered that two RNA-binding proteins called EloR and KhpA are part of a novel regulatory pathway controlling elongation in S. pneumoniae. EloR and KhpA form a complex that works closely with the Ser/Thr kinase StkP to regulate cell elongation. Here, we have further explored how this regulation occurs. EloR/KhpA is found at midcell, a localization fully dependent on EloR. Using a bacterial two-hybrid assay, we probed EloR against several elongasome proteins and found an interaction with the lytic transglycosylase homolog MltG. By using EloR as bait in immunoprecipitation assays, MltG was pulled down, confirming that they are part of the same protein complex. Fluorescence microscopy demonstrated that the Jag domain of EloR is essential for EloR’s midcell localization and its interaction with MltG. Since MltG is found at midcell independent of EloR, our results suggest that MltG is responsible for the recruitment of the EloR/KhpA complex to the division zone to regulate cell elongation.
IMPORTANCE Bacterial cell division has been a successful target for antimicrobial agents for decades. How different pathogens regulate cell division is, however, poorly understood. To fully exploit the potential for future antibiotics targeting cell division, we need to understand the details of how the bacteria regulate and construct the cell wall during this process. Here, we have revealed that the newly identified EloR/KhpA complex, regulating cell elongation in S. pneumoniae, forms a complex with the essential peptidoglycan transglycosylase MltG at midcell. EloR, KhpA, and MltG are conserved among many bacterial species, and the EloR/KhpA/MltG regulatory pathway is most likely a common mechanism employed by many Gram-positive bacteria to coordinate cell elongation and septation.
INTRODUCTION
In order to multiply, a bacterial cell splits into two daughter cells in an intricate process involving chromosome replication and segregation, the production of a new cell membrane, and the synthesis of a new cell wall. Streptococcus pneumoniae is a Gram-positive species, meaning that it produces a thick cell wall that surrounds and protects the cell. The major component of the cell wall is peptidoglycan (PG), which is made up of chains of polysaccharides that are cross-linked with short peptide bridges. The polysaccharides consist of alternating molecules of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc). The cross-links are made between pentapeptides attached to MurNAc (1).
S. pneumoniae has an ellipsoid shape resulting from the synthesis of the PG layer by two protein complexes: the elongasome and the divisome (2, 3). As the names suggest, the elongasome is responsible for producing PG in the peripheral direction, creating the elongated shape of pneumococci. The divisome, on the other hand, is responsible for synthesizing the septal disc that divides one cell into two. The precursor for PG is made inside the pneumococcal cell, transported to the outside, and incorporated into the growing PG through transglycosylation (TG) and transpeptidation (TP) reactions (1, 4). One group of enzymes performing this incorporation are the penicillin-binding proteins (PBPs). S. pneumoniae has six PBPs: three class A PBPs (PBP1a, PBP1b, and PBP2a) that harbor both TG and TP activities, two class B PBPs (PBP2b and PBP2x) that harbor only TP activity, and PBP3, a d,d-carboxypeptidase whose activity affects the number of cross-links in PG by removing the terminal d-Ala residues of pentapeptides (5–7). It is widely acknowledged that PBP2b is an essential part of the elongasome and that PBP2x is an essential part of the divisome (8–10). The shape elongation division and sporulation (SEDS) proteins RodA and FtsW have emerged as the main TG enzymes during PG production, working alongside the TP enzymes PBP2b and PBP2x, respectively. These essential protein pairs (PBP2b/RodA and PBP2x/FtsW) are the core PG-polymerizing units in S. pneumoniae (3, 11). The discovery that SEDS proteins are the primary TG enzymes in PG synthesis has prompted a reassessment of the role that class A PBPs have in PG synthesis. Rather than being essential in building the primary PG, recent data strongly indicate that the class A PBPs are essential for the maturation of newly synthesized PG, e.g., filling in gaps or mistakes left by the divisome and possibly the elongasome (12, 13). Other proteins considered to be part of the elongasome and divisome are found to be important for scaffolding, localization, and regulation of PG production. One newly identified member of the elongasome is the membrane-bound lytic transglycosylase MltG (14, 15). MltG in S. pneumoniae consists of a cytosolic domain, a transmembrane α-helix, and an extracellular catalytic domain. Several lines of evidence support that MltG is part of the elongasome: cells depleted of MltG have reduced lengths, MltG fused to superfolder green fluorescent protein (sfGFP-MltG) colocalizes with elongasome proteins throughout the cell cycle, and suppressor mutations have been found in mltG upon deletion of the essential elongasome transpeptidase pbp2b, demonstrating a functional link between these genes. The specific function of MltG in cell division is still unknown, but it has been proposed to release PG strands synthesized by PBP1a for cross-linking by RodA/PBP2b (15).
A particularly interesting aspect of cell wall synthesis is how PG production is regulated. By tracking the incorporation of new PG material using superresolution fluorescence microscopy, both the elongasome and divisome PG synthesis machineries have been shown to be organized in regularly spaced nodes in pneumococci (16), and the cells seem to elongate for a short time before septal PG synthesis is initiated (9, 17). Although several proteins are known to be involved in the regulation of PG synthesis (18–24), there is little knowledge about how the temporal and spatial control of elongation and division is achieved. The eukaryotic-type Ser/Thr kinase StkP appears to play a key role in coordinating these two events (25–27). StkP phosphorylates and thereby modulates the activity of several cell division proteins, i.e., DivIVA, GpsB, MapZ, MurC, MacP, and EloR (also known as Jag/KhpB) (18, 21, 24, 28–31). DivIVA and its paralog GpsB together with StkP are important for tuning septal and peripheral PG synthesis (18, 32), and phosphorylated MacP regulates the function of the class A PBP PBP2a (24). Furthermore, phosphorylation of MapZ (scaffolding protein for FtsZ) has been shown to be important for FtsZ ring constriction and splitting, while the effect of MurC (UDP‐N‐acetylmuramoyl l‐alanine ligase catalyzing the addition of alanine to UDP-MurNAc at an early step of the PG synthesis pathway) phosphorylation is still unclear. StkP is also important for the localization of PBP2x through interaction between StkP’s PASTA domains and the pedestal and/or the transpeptidase domain of PBP2x (33).
