There is an increasing understanding of the clinical correlations and potential mechanistic roles of specific members of the gut and tumoral microbiota in colorectal cancer (CRC) initiation, progression, and survival. However, we have yet to parlay this knowledge into better CRC outcomes through microbially informed diagnostic, preventive, or therapeutic approaches.
KEYWORDS: Fusobacterium nucleatum, aspirin, colon cancer
ABSTRACT
Aspirin is a chemopreventive agent for colorectal adenoma and cancer (CRC) that, like many drugs inclusive of chemotherapeutics, has been investigated for its effects on bacterial growth and virulence gene expression. Given the evolving recognition of the roles for bacteria in CRC, in this work, we investigate the effects of aspirin with a focus on one oncomicrobe—Fusobacterium nucleatum. We show that aspirin and its primary metabolite salicylic acid alter F. nucleatum strain Fn7-1 growth in culture and that aspirin can effectively kill both actively growing and stationary Fn7-1. We also demonstrate that, at levels that do not inhibit growth, aspirin influences Fn7-1 gene expression. To assess whether aspirin modulation of F. nucleatum may be relevant in vivo, we use the ApcMin/+ mouse intestinal tumor model in which Fn7-1 is orally inoculated daily to reveal that aspirin-supplemented chow is sufficient to inhibit F. nucleatum-potentiated colonic tumorigenesis. We expand our characterization of aspirin sensitivity across other F. nucleatum strains, including those isolated from human CRC tissues, as well as other CRC-associated microbes, enterotoxigenic Bacteroides fragilis, and colibactin-producing Escherichia coli. Finally, we determine that individuals who use aspirin daily have lower fusobacterial abundance in colon adenoma tissues, as determined by quantitative PCR performed on adenoma DNA. Together, our data support that aspirin has direct antibiotic activity against F. nucleatum strains and suggest that consideration of the potential effects of aspirin on the microbiome holds promise in optimizing risk-benefit assessments for use of aspirin in CRC prevention and management.
INTRODUCTION
The microbiome and specific members thereof are increasingly recognized for their potential contributions to the initiation and progression of colorectal cancer (CRC). CRC-associated microbes—including Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, and colibactin-producing Escherichia coli—influence carcinogenesis through a number of mechanisms, such as: inducing host cell DNA damage (1), shaping the tumor-immune microenvironment (2–4), and promoting metastasis (5, 6). Thus, these bacteria are an increasing focus for CRC diagnostics and therapeutics.
F. nucleatum is a Gram-negative anaerobe found commonly in the human oral cavity but rarely in stool. F. nucleatum is specifically enriched in colonic adenomas and CRC compared with normal colonic tissues (3, 7–11). In cell culture and animal models, F. nucleatum increases intestinal cancer cell proliferation (12, 13), localizes to CRC tissues (14), and influences immune responses within the tumor microenvironment (3, 4). Intratumoral fusobacterial burden is associated with poorer patient prognosis (15, 16) and CRC recurrence after treatment (17). These observations support the idea that F. nucleatum is a potential target for CRC prevention and treatment. A preclinical study using metronidazole demonstrated slower growth of mouse-implanted patient-derived xenografts harboring F. nucleatum (18), providing a proof of concept that modulating F. nucleatum levels could slow CRC growth. However, antibiotic resistance and antibiotic-induced dysbiosis highlight the need to identify alternative agents that might similarly be used without such concerns.
Aspirin, acetylsalicylic acid, is a nonsteroidal anti-inflammatory drug (NSAID) that targets cyclooxygenase-2 (COX-2; or prostaglandin-endoperoxide synthase 2) to inhibit prostaglandin biosynthesis (19, 20). Widely used for pain and inflammation, aspirin is recommended by the United States Preventive Services Task Force to prevent CRC and cardiovascular disease in certain populations (21). Meta-analyses support aspirin as a highly effective CRC chemopreventive treatment (22, 23). However, conflicting results in mouse models (24–26), as well as a recent work in which concurrent antibiotic treatment was necessary to observe aspirin’s antitumoral effects (27), suggest that part of aspirin’s efficacy as a CRC chemopreventive may be mediated by the microbiota. Aspirin and other NSAIDs cause shifts in the microbiota (28–30). Aspirin and salicylic acid, its primary bioactive metabolite, directly affect bacteria by inhibiting growth (31, 32) and altering virulence factor expression (33–37). While some bacteria have specific salicylic acid-responsive regulators (38, 39), how aspirin and salicylic acid drive these changes in other bacteria is less understood. Given these responses, the microbiota has been proposed as a potential mechanism for aspirin chemoprevention and a rich target for precision prevention biomarkers (40).
As aspirin is already employed for CRC prevention, although it is underutilized, and can influence other bacteria through both its antimicrobial and regulatory effects, we posited that aspirin might affect F. nucleatum growth or behavior. Here, we examine the effects of aspirin on F. nucleatum both in culture and during tumorigenesis to ascertain if aspirin holds potential for modulating F. nucleatum-associated CRC outcomes.
RESULTS
Aspirin and salicylic acid alter Fn7-1 growth in culture.
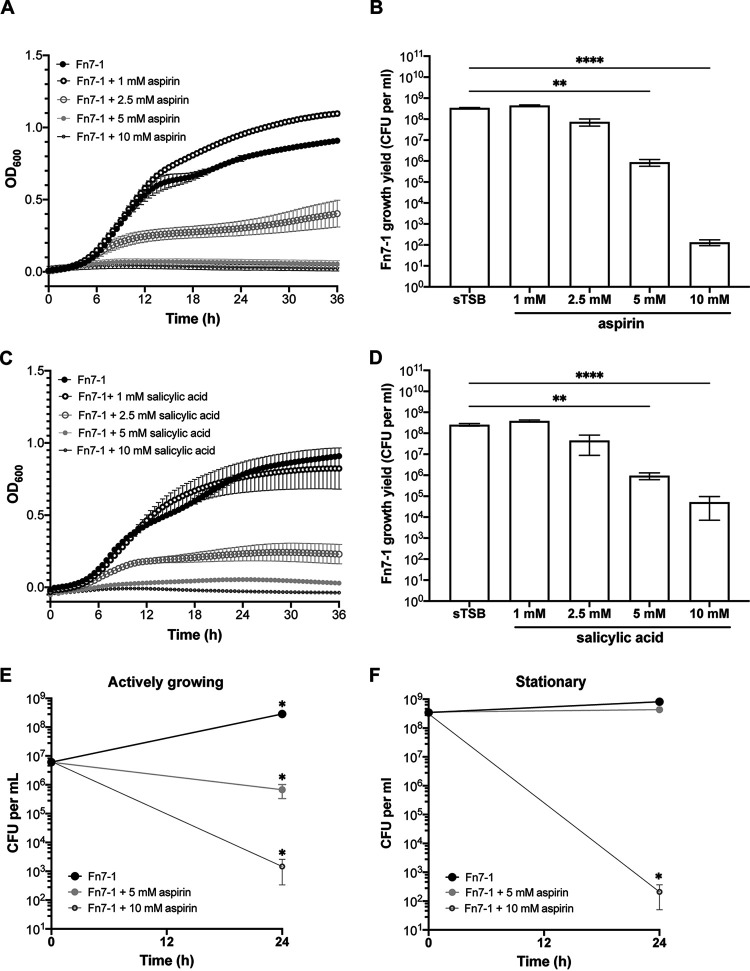
To probe whether aspirin affects the well-studied F. nucleatum strain Fn7-1, we assayed growth in media supplemented with increasing aspirin concentrations comparable to those used in prior studies (31, 33, 41) (Fig. 1A). We adjusted the pH of all media after aspirin or salicylic acid addition to match the control medium, to avoid conflating aspirin-specific and pH-based responses. In 1 mM aspirin, Fn7-1 growth mildly increased, as determined by maximum optical density (optical density at 600 nm [OD600]), despite indistinguishable log-phase growth from Fn7-1 grown in control medium (supplemented tryptic soy broth [sTSB]) (Fig. 1A). However, higher levels of aspirin slowed or entirely inhibited Fn7-1 growth. Because OD600 is a proxy for bacterial abundance and affected by aggregation and cell morphology, we determined growth yield by CFUs from cultures grown in the presence or absence of aspirin for 24 h (Fig. 1B). Beginning at 2.5 mM aspirin, we observed decreased growth yield (4-fold, but not statistically significant) compared with that in sTSB. In 5 mM and 10 mM aspirin, Fn7-1 growth yield was significantly reduced by 388-fold and over 106-fold, respectively. These data demonstrate that aspirin can inhibit Fn7-1 growth in culture.
FIG 1.
