Dichloroacetate (DCA) is ubiquitous in the environment due to natural formation via biological and abiotic chlorination processes and the turnover of chlorinated organic materials (e.g., humic substances). Additional sources include DCA usage as a chemical feedstock and cancer drug and its unintentional formation during drinking water disinfection by chlorination.
KEYWORDS: dichloroacetate, haloacid dehalogenase, fermentation, comparative proteomics, anaerobic catabolic pathways
ABSTRACT
Dichloroacetate (DCA) commonly occurs in the environment due to natural production and anthropogenic releases, but its fate under anoxic conditions is uncertain. Mixed culture RM comprising “Candidatus Dichloromethanomonas elyunquensis” strain RM utilizes DCA as an energy source, and the transient formation of formate, H2, and carbon monoxide (CO) was observed during growth. Only about half of the DCA was recovered as acetate, suggesting a fermentative catabolic route rather than a reductive dechlorination pathway. Sequencing of 16S rRNA gene amplicons and 16S rRNA gene-targeted quantitative real-time PCR (qPCR) implicated “Candidatus Dichloromethanomonas elyunquensis” strain RM in DCA degradation. An (S)-2-haloacid dehalogenase (HAD) encoded on the genome of strain RM was heterologously expressed, and the purified HAD demonstrated the cofactor-independent stoichiometric conversion of DCA to glyoxylate at a rate of 90 ± 4.6 nkat mg−1 protein. Differential protein expression analysis identified enzymes catalyzing the conversion of DCA to acetyl coenzyme A (acetyl-CoA) via glyoxylate as well as enzymes of the Wood-Ljungdahl pathway. Glyoxylate carboligase, which catalyzes the condensation of two molecules of glyoxylate to form tartronate semialdehyde, was highly abundant in DCA-grown cells. The physiological, biochemical, and proteogenomic data demonstrate the involvement of an HAD and the Wood-Ljungdahl pathway in the anaerobic fermentation of DCA, which has implications for DCA turnover in natural and engineered environments, as well as the metabolism of the cancer drug DCA by gut microbiota.
INTRODUCTION
Dichloroacetate (CHCl2-COO– [DCA]) is a naturally occurring compound produced through both biological and geochemical processes (1, 2). Marine algae, such as Asparagopsis spp., produce DCA, and algal blooms form extensive halogenated dissolved organic matter (chlorine- and iodine-containing metabolites) (3–5). Enzymatic chlorination (e.g., chloroperoxidases) results in substantial chlorination of decaying plant and humic materials leading to the formation of DCA (6–8). Reactive chlorine species (e.g., chlorine radicals generated in photochemical reactions) contribute to organic matter chlorination producing chloroacetates (9, 10). Photochemical degradation of chlorinated hydrocarbons generates DCA, and 1 to 5 μg liter−1 DCA has been detected in fog water and rainwater samples (11, 12). Detection of DCA in pristine Antarctic firn is seen as evidence for its natural formation (13, 14). DCA also has various anthropogenic sources, foremost as a consequence of drinking water sanitation. DCA is a common disinfection by-product of water chlorination and occurs broadly in drinking water systems with concentrations reported in the low μg liter−1 to hundreds of μg liter−1 range (15–17). DCA can also be detected in both bottled and tap water at low μg liter−1 levels (18–21). Swimming pool waters treated with chlorine contain DCA, and concentrations reaching 250 μg liter−1 have been reported (22). Its use as a therapeutic for a variety of diseases, including cancers and lactic acidosis (23–25), has triggered intense scrutiny by clinical scientists for decades, resulting in rigorous pharmacokinetic, biotransformation, and toxicological studies (26–28). Despite its therapeutic use, DCA is considered a hazardous chemical with cytotoxic and genotoxic effects (29), and it has been classified as an environmental pollutant (30).
In mammalian liver cells, glutathione S-transferase (GST) zeta 1 is the primary cytosolic enzyme that transforms DCA to glyoxylate, which is subsequently metabolized via the glyoxylate shunt pathway (31, 32). A novel rho (ρ) class of GST enzymes that catalyze the dehalogenation of DCA to glyoxylate has recently been identified and characterized in the cyanobacterium Synechocystis sp. strain PCC 6803 (33). The majority of aerobic bacteria, however, employ distinct enzymes belonging to the group of haloacid dehalogenases (HADs) to convert DCA to glyoxylate via hydrolytic dehalogenation (34–38). Aerobic bacterial degradation of DCA has been studied (39–41); however, the fate of DCA in anoxic environments and anaerobic microbial metabolism of DCA have remained elusive.
A microbial mixed culture, designated culture RM, was derived from pristine freshwater sediment enriched with dichloromethane (CH2Cl2 [DCM]) as the sole energy source under anoxic conditions (42). Acetate and methane were the final products, and H2 was an intermediate during DCM degradation (42–44). Phylogenetic, genomic, and physiological characterization identified the DCM degrader as “Candidatus Dichloromethanomonas elyunquensis” strain RM, representing a new genus and species affiliated with the Peptococcaceae family (45). Growth of strain RM was strictly dependent on DCM. Other chlorinated solvents, including chloroform, tetrachloroethene, trichloroethene, cis-1,2-dichloroethene, 1,1,1-trichloroethane, and 1,1-dichloroethane, did not support growth of strain RM (42, 43). The analysis of the metagenome-assembled genome (MAG) of strain RM identified two putative HADs (46, 47), which triggered the search for additional substrates, specifically chlorinated acetates, that could support growth of strain RM. Here, we report the utilization of DCA as a substrate supporting growth of strain RM, identify a novel HAD that enables the organism to convert DCA to glyoxylate via a glutathione-independent mechanism, and characterize the DCA catabolic pathway. Instead of utilizing DCA as an electron acceptor for reductive dehalogenation (i.e., organohalide respiration), strain RM employs a hydrolytic dechlorination mechanism and ferments DCA to acetate, CO2, and H2. The new findings advance understanding of the fate of DCA under anoxic conditions and have implications for the flow of carbon and electrons in electron acceptor-depleted environments and the human gut.
RESULTS
Dichloroacetate utilization by mixed culture RM.
When RM cultures that had completely consumed DCM were challenged with 2.5 mM DCA, DCA utilization commenced after a lag phase of about 3 weeks, and consumption was complete within 2 weeks (Fig. 1A). In parallel incubations, DCM-grown cultures rapidly consumed additional DCM (∼150 μmol per bottle) within 1 to 2 days (Fig. 1A). DCA concentrations remained constant in heat-inactivated and no-inoculum controls. Incubations using chloride-free medium demonstrated that the consumption of 43 ± 4 μmol DCA resulted in concomitant formation of 90 ± 7 μmol chloride, indicating that both chlorine substituents were released during DCA catabolism by mixed culture RM (Fig. 1B). During DCA utilization, formate was transiently produced, and up to 7 ± 2 μmol per bottle was observed (Fig. 1B). The terminal product was acetate, and 22 ± 2 μmol—about half the amount of the added DCA (i.e., 43 ± 4 μmol)—was formed (Fig. 1B). In contrast to DCM-grown cultures, DCA-fed cultures did not produce methane. Following the consumption of 2 mM DCA, the cultures were visibly turbid, with optical density at 600 nm (OD600) values less than 0.1 optical density unit. Quantitative real-time PCR (qPCR) targeting the 16S rRNA gene of strain RM revealed 141 ± 32-fold increases of cell abundances from (1.04 ± 0.56) × 106 per ml (cells introduced with the inoculum) to (1.46 ± 0.23) × 108 per ml following DCA consumption (Fig. 1C), demonstrating DCA catabolism by strain RM. Monochloroacetate (CH2Cl-COO– [MCA]) was never detected in cultures growing with DCA, and MCA could not replace DCA as a growth substrate.
FIG 1.

DCA and DCM degradation by mixed culture RM. (A) Utilization of DCA in RM cultures that had consumed an initial feeding of 93 ± 15 μmol of DCM. DCA utilization commenced after a lag phase of about 3 weeks, and consumption was complete within 2 weeks. Replicate DCM-grown cultures rapidly consumed additional DCM feedings without a lag phase. (B) Formation of inorganic chloride, acetate, and formate during DCA catabolism by mixed culture RM. (C) Increase in 16S rRNA gene copies of “Ca. Dichloromethanomonas elyunquensis” during DCA catabolism by mixed culture RM under anoxic conditions. The data represent the average from triplicate incubations, and the error bars represent the standard deviations.
During growth with DCA (538.8 ± 4.8 μmol of DCA per bottle), H2 was intermittently produced, and a maximum amount of 7.00 ± 0.06 μmol of H2 was observed (Fig. 2). H2 was slowly consumed to a threshold concentration of 1,500 ± 180 ppmv corresponding to 3.75 ± 0.45 μmol H2 per bottle (Fig. 2). H2 was previously identified as an intermediate of DCM metabolism in culture RM, which supported growth of hydrogenotrophic methanogens (e.g., Methanospirillum spp.) and homoacetogens (e.g., Acetobacterium spp.) to produce methane and acetate, respectively (43, 44). In addition to H2, carbon monoxide (CO) was detected as a transient intermediate during DCA metabolism and increased from 0.03 ± 0.003 μmol to a maximum of 0.24 ± 0.01 μmol per bottle (Fig. 2). The transient formation of H2 and CO only occurred in live cultures amended with DCA (see Fig. S1 in the supplemental material). CO formation was not observed in DCM-grown cultures (Fig. S1).