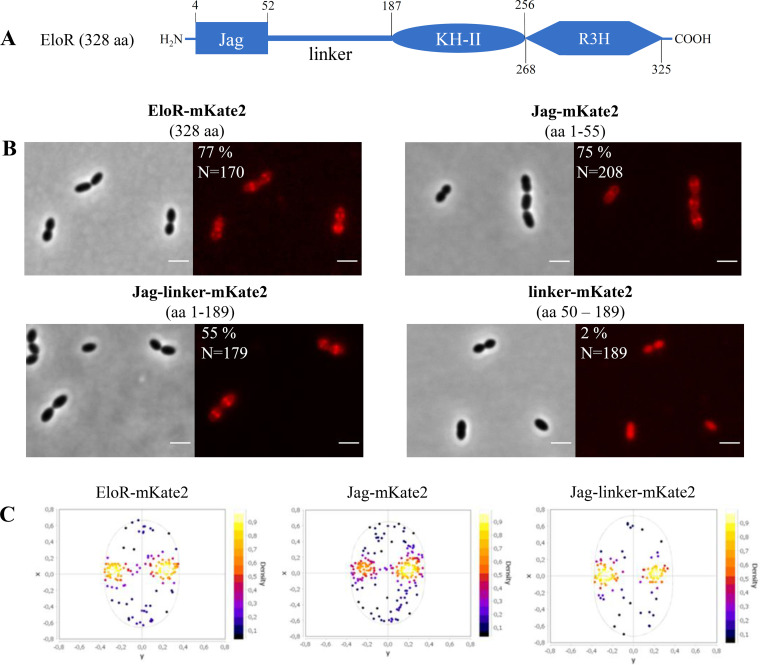
StkP-dependent phosphorylation of EloR has also been shown to be essential in the regulation of cell elongation in S. pneumoniae (19, 21). EloR (short for elongasome-regulating protein) is conserved in a range of Gram-positive genera, including Streptococcus, Bacillus, Clostridium, Listeria, Enterococcus, Lactobacillus, and Lactococcus. The protein is composed of three domains, (i) an N-terminal Jag domain, (ii) a KH-II domain, and (iii) an R3H domain at its C-terminal end (Fig. 1A), but no transmembrane segment. The KH-II and R3H domains are both single-stranded nucleic acid-binding domains that usually bind RNAs, while the Jag domain has an unknown function. EloR interacts with another RNA-binding protein, called KhpA (composed of one KH-II domain). If the EloR/KhpA complex is broken, cells become shorter, consistent with a loss of elongasome function, and are no longer dependent upon the essential PBP2b/RodA pair (19, 21). Point mutations inactivating the RNA-binding domains of EloR suggest that the phosphorylation of EloR by StkP leads to the release of bound RNA. This stimulates cell elongation in an unknown fashion (19). Interestingly, knockdown of eloR and khpA expression in the rod-shaped bacterium Lactobacillus plantarum also resulted in the shortening of cells, suggesting a conserved role for these proteins in regulating cell elongation (34). EloR and KhpA localize to the division zone of S. pneumoniae. While the midcell localization of KhpA depends on its interaction with the KH-II domain of EloR, it is not known what directs EloR to midcell.
FIG 1.
The Jag domain directs EloR to midcell. (A) Schematic representation of EloR, including predicted domains and domain borders. (B) Micrographs showing the subcellular localization of EloR-mKate2 (AW407), Jag-mKate2 (AW408), Jag-linker-mKate2 (AW409), and linker-mKate2 (AW410). Phase-contrast and corresponding fluorescence images are shown. The numbers above the micrographs indicate the amino acids (aa) of EloR utilized in the different constructs. The percentages of cells that displayed midcell localization of the mKate2 fusions are indicated, as are the numbers of cells included in the analyses. Bars, 2 μm. (C) Analysis of subcellular localization. For the strains where the majority of fusion proteins (EloR-mKate, Jag-mKate, and Jag-linker-mKate) displayed midcell localization, fluorescence maxima were detected and plotted in focus density plots using MicrobeJ (see Materials and Methods). x and y in the focus density plots denote the relative length and width axes, respectively.
In this study, we employed fluorescence microscopy and protein-protein interaction assays to further explore the EloR-mediated regulation of cell elongation in S. pneumoniae. We show that the Jag domain of EloR is critical for the midcell localization of this protein. Furthermore, EloR was shown to interact with the elongasome protein MltG via its Jag domain, suggesting a role of MltG in positioning EloR at midcell.
RESULTS AND DISCUSSION
The Jag domain is solely responsible for recruiting EloR to midcell.
EloR consists of an N-terminal Jag domain and two C-terminal RNA-binding domains, KH-II and R3H (Fig. 1A). We and others have previously shown that EloR localizes to the division zone, where it forms a complex with KhpA (20, 21). While KhpA depends on its interaction with EloR in order to localize to the division zone, it is not known how EloR finds midcell. We hypothesized that EloR must form an interaction(s) with other elongasome proteins to localize correctly. The Jag domain is connected to the KH-II domain by a large linker region (134 amino acids long) with an unknown structure and function. Since the KH-II and R3H domains bind RNA, our rationale was that the Jag-linker part of EloR would be important for its subcellular localization. We tested this by fusing full-length EloR, the Jag domain, the linker region, and the Jag-linker domains to the far-red fluorescent protein mKate2, creating the strains AW407, AW408, AW410, and AW409, respectively. These fusions were expressed ectopically from an inducible promoter using the ComRS system (35). The native eloR gene was kept unchanged in the genome. When the inducer (ComS) was supplied to the growth medium, we saw, as expected, that full-length EloR fused with mKate2 (EloR-mKate2) was concentrated at midcell for 77% of the cells investigated (Fig. 1B and C). It should be noted that we observed a background signal from the cytoplasm in most cells, suggesting that not all EloR proteins are midcell localized. We also found that the Jag-mKate2 and the Jag-linker-mKate2 fusions concentrated at midcell for 75% and 55% of the cells, respectively (Fig. 1B and C). The linker-mKate2 fusion, on the other hand, did not localize to midcell (Fig. 1B), as only 2% of the linker-mKate2 cells investigated displayed a midcell fluorescence signal. These results clearly suggest that the Jag domain is solely responsible for localizing EloR to midcell. The three-dimensional (3D) structure of the Jag domain has been solved for EloR in Clostridium symbiosum (PDB accession number 3GKU). It has a β-α-β-β fold with the α-helix laying on top of a three-stranded β-sheet. The conserved motif KKGFLG is found in the loop connecting the β2- and β3-strands (see Fig. S1A in the supplemental material). The same is true for the predicted structure of EloR from S. pneumoniae (Fig. S1B). We hypothesized that the conserved region (KKGFLG) could be involved in a protein-protein interaction possibly important for EloR localization. However, substitutions of several residues (K36A, K37A, F39A, and L40M) in this motif did not abrogate the midcell localization of EloR (Fig. S2).
StkP-mediated phosphorylation is not critical for EloR localization.
