The complexity of animal microbiomes presents challenges to defining signaling molecules within the microbial consortium and between the microbes and the host. By focusing on the binary symbiosis between Vibrio fischeri and Euprymna scolopes, we have combined genetic analysis with direct imaging to define and study small molecules in the intact symbiosis.
KEYWORDS: mass spectrometry, specialized metabolites, symbiosis
ABSTRACT
The lifelong relationship between the Hawaiian bobtail squid Euprymna scolopes and its microbial symbiont Vibrio fischeri represents a simplified model system for studying microbiome establishment and maintenance. The bacteria colonize a dedicated symbiotic light organ in the squid, from which bacterial luminescence camouflages the host in a process termed counterillumination. The squid host hatches without its symbionts, which must be acquired from the ocean amidst a diversity of nonbeneficial bacteria, such that precise molecular communication is required for initiation of the specific relationship. Therefore it is likely there are specialized metabolites used in the light organ microenvironment to modulate these processes. To identify small molecules that may influence the establishment of this symbiosis, we used imaging mass spectrometry to analyze metabolite production in V. fischeri with altered biofilm production, which correlates directly to colonization capability in its host. “Biofilm-up” and “biofilm-down” mutants were compared to a wild-type strain, and ions that were more abundantly produced by the biofilm-up mutant were detected. Using a combination of structural elucidation and synthetic chemistry, one such signal was determined to be a diketopiperazine, cyclo(d-histidyl-l-proline). This diketopiperazine modulated luminescence in V. fischeri and, using imaging mass spectrometry, was directly detected in the light organ of the colonized host. This work highlights the continued need for untargeted discovery efforts in host-microbe interactions and showcases the benefits of the squid-Vibrio system for identification and characterization of small molecules that modulate microbiome behaviors.
INTRODUCTION
Nearly all animals are hosts to microbial species (1). Communities of microbes can inhabit host organs intracellularly or extracellularly, may confer benefits to the host, and collectively are referred to as the microbiome (2). Some of these relationships have been explored by investigating the role that chemical communication plays between species for ascertaining whether to initiate interactions or to maintain them (3). For instance, the plant pathogen Ralstonia solanacearum produces a hybrid nonribosomal peptide synthetase-polyketide synthase (NRPS-PKS) lipopeptide that induces chlamydospore development in soil-dwelling filamentous fungi, which may help it persist in the soil environment. Conversely, the fungal species Fusarium fujikuroi and Botrytis cinerea produce bikaverin to inhibit invasion of ralsolamycin-producing strains of R. solanacearum (4, 5). In cheese rind-derived microbial interactions, small molecules like zinc-coproporphyrin III and other siderophores mediate trace metal acquisition between bacteria and fungi (6, 7). In both of these cases, specialized metabolites were central to understanding the observed phenotypes between the domains of life. Beyond antagonistic relationships, in many bacterial species, the density of the population is monitored by self-secreted “autoinducer” compounds, which include homoserine lactones in Gram-negative bacteria and small peptides in many Gram-positive organisms (8). Quorum sensing was discovered in Vibrio fischeri, a marine Gram-negative bacterium that colonizes squid and fish hosts (9). Studying V. fischeri in the context of its natural hosts provides an opportunity to expand our knowledge of biologically relevant compounds that influence microbial behaviors and microbe-host signaling in animal microbiomes.
To study the role of specialized metabolites in microbiome-host interactions, we have focused on the binary partnership between V. fischeri and the Hawaiian bobtail squid, Euprymna scolopes. Shortly after hatching, the squid host acquires the symbiont from the seawater, and the microbe proceeds to colonize the epithelial-lined crypts of the light organ. The bacteria gain a safe nutrient-rich habitat in which they can replicate (10). In turn, the bacteria in the aptly named “light organ” produce luminescence for the squid. The bacterial light camouflages the nocturnal predator via counterillumination, hiding the animal’s shadow in the moonlight so that it is less visible to predators and prey while the squid forages (11). Many aspects of the colonization process are comparable to bacterial colonization of human epithelial cells in the gut and on skin (3, 12, 13). As a naturally coevolved relationship where one host organ houses only one bacterial species, the simplified Vibrio-squid system allows for analysis of specialized metabolites involved in colonization of epithelial tissues. The colonization process is highly efficient and the host tissues are largely transparent, facilitating imaging approaches during the early stages of colonization. V. fischeri is amenable to genetic manipulation, including mimicking of symbiotic behaviors in culture. Furthermore, both partners can be raised separately and then mixed in a controlled fashion, enabling well-controlled experiments.
There is remarkable colonization specificity in that only V. fischeri—and only specific strains of V. fischeri—colonize the E. scolopes light organ (14). Part of this specificity lies in the exchange of chemical cues between the two partners that leads to maturation of the symbiosis, including the release of bacterial products peptidoglycan, the peptidoglycan monomer tracheal cytotoxin (TCT), lipopolysaccharide (LPS), while the host releases nitric oxide (NO) and chitin (15–20). A major checkpoint for this specificity is that bacterial biofilm production is required for bacterial aggregation and subsequent colonization (21, 22). Bacterial aggregates were first observed during confocal imaging of V. fischeri strain ES114 colonization of squid (23), and the genetic basis for aggregate formation was shown to require the hybrid histidine kinase RscS, hybrid histidine kinase SypF, response regulator SypG, and the 18-gene target symbiosis polysaccharide (syp) locus (21, 24, 25). Strains lacking any of the above regulators are unable to colonize squid robustly. Recent work has identified additional biofilm regulators, including BinK and HahK, which interface with the above pathway. BinK is a strong negative regulator of biofilm formation; in strains lacking BinK, increased biofilm formation is observed, whereas overexpression of BinK leads to reduced or absent biofilm development (26). HahK mediates NO signaling from the squid host via the HnoX NO sensor; in the presence of NO, HnoX inhibits HahK’s activity to promote biofilm formation, thus leading to enhanced dispersal of the symbiotic biofilm (27). The work on NO highlights the importance of small signaling molecules in the biofilm at the host interface.
