Summary
Positive selection in Europeans at the 2q21.3 locus harboring the lactase gene has been attributed to selection for the ability of adults to digest milk to survive famine in ancient times. However, the 2q21.3 locus is also associated with obesity and type 2 diabetes in humans, raising the possibility that additional genetic elements in the locus may have contributed to evolutionary adaptation to famine by promoting energy storage, but which now confer susceptibility to metabolic diseases. We show here that the miR-128-1 microRNA, located at the center of the positively selected locus, represents a crucial metabolic regulator in mammals. Antisense targeting and genetic ablation of miR-128-1 in mouse metabolic disease models result in increased energy expenditure and amelioration of high fat diet-induced obesity, and markedly improved glucose tolerance. A thrifty phenotype connected to miR-128-1-dependent energy storage may link ancient adaptation to famine and modern metabolic maladaptation associated with nutritional overabundance.
In Brief
A positively selected locus linked to ancient adaptation to milk consumption is also linked to metabolic disorders and contains a microRNA that controls energy expenditure, potentially connecting these two phenotypes and the role of selection in metabolic disease.
Graphical Abstract

Introduction
The extremely long (~1 Mb) haplotype on human chromosome 2 (2q21.3) represents a classic example of positive selection, where a large chromosome segment containing genetic variants has risen to high frequency among European populations over the last several thousand years, presumably due to selection for traits associated with a survival advantage (Bersaglieri et al., 2004; Field et al., 2016; Grossman et al., 2010; Grossman et al., 2013; Itan et al., 2009; Sabeti et al., 2002; Sabeti et al., 2007). The 2q21.3 locus harbors the lactase gene (LCT), encoding the enzyme lactase-phlorizin hydrolase, which hydrolyzes lactose and is expressed in the small intestine of infants allowing digestion of breast milk (Swallow, 2003). Intestinal expression of lactase is typically shut off after weaning in mammals. However, the long 2q21.3 haplotype in Europeans is strongly linked to expression of intestinal lactase into adulthood (termed lactase persistence, LP), allowing adult milk consumption (Gerbault et al., 2011; Itan et al., 2009; Segurel and Bon, 2017). The signature European LP single nucleotide polymorphism (SNP), rs4988235, is present in an intronic enhancer in the MCM6 gene, which is located just upstream of the LCT gene.
LP has been postulated to have provided a selective advantage in nutrient-deprived populations and at higher latitudes with decreased sunlight due to supplementation of calcium and vitamin D3 to support proper bone formation and calcification (Flatz and Rotthauwe, 1973). However, other dietary sources might have provided sufficient sources of such nutrients (Itan et al., 2009). The current prevailing theory has posited that adult consumption of fresh milk among herding populations would have allowed additional calorie intake to produce a survival advantage during famines (Sverrisdottir et al., 2014). However, fresh milk is highly susceptible to microbial degradation, and from archeological records it is clear that farmers and herders in ancient times knew well how to preserve milk products through the generation of butter, yogurt and cheese products, where much of the lactose is removed or consumed (Curry, 2013; Salque et al., 2013; Segurel and Bon, 2017). Studies of ancient DNA have also shown that steppe populations lacked the rs4988235 LP SNP for thousands of years, yet were known to be herders and exhibited evidence of consumption of milk products (Jeong et al., 2018; Mathiesen et al., 2015). Hence, individuals among herding populations, even those with moderate lactose intolerance, should have been able to consume stored milk products during most famines.
Challenged by severe and prolonged famines that would generate strong selective pressure, increased energy efficiency and fat storage among human populations might also have provided a selective advantage for survival and propagation, perhaps in cooperation with the ability of adults to consume milk. While potentially beneficial in times of famine frequent after the transition from hunting and gathering to agriculture, this “thrifty gene” trait may now represent a metabolic maladaptation predisposing to obesity, insulin resistance and type 2 diabetes (T2D) (Neel, 1962).
