Malaria remains a major public health and economic burden currently. The mutually exclusive expression of the virulence genes is associated with the pathogenesis and immune evasion of human malaria parasites in the host.
KEYWORDS: architectural factor, gene expression, genome organization, HMGB1, malaria
ABSTRACT
The three-dimensional (3D) genome organization plays a critical role in the regulation of gene expression in eukaryotic organisms. In the unicellular malaria parasite Plasmodium falciparum, the high-order chromosome organization has emerged as an important epigenetic pathway mediating gene expression, particularly for virulence genes, but the related architectural factors and underlying mechanism remain elusive. Herein, we have identified the high-mobility-group protein HMGB1 as a critical architectural factor for maintenance of genome organization in P. falciparum. Genome-wide occupancy analysis (chromatin immunoprecipitation sequencing [ChIP-seq]) shows that the HMGB1 protein is recruited mainly to centromeric regions likely via a DNA-binding-independent pathway. Chromosome conformation capture coupled with next-generation sequencing (Hi-C-seq) and 3D modeling analysis show that the loss of HMGB1 disrupts the integrity of centromere/telomere-based chromosome organization accompanied with diminished interaction frequency among centromere clusters. This triggers local chromatin alteration and dysregulated gene expression. Notably, the entire repertoire of the primary virulence genes (var) was completely silenced in the absence of P. falciparum HMGB1 (PfHMGB1). Furthermore, the disrupted nuclear organization was reconstituted by complementation of HMGB1, thereby rescuing the mutually exclusive expression of the var gene family. Collectively, these data demonstrate that the architectural factor HMGB1 is associated with gene expression via mediating the high-order structure of genome organization. This finding not only contributes better understanding of the epigenetic regulation of gene expression but may also provide novel targets for antimalarial strategies.
INTRODUCTION
Malaria parasites utilize various strategies to escape from host immune responses. One of these strategies involves the mutually exclusive expression of variant surface antigens encoded by multigene families (1). Perhaps among the most important of these multigene families are the ∼60 var genes that encode variants of the Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1s). These variant antigens on the surface of the infected red blood cells (iRBCs) have been shown to mediate antigenic variation, which underlies pathogenesis through the selective adhesion of iRBCs to various surface receptors of host cells (2, 3). Each individual malaria parasite only expresses a single var gene at a given time, with the other members of the multigene family remain silenced (4–6). Nevertheless, the complex mechanisms underlying the epigenetic control of the mutual expression of variant genes are not fully resolved.
The histone modification-associated chromatin microenvironment surrounding the upstream promoter regions determines the transcriptional activity of individual variant genes (7–9). For example, HP1-dependent heterochromatin marked by trimethylation of lysine 9 on histone 3 (H3K9me3) characterizes transcriptionally repressed regions, whereas euchromatin marked by H3K4me2/3, acetylated lysine 9 of histone 3 (H3K9ac), or H2A.Z is usually linked to active transcription (10–12). In particular, local chromatin alterations occurring within the promoter regions lead to silencing or activation of variant genes, suggesting that the dynamics of local chromatin structure control the transcriptional state of variant genes (13, 14). In addition, other mechanisms involving agents such as transcription factor (TF) ApiAP2, long noncoding RNAs (lncRNAs), and nascent RNAs are also involved in the singular expression of members of the variant gene family (15–18).
Another layer of the complex epigenetic mechanism regulating variant gene expression is the nuclear chromatin structure and perinuclear gene locus repositioning (19, 20). Precise organization of all chromosomes forms multiple intra- or interchromosome interaction events and constitutes the high-order chromatin structure in the nucleus (21). Recently, the application of chromosome conformation capture (Hi-C) to malaria parasites has made it feasible to capture the spatial chromosome interaction network through resolution of the three-dimensional (3D) genome organization (22, 23). Strikingly, the 3D genome model of P. falciparum shows that telomeres and centromeres cluster on two opposite perinuclear sides of the nucleus, and those silenced virulence genes colocalize with the telomere superdomain (22). The two separated superdomains were proposed to be linked to the transcriptional activity of specific gene families throughout the life cycle. Moreover, apparent dynamic relocation and interactions were observed for genes involved in multiple physiological processes, including immune evasion, pathogenesis, host cell invasion, and sexual commitment in P. falciparum (23).
Hence, it is speculated that high-order chromatin structure plays a role in the hierarchical control of mutually exclusive expression of variant genes via deposition of individual genes in centromere- or telomere-associated superdomains. However, little is known concerning the molecular basis of the configuration and regulation of the dynamic 3D genome. In malaria parasites, those architectural proteins such as CCCTC binding factor (CTCF) or lamins, which are known to be involved in chromosome organization in metazoans, have not yet been identified. Recently, two studies have reported that RecQ helicases (P. falciparum WRN [PfWRN] and PfBLM) targeting the noncanonical G-quadruplexes (G4s) DNA structures are associated with var gene expression (24, 25). In other eukaryotic organisms, nonhistone architectural chromosomal proteins, HMGB (high-mobility group B family), have been implicated in the control of gene expression through regulation of local nucleosome dynamics at the promoter regions of a variety of genes. They have also been found to regulate chromosome structure through the action of specific topologically associating domain (TAD) boundaries (26–28). For P. falciparum, four putative HMG proteins (PfHMGB1 to -4) have been predicted by sequence homology. Among which, PfHMGB1 and -2 belong to the HMGB/UB family with members that able to bind specific linear DNA. Moreover, PfHMGB1/2 can interact with four-way DNA junctions (4H) and induce DNA binding. Intriguingly, compared to higher protein abundance of PfHMGB2 in gametocytes, PfHMGB1 exhibits obvious higher protein abundance in mixed asexual stages (29), indicating a specific role of PfHMGB1 in the asexual stage. However, the biological functions of PfHMGB1 have yet to be investigated.
In this study, we provide experimental evidence for a critical role of the HMGB1 protein during the maintenance of the centromere/telomere-dependent genome organization. Moreover, the loss of HMGB1 results in complete silencing of the entire repertoire of the var gene family, which was rescued upon gene complementation. This finding provides a new insight into the complex epigenetic mechanism of virulence gene expression in malaria parasites and suggests an important role of the architectural factor HMGB1 during malaria pathogenesis.
RESULTS
Pfhmgb1 gene knockout dysregulates variant genes.
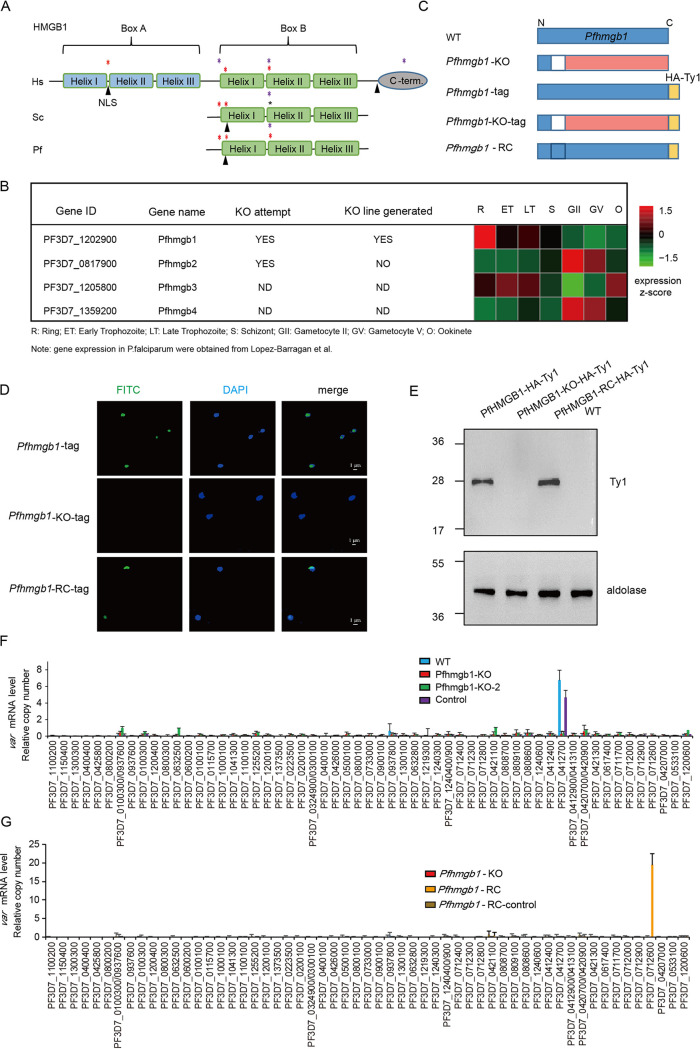
Although HMGB1 proteins are highly conserved in eukaryotes (see Fig. S1A and B in the supplemental material), their orthologues in the Plasmodium genus harbor different determinants reported for sequence-specific or structure-specific HMG box domains (Fig. 1A and Fig. S1C). As in yeast, Plasmodium HMGB1 contains neither the box-A domain nor the C-terminal hydrophobic tail, which is involved in the recognition of structural DNA targets (30). In the genome of human malaria parasite P. falciparum, four putative HMG box proteins have been predicted. Of these, PfHMGB1 (PF3D7_1202900) is preferentially expressed in asexual blood-stage parasites (29) (Fig. 1B).