We hypothesized that additional small molecule production may contribute to the propensity for V. fischeri to outcompete other bacteria. An experimental advantage offered by the Vibrio-squid system is the ability to investigate symbiotic phenotypes in culture-based assays that closely mirror the behavior in the host. Characterization of the biological roles for the positive regulator RscS and the negative regulator BinK were facilitated by culture-based assays that reflected the symbiotic phenotypes. Specifically, when RscS was overexpressed (using either a plasmid-based or chromosomal rscS* allele), V. fischeri produced large aggregates and outcompeted wild type in the squid. These same strains form visible biofilms on agar that manifest as wrinkled colonies. At the other end of the spectrum, strains that are interrupted in the gene encoding the RscS positive regulator are unable to form biofilm in culture or in the animal. Therefore, by using a set of genetically altered V. fischeri ES114 derivatives, we were able to examine a spectrum of biofilm phenotypes in culture that are relevant for the symbiotic behavior in the host. The three isogenic derivatives that were the focus for discovery in this paper are as follows. (i) A strain that does not form symbiotic biofilm in culture or in the animal (“biofilm-down”; i.e., ES114 rscS* rscS::Tnerm). This strain contains an interruption in rscS that blocks biofilm signaling. (ii) A strain that forms symbiotic biofilm in the animal but not in culture (“wild type” [WT]; i.e., unaltered ES114). (iii) A strain that forms symbiotic biofilm in culture and forms enhanced aggregates in the animal due to overexpression of the positive regulator RscS and the lack of the negative regulator BinK (“biofilm-up”; i.e., ES114 rscS* ΔbinK) (26). We have shown previously that a similar gradient of strains was useful for revealing biofilm phenotypes that are relevant in vivo (26), and here we integrated these defined strains with cutting edge analytical technologies for the discovery and characterization of small molecules connected to symbiotic behaviors.
One method that can rapidly determine differences in metabolite production of microbes is agar-based imaging mass spectrometry (IMS). This approach has been applied to decipher microbe-microbe chemical communication and changes in metabolite production between microbial colonies grown in different environmental conditions (6, 28, 29). IMS is especially valuable for the comparison of metabolites of defined strains because changes in production, as measured by ion intensity, can be specifically and rapidly detected and evaluated in the context of genetic differences (30). IMS analysis of WT V. fischeri compared to biofilm-up and biofilm-down strains revealed that several metabolites were significantly upregulated in the biofilm-up strain compared to the other strains. These metabolites were hypothesized to be important for the production of the symbiotic biofilm and/or the increased fitness of the biofilm-up mutant in the light organ environment. Additionally, because of the small size of E. scolopes hatchlings and the anatomical accessibility of the light organ, a whole-body imaging approach was optimized for in vivo investigation of the light organ (31). Together, these two IMS approaches were utilized to determine differences in specialized metabolism in V. fischeri mutants, both in vitro and in vivo, and led to the isolation and structural elucidation of a small molecule that increases V. fischeri luminescence, which may provide a fitness advantage in host-microbe recognition.
RESULTS AND DISCUSSION
Untargeted spatial metabolomics of V. fischeri biofilm mutants.
IMS was employed to generate a rapid screen of mass-to-charge ratios (m/z) in WT, biofilm-up, and biofilm-down V. fischeri strains to identify ions that differ between solid agar colonies of the three strains. A positive mode analysis in the small molecule range (100 to 1,000 Da) generated a panel of masses that were significantly more abundant (P < 0.1) in the biofilm-up mutant than in the other two samples, as determined by the “colocalization” function in SCiLS software (Bruker). Figure S1 in the supplemental material displays seven of these significant features that replicated at least twice across four biological replicates. One of these signals, m/z 257, was detected with high signal intensity in biofilm-up, and low signal intensity in WT and biofilm-down (Fig. 1). We proceeded to validate this compound using a different model of biofilm induction. It was recently shown that calcium in the medium can stimulate V. fischeri biofilm formation in strains lacking BinK, but without the need for overexpression of rscS* (32). This both provided an independent biofilm model in which we could test for production of the compound, and additionally allowed us to ask whether the compound was produced by other natural isolates, given that the rscS* overexpression approach does not apply in all backgrounds (33). In ES114, we observed elevated abundance of the compound on the calcium medium upon biofilm induction (i.e., in the strain lacking BinK), supporting the use of this model (Fig. S2). We also detected elevated compound in biofilm-induced strains MB11B1, MB15A4, and SR5 (Fig. S2). Additionally, we detected the compound in strain ES213 but no increase was detected upon biofilm induction (Fig. S2). These results therefore report that this compound is reliably detected in multiple biofilm models and can be detected in strains representing the three major phylogenetic groups of symbiotic V. fischeri (33). This signal was therefore prioritized for dereplication, the process of identifying known unknowns, because biofilm production is strongly correlated with a colonization advantage.
FIG 1.
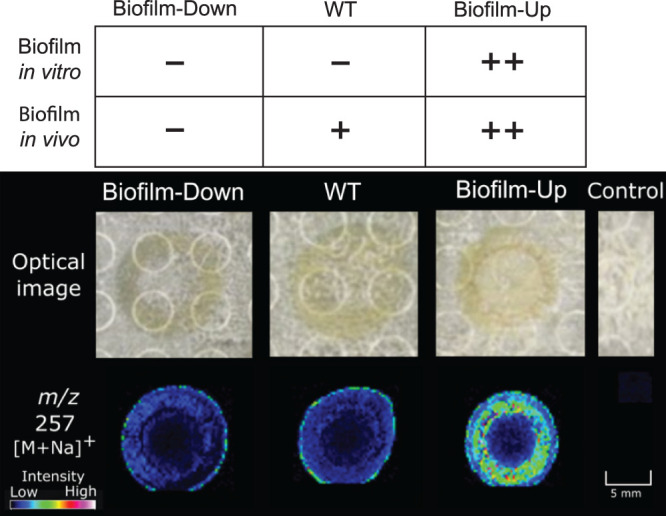
The m/z 257 from the mass panel in Fig. S1 was statistically more abundant in a biofilm-up strain compared to WT and biofilm-down strains (n = 4) (see the Materials and Methods for detailed genotypes). Using the “colocalization” function in SCiLS Lab (Bruker), the P value of m/z 257 in biofilm-up compared to WT and biofilm-down is 0.05 < P < 0.1, based on manual adjustment of the significance threshold.