Support for the possibility of a “thrifty” genotype-phenotype link comes from the fact that several genetic variants across the locus have also been linked to metabolic abnormalities and diseases associated with decreased energy expenditure, such as obesity and T2D, as well as abnormal blood metabolites and lipids (Albuquerque et al., 2013; Almon et al., 2012; Bauer et al., 2011; Camporez et al., 2018; Corella et al., 2011; Gupta et al., 2013; Heard-Costa et al., 2009; Kettunen et al., 2010; Knowles et al., 2015; Ma et al., 2010; Rung et al., 2009; Scherag et al., 2010; Shin et al., 2014; Sladek et al., 2007; Suhre et al., 2011; Willer et al., 2013). For example, a meta-analysis of 31,720 individuals from diverse European populations showed a statistically significant (P=7.9 X 10−5) elevation of body mass index (BMI, kg/m2) associated with the positively selected allele of the rs4988235 SNP (Kettunen et al., 2010). Increased ectopic fat storage in liver and skeletal muscle represent strong risk factors for the development of insulin resistance and T2D (Shulman, 2014). In support of a link of the 2q21.3 locus to T2D, two genome-wide association studies (GWAS) coupled to metabolomics also reported an association of SNPs across this locus with altered levels of 1,5-anhydroglucitol (1,5-AG), a metabolite linked to abnormal glucose homeostasis and T2D (Kim and Park, 2013; Shin et al., 2014; Suhre et al., 2011). Hence, the long haplotype at 2q21.3 is linked to both LP and metabolic aberrations, which may reflect positive selection for multiple cooperative traits (ability to consume milk in adulthood and increased fat storage) that together could have conferred a strong survival advantage during severe famines.
The genetic and molecular underpinnings for the association of the locus with obesity and T2D have remained obscure. It is possible that these additional associations with metabolic aberrations could be explained by one or multiple genetic elements present in the locus. While obesity has a clear genetic component in humans (Allison et al., 1996; Coleman, 1979; Maes et al., 1997; Rosenbaum and Leibel, 1998), meta-analysis of a number of genetic studies linking genetic loci to obesity failed to uncover strong support for single obesity genes producing a selective advantage (Speakman, 2013; Wang and Speakman, 2016), and there are no obvious candidate genes in the 2q21.3 locus. To date, the majority of genotype-phenotype studies in humans have focused on protein coding genes; however, microRNAs (miRNAs) have emerged as crucial regulators of the expression of hundreds of genes, often controlling the output of entire biological pathways (Ambros, 2004; Bartel, 2009, 2018). We and others have uncovered several miRNAs as crucial metabolic regulators (Goedeke et al., 2016; Najafi-Shoushtari et al., 2010; Rottiers and Näär, 2012). We previously employed an unbiased analysis of GWAS data from >188,000 individuals to identify miRNAs associated with abnormal blood lipids (Wagschal et al., 2015). These studies uncovered miR-128-1 as a critical regulator of circulating total cholesterol in vivo in mice (Wagschal et al., 2015). Importantly, miR-128-1 is an intronic miRNA present in the R3HDM1 gene, which is located at the center of the positively selected locus on human chromosome 2, approximately 200 kb from the LCT gene. Given the strong link of miR-128-1 to the GWAS association with cholesterol/lipid abnormalities (Wagschal et al., 2015), and the prediction that it might regulate expression of genes involved in metabolic control (www.targetscan.org), we sought to further evaluate whether miR-128-1 represents a metabolic miRNA that could have contributed to the association of the long haplotype on human chromosome 2 with metabolic diseases and positive selection.
