Mycobacterium abscessus infections in cystic fibrosis patients are challenging to treat due to widespread antibiotic resistance. The therapeutic use of lytic bacteriophages presents a new potential strategy, but the great variation among clinical M. abscessus isolates demands determination of phage susceptibility prior to therapy.
KEYWORDS: Mycobacterium abscessus, bacteriophage therapy, bacteriophages
ABSTRACT
Mycobacterium abscessus is an opportunistic pathogen whose treatment is confounded by widespread multidrug resistance. The therapeutic use of bacteriophages against Mycobacterium abscessus infections offers a potential alternative approach, although the spectrum of phage susceptibilities among M. abscessus isolates is not known. We determined the phage infection profiles of 82 M. abscessus recent clinical isolates and find that colony morphotype—rough or smooth—is a key indicator of phage susceptibility. None of the smooth strains are efficiently killed by any phages, whereas 80% of rough strains are infected and efficiently killed by at least one phage. The repertoire of phages available for potential therapy of rough morphotype infections includes those with relatively broad host ranges, host range mutants of Mycobacterium smegmatis phages, and lytically propagated viruses derived from integrated prophages. The rough colony morphotype results from indels in the glycopeptidolipid synthesis genes mps1 and mps2, negating reversion to smooth as a common route to phage resistance. Resistance is thus rare, and although mutations in polyketide synthesis, uvrD2, and rpoZ can confer resistance, these likely also impair survival in vivo. The expanded therapeutic repertoire and the resistance profiles show that small cocktails or single phages could be suitable for controlling infections with rough strains.
INTRODUCTION
Nontuberculous mycobacteria (NTM) are frequent pathogens of cystic fibrosis (CF) and bronchiectasis patients (1, 2). They are commonly refractory to treatment due to widespread antibiotic resistance and antibiotic toxicity over the required long treatment regimens (2–4). Among the NTMs, Mycobacterium abscessus is prevalent in CF patients, can greatly diminish lung function, and is a negative factor for lung transplantation (5, 6). There is an evident need for alternative treatments to control these infections.
There are three common subspecies of M. abscessus, subsp. abscessus, subsp. bolletii, and subsp. massiliense (7). M. abscessus subsp. abscessus is the most the common in CF patients, and genomic analysis shows that a high proportion of these strains form a clade of closely related strains that includes the ATCC 19977 type strain (8). Two distinct colony morphotypes of M. abscessus are observed, forming smooth (S) or rough (R) colonies on solid medium (9). The S strains generally are thought to be less virulent, due to the prevalence of surface glycopeptidolipids (GPLs) recognized by the host immune system (10–12). Rough strains are strongly depleted for GPLs, escape immune recognition more efficiently, and are more virulent (13). Smooth-to-rough transition occurs by interruption of GPL synthesis or localization—including mutations in mmpL4, which is required for GPL transport across the inner membrane—and mutation or silencing of mps1, mps2, and gap (9, 11, 14). Smooth-to-rough transitions are often nonreversible, although temperature-dependent variation has been reported for one variant (11, 15, 16).
The therapeutic use of bacteriophages may provide an alternative treatment strategy for NTM infections. A three-phage cocktail administered intravenously in a 15-year-old CF patient with a disseminated M. abscessus infection following a bilateral lung transplant showed substantial improvement and alleviation of infection (17). Whether phage interventions are useful in other patients with similar infections is unclear because of the extensive genetic variation among M. abscessus strains (18, 19).
RESULTS
Collection of Mycobacterium abscessus clinical isolates.
Following the successful treatment of a disseminated M. abscessus infection with a bacteriophage cocktail (17), we received 82 M. abscessus strains (designated GD01 to GD111; Table 1) from 78 different patients for characterization of phage susceptibilities; 54 (69%) of these are from within the United States and the others are from 10 different other countries (Table 1). We scored the colony morphotypes of each strain as being either rough (R) or smooth (S) (Table 1; Fig. 1, Fig. 2A); 48 (58.5%) have R morphotypes (Table 1). One-half of the strains are from patients with cystic fibrosis, and among these a similar proportion of strains (64%) are rough (Table 1). Smooth morphotype strains are less virulent than rough strains in some assays (20), but overall the smooth and rough profiles of these strains is similar to those reported previously (21, 22).
TABLE 1.
Properties of Mycobacterium abscessus clinical isolates
| Straina | Originb | CFc | Sspd | R/Se | Seqf | Prophagesg | Plasmidsh |
|---|---|---|---|---|---|---|---|
| GD01 | London, UK | Y | m | R | Com | None | None |
| GD02 | London, UK | Y | m | R | WGS | prophiGD02-1 (MabN), prophiGD02-2 (MabA2) | pGD02 (pF) |
| GD03 | Seattle, WA | N | m | S | WGS | prophiGD03-1 (MabG) | None |
| GD04 | Los Angeles, CA | Y | m | S | WGS | prophiGD04-1 (MabE1) | None |
| GD05 | Winston-Salem, NC | N | a | R | Com | prophiGD05-1 (MabD), prophiGD05-2 (MabH), prophiGD05-3 (MabM) | None |
| GD08 | Indianapolis, IN | Y | a | R | WGS | prophiGD08-1 (MabA1), prophiGD08-2 (MabB), prophiGD08-3 (MabF) | pGD08 (pA) |
| GD09 | Netherlands | Y | a | S | WGS | prophiGD09-1 (MabE1) | None |
| GD10 | Pittsburgh, PA | N | m | R | WGS | prophiGD10-1 (MabA1) | pGD10 (pSin) |
| GD11 | Pittsburgh, PA | Y | a | R | WGS | prophiGD11-1 (MabA1), prophiGD11-2 (MabB), phophiGD11-3 (MabF) | None |
| GD12 | Pittsburgh, PA | NA | a | R | WGS | prophiGD12-1 (MabA1), prophiGD12-2 (MabD) | None |
| GD13 | Pittsburgh, PA | NA | a | R | WGS | prophiGD13-1 (MabA1), prophiGD13-2 (MabC) | pGD13 (Sin) |
| GD14 | Pittsburgh, PA | NA | a | R | WGS | prophiGD14-1 (MabA1), prophiGD14-2 (MabD) | None |
| GD15 | Pittsburgh, PA | NA | a | R | WGS | prophiGD15-1 (MabA1) | None |
| GD16 | St Louis, MO | Y | m | S | WGS | prophiGD16-1 (MabB), prophiGD16-2 (MabA2) | pGD16 (pH) |
| GD17 | Durham, NC | Y | a | R | Com | prophiGD17-1 (MabD), prophiGD17-2 (MabA1) | None |
| GD18 | San Diego, CA | N | a | S | WGS | None | pGD18 (pB) |
| GD19 | San Diego, CA | Y | a | R | Com | None | pGD19 (pD) |
| GD20 | Durham, NC | Y | a | R | Com | prophiGD20-1 (MabA1) | None |
| GD21 | Baton Rouge, LA | N | a | S | Com | prophiGD21-1 (MabB), prophiGD21-2 (MabA1), prophiGD21-3 (MabG), prophiGD21-4 (MabJ) | pGD21-1 (pSin), pGD21-2 (pSin) |
| GD22 | Pittsburgh, PA | Y | a | R | Com | prophiGD22-1 (MabA1) | pGD22-1 (pSin), pGD22-2 (pC) |