Prioritized mass features detected using IMS. Download FIG S1, PDF file, 0.3 MB (353.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of natural V. fischeri isolates. Download FIG S2, PDF file, 0.4 MB (456.8KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Dereplication and structure elucidation of cyclo(histidyl-proline).
Masses identified via IMS were prioritized to a select list by evaluating their statistical significance. Because ionization capacities change with different modalities (e.g., matrix-assisted laser desorption ionization [MALDI] versus electrospray ionization [ESI]) liquid chromatography-tandem mass spectrometry (LC-MS/MS) data of crude V. fischeri extracts from each mutant were queried using Global Natural Products Social molecular networking (GNPS) to investigate signals detected in IMS, as well as to probe compounds that may only ionize in ESI. A spectral match from the V. fischeri biofilm-up extract was detected for a small molecule, cyclo(histidyl-proline) (cHP), with a molecular weight of 234 g/mol (Fig. S3) (34, 35). The precursor ion from GNPS (m/z 235) matched one of the statistically significant masses from IMS analysis; often, ions detected in IMS are sodiated adducts ([M+Na]+) because of salt added to the medium to support growth of marine bacteria. Therefore, the m/z 257 from Fig. 1 was likely the sodiated adduct of cyclo(His-Pro), and the protonated molecule was m/z 235 (Fig. S4). Of interest to this context, cyclo(His-Pro) is a member of the diketopiperazine (DKP) structural class. DKPs are formed from the cyclization of two amino acids and are prevalent natural products that play a variety of roles in microbial relationships (36).
GNPS match of m/z 235 to cyclo(His-Pro). Download FIG S3, PDF file, 0.3 MB (343.9KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MALDI-TOF dried droplet spectrum of m/z 235 in biofilm-up extract. Download FIG S4, PDF file, 0.3 MB (309.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
While GNPS identified cyclo(His-Pro) as a putative assignment, we employed direct infusion to detect and confirm the fragmentation patterns of the m/z 235 from the V. fischeri biofilm-up extract and a commercial cyclo(l-His-l-Pro) standard. Figure 2 depicts a near identical match between the fragmentation patterns of both samples using direct infusion, which was validated using high-resolution electrospray ionization mass spectrometry (HRESIMS) (Fig. S5). The protonated precursor ion (m/z 235.12) and all high-intensity fragments matched between the standard and extracted samples. These data provided strong evidence to support a level 2 identification, as defined by the chemical analysis working group (CAWG), of cyclo(His-Pro) in the V. fischeri biofilm-up extract (37).
FIG 2.
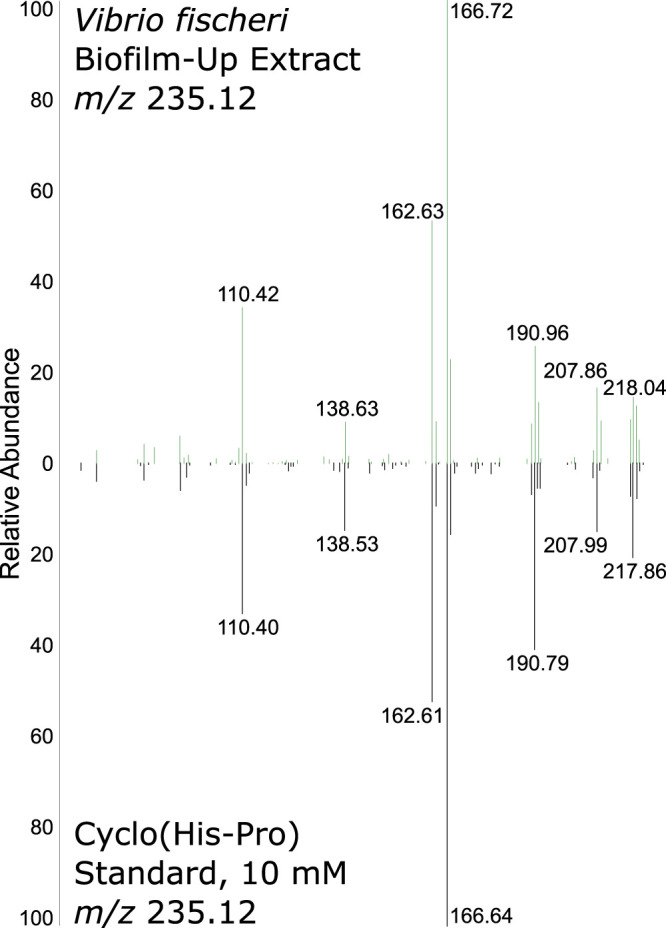
A standard of cyclo(His-Pro) mass fragmentation matched with the extracted molecule at m/z 235.12 using direct infusion.