FIG 2.
Transient formation of H2 and CO during DCA catabolism by mixed culture RM. The data represent the average from triplicate incubations, and the error bars represent the standard deviations.
Change of H2 and CO concentrations in abiotic and biotic control incubations and CO concentrations in RM cultures during growth with DCM. Download FIG S1, PDF file, 0.2 MB (207.6KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbial community response to enrichment with DCA.
16S rRNA gene amplicon sequencing revealed changes in microbial community structure in response to repeated transfers with DCA as the sole energy source. In the first transfer cultures with DCA, strain RM was the dominant population, accounting for approximately 65% of all sequences (Fig. 3; see Table S1 in the supplemental material). Following six consecutive transfers with DCA, the relative sequence abundance of amplicons representing strain RM increased to 87%, implying that strain RM is responsible for DCA degradation (Fig. 3). In DCM-grown cultures, Methanospirillum contributed about 2% to the total 16S rRNA gene amplicons (Fig. 3) and was implicated in methane formation (42, 43). Following repeated transfers in mineral salts medium with DCA, Methanospirillum sequences were no longer detected (Fig. 3), consistent with the loss of methane formation in cultures grown with DCA. Populations belonging to the genus Anaerolineae were also eliminated during repeated transfers with DCA (Fig. 3). The operational taxonomic units (OTUs) representing the genera Acetobacterium and Treponema, both known to comprise species capable of H2/CO2 reductive acetogenesis, were maintained at relative abundances of around 1% and 4%, respectively (Fig. 3). OTUs representing Bacteroidales were also maintained at a relative abundance of 4 to 5% during consecutive transfers with DCA. The abundances of OTUs representing Desulfovibrio, Dethiosulfovibrionaceae, Sulfuricurvum, and Veillonellaceae all declined after repeated transfers with DCA (Fig. 3).
FIG 3.
Microbial community structure responses to consecutive transfers of mixed culture RM with DCA as the sole energy source, as revealed by 16S rRNA gene amplicon sequencing. Taxa with relative abundances below 1% were categorized as “Others.” The operational taxonomic units (OTUs) representing bacteria and archaea are reported to the lowest taxonomic rank possible. “Ca. Dichloromethanomonas elyunquensis” was the dominant population in mixed culture RM, and continuous transfers with DCA resulted in further enrichment.
Taxonomy, relative abundance, and representative sequences of OTUs identified through 16S rRNA gene amplicon sequencing. Download Table S1, XLSX file, 0.01 MB (12.2KB, xlsx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Haloacid dehalogenases and DCA dehalogenation.
Examination of the genome of strain RM (46) revealed two genes (locus tags prokka_14346 and prokka_14344) encoding putative HADs (EC 3.8.1.2), designated HAD1 and HAD2, respectively. HAD1 and HAD2 shared 60.6% amino acid sequence identity with each other and clustered with biochemically characterized HADs from aerobic bacteria: e.g., Pseudomonas putida, Xanthobacter autotrophicus, Moraxella sp., and Burkholderia cepacia (34–36, 38) (Fig. 4).
FIG 4.
Amino acid sequence-based phylogenetic tree of select HADs. HAD1 (prokka_14346) and HAD2 (prokka_14344) of “Ca. Dichloromethanomonas elyunquensis” are shown in red font. Biochemically characterized HADs with demonstrated activity toward DCA are shown in blue font. The scale bar indicates the number of amino acid substitutions per site.
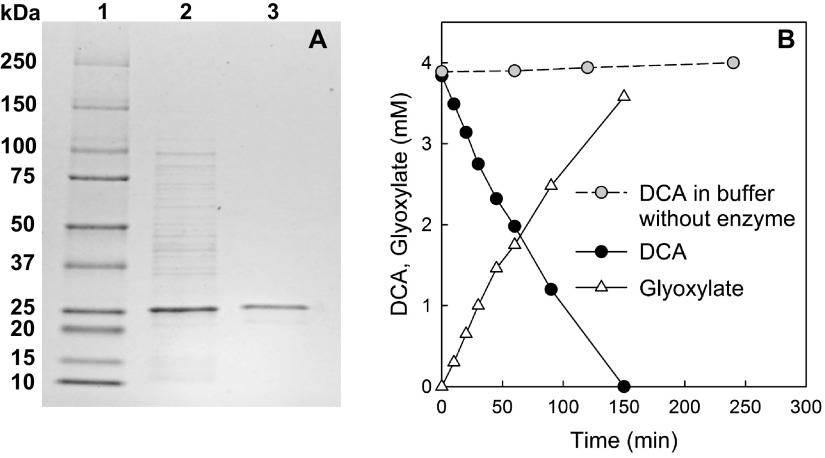
To functionally characterize the putative HADs of the strict anaerobe “Ca. Dichloromethanomonas elyunquensis” strain RM, the genes encoding HAD1 and HAD2 were cloned and heterologously expressed in Escherichia coli. Assays with cell extracts of the E. coli transformant carrying the had1 gene revealed the stoichiometric conversion of DCA to glyoxylate (see Fig. S2 in the supplemental material). Extracts of E. coli cells expressing HAD2 did not convert DCA to glyoxylate (Fig. S2). Similarly, cell extracts of an E. coli strain carrying the empty vector without an had gene did not catalyze the conversion of DCA to glyoxylate (Fig. S2). Based on these findings, we concluded that HAD1 was responsible for the initial attack on DCA in strain RM. After purification of the His-tagged HAD1 protein using a HisTrap Ni Sepharose column, a single protein band with a size of approximately 25 kDa was observed in SDS-PAGE (Fig. 5A), matching the expected size of the HAD1 protein (i.e., 221 amino acids with a calculated molecular mass of 25.59 kDa). In vitro assays demonstrated that the purified HAD1 stoichiometrically converted DCA to glyoxylate at a rate of 90 ± 4.6 nkat mg−1 protein (Fig. 5B). The purified HAD1 also converted MCA to glycolate, but at an approximately 70-fold lower rate of 1.3 nkat mg−1 protein (see Fig. S3 in the supplemental material). The purified HAD1 protein did not exhibit activity toward DCM, trichloroacetate, and mono- or difluoroacetate.
FIG 5.
Enzymatic activity of the heterologously expressed HAD1 protein of “Ca. Dichloromethanomonas elyunquensis.” (A) SDS-PAGE illustrating HAD1 purification. Lane 1, protein size markers; lane 2, soluble crude extract of E. coli strain FEL153 carrying the had1 gene (prokka_14346); lane 3, purified His-tagged HAD1 protein. (B) Enzymatic activity of heterologously expressed and purified HAD1 of “Ca. Dichloromethanomonas elyunquensis” showing the stoichiometric conversion of DCA to glyoxylate. In abiotic control incubations without protein, DCA was stable. The data shown are from a single experiment, and independent experiments yielded similar results.
Conversion of DCA to glyoxylate in cell-free extracts of E. coli transformants expressing the “Ca. Dichloromethanomonas elyunquensis” strain RM had1 or had2 gene. Download FIG S2, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro conversion of monochloroacetate (MCA) to glycolate using the purified HAD1 protein. Download FIG S3, PDF file, 0.08 MB (82.8KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparative proteome analysis.
The 3-week adaptation time required for DCM-grown cultures to commence DCA utilization (Fig. 1A) suggested that proteins involved in metabolizing DCA are inducible. A comparative global proteomic analysis between DCA- and DCM-grown cells was performed to elucidate differential abundance expression of proteins involved in DCA versus DCM metabolism in strain RM. A complete list of proteins identified under the different growth conditions at two sampling time points (i.e., immediately before the 2nd electron donor amendment [TP1] and near the end of electron donor consumption [TP2]; see Fig. S4 in the supplemental material) is presented in Table S2 in the supplemental material. HAD1 (prokka_14346) was detected in the proteomes of DCA- and DCM-grown cells; however, the expression was greater in DCA-grown cells at both time points, with log2 fold changes of +2.38 and +1.69. HAD2 (prokka_14344) was not detected in cultures grown with either substrate (Fig. 6), an observation consistent with the in vitro enzyme activity results (Fig. S2) and indicating that HAD2 is not involved in DCA metabolism.
FIG 6.