| GD23 | St. Louis, MO | NA | a | R | WGS | prophiGD23-1 (MabA1) | pGD23 (pB) |
| GD24 | Genoa, Italy | Y | a | R | WGS | prophiGD24-1 (MabA1), prophiGD24-2 (MabG), prophiGD24-3 (MabJ) | pGD24 (pC) |
| GD25 | Anchorage, AL | Y | a | R | Com | prophiGD25-1 (MabE1) | pGD25-1 (pF), pGD25-2 (pG), pGD25-3 (pSin) |
| GD26 | Pittsburgh, PA | Y | a | R | Com | prophiGD26-1 (MabA1) | None |
| GD27 | Boston, MA | Y | a | R | WGS | prophiGD27-1 (MabA1) | None |
| GD28 | Durham, NC | NA | a | S | WGS | None | None |
| GD30 | Durham, NC | NA | m | R | WGS | prophiGD30-1 (MabA1) | None |
| GD33 | Illes Balears, Spain | Y | a | S | WGS | prophiGD33-1 (MabC) | pGD33 (pE) |
| GD34 | Boston, MA | N | a | S | WGS | prophiGD34-1 (MabA1), prophiGD34-2 (MabB) | pGD34 (pC) |
| GD35 | New York, NY | Y | a | R | WGS | prophiGD35-1 (MabA1) | None |
| GD36 | Vancouver, BC, Canada | N | a | S | WGS | prophiGD36-1 (MabA1), prophiGD36-2 (MabH) | pGD36-1 (pB), pGD36-2 (pE) |
| GD37 | Vancouver, BC, Canada | N | b | S | WGS | None | None |
| GD38 | Hamilton, ON, Canada | NA | a | R | Com | None | None |
| GD39 | Hartford, CT | Y | a | S | WGS | prophiGD39-1 (MabA1), prophiGD39-2 (MabC) | pGD39 (pC) |
| GD40 | NSW, Australia | Y | a | R | WGS | prophiGD40-1 (MabA1) | None |
| GD41 | Pittsburgh, PA | Y | a | R | Com | prophiGD41-1 (MabA1) | None |
| GD42 | Pittsburgh, PA | Y | a | S | Com | prophiGD42-1 (MabA1), prophiGD42-2 (MabB), prophiGD42-3 (MabC) | pGD42-1 (pB), pGD42-2 (pA) |
| GD43A | NSW, Australia | Y | a | S | Com | prophiGD43A-1 (MabA1), prophiGD43A-2 (MabB), prophiGD43A-3 (MabC), prophiGD43A-4 (MabJ), prophiGD43A-5 (MabK), prophiGD43A-6 (MabL) | None |
| GD43B | NSW, Australia | Y | a | R | Com | prophiGD43B-1 (MabL), prophiGD43B-2 (MabK), prophiGD43B-3 (MabA1), prophiGD43B-4 (MabJ) | None |
| GD44 | Winnipeg, Canada | N | m | S | WGS | prophiGD44-1 (MabC) | None |
| GD45 | Miami, FL | Y | a | R | WGS | prophiGD45-1 (MabE1) | pGD45-1 (pG), pGD45-2 (pD) |
| GD47 | Dallas, TX | N | a | S | WGS | prophiGD47-1 (MabA1) | pGD47 (pB) |
| GD51 | Portland, OR | Y | a | R | WGS | prophiGD51-1 (MabC), prophiGD51-2 (MabP) | pGD51 (pSin) |
| GD52 | Sheffield, UK | Y | a | R | WGS | prophiGD52-1 (MabC), prophiGD52-2 (MabE1) | pGD52 (pSin) |
| GD53 | Lebanon, NH | Y | m | S | WGS | prophiGD53-1 (MabE1), prophiGD53-2 (MabK), prophiGD53-3 (MabN) | None |
| GD54 | NSW, Australia | Y | a | R | Com | prophiGD54-1 (MabE1), prophiGD54-2 MabI) | pGD54 (pF) |
| GD55 | Cincinnati, OH | Y | a | S | WGS | prophiGD55-1 (MabL) | pGD55 (pSin) |
| GD56 | San Jose, CA | NA | a | R | WGS | prophiGD56-1 (MabC), prophiGD56-2 (MabH) | None |
| GD57 | Barcelona, Spain | Y | a | R | Com | prophiGD57-1 (MabC), prophiGD57-2 (MabA1) | None |
| GD58 | Chapel Hill, NC | NA | m | S | WGS | prophiGD58-1 (MabG), prophiGD58-2 (MabA1) | pGD58 (pH) |
| GD59 | Chapel Hill, NC | NA | a | R | Com | prophiGD59-1 (MabA1) | None |
| GD60 | San Diego, CA | N | m | R | WGS | prophiGD60-1 (MabL) | None |
| GD61 | San Jose, CA | NA | a | S | WGS | prophiGD61-1 (MabC), prophiGD61-2 (MabH) | None |
| GD62 | Baton Rouge, LA | N | a | R | WGS | prophiGD62-1 (MabB), prophiGD62-2 (MabF), prophiGD62-3 (MabN) | pGD62-1 (pB), pGD62-2 (pC) |
| GD63 | Memphis, TN | Y | a | S | WGS | None | None |
| GD64 | Cambridge, UK | N | a | S | WGS | None | None |
| GD68A | Long Beach, CA | Y | m | R | Com | prophiGD68-1 (MabE1) | None |
| GD68B | Long Beach, CA | Y | m | S | Com | prophiGD68-1 (MabE1) | None |
| GD69A | Long Beach, CA | N | a | R | Com | prophiGD69-1 (MabN) | pGD69-1 (pB), pGD69-2 (pC) |
| GD69B | Long Beach, CA | N | a | S | Com | prophiGD69-1 (MabN) | pGD69-1 (pB), pGD69-2 (pC) |
| GD72 | San Jose, CA | N | a | S | WGS | prophiGD72-1 (MabC) | pGD72 (pB) |
| GD75 | Charleston, SC | N | a | S | WGS | prophiGD75-1 (MabJ), prophiGD75-2 (MabA1) | pGD75 (pC) |
| GD79 | Bethesda, MD | N | m | R | WGS | prophiGD79-1 (MabQ) | None |
| GD81 | Hartford, CT | Y | a | S | WGS | prophiGD81-1 (MabA1) | None |
| GD82 | Baltimore, MD | N | m | R | WGS | prophiGD82-1 (MabC) | None |
| GD84 | Dallas, TX | N | a | S | WGS | prophiGD84-1 (MabA1), prophiGD84-2 (MabD) | None |
| GD85 | Ankara, Turkey | Y | a | S | WGS | None | pGD85 (pD) |
| GD86 | Jerusalem, Israel | Y | a | R | WGS | prophiGD86-1 (MabI), prophiGD86-2 (MabE1) | pGD86-1 (pF), pGD86-2 (pG) |
| GD87 | Jerusalem, Israel | Y | a | R | WGS | None | pGD87 (pB) |
| GD88 | Jerusalem, Israel | Y | a | R | WGS | prophiGD88-1 (MabL) | None |
| GD89 | Jerusalem, Israel | Y | a | R | WGS | prophiGD89-1 (MabB) | None |
| GD90 | Jerusalem, Israel | Y | m | S | WGS | prophiGD90-1 (MabA2) | None |
| GD91 | Bordeaux, France | N | b | S | Com | prophiGD91-1 (MabC), prophiGD91-2 (MabA3), prophiGD91-3 (MabO), prophiGD91-4 (MabE2) | None |
| GD92 | Jacksonville, FL | N | a | R | WGS | None | None |
| GD95 | Dallas, TX | N | a | R | WGS | prophiGD95-1 (MabG), prophiGD95-2 (MabN) | pGD95-1 (pB), pGD95-2 (pC) |
| GD97 | Bizkaia, Spain | Y | a | S | WGS | None | None |
| GD100A | Philadelphia, PA | NA | a | S | Com | prophiGD100A-1 (MabA1), prophiGD100A-2 (MabC) | pGD100 (pC) |
| GD100B | Philadelphia, PA | NA | a | R | Com | prophiGD100A-1 (MabA1), prophiGD100A-2 (MabC) | pGD100 (pC) |
| GD102 | Finland | Y | a | R | WGS | prophiGD102-1 (MabA1), prophiGD102 (MabE1) | pGD102-1 (pF), pGD102-2 (pG) |
| GD104 | Knoxville, TN | N | m | R | WGS | prophiGD104-1 (MabA1), phiGD104-2 (MabC) | pGD104 (pSin) |
| GD108 | Dallas, TX | NA | a | S | WGS | prophiGD108-1 (MabN) | pGD108 (pB) |
| GD111 | Morton Grove, IL | N | m | R | WGS | prophiGD111-1 (MabE1) | None |
Strains are given a GDXX designation; multiple isolates from the same patient are designated GDXXA, GDXXB, etc.
Strain origin indicates the location of the hospital or laboratory from which the strain was sent.
Indication of whether strain was from a cystic fibrosis patient; Y, yes; N, no; NA, data not available.
Subspecies derived from genome sequencing; a, abscessus; b, bolletii; m, massiliense.
Colony morphotype is shown as R, rough or S, smooth.
Genome sequencing with Illumina whole-genome sequencing (WGS) or Illumina and Nanopore to completion (Com).
Prophages are designated prophiGDXX, with -1, -2-, etc. extensions denoting different prophages in the same strain.
Plasmids are designated pGDXX, with -1, -2-, etc. extensions if there is more than one plasmid in the same strain.
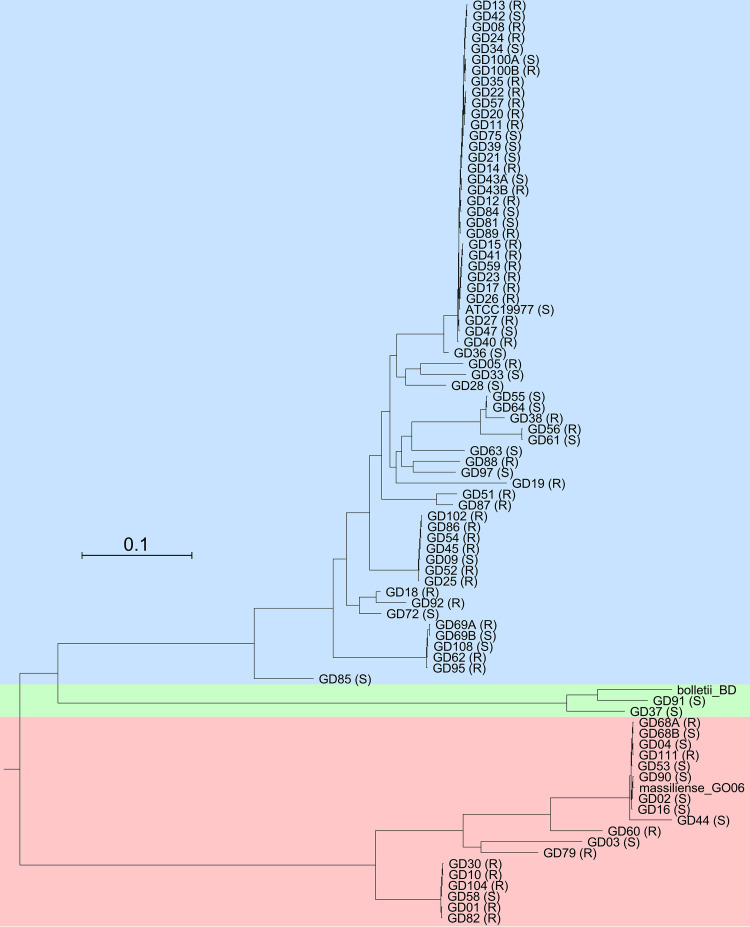
FIG 1.
M. abscessus phylogeny. Phylogenetic relationships of M. abscessus clinical isolates based on 3,682,630 (78%) conserved nucleotide positions in all strains. Subspecies are shaded blue, green, and red for M. abscessus subsp. abscessus, M. abscessus subsp. bolletii, and M. abscessus subsp. massiliense, respectively. The rough (R) or smooth (S) morphotype is indicated after the strain name. Scale corresponds to 0.1 substitutions per position.