Extracted ion chromatograms of cyclo(His-Pro) on the Q-ToF mass spectrometer. Download FIG S5, PDF file, 0.4 MB (415.5KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
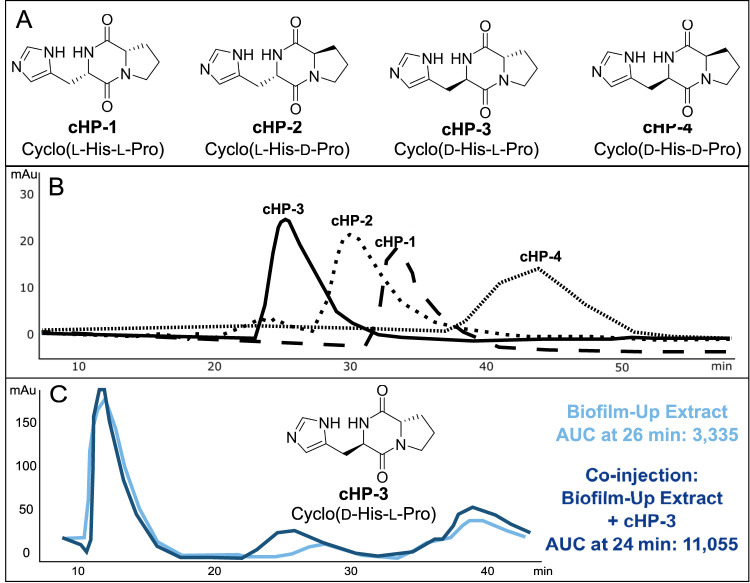
To elucidate the stereochemical configuration of the DKP, as MS/MS is considered to be largely stereochemically blind, we synthesized all four possible stereoisomers of cyclo(His-Pro): (i) cyclo(l-His-l-Pro) (cHP-1); (ii) cyclo(l-His-d-Pro) (cHP-2); (iii) cyclo(d-His-l-Pro) (cHP-3); and cyclo(d-His-d-Pro) (cHP-4). The four stereoisomers were synthesized for chemical comparison to the isolated compound from the V. fischeri biofilm-up extract (Fig. 3A) (38). To validate the configuration of the synthetic material, each stereoisomer was analyzed using both nuclear magnetic resonance (NMR) and optical rotation (OR). Kukla et al. reported optical rotation values, which were used for comparison and validation of each isomer (Text S1) (38).
FIG 3.
Retention time matching of V. fischeri biofilm-up extract on a chiral column indicates that the configuration of cyclo(His-Pro) in the microbial extract is stereoisomer cHP-3, cyclo(d-His-l-Pro). (A) Structures of all stereoisomers of cyclo(His-Pro). (B) Retention times of all synthesized stereoisomers: cHP-3 (24 min), cHP-2 (30 min), cHP-1 (33 min), and cHP-4 (44 min). A cellulose-B column was used to retain stereoisomers using 13:87 isopropyl alcohol (IPA):hexanes over 60 min at 2 ml/min. (C) A peak at 26 min was observed in the biofilm-up extract (light blue trace) and the area under the curve (AUC) measured 3,335. When coinjected with cHP-3 (dark blue trace), the peak at 26 min increased in AUC to 11,055, indicating the presence of cHP-3 in the biofilm-up extract. UV-vis was monitored at 214 nm.
General experimental, synthesis, and characterization of the different DKP stereoisomers. Download Text S1, PDF file, 1.0 MB (1,009KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chiral chromatography was used to match the retention time of the DKP in the V. fischeri extract to all four synthesized stereoisomers (Fig. 3B). After comparison with each stereoisomer, it was determined that the peak of stereoisomer cHP-3 was also detected in the V. fischeri biofilm-up extract. When coinjected, the peaks from both samples coalesced to one peak, indicating that their structures and configuration were identical (Fig. 3C). This retention time matching provides a level 1 identification for the m/z 235 from the V. fischeri biofilm-up, stereoisomer cHP-3 (cyclo[d-His-l-Pro]). The other stereoisomers, namely cHP-2, were eliminated as being produced in the biofilm-up extract because either their retention time did not match an existing peak in the biofilm-up extract, or the coinjection peak did not coalesce with an extract peak (Fig. S6). Quantification of the molecule in extracts of the biofilms produced by the mutants also indicated that the biofilm-up mutant produced a larger amount of cHP (measured as m/z 235.12) (Fig. S7). To attempt to validate the role of cHP-3 in the E. scolopes host, we aimed to capture production of cHP-3 in vivo.
Retention time matching of cyclo(d-His-l-Pro) to biofilm-up. Download FIG S6, PDF file, 0.3 MB (338.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantification of cyclo(d-His-l-Pro) in V. fischeri wild type and mutants. Download FIG S7, PDF file, 0.3 MB (327.8KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vivo detection in hatchlings.
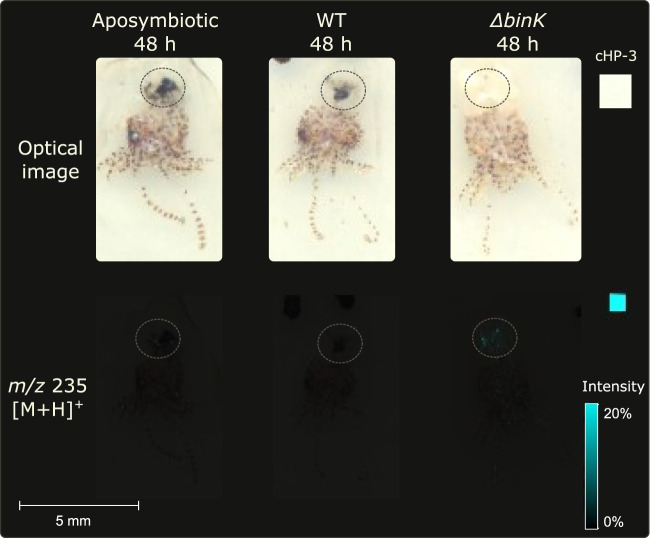
A recently developed sample preparation protocol for minimally manipulated whole-invertebrate-organism IMS was utilized to assess whether cHP-3 was being generated in vivo in the light organ of E. scolopes hatchlings (31). The presence of the ion in the light organ would indicate that cHP-3 has an ecological importance if it can be detected in vivo. Three conditions were evaluated: (i) no V. fischeri (aposymbiotic), (ii) V. fischeri WT, and (iii) V. fischeri WT ΔbinK (analogous to the biofilm-up strain, but instead of genetically inducing biofilm formation with the rscS* allele, relies on induction from native signals in the squid host). E. scolopes hatchlings were inoculated with each sample for 3 h, washed, and allowed to continue to colonize for 48 h, as described previously (39). At 48 h, m/z 235 was not detected in the aposymbiotic control, was produced weakly in the WT condition, and was produced strongly in the WT ΔbinK condition (Fig. 4). This was evidence that the molecule was produced by the WT strain in vivo and that the knockout of binK resulted in increased production of the ion.