Results
The 2q21.3 locus contains several selection signals in multiple species
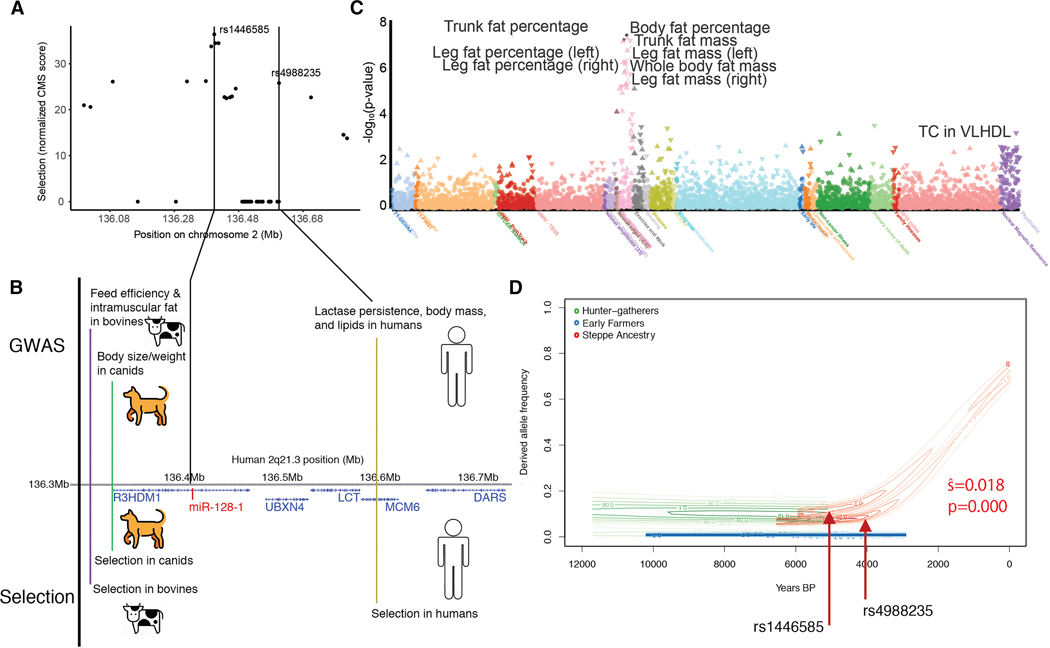
We began by examining the haplotype structure of 2q21.3 in Europeans, where we observed that there is a well-studied, strong selection signal across a long haplotype centered on the R3HDM1 gene (Figure 1A, Figure S1A). This selection signal is directly attributed to the LP phenotype associated with the previously reported tag SNP rs4988235. We note a strong metabolic association at the locus from our previous work detailing a lipid association with expression of miR-128-1 (Wagschal et al., 2015) present in the R3HDM1 gene. In light of this, along with the apparent selection signal associated with SNPs in R3HDM1 near miR-128-1 (e.g., rs1446585, Figure 1A), we examined the selection signal across species in more detail by performing a literature search to identify species exhibiting selection at the 2q21.3 syntenic locus, and if so, whether there is evidence for a metabolic association.
Figure 1. Selection at the 2q21.3 locus in mammals.
(A) A strong signal of positive selection is present at the 2q21.3 locus, evaluated using the Composite of Multiple Signals (CMS, Grossman et al., 2010). (B) Selection at 2q21.3 across mammals. There is evidence for selection on metabolic and anthropomorphic traits in dogs, cows, and humans. Relative locations annotated on the basis of lead SNPs in other species transferred to human GRCh37. (C) PheWAS of UK Biobank traits at rs1438307. Aggregated traits show a strong effect primarily on body composition and adiposity, with a smaller effect on cholesterol. (D) Ancient DNA from different ancient European populations reveals that that a shared selection event is present at the locus, starting in the Steppe ancestry around 4KY (Mathieson and Mathieson, 2018).
Interestingly, the 2q21.3 syntenic locus in cattle has been shown in GWAS to be associated with increased feed efficiency (thrifty phenotype) and increased intramuscular fat (both are associated with the miR-128-1 host gene R3HDM1), desirable traits in cattle breeding (Barendse et al., 2009; Bovine HapMap et al., 2009; Mei et al., 2018). There is also considerable genetic variation at the locus between breeds, potentially indicative of either artificial or natural selection (Bovine HapMap et al., 2009). Recent work indicates a selection signal in canids as well, centered on the promoter of R3HDM1 (XP-EHH 1.02, p = 0.0056; Plassais et al., 2019). This selection signal on the derived allele is also associated with a 14.5 kg increase in body weight (p = 2.17 × 10−13) and is observed exclusively in large breeds with the “bulky” phenotype. Together, these data suggest a complex history of shared natural and artificial selection on multiple metabolic phenotypes at the 2q21.3 locus (Figure 1B).