Fn7-1 responses to aspirin and salicylic acid in culture. (A and C) Growth curve as determined by OD600 for Fn7-1 grown in sTSB supplemented with different concentrations of aspirin or salicylic acid. (B and D) Fn7-1 growth after 24 h in indicated media, as determined by final CFUs per ml of culture. (E and F) Fn7-1 growth at t0 and after 24 h in indicated media, as determined by CFU per ml of culture. For actively growing cells (E), cultures were inoculated 1:100 from an overnight preculture into indicated media. For stationary-phase cells (F), cells were pelleted and washed into fresh, prereduced indicated media. All data represent the mean ± SEM for at least 6 cultures. For A and C, data were analyzed by two-way repeated measures analysis of variance (ANOVA) with post hoc Dunnett’s test. For aspirin, samples are significantly different from sTSB at P values of <0.05 beginning at 7.5 h (5 mM and 10 mM), 9.5 h (2.5 mM), or 17 h (1 mM). For salicylic acid, samples are significantly different from sTSB at P values of <0.05 beginning at 8 h (10 mM), 8.5 h (5 mM), or 11.5 h (2.5 mM). For B and D, ** indicates P values of <0.01 and **** indicates P values of <0.0001 as determined by Kruskal-Wallis test with post hoc Dunn’s test for multiple comparisons to the control medium sample. For E and F, Wilcoxon signed-rank test was performed for each condition at 24 h compared with t0. *, P < 0.05.
We next probed whether salicylic acid, aspirin’s primary metabolite, affected Fn7-1 growth. In OD600 and growth yield experiments (Fig. 1C and D), we observed similar inhibition of Fn7-1 growth. While there are subtle differences between the responses to aspirin and salicylic acid, such as greater sensitivity to 10 mM aspirin than to 10 mM salicylic acid, the responses to both compounds were largely similar. Because of the clinical relevance of aspirin in contrast to salicylic acid, we focused on aspirin in further studies.
We next investigated if aspirin could efficiently kill Fn7-1. We tested actively growing cells by subculturing Fn7-1 into sTSB or media supplemented with 5 mM or 10 mM aspirin (Fig. 1E). There was a significant decrease in CFU for both aspirin media, suggesting that aspirin was not only inhibiting growth but also reducing culture viability. In 10 mM aspirin, cell viability was reduced by ∼3 logs from the time of inoculation (t0). To address if aspirin could kill stationary cells, we grew Fn7-1 overnight in the absence of aspirin and then transferred cells into media containing aspirin (Fig. 1F). Under these conditions, Fn7-1 growth yield doubled only over 24 h in sTSB, supporting the conclusion that these cells were not rapidly growing and were likely in stationary phase, despite being washed into fresh medium to allow pH matching with the aspirin-supplemented media. In 5 mM aspirin, there was effectively no change in CFU over 24 h. However, when cells were washed into 10 mM aspirin, there was a 6-log decrease in Fn7-1 CFUs compared with those of t0, a reproducible but surprising result given the results for 5 mM. These data support that, in addition to inhibiting Fn7-1 growth, aspirin is able to reduce viable Fn7-1 cells, under both actively growing and stationary states.
Subinhibitory concentrations of aspirin affect Fn7-1 gene expression.
We observed subtle changes in phenotypes associated with autoaggregation when Fn7-1 was grown in subinhibitory levels of aspirin. In 1 mM aspirin, cells still formed aggregates, as Fn7-1 does under these conditions, but they appeared looser (see Fig. S1A in the supplemental material). Unlike Fn7-1 grown in the sTSB where clumping leads to large fluctuations in OD600 during stationary phase (Fig. S1B), the OD600 of Fn7-1 grown in 1 mM aspirin remained more consistent (Fig. S1C). As aspirin and salicylic downregulate surface proteins in some bacteria (35, 36), we posited that aspirin may have similar effects on Fn7-1 gene expression, as suggested by these autoaggregation observations.
Changes to Fn7-1 autoaggregation when grown in 1 mM aspirin. (A) Fn7-1 grown in sTSB or sTSB supplemented with 1 mM aspirin demonstrates different clumping behaviors. (B and C) OD600 data from two individual wells of Fn7-1 grown in either sTSB or sTSB with 1 mM aspirin. For this experiment, Fn7-1 was grown in a 96-well plate to better visualize autoaggregation phenotypes. Download FIG S1, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To address this possibility, we performed RNA sequencing (RNA-seq) on Fn7-1 cells grown overnight in sTSB and sTSB supplemented with 1 mM aspirin (Fig. 2A; Data Set S1 in the supplemental material). Consistent with prior studies (33, 42), we observed that this subinhibitory level of aspirin led to a largely downregulatory shift in gene expression. Using a cutoff P value of <0.05 and a 2-fold change, we observed 53 genes with significantly lower expression and only 2 genes (both encoding proteins of unknown function) with significantly higher expression when Fn7-1 was grown in 1 mM aspirin. Genes with reduced expression in 1 mM aspirin include fap2, encoding an autotransporter with important roles in CRC and other host interactions (4, 14, 43, 44), as well as FSDG_01349 and FSDG_01370, also encoding autotransporters, a class of potential virulence factors abundant in F. nucleatum genomes (45). In Fn23726, the FSDG_01349 homolog (Gene_2067) is also genomically linked to fap2 (45). Another downregulated gene, FSDG_00378, encodes a β-barrel protein, like the autotransporters, and is similarly predicted to localize to the outer membrane. Given their subcellular localization, one of these genes may be responsible for the altered autoaggregation, but Fn7-1’s lack of genetic tractability makes it difficult to functionally test. Expression of housekeeping genes involved in transcription (rpoC), translation (rpsE and rplF), and cell division (mreB) were also reduced in response to aspirin, which may provide insight into how aspirin reduces Fn7-1 growth. Other downregulated genes include genes in the ϕFunu1 phage locus (46), a gene upregulated during Caco2 infection (oppD5) (47), and macB, encoding an efflux pump predicted to play a role in macrolide export and consistent with aspirin and salicylic acid altering antibiotic sensitivity in other bacteria (48–51). Overall, these results support a conclusion that subinhibitory aspirin alters Fn7-1 gene expression; however, our interpretation is limited by homology-based inferred functions and an overrepresentation of genes of unknown function.
FIG 2.
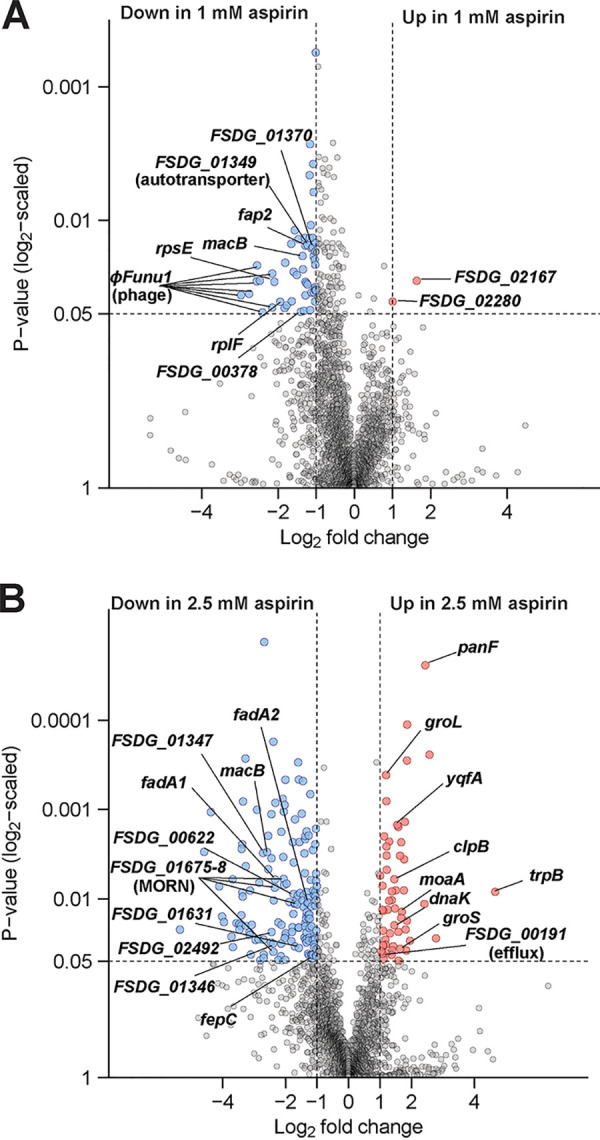
Downregulation of Fn7-1 gene expression in response to aspirin in culture. Volcano plots of RNA-seq expression data from Fn7-1 grown in sTSB compared with media supplemented with 1 mM aspirin (A) or 2.5 mM aspirin (B) for 24 h. Data represent 2 replicates for Fn7-1 grown in sTSB and 3 replicates each for Fn7-1 grown in 1 mM or 2.5 mM aspirin. Cutoffs indicate 2-fold changes in expression and P values <0.05 as described in the Materials and Methods. A full list of differentially expressed genes can be found in Data Set S1 and S2.