FIG 4.
Representative replicates depicting m/z 235 (cHP-3) is detected in vivo in the light organ of E. scolopes hatchlings colonized both when inoculated with V. fischeri WT and (strongly) with WT ΔbinK (n = 3). Dotted circles surround the light organ region in each hatchling. Colors in the light organ regions vary depending on level of ink removal. Replicates can be found in Fig. S8 in the supplemental material.
Detection of cHP-3 in light organ replicates. Download FIG S8, PDF file, 0.3 MB (319.6KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The consistency in detection of cHP-3 production in vitro and in vivo is significant in light of the bacterial strains that were used. The WT strain is the same for both assays, whereas the comparative strain in each case lacks BinK. However, in the in vitro assay, biofilms are genetically induced with an rscS* allele (i.e., an overexpressed biofilm regulator), while this allele is not present for the strain used in vivo (Fig. 4). The rationale for this difference is that the activated RscS allele is used to mimic behavior in culture that is stimulated by the host conditions. Given that cHP-3 production is similarly induced in the host, we conclude that its production is a bona fide output of the symbiotic biofilm pathway and is induced in the host in both WT and in a ΔbinK background, independent of artificial RscS activation.
Role in stimulating bioluminescence.
Quorum sensing (QS) is the activity of bacterial cells engaging in group behavior facilitated by the production of QS molecules by individual cells until a concentration is reached to activate a particular pathway. In the case of V. fischeri, the LuxR pathway is activated and bioluminescent activity occurs. The most well studied QS molecules in the V. fischeri system are acyl-homoserine lactones (AHL), such as N-3-oxohexanoyl-l-homoserine lactone (OHHL). In other bacterial QS systems, proline-containing DKPs are responsible for activating the relative pathways. For example, cyclo(l-Pro-l-Leu) produced by Cronobacter sakazakii, cyclo(l-Pro-l-Tyr) and cyclo(l-Phe-l-Pro) produced by Pseudomonas aeruginosa, and cyclo(l-Pro-l-Val) isolated from Haloterrigena halophilus all influence the QS systems of other microbes (40–42). With so many structural combinations possible, there are many more examples of DKPs that influence microbial QS systems, some in biofilm formation, and some even in Vibrio spp. (43–46). For example, cyclo(l-Phe-l-Pro), which was originally isolated from V. vulnificus, was shown to activate the V. fischeri luminescence locus (47).
Because of the relationship between biofilm production potential and appearance of cHP-3 in the colonized host, we sought to query whether cHP-3 affected the bioluminescence of V. fischeri. An increase in luminescence was observed at low cell densities in the WT strain (ES114) (Fig. 5). Since ES114 has low luminescence compared to other V. fischeri isolates, we tested the effect of cHP-3 in another strain, EM17, which is a much brighter isolate. This strain also exhibited an increase in luminescence with the addition of exogenous cHP-3 and the effect is much more pronounced than in ES114 (Fig. 5). The greatest effect on luminescence in both strains was seen with concentrations of 100 and 250 μM cHP-3. The effect of cHP-3 at low cell densities is similar for other quorum-sensing molecules, as there is a concentration threshold in these systems after which the addition of more compound does not increase activity (48). Given the optimal response at 250 μM for ES114, we asked whether exogenous addition of cHP-3 at this concentration would affect squid colonization. It did not impact colonization level or luminescence for the WT symbiont (Fig. S9). Given the high level of luminescence in vivo, we suspect that light production is saturated, and it will be necessary in future work to interrupt biosynthesis of cHP-3 to further investigate its role in the host.
FIG 5.
cHP-3 stimulates V. fischeri luminescence. Line graphs show cHP-3 increases relative light units in V. fischeri cultures at low concentrations in both a low-luminescence strain (ES114) and in a high-luminescence strain (EM17). Bar graphs show luminescence levels at a specific OD (OD600 = ∼0.3) to illustrate the concentration-dependent effect. Graphs are representative of three independent experiments.
Colonization of squid hatchlings with exogenous cHP-3. Download FIG S9, PDF file, 0.3 MB (313.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Roles of microbial diketopiperazines.
Microbial DKPs have been detailed in a number of biological contexts, including production in sexual pheromone signaling, as enzyme inhibitors, and in quorum sensing (46, 49–51). Their structures vary widely, as there are many possible amino acid combinations and side chain modifications (36). A thorough understanding of the ecology of the squid-Vibrio system provides context for the potential role of cHP-3. With an observable increase in production of cHP-3 in a biofilm-up mutant, it is possible to surmise that cHP-3 may be critical for biofilm development, either as an end goal or as a preliminary step for a downstream process, or even as a quorum-sensing molecule. Bacterial DKPs have been implicated in a variety of roles. As an example, cyclo(l-Phe-l-Pro) was isolated from Vibrio vulnificus due to its ability to activate a V. fischeri luminescence locus (47). The compound is produced widely among Vibrio species, can affect production of toxin genes, and can influence host innate immunity (52–54). Several bacterially derived DKPs have been described as chitinase inhibitors: cyclo(l-Arg-l-Pro), cyclo(Gly-l-Pro), cyclo(l-His-l-Pro) (cHP-1), and cyclo(l-Tyr-l-Pro) (51, 55, 56). While we have not explored the chitinase inhibition activity of cHP-3, if the stereoselectivity is not important in other activities of DKPs, it is possible that cHP-3 displays the same inhibitory activity in the squid-Vibrio system, especially if cHP-1 is known to be inhibitory. Because E. scolopes generates chitin as a source of nutrients for V. fischeri, presumably inhibition of chitinase by the bacterium would result in fewer monomers for consumption which would be thought to decrease fitness. Although this hypothesis has not yet been tested, there is precedent for the regulation of chitin and chitinase activity in this system (57).