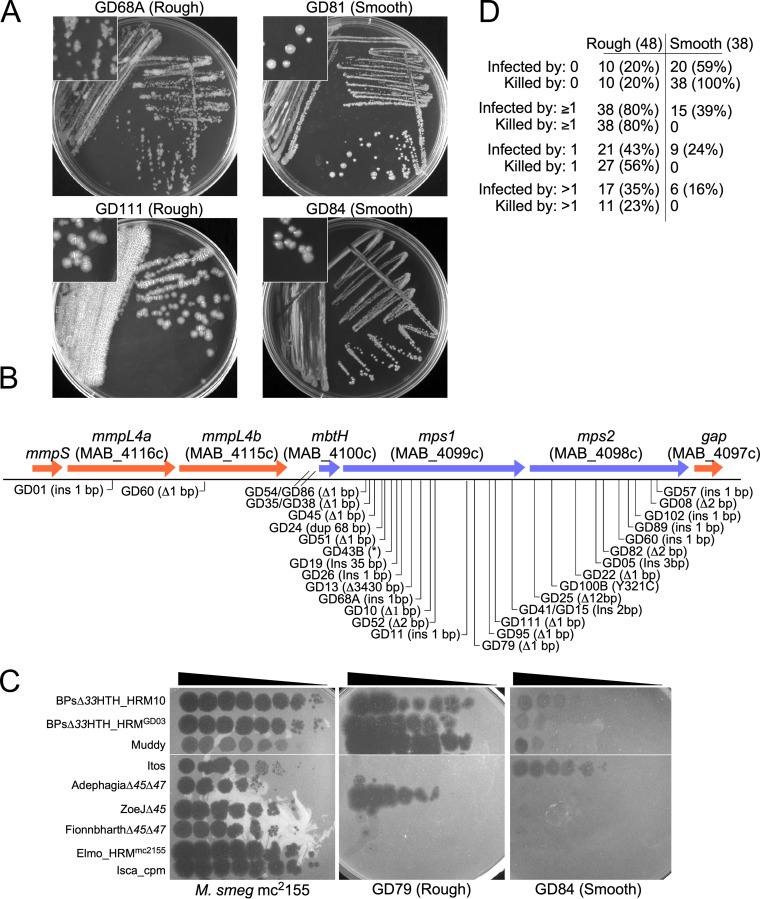
FIG 2.
Smooth and rough M. abscessus morphotypes. (A) Examples of strain colony morphotypes showing strains GD68A and GD111 (both rough) and GD81 and GD84 (both smooth) growing on solid medium. Insets show magnified view of colonies. (B) Mutations contributing to rough colony morphotypes. Rough strain sequences were compared with ATCC 19977 genes involved in GPL synthesis (as shown) and those with small (1 to 2 bp) insertions or deletions are indicated. One mutation (GD100B) is a single base substitution in mps2 and is the sole difference from its smoother counterpart, GD100A. A large spectrum of mutations is observed, although three pairs of rough strains (e.g., GD35 and GD38) have the same mutations. See Table S2 for details. (C) Plaque assay showing infection of M. smegmatis mc2155, M. abscessus GD79 (rough), and M. abscessus GD84 (smooth), as indicated. Ten-fold serial dilutions of phages were spotted from left to right on lawns of each strain, as indicated. (D) Phage susceptibilities of rough and smooth morphotype strains. The numbers of each strain morphotype that are not infected or killed by any phage tested (0), by at least one phage (≥1), by only a single phage (1), or by 2 or more phages (>1), as indicated, are shown. See Fig. 3 legend for details. The total number of strains (86) includes four strain pairs (e.g., A, B) each from a single patient source.
Genomic differences in smooth and rough Mycobacterium abscessus clinical isolates. Download Table S2, PDF file, 0.06 MB (61.3KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The M. abscessus genomes were sequenced either by Illumina technology to give whole-genome sequences (WGS) or to completion by the addition of Nanopore sequencing reads (Table 1; Table S1 in the supplemental material). Phylogenetic analysis of these strains together with the type-strains [M. abscessus subsp. abscessus ATCC 19977 (23), M. abscessus subsp. bolletii BDT (24), and M. abscessus subsp. massiliense GO06 (25)], shows that 62 of the strains are subspecies abscessus, 18 are massiliense, and 2 are bolletii (Table 1, Fig. 1). The substantial genetic diversity is not surprising (8), and 50% of the strains form a closely related clade that includes the ATCC 19977 type strain (Fig. 1); the prevalence of this clade was noted previously (8). There are two smaller massiliense clades, one of which includes the previously characterized strain GO06 (25), and one that includes GD01, the strain from the previously phage-treated patient (17) (Fig. 1). Both of the subsp. boletii strains have S morphotypes, but R and S morphotypes are distributed throughout the rest of the phylogenetic spectrum (Table 1, Fig. 1, Fig. 2A). All of the strains are CRISPR-free.
Sequencing details of Mycobacterium abscessus clinical isolates. Download Table S1, PDF file, 0.06 MB (99.1KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genotypes of smooth and rough colony morphotypes.
For four strains (GD43, GD68, GD69, and GD100), both S and R variants were recovered from the same sample and were sequenced to completion (Table 1). The R variants generally have several differences from the S variants, but these include mutations in GPL synthesis implicated in the rough morphotype (14). For example, rough strains GD43B and GD68A have single base differences in mps1 (MAB_4099) relative to their smooth variants GD43A and GD68B, introducing nonsense and single base insertions, respectively (Fig. 2B; Table S2). Although the other R strains do not have a smooth counterpart, over 75% of them have mutations in mps1 and mps2 (MAB_4098), most commonly 1- to 2-bp insertions or deletions. One (GD60) also has a frameshift mutation in mmpL4b (MAB_4115) (Fig. 2B, Table S2). Only three instances of the same mutation in different strains were identified.
Phage susceptibility profiles of M. abscessus clinical isolates.
We previously reported that M. abscessus strain GD01 is efficiently infected and killed by the phages Muddy, BPs, and ZoeJ, although these were the only phages identified from a screen of about 100 individual phages isolated on M. smegmatis (17). Few phages have been isolated on M. abscessus strains directly (17), and initial evaluation of several dozen M. smegmatis phages confirmed that many do not infect any of the clinical isolates; we therefore focused on eight of the most promising candidates (Fig. 1, Fig. 2C, Fig. 3): Muddy, BPs, ZoeJ, Itos, Faith1, Fionnbharth, D29, and Elmo (members of clusters/subclusters AB, G1, K1, L2, L2, K4, A2, and A3, respectively) (26, 27) or their derivatives, as described below.
FIG 3.
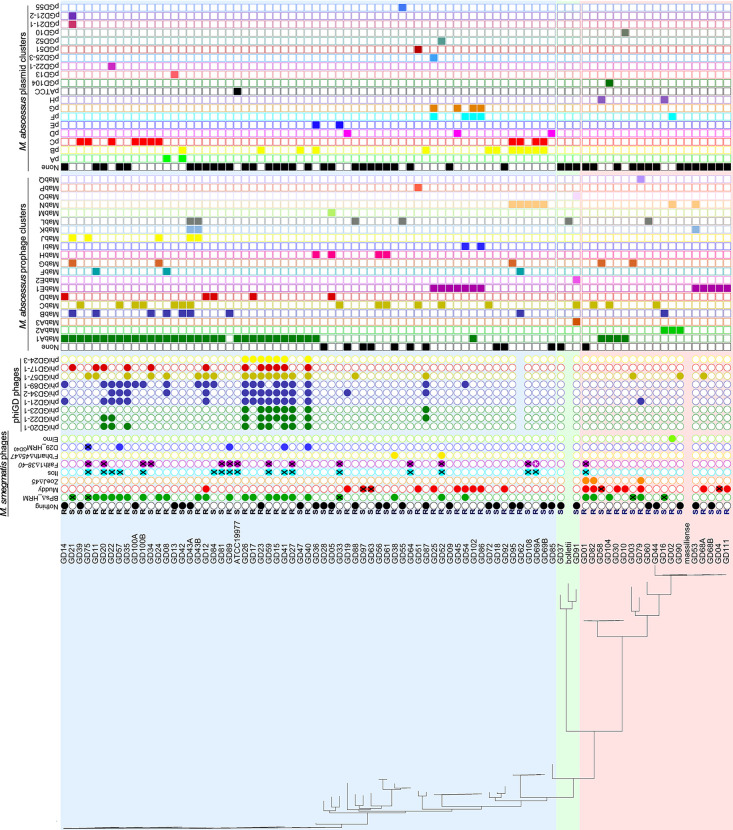
Properties and behaviors of M. abscessus clinical isolates. M. abscessus clinical isolates are shown with their phylogenetic relationships (as in Fig. 1), colony morphotype (R, rough; S, smooth) and susceptibilities to M. smegmatis phages (leftmost circles). Filled circles indicate either no phage infection (black) or efficiency of plating (EOP) relative to M. smegmatis of >0.1. Empty circles indicate no infection or an EOP relative to M. smegmatis of <0.1. Filled circles with an “X” reflect efficient infection (EOP >0.1) but poor killing; absence of circle, not tested. Filled circles with a white asterisk denote strains infected by HRM, but not the parental phage. The rightmost circles indicate susceptibilities to phiGD phages. Empty circles indicate no infection and filled circles reflect efficiency of plating relative to the original recipient strain of >0.01. To the right are squares indicating the presence of prophages and plasmids, with the cluster indicated at the top. Filled squares, presence of prophage or plasmid; empty squares, absence of prophage or plasmid.
The substantial variation in the infection profiles is striking (Fig. 3). Many different susceptibility combinations are observed and there is no evident correlation with the whole-genome phylogenies. Approximately 34% of the strains are not susceptible to any of the phages tested, spanning subspecies and clades (Fig. 3). There are several instances in which closely related strains of different origins have similar profiles, such as GD01 and GD82 from patients in the UK and US, respectively, which differ by only a few dozen nucleotide polymorphisms and have similar susceptibility profiles (Table 1, Fig. 3).