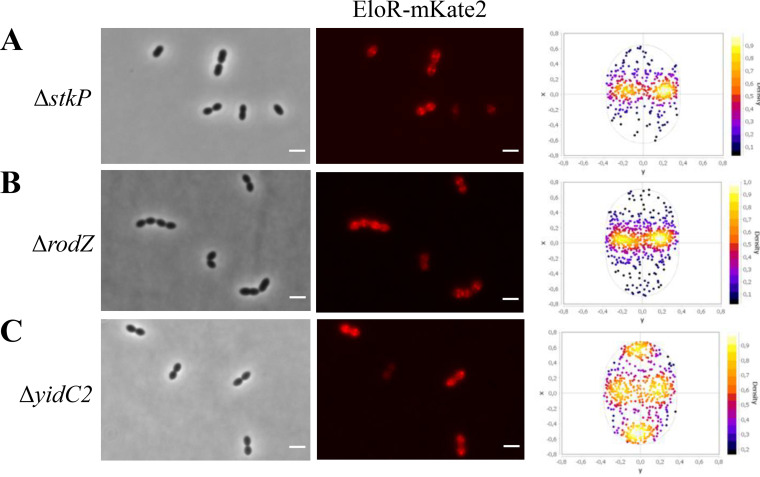
The results described above clearly suggest that the Jag domain targets EloR to the division zone independently of the linker domain. Nevertheless, the fact that the conserved threonine (threonine 89 in S. pneumoniae) phosphorylated by StkP to modulate EloR activity is located in the linker domain suggests that the linker could be involved in conformational rearrangements of the EloR protein between the active and inactive forms. The StkP kinase is located at midcell in S. pneumoniae, and to explore whether this protein or the phosphorylation state affected the localization of EloR (36, 37), we analyzed the localization of EloR-mKate2 in a genetic background lacking stkP (ΔstkP::janus). However, this demonstrated that StkP is not the reason why EloR-mKate2 is concentrated at midcell since the protein retained its localization in cells lacking stkP (Fig. 2A).
FIG 2.
EloR-mKate2 localization in different genetic backgrounds. The subcellular localization of EloR-mKate2 in ΔstkP (n = 1,155) (A), ΔrodZ (n = 1,681) (B), and ΔyidC2 (n = 1,280) (C) mutants is shown by microscopy images and corresponding focus density plots of detected foci. n indicates the number of cells analyzed for each strain. x and y in the focus density plots denote the relative length and width axes, respectively. Bars, 2 μm.
Based on our localization results (Fig. 1), the linker region is not crucial for recruiting EloR to midcell. Interestingly, when aligning the amino acid sequences of EloR homologs from different Gram-positive species, the length of the linker region varies from approximately 135 amino acid residues in S. pneumoniae to approximately 10 residues in Bacillus subtilis (Fig. S3). The reason for these variations is not clear, but if the linker domain is involved in protein-protein interactions, the larger linker region in pneumococcal EloR could accommodate more interaction partners and, hence, more regulatory possibilities. Why pneumococci would need this is not clear. The structural fold of the EloR linker in S. pneumoniae is unknown, making it particularly interesting for future studies to explore its 3D structure and the rearrangements occurring between phosphorylated EloR and the nonphosphorylated form.
EloR interacts with several proteins known to be part of the elongasome.
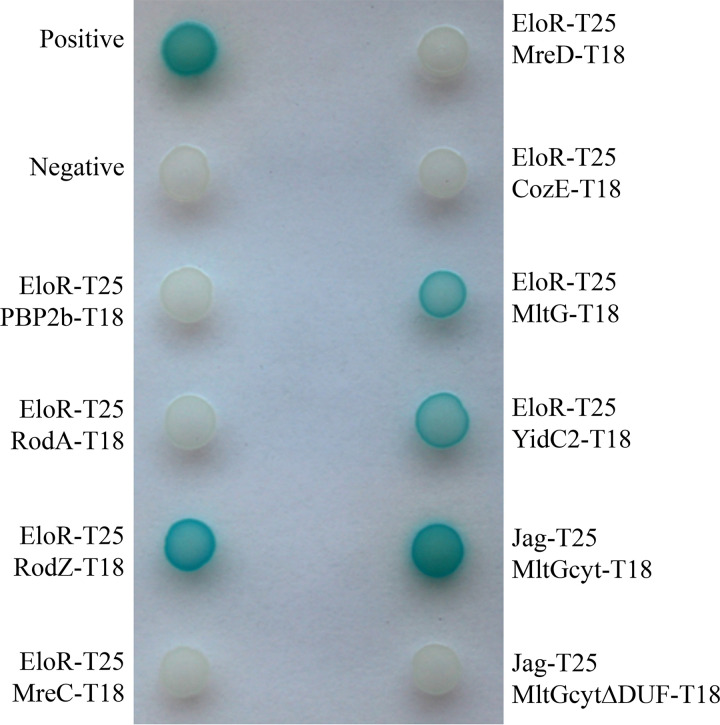
EloR has been shown to interact with the midcell-localized proteins StkP and KhpA, but these interactions did not affect the localization of EloR. In order to investigate how EloR localizes at midcell and to understand its regulatory function in cell elongation, we wanted to explore what other protein interactions EloR forms in addition to the one with KhpA and StkP. We screened our bacterial two-hybrid (BACTH) library in Escherichia coli for possible interaction partners for EloR. The BACTH assay is based on blue (positive) and white (negative) color selection, where the blue color comes from the cleavage of X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) in the medium by β-galactosidase. Briefly, the two proteins that are tested for interaction are fused to either the T18 or T25 domain. If an interaction between the two proteins occurs, T18 and T25 reconstitute an adenylate cyclase producing cAMP, which induces the expression of β-galactosidase (38). EloR was probed against a range of known cell division proteins, namely, PBP2b, RodA, RodZ, MreC, MreD, CozE, and MltG (Fig. 3). We also tested YidC2, whose gene shares an operon with eloR. YidC2 is an insertase that assists in the insertion of membrane proteins into the lipid bilayer, working together with the SecYEG translocon, the signal recognition particle (SRP), and the SRP receptor FtsY (39, 40). The presence of yidC2 and eloR in one operon seems to be conserved in several species, e.g., S. pneumoniae, Streptococcus mitis, Streptococcus oralis, B. subtilis, and Listeria monocytogenes, indicating a functional link between the two.
FIG 3.