We note that cattle and dogs are weaned and do not consume milk in adulthood. This makes it unlikely that LP would have contributed to the observed differences in body mass among bovine breeds and modern canids, which suggests other mechanisms might be at play. Given this, we performed a phenome-wide association study (PheWAS) using data from the UK Biobank, and found that European long haplotype 2q21.3 variants are associated with diverse body fat-related traits (Figure 1C), including overall body fat percentage/mass, trunk fat percentage/mass, and leg fat percentage/mass. This is supported by independent (non-UKBB) datasets, where we found robust associations at this locus with a number of metabolic traits, including strong associations with total cholesterol, LDL cholesterol, and 1,5-AG, and we verified the positive selection at the locus using the singleton density score (Field et al., 2016) (Figure S1B). We also examined other LP selection signals and found lipid associations consistent with the effects observed on the European haplotype (Figure S1C). We found tight linkage across ancient DNA samples in Europe, confirming the haplotype was selected as a whole (Mathieson and Mathieson, 2018) (Figure 1D), and consistent with the observed shift in allele frequencies across the locus in present-day Europeans (Figure S1D).
To explore which cell types are most likely responsible for the association with metabolic traits at this locus, we considered variants at the whole extended haplotype. Analyzing chromatin state maps across 127 human cell types from the Roadmap (Roadmap Epigenomics et al., 2015) and ENCODE (Hoffman et al., 2013) consortia (Figure 2A), we found that the 2q21.3 locus is characterized by enhancer-associated marks in blood, fetal brain, and mesenchymal lineages, including adipocytes and their progenitor cells. Using chromatin conformation data in lymphoblastoid cells (Mumbach et al., 2016; Rao et al., 2014), we discovered that the rs1438307 variant is located on the edge of a topological associated domain (TAD) which contains the miR-128-1, ZRANB3, and R3HDM1 gene promoters (Figure 2B; Figure S2B), as well as a larger domain further containing RAB3GAP1 (Figure 2C). We thus considered the possibility that the positive selection haplotype might be associated with altered chromatin accessibility and expression across the locus (Flavahan et al., 2016; Hnisz et al., 2016).
Figure 2. The 2q21.3 regulatory circuitry in humans.
(A) Chromatin state maps from 127 cell types for the region surrounding miR-128-1. (B) Hi-C (GM12878) indicates a stable chromatin domain whose boundary coincides with rs1438307. (C) Wider view of the topological domain reveals a larger domain structure also encompassing RAB3GAP1. A further domain contains DARS and CXCR4. (D) The intensity of the CTCF peak of interest and the expression of R3HDM1 gene are strongly increased in the ENCODE cell lines with the variant locus rs1438307. Boxplots of the distributions of CTCF ChIP-seq enrichment near the TSS of UBXN4 gene (left) and the expression of R3HDM1 (right) for the ENCODE samples with two different variants of rs1438307 (T, red vs G, blue). (E) Data from CellMinerCDB, derived from GDSC, show that expression of genes in the locus, spanned by R3HDM1 and MCM6, are highly correlated. (F) The expression of miR-128-1 is highly correlated with the expression of the host gene R3HDM1 (CCLE).
We tested for differences of binding of the chromatin boundary factor CTCF to the haplotype and found that CTCF indeed preferentially binds the derived T allele of rs1438307 (Figure 2D, S2A). Because of the relatively low read depth at the canonical rs4988235, we were unable to test for allelic bias at this particular position, and our findings should only indicate that the haplotype as a whole alters CTCF binding. The T allele further associated with increased expression of R3HDM1 as well as UBXN4, ZRANB3, and RAB3GAP1, but not flanking genes outside the positively selected region such as CXCR4 (Figure 2D). In addition, we found a substantial allelic bias of rs1438307 for histone H3 lysine 4 trimethylation (H3K4me3; a histone mark associated with open chromatin and active transcription) across cell types (Figure S2C), supporting the notion that increased accessibility was associated with the T allele.
Consistent with our hypothesis that chromatin accessibility and gene expression across the locus is programmed by the genetic architecture, we found that expression of the miR-128-1 host gene R3HDM1 is strongly linked to the expression of the MCM6 gene in 984 cell lines in the GDSC database (GDSC, CellMinerCDB; https://discover.nci.nih.gov/cellminercdb/) (r = 0.488, p = 3.44e-60, Figure 2E), indicative of concerted differential gene regulation at the locus. Furthermore, using expression data from 940 cell lines in the CCLE database (CCLE, CellMinerCDB), we found that miR-128-1 expression also correlated with the expression of the host gene R3HDM1 (r = 0.337, p = 2.05e-26, Figure 2F). Lastly, we confirmed that the rs1438307 T allele associates with higher DNase I accessibility compared to the G allele at the miR-128-1 promoter, just upstream of the sequence encoding the miRNA itself (Figure S2D). These results link rs1438307 to altered chromatin architecture and accessibility leading to coordinated expression changes across the locus through epigenetic regulation.