Proposed anaerobic catabolic pathway for DCA in “Ca. Dichloromethanomonas elyunquensis” strain RM. The shaded boxes indicate the log2 fold change of normalized protein abundance values in DCA- versus DCM-grown cells at TP1 (dashed line boxes) and TP2 (solid line boxes). The protein abundance values represent average from three biological replicate cultures for each growth condition. Boxes marked with “+” signs indicate that the fold changes were statistically significant (P < 0.05) in the pairwise comparisons of DCA- versus DCM-grown cells at TP1 and/or TP2. Gene locus tags of each protein are depicted below protein names. Abbreviations: HAD1, haloacid dehalogenase 1; Gcl, glyoxylate carboligase; Hyi, hydroxypyruvate isomerase; Ghr, glyoxylate/hydroxypyruvate reductase; GlxR, 2-hydroxy-3-oxopropionate reductase; Gck, glycerate 2-kinase; Eno, enolase; Pyk, pyruvate kinase; POR, pyruvate-flavodoxin oxidoreductase; PTA, phosphate acetyltransferase; ACK, acetate kinase; ACS/CODH, acetyl coenzyme A synthase/carbon monoxide dehydrogenase; CFeSP, corrinoid iron-sulfur protein; MeTr, methyltransferase; MetF, methylene-tetrahydrofolate (H4folate) reductase; MTHFD, methylene-H4folate dehydrogenase; FolD, formyl-H4folate cyclohydrolase; Fhs, formyl-H4folate synthase; Fdh, formate dehydrogenase. WLP proteins are depicted in green font, and proteins involved in DCA reduction to acetate are shown in blue font. The fold change values are shown in Table S2.
Growth of “Ca. Dichloromethanomonas elyunquensis” during DCA and DCM catabolism and collection of samples for comparative proteome analysis. Download FIG S4, PDF file, 2.1 MB (2.1MB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of proteins identified in RM cultures grown with DCA or DCM at two sampling points. Samples were collected immediately before the 2nd electron donor amendment (TP1) and near the end of electron donor consumption (TP2); approximately 95% of the initial amount of electron donor was consumed (Fig. S4). Imputed values are shown in orange. Download Table S2, XLSX file, 0.6 MB (643.7KB, xlsx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A glyoxylate carboligase (Gcl; prokka_21461), which catalyzes the decarboxylation of glyoxylate and the ligation to a second molecule of glyoxylate to form the three-carbon compound tartronate semialdehyde, was among the most highly abundant proteins in DCA-grown cells. The log2 fold changes in Gcl in DCA- versus DCM-grown cells were +8.09 and +9.26 at the TP1 and TP2 time points, respectively, and both log2 fold changes were above the statistically significant level (P < 0.05 [Fig. 6]). A series of enzymes involved in the stepwise conversion of tartronate semialdehyde to acetyl coenzyme A (acetyl-CoA), including 2-hydroxy-3-oxopropionate reductase (GlxR; prokka_14716), hydroxypyruvate isomerase (Hyi; prokka_14715), glyoxylate/hydroxypyruvate reductase (Ghr; prokka_20728), glycerate 2-kinase (Gck; prokka_14717), and pyruvate kinase (Pyk, prokka_14576), were significantly more abundant in DCA-grown cells at both time points (Fig. 6). Other predicted pathway enzymes, such as enolase (Eno; prokka_20734), a second Pyk (prokka_17967), and pyruvate-flavodoxin oxidoreductase (POR; prokka_27155, _20890, and _11715), did not show statistically significant abundance fold changes in DCA-grown versus DCM-grown cells (Fig. 6); however, they were all detected in the proteome, with potential roles in the transformation of 2-phospho-d-glycerate to acetyl-CoA. The presence of phosphate acetyltransferase (PTA; prokka_76015) and acetate kinase (ACK; prokka_39944) enzymes, both of which are encoded on the genome, was confirmed in the analyses of both DCA- and DCM-grown cells. Interestingly, PTA was less abundant in DCA-grown cells, while the ACK abundance was higher than that in DCM-grown cells. Although more information is required to understand the regulatory controls of these enzymes, their detection in cultures growing with DCA suggests an acetyl transfer reaction followed by a dephosphorylation reaction to convert acetyl-CoA to acetate, which was measured as a terminal product in DCA-grown cultures (Fig. 6). In addition, all of the Wood-Ljungdahl pathway (WLP) proteins encoded on the genome of strain RM were detected in the proteome of DCA-grown cells, suggesting the involvement of WLP enzymes in DCA catabolism (Fig. 6).
DISCUSSION
Previous studies focused on DCA catabolism in aerobes; however, the fate of DCA under anoxic conditions remained obscure. The anaerobic mixed culture RM could be maintained with DCA as the sole source of energy, and the molecular analyses implicated the DCM-degrading bacterium “Ca. Dichloromethanomonas elyunquensis” strain RM in DCA catabolism. The findings demonstrate that specialized anaerobes metabolize DCA and illustrate that the range of substrates strain RM can utilize is not limited to DCM.
Initial enzymatic attack on DCA.
Both chlorine substituents were released during DCA degradation, and acetate was an end product (Fig. 1B). The stepwise reductive dechlorination of trichloroacetate (CCl3-COO– [TCA]), DCA, and MCA via reductive dechlorination is thermodynamically favorable, with Gibbs free energy changes of –171.2, –154.0, and –152.0 kJ per reaction, respectively, under standard conditions with H2 as electron donor (48). The transformation of TCA to DCA has been observed when TCA was incubated with mouse or rat gut microflora under anoxic conditions (49). Reductive dechlorination of TCA to DCA was explicitly demonstrated in an axenic culture of Geobacter thiogenes (formerly Trichlorobacter thiogenes) strain K1; however, a cryptic sulfur-sulfide redox cycle was involved in dechlorination, and the organism apparently does not perform organohalide respiration (50, 51). The genome of strain RM encodes three putative reductive dehalogenases (RDases), and two of them were expressed during growth with DCM (47). One of these RDases (prokka_14638) was detected during growth with DCA, albeit at very low abundance. The experimental efforts did not generate any evidence for reductive dechlorination. MCA, the product of a single reductive dechlorination (hydrogenolysis) reaction, was neither detected as an intermediate nor supported growth of mixed culture RM. Furthermore, the utilization of DCA as an electron acceptor in organohalide respiration should result in the formation of stoichiometric amounts of acetate; however, only about 50% of the initial amount of DCA was recovered as acetate in RM cultures. The experimental data indicate that strain RM ferments DCA to acetate, H2, and CO2 and does not utilize DCA as an electron acceptor in organohalide respiration (Fig. 6).
The integrated physiologic, proteogenomic, and enzymatic studies pinpoint a novel HAD involved in converting DCA to glyoxylate in a strictly anaerobic bacterium. HADs belong to a large superfamily of hydrolases with diverse substrate specificities and catalyze the hydrolytic dehalogenation of 2-haloalkanoic acid to the corresponding 2-hydroxyalkanoic acids (38, 52). Many aerobic bacteria, including members of the genera Pseudomonas, Xanthobacter, and Moraxella, possess HADs that convert DCA to glyoxylate to initiate DCA metabolism and growth (34–36, 38). HADs are not sensitive to O2 and do not require any cofactors such as O2 or glutathione for activity. In contrast, mammalian liver cells employ glutathione S-transferases (GSTs) to convert DCA to glyoxylate (31–33). GSTs play central roles for detoxification of various groups of harmful compounds, such as halogenated nitrobenzenes, arene oxides, and quinones (53). DCM dehalogenases of aerobic and facultative aerobic methylotrophic bacteria, which catalyze the conversion of DCM to formaldehyde, also belong to GSTs (54, 55). The active site of GSTs is the thiol group of the glutathione cofactor, which in its reduced form performs a nucleophilic attack on nonpolar compounds containing an electrophilic carbon, nitrogen, or sulfur atom (53). GSTs strictly require glutathione as a cofactor and have been found in eukaryotes, some aerobic and facultative aerobic methylotrophic bacteria, and recently in a cyanobacterium (33), but never in strict anaerobes (53). HADs, in contrast, employ the carboxyl group of an aspartate residue in the active center to carry out an SN2 nucleophilic attack on the α-carbon atom of the halogenated carboxylate substrate to displace a halogen atom and produce an enzyme-bound ester intermediate. The ester bond is subsequently hydrolyzed to produce the corresponding d-2-hydroxyalkanoate and regenerate the aspartate residue (56, 57). Although both HADs (EC 3.8.1.2) and GSTs (EC 2.5.1.18) are capable of removing chlorine substituents from DCA, yielding the same products (i.e., glyoxylate and inorganic chloride), their reaction mechanisms are fundamentally different and belong to distinct enzyme classes.
DCA fermentation pathway.
Based on 16S rRNA gene sequence analysis, “Ca. Dichloromethanomonas elyunquensis” strain RM is phylogenetically related to Syntrophobotulus glycolicus (45), an isolate capable of fermenting glyoxylate to glycolate, CO2, and H2 (58, 59). Based on physiological and enzymatic evidence, S. glycolicus strain FlGlyRT (DSM 8271) was proposed to metabolize glyoxylate via malyl-CoA to glycolate, CO2, and H2 under anoxic conditions without an external electron acceptor (58). Genes encoding HADs were not found on the genome of S. glycolicus (60), and strain FlGlyRT could not utilize DCA as a growth substrate. The HAD-catalyzed DCA dehalogenation leads to the formation of glyoxylate, which is subsequently fermented by strain RM, generating acetate, CO2, and H2. In addition to DCA, strain RM is also able to ferment glyoxylate (see Fig. S5 in the supplemental material), consistent with the observation that glyoxylate is an intermediate of DCA metabolism. In contrast to S. glycolicus, strain RM does not possess the canonical genes for malyl-CoA lyase and malate dehydrogenase, and we never detected glycolate in culture supernatant. Instead, the comparative proteome analysis revealed high expression of glyoxylate carboligase (Gcl; prokka_21461) in DCA-grown cells (Fig. 6). This enzyme catalyzes the condensation of two molecules of glyoxylate to form tartronate semialdehyde, suggesting this C3 compound is a pathway intermediate. The comparative proteome analysis further revealed the abundance of proteins (i.e., GlxR, Hyi, Ghr, Gck, Eno, Pyk, and POR) potentially involved in converting tartronate semialdehyde to acetyl-CoA via glycerate and pyruvate (Fig. 6). Glyoxylate metabolism through tartronate semialdehyde and the glycerate pathway was proposed previously in the oxalate-degrading anaerobic bacterium Oxalobacter formigenes based on the detection of enzymatic activities in the cell-free crude extract, specifically the activities of glyoxylate carboligase (Gcl), tartronic semialdehyde reductase (GlxR), and glycerate kinase (Gck) (61).