FIG 1.
Pfhmgb1-knockout dysregulates variant gene expression. (A) Schematic representation of HMGB1 proteins of Homo sapiens (Hs), Saccharomyces cerevisiae (Sc), and P. falciparum (Pf). The two boxes and individual α-helix domains are shown in parallel. The purple asterisks indicate the determinants for structure-specific recognition, and red asterisks indicate critical intercalating sites. The sites of putative nuclear localization signals (NLS) are indicated with each protein. (B) Transcriptional profiles and CRISPR-Cas9 knockout attempts of the four Pfhmgb genes in the 3D7 strain. Transcriptional abundance data were obtained from Lopez-Barragan et al. (51). (C) Schematic representation of generation of Pfhmgb1-KO, Pfhmgb1-HA-Ty1, Pfhmgb1-KO-HA-Ty1, and Pfhmgb1-RC transgenic parasites by CRISPR-Cas9. The blank region represents the deleted fragment and the pink region represents the disrupted ORF of the Pfhmgb1 gene. (D) Subcellular location of PfHMGB1 in transgenic ring-stage parasites detected by IFA with anti-Ty1 antibody (green). The nuclei are stained by 4′,6-diamidino-2-phenylindole (DAPI). (E) Western blot assay of PfHMGB1 protein in different clones. (F) RT-qPCR result for var genes in Pfhmgb1-KO (two clones from independent transfections), vector control, and WT clones. Error bars represent the standard errors of the means (SEMs) from two biological replicates. (G) RT-qPCR result for var genes in Pfhmgb1-KO, Pfhmgb1-RC, and transfection control (vector) clones. Error bars represent the SEMs for two biological replicates.
Evolutionally conserved HMGB1 family in eukaryotes. (A) Phylogenetic tree of the HMGB family in various organisms. (B) Multiple alignment of HMG box domains of HMG proteins in diverse species. Dashes represent gaps and missing sequences. The bar plot (bottom) indicates the level of conservation for each amino acid. The amino acids were colored according to default ClustalX (v2.1) color palette. (C) Modeling of crystal structure of the HMGB1 proteins of human (GenBank CAG33144.1), yeast (GenBank KZV07568.1), and P. falciparum (GenBank CZT99193.1). The residues associated with structure- or sequence-specific recognition of HMGB1-DNA are indicated individually corresponding to Fig. 1A. The PDB files of each protein sequence were produced by SWISS-MODEL online (https://swissmodel.expasy.org/) and were viewed and modified in PyMol. Download FIG S1, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To explore the biological function of PfHMGB1 during asexual development, we disrupted the open reading frame (ORF) by deleting a fragment of the P. falciparum hmgb1 (Pfhmgb1) gene in the wild-type (WT) 3D7-G7 strain (18) through the CRISPR-Cas9 gene editing system (31). The knockout (KO) strain represented the disruption of the original open reading frame gene from the 17th codon (Fig. 1C, Fig. S2A and B, and Table S1A). An empty plasmid without a single guide RNA (sgRNA) expression cassette was used as a transfection control. Following drug selection and cloning of transgenic parasites, we successfully obtained both Pfhmgb1-KO and control lines. To confirm the loss of PfHMGB1 expression in the Pfhmgb1-KO transgenic line, we performed a second-round transfection to fuse an HA×3-Ty1×3 sequence (hemagglutinin [HA]-Ty1 tag) to the 3′ end of the coding region of the Pfhmgb1 gene of the WT and Pfhmgb1-KO lines (Fig. 1C). Immunofluorescence assay (IFA) and Western blot analyses validated the knockout in the Pfhmgb1-KO line (Fig. 1D and E). No apparent growth retardation was observed for the Pfhmgb1-KO line during the blood-stage development of parasites (Fig. S2C and D).
Generation of Pfhmgb1-KO line. (A) Schematic representation of generation of Pfhmgb1-KO transfectant line by CRISPR-Cas9 technique. (B) Nucleotide sequencing of the mutation and reverse mutation events in Pfhmgb1-KO and Pfhmgb1-RC lines. (C) Growth curves of synchronized parasites of Pfhmgb1-KO and WT control. Error bars represent SEMs from three biological replicates. (D) Flow cytometry analysis of the blood-stage growth of Pfhmgb1-KO and WT lines in parallel. Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of oligonucleotide sequences and RNA-seq, ChIP-seq, and Hi-C-seq analyses. Download Table S1, XLSX file, 0.06 MB (60.3KB, xlsx) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we harvested synchronized ring-stage parasites of the Pfhmgb1-KO and WT clones and performed transcriptome sequencing (RNA-seq) analysis with high reproducibility between replicates (see Fig. S3A). We examined the changes in transcriptome upon Pfhmgb1 KO at ring, trophozoite, and schizont stages and found that most differentially expressed genes (DEGs) were structured noncoding RNAs (ncRNAs) such as tRNA, rRNA, and snoRNA and heterochromatin-associated genes (10, 11), such as genes encoding exported proteins, variant genes of var, rifin, stevor, and Pfmc-2tm, and var-associated noncoding ruf6 genes (14, 17) (Fig. S3B and C and Table S1B). Strikingly, the active var gene (PF3D7_0412700; var0412700) originally expressed in WT parasites was completely silenced in the Pfhmgb1-KO clone, whereas it maintained predominant expression in the transfection control line (Fig. S3D). This finding was validated by quantitative reverse transcription-PCR (RT-qPCR) analysis with another clone of an independent knockout transfection (Pfhmgb1-KO), which enabled exclusion of the accidental side effect of transfection itself (Fig. 1F). Notably, most other members of variant gene families were also significantly downregulated, including those “silenced” in the WT clone. In particular, all stevor and Pfmc-2tm genes were completely silenced (Fig. S3E).
Pfhmgb1 knockout silenced variant genes. (A) Correlation of transcript abundance in samples under different conditions. (B) Comparative transcriptome analysis between Pfhmgb1-KO (y axis) and WT (x axis) clones at different stages. All the significantly up- and downregulated genes (≥3-fold change) are indicated with red and green circles, respectively. Dashed lines indicate the fold change cutoff. (C) Gene function classification of upregulated and downregulated genes in Pfhmgb1-KO versus WT. The numbers of genes in each category are indicated in the figure. (D) Transcriptional level of var genes in Pfhmgb1-KO, WT 3D7-G7, and transfection control clones measured by RNA-seq assays. Error bars represent SEMs for two biological replicates. (E) Transcription levels of variant genes, including rifin, stevor, and Pfmc-2tm in Pfhmgb1-KO and WT clones at ring measured by RNA-seq assays. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To test whether the disrupted mutually exclusive expression of variant genes in Pfhmgb1-KO parasites could be rescued by the complementation of the Pfhmgb1 gene, we performed reverse mutation experiments to recover the deleted fragment by genetic editing using CRISPR-Cas9 transfection with a new sgRNA sequence (Fig. 1C to E and see Fig. S2B and S4A and Table S1A). We performed two highly correlated replicates of RNA-seq with the resulting Pfhmgb1-RC clone at the ring stage (Fig. S4B). Comparative transcriptome analysis of the Pfhmgb1-RC and WT clones revealed a high correlation of genome-wide gene expression in the two clones (Fig. S4C). As expected, the transcriptional abundance of most DEGs was reversed in the Pfhmgb1-RC line (Fig. S4D). Importantly, var gene expression was reversed back to the WT pattern, although the predominantly transcribed var gene had switched to another one located on chromosome 7 (PF3D7_0712600; var0712600) (Fig. S4E), which was validated by RT-qPCR with a vector control (Pfhmgb1-RC-control) (Fig. 1G). Taken together, our data strongly suggests that the PfHMGB1 is required for mutually exclusive expression of variant genes in P. falciparum.