In conclusion, application of MALDI-IMS across a gradient of V. fischeri ES114 biofilm-producing derivatives identified a small molecule, cHP-3, that is produced in significant quantities in the biofilm-up strain. cHP-3 is a member of the DKP molecular class, members of which are increasingly ubiquitous across microbial species and have various and dedicated activities, including signaling, quorum sensing, and enzyme inhibition. The effect we observed on bacterial luminescence suggests that this DKP may link the early developmental process of biofilm formation with the later light production.
The biosynthetic origin of cHP-3 is an immediate focus for our future work. Oftentimes, DKPs are shunt products to larger nonribosomal peptide synthetase (NRPS)-derived molecules; however, in our case, no larger peptidic masses (based on MS/MS) have been detected in the culture broth as of yet, which makes this biosynthetic route unlikely. Future studies will also focus on the protein target for luminescence activity, as well as the role in the light organ. The investigation of differentially produced small molecules in this controlled, two-partner system provides a platform for an increased understanding of the mechanisms that may be responsible for animal-microbe partners to recognize one another and to maintain a specific, lifelong relationship.
MATERIALS AND METHODS
V. fischeri strains.
All strains for the initial screen and squid colonizations are derivatives of the MJM1100 isolate of V. fischeri ES114, an E. scolopes light organ isolate (58, 59). MJM1776 is the “biofilm-down” isolate, with genotype MJM1100 rscS* rscS::Tnerm. It was isolated as a mariner transposon insertion from pMarVF1 in the rscS gene in strain MJM1198 (Mattias Gyllborg and M.J.M., unpublished) (60). MJM2255 is the “biofilm-up” isolate with genotype MJM1100 rscS* ΔbinK, which was described previously (26). MJM2251 is the ΔbinK strain used in Fig. 4, with genotype MJM1100 ΔbinK (26).
For the broader natural isolate assessment, wild type and ΔbinK derivatives are listed, respectively, for each strain: ES114 (MJM1100, MJM2251 [26]); MB11B1 (MJM1130 [22], MJM3084 [33]); MB15A4 (MJM2114 [33], MJM3087); ES213 (MJM1117 [22], MJM3551); and SR5 (MJM1125 [22], MJM4037). Strains MJM3087 and MJM3551 were constructed by allelic exchange, with the resulting strains bearing an unmarked ΔbinK allele. Strain MJM4037 is the ΔbinK::bar derivative of MJM3751, which carries the ΔbinK::erm-bar allele constructed via tfoX-based transformation (33, 61).
Bioluminescence assays.
Two Vibrio fischeri strains, ES114 (WT) and Euprymna morsei symbiont EM17, were grown overnight in LBS at 25°C. Cultures were diluted 1/1,000 in seawater tryptone with adjusted osmolarity (SWTO) with various concentrations of cHP-3 from 0 to 500 μM (62). Samples were then transferred in triplicate to a Nunc clear bottom plate and measurements were taken by Biotek Synergy Neo2 plate reader. Relative luminescence (RLU) and optical density at 600 nm (OD600) were measured every 30 min for 22 h. Triplicates were averaged and the specific luminescence (RLU/OD600) was plotted as a function of the OD600.
Data availability.
Data files for all IMS and LS-MS/MS analyses can be found in the MassIVE database under ID MSV000085327.
ACKNOWLEDGMENTS
We gratefully acknowledge Ella Rotman and Katherine Bultman for bacterial mutant strains.
This publication was funded in part by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust (to L.M.S. and M.J.M.). Studies in the lab of L.M.S. were supported by NIH grant R01GM125943-02S2 and UIC startup funds. Studies in the lab of M.J.M. are supported by NIH grant R35GM119627. K.E.Z. was supported by NIH grant F31 CA236237.
Footnotes
Citation Zink KE, Ludvik DA, Lazzara PR, Moore TW, Mandel MJ, Sanchez LM. 2021. A small molecule coordinates symbiotic behaviors in a host organ. mBio 12:e03637-20. https://doi.org/10.1128/mBio.03637-20.
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kostic AD, Howitt MR, Garrett WS. 2013. Exploring host-microbiota interactions in animal models and humans. Genes Dev 27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cleary JL, Condren AR, Zink KE, Sanchez LM. 2017. Calling all hosts: bacterial communication in situ. Chem 2:334–358. doi: 10.1016/j.chempr.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spraker JE, Sanchez LM, Lowe TM, Dorrestein PC, Keller NP. 2016. Ralstonia solanacearum lipopeptide induces chlamydospore development in fungi and facilitates bacterial entry into fungal tissues. ISME J 10:2317–2330. doi: 10.1038/ismej.2016.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spraker JE, Wiemann P, Baccile JA, Venkatesh N, Schumacher J, Schroeder FC, Sanchez LM, Keller NP. 2018. Conserved responses in a war of small molecules between a plant-pathogenic bacterium and fungi. mBio 9:e00820-18. doi: 10.1128/mBio.00820-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary JL, Kolachina S, Wolfe BE, Sanchez LM. 2018. Coproporphyrin III produced by the bacterium Glutamicibacter arilaitensis binds zinc and is upregulated by fungi in cheese rinds. mSystems 3:e00036-18. doi: 10.1128/mSystems.00036-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierce EC, Morin M, Little JC, Liu RB, Tannous J, Keller NP, Pogliano K, Wolfe BE, Sanchez LM, Dutton RJ. 2021. Bacterial-fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. Nat Microbiol 6:87–102. doi: 10.1038/s41564-020-00800-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteley M, Diggle SP, Greenberg EP. 2017. Progress in and promise of bacterial quorum sensing research. Nature 551:313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104:313–322. doi: 10.1128/JB.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nealson KH, Hastings JW. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev 43:496–518. doi: 10.1128/MR.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones BW, Nishiguchi MK. 2004. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda). Mar Biol 144:1151–1155. doi: 10.1007/s00227-003-1285-3. [DOI] [Google Scholar]