Growth of “Ca. Dichloromethanomonas elyunquensis” in mixed culture RM with glyoxylate as the sole electron donor as determined by 16S rRNA gene-targeted qPCR. Download FIG S5, PDF file, 0.2 MB (227.9KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The genome of strain RM encodes a complete WLP, and the corresponding proteins were detected in DCA-grown cells, indicating the involvement of the WLP in DCA metabolism. Based on gene content, the tricarboxylic acid (TCA) cycle is incomplete, and the oxidation of acetyl-CoA through the TCA cycle is not possible. Half of the acetyl-CoA formed during DCA metabolism is likely oxidized to CO2 via the reverse WLP generating reducing equivalents (i.e., electrons and protons) (Fig. 6). The WLP has also been implicated in glyoxylate metabolism in the thermophilic homoacetogenic bacterium Moorella sp. strain HUC22-1, which ferments glyoxylate to acetate and CO2 via malyl-CoA rather than tartronate semialdehyde and glycerate (62). Anaerobic DCM metabolism also proceeds via the WLP (43, 63), and all WLP proteins were highly expressed in strain RM cells during growth with DCM (47). To facilitate direct comparisons, the pathway postulated for anaerobic DCM metabolism in strain RM is shown in Fig. S6 in the supplemental material. The detection of formate and CO as intermediates during DCA metabolism (Fig. 1 and 2) lends further support for the involvement of the WLP in DCA metabolism. CO is an obligatory intermediate of the WLP, generated by the bifunctional enzyme CO dehydrogenase/acetyl-CoA synthase (CODH/ACS) during the reduction of CO2 (64). CO was not detected in cultures grown with DCM, which may be explained by the direction of the CODH/ACS reaction during DCA versus DCM metabolism, viz., the oxidative direction during DCA degradation versus the reductive direction during DCM catabolism. Another possible explanation is the mineralization of DCM to CO2 and H2 via the oxidative WLP, with a small fraction of the DCM carbon being assimilated through anabolic reactions (i.e., via the reductive route of the WLP) with CO as an intermediate (Fig. S6). Although strain RM metabolizes both DCM and DCA via the WLP, the proteomic data indicate relative higher expression levels of WLP proteins (with the exception of MetF) in DCM-grown cells.
Postulated anaerobic DCM metabolic pathway in “Ca. Dichloromethanomonas elyunquensis” strain RM. DCM is metabolized via the Wood-Ljungdahl pathway (WLP). Download FIG S6, PDF file, 0.04 MB (45.8KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
H2 was detected as an intermediate during DCA degradation in culture RM, similar to what has been observed in S. glycolicus cultures fermenting glyoxylate. The genes encoding two putative group 4 H2-evolving [NiFe]-hydrogenases (prokka_18969 and prokka_18970) are present on the genome of strain RM, and both [NiFe]-hydrogenases were highly expressed during growth with DCA (Fig. 6). Very likely, one or both [NiFe]-hydrogenases is involved in H2 formation during DCA metabolism by catalyzing the reduction of protons generated from the oxidation of acetyl-CoA.
Degradation of both DCA and DCM generated H2, but methanogenesis only occurred in DCM-grown cultures. Repeated transfers with DCA eliminated methanogens (Fig. 3), hinting at possible toxic effects of DCA on methanogens (65). H2 consumption in DCA-grown mixed culture RM was attributed to bacteria performing H2/CO2 reductive acetogenesis, e.g., Acetobacterium and Treponema (66, 67). H2 was eventually scavenged to 1,500 ± 180 ppmv during growth on DCA (Fig. 2), which is consistent with the H2 consumption threshold concentration range reported for H2/CO2 reductive acetogenesis as the terminal electron accepting process (68, 69).
“Ca. Dichloromethanomonas elyunquensis” strain RM has resisted isolation—presumably due to the requirement for a hydrogenotrophic partner population to remove H2 (Fig. 3). Elevated H2 partial pressures inhibit DCM (43) and DCA degradation (see Fig. S7 in the supplemental material) indicative of strict syntrophy, and strain RM relies on H2-scavenging populations to metabolize DCM and DCA. Based on the physiological observations and proteomic data, DCA metabolism in strain RM generates acetate, CO2, H2, chloride (Cl–), and biomass (Fig. 1) and proceeds according to equation 1:
| (1) |
The generated H2 is consumed in H2/CO2 reductive acetogenesis leading to acetate formation according to equation 2:
| (2) |
Therefore, DCA catabolism in mixed culture RM proceeds according to equation 3, with half of the DCA being reduced to acetate and the other half being oxidized to CO2, which is consistent with the experimentally measured stoichiometry, viz., the ratio of DCA degraded versus acetate generated was 2.15 ± 0.05 to 1 (Fig. 1B). Based on these observations, half of the acetate formed is directly derived from DCA, and the other half is generated via reductive acetogenesis by utilizing H2 generated during DCA catabolism:
| (3) |
Inhibitory effect of H2 on DCA degradation in mixed culture RM. Download FIG S7, PDF file, 0.9 MB (949.2KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Implications.
Natural and anthropogenic processes introduce DCA into the environment, and the prevalence of had genes in the genomes of aerobic bacteria can be viewed as a consequence of ubiquitously present DCA (6–8, 21). DCA formation has been reported in various terrestrial environments, as well as marine and peat bog ecosystems (7, 8). Information about DCA pool sizes is only available from coniferous forest soils, which contain approximately 300 ng g−1 soil (6). Based on this information, we calculate that the global DCA amount in coniferous forest soils alone exceeds 8 × 109 kg (assuming an area of 40 × 106 km2, a surface soil depth [A horizon] of 40 cm, and a soil bulk density of 1.6 g/cm3) (70). Because information about DCA fluxes in environmental systems is lacking, DCA turnover may be substantial even in the absence of measurable DCA pools. The heretofore unrecognized anaerobic DCA degradation pathway via glyoxylate likely constitutes the dominant route of DCA catabolism under electron acceptor-depleted conditions, with implications for carbon and electron flow in anoxic environments. DCA fermentation generates acetate and H2, both of which are central intermediates during carbon cycling and can fuel anaerobic food webs. Therefore, the abiotic and biotic formation and subsequent fermentation of DCA may be relevant processes for sustaining microbial activity in energy-depleted environments such as the deep subsurface. The findings also have bearing on the clinical use of DCA as a drug and future studies should explore if members of the gut microbiota have the ability to ferment DCA and assess the responses of the gut microbiome to DCA treatment (71, 72).
MATERIALS AND METHODS
Chemicals.
DCA (purity, >99.8%) and DCM (>99.95%) were purchased from Sigma-Aldrich Co. (St. Louis, MO) and Acros Organics (Fair Lawn, NJ), respectively. Gas mixtures with H2 partial pressures of 10, 50, and 100 ppmv were purchased from Airgas (Radnor, PA), and CO gas (>99.0%) was purchased from Sigma-Aldrich Co. and used for standard curve preparation. All other chemicals used were analytical reagent grade or higher.
Microorganisms and cultivation.
Mixed culture RM was derived from pristine freshwater sediment and maintained with DCM as the sole energy source for 8 years (42, 45). Culture RM was routinely grown in 160-ml glass serum bottles containing 100 ml of anoxic, bicarbonate-buffered (30 mM, pH 7.3) basal salts medium reduced with 0.2 mM sulfide and 0.2 mM l-cysteine (73). The vessels were sealed with black butyl rubber stoppers (Bellco Glass, Inc., Vineland, NJ) under a headspace of N2/CO2 (80/20 [vol/vol]), with 5 to 10 μl neat DCM (78 to 156 μmol) provided as the sole electron donor prior to inoculation from a DCM-grown culture (5% [vol/vol]). Cultures that had consumed the initial dose of DCM received 1 to 2 mM DCA to examine its potential utilization as an energy source. Following the consumption of DCA, RM cultures were repeatedly transferred (3% [vol/vol]) with DCA as the sole energy source before the experiments reported herein were initiated. All culture vessels were incubated at 30°C in the dark without agitation. To quantitatively measure inorganic chloride release during DCA degradation, incubations were conducted in chloride-free medium with bromide salts substituting for chloride salts.