Recovery of gene expression in the Pfhmgb1-RC clone. (A) Schematic representation of generation of Pfhmgb1-RC transgenic parasites by CRISPR-Cas9 technique. (B) Correlation of global gene expression between two biological replicates of Pfhmgb1-RC clones at ring. (C) Comparative transcriptome analysis of WT and Pfhmgb1-RC clones by RNA-seq at ring. (D) Boxplots show transcript abundance of dysregulated genes upon Pfhmgb1-KO in WT, Pfhmgb1-KO, and Pfhmgb1-RC clones. Lines and error bars represent medians with 95% CIs. Wilcoxon test, ***, P ≤ 0.001; **, P ≤ 0.01. (E) Transcriptional level of var genes in Pfhmgb1-KO and Pfhmgb1-RC clones measured by RNA-seq assays. Error bars represent SEMs for two biological replicates. Download FIG S4, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PfHMGB1 interacts with centromere clusters.
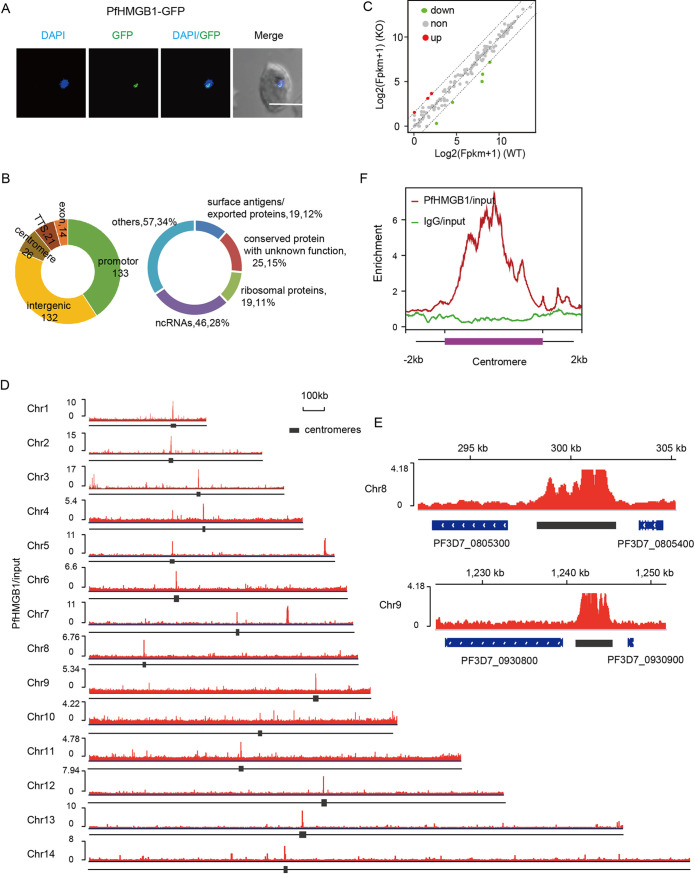
To explore the underlying mechanism of PfHMGB1-mediated virulence gene expression, we next attempted to resolve the genome-wide distribution of PfHMGB1 and identified its target genes. To this end, we constructed a Pfhmgb1-gfp transgenic line with the C terminus of the endogenous PfHMGB1 tagged with a green fluorescent protein (GFP) for chromatin immunoprecipitation sequencing (ChIP-seq) analysis (see Fig. S5A and Fig. 2A). As it has been reported that the conventional formaldehyde cross-linking may fail to capture HMG complex bound to chromatin in other eukaryotes (32), here we adopted a modified dual-cross-linking strategy (27) to identify the binding targets of PfHMGB1 in ring-stage parasites.
FIG 2.
PfHMGB1 is preferentially enriched in centromeric regions. (A) Live-cell fluorescence assay of PfHMGB1-GFP. The nuclei are stained by DAPI. Bar, 5 μm. (B, left) Distribution of PfHMGB1 peaks at the whole-genome level. The numbers indicate the number of the enriched peaks for each region (see Table S1C in the supplemental material). (Right) Detailed classification of PfHMGB1 binding genes. (C) Transcriptomic changes of PfHMGB1 binding genes in WT and Pfhmgb1-KO clones. All the significantly up- and downregulated genes (≥3-fold change) are indicated with red and green circles, respectively. Dashed lines indicate the fold change cutoff. (D) IGV snapshots of PfHMGB1 binding sites at 14 chromosomes. Black boxes indicate the location of centromeres. The enrichment of PfHMGB1 signal was normalized with input. (E) Integrative Genomics Viewer (IGV) snapshots of PfHMGB1 binding sites at centromere regions of indicated chromosomes. (F) ChIP-seq enrichment profile of PfHMGB1 (red line) and IgG control (green line) at the region of centromeres. Both PfHMGB1 (red line) and IgG were normalized with genomic input signal.
Reproducibility of PfHMGB1 ChIP-seq experiments (A) Schematic representation of generation of Pfhmgb1-GFP transgenic parasites by CRISPR-Cas9 modification. (B) Correlation analysis for biological replicates of PfHMGB1 ChIP-seq data. Download FIG S5, TIF file, 0.2 MB (245KB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ChIP-seq analysis on two highly correlated biological replicates (Fig. S5B) showed that PfHMGB1 mainly binds to 132 intergenic sites and upstream regions of 133 protein-coding or noncoding genes encoding surface antigens, exported proteins, ribosomal proteins, and structured ncRNAs (Fig. 2B and Table S1C). However, the majority of these target genes did not show any differential expression between the Pfhmgb1-KO and WT lines (Fig. 2C). This suggests that PfHMGB1 binding to these gene loci, e.g., tRNAs (33), may be associated with nuclear organization rather than acting as a transcription factor directly involved in transcription activity.
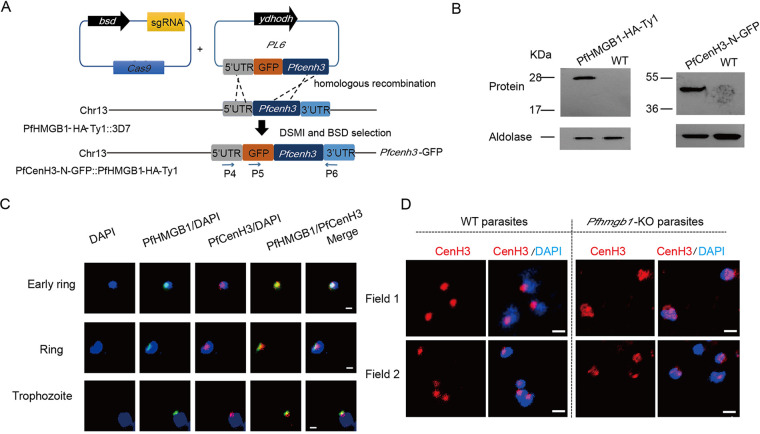
Strikingly, a strong selective enrichment of PfHMGB1 signals was detected within the centromere regions of all 14 chromosomes, indicative of an interaction between PfHMGB1 and centromeres (Fig. 2D to F). The core regions of P. falciparum centromeres are extremely AT rich (∼98%) and contain multiple repetitive sequences with various sizse and copy numbers (34), and this may cause amplification bias in DNA library preparation for high-throughput sequencing. To address this issue, we constructed another dual-gene-tagging transfectant (GFP-PfCenH3::PfHMGB1-HA-Ty1) by integrating the gfp gene sequence into the PfCenH3 gene locus with the Pfhmgb1-HA-Ty1 transgenic line as a parental line for transfection (Fig. 3A). Western blot analysis confirmed the endogenous coexpression of GFP-PfCenH3 and PfHMGB1-HA-Ty1 fusion proteins (Fig. 3B). Thus, the centromeres could be labeled by GFP-PfCenH3 histone specifically. Co-IFA assays confirmed the colocalization of PfHMGB1 and the centromere superdomain (Fig. 3C). They mainly formed a single focus at a unique site on the nuclear periphery, which is consistent with a previous description of centromere distribution in the nucleus of P. falciparum (35, 36) and in fission yeast (34, 37, 38). We observed that, to some extent, the size of a single centromeric focus was larger in the nucleus of Pfhmgb1-KO parasites (Fig. 3D), supportive of a reduced interactome within the centromere superdomain which was previously described by 3D genome modeling (22).