Bacterial two-hybrid assay probing EloR against other elongasome proteins. PBP2b, RodA, MreC, MreD, and CozE (CozEa) probed against EloR gave colorless spots of bacteria, complying with no interaction between the two proteins. RodZ, MltG, and YidC2, on the other hand, gave blue spots when probed against EloR, suggesting that an interaction occurs. Positive and negative controls were supplied by the manufacturer (Euromedex). The Jag domain of EloR was tested against the cytosolic domain of MltG with and without the DUF. The interactions between the two domains were lost in the absence of the DUF in the cytosolic part of MltG. This indicates that the Jag domain of EloR interacts with the DUF of MltG.
Of all the proteins tested using the BACTH assay, the positive hits were RodZ, YidC2, and MltG. MltG is a membrane protein predicted to be a lytic transglycosylase and is essential in S. pneumoniae (14). RodZ, similar to EloR, is considered to be part of the elongasome, and studies in E. coli indicate that RodZ is important for the elongated cell shape (41). To test whether the interactions with these proteins were important for EloR localization, we analyzed EloR-mKate2 localization in cells devoid of these genes. Deletions of either rodZ or yidC2, however, did not abrogate the midcell localization of EloR-mKate2 (Fig. 2B and C). Interestingly, however, in the genetic background lacking yidC2, we now detected an accumulation of EloR-mKate2 at the poles of the cells as well as at midcell. Further investigations of the polar localization of EloR-mKate2 revealed that the polar foci of EloR-mKate2 were found in old cellular poles (Fig. S4).
EloR and MltG are part of the same complex.
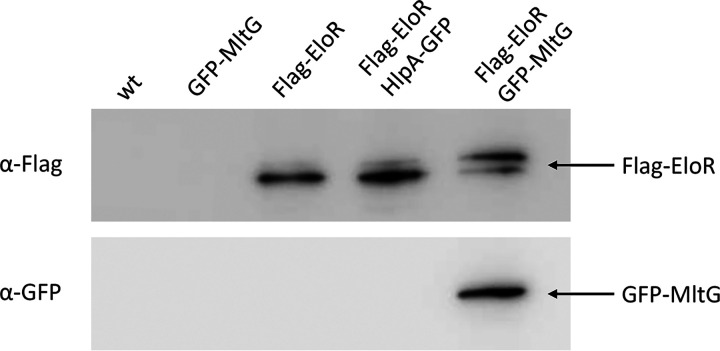
Since EloR was still localized at midcell when rodZ or yidC2 was deleted, we hypothesized that MltG, which also has a midcell localization (15), could be important for this matter. However, MltG is essential in wild-type cells, making it impossible to track EloR-mKate2 in a ΔmltG mutant. Furthermore, we did not succeed in making an mltG depletion strain having EloR-mKate2 in the native eloR locus. Instead, to confirm the interaction between EloR and MltG in vivo in S. pneumoniae, we attempted to use EloR as bait to pull down MltG. In order to do so, we constructed a mutant expressing Flag-tagged EloR and sfGFP-tagged MltG (strain AW447). By using resin beads tethered with anti-Flag antibodies, we pulled out Flag-EloR from the cell lysate as previously described by Stamsås et al. (19). Next, we looked for both Flag-EloR and sfGFP-MltG among the immunoprecipitated proteins using immunodetection and anti-Flag and anti-GFP antibodies (Fig. 4). Indeed, when pulling out Flag-EloR using the anti-Flag resin, we found that sfGFP-MltG followed in the same fraction. Strain ds515 expressing only sfGFP-MltG was used as a negative control for a possible GFP/anti-Flag interaction. In addition, to exclude a possible GFP/Flag-EloR unspecific interaction, we coexpressed Flag-tagged EloR and GFP-tagged HlpA (DNA-binding protein [42]) in strain AW459. When performing anti-Flag immunoprecipitation on lysates from this strain, no HlpA-GFP was pulled down together with Flag-EloR.
FIG 4.
Immunoblot analysis confirming the EloR-MltG interaction. Lysates from strains RH425 (wild type [wt]), ds515 (sfgfp-mltG), AW98 (flag-eloR), AW459 (flag-eloR hlpA-gfp), and AW447 (flag-eloR sfgfp-mltG) were incubated with resin beads tethered with anti-Flag antibodies to pull down Flag-EloR. As expected, immunoprecipitated Flag-EloR was found in strains AW98, AW459, and AW447 but not in strain ds515. sfGFP-MltG was found in immunoprecipitated fractions only when it was coexpressed with Flag-tagged EloR. The two EloR bands visible in the blot represent the phosphorylated and unphosphorylated forms of the protein (19). All fusion proteins used in the co-IP (HlpA-GFP, Flag-EloR, and GFP-MltG) have previously been shown to be stable when expressed in S. pneumoniae (19, 42). Uncropped versions of the immunoblots can be found in Fig. S6 in the supplemental material.
The Jag domain of EloR interacts with the intracellular DUF1346 domain of MltG.
The immunoprecipitation result proved that EloR is in complex with MltG in vivo in S. pneumoniae. Our BACTH results suggested that the EloR/MltG interaction is direct. To further pinpoint the EloR-MltG interaction, we performed BACTH assays with the Jag domain of EloR (which is targeted to midcell) and the cytoplasmic part of MltG (Fig. 5). Indeed, the sole Jag domain interacted with the cytosolic part of MltG (Fig. 3). The cytosolic part of MltG mainly consists of a DUF1346 (domain of unknown function 1346) domain. When we tested the cytosolic part of MltG lacking the DUF against the Jag domain of EloR, the interaction between the two was lost (Fig. 3). Since MltG is located at the division zone of S. pneumoniae, it is plausible that EloR is recruited to midcell through its interaction with MltG. Nevertheless, we cannot completely exclude the possibility that MltG is pulled down with EloR because both EloR and MltG interact with a common third protein. Further investigations (BACTH assays, coimmunoprecipitations [co-IPs], and cross-linking) are required to rule out this possibility.
FIG 5.
Schematic representation of MltG showing the N-terminal cytosolic domain, the transmembrane helix, and the C-terminal extracellular domain. The domain borders are indicated.
YidC2 does not affect MltG localization.