To assess whether the miR-128-1 haplotype has an effect on miR-128-1 expression, we conducted an expression quantitative trait locus (eQTL) analysis in subcutaneous and visceral white adipose tissues (WAT) from 64 derived homozygotes, 76 heterozygotes, and 39 ancestral homozygotes from a European cohort. We found a subtle haplotype-specific differential expression of miR-128-1 (F-test p = 0.06; Figure S3A, B). We observed this trend to be depot-specific, with visceral WAT expression being somewhat higher than in subcutaneous WAT in both our discovery and replication cohorts (1.8-fold, p = 1.2e-4 and 1.8-fold, p = 4.7e-3, Figure S3A, B). When evaluating the effect of the haplotype tagging variant rs1446585, there was a trend towards the G allele lowering miR-128-1 expression in three cohorts (Figure S3C). We further noted a significant association of miR-128-1 levels with elevated fasting glucose (beta = 0.345, p = 0.0015, Figure S3D), HbA1c levels (beta = 0.135, p = 0.028, Figure S3E), and triacylglycerol (TAG)/high-density lipoprotein (HDL) ratio (all associated with the Metabolic Syndrome; beta = 0.071, p = 0.044, Figure S3F) in our well-annotated adipose tissue replication cohort (n = 31). Using data from the METSIM study (n = 200, Civelek et al., 2013), where mature miR-128 expression was measured with high-throughput sequencing, we confirmed the significant association between log-transformed miR-128 expression and serum fasting glucose levels (beta = 0.47, p = 0.01; Figure S3G). When examining a time course of human adipocyte differentiation, we found that miR-128-1 expression increases steadily throughout differentiation (Figure S3H), consistent with a mechanism active in mature adipocytes. Finally, we examined the FINEMAP-derived posterior estimates (https://www.finucanelab.org/data) and found an association block next to rs4988235 for which the derived allele of rs1446585 was a strong causal candidate for decreased ApoA1 and HDL-cholesterol levels (Figure S3I), but which was not associated with other metabolic traits.
Together, we show that the European long haplotype at 2q21.3 is associated with a locus-wide increased chromatin accessibility, which leads to coordinated elevated expression of miR-128-1 and co-expressed genes at the locus. Given the increased expression of miR-128-1 through differentiation and depot-specific expression, we implicate adipose and possibly other metabolic tissues in this effect. Altogether, our data provide evidence that the 2q21.3 locus associates with metabolic traits.