Differentially expressed Fn7-1 genes in response to 1 mM aspirin. Download Data Set S1, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed Fn7-1 genes in response to 2.5 mM aspirin. Download Data Set S2, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To further probe how aspirin inhibits Fn7-1 growth, we assessed gene expression in 2.5 mM aspirin, a dosage at which Fn7-1 growth was reduced (Fig. 1A) while providing sufficient RNA yield. In response to 2.5 mM aspirin, 55 genes were significantly upregulated and 155 genes significantly downregulated using the same cutoff (Fig. 2B; see Data Set S2 in the supplemental material). These changes represent over 8% of the predicted coding sequences in Fn7-1, suggesting that global changes in response to this higher dosage are indicative of a general stress response. Consistent with this hypothesis, we observed upregulation of several chaperone-related genes, including groS, groL, clpB, and dnaK. Of the genes downregulated in 1 mM aspirin, 15 are also downregulated in 2.5 mM and may represent an aspirin-specific response as opposed to a general stress or death response. These genes include macB and its neighboring genes FSDG_01346 and FSDG_01347, as well as fepC, predicted to be involved in iron transport. In contrast to the downregulation of macB, another gene predicted to encode an efflux pump protein, FSDG_00191, is upregulated in 2.5 mM aspirin, which together could conversely affect how aspirin might alter antibiotic sensitivity in F. nucleatum. As with 1 mM aspirin, we observed downregulation of potential virulence factors in 2.5 mM aspirin, including two genes encoding FadA-like domains (12, 52) (FSDG_02507 and FSDG_02530), multiple autotransporters (FSDG_00622, FSDG_01631, and FSDG_02492), and a locus of genes (FSDG_01675-01678) predicted to encode MORN domain-containing proteins that are overrepresented in invasive F. nucleatum strains (52). These data support that aspirin alters expression of Fn7-1 genes that have the potential to affect how Fn7-1 behaves and interacts with host cells.
Aspirin treatment reduces Fn7-1-potentiated tumorigenesis in the ApcMin/+ model.
We next sought to determine if aspirin might affect Fn7-1-potentiated intestinal tumorigenesis, given (i) the pleiotropic effects of different levels of aspirin on Fn7-1 in culture, (ii) the difficulty of determining colonic intraluminal aspirin concentration to investigate in culture-based experiments, and (iii) noted discrepancies in how bacteria respond to antimicrobial compounds in vivo and in vitro (53, 54). Using the ApcMin/+ mouse model of intestinal tumorigenesis in which Fn7-1 is orally inoculated into conventionally reared mice daily beginning at 6 weeks of age (3), we concurrently transitioned mice to either a control chow or one supplemented with 200 ppm aspirin, a dosage used in previous studies that demonstrated mild, if any, effects on tumorigenesis (24, 25). After 8 weeks of treatment, consistent with our prior study (3), we observed that Fn7-1 treatment led to a significant increase in colonic adenoma burden in mice on the control chow, compared with mice orally inoculated with medium alone (sham mice) on the control chow that rarely develop colonic adenomas in our facility (Fig. 3). Similarly, sham mice placed on the aspirin chow seldomly developed colonic adenomas. In contrast, when mice treated with Fn7-1 were placed on the aspirin-supplemented chow, we saw complete abrogation of colonic adenoma development. These in vivo results suggest a specific role for aspirin in blocking Fn7-1-potentiated intestinal tumorigenesis.
FIG 3.
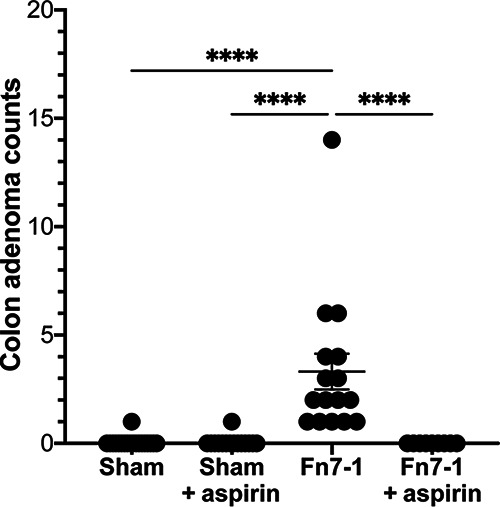
Colonic adenoma burden in the murine ApcMin/+ model in response to daily Fn7-1 instillation and aspirin supplementation. Conventional specific-pathogen-free ApcMin/+ mice were orally inoculated with Fn7-1 or a sham control (sTSB) daily and simultaneously maintained on either a control chow or a chow supplemented with 200 ppm aspirin, with both treatments beginning at 6 weeks of age and continuing until 14 weeks. Mice were then sacrificed, and the colons were prepared for histological analysis for enumeration of colon adenomas. Data points represent adenoma counts from individual mice, bars indicate the mean ± SEM, and statistical analysis was performed by Kruskal-Wallis test with post hoc Dunn’s test for multiple comparisons. ****, P < 0.0001.
Comparative survey of aspirin sensitivity in F. nucleatum ATCC strains and clinical tumor isolates.
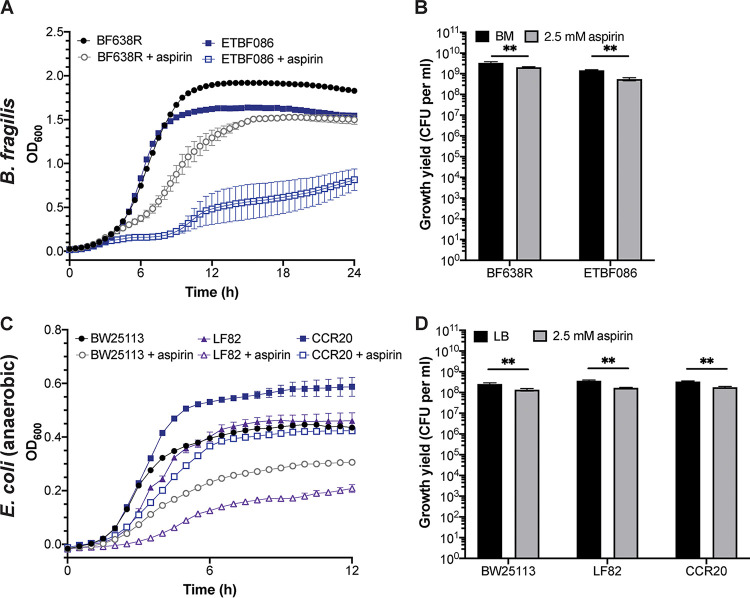
Given our results in culture and in mice, we next explored how aspirin might affect F. nucleatum as it relates to human CRC by assessing the sensitivity of other F. nucleatum strains, including isolates from human CRC tissues. We first examined how aspirin affects the ATCC strains Fn23726 (F. nucleatum subsp. nucleatum) and Fn10953 (F. nucleatum subsp. polymorphum). We performed growth curve assays for these strains in sTSB or in the presence of 1 mM and 2.5 mM aspirin (Fig. 4A). Unlike Fn7-1 which exhibited a slight growth increase at this dosage (Fig. 1A), even 1 mM aspirin was sufficient to fully inhibit Fn23726 growth. Similarly, Fn10953 demonstrated reduced growth at 1 mM and no appreciable growth at 2.5 mM aspirin. These observations were supported by strong decreases in yield (>5 logs lower CFU/ml than that in sTSB) for both Fn23276 and Fn10953 in 2.5 mM aspirin (Fig. 4B). These phenotypes demonstrated that these F. nucleatum strains are more sensitive to aspirin in culture than Fn7-1, for which we observed only ∼4-fold reduction in CFU in 2.5 mM aspirin (Fig. 1B).
FIG 4.