- 12.Cundell AM. 2018. Microbial ecology of the human skin. Microb Ecol 76:113–120. doi: 10.1007/s00248-016-0789-6. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt TSB, Raes J, Bork P. 2018. The human gut microbiome: from association to modulation. Cell 172:1198–1215. doi: 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 14.Mandel MJ. 2010. Models and approaches to dissect host-symbiont specificity. Trends Microbiol 18:504–511. doi: 10.1016/j.tim.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Altura MA, Stabb E, Goldman W, Apicella M, McFall-Ngai MJ. 2011. Attenuation of host NO production by MAMPs potentiates development of the host in the squid-vibrio symbiosis. Cell Microbiol 13:527–537. doi: 10.1111/j.1462-5822.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 17.Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ. 2009. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol 11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremer N, Philipp EER, Carpentier M-C, Brennan CA, Kraemer L, Altura MA, Augustin R, Häsler R, Heath-Heckman EAC, Peyer SM, Schwartzman J, Rader BA, Ruby EG, Rosenstiel P, McFall-Ngai MJ. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. 2004. NO means “yes” in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol 6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 20.Visick KL, Ruby EG. 2006. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol 9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Visick KL. 2009. An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol 74:782–789. doi: 10.1111/j.1365-2958.2009.06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yip ES, Grublesky BT, Hussa EA, Visick KL. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 25.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brooks JF, 2nd, Mandel MJ. 2016. The histidine kinase BinK is a negative regulator of biofilm formation and squid colonization. J Bacteriol 198:2596–2607. doi: 10.1128/JB.00037-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson CM, Tischler AH, Tarnowski DA, Mandel MJ, Visick KL. 2019. Nitric oxide inhibits biofilm formation by Vibrio fischeri via the nitric oxide sensor HnoX. Mol Microbiol 111:187–203. doi: 10.1111/mmi.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang JY, Phelan VV, Simkovsky R, Watrous JD, Trial RM, Fleming TC, Wenter R, Moore BS, Golden SS, Pogliano K, Dorrestein PC. 2012. Primer on agar-based microbial imaging mass spectrometry. J Bacteriol 194:6023–6028. doi: 10.1128/JB.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Condren AR, Kahl LJ, Boelter G, Kritikos G, Banzhaf M, Dietrich LE, Sanchez LM. 2019. Biofilm inhibitor taurolithocholic acid alters colony morphology, specialized metabolism, and virulence of Pseudomonas aeruginosa. ACS Infect Dis 6:603–612. doi: 10.1021/acsinfecdis.9b00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minakshi P, Ghosh M, Kumar R, Patki HS, Saini HM, Ranjan K, Brar B, Prasad G. 2019. Single-cell metabolomics: technology and applications, p 319–353. In Barh D, Azevedo V (ed), Single-cell omics. Academic Press, Cambridge, MA. [Google Scholar]
- 31.Zink KE, Tarnowski DA, Mandel MJ, Sanchez LM. 2020. Optimization of a minimal sample preparation protocol for imaging mass spectrometry of unsectioned juvenile invertebrates. J Mass Spectrom 55:e4458. doi: 10.1002/jms.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tischler AH, Lie L, Thompson CM, Visick KL. 2018. Discovery of calcium as a biofilm-promoting signal for Vibrio fischeri reveals new phenotypes and underlying regulatory complexity. J Bacteriol 200:e00016-18. doi: 10.1128/JB.00016-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rotman ER, Bultman KM, Brooks JF, 2nd, Gyllborg MC, Burgos HL, Wollenberg MS, Mandel MJ. 2019. Natural strain variation reveals diverse biofilm regulation in squid-colonizing Vibrio fischeri. J Bacteriol 201:e00033-19. doi: 10.1128/JB.00033-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, et al. 2016. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang JY, Sanchez LM, Rath CM, Liu X, Boudreau PD, Bruns N, Glukhov E, Wodtke A, de Felicio R, Fenner A, Wong WR, Linington RG, Zhang L, Debonsi HM, Gerwick WH, Dorrestein PC. 2013. Molecular networking as a dereplication strategy. J Nat Prod 76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borthwick AD. 2012. 2,5-Diketopiperazines: synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem Rev 112:3641–3716. doi: 10.1021/cr200398y. [DOI] [PubMed] [Google Scholar]
- 37.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW-M, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, Reily MD, Thaden JJ, Viant MR. 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kukla MJ, Breslin HJ, Bowden CR. 1985. Synthesis, characterization, and anorectic testing of the four stereoisomers of cyclo(histidylproline). J Med Chem 28:1745–1747. doi: 10.1021/jm00149a035. [DOI] [PubMed] [Google Scholar]
- 39.Naughton LM, Mandel MJ. 2012. Colonization of Euprymna scolopes squid by Vibrio fischeri. J Vis Exp e3758. doi: 10.3791/3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bofinger MR, de Sousa LS, Fontes JEN, Marsaioli AJ. 2017. Diketopiperazines as cross-communication quorum-sensing signals between Cronobacter sakazakii and Bacillus cereus. ACS Omega 2:1003–1008. doi: 10.1021/acsomega.6b00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holden MTG, Ram Chhabra S, De Nys R, Stead P, Bainton NJ, Hill PJ, Manefield M, Kumar N, Labatte M, England D, Rice S, Givskov M, Salmond GPC, Stewart GSAB, Bycroft BW, Kjelleberg S, Williams P. 1999. Quorum-sensing cross talk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol Microbiol 33:1254–1266. doi: 10.1046/j.1365-2958.1999.01577.x. [DOI] [PubMed] [Google Scholar]
- 42.Tommonaro G, Abbamondi GR, Iodice C, Tait K, De Rosa S. 2012. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb Ecol 63:490–495. doi: 10.1007/s00248-011-9980-y. [DOI] [PubMed] [Google Scholar]