FIG 3.
PfHMGB1 interacts with perinuclear centromeres. (A) Schematic representation of generation of dual-gene-tagging transfectant (PfHMGB1-HA-Ty1::GFP-PfCenH3) by CRISPR-Cas9 with the Pfhmgb1-HA-Ty1 strain as the parent line for transfection. (B) Western blot for PfHMGB1-HA-Ty1 (left) and PfCenH3-N-GFP (right). The aldolase signals were used as the internal control. (C) Co-IFA assay of PfHMGB1-HA-Ty1 (mouse anti-Ty1 antibody, green) and GFP-PfCenH3 (rabbit anti-GFP antibody, red) in the ring or trophozoite-stage parasites of PfHMGB1-HA-Ty1::GFP-PfCenH3 line. Nuclear DNA was stained by DAPI (blue). Bars, 1 μm. (D) IFA assay of GFP-PfCenH3 with anti-GFP antibody in the ring-stage parasites of Pfhmgb1-KO and WT control. Nuclear DNA was stained by DAPI (blue). Bars, 1 μm.
Absence of PfHMGB1 reduces centromere interactome.
In other eukaryotes, HMG box proteins have been shown to bind to non-B-type DNA structures such as cruciform or distorted AT-rich DNA sequences in a non-sequence-specific manner (39). An in vitro DNA binding assay also demonstrated that the recombinant Plasmodium HMGB1 preferentially interacts with structured DNA probes such as 4H instead of linear DNA targets (29), suggesting that PfHMGB1 may be involved in spatial chromosome organization via binding to the junction sites of different chromosomes.
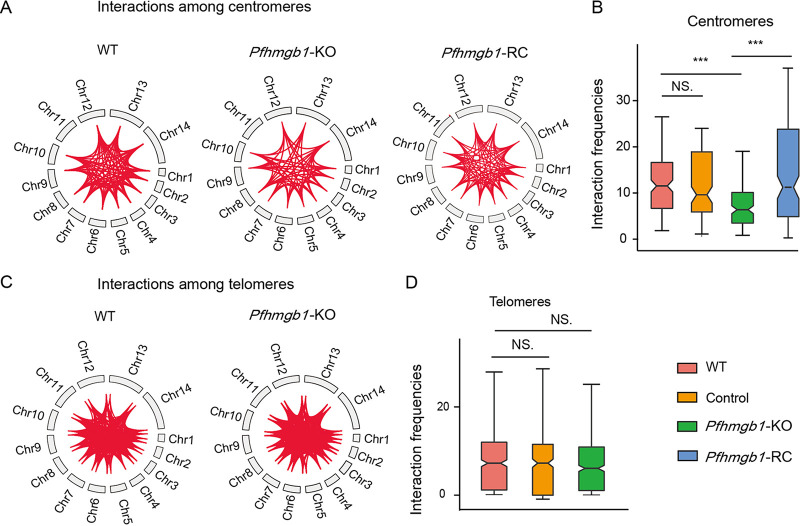
To examine this hypothesis, we utilized Hi-C coupled with next-generation sequencing (Hi-C-seq) analysis to examine the potential impact of PfHMGB1 knockout on the 3D nuclear organization, particularly, the reposition of variant genes. For each of the Pfhmgb1-KO, Pfhmgb1-RC, and WT clones, we performed Hi-C experiments with ring-stage parasites for at least two independent replicates of high reproducibility (see Fig. S6A). We then combined our Hi-C data replicates and normalized the sum of all valid interaction frequencies of each clone for downstream analysis (Table S1D). We observed an obvious reduction of interaction frequencies among centromeres (Fig. 4A), which is further confirmed by our statistical comparison of interaction frequencies between the Pfhmgb1-KO and WT clones (Fig. 4B). Importantly, the reduced interactome within the centromere superdomain had been rescued by the complementation of PfHMGB1 in the Pfhmgb1-RC line. We also examined telomere clustering and observed no significant difference for the interaction frequencies among telomeres between Pfhmgb1-KO and WT clones (Fig. 4C and D). To exclude the effect of manipulations of transfection or drug selection, we also performed a Hi-C-seq assay with the ring-stage parasites of the vector control line. While the sequencing depth and total valid interactions were comparable among them, no significant difference for the interactions among either centromeres or telomeres was found between the control and WT clones (Fig. S6B).
FIG 4.
Loss of PfHMGB1 diminishes the centromere interactome. (A) Significant interactions among centromeres in WT, Pfhmgb1-KO, and Pfhmgb1-RC clones. Red lines represent interactions among genomic features. Interactions among centromeres were obviously reduced in Pfhmgb1-KO strain and back to the wild-type level in the Pfhmgb1-RC strain. (B) Statistical chart for interaction frequency among centromeres. Lines and error bars represent medians and 95% confidence intervals (CIs), respectively. Wilcoxon test, ***, P ≤ 0.001; NS, no significant change. (C) Significant interactions among telomeres in the WT and Pfhmgb1 KO. Red lines represent interactions among genomic features. There is no significant change in interactions among telomeres between WT and Pfhmgb1-KO strains. (D) Statistical chart for interaction frequency among telomeres. Lines and error bars represent medians with 95% CIs. Wilcoxon test, NS, no significant change.
Quality control of Hi-C-seq data between replicates and clones. (A) The correlation between replicates by HiCExplorer are shown for WT, Pfhmgb1-KO, and Pfhmgb1-RC lines in ring-stage parasites. (B) Significant interactions among centromeres (left) or telomeres (right) in WT and vector control clones, respectively. Red lines represent interactions among genomic features. There is no significant change in interactions among centromeres or telomeres between WT and vector control strains. Download FIG S6, TIF file, 0.9 MB (897.4KB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
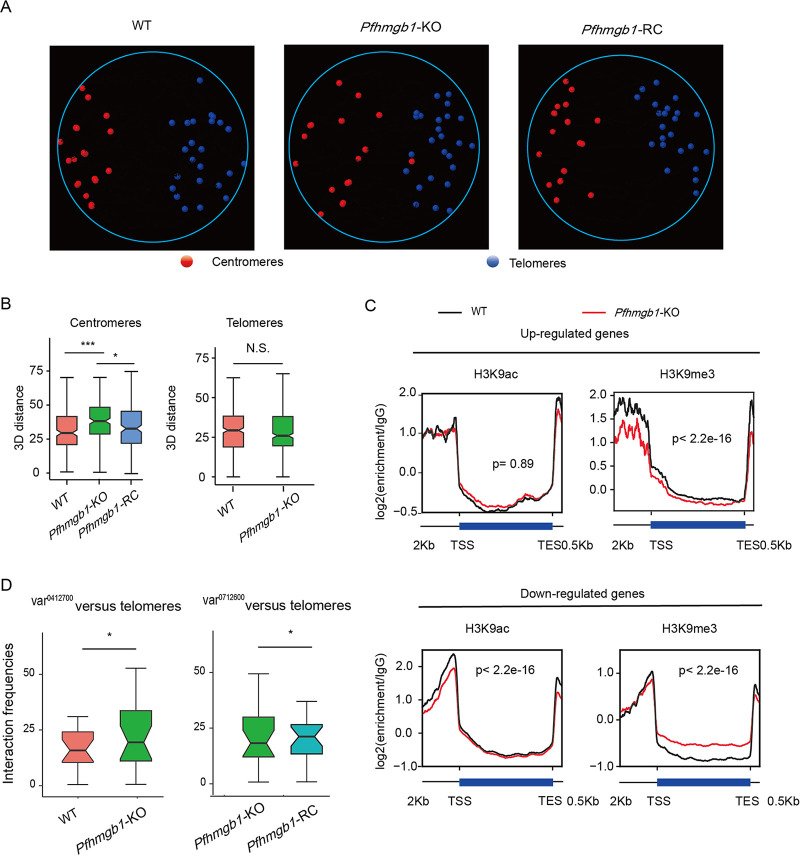
Next, we constructed 3D models of genome organization to evaluate the mutual positions of telomeres, centromeres, and var gene loci in the nuclei of Pfhmgb1-KO, Pfhmgb1-RC, and WT clones. Similar to a previous report (22), all telomeres and centromeres occupied two distinct spaces at perinuclear sides in the nucleus of WT parasites (Fig. 5A, left, and see Video S1). The clustering of centromeres was conserved (P = 0.001, Witten-Noble colocalization test), and the telomeres also colocalized significantly (P < 0.001). Strikingly, we observed significant increased 3D distances among the centromeres in the Pfhmgb1-KO line, which results in an enlarged centromere superdomain at the nuclear periphery. No significant difference in 3D distances was observed for telomeres (Fig. 5A and B and Video S1). These results were further confirmed by a Witten-Noble colocalization test in the Pfhmgb1-KO clone that showed the clustering of centromeres lost colocalization (P = 0.759), while the clustering of telomeres remained conserved (P = 0.003). Finally, we repeated the Hi-C-seq analysis to examine the genome organization in ring-stage Pfhmgb1-RC parasites. As expected, the interaction frequencies among centromeres had been recovered (Fig. 4A). Consistent with this, the clustering of centromeres reverted to colocalization (P < 0.001, Witten-Noble colocalization test), followed by a significant decrease in 3D distances among centromeres in the Pfhmgb1-RC clone (Fig. 5A and B and Video S1).
