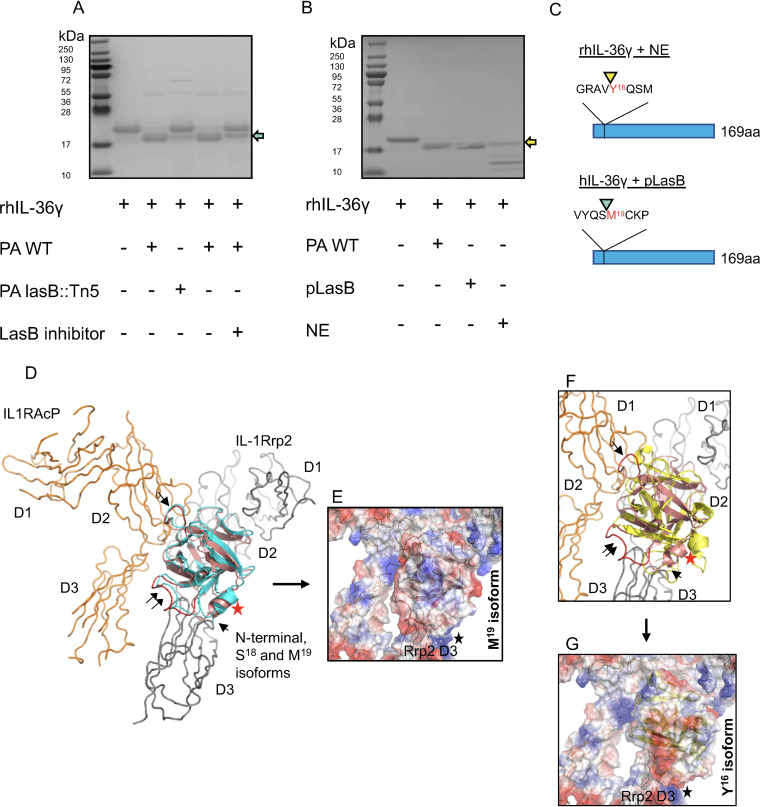
FIG 3.
PA14 LasB cleaves IL-36γ proximally to M19, and sequential N-terminal truncation models in silico predict the bioactivity of the M19 isoform. (A) Full-length IL-36γ was incubated with wild-type PA14 (PA WT) or lasB::Tn5 mutant supernatant in the presence or absence of LasB inhibitor. (B) Full-length IL-36γ was incubated with PA WT, purified LasB (pLasB), or recombinant NE and visualized by SDS-PAGE. Teal arrow points to M19 IL-36γ, and yellow arrows point to Y16 IL-36γ. (C) N-terminal sequencing of IL-36γ NE and IL-36γ LasB were analyzed by Edman degradation, with arrowheads indicating the site of cleavage. (D) IL-36γ S18 (salmon) and M19 (teal) isoforms associate in a similar orientation to a model of the IL-1Rrp2/IL1RAcP receptor complex (gray and orange, respectively). Parts of D2 and D3 of IL-1Rrp2 and the D2 domain of IL1RAcP contribute to the identified cytokine-binding site. Helix I104-G109 (red star) is close to the N termini of both isoforms pointed toward the IL-1Rrp2 D3 domain. The single black arrow indicates loop L155-N160 (red), which makes favorable electrostatic contact with the IL1RAcP D2 domain. Loop T61-D72 (double black arrow, red loop) faces the D3 domain of the accessory protein. (E) The electrostatic pattern of M19 isoform binding may influence the IL-1Rrp2 D3 domain. The black star denotes a predicted concentration of electrostatic repulsions (basic charge in blue) exist at the lower contact interface between the isoform and the IL-1Rrp2 D3 domain. (F) Y16 isoform (yellow) binds in the upside-down fashion, but there are differences compared to the M19 isoform (see position shift of the landmark helix [red star]). For reference, the single and double black arrows again indicate the loops shown in in panel D. (G) The Y16 isoform binds in the upside-down arrangement, but the intermolecular packing is less efficient between the cytokine and IL-36R complex. The figures were prepared and electrostatic potential surfaces calculated using PyMol Molecular Graphics System v1.3 (Schrodinger, LLC).