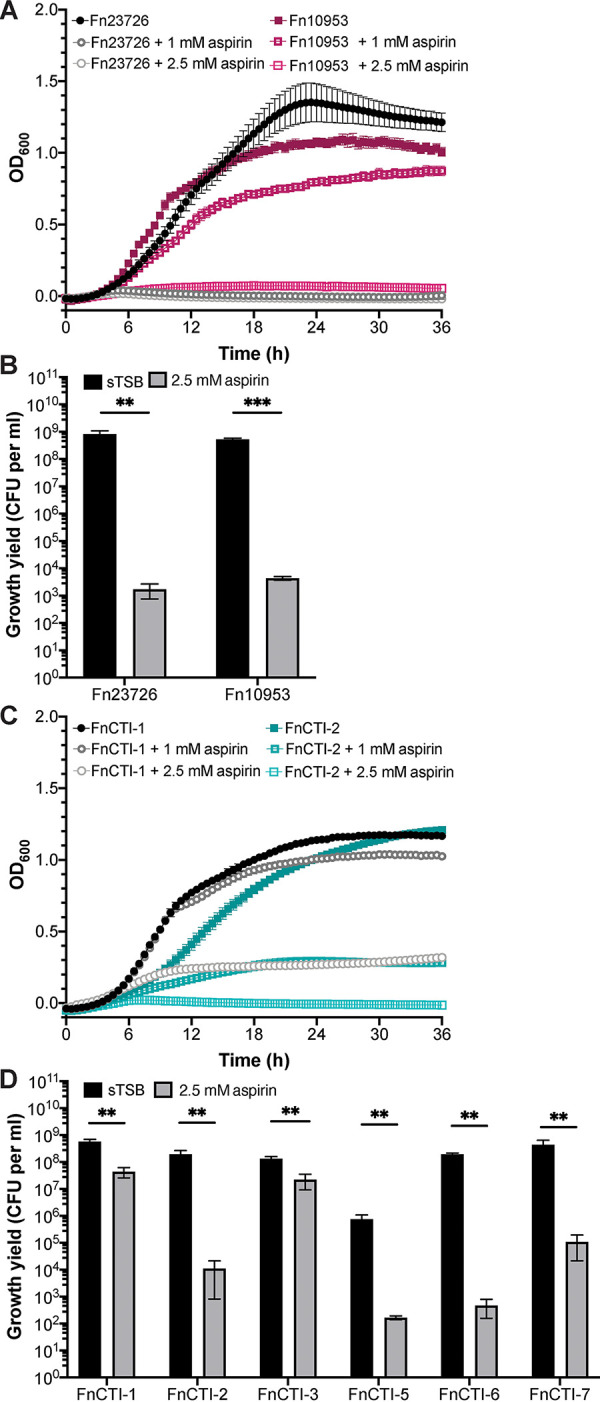
Growth inhibition of aspirin on ATCC and CRC isolates of F. nucleatum. Growth of ATCC strains Fn23726 and Fn10953 in response to aspirin as determined by optical density (A) and growth yield (CFU per ml) at 24 h (B). (C) Growth of the CRC F. nucleatum isolates FnCTI-1 and FnCTI-2 in response to aspirin as determined by optical density. (D) Growth yield of FnCTI-1, -2, -3, -5, -6, and -7 in response to 2.5 mM aspirin after 24 h as determined by CFU per ml. All data represent the mean ± SEM for at least 6 cultures. Growth curves were analyzed by two-way repeated measures ANOVA with post hoc Dunnett’s test. Growth was significantly different from sTSB at P values of <0.05 for each of the following strains at the indicated time points and concentrations: Fn23762 (1 mM and 2.5 mM at 7.5 h), Fn10953 (1 mM at 6.5 h and 2.5 mM at 5.5 h), FnCTI-1 (1 mM at 17.5 h and 2.5 mM at 7 h), and FnCTI-2 (1 mM at 9 h and 2.5 mM at 7 h). Analysis of growth yield data was performed by Mann-Whitney test. **, P < 0.01; ***, P < 0.001.
Given this observation, we asked if F. nucleatum strains isolated from human CRC tissues (FnCTI-1, -2, -3, -5, -6, and -7) (4) would behave more like Fn7-1, a clinical isolate from a patient with inflammatory bowel disease (55), or like the extraintestinal and lab-adapted ATCC strains Fn23726 and Fn10953. In growth curve assays, we observed varied phenotypes. FnCTI-1 exhibited only mildly reduced growth in the presence of 1 mM aspirin and was able to grow slightly in 2.5 mM, whereas the growth of FnCTI-2 was strongly reduced in 1 mM aspirin and entirely abrogated in 2.5 mM (Fig. 4C). The greater aspirin sensitivity of FnCTI-2 was supported by larger reductions in viable CFU in the presence of 2.5 mM aspirin for FnCTI-2 (>104-fold) than for FnCTI-1 (∼10-fold) (Fig. 4D). FnCTI-3 was the least aspirin-sensitive isolate, as determined by both minimal effects on growth yield (Fig. 4D) and OD600 (see Fig. S2A in the supplemental material). FnCTI-5, FnCTI-6, and FnCTI-7 all showed multilog decreases in growth yield by CFU in 2.5 mM aspirin (Fig. 4D) and intermediate OD600 phenotypes for FnCTI-6 and FnCTI-7 (Fig. S2B and C). FnCTI-5 exhibits strong clumping and cannot be accurately measured by OD600. Together, these data demonstrate that both CRC tumor isolates and ATCC strains of F. nucleatum are sensitive to aspirin in culture, frequently more than we observed for Fn7-1 (Fig. 1A and B), supporting the relevance of F. nucleatum aspirin sensitivity as we consider approaches to reduce fusobacterial burden.
Differential growth responses of F. nucleatum CTI strains in the presence of aspirin. Growth of FnCTI-3 (A), FnCTI-6 (B), and FnCTI-7 (C) in sTSB or media supplemented with 1 mM or 2.5 mM aspirin. All data represent the mean ± SEM for at least 6 cultures. Growth curves were analyzed by two-way repeated measures ANOVA with post hoc Dunnett’s test. Growth was significantly different from sTSB at a P value of <0.05 for each of the following strains beginning at the indicated timepoints and concentrations: CTI-3 (2.5 mM at 11.5 h), CTI-5 (1 mM at 8 h and 2.5 mM at 6 h), and FnCTI-7 (2.5 mM at 11 h). Download FIG S2, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effect of aspirin on growth of CRC-associated enterotoxigenic B. fragilis and colibactin-producing E. coli.
Beyond F. nucleatum, two of the best-characterized CRC-associated microbes are enterotoxigenic B. fragilis (ETBF), which drives a strong proinflammatory and protumorigenic response, and colibactin-producing E. coli, which induces DNA damage and a specific mutational signature that can be found in CRC tissues and increases tumor multiplicity in preclinical mouse models (1, 2, 5, 56–59). We used growth assays on ETBF and colibactin-producing E. coli, as well as additional B. fragilis and E. coli strains, to ascertain if these CRC-associated bacteria are similarly sensitive to aspirin and could represent additional targets for aspirin modulation. For B. fragilis, we investigated ETBF086, an enterotoxigenic isolate, and BF638R, a nontoxigenic isolate. Both B. fragilis strains exhibited reduced growth in 2.5 mM aspirin (Fig. 5A and B). However, while statistically significant, the effects were far milder than we observed for many of the F. nucleatum strains, and represented only less than 3-fold decreases in growth yield.
FIG 5.
CRC-associated microbe B. fragilis and E. coli responses to aspirin. Growth of B. fragilis strains BF638R and ETBF086 in basal medium (BM) or BM supplemented with 2.5 mM aspirin as determined by optical density (A) and growth yield (CFU per ml; B) after anaerobic growth for 24 h at 37°C. Growth of E. coli strains BW25113, LF82, and CCR20 and ETBF086 in LB or LB supplemented with 2.5 mM aspirin as determined by optical density (C) and growth yield (CFU per ml; D) after anaerobic growth for 24 h at 37°C. All data represent the mean ± SEM for at least 6 cultures. Growth curves were analyzed by two-way repeated measures ANOVA with post hoc Dunnett’s test, and growth in 1 mM aspirin was significantly different from control medium at P values of <0.05 for each of the following strains beginning at the indicated time points: BF638R (6 h), ETBF086 (5.5 h), BW25113 (3 h), LF82 (3.5 h), and CCR20 (3.5 h). Growth yield analysis was performed by Mann-Whitney test; **, P < 0.01.
For E. coli, we investigated the following: BW25113, a K-12 strain; LF82, an adherent-invasive strain; and CCR20, a colibactin-producing strain isolated from CRC tissues. All strains exhibited reduced growth in 2.5 mM aspirin compared with that in Luria-Bertani (LB) (Fig. 5C and D). Much like B. fragilis, however, the effects were statistically significant but mild, reducing growth yield by ∼2-fold. Because we performed the growth assays under anaerobic conditions, even for E. coli, we also determined growth yield for the E. coli strains in 2.5 mM aspirin under aerobic conditions and again observed minimal aspirin sensitivity (see Fig. S3 in the supplemental material). Taken together, these data suggest that, although both B. fragilis and E. coli exhibited slight growth inhibition in response to aspirin, many F. nucleatum strains are far more aspirin sensitive, at least under the conditions of our assays. Therefore, the observations we make using our mouse models and human study regarding the effects of aspirin on F. nucleatum in vivo may not extend to these other CRC-associated microbes.
Aspirin minimally affects E. coli strains grown under aerobic conditions. Growth yield (CFU per ml) of E. coli strains BW25113, LF82, and CCR20 in LB or LB supplemented with 2.5 mM aspirin for 18 h at 37°C under aerobic conditions. All data represent the mean ± SEM for at least 6 cultures. Growth yield analysis was performed by Mann-Whitney test and ** indicates a P value of <0.01. Download FIG S3, TIF file, 0.3 MB (316KB, tif) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Fusobacterial abundance is reduced in colonic adenomas from patients who use aspirin.