Phages forming plaques on clinical isolates may not necessarily kill them efficiently, and we therefore determined this using challenge assays to measure bacterial survival (Fig. 4C and E, Table S3). There is a striking correlation between phage killing efficiency and strain colony morphotype. Of the 48 R strains, 38 (80%) are efficiently killed by at least one phage, 17 of which (35%) are killed by more than one (Fig. 2D). In contrast, none of the 38 S strains are killed, although a subset (21%) is infected by phages Faith1 and/or Itos, but neither kills the strains efficiently (Fig. 1, Fig. 2D). Surface GPLs on the smooth strains may be responsible for the poor phage infection, but it is unclear why lytic derivatives of Faith1 and Itos (see below) do not efficiently kill the S strains they infect (Fig. 2D, Fig. 3). This poor-killing phenotype is not constrained to the subcluster L2 phages and is also observed in some R and S strains with phages BPs, Muddy, and D29 (Fig. 2D, Fig. 3).
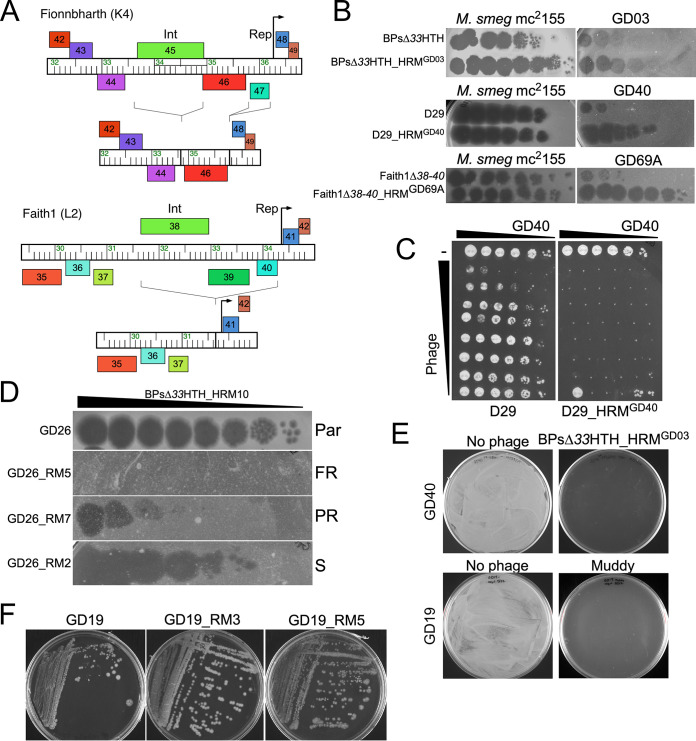
FIG 4.
M. abscessus phage mutants and resistance. (A) Engineering lytic derivatives of phage Fionnbharth and Faith1. Central genomic regions are shown for each phage with rightward- and leftward-transcribed genes shown above and below the genome ruler, respectively. The subcluster K4 phage Fionnbharth genome was edited to remove the integrase (int) and repressor (rep) genes as indicated. The subcluster L2 phage Faith1 was edited to remove genes 38 to 40 including int and rep. (B) Ten-fold serial dilutions of host range mutants (HRM) of BPsΔ33HTH, D29, and Faith1Δ38-40 were plated from left to right on M. smegmatis and the M. abscessus strain on which the HRM was isolated. (C) Killing assay of M. abscessus strain GD40 with phages D29 (left) and D29_HRMGD40 (right). Each column has 10-fold serial dilutions of M. abscessus GD40 (leftmost column, 3 × 105 CFU), and each row has 10-fold serial dilutions of either phage D29 or D29_HRMGD40 (topmost row, 107 PFU). Cells and phage were mixed and incubated for 48 h prior to plating on solid medium. (D) Phage susceptibility assay illustrating that strains recovered from BPsΔ33HTH_HRM10 challenge of GD26: Par, parental; FR, fully resistant GD26_RM5; PR, partially resistant GD26_RM7; and S, fully sensitive GD26_RM2. Spots are 10-fold serial dilutions of BPsΔ33HTH_HRM10 from left to right. (E) Efficient killing of M. abscessus strains GD40 and GD19 (as indicated) with phages BPsΔ33HTH_HRMGD03 or Muddy. Aliquots of 107 CFU were incubated with or without phage (multiplicity of infection [MOI] 10) for 48 h and plated on solid medium. (F) Colony morphotypes of GD19 and Muddy-resistant mutants GD19_RM3 and GD19_RM5. GD19 and GD19_RM3 are rough, and GD19_RM5 is smooth.
M. abscessus survival following phage challenge. Download Table S3, PDF file, 0.1 MB (118.1KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expanding the phage repertoire by genome engineering.
Several of the potentially useful phages are temperate and form stable lysogens in M. smegmatis. Muddy is not evidently temperate, and efficiently kills most of the M. abscessus strains it infects. Itos, D29, and Elmo are not temperate, but are naturally lytic derivatives of temperate phages that lack the repressor gene, which is responsible for establishing and maintaining lysogeny. We previously described the engineering of both BPs and ZoeJ to construct lytic derivatives (17, 28), and we used similar approaches to construct lytic versions of Fionnbharth (i.e., FionnbharthΔ45Δ47) and Faith1 (i.e., Faith1Δ38-40) (Fig. 4A).
Expanding the phage repertoire using host range mutants.
In screening strains, several different phenotypes of M. abscessus phage infection were observed. These include an absence of infection, an efficiency of plaquing (e.o.p.) of one relative to M. smegmatis, and killing of cells only at high phage concentrations (Fig. 2C). Sometimes, individual plaques are observed at high phage titers, arising from phenotypic escape of defense systems, such as restriction or genetic alteration that promotes efficient infection. We previously described a host range mutant (HRM) of phage BPsΔ33HTH (BPsΔ33HTH_HRM10) (17), and we successfully isolated another HRM of BPsΔ33HTH (BPsΔ33HTH_HRMGD03) using M. abscessus GD03 as the host (Fig. 4B). We similarly isolated HRMs of phages D29 and Faith1Δ38-40 (Fig. 4B).
The lytic subcluster A2 phage D29 (29, 30) does not efficiently infect any M. abscessus strain, but an HRM was isolated on M. abscessus GD40, purified, and shown to efficiently infect GD57, GD89, and GD41, in addition to GD40 (Fig. 3, Fig. 4B). D29_HRMGD40 has three mutations relative to its immediate precursor, C12442A, T31726C, and A43657T, but reconstruction of the individual mutants demonstrated that the C12442A mutation conferring a T116N substitution in the capsid protein is responsible for the host range phenotype (Fig. 4B). Capsid mutations associated with phage host range are unusual but have been described to influence adsorption of ϕX174 (31). The potential therapeutic utility of D29_HRMGD40 is illustrated by its enhanced killing of strain GD40 relative to the D29 parent phage (Fig. 4C).
Faith1 does not efficiently infect M. abscessus GD69A but an HRM was isolated (Faith1Δ38-40_HRMGD69A) that does (Fig. 4B), although the HRM does not change its infection of other strains. Faith1Δ38-40_HRMGD69A has a single mutation at G34936T conferring an early termination codon in gene 43 (E11*), whose function is unknown, but it is highly expressed in early lytic growth of its relative Crossroads (32). Faith1 gp43 is unlikely to be involved in cell surface interactions and may be triggering a host defense system to which the Faith1Δ38-40_HRMGD69A mutant is able to escape.
M. abscessus phage resistance.
Phage-susceptible strains were challenged with phage(s) in liquid cultures and 107 cells plated on solid medium to recover survivors. Robust growth was observed for all of the smooth strains (>50% survival) reflecting very poor killing (Table S3). In contrast, 70% of the 75 combinations of phage and rough strains, using a total of 28 strains, yielded either no or very few survivors, which upon retesting were either fully or partially phage sensitive (Fig. 4D and E; Table S3). Only 12 of the 28 strains (43%) yielded any resistant mutants (Table S3). With the exception of mutant GD19_RM5, which converted to S, all of the resistant mutants retained their R morphotype (Fig. 4F). This is notable because reversion or suppression to smooth could be a simple route to resistance, but evidently is relatively uncommon due to the type of mutations conferring rough morphotypes (Fig. 2B). We note that no resistant strains for GD43B were recovered (Table S3), suggesting that reversion or suppression of the mps1 nonsense mutation occurs below the detection limit of the assay.
Nine phage-resistant strains were sequenced and compared to their phage-sensitive parents (Table S4). GD19_RM5 (Fig. 4F) contains a wild-type allele of mps1 (MAB_4099), and direct reversion of the 35-bp insertion in mps1 (MAB_4099) has given both the smooth morphotype and resistance to Muddy (Fig. 2B, Fig. 4F). Two mutants (GD17_RM1 and GD22_RM4) have mutations in a type I polyketide synthase (MAB_0939), implicated in synthesis of trehalose polyphleates (33); related proteins are nonessential for M. tuberculosis growth in vitro but are important for virulence (34). Mutants GD22_RM1 and GD22_RM2 have mutations in the C-terminal HRDC domain of UvrD2 (MAB_3511), which is nonessential for M. tuberculosis growth (35). For GD26_RM4 and GD25_RM2, it is unclear which genes are implicated in resistance, either a 28.5-kbp deletion or multiple changes from its parent; however, both include genes implicated in virulence (Table S4). GD22_RM3 has lost plasmid pGD22-1 (36), which could impact phage infection.
Genomic differences in phage-resistant mutants (RMs) of Mycobacterium abscessus clinical isolates. Download Table S4, PDF file, 0.04 MB (40.4KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mutant GD19_RM3 (Fig. 4F) has a frameshift mutation (4-bp deletion) in rpoZ coding for the RNA polymerase omega subunit, and is the sole sequence difference from GD19; rpoZ is clearly not essential for M. abscessus growth, as reported for M. smegmatis (37). Deletion of rpoZ in M. smegmatis does not alter the GPL profile but it reduces both sliding motility and biofilm formation, and has a notable reduction in short-chain mycolates on the cell surface (37). Similar surface changes in GD19_RM3 may be responsible for the inability of phage Muddy to infect. None of the other strains described here have rpoZ mutations, and this RM characterization illustrates the multitude of mechanisms that alter phage susceptibility.
Isolation and propagation of spontaneously released lytic phages.
