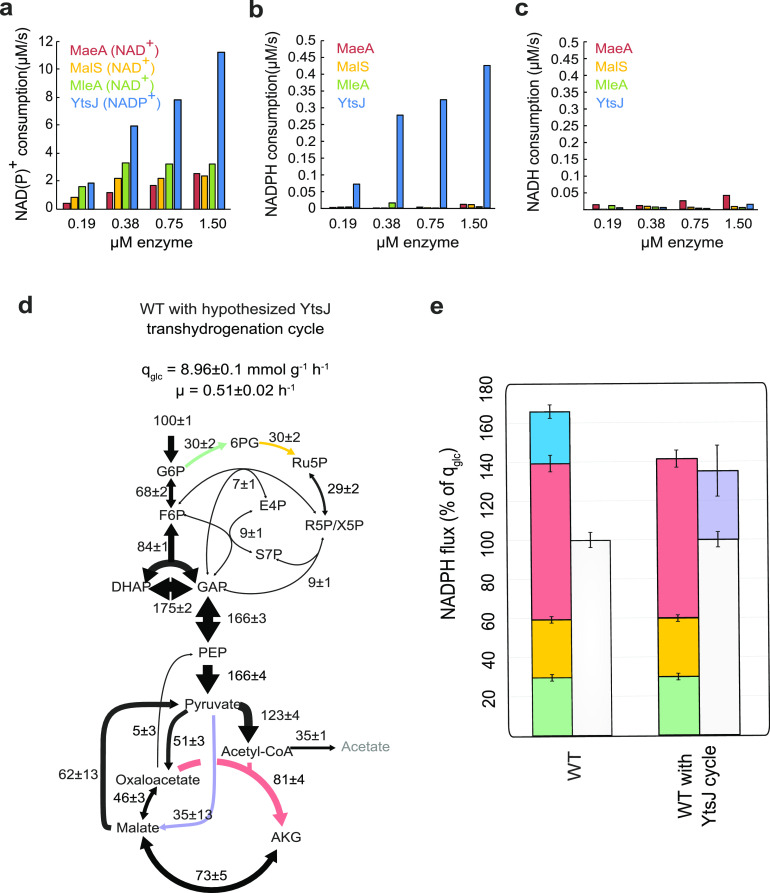
FIG 2.
NADPH oxidation by malic enzyme. (a to c) Initial reaction rates of purified MaeA, MalS, MleA, and YtsJ at different enzyme concentrations. (a) Oxidative malate decarboxylation rate in the presence of each enzymes’ preferred cofactor. (b and c) Reductive pyruvate carboxylation rate with NADPH (b) and NADH as cofactor (c). (d) Metabolic flux distribution in B. subtilis wild type with hypothetical NADPH-oxidizing cycle, estimated from stationary and nonstationary 13C flux data. (e) NADPH balance in B. subtilis wild type with and without the hypothetical, NADPH-oxidizing cycle. Glucose-6-phosphate dehydrogenase (green), 6-phosphogluconate dehydrogenase (yellow), isocitrate dehydrogenase (red), and NADP+ specific malic enzyme (blue) compared to the normalized growth-dependent NADPH consumption (white) and YtsJ cycle (purple).