Given the aspirin sensitivity of F. nucleatum, including isolates from human CRC tissues, that we observed in culture, we posited that regular aspirin use may affect fusobacterial load in human tissues. As F. nucleatum is infrequently detected in healthy human stool samples, we used human colonic adenoma tissues, in which F. nucleatum is enriched (3, 10, 11), to determine if aspirin affects fusobacterial burden. We isolated DNA from 36 human adenoma samples and determined Fusobacterium sp. abundance, normalized to human PGT gene copies as a reference for adenoma DNA abundance, by quantitative PCR (qPCR) as previously described (8, 16). After data analysis when we were no longer blind to aspirin use, we compared the relative adenoma fusobacterial abundance across patients who reported taking aspirin daily and those whose reported no aspirin use (Fig. 6). We observed a 2.3-fold lower mean fusobacterial burden for patients taking aspirin. However, if we instead categorized adenomas as Fusobacterium high or Fusobacterium low/negative, as often done in the epidemiological literature, we observed 7 of 14 adenomas as Fusobacterium high in the control group and only 6 of 22 adenomas as Fusobacterium high in the aspirin group, using a relative fusobacterial load cutoff of 0.0001. We further analyzed the data comparing patients reporting low-dose (<325 mg daily) or high-dose (≥325 mg daily) aspirin use and found no dose-related differences between the two groups (Mann-Whitney test, P > 0.9999). These data demonstrate an association between aspirin use and reduced Fusobacterium sp. load in precancerous adenomas and suggest that the aspirin modulation of F. nucleatum we have described in culture may also occur in humans. Unfortunately, we were unable to determine the carriage of ETBF or colibactin-producing E. coli in the human adenoma samples we examined, as growth to enrich for either B. fragilis or E. coli isolates is required to assess their presence (58, 60) rather than direct lysis of the tissue, as performed for Fusobacterium sp. quantification. Thus, we could not test if these bacteria are less affected by aspirin use as our in vitro data might suggest.
FIG 6.
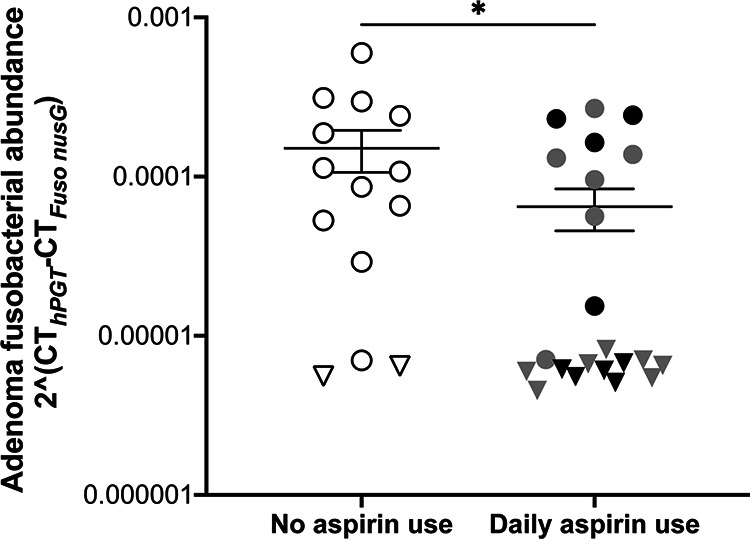
Fusobacterial abundance in human colonic adenomas of patients who self-reported aspirin use. qPCR was performed on DNA isolated from human colonic adenomas using primers targeting the nusG gene of Fusobacterium spp. and the human PGT gene to normalize for human DNA abundance. Samples were grouped based on patients who reported daily aspirin use and those who reported no aspirin use (white data points). For patients reporting daily aspirin use, gray data points indicate low-dose aspirin (<325 mg daily) and black data points indicate those reporting high-dose aspirin (≥325 mg daily). Data points (circles) represent individual adenomas, and triangles represent adenomas for which Fusobacterium sp. abundance was below the limit of detection and marked at the limit of detection based on the cycle threshold (CT) for hPGT in that individual sample. Bars indicate the mean ± SEM. Analysis was performed by Mann-Whitney test, with * indicating P values of <0.05.
DISCUSSION
In this work, we have coupled complementary culture-based experiments, a murine model of intestinal tumorigenesis, and human tissues to reveal that aspirin, a CRC chemopreventive drug, has antibacterial activity against F. nucleatum. More broadly, the effects of aspirin on F. nucleatum we observe demonstrate the merit of reconsidering existing preventative and therapeutic options and investigating how they may also be affecting the CRC-associated microbiome beyond their more well-known mechanisms.
In culture, we demonstrated the sensitivity of F. nucleatum strains and other CRC-associated bacteria to millimolar levels of aspirin. Although relatively high compared with more canonical antimicrobial compounds, these levels of aspirin are comparable to those used to study the effect of aspirin on other bacteria and colon cancer cell lines and those observed in human serum in some studies, supporting their relevance (31, 41, 61–63). At these levels, all F. nucleatum strains demonstrated aspirin sensitivity. The responses of Fn7-1 to aspirin at these doses were often surprisingly stark, such as a 6-log reduction in stationary Fn7-1 CFU in 10 mM aspirin compared with no reduction in 5 mM aspirin. Similarly, the range of aspirin levels used in our study often covered the inflection point for sensitivity of other tested strains, which may partially explain why we were able to observe differences across F. nucleatum strains. The underlying basis for why some F. nucleatum strains exhibit stronger aspirin sensitivity may inform the method of action for how aspirin affects F. nucleatum and help explain the results we observed for Fn7-1. An experimental evolution study of benzoate tolerance in E. coli observed concomitant salicylic acid resistance in the benzoate-adapted strains, whose mutations largely mapped to multidrug efflux systems (64). Variation in either the presence or expression of these systems, such as the MacB macrolide transporter whose expression in Fn7-1 was downregulated in response to aspirin, across F. nucleatum strains may be one explanation for different aspirin sensitivity. Isolation and characterization of F. nucleatum mutants that exhibit improved growth directly in aspirin or salicylic acid, while beyond the scope of this project, might reveal specific mechanisms underlying fusobacterial aspirin sensitivity and resistance. In addition to directly inhibiting bacterial growth, aspirin can alter sensitivity to antibiotics (48–51), which could synergize to further reduce F. nucleatum levels in vivo, although it is yet to be empirically tested in F. nucleatum.
Beyond culture-based experiments, we investigated how aspirin influences F. nucleatum in the setting of CRC using a genetically driven mouse model to show that aspirin abrogated F. nucleatum-potentiated tumorigenesis. Experimental limitations of this model—including the lack of colonic adenomas in either the absence of Fn7-1 or in Fn7-1 mice on aspirin chow and the difficulty in isolating viable Fn7-1 from the stool of conventional (specific pathogen free) mice—left us unable to fully ascertain if this observation was due to aspirin directly altering Fn7-1 viability or behavior or dominant effects of aspirin on inflammation and colonic tumor development independent of Fn7-1. However, aspirin leads to only mild, if any, effects on spontaneous intestinal adenoma formation in ApcMin/+ mice at this dosage (24, 25). Furthermore, a recent study proposed a role for the microbiota in mediating the response to aspirin in this model, as intestinal tumor burdens were reduced only when the microbiota was also disrupted by antibiotics (27). Taken together, these studies suggest that it is unlikely that the anti-inflammatory effects of aspirin alone are sufficient for the strong inhibition of Fn7-1-potentiated tumorigenesis we observed, and therefore, there likely may be a role for aspirin modulation of Fn7-1 in this phenotype. F. nucleatum aspirin-resistant mutants, as proposed earlier, would also prove useful in more definitively probing the role of aspirin-sensitivity in blocking F. nucleatum-potentiating tumorigenesis in the ApcMin/+ model, representing a potential future direction for in vivo experimentation. Beyond aspirin sensitivity, our observations could also be influenced by changes in Fn7-1 gene expression in response to lower levels of aspirin, including downregulation of important virulence factors like fap2, required to mediate CRC tissue localization and anti-tumor immunity (4, 14), and fadA, encoding a fusobacterial adhesin that engages E-cadherin to promote Wnt/β‐catenin signaling and drive cancer cell proliferation (12).
Using human samples, we demonstrated reduced fusobacterial load in colonic adenomas for individuals who self-reported daily aspirin use, by both mean fusobacterial burden and categorization as Fusobacterium high or Fusobacterium low/negative. The poorer prognosis and development of chemoresistance associated with fusobacterial load were determined using analyses with stratification of Fusobacterium-high and Fusobacterium-low/negative CRC tissues (15, 17) rather than based on correlation to a continuum of fusobacterial load. Therefore, the categorical threshold differences in fusobacterial load in response to aspirin use may indicate a greater potential to shape CRC outcomes than the 2.3-fold difference in mean adenoma fusobacterial abundance may suggest. Important remaining questions include where in the body (e.g., the oral cavity, the intestinal tract, or within the tumor microenvironment) and when (relative to tumor development) F. nucleatum exposure is occurring so that aspirin can be used most effectively.