Since EloR and MltG are part of the same complex, and since EloR localization was somewhat altered in the ΔyidC2 mutant (localization to old poles [Fig. 2C and Fig. S4]), we wondered whether MltG, similar to EloR, would concentrate at the cell poles as well as at midcell in the ΔyidC2 mutant. We therefore deleted yidC2 in the strain expressing sfGFP-MltG from the native locus. However, like in wild-type cells, sfGFP-MltG was found at midcell in the ΔyidC2 mutant, and no polar foci were observed (Fig. S5A and B). Thus, in contrast to EloR, the deletion of yidC2 did not affect the localization of MltG. This suggests that EloR may have additional interaction partners that need to be identified in the future or possibly that the RNA molecules that it binds are concentrated at the old poles in the ΔyidC2 mutant. The fact that EloR displayed interaction with YidC2 in BACTH assays and the fact that the absence of YidC2 induces an altered EloR localization pattern suggest a functional role of YidC2 in the EloR/KhpA regulatory pathway. Since YidC proteins assist with the insertion of membrane proteins during translation, it is easy to imagine that the RNA-binding protein EloR is functionally linked to this process, e.g., controlling the expression of one or several elongasome proteins. Unraveling this would require additional research. The localization of sfGFP-MltG was not affected by the loss of eloR, as previously reported (19) and as shown in Fig. S5C in the supplemental material.
Concluding remarks.
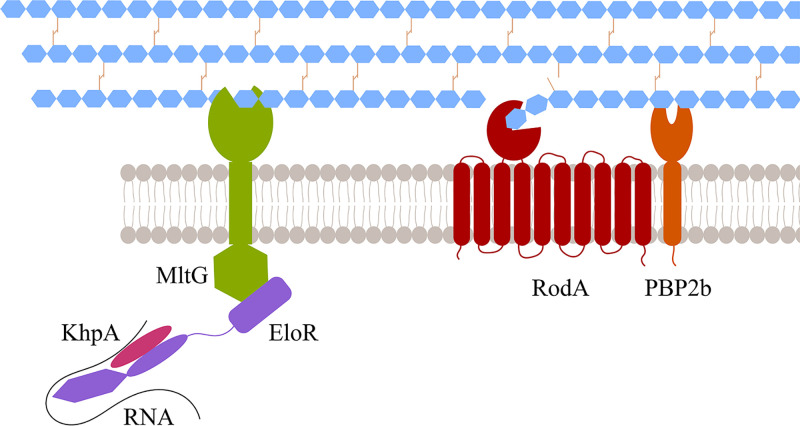
It has previously been shown that knocking out the essential pbp2b gene results in suppressor mutations in mltG, eloR, or khpA relieving the requirement of the elongasome in S. pneumoniae (15, 19). The current finding that EloR and MltG interact therefore corroborates that MltG and EloR are part of the same regulatory pathway. KhpA is also part of this complex since it has been shown previously to interact directly with EloR at the division zone (20, 21). In sum, we can conclude that MltG, EloR, and KhpA form a protein complex at the division zone, which regulates the elongasome on command from StkP. The relationship between EloR/KhpA and MltG is unknown. We have previously speculated that the expression level of MltG is controlled via the RNA-binding capacity of EloR/KhpA. This, however, turned out to be wrong as the amounts of MltG in a wild-type background and in a ΔeloR mutant are similar (19). Another hypothesis is that the EloR/KhpA complex regulates the activity of MltG. MltG in E. coli has been shown to possess endolytic transglycosylase activity, i.e., breaking glycosidic bonds within a glycan strand (14). Structural modeling and site-directed mutagenesis of the active site of pneumococcal MltG suggest that it has the same muralytic activity (15). Interestingly, it seems that MltG is not tolerated in either E. coli or S. pneumoniae mutants with compromised PG synthases (14, 15, 43), suggesting that the muralytic activity of MltG becomes lethal if a weaker PG layer is produced due to inefficient PG synthesis. In S. pneumoniae, MltG is most likely activated by EloR/KhpA since deletions of EloR and KhpA suppress the toxic effect that MltG has on Δpbp2b and ΔrodA mutants. E. coli, on the other hand, does not express EloR, and it has an MltG lacking the cytoplasmic domain found to interact with EloR in S. pneumoniae. Hence, MltG must be regulated through a different mechanism in this species. It has been hypothesized that MltG releases glycan strands made by both class A and B PBPs so that they can be cross-linked to new PG by the divisome and elongasome (15, 43). While this intriguing model might be proven correct, recent discoveries suggesting that class A PBPs are not involved in primary PG synthesis (elongasome and divisome) but rather function to repair, mature, or strengthen newly synthesized PG, combined with the fact that MltG is associated with the elongasome (12, 13, 15), lead us to suggest an alternative model: MltG may work together with amidases to open the PG layer so that PBP2b/RodA can add new PG to the existing layer and hence elongate the dividing cell (Fig. 6). MltG must therefore be strictly regulated to avoid uncontrolled damage to the PG layer when PG synthase activity is reduced. The present study has shown just how important the MltG levels in the cells are: adjusting the expression level of MltG from an inducible promoter proved to be difficult, and hence, subsequent genetic changes to eloR, which is part of the same pathway, are not possible. Based on the data presented here, the EloR/KhpA complex appears to play a key role in the regulation of MltG. Since the inactivation of the RNA-binding domains of EloR gives the same phenotype as the inactivation of the catalytic domain of MltG (PBP2b/RodA becomes redundant) (15, 19), one could speculate that the EloR/KhpA complex modulates the activity of MltG via the RNA-binding domains. Another possibility is that EloR regulates the MltG activity directly by interacting with other cell division proteins. This must be confirmed or rejected by further experimental evidence.
FIG 6.
Model of MltG/EloR/KhpA function. The communication between EloR/KhpA/RNA and MltG allows for a controlled opening of the PG. These openings are utilized by PBP2b/RodA to insert new PG in the lateral direction of the cell and in this way elongate the cell.
MATERIALS AND METHODS
Bacterial strains, cultivation, and transformation.