Syntrophobotulus glycolicus strain FlGlyRT (DSM 8271) was purchased from DSMZ-German Collection of Microorganisms and Cell Cultures GmbH (Braunschweig, Germany) and was grown in the anoxic, bicarbonate-buffered (30 mM, pH 7.3) basal salts medium as described above, with glyoxylate (5 mM) as the sole energy source. To test DCA as a potential substrate for S. glycolicus strain FlGlyRT, 2 and 5 mM DCA replaced glyoxylate in medium inoculated from a glyoxylate-grown S. glycolicus culture (3% [vol/vol]).
DNA extraction and quantitative real-time PCR.
For DNA extraction, 5 ml of culture suspension was periodically collected during a growth cycle on DCA or DCM and filtered onto 0.22-μm-pore Durapore membranes (Millipore, Cork, Ireland). DNA was extracted using the DNeasy PowerSoil DNA isolation kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. 16S rRNA gene-targeted qPCR was used to monitor growth of strain RM in cultures grown with DCA or DCM. qPCR assays used primers and a probe specifically targeting the 16S rRNA gene of strain RM (45) and were conducted using an ABI ViiA7 real-time PCR system (43, 45).
16S rRNA gene amplicon sequencing.
To monitor the microbial community response to repeated transfers with DCA, 16S rRNA gene-based amplicon sequencing targeting the V4 region of both bacterial and archaeal 16S rRNA genes was performed following established procedures (74, 75). The amplicons were sequenced on the Illumina MiSeq platform (San Diego, CA), and data analysis was performed with the QIIME v.1.9.1 software package (76). Raw sequencing reads were jointly paired, demultiplexed, and trimmed to a length of 250 bp, and chimeric reads were removed. After quality control, over 100,000 individual sequences were obtained for each library generated with DNA samples collected from consecutive transfers with DCM or DCA. Operational taxonomic units (OTUs) were picked via the default UCLUST pipeline (77) and filtered at a 0.005% threshold. Taxonomic assignments were performed using the RDP classifier trained against the Greengenes 16S rRNA gene database (version 13.8) (78). Taxonomy, relative abundance, and sequences of representative OTUs are shown in Table S1. The most abundant sequence within each taxon was chosen as the representative sequence for each OTU.
Heterologous had gene expression, purification, and in vitro activity testing.
The pET-28a(+) expression vector backbone was used to clone and express had genes carrying an N-terminal His tag. “Ca. Dichloromethanomonas elyunquensis” had genes (locus tags prokka_14344 and prokka_14346) were amplified from DNA extracted from DCA-grown cells using Phusion Flash High-Fidelity PCR master mix (Thermo Fisher) and primer sets NJ762 (5′-CTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACCATGATTAGAGCTGTAGTCTTTGATGCC-3′) and NJ763 (5′-AGCAGCCGGATCTCAGTGGTGGTGGTGGTGGTGCTCGAGTTCAAATATTCTTAGTTTTGAGGGCCAAC-3′) and NJ764 (5′-CTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACCATGATTAAGGCATGCGCATTTGATG-3′) and NJ765 (5′-AGCAGCCGGATCTCAGTGGTGGTGGTGGTGGTGCTCGAGTTTATAGGCCTTTAACTTTTTGAGCC-3′), respectively. PCR products were cleaned using the UltraClean PCR Clean-Up kit (MO BIO Laboratories, Inc., or Zymo DNA Clean and Concentrator). The pET-28a(+) vector backbone was digested with the restriction endonucleases BamHI, NcoI, NdeI, and NotI for untagged constructs and BamHI, NdeI, and NotI for tagged constructs and gel extracted to remove any remaining supercoiled plasmid. The linearized vector and had gene inserts were then cotransformed into electrocompetent E. coli strain BW25113 cells with preinduced λ Red recombinase from plasmid pKD46 (79) to allow for homologous recombination. All PCR amplicons in recombinant vectors were sequence verified using Sanger sequencing. Following sequence verification, recombinant vectors pNJ100 (carrying prokka_14344) and pNJ101 (carrying prokka_14346) were introduced into E. coli strain BL21(DE3) (New England BioLabs) for overexpression and purification. The supplemental material provides additional information about primers (Table S3A), plasmids (Table S3B), and E. coli strains (Table S3C) used for the heterologous expression of had genes.
Primers, plasmids, and E. coli strains used for cloning and overexpression of had genes. Download Table S3, XLSX file, 0.01 MB (14KB, xlsx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
E. coli strain BL21(DE3) carrying an had expression plasmid was grown in 300 ml of Terrific Broth (Thermo Fisher) with 50 μg ml−1 kanamycin at 37°C and 150 rpm to an optical density at 600 nm (OD600) of 1.0. Cultures were then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Thermo Fisher) and incubated overnight at room temperature at 150 rpm. Cells were collected by centrifugation at 9,000 × g for 25 min, suspended in 25 mM Tris-HCl buffer (pH 7.5), and sonicated at 50% amplitude (Branson Sonifier 250; Branson Sonifiers, Danbury, CT) in an ice bath for 8 min with a 50% duty cycle (30 s on and 30 s off). The lysate was centrifuged at 38,000 × g for 20 min, and the supernatant was passed through a 5-ml HisTrap Ni Sepharose column (GE Healthcare) using an ÄKTA Prime fast protein liquid chromatography (FPLC) system (GE Healthcare, Pittsburgh, PA). Proteins were eluted with 25 mM Tris-HCl buffer (pH 7.5) containing 100 mM NaCl and 300 mM imidazole. Fractions containing protein were combined, and the elution buffer was exchanged with 4 ml of 100 mM Tris-HCl (pH 8.0) by using a 10-kDa-cutoff Amicon Ultra-4 filter unit (Millipore). Protein concentrations were quantified using the Bradford assay (80), and protein purity was examined by SDS-PAGE and Coomassie blue staining. A standard protein marker (Bio-Rad, Hercules, CA) with protein sizes ranging from 10 to 250 kDa allowed size estimations. The purified HAD1 protein stock solutions (0.6 mg ml−1) were frozen immediately and stored at −80°C. The N-terminal hexahistidine tag was retained for all experiments.
Enzyme assays were conducted in 100 mM Tris-HCl buffer (pH 8.0) in Eppendorf tubes in a total assay volume of 0.5 ml. DCA was added at a concentration of 4 mM, and the reactions were started by adding 3 μg of HAD protein. The tubes were agitated with 150 rpm at 30°C. Aliquots of 50 μl were collected over time, acidified with 1 μl of 1 M H2SO4 to quench the reaction, centrifuged at 17,000 × g for 5 min at room temperature, and analyzed by high-performance liquid chromatography (HPLC) to determine the concentrations of DCA and glyoxylate. HAD enzyme activity was calculated based on the formation of glyoxylate: 1 nkat is the amount of enzyme that generates 1 nmol of glyoxylate per second.
Global proteomics of RM cultures grown with DCA versus DCM.
Prior to proteomic analysis, cultures were consecutively passaged at least three times on the same substrate (i.e., DCA or DCM). Following the consumption of 361.0 ± 19.5 μmol of DCA and 373.2 ± 5.8 μmol of DCM, the respective cultures received one additional feeding of the respective substrate. Samples for proteomic analysis were collected upon the consumption of the first (time point 1 [TP1]) and the second (TP2) substrate feedings (Fig. S4). Cells grown with DCA or DCM were collected from triplicate cultures by passing 100 ml of culture suspension through Sterivex 0.22-μm-pore filter units (EMD Millipore Corporation, Billerica, MA). The outlet of a filter unit was capped, and 1.5 ml of boiling SDS lysis buffer (4% SDS [wt/wt] in 100 mM Tris-HCl buffer, pH 8.0) was added. Following gentle agitation on a shaker with three-dimensional (3D) gyratory action for 1 h at room temperature, a 3-ml plastic syringe was connected to the cartridge’s inlet, the unit was inverted, and as much lysate as possible was transferred into the syringe. The filter units were rinsed once with 500 μl of fresh lysis buffer at room temperature, and the recovered volumes were combined. The cell lysates were then subjected to trichloroacetic acid precipitation followed by urea denaturation, reduction, blocking of disulfide bonds, and tryptic digestion (trypsin/protein ratio of 1:50 [wt/wt]) as described previously (81). The protein contents in crude and peptide extracts were quantified using the bicinchoninic acid (BCA) assay (Pierce Biotechnology, Waltham, MA). Peptide extracts were stored at −80°C until liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Proteomics data sets from culture RM were obtained with an Orbitrap Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA) equipped with an electrospray ionization (ESI) source and interfaced with a Proxeon EASY-nLC 1200 system. Peptides (2 μg) from each sample were suspended in solvent A (2% acetonitrile, 98% water, 0.1% formic acid) and injected onto a 75-μm-inner-diameter microcapillary column packed with 35 cm of Kinetex C18 resin (1.7 μm, 100 Å; Phenomenex). Peptides were separated using a 90-min gradient from 2% to 30% solvent B (80% acetonitrile, 20% water, 0.1% formic acid), followed by an increase to 40% solvent B within 10 min and a 10-min wash with 98% solvent A. The flow rate was kept at 250 nl min−1. MS data were acquired with the Thermo Xcalibur software version 4.27.19, with a topN method, where N was capped to 15. Other scanning and spectral data collection parameters were similar to those reported previously (82). All spectral data collected in this study have been deposited in the MASSIVE and ProteomeXchange repositories with identifiers MSV000086520 and PXD022742, respectively (ftp://massive.ucsd.edu/MSV000086520/).