The effect of aspirin on F. nucleatum growth and behavior is only one aspect of the interactions between aspirin and the microbiota in the context of colorectal tumorigenesis. For example, we previously demonstrated that F. nucleatum drives expression of Ptgs2 (3), which encodes COX-2, the predominant cellular target of aspirin in its anti-inflammatory role. Furthermore, a recent study suggested a role of the microbiota in influencing aspirin bioavailability in intestinal tumor models (27). Thus, the interactions among aspirin, F. nucleatum, other microbiota constituents, and inflammation in CRC tissues likely represent a highly complex interplay rather than the reductionist focus here. The critical question is how we use our understanding of these interactions to inform personalized microbiota-based medicine, such as more strongly recommending aspirin to individuals who harbor detectable fecal F. nucleatum and therefore are at a higher risk for CRC in a microbiota-based screening (65, 66).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
F. nucleatum strains Fn7-1 (55); FnCTI-1, -2, -3, -5, -6, and -7 (4); Fn23726; and Fn10953 (ATCC; Manassas, VA) were grown in Columbia broth or tryptic soy broth supplemented with hemin (5 μg/ml) and menadione (1 μg/ml) (sTSB) at 37°C, under anaerobic conditions using a vinyl chamber (Coy Lab Products, Grass Lake, MI). Fastidious anaerobe agar (FAA; Neogen, Lansing, MI) supplemented with 5% defibrinated sheep blood was used for plating. E. coli strains BW25113 (67), LF82 (68), and CCR20 (59, 69) were grown in Luria-Bertani (LB) broth at 37°C under aerobic or anaerobic conditions, as indicated, and plated onto either LB or MacConkey agar. B. fragilis strains BF638R (70) and ETBF 086-54443-2-2 (ETBF086) (71) were grown in supplemented basal medium (BM; 20 g/liter proteose peptone, 5 g/liter yeast extract, 5 g/liter sodium chloride, 5 g/liter glucose, 5 g/liter dipotassium phosphate, 0.5 g/liter l-cysteine, 5 μg/ml hemin, 2.5 μg/ml vitamin K1; modified from reference 72) at 37°C under anaerobic conditions and plated onto FAA supplemented with 5% defibrinated sheep blood.
All media supplemented with aspirin or salicylic acid were pH adjusted to match the control medium. For growth experiments, bacteria were grown overnight and then subcultured 1:100 into indicated media for 24 h at 37°C, unless otherwise noted. OD600 was measured in 48-well plates, unless otherwise indicated, using a Biotek Eon plate reader (Winooski, VT), located within the anaerobic chamber.
Gene expression studies.
Fn7-1 was grown for 24 h under the conditions described. RNA was extracted from the cultures by using a Directzol RNA Miniprep kit (Zymo Research, Irvine, CA), followed by Turbo DNA-free treatment (Invitrogen, Carlsbad, CA) and concentration with an RNeasy MinElute cleanup kit (Qiagen, Germantown, MD). RNA quality assessment (2200 Tapestation; Agilent, Santa Clara, CA), rRNA depletion (Ribo-Zero rRNA removal kit for bacteria; Illumina, San Diego, CA), library construction (Wafergen PrepX directional RNA-seq library kit; TaKaRa Bio, Mountain View, CA), and sequencing (HiSeq 2500; Illumina) were performed by the Harvard Medical School (HMS) Biopolymers Core Facility using standard techniques to generate 75-base pair paired-end reads.
Raw sequencing reads were quality trimmed using Trimmomatic (73) (v0.39) configured to perform sliding window scan with the following settings: “ILLUMINACLIP:${adapter_library_FASTA}:2:36:7:1: keepBothReads LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36.” Optical and PCR duplicate sequences were then identified and removed using Clumpify with the option “dedupe optical spany adjacent subs = 0” from the BBTools bioinformatic suite (http://sourceforge.net/projects/bbmap). We indexed 2,419 Fn7-1 genomic loci identified by the GenBank database (assembly accession GCA_000158275.2) using Burrows-Wheeler Aligner (BWA; v0.7.17) (74) to create a gene database for short read alignment. Low-complexity gene subsequences were hard masked with the DUST program (75) prior to read mapping with the maximal exact match (mem) algorithm and default penalty scoring strategy (76). To maximize the biological interpretation of gene expression data, we used Prokka (77) to reannotate genome loci with protein families by incorporating profile hidden Markov model (pHMM) databases from Pfam (78) (prioritized; v33), EggNOG bacteria archaea viruses (79) (v5), KOfam (80) (v96), TIGRFAMs (81) (v15), and PSORTb (82) (v3.0.2) to predict subcellular localization of proteins in Gram-negative bacteria. A gene expression matrix was constructed by counting only reads that mapped to a single gene (i.e., unambiguous reads) using the htseq-count script included with the HTSeq python library (83). Read counts were normalized to transcripts per kilobase million (TPM) to equalize variable sequencing depth across samples for differential gene expression analysis by least square linear regression as implemented in the limma R/Bioconductor package (84). Fold change between treatment conditions for each gene was calculated by determining the mean value of all combinatorial pairs of samples that fell within the interquartile range (85). To ensure robust statistical analysis, we aligned reads against a fusobacterial clade-specific marker gene collection using the ChocoPhlAn pangenome database (v30) (86).
Animal studies.
Beginning at 6 weeks of age and continuing daily for 8 weeks, male and female ApcMin/+ (sourced from Jackson and bred in-house in a barrier facility) mice were orally instilled with either 108 CFU of Fn7-1 (in <100-μl volume) or medium control (sTSB). Concurrently, animals were placed on either an AIN-76A diet or AIN-76A supplemented with 200 ppm aspirin (Research Diets, Inc., New Brunswick, NJ). At 14 weeks, mice were euthanized and colons were excised for histological analysis conducted in a blind manner by J.N.G., as previously described (3). All experiments were approved and carried out in accordance with HMS’s Standing Committee on Animals and the National Institutes of Health guidelines for animal use and care.
Determination of Fusobacterium sp. abundance in human colon adenomas.
Human colonic adenomas were acquired from the Pitt Biospecimen Core (PBC) at the University of Pittsburgh. Acquisition of the samples was institutional review board (IRB) approved, and informed consent was received from all participants, who also completed questions about NSAID and aspirin use. All adenomas were ≥1 cm in size. Upon endoscopic removal, they were placed into a saline ice bath and transported to pathology for sectioning. The pathologist allocated tissue for clinical diagnosis and research purposes, which was flash frozen and stored at −80°C.
DNA was isolated from adenoma tissues (∼2 mm by 2 mm by 2 mm) by overnight lysis (100 mM Tris-HCl [pH 8.5], 5 mM EDTA [pH 8.0], 0.2% sodium dodecyl sulfate, 200 mM NaCl, and 1 mg/ml proteinase K; rotating at 55°C), followed by a standard phenol-chloroform extraction. qPCR was performed on tumoral DNA (160 ng per reaction, technical duplicates) using the Kapa ProbeFast Rox low kit (Wilmington, MA) on an Agilent Mx3005P cycler. Primers targeted Fusobacterium spp. (forward, 5′-CAACCATTACTTTAACTCTACCATGTTCA-3′; reverse, 5′-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3′; probe, 5′-6-6-carboxyfluorescein [FAM]-TCAGCAACTTGTCCTTCTTGATCTTTAAATGAACC-black hole quencher [BHQ]1-3′) and human PGT (forward, 5′-ATCCCCAAAGCACCTGGTTT-3′; reverse, 5′-AGAGGCCAAGATAGTCCTGGTAA-3′; probe, 5′-6-FAM-CCATCCATGTCCTCATCTC-BHQ1-3′) as previously described (8, 16). We were blind to aspirin use until after data analysis.
Statistical analysis.
Graphs and statistical analysis were generated using Prism 9 (GraphPad Software, San Diego, CA). Statistical tests used for each analysis are described in figure legends.
Data availability.
RNA-seq data used in this study have been deposited in the NCBI SRA database under the BioProject identifier (ID) PRJNA701284.
ACKNOWLEDGMENTS
We thank the Garrett lab for thoughtful discussions and contributions.
These studies were supported by NIH RO1CA154426 and the Cancer Research UK Grand Challenge Initiative C10674/A27140, which were both awarded to W.S.G.; NCI R01 CA202704 to W.S.G. and A.T.C; and NCI R35 CA253185 to A.T.C. Adenoma tissue collection was supported by the Early Detection Research Network grant U01-CA152753 to R.E.S. C.A.B. was the Dennis and Marsha Dammerman fellow of the Damon Runyon Cancer Research Foundation (DRG-2205-14) and funded by the DFCI Cancer Immunology Training Program (NIH T32CA207021). D.A.D. is supported by NIH K01DK120742. A.T.C. is the Stuart and Suzanne Steele MGH research scholar.
We greatly thank Cynthia Sears (The Johns Hopkins University School of Medicine), Dennis Kasper (Harvard Medical School), and Emily Balskus (Harvard University) for strains. We thank the HMS Biopolymers Facility for their contributions to the RNA-seq studies.
Footnotes
This article is a direct contribution from Wendy S. Garrett, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Kim Lewis, Northeastern University, and Daniel Slade, Virginia Polytechnic Institute and State University.
Citation Brennan CA, Nakatsu G, Gallini Comeau CA, Drew DA, Glickman JN, Schoen RE, Chan AT, Garrett WS. 2021. Aspirin modulation of the colorectal cancer-associated microbe Fusobacterium nucleatum. mBio 12:e00547-21. https://doi.org/10.1128/mBio.00547-21.