FIG 5.
Defects of genomic organization alter local chromatin environment. (A) 3D genome modeling of WT, Pfhmgb1-KO, and Pfhmgb1-RC clones. Centromeres are indicated with red spheres and telomeres with blue spheres (See Video S1 for panoramic view). (B) Statistical charts for 3D distance among centromeres (left) and telomeres (right) in different clones. Lines and error bars represent medians with 95% CIs. Wilcoxon test, ***, P ≤ 0.001; *, P ≤ 0.05; NS, not significant. (C) H3K9ac and H3K9me3 ChIP signal at upregulated gene loci and downregulated gene loci in Pfhmgb1-KO versus WT strains at ring. P values are for the Wilcoxon test. (D) Interaction frequencies of the telomere cluster with var0412700, which is originally actively transcribed in WT clone (left), or with var0712600, which is activated in the Pfhmgb1-RC clone (right). Lines and error bars represent medians with 95% CIs. Wilcoxon test, *, P ≤ 0.05.
Video for 3D model in WT, Pfhmgb1-KO, and Pfhmgb1-RC clones at ring with various genomic features. Chromosomes are indicated with white ribbons. Centromeres are indicated with red spheres and telomeres with blue spheres. Download Video S1, MOV file, 2.1 MB (2.1MB, mov) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Defect of nuclear architecture alters local chromatin environment.
Recently, by 3D genome modeling analysis, nuclear gene expression has been assumed to occur in a gradient of increasing transcriptional activity from the transcriptionally repressive heterochromatic telomeres to the opposite side, and the genes nearest to the centroid of telomeres exhibit the lowest expression levels in P. falciparum and other apicomplexan parasites (22, 23). Similarly, we observed the same gene expression pattern in WT parasites, and the global profile remained largely unchanged upon Pfhmgb1 KO, which is consistent with the observation that the development of asexual-stage parasites was not significantly arrested in Pfhmgb1-KO parasites. Interestingly, we observed variations of this expression pattern at some local genomic locations in the Pfhmgb1-KO line compared to that in the WT line (see Fig. S7A), which may correspond to those dysregulated genes upon Pfhmgb1 KO (Fig. S3B). The differential transcriptome analysis has shown that variant genes, including most “silent” members, were also downregulated upon the loss of PfHMGB1 (Fig. S3D and E). Nevertheless, the HMGB1-bound genes did not correlate with their expression (Fig. 2C). Collectively, these data are supportive of the speculation that the loss of PfHMGB1 releases the centromere superdomain, which, to some extent, disrupts the strictly organized centromere/telomere-dependent nuclear architecture. Such a defect may trigger alteration of local chromatin environment and gene expression, particularly for those genes which are highly dependent on the local chromatin structure, i.e., variant genes (13, 14).
PfHMGB1 knockout induces local histone modifications alteration in the 3D genome. Relation between gene expression (A) and histone modification (HM) level of H3K9me3 (B) or H3K9ac (C), and distance from the centroid of the telomeres (D). Windows were divided into 30 bins (for more information, see Text S1). For each bin, the median value is plotted as a line plot, and bar plots display gene expression values or HM signals within each bin. (C) IGV snapshot shows dynamic of histone modifications (H3K9ac and H3K9me3) at the surrounding loci of var0412700. Yellow box indicates promoter region of var0412700. Download FIG S7, TIF file, 0.6 MB (626.5KB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To address whether the different transcriptomes of Pfhmgb1-KO parasites are the result of histone modification (HM) alteration upon the loss of PfHMGB1, we first examined the changes of HM signals at a three-dimensional organization. Similar to our result of gene expression, both H3K9ac and H3K9me3 remained largely at the same distribution profile along the centroid of telomeres upon Pfhmgb1 KO (Fig. S7B and C). However, there were variations in HM signal at some local genomic regions. Based on this result, we then examined the changes in HM signal at the promoter regions of those dysregulated genes. The upregulated genes displayed a significant reduction in H3K9me3 enrichment at promoter regions, whereas the downregulated genes showed a significant decrease in H3K9ac signals at the promoter region in the Pfhmgb1-KO clone (Fig. 5C). For instance, the H3K9ac level was dramatically downregulated at the upstream region of the var0412700 gene locus in the Pfhmgb1-KO clone (Fig. S7D). These results suggest that the chromatin microenvironment accounted for the changes of transcriptional activities of these genes, while the global histone modification profile remained largely unchanged. Consistently, this originally expressed var gene in the WT 3D7-G7 clone exhibited elevated interactions with the telomere cluster when it was silenced in the Pfhmgb1-KO clone (Fig. 5D, left). On the contrary, the newly expressed var gene (var0712600) in the Pfhmgb1-RC clone exhibited a significant reduction in interactions with the telomere cluster compared to that in the Pfhmgb1-KO clone (Fig. 5D, right).
DISCUSSION
In addition to those general transcriptional factors such as the TATA-binding protein (TBP) and RNA polymerase-associated TFs, there are two groups of DNA-interaction factors associated with either specific DNA sequences or DNA structures involved in transcriptional regulation in eukaryotic organisms (40, 41). In the human malaria parasites, few DNA-binding TFs have been identified so far (35, 42). The major class of transcriptional regulators is the apicomplexan AP2 (ApiAP2) protein family. Among these, a small number of ApiAP2 factors (PfSIP2, AP2-exp, and AP2-Tel) have been shown to be involved in the formation of telomeric heterochromatin structures or in the mutually exclusive expression of variant genes (36). Here, we describe another layer in the complex epigenetic mechanism of variant gene expression in addition to histone modification-based local chromatin alteration. PfHMGB1 is, to our knowledge, the first architectural factor regulating virulence gene expression identified and offers mechanistic insight to the biological role of the 3D genome structure of the malaria parasite described recently.
In yeast and metazoan cells, the high-mobility-group (HMG) superfamily is a class of abundant nonhistone proteins involved in gene regulation via interaction with the AT hook (HMGA subgroup), with the nucleosomes (HMGN subgroup), or with HMG box DNA-binding domains (HMGB subgroup) (43). These HMG proteins are able to regulate gene transcription directly by binding to cruciform or distorted DNA sequences as DNA chaperones and through the induction of DNA bending, thereby influencing the accessibility of nucleosomes by TFs (44). The orthologue of HMGB1 in yeast, Nhp6A, affects nucleosome dynamics and gene transcription by binding to the promoters of discrete gene groups. Moreover, it has recently been reported that HMGB2 regulates an early event on the path to replicative senescence by binding chromatin at TAD boundaries to control heterochromatic and transcriptional remodeling in primary human cells, revealing another pathway by which 3D nuclear organization regulates gene expression (27). However, although we have identified a panel of PfHMGB1-binding target genes at their upstream regions in P. falciparum, the transcriptional activities of these genes are not directly controlled by this factor. A recent study also shows that PfHMGB1 is not present in the protein complex binding to var gene loci (45). Therefore, differential expression of those genes in the Pfhmgb1-KO line is a consequence of nuclear reorganization. Most of them belong to heterochromatic genes and experience local chromatin alteration at their promoter regions. These data strongly suggest that PfHMGB1 is an architectural factor involved in the dynamics of nuclear structure instead of a transcription factor.