Peptide and protein identification by database searching.
MS/MS raw data files from culture RM were searched against a database of sequences annotated from the draft genome of “Ca. Dichloromethanomonas elyunquensis” (accession no. LNDB00000000), to which common contaminant proteins were appended (www.thegpm.org/crap). The MyriMatch v2.2 algorithm was used for standard database searching and was set to the same parameters described previously (82, 83). Confidently identified peptides at a false-discovery rate below 1% were assembled into proteins using the IDPicker v.3.1 software (84). Every protein in the data set was identified with at least two unique peptide sequences. For label-free quantification, the MS1-level peptide precursor intensities were extracted from IDPicker with IDPQuantify, summed by protein, and then divided by the sequence length of the protein to which they matched (85). Protein abundance values were log2 transformed and then normalized by mean central tendency analysis with Inferno RDN (https://omics.pnl.gov/software/infernordn). The Perseus software (86) was then used to filter proteins with non-zero abundance values in two out of three biological replicates in at least one growth condition and time point. After data filtering, undetected proteins (i.e., proteins with missing abundance values) were imputed with a simulated Gaussian distribution of low-abundance values to provide non-zero abundance metrics at the detection threshold. This approach enables statistical analyses across the entire data set, as is commonly done in proteome measurements (87, 88). Pairwise t test comparisons were conducted to identify proteins having statistically significant abundance changes (P < 0.05) between growth conditions at each respective substrate feeding time point.
Analytical methods.
DCA, glyoxylate, formate, and acetate were analyzed on an Agilent 1200 series HPLC system equipped with an Aminex HPX-87H column (Bio-Rad, Hercules, CA) operated at 30°C and a multiple-wavelength detector set to 210 nm. Operation was isocratic using 4 mM H2SO4 as the eluent at a flow rate of 0.6 ml min−1. Aqueous samples (200 μl) were acidified with 4 μl 1 M H2SO4 and filtered prior to HPLC analysis. The identification of peaks was based on retention times of authentic standards, and quantification was achieved using external calibration curves. DCM was measured by manually injecting 0.1-ml headspace samples into an Agilent 7890A gas chromatograph (Santa Clara, CA) equipped with a DB-624 column (60-m length, 0.32-mm inside diameter, 1.8-μm film thickness) and a flame ionization detector as described previously (89). Chloride ions were measured with an ion chromatograph using a Dionex ICS-2100 system equipped with a 4- by 250-mm IonPac AS18 hydroxide-selective anion-exchange column (Thermo Fisher Scientific, Waltham, MA) operated at 30°C. The 10 mM KOH eluent was delivered at a flow rate of 1 ml min−1, and an ERS 500 suppressor (4 mm) was set at a current of 57 mA. To follow the formation of H2 and CO, 0.5 ml of culture headspace samples was injected into a Peak Performer 1 gas chromatograph coupled with a reducing compound photometer (Peak Laboratories, Mountain View, CA) with detection limits for H2 and CO below 8 ppb by volume (ppbv).
ACKNOWLEDGMENTS
This work was partly supported by The Chemours Company.
We acknowledge David Graham, Oak Ridge National Laboratory, for providing access to an ÄKTA chromatography system.
Footnotes
This article is a direct contribution from Frank E. Löffler, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Max Häggblom, Rutgers, The State University of New Jersey, and Elizabeth Edwards, University of Toronto.
Citation Chen G, Jiang N, Villalobos Solis MI, Kara Murdoch F, Murdoch RW, Xie Y, Swift CM, Hettich RL, Löffler FE. 2021. Anaerobic microbial metabolism of dichloroacetate. mBio 12:e00537-21. https://doi.org/10.1128/mBio.00537-21.
REFERENCES
- 1.Gribble GW. 2010. Progress in the chemistry of organic natural products, vol 91, p 12–13. Springer, Vienna, Austria. [Google Scholar]
- 2.Matucha M, Gryndler M, Forczek ST, Uhlířová H, Fuksová K, Schröder P. 2003. Chloroacetic acids in environmental processes. Environ Chem Lett 1:127–130. doi: 10.1007/s10311-003-0030-y. [DOI] [Google Scholar]
- 3.McConnell O, Fenical W. 1977. Halogen chemistry of the red alga Asparagopsis. Phytochemistry 16:367–374. doi: 10.1016/0031-9422(77)80067-8. [DOI] [Google Scholar]
- 4.Moore RE. 1977. Volatile compounds from marine algae. Acc Chem Res 10:40–47. doi: 10.1021/ar50110a002. [DOI] [Google Scholar]
- 5.Kuhlisch C, Schleyer G, Shahaf N, Vincent F, Schatz D, Vardi A. 2020. Viral infection of algal blooms leaves a halogenated footprint on the dissolved organic matter in the ocean. bioRxiv https://www.biorxiv.org/content/10.1101/2020.09.08.287805v1. [DOI] [PMC free article] [PubMed]
- 6.Fahimi IJ, Keppler F, Schöler HF. 2003. Formation of chloroacetic acids from soil, humic acid and phenolic moieties. Chemosphere 52:513–520. doi: 10.1016/S0045-6535(03)00212-1. [DOI] [PubMed] [Google Scholar]
- 7.Bond T, Goslan EH, Parsons SA, Jefferson B. 2012. A critical review of trihalomethane and haloacetic acid formation from natural organic matter surrogates. Environ Technol Rev 1:93–113. doi: 10.1080/09593330.2012.705895. [DOI] [Google Scholar]
- 8.Laturnus F, Fahimi I, Gryndler M, Hartmann A, Heal MR, Matucha M, Schöler HF, Schroll R, Svensson T. 2005. Natural formation and degradation of chloroacetic acids and volatile organochlorines in forest soil—challenges to understanding. Environ Sci Pollut Res Int 12:233–244. doi: 10.1065/espr2005.06.262. [DOI] [PubMed] [Google Scholar]
- 9.Parker KM, Mitch WA. 2016. Halogen radicals contribute to photooxidation in coastal and estuarine waters. Proc Natl Acad Sci U S A 113:5868–5873. doi: 10.1073/pnas.1602595113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, Parker KM. 2018. Halogen radical oxidants in natural and engineered aquatic systems. Environ Sci Technol 52:9579–9594. doi: 10.1021/acs.est.8b02219. [DOI] [PubMed] [Google Scholar]
- 11.Reimann S, Grob K, Frank H. 1996. Chloroacetic acids in rainwater. Environ Sci Technol 30:2340–2344. doi: 10.1021/es9507776. [DOI] [Google Scholar]
- 12.Rompp A, Klemm O, Fricke W, Frank H. 2001. Haloacetates in fog and rain. Environ Sci Technol 35:1294–1298. doi: 10.1021/es0012220. [DOI] [PubMed] [Google Scholar]
- 13.von Sydow L, Bordn H, Grimvall A. 1999. Chloroacetates in snow, firn and glacier ice. Chemosphere 39:2479–2488. doi: 10.1016/S0045-6535(99)00160-5. [DOI] [Google Scholar]
- 14.von Sydow LM, Nielsen AT, Grimvall AB, Borén HB. 2000. Chloro- and bromoacetates in natural archives of firn from Antarctica. Environ Sci Technol 34:239–245. doi: 10.1021/es9812150. [DOI] [Google Scholar]
- 15.Malliarou E, Collins C, Graham N, Nieuwenhuijsen MJ. 2005. Haloacetic acids in drinking water in the United Kingdom. Water Res 39:2722–2730. doi: 10.1016/j.watres.2005.04.052. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Collins C, Graham N, Templeton MR, Huang J, Nieuwenhuijsen M. 2010. Speciation and variation in the occurrence of haloacetic acids in three water supply systems in England. Water Environ J 24:237–245. doi: 10.1111/j.1747-6593.2009.00200.x. [DOI] [Google Scholar]
- 17.Liu W, Zhao Y, Chow CWK, Wang D. 2011. Formation of disinfection byproducts in typical Chinese drinking water. J Environ Sci 23:897–903. doi: 10.1016/S1001-0742(10)60493-7. [DOI] [PubMed] [Google Scholar]
- 18.Al-Shatri MA, Nuhu AA, Basheer C. 2014. Determination of haloacetic acids in bottled and tap water sources by dispersive liquid-liquid microextraction and GC-MS analysis. ScientificWorldJournal 2014:695049. doi: 10.1155/2014/695049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CY, Chang SN, Wang GS. 2009. Determination of ten haloacetic acids in drinking water using high-performance and ultra-performance liquid chromatography-tandem mass spectrometry. J Chromatogr Sci 47:67–74. doi: 10.1093/chromsci/47.1.67. [DOI] [PubMed] [Google Scholar]
- 20.Xie YF. 2004. Disinfection byproducts in drinking water: formation, analysis, and control. Lewis Publishers, Boca Raton, FL. [Google Scholar]