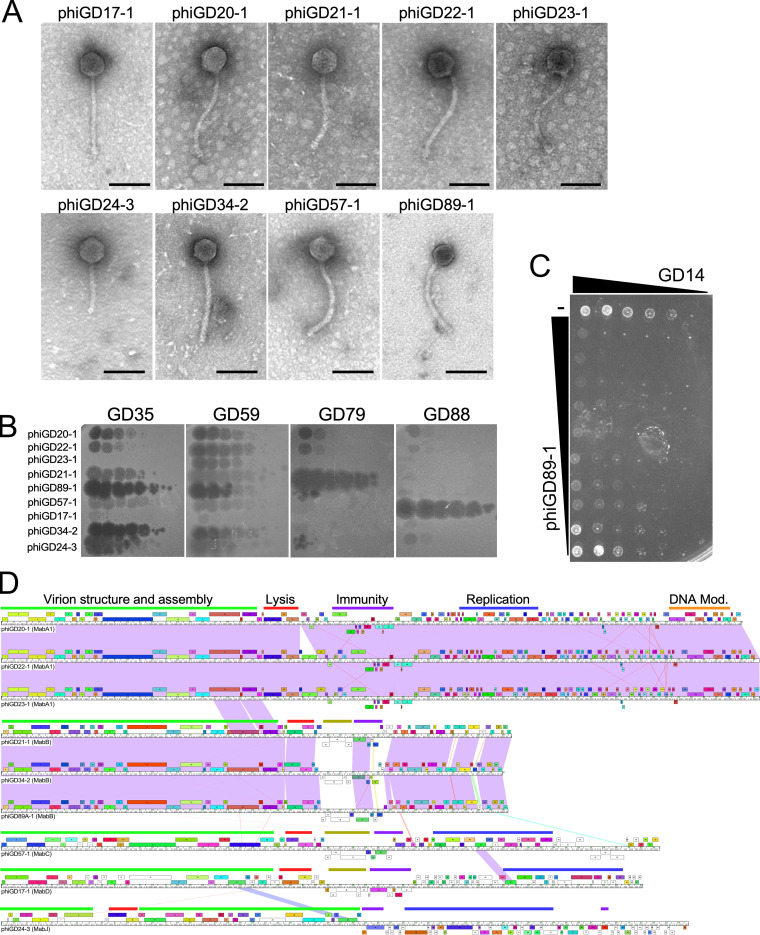
To expand the suite of phages with therapeutic potential, we searched for phages that are spontaneously released from the M. abscessus strains that form plaques on other strains. A screen of over 1,200 pairwise tests identified nine distinct phages (designated phiGDxx), corresponding to nine different donor and five recipient strains (Table 2). Each phage was purified on its recipient strain, sequenced, and annotated (Fig. 5, Table 2, Fig. S1 to S5, Fig. S6 to S9) (https://phagesdb.org/documents/categories/15/). All have features typical of temperate phages and their organizations and virion genes are consistent with their siphoviral morphologies (Fig. 5A). None grow on M. smegmatis, and none are closely related to previously described M. smegmatis phages (26, 27). Electron microscopy showed all nine phages have siphoviral morphologies (Fig. 5A). The phiGDxx phages are expected to be temperate and form turbid plaques, as they are derived from resident prophages. This is observed for most of the phages, but not for phiGD89-1, which forms clear plaques (Fig. 5B). phiGD89-1 differs from its close relatives phiGD21-1 and phiGD34-2 by several nucleotide differences immediately upstream of the early lytic genes, which may confer the clear-plaque phenotype. phiGD34-2 forms turbid plaques on most strains, but not on all, and the plaques appear clear on strains such as GD35 (Fig. 5B).
TABLE 2.
Lytically propagated phages derived from Mycobacterium abscessus
| Namea | Recipientb | Clusterc | Length (bp)d | ORFse | Equivalent prophagef | Accession no. |
|---|---|---|---|---|---|---|
| phiGD20-1 | GD35 | MabA1 | 59,055 | 118 | prophiGD20-1 (MabA1) | MW314858 |
| phiGD22-1 | GD40 | MabA1 | 60,512 | 120 | prophiGD22-1 (MabA1) | MW314856 |
| phiGD23-1 | GD40 | MabA1 | 60,535 | 120 | prophiGD23-1 (MabA1) | MW314855 |
| phiGD21-1 | GD41 | MabB | 40,735 | 63 | prophiGD21-1 (MabB) | MW314857 |
| phiGD34-2 | GD41 | MabB | 40,005 | 63 | prophiGD34-2 (MabB) | MW314853 |
| phiGD89-1 | GD89B | MabB | 40,450 | 63 | prophiGD89A-1 (MabB) | MW314851 |
| phiGD57-1 | GD35 | MabC | 52,563 | 72 | prophiGD57-1 (MabC) | MW314852 |
| phiGD17-1 | GD40 | MabD | 51,185 | 88 | prophiGD17-1 (MabD) | MW314859 |
| phiGD24-3 | GD17 | MabJ | 54,877 | 94 | prophiGD24-3 (MabJ) | MW314854 |
Phages are named according to the donor strains from which they were isolated.
Recipient is the sensitive strain on which the phage was isolated on.
Cluster designation (MabA, MabB, etc.), corresponds to the prophage cluster assignment.
Phage genome length in base pairs (bp).
Predicted numbers of open reading frames (ORFs).
Name of prophage present in the donor strain corresponding to the lytically propagated phage. Cluster designation is shown in parentheses.
FIG 5.
Lytically propagating phiGDxx phages. (A) Electron micrographs of nine lytically growing phages released by spontaneous prophage induction. Scale bar is 100 nm. (B) The nine lytically growing phages recovered from M. abscessus GD strains (as shown) were 10-fold serially diluted and spotted onto lawns of GD35, GD59, GD79, and GD88 to illustrate infection profiles. Full infection profiles are shown in Fig. 3. (C) Killing assay showing reduction in viability of M. abscessus GD14 by phiGD89-1. Configuration is as shown in Fig. 4C. (D) Genome maps of the nine lytically growing phages recovered from M. abscessus strains, with pairwise nucleotide sequence similarity shown as spectrum colored shading, with violet indicating closest similarity and red the least above a threshold BLASTN E value of 10−4. Genes are shown as boxes above (transcribed rightward) and below (transcribed leftward) each genome; boxes are colored according to the gene phamilies they are assigned (50). Slim bars above genome sequence identify categories of gene functions: green bar, structural genes; red bar, lysis cassette; purple bar, immunity cassette; blue bar, replication genes, orange bar, DNA modification genes; and olive bars, polymorphic toxin genes. Detailed genome maps for all phages are in Fig. S1 to S5 and Fig. S6 to S9 in the supplemental material (https://phagesdb.org/documents/categories/15/).
Each of the nine phages was tested for infection of the GDxx strains (Fig. 3). Phage phiGD89-1 infects about one-third of the R strains, most of which are in the large M. abscessus subsp. abscessus clade (Fig. 3, Fig. 5B). The closely related phiGD21-1 has a similar profile, although some strains, such as GD08, distinguish between the two phages (Fig. 3). Seven strains—four S type (GD34, GD84 GD90, and GD100A) and three R type (GD14, GD23, and GD88)—that are not killed efficiently by other phages are efficiently infected by at least one phiGD phage (Fig. 3), increasing the proportion of efficiently infected R strains to 85%. They also add a potential second phage to 10 other R strains that are only killed by a single phage. This expansion of the phage repertoire is illustrated by the ability of phiGD89-1 to kill GD14, for which no other phages have been identified (Fig. 3, Fig. 5C). The temperate phiGDxx phages will need to be engineered for obligatory lytic growth prior to therapeutic consideration. The phiGDxx genomes vary in length from 40 to 60 kbp and represent five distinct groups based on overall sequence relationships (Fig. 5D, Table 2); detailed genome maps are shown in Fig. S1 to S5 and Fig. S6 to S9 (https://phagesdb.org/documents/categories/15/).
Genome organization of phiGD20-1 (MabA1). The genome of phiGD20-1 is shown with predicted genes shown as boxes either above or below the genome indicating rightward- and leftward-transcription, respectively. Gene numbers are shown within each gene box. Phamily designations are shown above or below each gene, with the numbers of phamily members in parentheses; genes are colored according to the phamily designations. White boxes represent “orphams,” i.e., genes with no close relatives in this dataset. Phamily assignments were determined using Phamerator and database Actino_prophage (version 5). Predicted gene functions are indicated. Download FIG S1, PDF file, 0.4 MB (405.5KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD21-1 (MabB). See Fig. S1 for details. Download FIG S2, PDF file, 0.2 MB (211.2KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD57-1 (MabC). See Fig. S1 for details. Download FIG S3, PDF file, 0.2 MB (213.6KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD17-1 (MabD). See Fig. S1 for details. Download FIG S4, PDF file, 0.3 MB (296.9KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD24-3 (MabJ). See Fig. S1 for details. Download FIG S5, PDF file, 0.3 MB (312.9KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