All bacterial strains used in this work are listed in Table 1. All E. coli strains were grown in liquid LB broth with shaking or on LB agar plates at 30°C or 37°C. When necessary, the following antibiotics were used: 100 μg/ml ampicillin and 50 μg/ml kanamycin. Transformation of E. coli was performed with heat shock at 42°C for 30 s. All S. pneumoniae strains were grown in C-medium (44) without shaking or on Todd-Hewitt (TH) agar plates in an oxygen-depleted chamber using AnaeroGen bags from Oxoid at 37°C. A concentration of 200 μg/ml streptomycin, 400 μg/ml kanamycin, or 2 μg/ml chloramphenicol was employed when necessary. When introducing genetic changes, natural transformation was utilized. Exponentially growing cells were diluted to an optical density at 550 nm (OD550) of 0.05 to 0.1 and grown for 2 h with 100 to 200 ng of the transforming DNA and 250 ng/ml (final concentration) competence-stimulating peptide (CSP) added to the growth medium. Thirty microliters of the transformed cell cultures was plated on TH agar plates with the appropriate antibiotic and incubated at 37°C overnight.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| S. pneumoniae | ||
| R704 | R6 derivative; comA::ermAM Eryr | J. P. Claverys |
| RH425 | R704 but streptomycin resistant Eryr Smr | 49 |
| SPH131 | ΔcomA P1::PcomR::comR PcomX::Janus Eryr Kanr | 35 |
| AW407 | ΔcomA P1::PcomR::comR PcomX::eloR-mKate2 Eryr Smr | This work |
| AW408 | ΔcomA P1::PcomR::comR PcomX::jag-mKate2 Eryr Smr | This work |
| AW409 | ΔcomA P1::PcomR::comR PcomX::jag-linker-mKate2 Eryr Smr | This work |
| AW410 | ΔcomA P1::PcomR::comR PcomX::linker-mKate2 Eryr Smr | This work |
| AW420 | ΔcomA P1::PcomR::comR PcomX::eloRK36A-mKate2 Eryr Smr | This work |
| AW424 | ΔcomA P1::PcomR::comR PcomX::eloRK37A-mKate2 Eryr Smr | This work |
| AW425 | ΔcomA P1::PcomR::comR PcomX::eloRF39A-mKate2 Eryr Smr | This work |
| AW426 | ΔcomA P1::PcomR::comR PcomX::eloRL40M-mKate2 Eryr Smr | This work |
| AW453 | ΔcomA P1::PcomR::comR PcomX::eloR-mKate2 ΔstkP::janus Eryr Kmr | This work |
| AW415 | ΔcomA P1::PcomR::comR PcomX::eloR-mKate2 ΔyidC2::janus Eryr Kmr | This work |
| AW417 | ΔcomA P1::PcomR::comR PcomX::eloR-mKate2 ΔrodZ::janus Eryr Kmr | This work |
| AW447 | ΔcomA m(sf)gfp-mltG flag-eloR Eryr Smr | This work |
| AW459 | ΔcomA flag-eloR hlpA-gfp-chloramphenicol Eryr Smr Camr | This work |
| DS515 | ΔcomA m(sf)gfp-mltG Eryr Smr | 19 |
| AW98 | ΔcomA flag-eloR Eryr Smr | 19 |
| MH43 | ΔcomA m(sf)gfp-mltG; ΔyidC2::janus Eryr Kmr | This work |
| SPH472 | ΔcomA m(sf)gfp-mltG; ΔeloR::janus Eryr Kmr | 19 |
| E. coli | ||
| XL1-Blue | Host strain | Agilent Technologies |
| BTH101 | BACTH expression strain; lacks cya | Euromedex |
| Plasmids | ||
| pKT25 | Plasmid used in BACTH analysis | Euromedex |
| pKNT25 | Plasmid used in BACTH analysis | Euromedex |
| pUT18C | Plasmid used in BACTH analysis | Euromedex |
| pUT18 | Plasmid used in BACTH analysis | Euromedex |
| pKNT25-eloR | T25 domain fused to the C terminus of EloR | 19 |
| pKT25-jag | T25 domain fused to the N terminus of the Jag domain of EloR | This work |
| pUT18C-mltG | T18 domain fused to the N terminus of MltG | 19 |
| pUT18C-mltGcyt | T18 domain fused to the N terminus of the cytoplasmic domain of MltG | This work |
| pUT18C-mltGcytΔDUF | T18 domain fused to the N terminus of the cytoplasmic domain of MltG without DUF | This work |
| pUT18C-pbp2b | T18 domain fused to the N terminus of PBP2b | 50 |
| pUT18C-rodA | T18 domain fused to the N terminus of RodA | 50 |
| pUT18C-rodZ | T18 domain fused to the N terminus of RodZ | 19 |
| pUT18C-mreC | T18 domain fused to the N terminus of MreC | 19 |
| pUT18C-mreD | T18 domain fused to the N terminus of MreD | This work |
| pUT18-cozE | T18 domain fused to the C terminus of CozE | 50 |
| pUT18-yidC2 | T18 domain fused to the C terminus of YidC2 | This work |
DNA constructs.
All primers used in this study are listed in Table S1 in the supplemental material. DNA constructs used to transform S. pneumoniae were made using overlap extension PCR (45). In short, in order to create deletion mutants, ∼1,000-bp sequences upstream and downstream of the gene in question were amplified and fused with the 5′ end and the 3′ end of the Janus cassette (46), respectively. The same flanking regions were then used to replace the Janus cassette with an alternative DNA sequence (47). Constructs used to produce bacterial two-hybrid (BACTH) plasmids were amplified from S. pneumoniae, cleaved with restriction enzymes (XbaI and EcoRI from New England BioLabs), and ligated into the preferred plasmid using Quick ligase (New England BioLabs). The plasmids used in this study are listed in Table 1. All constructs were verified by DNA sequencing.
Bacterial two-hybrid assay.
BACTH assays are based on the two tags T18 and T25 that make up the catalytic domain of Bordetella pertussis adenylate cyclase (CyaA). In order to test whether two proteins interact, their genes are cloned in frame with one tag each and coexpressed in E. coli BTH101 cells (lacking cyaA). If the two proteins interact, T18 and T25 are brought into proximity to make up an active CyaA catalytic domain. This results in cAMP production, which induces the expression of lacZ (β-galactosidase). β-Galactosidase cleaves X-gal, resulting in blue bacteria on X-gal-containing agar plates. In instances where the two tested proteins do not interact, no β-galactosidase is expressed, and the bacteria remain white. The BACTH experiments were performed as described by the manufacturer (Euromedex). The genes encoding our proteins of interest were cloned in frame with either the T18- or T25-encoding gene in the plasmids pUT18, pUT18C, pKNT25, and pKT25. The plasmids were then transformed into E. coli XL1-Blue cells, isolated, and sequenced. In order to test the interaction between two proteins, they were coexpressed with one tag (T18 or T25) each in E. coli BTH101 cells. After overnight incubation of transformants, five random colonies were picked, grown to exponential phase, and spotted (2 μl) onto LB agar plates containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM), and X-gal (40 μg/ml). After overnight incubation at 30°C, the results were documented.
Coimmunoprecipitation and Western blotting.