- 21.Villanueva CM, Kogevinas M, Grimalt JO. 2003. Haloacetic acids and trihalomethanes in finished drinking waters from heterogeneous sources. Water Res 37:953–958. doi: 10.1016/S0043-1354(02)00411-6. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Jun M-J, Lee M-H, Lee M-H, Eom S-W, Zoh K-D. 2010. Production of various disinfection byproducts in indoor swimming pool waters treated with different disinfection methods. Int J Hyg Environ Health 213:465–474. doi: 10.1016/j.ijheh.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Stacpoole PW, Harman EM, Curry SH, Baumgartner TG, Misbin RI. 1983. Treatment of lactic acidosis with dichloroacetate. N Engl J Med 309:390–396. doi: 10.1056/NEJM198308183090702. [DOI] [PubMed] [Google Scholar]
- 24.Stacpoole PW, Kurtz TL, Han Z, Langaee T. 2008. Role of dichloroacetate in the treatment of genetic mitochondrial diseases. Adv Drug Deliv Rev 60:1478–1487. doi: 10.1016/j.addr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michelakis ED, Webster L, Mackey JR. 2008. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br J Cancer 99:989–994. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stacpoole PW. 1989. The pharmacology of dichloroacetate. Metabolism 38:1124–1144. doi: 10.1016/0026-0495(89)90051-6. [DOI] [PubMed] [Google Scholar]
- 27.Stacpoole PW, Henderson GN, Yan ZM, James MO. 1998. Clinical pharmacology and toxicology of dichloroacetate. Environ Health Perspect 106:989–994. doi: 10.1289/ehp.98106s4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammini CV, Stacpoole PW. 2003. Biotransformation, toxicology and pharmacogenomics of dichloroacetate, p 215–234. In Gribble G (ed), Natural production of organohalogen compounds. Springer, Berlin, Germany. [Google Scholar]
- 29.Plewa MJ, Kargalioglu Y, Vankerk D, Minear RA, Wagner ED. 2002. Mammalian cell cytotoxicity and genotoxicity analysis of drinking water disinfection by-products. Environ Mol Mutagen 40:134–142. doi: 10.1002/em.10092. [DOI] [PubMed] [Google Scholar]
- 30.Stacpoole PW. 2011. The dichloroacetate dilemma: environmental hazard versus therapeutic goldmine—both or neither? Environ Health Perspect 119:155–158. doi: 10.1289/ehp.1002554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong Z, Board PG, Anders MW. 1998. Glutathione transferase zeta catalyses the oxygenation of the carcinogen dichloroacetic acid to glyoxylic acid. Biochem J 331:371–374. doi: 10.1042/bj3310371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong Z, Board PG, Anders MW. 1998. Glutathione transferase zeta-catalyzed biotransformation of dichloroacetic acid and other alpha-haloacids. Chem Res Toxicol 11:1332–1338. doi: 10.1021/tx980144f. [DOI] [PubMed] [Google Scholar]
- 33.Pandey T, Chhetri G, Chinta R, Kumar B, Singh DB, Tripathi T, Singh AK. 2015. Functional classification and biochemical characterization of a novel rho class glutathione S-transferase in Synechocystis PCC 6803. FEBS Open Bio 5:1–7. doi: 10.1016/j.fob.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motosugi K, Esaki N, Soda K. 1982. Purification and properties of a new enzyme, dl-2-haloacid dehalogenase, from Pseudomonas sp. J Bacteriol 150:522–527. doi: 10.1128/JB.150.2.522-527.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Ploeg J, van Hall G, Janssen DB. 1991. Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene. J Bacteriol 173:7925–7933. doi: 10.1128/jb.173.24.7925-7933.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diez A, Prieto MI, Alvarez MJ, Bautista JM, Puyet A, Pertierra G. 1996. Purification and properties of a high-affinity l-2-haloacid dehalogenase from Azotobacter sp. strain RC26. Lett Appl Microbiol 23:279–282. doi: 10.1111/j.1472-765X.1996.tb00189.x. [DOI] [Google Scholar]
- 37.Novak HR, Sayer C, Isupov MN, Paszkiewicz K, Gotz D, Mearns Spragg A, Littlechild JA. 2013. Marine Rhodobacteraceae l-haloacid dehalogenase contains a novel His/Glu dyad that could activate the catalytic water. FEBS J 280:1664–1680. doi: 10.1111/febs.12177. [DOI] [PubMed] [Google Scholar]
- 38.Adamu A, Wahab RA, Huyop F. 2016. l-2-Haloacid dehalogenase (DehL) from Rhizobium sp. RC1. SpringerPlus 5:695. doi: 10.1186/s40064-016-2328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinze U, Rehm H-J. 1993. Biodegradation of dichloroacetic acid by entrapped and adsorptive immobilized Xanthobacter autotrophicus GJ10. Appl Microbiol Biotechnol 40:158–164. doi: 10.1007/BF00170445. [DOI] [Google Scholar]
- 40.Zhang P, LaPara TM, Goslan EH, Xie Y, Parsons SA, Hozalski RM. 2009. Biodegradation of haloacetic acids by bacterial isolates and enrichment cultures from drinking water systems. Environ Sci Technol 43:3169–3175. doi: 10.1021/es802990e. [DOI] [PubMed] [Google Scholar]
- 41.Berthiaume C, Gilbert Y, Fournier-Larente J, Pluchon C, Filion G, Jubinville E, Sérodes J-B, Rodriguez M, Duchaine C, Charette SJ. 2014. Identification of dichloroacetic acid degrading Cupriavidus bacteria in a drinking water distribution network model. J Appl Microbiol 116:208–221. doi: 10.1111/jam.12353. [DOI] [PubMed] [Google Scholar]
- 42.Justicia-Leon SD, Ritalahti KM, Mack EE, Löffler FE. 2012. Dichloromethane fermentation by a Dehalobacter sp. in an enrichment culture derived from pristine river sediment. Appl Environ Microbiol 78:1288–1291. doi: 10.1128/AEM.07325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen G, Kleindienst S, Griffiths D, Erin Mack E, Seger E, Löffler F. 2017. Mutualistic interaction between dichloromethane- and chloromethane-degrading bacteria in an anaerobic mixed culture. Environ Microbiol 19:4784–4796. doi: 10.1111/1462-2920.13945. [DOI] [PubMed] [Google Scholar]
- 44.Chen G, Fisch AR, Gibson CM, Erin Mack E, Seger ES, Campagna SR, Löffler FE. 2020. Mineralization versus fermentation: evidence for two distinct anaerobic bacterial degradation pathways for dichloromethane. ISME J 14:959–970. doi: 10.1038/s41396-019-0579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleindienst S, Higgins SA, Tsementzi D, Chen G, Konstantinidis KT, Mack EE, Löffler FE. 2017. ‘Candidatus Dichloromethanomonas elyunquensis’ gen. nov., sp. nov., a dichloromethane-degrading anaerobe of the Peptococcaceae family. Syst Appl Microbiol 40:150–159. doi: 10.1016/j.syapm.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Kleindienst S, Higgins SA, Tsementzi D, Konstantinidis KT, Mack EE, Löffler FE. 2016. Draft genome sequence of a strictly anaerobic dichloromethane-degrading bacterium. Genome Announc 4:e00037-16. doi: 10.1128/genomeA.00037-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleindienst S, Chourey K, Chen G, Murdoch RW, Higgins SA, Iyer R, Campagna SR, Mack EE, Seger ES, Hettich RL, Löffler FE. 2019. Proteogenomics reveals novel reductive dehalogenases and methyltransferases expressed during anaerobic dichloromethane metabolism. Appl Environ Microbiol 85:e02768-18. doi: 10.1128/AEM.02768-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolfing J, Janssen DB. 1994. Estimates of Gibbs free energies of formation of chlorinated aliphatic compounds. Biodegradation 5:21–28. [Google Scholar]
- 49.Moghaddam AP, Abbas R, Fisher J, Stavrou S, Lipscomb J. 1996. Formation of dichloroacetic acid by rat and mouse gut microflora, an in vitro study. Biochem Biophys Res Commun 228:639–645. doi: 10.1006/bbrc.1996.1709. [DOI] [PubMed] [Google Scholar]
- 50.De Wever H, Cole JR, Fettig MR, Hogan DA, Tiedje JM. 2000. Reductive dehalogenation of trichloroacetic acid by Trichlorobacter thiogenes gen. nov., sp. nov. Appl Environ Microbiol 66:2297–2301. doi: 10.1128/aem.66.6.2297-2301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nevin KP, Holmes DE, Woodard TL, Covalla SF, Lovley DR. 2007. Reclassification of Trichlorobacter thiogenes as Geobacter thiogenes comb. nov. Int J Syst Evol Microbiol 57:463–466. doi: 10.1099/ijs.0.63408-0. [DOI] [PubMed] [Google Scholar]
- 52.Chan WY, Wong M, Guthrie J, Savchenko AV, Yakunin AF, Pai EF, Edwards EA. 2010. Sequence- and activity-based screening of microbial genomes for novel dehalogenases. Microb Biotechnol 3:107–120. doi: 10.1111/j.1751-7915.2009.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayes JD, Flanagan JU, Jowsey IR. 2005. Glutathione transferases. Annu Rev Pharmacol Toxicol 45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 54.Leisinger T, Braus-Stromeyer SA. 1995. Bacterial growth with chlorinated methanes. Environ Health Perspect 103:33–36. doi: 10.1289/ehp.95103s433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vuilleumier S, Pagni M. 2002. The elusive roles of bacterial glutathione S-transferases: new lessons from genomes. Appl Microbiol Biotechnol 58:138–146. doi: 10.1007/s00253-001-0836-0. [DOI] [PubMed] [Google Scholar]
- 56.Kurihara T, Esaki N, Soda K. 2000. Bacterial 2-haloacid dehalogenases: structures and reaction mechanisms. J Mol Catal B Enzym 10:57–65. doi: 10.1016/S1381-1177(00)00108-9. [DOI] [Google Scholar]