M. abscessus prophages.
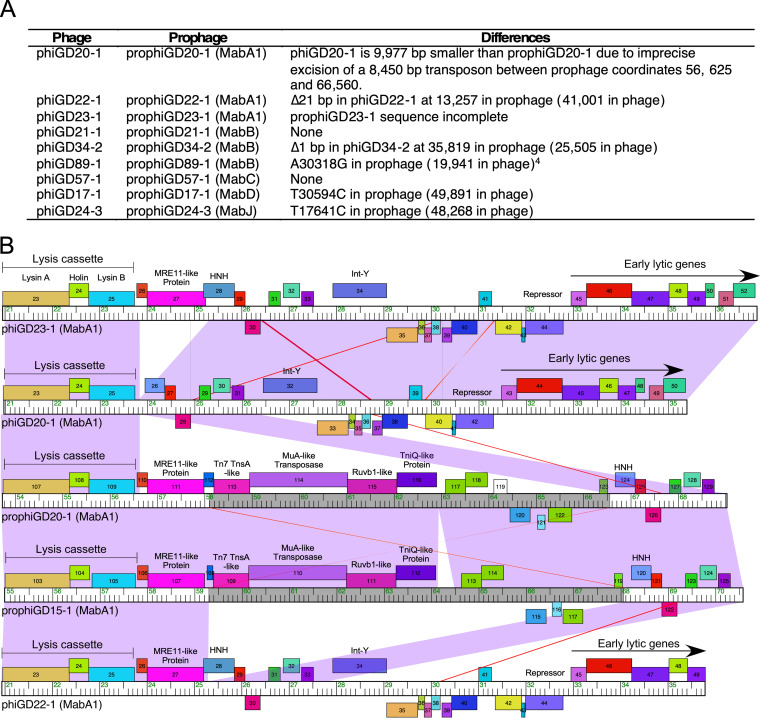
Using the phiGDxx sequences, we identified the cognate prophages (designated prophiGDxx) in the donor strains and extracted their complete prophage sequences (Table 1, Fig. 6). Genome comparisons showed that phages phiGD21-1 and phiGD57-1 are identical to their cognate prophages, and spontaneous induction involves simple excisive site-specific recombination between the attL and attR junctions. However, six of the phages differ from their prophages by one or more base substitutions and/or insert/deletions, which presumably occurred during induction or subsequent lytic growth (Fig. 6A). Curiously, prophiGD20-1 is 9,977 bp larger than phiGD20-1 and contains an 8,452-bp transposon insertion in an HNH-like gene, which is defined by 25-bp imperfect inverted repeats (IRs) and flanked by a 5-bp target duplication (Fig. 6B). Loss of the transposon—presumably a requirement for lytic growth due to DNA packaging constraints—has occurred by transposase-mediated imprecise excision between the rightmost IR junction and a location within prophiGD21-1 gene 110; excision also removes an Mre-11-like gene which is evidently not required for lytic growth (Fig. 6B). A second closely related prophage (prophiGD15-1; see below) has a similar transposon inserted within the same HNH gene, 25 bp to the left of that in prophiGD20-1. Moreover, two additional prophages, prophiGD41-1 and prophiGD59-1, are identical to prophiGD15-1 with the same transposon insertions. The transposon is also present in M. abscessus subsp. bolletii CCUG, M. abscessus FLAC013, and M. abscessus UC22 (GenBank accession numbers AP014547, CP014955, and CP012044).
FIG 6.
Comparison of induced phages and their cognate prophages. (A) Comparisons of phage (i.e., phiGDxx) and their cognate prophages (i.e., prophiGDxx) showing genomic differences; phages and prophages are named after the strain they were isolated from. The prophiGD23-1 sequence is incomplete in the WGS assembly and thus cannot be fully compared with phiGD23-1. In phiGD89-1, there are several differences in a region that is well conserved in other MabB phages, including phiGD21-1. Specifically, the sequence 5′-TGGACTACGGCTGAGCAGCA-TGCT (coordinates 30,519 to 30,542) replaces 5′-TGGGCTACGGCTGAGCAGTAGAACT. This is in the location of a predicted rightward early promoter and operator site, and likely inactivates lysogeny. (B) Comparison of phage and prophage genome segments, illustrating the transposon insertions (gray areas on genome rulers) in prophages prophiGD20-1 and prophiGD15-1. Genome annotations and comparisons are illustrated as in Fig. 5D. The alignment illustrates the transposon insertion (comparing phiGD22-1 and prophiGD15-1), and excision of the transposon and flanking gene in phiGD20-1. The transposons in prophiGD20-1 and prophiGD15-1 are closely related but inserted at targets 25 bp apart.
Bioinformatic analyses shows that prophages are abundant in the GD strains, as has been reported for other M. abscessus strains (38). These prophages, along with plasmids from these strains, are described in detail elsewhere (36) (Tables S5 and S6) (https://phagesdb.org/documents/categories/15/), and their inclusion in each strain is indicated in Fig. 3. Only 12 strains are prophage-free, and the 122 prophages and nine lytically growing phages (Table 2) are grouped into clusters (MabA to MabQ) according to their sequence relationships (Fig. 6A, Table S5). Because the phage susceptibility profiles do not correlate closely with nucleotide sequence-based phylogeny (Fig. 3), the prophage and plasmid contents are strong candidates for contributing to the phage infection profiles. The abundance and diversity of the M. abscessus mobilome confounds any simple elucidation of their roles. We note, however, that many of the prophages and plasmids code for type II toxin-antitoxin systems which can confer viral defense (36).
DISCUSSION
The phage infection profiles of these M. abscessus strains illuminate both the opportunities and challenges in their therapeutic prospects. Smooth morphotype strains are problematic and not efficiently killed by any of the phages, and a surprisingly high proportion (36%) of strains are smooth, suggesting that they are important pathogens, but perhaps with pathologies distinct from rough strains. The observation that smooth strains with abundant surface GPLs are not efficiently killed by phages is in contrast to the finding that GPLs are required for M. smegmatis infection by phage I3 (39). Through extensive phage screening, genome engineering, host range mutant identifications, and induction of prophages, a relatively small set of phages were identified that can infect and kill 80% of the rough strains. The induced prophages require further engineering to remove genes for lysogeny and potential virulence genes, but are a promising source of new phages, increasing the number of rough strains targeted to about 85%. Until additional phages are identified that expand the repertoire to kill all of the rough strains, screening of individual strains is still required before therapeutic intervention because of the variable and unpredictable phage susceptibility profiles. The variability in phage infection is likely determined in large part by the impressive array of diverse prophages and plasmids in these M. abscessus clinical isolates.
With only a rather limited set of phages available for therapy—and a substantial proportion of rough strains are killed by only a single phage—the frequencies and mechanisms of phage resistance are key concerns. For most M. abscessus phage infections, no stably resistant survivors were isolated, and when resistant mutants were recovered, the mutations suggest there may be a fitness cost for in vivo growth. Furthermore, although reversion to a smooth morphotype is expected to contribute to resistance, this occurs only rarely, reflecting the predominance of mutations leading to rough morphotypes that occur only infrequently. In general, use of a single phage for therapy when antibiotic treatments are no longer available would seem a reasonable strategy. We recognize that some infections contain both rough and smooth M. abscessus variants and can vary by anatomical location and duration (14, 15, 22), raising the question as to whether phage-mediated killing of the resistant strain alone will provide any therapeutic effect. This is likely to vary considerably among patients, depending on the virulence capacity of the smooth variant.
Challenges in the therapeutic use of phages arise for many other bacterial pathogens other than M. abscessus, including Mycobacterium avium and Burkholderia spp. infections. The strategies deployed here, including exploitation of phage libraries of related hosts, phage genome engineering, host range expansion, and growth of induced prophages, can be applied to these and other pathogens to expand the repertoire of therapeutically useful phages. Although the number of useful phages may remain relatively small for some infections, infrequent phage resistance and tradeoffs for in vivo fitness should improve their therapeutic utility.
MATERIALS AND METHODS
Bacterial strains.
M. smegmatis mc2155 is a laboratory stock strain and was grown as previously described (40). M. abscessus ATCC 19977 was obtained from the American Type Culture Collection. M. abscessus clinical isolates were received on Lowenstein-Jensen slants and streaked out on Middlebrook 7H10 agar (Difco) supplemented with oleic acid-albumin-dextrose-catalase (OADC) and 1 mM CaCl2 and were grown for 5 to 7 days at 37°C. Liquid cultures were inoculated from a single colony and grown in Middlebrook 7H9 medium with OADC and 1 mM CaCl2 for 4 to 5 days at 37°C, with shaking. For plaque assays, M. abscessus cultures were sonicated briefly in a cup-horn sonicator (Q-sonica) as described previously (17). The GD54 strain sequenced and analyzed was GD54H, one of several isolates from the same source. Similarly, the GD35 strain sequenced and analyzed was otherwise designated GD35B. Strains GD43A and B have different numbers of prophages and were therefore analyzed as separate strains. Strain GD43A has six prophages (in clusters MabA, MabB, MabC, MabJ, MabK, and MabL) and GD43B has only four of these (MabA, MabL, MabK, and MabJ). There are no plasmids in either GD43A or GD43B.
Genome sequencing.
Bacterial and phage genomes were sequenced by Illumina alone or supplemented with Nanopore reads as described previously (41, 42) and details are provided in Table S1. For all strains, Illumina sequencing libraries were prepared from genomic DNA using NEB Ultra II FS kits with dual-indexed barcoding. Libraries were pooled and run on an Illumina MiSeq, yielding 300-base paired-end reads. In some cases, Oxford Nanopore libraries were also constructed from genomic DNA using Rapid Sequencing Barcoding kits, then pooled and run on a MinION device using FLO-MIN106D flowcells. Illumina reads for each strain were trimmed and quality-controlled using Skewer (43). Trimmed Illumina reads were then assembled using Unicycler (44), incorporating Nanopore reads when available.
In the case of complete genomes, assemblies were viewed, stitched, corrected, and finalized using Consed version 29 (45, 46). GraphMap (47) was used to align long Nanopore reads to provisional assemblies and resolve repetitive regions. The first base and orientation of each complete circular chromosome was chosen to match those of the ATCC 19977 strain and/or to align with the first base of the dnaA gene. Complete circular plasmids were similarly oriented and cut so that base 1 was the first base of a predicted repA gene.
Phylogenetic trees.
Phylogenies were created using CSI Phylogeny 1.4, a SNP-based concatenated alignment, available on the DTU server (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) (48). Complete and multiple sequence fasta files for 84 genomes were aligned to reference genome (ATCC 19977, CU458896), snp pruning disabled. Newick files were viewed in FigTree v1.4.4 and imported into iTOL (https://itol.embl.de/).
Prophage identification.
Identification of additional prophages was accomplished by searching with PHASTER (49) for phage-like regions followed by careful manual inspection, identification of attL and attR attachment sites, and confirmation of the predicted attB sequences in the type-strains of either M. abscessus subsp. abscessus ATCC 19977, M. abscessus subsp. massiliense, or M. abscessus subsp. bolletii BDT (23–25). Phamerator (50) databases “Actino_prophage_15” and “Abscessus_prophages_5” were constructed for comparative genomic analyses.