Co-IP was performed using anti-Flag M2 affinity gel (Sigma-Aldrich). S. pneumoniae strains were grown to an OD550 of 0.3 and lysed with 1 ml lysis buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) by triggering the endogenous pneumococcal autolysin LytA at 37°C for 5 min. The lysate was incubated with 40 μl anti-Flag M2 affinity gel with gentle rotation at 4°C overnight. After washing the affinity gel three times with 500 μl Tris-buffered saline (TBS), SDS sample buffer was added, and the samples were incubated at 95°C for 10 min. Proteins from 8 μl of each sample were separated in a 12% SDS-PAGE gel. After electrophoresis, the separated proteins were electroblotted onto a polyvinylidene difluoride (PVDF) membrane using a Trans-Blot Turbo transfer system (Bio-Rad) with a standard protocol for 7 min. Finally, Flag-tagged proteins were detected as previously described by Stamsås et al. (19). GFP-tagged proteins were detected with Chromotek rabbit polyclonal antibody for GFP, using the same protocol as the one described above and dilutions as recommended by the manufacturer.
Phase-contrast and fluorescence microscopy.
Cells were prepared for microscopic imaging by growing them to an OD550 of 0.4 and then diluting the culture to an OD550 of 0.1 and grown for another hour prior to microscopy. When relevant, 2 μM ComS inducer was added. Proteins fused with fluorescent mKate2 were visualized as previously described (19), using a Zeiss AxioObserver with ZEN Blue software, an Orca-Flash 4.0 V2 digital complementary metal-oxide semiconductor (CMOS) camera (Hamamatsu Photonics), and a 100× phase-contrast objective. An HXP 120 Illuminator (Zeiss) was used as a fluorescence light source. Images were prepared and analyzed using ImageJ software with the MicrobeJ plug-in (48). For subcellular localization analysis, the Maxima function in MicrobeJ was used to define fluorescence maxima within cells, and the average subcellular localizations of these maxima were plotted using the XYCellDensity plot (focus density plots) in MicrobeJ.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Research Council of Norway (project numbers 296906 and 250976).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Vollmer W, Blanot D, De Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Pinho MG, Kjos M, Veening J-W. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11:601–614. 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 3.Cho H, Wivagg CN, Kapoor M, Barry Z, Rohs PD, Suh H, Marto JA, Garner EC, Bernhardt TG. 2016. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol 1:16172. 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 6.Zapun A, Vernet T, Pinho MG. 2008. The different shapes of cocci. FEMS Microbiol Rev 32:345–360. 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- 7.Morlot C, Pernot L, Le Gouellec A, Di Guilmi AM, Vernet T, Dideberg O, Dessen A. 2005. Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae. J Biol Chem 280:15984–15991. 10.1074/jbc.M408446200. [DOI] [PubMed] [Google Scholar]
- 8.Berg KH, Stamsås GA, Straume D, Håvarstein LS. 2013. Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J Bacteriol 195:4342–4354. 10.1128/JB.00184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsui H-CT, Boersma MJ, Vella SA, Kocaoglu O, Kuru E, Peceny JK, Carlson EE, VanNieuwenhze MS, Brun YV, Shaw SL, Winkler ME. 2014. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol Microbiol 94:21–40. 10.1111/mmi.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez AJ, Cesbron Y, Shaw SL, Bazan Villicana J, Tsui H-CT, Boersma MJ, Ye ZA, Tovpeko Y, Dekker C, Holden S, Winkler ME. 2019. Movement dynamics of divisome proteins and PBP2x: FtsW in cells of Streptococcus pneumoniae. Proc Natl Acad Sci U S A 116:3211–3220. 10.1073/pnas.1816018116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meeske AJ, Riley EP, Robins WP, Uehara T, Mekalanos JJ, Kahne D, Walker S, Kruse AC, Bernhardt TG, Rudner DZ. 2016. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537:634–638. 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Straume D, Piechowiak KW, Olsen S, Stamsås GA, Berg KH, Kjos M, Heggenhougen MV, Alcorlo M, Hermoso JA, Håvarstein LS. 2020. Class A PBPs have a distinct and unique role in the construction of the pneumococcal cell wall. Proc Natl Acad Sci U S A 117:6129–6138. 10.1073/pnas.1917820117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vigouroux A, Cordier B, Aristov A, Alvarez L, Özbaykal G, Chaze T, Oldewurtel ER, Matondo M, Cava F, Bikard D, van Teeffelen S. 2020. Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 9:e51998. 10.7554/eLife.51998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yunck R, Cho H, Bernhardt TG. 2016. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol Microbiol 99:700–718. 10.1111/mmi.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsui HCT, Zheng JJ, Magallon AN, Ryan JD, Yunck R, Rued BE, Bernhardt TG, Winkler ME. 2016. Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol Microbiol 100:1039–1065. 10.1111/mmi.13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez AJ, Boersma MJ, Bruce KE, Lamanna MM, Shaw SL, Tsui H-CT, Taguchi A, Carlson EE, VanNieuwenhze MS, Winkler ME. 2 December 2020. Organization of peptidoglycan synthesis in nodes and separate rings at different stages of cell division of Streptococcus pneumoniae. Mol Microbiol 10.1111/mmi.14659. [DOI] [PMC free article] [PubMed]
- 17.Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. 2011. Super‐resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol 82:1096–1109. 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- 18.Fleurie A, Manuse S, Zhao C, Campo N, Cluzel C, Lavergne J-P, Freton C, Combet C, Guiral S, Soufi B, Macek B, Kuru E, VanNieuwenhze MS, Brun YV, Di Guilmi A-M, Claverys J-P, Galinier A, Grangeasse C. 2014. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet 10:e1004275. 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamsås GA, Straume D, Ruud Winther A, Kjos M, Frantzen CA, Håvarstein LS. 2017. Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae. Mol Microbiol 105:954–967. 10.1111/mmi.13748. [DOI] [PubMed] [Google Scholar]
- 20.Winther AR, Kjos M, Stamsås GA, Håvarstein LS, Straume D. 2019. Prevention of EloR/KhpA heterodimerization by introduction of site-specific amino acid substitutions renders the essential elongasome protein PBP2b redundant in Streptococcus pneumoniae. Sci Rep 9:3681. 10.1038/s41598-018-38386-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng JJ, Perez AJ, Tsui HCT, Massidda O, Winkler ME. 2017. Absence of the KhpA and KhpB (JAG/EloR) RNA‐binding proteins suppresses the requirement for PBP2b by overproduction of FtsA in Streptococcus pneumoniae D39. Mol Microbiol 106:793–814. 10.1111/mmi.13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenton AK, El Mortaji L, Lau DT, Rudner DZ, Bernhardt TG. 2017. Erratum: CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat Microbiol 2:17011. 10.1038/nmicrobiol.2017.11. [DOI] [PubMed] [Google Scholar]