- 57.Kurihara T, Esaki N. 2008. Bacterial hydrolytic dehalogenases and related enzymes: occurrences, reaction mechanisms, and applications. Chem Rec 8:67–74. doi: 10.1002/tcr.20141. [DOI] [PubMed] [Google Scholar]
- 58.Friedrich M, Schink B. 1995. Electron transport phosphorylation driven by glyoxylate respiration with hydrogen as electron donor in membrane vesicles of a glyoxylate-fermenting bacterium. Arch Microbiol 163:268–275. doi: 10.1007/BF00393379. [DOI] [PubMed] [Google Scholar]
- 59.Friedrich M, Springer N, Ludwig W, Schink B. 1996. Phylogenetic positions of Desulfofustis glycolicus gen. nov., sp. nov. and Syntrophobotulus glycolicus gen. nov., sp. nov., two new strict anaerobes growing with glycolic acid. Int J Syst Evol Microbiol 46:1065–1069. [DOI] [PubMed] [Google Scholar]
- 60.Han C, Mwirichia R, Chertkov O, Held B, Lapidus A, Nolan M, Lucas S, Hammon N, Deshpande S, Cheng J-F, Tapia R, Goodwin L, Pitluck S, Huntemann M, Liolios K, Ivanova N, Pagani I, Mavromatis K, Ovchinikova G, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Brambilla E-M, Rohde M, Spring S, Sikorski J, Göker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk H-P, Detter JC. 2011. Complete genome sequence of Syntrophobotulus glycolicus type strain (FlGlyRT). Stand Genomic Sci 4:371–380. doi: 10.4056/sigs.2004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cornick NA, Allison MJ. 1996. Anabolic incorporation of oxalate by Oxalobacter formigenes. Appl Environ Microbiol 62:3011–3013. doi: 10.1128/AEM.62.8.3011-3013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakai S, Inokuma K, Nakashimada Y, Nishio N. 2008. Degradation of glyoxylate and glycolate with ATP synthesis by a thermophilic anaerobic bacterium, Moorella sp. strain HUC22-1. Appl Environ Microbiol 74:1447–1452. doi: 10.1128/AEM.01421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mägli A, Messmer M, Leisinger T. 1998. Metabolism of dichloromethane by the strict anaerobe Dehalobacterium formicoaceticum. Appl Environ Microbiol 64:646–650. doi: 10.1128/AEM.64.2.646-650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Menon S, Ragsdale SW. 1996. Evidence that carbon monoxide is an obligatory intermediate in anaerobic acetyl-CoA synthesis. Biochemistry 35:12119–12125. doi: 10.1021/bi961014d. [DOI] [PubMed] [Google Scholar]
- 65.Egli C, Thüer M, Suter D, Cook AM, Leisinger T. 1989. Monochloro- and dichloroacetic acids as carbon and energy-sources for a stable, methanogenic mixed culture. Arch Microbiol 152:218–223. doi: 10.1007/BF00409654. [DOI] [Google Scholar]
- 66.Balch WE, Schoberth S, Tanner RS, Wolfe RS. 1977. Acetobacterium, a new genus of hydrogen-oxidizing, carbon dioxide-reducing, anaerobic bacteria. Int J Syst Evol Microbiol 27:355–361. doi: 10.1099/00207713-27-4-355. [DOI] [Google Scholar]
- 67.Leadbetter JR, Schmidt TM, Graber JR, Breznak JA. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686–689. doi: 10.1126/science.283.5402.686. [DOI] [PubMed] [Google Scholar]
- 68.Cord-Ruwisch R, Seitz H-J, Conrad R. 1988. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch Microbiol 149:350–357. doi: 10.1007/BF00411655. [DOI] [Google Scholar]
- 69.Löffler FE, Tiedje JM, Sanford RA. 1999. Fraction of electrons consumed in electron acceptor reduction and hydrogen thresholds as indicators of halorespiratory physiology. Appl Environ Microbiol 65:4049–4056. doi: 10.1128/AEM.65.9.4049-4056.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lladó S, López-Mondéjar R, Baldrian P. 2017. Forest soil bacteria: diversity, involvement in ecosystem processes, and response to global change. Microbiol Mol Biol Rev 81:e00063-16. doi: 10.1128/MMBR.00063-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin N-R, Whon TW, Bae J-W. 2015. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 73.Löffler FE, Sanford RA, Ritalahti KM. 2005. Enrichment, cultivation, and detection of reductively dechlorinating bacteria. Methods Enzymol 397:77–111. doi: 10.1016/S0076-6879(05)97005-5. [DOI] [PubMed] [Google Scholar]
- 74.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 78.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 81.Yang S, Giannone RJ, Dice L, Yang ZK, Engle NL, Tschaplinski TJ, Hettich RL, Brown SD. 2012. Clostridium thermocellum ATCC27405 transcriptomic, metabolomic and proteomic profiles after ethanol stress. BMC Genomics 13:336. doi: 10.1186/1471-2164-13-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson CW, Abraham PE, Linger JG, Khanna P, Hettich RL, Beckham GT. 2017. Eliminating a global regulator of carbon catabolite repression enhances the conversion of aromatic lignin monomers to muconate in Pseudomonas putida KT2440. Metab Eng Commun 5:19–25. doi: 10.1016/j.meteno.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tabb DL, Fernando CG, Chambers MC. 2007. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res 6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma Z-Q, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, Tabb DL. 2009. IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J Proteome Res 8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y-Y, Chambers MC, Li M, Ham A-JL, Turner JL, Zhang B, Tabb DL. 2013. IDPQuantify: combining precursor intensity with spectral counts for protein and peptide quantification. J Proteome Res 12:4111–4121. doi: 10.1021/pr400438q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein M, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 87.Karpievitch YV, Dabney AR, Smith RD. 2012. Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics 13:S5. doi: 10.1186/1471-2105-13-S16-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lazar C, Gatto L, Ferro M, Bruley C, Burger T. 2016. Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J Proteome Res 15:1116–1125. doi: 10.1021/acs.jproteome.5b00981. [DOI] [PubMed] [Google Scholar]
- 89.Amos BK, Christ JA, Abriola LM, Pennell KD, Löffler FE. 2007. Experimental evaluation and mathematical modeling of microbially enhanced tetrachloroethene (PCE) dissolution. Environ Sci Technol 41:963–970. doi: 10.1021/es061438n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Change of H2 and CO concentrations in abiotic and biotic control incubations and CO concentrations in RM cultures during growth with DCM. Download FIG S1, PDF file, 0.2 MB (207.6KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Taxonomy, relative abundance, and representative sequences of OTUs identified through 16S rRNA gene amplicon sequencing. Download Table S1, XLSX file, 0.01 MB (12.2KB, xlsx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Conversion of DCA to glyoxylate in cell-free extracts of E. coli transformants expressing the “Ca. Dichloromethanomonas elyunquensis” strain RM had1 or had2 gene. Download FIG S2, PDF file, 1.1 MB (1.1MB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro conversion of monochloroacetate (MCA) to glycolate using the purified HAD1 protein. Download FIG S3, PDF file, 0.08 MB (82.8KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of “Ca. Dichloromethanomonas elyunquensis” during DCA and DCM catabolism and collection of samples for comparative proteome analysis. Download FIG S4, PDF file, 2.1 MB (2.1MB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Summary of proteins identified in RM cultures grown with DCA or DCM at two sampling points. Samples were collected immediately before the 2nd electron donor amendment (TP1) and near the end of electron donor consumption (TP2); approximately 95% of the initial amount of electron donor was consumed (Fig. S4). Imputed values are shown in orange. Download Table S2, XLSX file, 0.6 MB (643.7KB, xlsx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Growth of “Ca. Dichloromethanomonas elyunquensis” in mixed culture RM with glyoxylate as the sole electron donor as determined by 16S rRNA gene-targeted qPCR. Download FIG S5, PDF file, 0.2 MB (227.9KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Postulated anaerobic DCM metabolic pathway in “Ca. Dichloromethanomonas elyunquensis” strain RM. DCM is metabolized via the Wood-Ljungdahl pathway (WLP). Download FIG S6, PDF file, 0.04 MB (45.8KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inhibitory effect of H2 on DCA degradation in mixed culture RM. Download FIG S7, PDF file, 0.9 MB (949.2KB, pdf) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers, plasmids, and E. coli strains used for cloning and overexpression of had genes. Download Table S3, XLSX file, 0.01 MB (14KB, xlsx) .
Copyright © 2021 Chen et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.