Other DNA-binding factors such as CenH3 may cooperate with PfHMGB1 to regulate this centromere structure. It is also possible that HP1 and other telomere-associated factors such as PfAP2tel, PfSIP2, and PfTRZ may contribute to the formation of the heterochromatic telomeric superdomain (10, 36, 46). A coordinated regulatory network is required to secure the nuclear organization. Although the two superdomains occupy opposite zones of the nuclear periphery, reorganization of centromere clusters can trigger the spread of the heterochromatin environment. Previously, an opposite phenomenon was observed in studies of the deacetylase PfSir2 protein. Knockout of this protein results in chromatin alteration at the telomere-proximal regions, i.e., the spread of histone acetylation modification, and repositioning of var gene loci, thereby activating multiple subtelomere virulence genes (7, 8). In this case, PfSir2 silences virulence genes via direct binding to the subtelomeric regions. It maintains the gradated chromatin structure at the telomere-associated regions as a boundary to acetylation modification. PfHMGB1 is involved in virulence gene expression in a distinct way. In this case, dynamic chromosome organization and chromatin structure are highly associated and coregulated, which secures the strict singular expression of virulence genes. This may partially explain the conversion of the activated var gene in the Pfhmgb1-RC clone. The highly organized genomic structure provides a supportive environment for var gene expression, whose disruption silences the var gene family upon Pfhmgb1 KO. However, activation of the exact var gene requires a more-specific regulating mechanism such as Ruf6 or PfSir2, etc., which shall be further investigated. In addition, PfHMGB1 functions in gene regulation as an architectural regulator and is mainly associated with virulence genes. This gene expression-related regulatory pathway is not essential for parasite development but is critical for immune evasion in the human host.
We used the 3D7-G7 clone to generate the Pfhmgb1 knockout parasite. This clone expresses relatively stable central var genes. This raises the question regarding whether all active var genes in other parasite line clones maintain their subnuclear position at the transition boundary between the centromere and telomere clusters. While upsC- and upsBC-type var genes are located in the central regions of chromosomes, upsA- and upsB-type var genes, which constitute more than half of the var gene family, are located in subtelomeric regions. One clue comes from the PfHMGB1-complementary line (Pfhmgb1-RC) in which the newly activated var gene is still located at the central site of another chromosome. It is of interest, therefore, to consider how these var genes are released from the transcriptionally repressive telomere superdomain through chromosome reorganization. One possible explanation is that chromosomally central var genes may be preferentially activated, as their chromosomal positions confer them priority for nuclear reposition compared with those of telomere-associated members. This hypothesis is perhaps supported by the observation that chromosomally central var genes are more likely to be activated and undergo lower switching rates in cultures ex vivo and in vitro (47, 48).
MATERIALS AND METHODS
Parasite cultivation and transfection.
P. falciparum parasites were cultured as described previously (18). The genetic manipulation (knock-in or knockout) of Pfhmgb1 or PfCenh3 genes was achieved using the CRISPR-Cas9 gene editing system (31) and was validated by PCR, sequencing, Western blotting, and IFA. For tagging, the HA-Ty1 or GFP sequences were fused to the C terminus of Pfhmgb1 or N terminus of PfCenh3, respectively. The resulting vectors were transfected into the 3D7-G7 parasites as described previously (49). All the nucleotide sequences of sgRNAs or primers are listed in Table S1A in the supplemental material.
Flow cytometry.
Ring-stage parasites were diluted to 0.5% parasitemia for sampling after every 4 h in culture. DNA content analysis was carried out for three intraerythrocytic developmental cycles. Samples (10 μl) were fixed in 4% formaldehyde-0.015% glutaraldehyde for 20 min. The nuclei were labeled by using Hoechst 33342 (Invitrogen, H3570) for 30 min. At least 500,000 cells were gated using forward- and side-scatter parameters, and then a second gate was implemented to select infected cells, according to Hoechst-positive cells. Parasites in different phases were distinguished on the basis of fluorescence intensity. Fluorescence signal data were obtained with a flow cytometer (BD FACS AriaII) and analyzed by using FlowJo software version 10.2.
Western blotting.
RBC-free parasites were isolated with 0.15% saponin treatment and resuspended in 1× SDS-loading buffer (Bio-Rad) for protein extracts. Proteins were separated by gel electrophoresis, transferred to a polyvinylidene difluoride (PVDF) membrane, and visualized by exposure to an imaging device. The antibodies used in this study were mouse anti-Pfaldolase (Abcam), mouse anti-Ty1 (Sigma), rabbit anti-HA (Abcam), mouse anti-GFP (Abcam) antibodies. Membranes were developed with an enhanced chemiluminescence (ECL) Western blot kit (GE health care).
Immunofluorescence assays.
Immunofluorescence assays were performed as described previously (18). Parasites at different stages were fixed with 4% paraformaldehyde and deposited on microscope slides. The dilution for the primary mouse anti-Ty1 antibody was 1:1,000 and for rabbit anti-GFP was 1:5,000, and the second antibodies of Alexa-Fluor-488-conjugated anti-mouse and Alexa-Fluor-568-conjugated anti-rabbit were diluted by 1:1,000. Images were captured by using a Nikon A1R microscope at ×100 magnification.
Quantitative reverse transcription-PCR.
cDNA was synthesized with random hexamer primers according to the manufacturer’s instructions (TaKaRa, number 2641A). Primer sequences for var genes were designed as in previous methods (50). PCR conditions were initial denaturing at 95°C for 30 s, followed by 40 cycles of 5 s at 95°C, 20 s at 54°C, 7 s at 56°C, 7 s at 59°C, and 27 s at 62°C. The seryl-tRNA synthetase gene (PF3D7_0717700) was used as an internal control.
High-throughput sequencing-associated analysis.
The technical details of ChIP-seq, RNA-seq, and Hi-C-seq assays and related detailed bioinformatics analysis are available in Text S1.
Detailed materials and methods for ChIP-seq, RNA-seq, and Hi-C-seq assays and related detailed bioinformatics analysis. Download Text S1, DOCX file, 0.04 MB (41.9KB, docx) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data availability.
All raw sequence data reported in this paper have been deposited in Gene Expression Omnibus (GEO) and are publicly accessible under GEO accession GSE141762.
ACKNOWLEDGMENTS
We thank Artur Scherf (Institut Pasteur, Paris) for technical and material support.
This work was supported by funding from the National Natural Science Foundation of China (NSFC; 81630063 and 81971959) and National Key R&D Program of China (2018YFA0507300) to Q.Z., NSFC (31972882, 31771419, and 31721003) to C.J., NSFC (31771431) to G.W.
Q.Z. and G.W. conceived and designed experiments. B.L., F.W., and Y.Z. generated transgenic parasite lines. B.L., G.G., X.H., and C.J. performed transcriptome analysis, IFA, and co-immunoprecipitation assays. B.L., and X.S performed ChIP-seq assay. B.L., Y.L., and G.W. performed the Hi-C assay. M.L., S.S., L.W., G.Y., L.G., and Q.Z. performed all the bioinformatics analyses. Q.Z. and G.W. wrote the manuscript with contributions from M.L., B.L., R.C., and J.C.
We declare no competing interests.
Footnotes
Citation Lu B, Liu M, Gu L, Li Y, Shen S, Guo G, Wang F, He X, Zhao Y, Shang X, Wang L, Yang G, Zhu Q, Cao J, Jiang C, Culleton R, Wei G, Zhang Q. 2021. The architectural factor HMGB1 is involved in genome organization in the human malaria parasite Plasmodium falciparum. mBio 12:e00148-21. https://doi.org/10.1128/mBio.00148-21.