REFERENCES
- 1.Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 2.Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med 15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. 2013. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. 2015. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, Destefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, Tam AJ, McAllister F, Fan H, Wu X, Ganguly S, Lebid A, Metz P, Van Meerbeke SW, Huso DL, Wick EC, Pardoll DM, Wan F, Wu S, Sears CL, Housseau F. 2018. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 23:203–214.e5. doi: 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umana A, Zhang Y, Peng H, Duncan AJ, Wang Y, Li L, Verbridge SS, Slade DJ. 2020. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal 13:eaba9157. doi: 10.1126/scisignal.aba9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano H-O, Sugai T, An B, Shureiqi I, Toyota M, Kondo Y, Estécio MRH, Issa J-PJ. 2014. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. 2013. Fusobacterium is associated with colorectal adenomas. PLoS One 8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. 2013. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, Li H, Guo B, Zhu Q, Wei Q, Moyer M-P, Wang P, Cai S, Goel A, Qin H, Ma Y. 2017. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology 152:851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. 2016. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, Milner DA, Baba H, Giovannucci EL, Garraway LA, Freeman GJ, Dranoff G, Garrett WS, Huttenhower C, Meyerson M, Meyerhardt JA, Chan AT, Fuchs CS, Ogino S. 2016. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut Gutjnl 65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JHM, Hughes DJ. 2014. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 17.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang J-Y. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. 2017. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vane JR. 1971. Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat New Biol 231:232–235. doi: 10.1038/newbio231232a0. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell JA, Akarasereenont P, Thiemermann C, Flower RJ, Vane JR. 1993. Selectivity of nonsteroidal antiinflammatory drugs as inhibitors of constitutive and inducible cyclooxygenase. Proc Natl Acad Sci U S A 90:11693–11697. doi: 10.1073/pnas.90.24.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bibbins-Domingo K, U.S. Preventive Services Task Force . 2016. Aspirin use for the primary prevention of cardiovascular disease and colorectal cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 164:836. doi: 10.7326/M16-0577. [DOI] [PubMed] [Google Scholar]
- 22.Nan H, Hutter CM, Lin Y, Jacobs EJ, Ulrich CM, White E, Baron JA, Berndt SI, Brenner H, Butterbach K, Caan BJ, Campbell PT, Carlson CS, Casey G, Chang-Claude J, Chanock SJ, Cotterchio M, Duggan D, Figueiredo JC, Fuchs CS, Giovannucci EL, Gong J, Haile RW, Harrison TA, Hayes RB, Hoffmeister M, Hopper JL, Hudson TJ, Jenkins MA, Jiao S, Lindor NM, Lemire M, Le Marchand L, Newcomb PA, Ogino S, Pflugeisen BM, Potter JD, Qu C, Rosse SA, Rudolph A, Schoen RE, Schumacher FR, Seminara D, Slattery ML, Thibodeau SN, Thomas F, Thornquist M, Warnick GS, Zanke BW, Gauderman WJ, Peters U, Hsu L, Chan AT, CCFR, GECCO . 2015. Association of aspirin and NSAID use with risk of colorectal cancer according to genetic variants. JAMA 313:1133–1142. doi: 10.1001/jama.2015.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothwell PM, Wilson M, Elwin C-E, Norrving B, Algra A, Warlow CP, Meade TW. 2010. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet 376:1741–1750. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- 24.Mahmoud NN, Dannenberg AJ, Mestre J, Bilinski RT, Churchill MR, Martucci C, Newmark H, Bertagnolli MM. 1998. Aspirin prevents tumors in a murine model of familial adenomatous polyposis. Surgery 124:225–231. doi: 10.1016/S0039-6060(98)70124-2. [DOI] [PubMed] [Google Scholar]
- 25.Sansom OJ, Stark LA, Dunlop MG, Clarke AR. 2001. Suppression of intestinal and mammary neoplasia by lifetime administration of aspirin in Apc(Min/+) and Apc(Min/+), Msh2(−/−) mice. Cancer Res 61:7060–7064. [PubMed] [Google Scholar]
- 26.Reuter BK, Zhang X-J, Miller MJS. 2002. Therapeutic utility of aspirin in the ApcMin/+ murine model of colon carcinogenesis. BMC Cancer 2:19. doi: 10.1186/1471-2407-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao R, Coker OO, Wu J, Zhou Y, Zhao L, Nakatsu G, Bian X, Wei H, Chan AWH, Sung JJY, Chan FKL, El-Omar E, Yu J. 2020. Aspirin reduces colorectal tumor development in mice and gut microbes reduce its bioavailability and chemopreventive effects. Gastroenterology 159:969–983.e4. doi: 10.1053/j.gastro.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Liang X, Bittinger K, Li X, Abernethy DR, Bushman FD, FitzGerald GA. 2015. Bidirectional interactions between indomethacin and the murine intestinal microbiota. Elife 4:e08973. doi: 10.7554/eLife.08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maseda D, Zackular JP, Trindade B, Kirk L, Roxas JL, Rogers LM, Washington MK, Du L, Koyama T, Viswanathan VK, Vedantam G, Schloss PD, Crofford LJ, Skaar EP, Aronoff DM. 2019. Nonsteroidal anti-inflammatory drugs alter the microbiota and exacerbate Clostridium difficile colitis while dysregulating the inflammatory response. mBio 10:S65. doi: 10.1128/mBio.02282-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prizment AE, Staley C, Onyeaghala GC, Vivek S, Thyagarajan B, Straka RJ, Demmer RT, Knights D, Meyer KA, Shaukat A, Sadowsky MJ, Church TR. 2020. Randomised clinical study: oral aspirin 325 mg daily vs placebo alters gut microbial composition and bacterial taxa associated with colorectal cancer risk. Aliment Pharmacol Ther 52:976–987. doi: 10.1111/apt.16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang WH, Wong WM, Dailidiene D, Berg DE, Gu Q, Lai KC, Lam SK, Wong BCY. 2003. Aspirin inhibits the growth of Helicobacter pylori and enhances its susceptibility to antimicrobial agents. Gut 52:490–495. doi: 10.1136/gut.52.4.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cederlund H, Mårdh PA. 1993. Antibacterial activities of non-antibiotic drugs. J Antimicrob Chemother 32:355–365. doi: 10.1093/jac/32.3.355. [DOI] [PubMed] [Google Scholar]
- 33.Afzal M, Shafeeq S. 2017. Impact of aspirin on the transcriptome of Streptococcus pneumoniae D39. Genom Data 12:38–40. doi: 10.1016/j.gdata.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kupferwasser LI, Yeaman MR, Nast CC, Kupferwasser D, Xiong Y-Q, Palma M, Cheung AL, Bayer AS. 2003. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J Clin Invest 112:222–233. doi: 10.1172/JCI16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunin CM, Hua TH, Guerrant RL, Bakaletz LO. 1994. Effect of salicylate, bismuth, osmolytes, and tetracycline resistance on expression of fimbriae by Escherichia coli. Infect Immun 62:2178–2186. doi: 10.1128/IAI.62.6.2178-2186.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosner JL, Chai TJ, Foulds J. 1991. Regulation of ompF porin expression by salicylate in Escherichia coli. J Bacteriol 173:5631–5638. doi: 10.1128/jb.173.18.5631-5638.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerner E, Almqvist S, Werthén M, Trobos M. 2020. Sodium salicylate interferes with quorum-sensing-regulated virulence in chronic wound isolates of Pseudomonas aeruginosa in simulated wound fluid. J Med Microbiol 69:767–780. doi: 10.1099/jmm.0.001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alekshun MN, Levy SB. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J Bacteriol 181:4669–4672. doi: 10.1128/JB.181.15.4669-4672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones RM, Pagmantidis V, Williams PA. 2000. sal Genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobacter sp. strain ADP1. J Bacteriol 182:2018–2025. doi: 10.1128/jb.182.7.2018-2025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drew DA, Chan AT. 2021. Aspirin in the prevention of colorectal neoplasia. Annu Rev Med 72:415–430. doi: 10.1146/annurev-med-060319-120913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomposiello PJ, Bennik MH, Demple B. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol 183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denkin S, Byrne S, Jie C, Zhang Y. 2005. Gene expression profiling analysis of Mycobacterium tuberculosis genes in response to salicylate. Arch Microbiol 184:152–157. doi: 10.1007/s00203-005-0037-9. [DOI] [PubMed] [Google Scholar]