Of the 122 prophages identified, 80 were either in completely sequenced genomes or were wholly within one contig in WGS assemblies (Table S5), and complete genome sequences were extracted. The other 42 were in multiple contigs (Table S5) and, although complete prophage sequences were not available, sufficiently large segments were available to reveal their relationships to other prophages.
Several of the clusters contained only a single prophage member, although database searches suggested that all of these have relatives in other sequenced M. abscessus genomes.
The very extensive diversity of the plasmid genes is reflected in the finding that when sorted into protein phamilies (as described previously), 58% of the genes are “orphams” without closely related genes in this data set.
Phage infections.
Mycobacterial strains were grown and tested for phage susceptibility as described previously (17). Twenty-three mycobacteriophages were used for the initial screening of M. abscessus clinical isolates. These phages were chosen from clusters known to have expanded host range (40) and others were chosen based on screening completed previously on M. abscessus clinical isolates (17). Phage lysates were 10-fold serially diluted and plated on M. smegmatis mc2155 and each M. abscessus clinical isolate. The phiGDxx series of phages were isolated by plating clarified lysates of M. abscessus strains on M. abscessus strains, purifying phages from cleared areas on the sensitive strains, and growing to high titer.
Phage engineering.
Bacteriophage recombineering of electroporated DNA (BRED) was used to generate the three single base mutations (C12442A, T31726C, and A43657T) of D29, as well as FionnbharthΔ45Δ47 and Faith1Δ38-40 (51). Oligonucleotides are shown in Table S8 (https://phagesdb.org/documents/categories/15/). For D29, MAMA PCR was used to screen potential mutants. Once pure, they were serially diluted and plated on M. smegmatis mc2155 and GD40. For Fionnbharth and Faith1, PCR using flanking primers was used to screen plaques for homogeneous deletion derivatives. All phage mutants were sequenced. See Table S8 (https://phagesdb.org/documents/categories/15/) for a list of oligonucleotides.
Phage-resistant M. abscessus mutants.
Survival assays of M. abscessus strains sensitive to phages were set up to isolate resistant mutants. An aliquot of 1 ml of ∼1 × 108 CFU/ml of culture was mixed with, or without, one log higher concentration (∼1 × 109 PFU/ml) of phage. These were incubated at 37°C with shaking, and 100 μl was plated after 2 and 5 days. Plates were grown at 37°C for 5 to 7 days and then photographed. Colonies from these plates were picked, streaked twice, and then grown in liquid culture to be used in top agar overlays for phage-resistance screening. Top agar overlays were spotted with serial dilutions of the phage of interest and incubated at 37°C for 5 to 7 days.
phiGDxx phages.
Log phase liquid cultures of clinical isolates were grown, and one ml of each culture was centrifuged at 14,000 × g for 2 min. The supernatant (from each “donor” strain) was saved and used for spotting on top agar overlays of other clinical isolates (the “recipient” strain). These plates were incubated at 37°C for 5 to 7 days. Any clearing on the overlay where a supernatant was spotted was picked, using a sterile pipet tip, into phage buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgSO4, and 68 mM NaCl). The phiGDxx phages were purified and then amplified on the receiving strain. DNA was extracted from lysates of phiGDxx phages using a standard phenol-chloroform/EtOH precipitation protocol.
Data availability.
The completed and WGS genome sequencing data for M. abscessus clinical isolates have been submitted to GenBank and accession numbers are listed in Table S1. The phiGDxx genomes have the following GenBank and accession numbers: phiGD89-1 (MW314851), phiGD57-1 (MW314852), phiGD34-2 (MW314853), phiGD24-3 (MW314854), phiGD23-1 (MW314855), phiGD22-1 (MW314856), phiGD21-1 (MW314857), phiGD20-1 (MW314858), phiGD17-1 (MW314859).
ACKNOWLEDGMENTS
We are very grateful to the many colleagues who sent us M. abscessus strains.
This work was supported by grants 1R35 GM131729 and 1R21AI151264 from the National Institutes of Health, GT12053 from the Howard Hughes Medical Institute, HATFUL19GO from the Cystic Fibrosis Foundation, and a kind donation from The Fowler Fund for Phage Research.
R.M.D., C.A.G.-B., and G.F.H. conceived and designed the experiments; R.M.D., B.E.S., H.G.A., C.A.G.-B., A.M.D., V.M., R.A.G., D.A.R., K.M.Z., and L.A. performed the experiments; R.M.D., B.E.S., H.G.A., C.A.G.-B., A.M.D., V.M., D.J.-S., R.A.G., D.A.R., L.A., C.H.G., and G.F.H. analyzed the data; R.M.D. and G.F.H. wrote the paper.
G.F.H. is a consultant for Janssen Pharmaceuticals.
Footnotes
Citation Dedrick RM, Smith BE, Garlena RA, Russell DA, Aull HG, Mahalingam V, Divens AM, Guerrero-Bustamante CA, Zack KM, Abad L, Gauthier CH, Jacobs-Sera D, Hatfull GF. 2021. Mycobacterium abscessus strain morphotype determines phage susceptibility, the repertoire of therapeutically useful phages, and phage resistance. mBio 12:e03431-20. https://doi.org/10.1128/mBio.03431-20.
REFERENCES
- 1.Johansen MD, Herrmann JL, Kremer L. 2020. Non-tuberculous mycobacteria and the rise of Mycobacterium abscessus. Nat Rev Microbiol 18:392–407. doi: 10.1038/s41579-020-0331-1. [DOI] [PubMed] [Google Scholar]
- 2.Floto RA, Haworth CS. 2015. The growing threat of nontuberculous mycobacteria in CF. J Cyst Fibros 14:1–2. doi: 10.1016/j.jcf.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Story-Roller E, Maggioncalda EC, Cohen KA, Lamichhane G. 2018. Mycobacterium abscessus and beta-lactams: emerging insights and potential opportunities. Front Microbiol 9:2273. doi: 10.3389/fmicb.2018.02273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greendyke R, Byrd TF. 2008. Differential antibiotic susceptibility of Mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria. Antimicrob Agents Chemother 52:2019–2026. doi: 10.1128/AAC.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavaliunaite E, Harris KA, Aurora P, Dixon G, Shingadia D, Muthialu N, Spencer H. 2020. Outcome according to subspecies following lung transplantation in cystic fibrosis pediatric patients infected with Mycobacterium abscessus. Transpl Infect Dis 22:e13274. doi: 10.1111/tid.13274. [DOI] [PubMed] [Google Scholar]
- 6.Degiacomi G, Sammartino JC, Chiarelli LR, Riabova O, Makarov V, Pasca MR. 2019. Mycobacterium abscessus, an emerging and worrisome pathogen among cystic fibrosis patients. Int J Mol Sci 20:5868. doi: 10.3390/ijms20235868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tortoli E, Kohl TA, Brown-Elliott BA, Trovato A, Leao SC, Garcia MJ, Vasireddy S, Turenne CY, Griffith DE, Philley JV, Baldan R, Campana S, Cariani L, Colombo C, Taccetti G, Teri A, Niemann S, Wallace RJ, Jr, Cirillo DM. 2016. Emended description of Mycobacterium abscessus, Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii and designation of Mycobacterium abscessus subsp. massiliense comb. nov. Int J Syst Evol Microbiol 66:4471–4479. doi: 10.1099/ijsem.0.001376. [DOI] [PubMed] [Google Scholar]
- 8.Davidson RM, Hasan NA, Reynolds PR, Totten S, Garcia B, Levin A, Ramamoorthy P, Heifets L, Daley CL, Strong M. 2014. Genome sequencing of Mycobacterium abscessus isolates from patients in the United States and comparisons to globally diverse clinical strains. J Clin Microbiol 52:3573–3582. doi: 10.1128/JCM.01144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernut A, Viljoen A, Dupont C, Sapriel G, Blaise M, Bouchier C, Brosch R, de Chastellier C, Herrmann JL, Kremer L. 2016. Insights into the smooth-to-rough transitioning in Mycobacterium bolletii unravels a functional Tyr residue conserved in all mycobacterial MmpL family members. Mol Microbiol 99:866–883. doi: 10.1111/mmi.13283. [DOI] [PubMed] [Google Scholar]
- 10.Howard ST, Rhoades E, Recht J, Pang X, Alsup A, Kolter R, Lyons CR, Byrd TF. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology (Reading) 152:1581–1590. doi: 10.1099/mic.0.28625-0. [DOI] [PubMed] [Google Scholar]
- 11.Pawlik A, Garnier G, Orgeur M, Tong P, Lohan A, Le Chevalier F, Sapriel G, Roux AL, Conlon K, Honore N, Dillies MA, Ma L, Bouchier C, Coppee JY, Gaillard JL, Gordon SV, Loftus B, Brosch R, Herrmann JL. 2013. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol Microbiol 90:612–629. doi: 10.1111/mmi.12387. [DOI] [PubMed] [Google Scholar]
- 12.Ripoll F, Deshayes C, Pasek S, Laval F, Beretti JL, Biet F, Risler JL, Daffe M, Etienne G, Gaillard JL, Reyrat JM. 2007. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genomics 8:114. doi: 10.1186/1471-2164-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernut A, Herrmann JL, Kissa K, Dubremetz JF, Gaillard JL, Lutfalla G, Kremer L. 2014. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A 111:E943–52. doi: 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez AV, Viljoen A, Ghigo E, Herrmann JL, Kremer L. 2018. Glycopeptidolipids, a double-edged sword of the Mycobacterium abscessus complex. Front Microbiol 9:1145. doi: 10.3389/fmicb.2018.01145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhoades ER, Archambault AS, Greendyke R, Hsu FF, Streeter C, Byrd TF. 2009. Mycobacterium abscessus Glycopeptidolipids mask underlying cell wall phosphatidyl-myo-inositol mannosides blocking induction of human macrophage TNF-alpha by preventing interaction with TLR2. J Immunol 183:1997–2007. doi: 10.4049/jimmunol.0802181. [DOI] [PubMed] [Google Scholar]