REFERENCES
- 1.Deitsch KW, Dzikowski R. 2017. Variant gene expression and antigenic variation by malaria parasites. Annu Rev Microbiol 71:625–641. doi: 10.1146/annurev-micro-090816-093841. [DOI] [PubMed] [Google Scholar]
- 2.Avril M, Tripathi AK, Brazier AJ, Andisi C, Janes JH, Soma VL, Sullivan DJ, Jr, Bull PC, Stins MF, Smith JD. 2012. A restricted subset of var genes mediates adherence of Plasmodium falciparum-infected erythrocytes to brain endothelial cells. Proc Natl Acad Sci U S A 109:E1782–E1790. doi: 10.1073/pnas.1120534109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner L, Lavstsen T, Berger SS, Wang CW, Petersen JEV, Avril M, Brazier AJ, Freeth J, Jespersen JS, Nielsen MA, Magistrado P, Lusingu J, Smith JD, Higgins MK, Theander TG. 2013. Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature 498:502–505. doi: 10.1038/nature12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, Fernandez V, Sundstrom A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. 1998. Developmental selection of var gene expression in Plasmodium falciparum. Nature 394:392–395. doi: 10.1038/28660. [DOI] [PubMed] [Google Scholar]
- 5.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J 17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duraisingh MT, Skillman KM. 2018. Epigenetic variation and regulation in malaria parasites. Annu Rev Microbiol 72:355–375. doi: 10.1146/annurev-micro-090817-062722. [DOI] [PubMed] [Google Scholar]
- 7.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. 2005. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. 2005. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 9.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. 2009. Genome-wide Analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe 5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Brancucci NMB, Bertschi NL, Zhu L, Niederwieser I, Chin WH, Wampfler R, Freymond C, Rottmann M, Felger I, Bozdech Z, Voss TS. 2014. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe 16:165–176. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Fraschka SA, Filarsky M, Hoo R, Niederwieser I, Yam XY, Brancucci NMB, Mohring F, Mushunje AT, Huang XM, Christensen PR, Nosten F, Bozdech Z, Russell B, Moon RW, Marti M, Preiser PR, Bartfai R, Voss TS. 2018. Comparative heterochromatin profiling reveals conserved and unique epigenome signatures linked to adaptation and development of malaria parasites. Cell Host Microbe 23:407.e8–420.e8. doi: 10.1016/j.chom.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartfai R, Hoeijmakers WAM, Salcedo-Amaya AM, Smits AH, Janssen-Megens E, Kaan A, Treeck M, Gilberger TW, Francoijs KJ, Stunnenberg HG. 2010. H2A.Z demarcates intergenic regions of the Plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog 6:e1001223. doi: 10.1371/journal.ppat.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss TS, Tonkin CJ, Marty AJ, Thompson JK, Healer J, Crabb BS, Cowman AF. 2007. Alterations in local chromatin environment are involved in silencing and activation of subtelomeric var genes in Plasmodium falciparum. Mol Microbiol 66:139–150. doi: 10.1111/j.1365-2958.2007.05899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan YT, Shen SJ, Wei GY, Tang JX, Zhao YM, Wang F, He XH, Guo GQ, Shang XM, Yu XY, Ma ZL, He XQ, Liu M, Zhu QS, Le Z, Wei G, Cao J, Jiang CZ, Zhang QF. 2020. Rrp6 regulates heterochromatic gene silencing via ncRNA RUF6 decay in malaria parasites. mBio 11:e01110-20. doi: 10.1128/mBio.01110-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martins RM, Macpherson CR, Claes A, Scheidig-Benatar C, Sakamoto H, Yam XY, Preiser P, Goel S, Wahlgren M, Sismeiro O, Coppee JY, Scherf A. 2017. An ApiAP2 member regulates expression of clonally variant genes of the human malaria parasite Plasmodium falciparum. Sci Rep 7:14042. doi: 10.1038/s41598-017-12578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amit-Avraham I, Pozner G, Eshar S, Fastman Y, Kolevzon N, Yavin E, Dzikowski R. 2015. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc Natl Acad Sci U S A 112:E982–E991. doi: 10.1073/pnas.1420855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barcons-Simon A, Cordon-Obras C, Guizetti J, Bryant JM, Scherf A. 2020. CRISPR interference of a clonally variant GC-Rich noncoding RNA family leads to general repression of var genes in Plasmodium falciparum. mBio 11:e03054-19. doi: 10.1128/mBio.03054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q, Siegel TN, Martins RM, Wang F, Cao J, Gao Q, Cheng X, Jiang L, Hon CC, Scheidig-Benatar C, Sakamoto H, Turner L, Jensen AT, Claes A, Guizetti J, Malmquist NA, Scherf A. 2014. Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature 513:431–435. doi: 10.1038/nature13468. [DOI] [PubMed] [Google Scholar]
- 19.Ralph SA, Scheidig-Benatar C, Scherf A. 2005. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci U S A 102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. 2006. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 439:1004–1008. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- 21.Batugedara G, Lu XM, Bunnik EM, Le Roch KG. 2017. The role of chromatin structure in gene regulation of the human malaria parasite. Trends Parasitol 33:364–377. doi: 10.1016/j.pt.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bunnik EM, Venkat A, Shao J, McGovern KE, Batugedara G, Worth D, Prudhomme J, Lapp SA, Andolina C, Ross LS, Lawres L, Brady D, Sinnis P, Nosten F, Fidock DA, Wilson EH, Tewari R, Galinski MR, Ben Mamoun C, Ay F, Le Roch KG. 2019. Comparative 3D genome organization in apicomplexan parasites. Proc Natl Acad Sci U S A 116:3183–3192. doi: 10.1073/pnas.1810815116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunnik EM, Cook KB, Varoquaux N, Batugedara G, Prudhomme J, Cort A, Shi L, Andolina C, Ross LS, Brady D, Fidock DA, Nosten F, Tewari R, Sinnis P, Ay F, Vert JP, Noble WS, Le Roch KG. 2018. Changes in genome organization of parasite-specific gene families during the Plasmodium transmission stages. Nat Commun 9:1910. doi: 10.1038/s41467-018-04295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claessens A, Harris LM, Stanojcic S, Chappell L, Stanton A, Kuk N, Veneziano-Broccia P, Sterkers Y, Rayner JC, Merrick CJ. 2018. RecQ helicases in the malaria parasite Plasmodium falciparum affect genome stability, gene expression patterns and DNA replication dynamics. PLoS Genet 14:e1007490. doi: 10.1371/journal.pgen.1007490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Yin SG, Sun MX, Cheng X, Wei JQ, Gilbert N, Miao J, Cui LW, Huang ZG, Dai XY, Jiang LB. 2019. DNA helicase RecQ1 regulates mutually exclusive expression of virulence genes in Plasmodium falciparum via heterochromatin alteration. Proc Natl Acad Sci U S A 116:3177–3182. doi: 10.1073/pnas.1811766116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowell NL, Sperling AS, Mason MJ, Johnson RC. 2010. Chromatin-dependent binding of the S. cerevisiae HMGB protein Nhp6A affects nucleosome dynamics and transcription. Genes Dev 24:2031–2042. doi: 10.1101/gad.1948910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, Drechsel O, Becker C, Altmuller J, Josipovic N, Georgomanolis T, Brant L, Franzen J, Koker M, Gusmao EG, Costa IG, Ullrich RT, Wagner W, Roukos V, Nurnberg P, Marenduzzo D, Rippe K, Papantonis A. 2018. HMGB2 loss upon senescence entry disrupts genomic organization and induces CTCF clustering across cell types. Mol Cell 70:730.e6–744.e6. doi: 10.1016/j.molcel.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 28.McCauley MJ, Huo R, Becker N, Holte MN, Muthurajan UM, Rouzina I, Luger K, Maher LJ, Israeloff NE, Williams MC. 2019. Single and double box HMGB proteins differentially destabilize nucleosomes. Nucleic Acids Res 47:666–678. doi: 10.1093/nar/gky1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briquet S, Boschet C, Gissot M, Tissandie E, Sevilla E, Franetich JF, Thiery I, Hamid Z, Bourgouin C, Vaquero C. 2006. High-mobility-group box nuclear factors of Plasmodium falciparum. Eukaryot Cell 5:672–682. doi: 10.1128/EC.5.4.672-682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy FV, Sweet RM, Churchill MEA. 1999. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J 18:6610–6618. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. 2014. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 32.Redmond AM, Byrne C, Bane FT, Brown GD, Tibbitts P, O'Brien K, Hill ADK, Carroll JS, Young LS. 2015. Genomic interaction between ER and HMGB2 identifies DDX18 as a novel driver of endocrine resistance in breast cancer cells. Oncogene 34:3871–3880. doi: 10.1038/onc.2014.323. [DOI] [PubMed] [Google Scholar]
- 33.Noma KI, Cam HP, Maraia RJ, Grewal SIS. 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 34.Hoeijmakers WAM, Flueck C, Francoijs KJ, Smits AH, Wetzel J, Volz JC, Cowman AF, Voss T, Stunnenberg HG, Bartfai R. 2012. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol 14:1391–1401. doi: 10.1111/j.1462-5822.2012.01803.x. [DOI] [PubMed] [Google Scholar]