- 43.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, Shi W. 2010. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun 78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coppenhagen-Glazer S, Sol A, Abed J, Naor R, Zhang X, Han YW, Bachrach G. 2015. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun 83:1104–1113. doi: 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Umaña A, Sanders BE, Yoo CC, Casasanta MA, Udayasuryan B, Verbridge SS, Slade DJ. 2019. Utilizing whole Fusobacterium genomes to identify, correct, and characterize potential virulence protein families. J Bacteriol 201:3703. doi: 10.1128/JB.00273-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cochrane K, McGuire AM, Priest ME, Abouelleil A, Cerqueira GC, Lo R, Earl AM, Allen-Vercoe E. 2016. Complete genome sequences and analysis of the Fusobacterium nucleatum subspecies animalis 7-1 bacteriophage ϕFunu1 and ϕFunu2. Anaerobe 38:125–129. doi: 10.1016/j.anaerobe.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cochrane K, Robinson AV, Holt RA, Allen-Vercoe E. 2020. A survey of Fusobacterium nucleatum genes modulated by host cell infection. Microb Genom 6:e000300. doi: 10.1099/mgen.0.000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen Z, Pu X-Y, Zhang Q. 2011. Salicylate functions as an efflux pump inducer and promotes the emergence of fluoroquinolone-resistant Campylobacter jejuni mutants. Appl Environ Microbiol 77:7128–7133. doi: 10.1128/AEM.00763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aumercier M, Murray DM, Rosner JL. 1990. Potentiation of susceptibility to aminoglycosides by salicylate in Escherichia coli. Antimicrob Agents Chemother 34:786–791. doi: 10.1128/aac.34.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price CT, O'Brien FG, Shelton BP, Warmington JR, Grubb WB, Gustafson JE. 1999. Effects of salicylate and related compounds on fusidic acid MICs in Staphylococcus aureus. J Antimicrob Chemother 44:57–64. doi: 10.1093/jac/44.1.57. [DOI] [PubMed] [Google Scholar]
- 51.Burns JL, Clark DK. 1992. Salicylate-inducible antibiotic resistance in Pseudomonas cepacia associated with absence of a pore-forming outer membrane protein. Antimicrob Agents Chemother 36:2280–2285. doi: 10.1128/aac.36.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manson McGuire A, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, Nice JB, MacDonald H, Birren BW, Berger BW, Allen-Vercoe E, Earl AM. 2014. Evolution of invasion in a diverse set of Fusobacterium species. mBio 5:e01864. doi: 10.1128/mBio.01864-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fantin B, Leggett J, Ebert S, Craig WA. 1991. Correlation between in vitro and in vivo activity of antimicrobial agents against gram-negative bacilli in a murine infection model. Antimicrob Agents Chemother 35:1413–1422. doi: 10.1128/aac.35.7.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 55.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. 2011. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 56.Wilson MR, Jiang Y, Villalta PW, Stornetta A, Boudreau PD, Carrá A, Brennan CA, Chun E, Ngo L, Samson LD, Engelward BP, Garrett WS, Balbo S, Balskus EP. 2019. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363:eaar7785. doi: 10.1126/science.aar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, Paganelli FL, Geurts MH, Beumer J, Mizutani T, Miao Y, van der Linden R, van der Elst S, Garcia KC, Top J, Willems RJL, Giannakis M, Bonnet R, Quirke P, Meyerson M, Cuppen E, van Boxtel R, Clevers H, Genomics England Research Consortium . 2020. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580:269–273. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L, Sauvanet P, Darcha C, Déchelotte P, Bonnet M, Pezet D, Wodrich H, Darfeuille-Michaud A, Bonnet R. 2014. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 60.Boleij A, van Gelder MMHJ, Swinkels DW, Tjalsma H. 2011. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis 53:870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 61.Kim KM, Song JJ, An JY, Kwon YT, Lee YJ. 2005. Pretreatment of acetylsalicylic acid promotes tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by down-regulating BCL-2 gene expression. J Biol Chem 280:41047–41056. doi: 10.1074/jbc.M503713200. [DOI] [PubMed] [Google Scholar]
- 62.Pathi S, Jutooru I, Chadalapaka G, Nair V, Lee S-O, Safe S. 2012. Aspirin inhibits colon cancer cell and tumor growth and downregulates specificity protein (Sp) transcription factors. PLoS One 7:e48208. doi: 10.1371/journal.pone.0048208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Juárez Olguín H, Flores Pérez J, Lares Asseff I, Loredo Abdalá A, Carbajal Rodríguez L. 2004. Comparative pharmacokinetics of acetyl salicylic acid and its metabolites in children suffering from autoimmune diseases. Biopharm Drug Dispos 25:1–7. doi: 10.1002/bdd.379. [DOI] [PubMed] [Google Scholar]
- 64.Creamer KE, Ditmars FS, Basting PJ, Kunka KS, Hamdallah IN, Bush SP, Scott Z, He A, Penix SR, Gonzales AS, Eder EK, Camperchioli DW, Berndt A, Clark MW, Rouhier KA, Slonczewski JL. 2017. Benzoate- and salicylate-tolerant strains of Escherichia coli K-12 lose antibiotic resistance during laboratory evolution. Appl Environ Microbiol 83:65. doi: 10.1128/AEM.02736-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo S, Li L, Xu B, Li M, Zeng Q, Xiao H, Xue Y, Wu Y, Wang Y, Liu W, Zhang G. 2018. A simple and novel fecal biomarker for colorectal cancer: ratio of Fusobacterium Nucleatum to probiotics populations, based on their antagonistic effect. Clin Chem 64:1327–1337. doi: 10.1373/clinchem.2018.289728. [DOI] [PubMed] [Google Scholar]
- 66.Young C, Wood HM, Seshadri RA, Van Nang P, Vaccaro C, Melendez LC, Bose M, Van Doi M, Piñero TA, Valladares CT, Arguero J, Balaguer AF, Thompson KN, Yan Y, Huttenhower C, Quirke P. 2021. The colorectal cancer-associated faecal microbiome of developing countries resembles that of developed countries. Genome Med 13:27. doi: 10.1186/s13073-021-00844-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 115:1405–1413. doi: 10.1016/S0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 69.Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R. 2013. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8:e56964. doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Privitera G, Dublanchet A, Sebald M. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J Infect Dis 139:97–101. doi: 10.1093/infdis/139.1.97. [DOI] [PubMed] [Google Scholar]
- 71.Wu S, Dreyfus LA, Tzianabos AO, Hayashi C, Sears CL. 2002. Diversity of the metalloprotease toxin produced by enterotoxigenic Bacteroides fragilis. Infect Immun 70:2463–2471. doi: 10.1128/IAI.70.5.2463-2471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. 1991. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun 59:2075–2082. doi: 10.1128/IAI.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. 2006. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol 13:1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 76.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997v2. [q-bio.GN]. https://arxiv.org/abs/1303.3997.
- 77.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 78.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, Mering von C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, Ogata H. 2020. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res 31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma Powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dembélé D, Kastner P. 2014. Fold change rank ordering statistics: a new method for detecting differentially expressed genes. BMC Bioinformatics 15:14–15. doi: 10.1186/1471-2105-15-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beghini F, McIver LJ, Blanco-Miguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Thomas AM, Manghi P, Valles-Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N. 2020. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. bioRxiv https://www.biorxiv.org/content/10.1101/2020.11.19.388223v1. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Changes to Fn7-1 autoaggregation when grown in 1 mM aspirin. (A) Fn7-1 grown in sTSB or sTSB supplemented with 1 mM aspirin demonstrates different clumping behaviors. (B and C) OD600 data from two individual wells of Fn7-1 grown in either sTSB or sTSB with 1 mM aspirin. For this experiment, Fn7-1 was grown in a 96-well plate to better visualize autoaggregation phenotypes. Download FIG S1, TIF file, 2.7 MB (2.7MB, tif) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed Fn7-1 genes in response to 1 mM aspirin. Download Data Set S1, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed Fn7-1 genes in response to 2.5 mM aspirin. Download Data Set S2, XLSX file, 1.0 MB (1MB, xlsx) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential growth responses of F. nucleatum CTI strains in the presence of aspirin. Growth of FnCTI-3 (A), FnCTI-6 (B), and FnCTI-7 (C) in sTSB or media supplemented with 1 mM or 2.5 mM aspirin. All data represent the mean ± SEM for at least 6 cultures. Growth curves were analyzed by two-way repeated measures ANOVA with post hoc Dunnett’s test. Growth was significantly different from sTSB at a P value of <0.05 for each of the following strains beginning at the indicated timepoints and concentrations: CTI-3 (2.5 mM at 11.5 h), CTI-5 (1 mM at 8 h and 2.5 mM at 6 h), and FnCTI-7 (2.5 mM at 11 h). Download FIG S2, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Aspirin minimally affects E. coli strains grown under aerobic conditions. Growth yield (CFU per ml) of E. coli strains BW25113, LF82, and CCR20 in LB or LB supplemented with 2.5 mM aspirin for 18 h at 37°C under aerobic conditions. All data represent the mean ± SEM for at least 6 cultures. Growth yield analysis was performed by Mann-Whitney test and ** indicates a P value of <0.01. Download FIG S3, TIF file, 0.3 MB (316KB, tif) .
Copyright © 2021 Brennan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
RNA-seq data used in this study have been deposited in the NCBI SRA database under the BioProject identifier (ID) PRJNA701284.