- 16.Ryan K, Byrd TF. 2018. Mycobacterium abscessus: shapeshifter of the mycobacterial world. Front Microbiol 9:2642. doi: 10.3389/fmicb.2018.02642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dedrick R, Guerrero Bustamante C, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choo SW, Wee WY, Ngeow YF, Mitchell W, Tan JL, Wong GJ, Zhao Y, Xiao J. 2014. Genomic reconnaissance of clinical isolates of emerging human pathogen Mycobacterium abscessus reveals high evolutionary potential. Sci Rep 4:4061. doi: 10.1038/srep04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapriel G, Konjek J, Orgeur M, Bouri L, Frezal L, Roux AL, Dumas E, Brosch R, Bouchier C, Brisse S, Vandenbogaert M, Thiberge JM, Caro V, Ngeow YF, Tan JL, Herrmann JL, Gaillard JL, Heym B, Wirth T. 2016. Genome-wide mosaicism within Mycobacterium abscessus: evolutionary and epidemiological implications. BMC Genomics 17:118. doi: 10.1186/s12864-016-2448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halloum I, Carrere-Kremer S, Blaise M, Viljoen A, Bernut A, Le Moigne V, Vilcheze C, Guerardel Y, Lutfalla G, Herrmann JL, Jacobs WR, Jr, Kremer L. 2016. Deletion of a dehydratase important for intracellular growth and cording renders rough Mycobacterium abscessus avirulent. Proc Natl Acad Sci U S A 113:E4228–E4237. doi: 10.1073/pnas.1605477113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruger K, Hampel A, Billig S, Rucker N, Suerbaum S, Bange FC. 2014. Characterization of rough and smooth morphotypes of Mycobacterium abscessus isolates from clinical specimens. J Clin Microbiol 52:244–250. doi: 10.1128/JCM.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw LP, Doyle RM, Kavaliunaite E, Spencer H, Balloux F, Dixon G, Harris KA. 2019. Children With cystic fibrosis are infected with multiple subpopulations of Mycobacterium abscessus with different antimicrobial resistance profiles. Clin Infect Dis 69:1678–1686. doi: 10.1093/cid/ciz069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann JL, Daffe M, Brosch R, Risler JL, Gaillard JL. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi GE, Cho YJ, Koh WJ, Chun J, Cho SN, Shin SJ. 2012. Draft genome sequence of Mycobacterium abscessus subsp. bolletii BD(T). J Bacteriol 194:2756–2757. doi: 10.1128/JB.00354-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raiol T, Ribeiro GM, Maranhão AQ, Bocca AL, Silva-Pereira I, Junqueira-Kipnis AP, Brigido M.dM, Kipnis A. 2012. Complete genome sequence of Mycobacterium massiliense. J Bacteriol 194:5455. doi: 10.1128/JB.01219-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatfull GF. 2020. Actinobacteriophages: genomics, dynamics, and applications. Annu Rev Virol 7:37–61. doi: 10.1146/annurev-virology-122019-070009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell DA, Hatfull GF. 2017. PhagesDB: the actinobacteriophage database. Bioinformatics 33:784–786. doi: 10.1093/bioinformatics/btw711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dedrick RM, Guerrero Bustamante CA, Garlena RA, Pinches RS, Cornely K, Hatfull GF. 2019. Mycobacteriophage ZoeJ: a broad host-range close relative of mycobacteriophage TM4. Tuberculosis (Edinb) 115:14–23. doi: 10.1016/j.tube.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford ME, Sarkis GJ, Belanger AE, Hendrix RW, Hatfull GF. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J Mol Biol 279:143–164. doi: 10.1006/jmbi.1997.1610. [DOI] [PubMed] [Google Scholar]
- 30.Froman S, Will DW, Bogen E. 1954. Bacteriophage active against Mycobacterium tuberculosis I. Isolation and activity. Am J Public Health Nations Health 44:1326–1333. doi: 10.2105/ajph.44.10.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Y, Roznowski AP, Tokuda JM, Klose T, Mauney A, Pollack L, Fane BA, Rossmann MG. 2017. Structural changes of tailless bacteriophage PhiX174 during penetration of bacterial cell walls. Proc Natl Acad Sci U S A 114:13708–13713. doi: 10.1073/pnas.1716614114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentile GM, Wetzel KS, Dedrick RM, Montgomery MT, Garlena RA, Jacobs-Sera D, Hatfull GF. 2019. More evidence of collusion: a new prophage-mediated viral defense system encoded by mycobacteriophage Sbash. mBio 10:e00196-19. doi: 10.1128/mBio.00196-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burbaud S, Laval F, Lemassu A, Daffe M, Guilhot C, Chalut C. 2016. Trehalose polyphleates are produced by a glycolipid biosynthetic pathway conserved across phylogenetically distant mycobacteria. Cell Chem Biol 23:278–289. doi: 10.1016/j.chembiol.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, de la Paz Santangelo M, Cataldi AA, Bigi F. 2013. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 4:3–66. doi: 10.4161/viru.22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams A, Guthlein C, Beresford N, Bottger EC, Springer B, Davis EO. 2011. UvrD2 is essential in Mycobacterium tuberculosis, but its helicase activity is not required. J Bacteriol 193:4487–4494. doi: 10.1128/JB.00302-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dedrick RM, Aull HG, Jacobs-Sera D, Garlena RA, Russell DA, Smith BE, Mahalingam V, Abad L, Gauthier CH, Hatfull GF. 2021. The prophage and plasmid mobilome as a likely driver of Mycobacterium abscessus diversity. mBio 12:e03441-20. doi: 10.1128/mBio.03441-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew R, Mukherjee R, Balachandar R, Chatterji D. 2006. Deletion of the rpoZ gene, encoding the omega subunit of RNA polymerase, results in pleiotropic surface-related phenotypes in Mycobacterium smegmatis. Microbiology (Reading) 152:1741–1750. doi: 10.1099/mic.0.28879-0. [DOI] [PubMed] [Google Scholar]
- 38.Glickman C, Kammlade SM, Hasan NA, Epperson LE, Davidson RM, Strong M. 2020. Characterization of integrated prophages within diverse species of clinical nontuberculous mycobacteria. Virol J 17:124. doi: 10.1186/s12985-020-01394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Kriakov J, Singh A, Jacobs WR, Jr, Besra GS, Bhatt A. 2009. Defects in glycopeptidolipid biosynthesis confer phage I3 resistance in Mycobacterium smegmatis. Microbiology (Reading) 155:4050–4057. doi: 10.1099/mic.0.033209-0. [DOI] [PubMed] [Google Scholar]
- 40.Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Modlin RL, Hendrix RW, Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science Sea-Phages Program. 2012. On the nature of mycobacteriophage diversity and host preference. Virology 434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell DA. 2018. Sequencing, assembling, and finishing complete bacteriophage genomes. Methods Mol Biol 1681:109–125. doi: 10.1007/978-1-4939-7343-9_9. [DOI] [PubMed] [Google Scholar]
- 42.Russell DA, Hatfull GF. 2016. Complete genome sequence of Arthrobacter sp. ATCC 21022, a host for bacteriophage discovery. Genome Announc 4:e00168-16. doi: 10.1128/genomeA.00168-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang H, Lei R, Ding SW, Zhu S. 2014. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon D, Abajian C, Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res 8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 46.Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29:2936–2937. doi: 10.1093/bioinformatics/btt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sovic I, Sikic M, Wilm A, Fenlon SN, Chen S, Nagarajan N. 2016. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun 7:11307. doi: 10.1038/ncomms11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marinelli LJ, Piuri M, Swigonova Z, Balachandran A, Oldfield LM, van Kessel JC, Hatfull GF. 2008. BRED: a simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS One 3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic differences in smooth and rough Mycobacterium abscessus clinical isolates. Download Table S2, PDF file, 0.06 MB (61.3KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequencing details of Mycobacterium abscessus clinical isolates. Download Table S1, PDF file, 0.06 MB (99.1KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
M. abscessus survival following phage challenge. Download Table S3, PDF file, 0.1 MB (118.1KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genomic differences in phage-resistant mutants (RMs) of Mycobacterium abscessus clinical isolates. Download Table S4, PDF file, 0.04 MB (40.4KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD20-1 (MabA1). The genome of phiGD20-1 is shown with predicted genes shown as boxes either above or below the genome indicating rightward- and leftward-transcription, respectively. Gene numbers are shown within each gene box. Phamily designations are shown above or below each gene, with the numbers of phamily members in parentheses; genes are colored according to the phamily designations. White boxes represent “orphams,” i.e., genes with no close relatives in this dataset. Phamily assignments were determined using Phamerator and database Actino_prophage (version 5). Predicted gene functions are indicated. Download FIG S1, PDF file, 0.4 MB (405.5KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD21-1 (MabB). See Fig. S1 for details. Download FIG S2, PDF file, 0.2 MB (211.2KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD57-1 (MabC). See Fig. S1 for details. Download FIG S3, PDF file, 0.2 MB (213.6KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD17-1 (MabD). See Fig. S1 for details. Download FIG S4, PDF file, 0.3 MB (296.9KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome organization of phiGD24-3 (MabJ). See Fig. S1 for details. Download FIG S5, PDF file, 0.3 MB (312.9KB, pdf) .
Copyright © 2021 Dedrick et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The completed and WGS genome sequencing data for M. abscessus clinical isolates have been submitted to GenBank and accession numbers are listed in Table S1. The phiGDxx genomes have the following GenBank and accession numbers: phiGD89-1 (MW314851), phiGD57-1 (MW314852), phiGD34-2 (MW314853), phiGD24-3 (MW314854), phiGD23-1 (MW314855), phiGD22-1 (MW314856), phiGD21-1 (MW314857), phiGD20-1 (MW314858), phiGD17-1 (MW314859).