- 43.De Rosa S, Mitova M, Tommonaro G. 2003. Marine bacteria associated with sponge as source of cyclic peptides. Biomol Eng 20:311–316. doi: 10.1016/s1389-0344(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 44.Klose KE. 2006. Increased chatter: cyclic dipeptides as molecules of chemical communication in Vibrio spp. J Bacteriol 188:2025–2026. doi: 10.1128/JB.188.6.2025-2026.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degrassi G, Aguilar C, Bosco M, Zahariev S, Pongor S, Venturi V. 2002. Plant growth-promoting Pseudomonas putida WCS358 produces and secretes four cyclic dipeptides: cross-talk with quorum sensing bacterial sensors. Curr Microbiol 45:250–254. doi: 10.1007/s00284-002-3704-y. [DOI] [PubMed] [Google Scholar]
- 46.Wang J-H, Yang C-Y, Fang S-T, Lu J, Quan C-S. 2016. Inhibition of biofilm in Bacillus amyloliquefaciens Q-426 by diketopiperazines. World J Microbiol Biotechnol 32:143. doi: 10.1007/s11274-016-2106-4. [DOI] [PubMed] [Google Scholar]
- 47.Park D-K, Lee K-E, Baek C-H, Kim IH, Kwon J-H, Lee WK, Lee K-H, Kim B-S, Choi S-H, Kim K-S. 2006. Cyclo(Phe-Pro) modulates the expression of ompU in Vibrio spp. J Bacteriol 188:2214–2221. doi: 10.1128/JB.188.6.2214-2221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deep A, Chaudhary U, Gupta V. 2011. Quorum sensing and bacterial pathogenicity: from molecules to disease. J Lab Physicians 3:4–11. doi: 10.4103/0974-2727.78553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lembke C, Stettin D, Speck F, Ueberschaar N, De Decker S, Vyverman W, Pohnert G. 2018. Attraction pheromone of the benthic diatom Seminavis robusta: studies on structure-activity relationships. J Chem Ecol 44:354–363. doi: 10.1007/s10886-018-0944-2. [DOI] [PubMed] [Google Scholar]
- 50.Frenkel J, Vyverman W, Pohnert G. 2014. Pheromone signaling during sexual reproduction in algae. Plant J 79:632–644. doi: 10.1111/tpj.12496. [DOI] [PubMed] [Google Scholar]
- 51.Houston DR, Synstad B, Eijsink VGH, Stark MJR, Eggleston IM, van Aalten DMF. 2004. Structure-based exploration of cyclic dipeptide chitinase inhibitors. J Med Chem 47:5713–5720. doi: 10.1021/jm049940a. [DOI] [PubMed] [Google Scholar]
- 52.Kim K, Kim N-J, Kim SY, Kim IH, Kim K-S, Lee GR. 2015. Cyclo(Phe-Pro) produced by the human pathogen Vibrio vulnificus inhibits host innate immune responses through the NF-κB pathway. Infect Immun 83:1150–1161. doi: 10.1128/IAI.02878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bina XR, Bina JE. 2010. The cyclic dipeptide cyclo(Phe-Pro) inhibits cholera toxin and toxin-coregulated pilus production in O1 El Tor Vibrio cholerae. J Bacteriol 192:3829–3832. doi: 10.1128/JB.00191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bina XR, Taylor DL, Vikram A, Ante VM, Bina JE. 2013. Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro). mBio 4:e00366–13–e00313. doi: 10.1128/mBio.00366-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izumida H, Nishijima M, Takadera T, Nomoto AM, Sano H. 1996. The effect of chitinase inhibitors, cyclo (Arg-Pro) against cell separation of Saccharomyces cerevisiae and the morphological change of Candida albicans. J Antibiot (Tokyo) 49:829–831. doi: 10.7164/antibiotics.49.829. [DOI] [PubMed] [Google Scholar]
- 56.de Carvalho MP, Abraham W-R. 2012. Antimicrobial and biofilm inhibiting diketopiperazines. Curr Med Chem 19:3564–3577. doi: 10.2174/092986712801323243. [DOI] [PubMed] [Google Scholar]
- 57.Heath-Heckman EAC, McFall-Ngai MJ. 2011. The occurrence of chitin in the hemocytes of invertebrates. Zoology (Jena) 114:191–198. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mandel MJ, Stabb EV, Ruby EG. 2008. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics 9:138. doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh P, Brooks JF, 2nd, Ray VA, Mandel MJ, Visick KL. 2015. CysK plays a role in biofilm formation and colonization by Vibrio fischeri. Appl Environ Microbiol 81:5223–5234. doi: 10.1128/AEM.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burgos HL, Burgos EF, Steinberger AJ, Suen G, Mandel MJ. 2020. Multiplexed competition in a synthetic squid light organ microbiome using barcode-tagged gene deletions. mSystems 5:e00846-20. doi: 10.1128/mSystems.00846-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stabb EV, Butler MS, Adin DM. 2004. Correlation between osmolarity and luminescence of symbiotic Vibrio fischeri strain ES114. J Bacteriol 186:2906–2908. doi: 10.1128/jb.186.9.2906-2908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prioritized mass features detected using IMS. Download FIG S1, PDF file, 0.3 MB (353.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of natural V. fischeri isolates. Download FIG S2, PDF file, 0.4 MB (456.8KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GNPS match of m/z 235 to cyclo(His-Pro). Download FIG S3, PDF file, 0.3 MB (343.9KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MALDI-TOF dried droplet spectrum of m/z 235 in biofilm-up extract. Download FIG S4, PDF file, 0.3 MB (309.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extracted ion chromatograms of cyclo(His-Pro) on the Q-ToF mass spectrometer. Download FIG S5, PDF file, 0.4 MB (415.5KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
General experimental, synthesis, and characterization of the different DKP stereoisomers. Download Text S1, PDF file, 1.0 MB (1,009KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Retention time matching of cyclo(d-His-l-Pro) to biofilm-up. Download FIG S6, PDF file, 0.3 MB (338.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantification of cyclo(d-His-l-Pro) in V. fischeri wild type and mutants. Download FIG S7, PDF file, 0.3 MB (327.8KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detection of cHP-3 in light organ replicates. Download FIG S8, PDF file, 0.3 MB (319.6KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Colonization of squid hatchlings with exogenous cHP-3. Download FIG S9, PDF file, 0.3 MB (313.1KB, pdf) .
Copyright © 2021 Zink et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Data files for all IMS and LS-MS/MS analyses can be found in the MassIVE database under ID MSV000085327.