- 35.Buendia-Orozco J, Guerrero A, Pastor N. 2005. Model of the TBP-TFIIB complex from Plasmodium falciparum: interface analysis and perspectives as a new target for antimalarial design. Arch Med Res 36:317–330. doi: 10.1016/j.arcmed.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 36.Jeninga MD, Quinn JE, Petter M. 2019. ApiAP2 transcription factors in apicomplexan parasites. Pathogens 8:47. doi: 10.3390/pathogens8020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verma G, Surolia N. 2013. Plasmodium falciparum CENH3 is able to functionally complement Cse4p and its, C-terminus is essential for centromere function. Mol Biochem Parasitol 192:21–29. doi: 10.1016/j.molbiopara.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Dong QH, Yin FX, Gao F, Shen Y, Zhang FB, Li Y, He HJ, Gonzalez M, Yang JP, Zhang S, Su M, Chen YH, Li F. 2016. Ccp1 homodimer mediates chromatin integrity by antagonizing CENP-A loading. Mol Cell 64:79–91. doi: 10.1016/j.molcel.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustin M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol 19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas MC, Chiang CM. 2006. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol 41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 41.Thapar R. 2015. Structure-specific nucleic acid recognition by L-motifs and their diverse roles in expression and regulation of the genome. Biochim Biophys Acta 1849:677–687. doi: 10.1016/j.bbagrm.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gissot M, Briquet S, Refour P, Boschet C, Vaquero C. 2005. PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J Mol Biol 346:29–42. doi: 10.1016/j.jmb.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 43.Stros M. 2010. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 1799:101–113. doi: 10.1016/j.bbagrm.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Reeves R. 2015. High mobility group (HMG) proteins: modulators of chromatin structure and DNA repair in mammalian cells. DNA Repair (Amst) 36:122–136. doi: 10.1016/j.dnarep.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 45.Bryant JM, Baumgarten S, Dingli F, Loew D, Sinha A, Claes A, Preiser PR, Dedon PC, Scherf A. 2020. Exploring the virulence gene interactome with CRISPR/dCas9 in the human malaria parasite. Mol Syst Biol 16:e9569. doi: 10.15252/msb.20209569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bertschi NL, Toenhake CG, Zou A, Niederwieser I, Henderson R, Moes S, Jenoe P, Parkinson J, Bartfai R, Voss TS. 2017. Malaria parasites possess a telomere repeat-binding protein that shares ancestry with transcription factor IIIA. Nat Microbiol 2:17033. doi: 10.1038/nmicrobiol.2017.33. [DOI] [PubMed] [Google Scholar]
- 47.Zhang QF, Zhang YL, Huang YF, Xue XY, Yan H, Sun XD, Wang J, McCutchan TF, Pan WQ. 2011. From in vivo to in vitro: dynamic analysis of Plasmodium falciparum var gene expression patterns of patient isolates during adaptation to culture. PLoS One 6:e20591. doi: 10.1371/journal.pone.0020591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank M, Dzikowski R, Amulic B, Deitsch K. 2007. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol 64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fidock DA, Wellems TE. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A 94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salanti A, Staalsoe T, Lavstsen T, Jensen ATR, Sowa MPK, Arnot DE, Hviid L, Theander TG. 2003. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol 49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 51.Lopez-Barragan MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ. 2011. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evolutionally conserved HMGB1 family in eukaryotes. (A) Phylogenetic tree of the HMGB family in various organisms. (B) Multiple alignment of HMG box domains of HMG proteins in diverse species. Dashes represent gaps and missing sequences. The bar plot (bottom) indicates the level of conservation for each amino acid. The amino acids were colored according to default ClustalX (v2.1) color palette. (C) Modeling of crystal structure of the HMGB1 proteins of human (GenBank CAG33144.1), yeast (GenBank KZV07568.1), and P. falciparum (GenBank CZT99193.1). The residues associated with structure- or sequence-specific recognition of HMGB1-DNA are indicated individually corresponding to Fig. 1A. The PDB files of each protein sequence were produced by SWISS-MODEL online (https://swissmodel.expasy.org/) and were viewed and modified in PyMol. Download FIG S1, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of Pfhmgb1-KO line. (A) Schematic representation of generation of Pfhmgb1-KO transfectant line by CRISPR-Cas9 technique. (B) Nucleotide sequencing of the mutation and reverse mutation events in Pfhmgb1-KO and Pfhmgb1-RC lines. (C) Growth curves of synchronized parasites of Pfhmgb1-KO and WT control. Error bars represent SEMs from three biological replicates. (D) Flow cytometry analysis of the blood-stage growth of Pfhmgb1-KO and WT lines in parallel. Download FIG S2, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of oligonucleotide sequences and RNA-seq, ChIP-seq, and Hi-C-seq analyses. Download Table S1, XLSX file, 0.06 MB (60.3KB, xlsx) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pfhmgb1 knockout silenced variant genes. (A) Correlation of transcript abundance in samples under different conditions. (B) Comparative transcriptome analysis between Pfhmgb1-KO (y axis) and WT (x axis) clones at different stages. All the significantly up- and downregulated genes (≥3-fold change) are indicated with red and green circles, respectively. Dashed lines indicate the fold change cutoff. (C) Gene function classification of upregulated and downregulated genes in Pfhmgb1-KO versus WT. The numbers of genes in each category are indicated in the figure. (D) Transcriptional level of var genes in Pfhmgb1-KO, WT 3D7-G7, and transfection control clones measured by RNA-seq assays. Error bars represent SEMs for two biological replicates. (E) Transcription levels of variant genes, including rifin, stevor, and Pfmc-2tm in Pfhmgb1-KO and WT clones at ring measured by RNA-seq assays. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Recovery of gene expression in the Pfhmgb1-RC clone. (A) Schematic representation of generation of Pfhmgb1-RC transgenic parasites by CRISPR-Cas9 technique. (B) Correlation of global gene expression between two biological replicates of Pfhmgb1-RC clones at ring. (C) Comparative transcriptome analysis of WT and Pfhmgb1-RC clones by RNA-seq at ring. (D) Boxplots show transcript abundance of dysregulated genes upon Pfhmgb1-KO in WT, Pfhmgb1-KO, and Pfhmgb1-RC clones. Lines and error bars represent medians with 95% CIs. Wilcoxon test, ***, P ≤ 0.001; **, P ≤ 0.01. (E) Transcriptional level of var genes in Pfhmgb1-KO and Pfhmgb1-RC clones measured by RNA-seq assays. Error bars represent SEMs for two biological replicates. Download FIG S4, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reproducibility of PfHMGB1 ChIP-seq experiments (A) Schematic representation of generation of Pfhmgb1-GFP transgenic parasites by CRISPR-Cas9 modification. (B) Correlation analysis for biological replicates of PfHMGB1 ChIP-seq data. Download FIG S5, TIF file, 0.2 MB (245KB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quality control of Hi-C-seq data between replicates and clones. (A) The correlation between replicates by HiCExplorer are shown for WT, Pfhmgb1-KO, and Pfhmgb1-RC lines in ring-stage parasites. (B) Significant interactions among centromeres (left) or telomeres (right) in WT and vector control clones, respectively. Red lines represent interactions among genomic features. There is no significant change in interactions among centromeres or telomeres between WT and vector control strains. Download FIG S6, TIF file, 0.9 MB (897.4KB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Video for 3D model in WT, Pfhmgb1-KO, and Pfhmgb1-RC clones at ring with various genomic features. Chromosomes are indicated with white ribbons. Centromeres are indicated with red spheres and telomeres with blue spheres. Download Video S1, MOV file, 2.1 MB (2.1MB, mov) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PfHMGB1 knockout induces local histone modifications alteration in the 3D genome. Relation between gene expression (A) and histone modification (HM) level of H3K9me3 (B) or H3K9ac (C), and distance from the centroid of the telomeres (D). Windows were divided into 30 bins (for more information, see Text S1). For each bin, the median value is plotted as a line plot, and bar plots display gene expression values or HM signals within each bin. (C) IGV snapshot shows dynamic of histone modifications (H3K9ac and H3K9me3) at the surrounding loci of var0412700. Yellow box indicates promoter region of var0412700. Download FIG S7, TIF file, 0.6 MB (626.5KB, tif) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detailed materials and methods for ChIP-seq, RNA-seq, and Hi-C-seq assays and related detailed bioinformatics analysis. Download Text S1, DOCX file, 0.04 MB (41.9KB, docx) .
Copyright © 2021 Lu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All raw sequence data reported in this paper have been deposited in Gene Expression Omnibus (GEO) and are publicly accessible under GEO accession GSE141762.