- 23.Stamsås GA, Restelli M, Ducret A, Freton C, Garcia PS, Håvarstein LS, Straume D, Grangeasse C, Kjos M. 2020. A CozE homolog contributes to cell size homeostasis of Streptococcus pneumoniae. mBio 11:e02461-20. 10.1128/mBio.02461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenton AK, Manuse S, Flores-Kim J, Garcia PS, Mercy C, Grangeasse C, Bernhardt TG, Rudner DZ. 2018. Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proc Natl Acad Sci U S A 115:2812–2817. 10.1073/pnas.1715218115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manuse S, Fleurie A, Zucchini L, Lesterlin C, Grangeasse C. 2016. Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol Rev 40:41–56. 10.1093/femsre/fuv041. [DOI] [PubMed] [Google Scholar]
- 26.Fleurie A, Cluzel C, Guiral S, Freton C, Galisson F, Zanella‐Cleon I, Di Guilmi AM, Grangeasse C. 2012. Mutational dissection of the S/T‐kinase StkP reveals crucial roles in cell division of Streptococcus pneumoniae. Mol Microbiol 83:746–758. 10.1111/j.1365-2958.2011.07962.x. [DOI] [PubMed] [Google Scholar]
- 27.Beilharz K, Nováková L, Fadda D, Branny P, Massidda O, Veening J-W. 2012. Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc Natl Acad Sci U S A 109:E905–E913. 10.1073/pnas.1119172109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleurie A, Lesterlin C, Manuse S, Zhao C, Cluzel C, Lavergne J-P, Franz-Wachtel M, Macek B, Combet C, Kuru E, VanNieuwenhze MS, Brun YV, Sherratt D, Grangeasse C. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516:259–262. 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holečková N, Doubravová L, Massidda O, Molle V, Buriánková K, Benada O, Kofroňová O, Ulrych A, Branny P. 2015. LocZ is a new cell division protein involved in proper septum placement in Streptococcus pneumoniae. mBio 6:e01700-14. 10.1128/mBio.01700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falk SP, Weisblum B. 2013. Phosphorylation of the Streptococcus pneumoniae cell wall biosynthesis enzyme MurC by a eukaryotic-like Ser/Thr kinase. FEMS Microbiol Lett 340:19–23. 10.1111/1574-6968.12067. [DOI] [PubMed] [Google Scholar]
- 31.Ulrych A, Holečková N, Goldová J, Doubravová L, Benada O, Kofroňová O, Halada P, Branny P. 2016. Characterization of pneumococcal Ser/Thr protein phosphatase phpP mutant and identification of a novel PhpP substrate, putative RNA binding protein Jag. BMC Microbiol 16:247. 10.1186/s12866-016-0865-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rued BE, Zheng JJ, Mura A, Tsui H-CT, Boersma MJ, Mazny JL, Corona F, Perez AJ, Fadda D, Doubravová L, Buriánková K, Branny P, Massidda O, Winkler ME. 2017. Suppression and synthetic‐lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin‐binding protein interactions in Streptococcus pneumoniae D39. Mol Microbiol 103:931–957. 10.1111/mmi.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morlot C, Bayle L, Jacq M, Fleurie A, Tourcier G, Galisson F, Vernet T, Grangeasse C, Di Guilmi AM. 2013. Interaction of penicillin‐binding protein 2x and Ser/Thr protein kinase StkP, two key players in Streptococcus pneumoniae R6 morphogenesis. Mol Microbiol 90:88–102. 10.1111/mmi.12348. [DOI] [PubMed] [Google Scholar]
- 34.Myrbråten IS, Wiull K, Salehian Z, Håvarstein LS, Straume D, Mathiesen G, Kjos M. 2019. CRISPR interference for rapid knockdown of essential cell cycle genes in Lactobacillus plantarum. mSphere 4:e00007-19. 10.1128/mSphere.00007-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg KH, Biørnstad TJ, Straume D, Håvarstein LS. 2011. Peptide-regulated gene depletion system developed for use in Streptococcus pneumoniae. J Bacteriol 193:5207–5215. 10.1128/JB.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirschfeld C, Gómez-Mejia A, Bartel J, Hentschker C, Rohde M, Maaß S, Hammerschmidt S, Becher D. 2019. Proteomic investigation uncovers potential targets and target sites of pneumococcal serine-threonine kinase StkP and phosphatase PhpP. Front Microbiol 10:3101. 10.3389/fmicb.2019.03101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X, Ge F, Xiao C-L, Yin X-F, Ge R, Zhang L-H, He Q-Y. 2010. Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J Proteome Res 9:275–282. 10.1021/pr900612v. [DOI] [PubMed] [Google Scholar]
- 38.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu ZC, de Keyzer J, Berrelkamp-Lahpor GA, Driessen AJ. 2013. Interaction of Streptococcus mutans YidC1 and YidC2 with translating and nontranslating ribosomes. J Bacteriol 195:4545–4551. 10.1128/JB.00792-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg R, Knüpffer L, Origi A, Asti R, Koch H-G. 2018. Co-translational protein targeting in bacteria. FEMS Microbiol Lett 365:fny095. 10.1093/femsle/fny095. [DOI] [PubMed] [Google Scholar]
- 41.Shiomi D, Sakai M, Niki H. 2008. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27:3081–3091. 10.1038/emboj.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kjos M, Aprianto R, Fernandes VE, Andrew PW, van Strijp JA, Nijland R, Veening J-W. 2015. Bright fluorescent Streptococcus pneumoniae for live-cell imaging of host-pathogen interactions. J Bacteriol 197:807–818. 10.1128/JB.02221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohrhunter JL, Rohs PDA, Torres G, Yunck R, Bernhardt TG. 5 December 2020. MltG activity antagonizes cell wall synthesis by both types of peptidoglycan polymerases in Escherichia coli. Mol Microbiol 10.1111/mmi.14660. [DOI] [PMC free article] [PubMed]
- 44.Lacks S, Hotchkiss RD. 1960. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta 39:508–518. 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 45.Higuchi R, Krummel B, Saiki R. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16:7351–7367. 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sung C, Li H, Claverys J, Morrison D. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnsborg O, Eldholm V, Bjørnstad ML, Håvarstein LS. 2008. A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol Microbiol 69:245–253. 10.1111/j.1365-2958.2008.06288.x. [DOI] [PubMed] [Google Scholar]
- 48.Ducret A, Quardokus EM, Brun YV. 2016. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 1:16077. 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnsborg O, Håvarstein LS. 2009. Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J Bacteriol 191:5859–5864. 10.1128/JB.00724-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straume D, Stamsås GA, Berg KH, Salehian Z, Håvarstein LS. 2017. Identification of pneumococcal proteins that are functionally linked to penicillin‐binding protein 2b (PBP2b). Mol Microbiol 103:99–116. 10.1111/mmi.13543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.