Anti-miR oligonucleotides promote increased energy expenditure and protection from diet-induced obesity
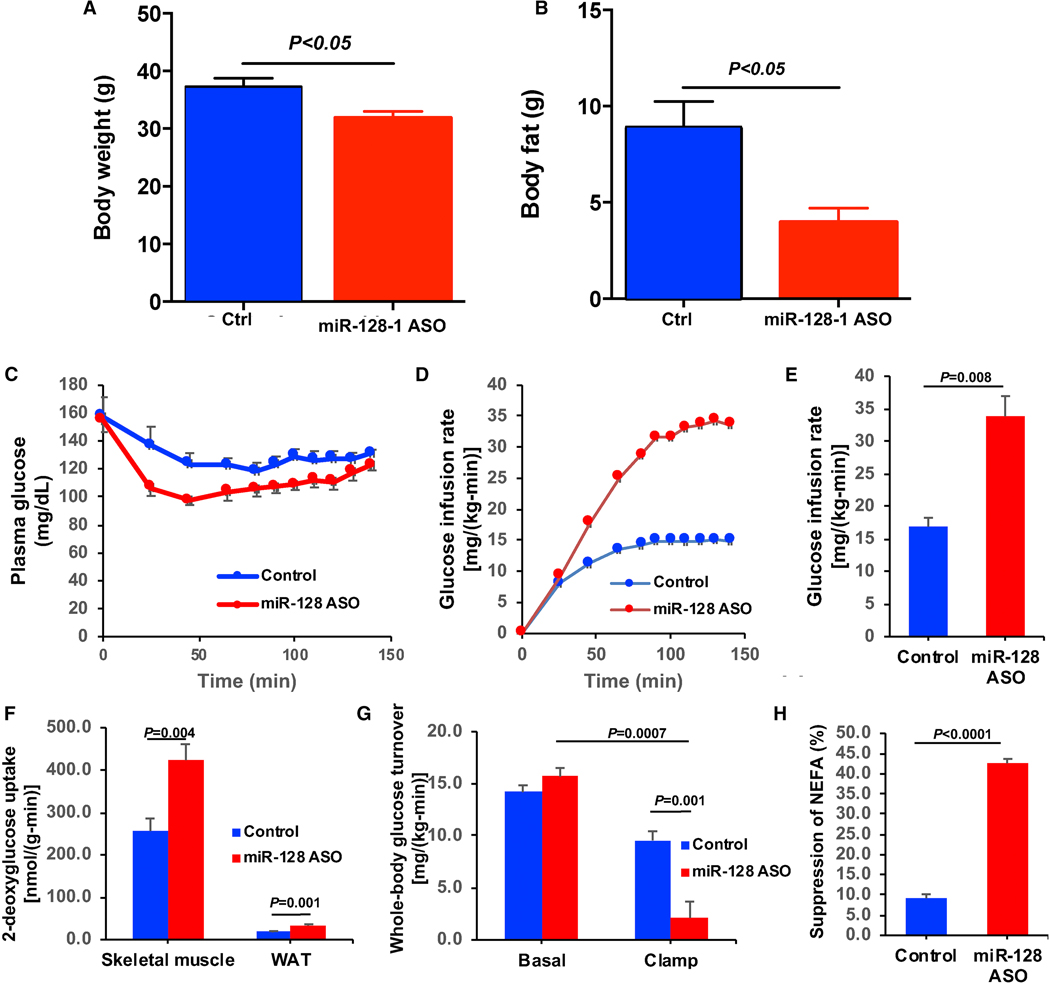
To explore whether miR-128-1 plays a role in regulating energy expenditure and metabolic control, we initially tested miR-128-1 inhibition by anti-miR oligonucleotides in high-fat diet (HFD)-fed male C57BL/6J mice, an established diet-induced obesity (DIO) model (Lee et al., 2014). Treatment with once-weekly subcutaneous injections of 10 mg/kg of a locked nucleic acid (LNA)-modified anti-miR targeting miR-128-1 (miR-128-1 ASO) over 16 weeks in male DIO mice potently reduced miR-128-1 levels in metabolic tissues (e.g., liver; Figure S4A), which was accompanied by strongly reduced weight gain on the HFD, being apparent already two weeks after treatment initiation (Figure 3A, B). This was not due to decreased food intake; if anything, the mice, at least temporarily, consumed more food in response to anti-miR-128-1 treatment (Figure S4B), possibly related to compensatory mechanisms (Soukas et al., 2000). In agreement with our previous studies in Apoe−/− mice (Wagschal et al., 2015), treatment with anti-miR-128-1 resulted in markedly lowered circulating total cholesterol and TAG levels (Figure S4C, D). Body composition studies with 1H nuclear magnetic resonance (EchoMRI) revealed substantially decreased fat stores (>25%; P<0.05) after 15 weeks of treatment, but no significant effect on lean mass (Figure 3C). Accordingly, the visceral (epididymal) and subcutaneous (inguinal) fat pads were markedly smaller in anti-miR-128-1 treated animals as compared with control anti-miR treated mice (Figure 3D). Livers of anti-miR-128-1 treated mice also exhibited smaller size and were less pale, indicative of decreased hepatic lipid accumulation as compared to controls (Figure 3D). In contrast, the inter-scapular brown adipose tissue (BAT) depot, which plays a key role in thermogenesis and regulation of energy expenditure, exhibited increased brown adipose appearance (less fat accumulation/hypertrophy and increased mitochondrial content) in response to anti-miR-128-1 treatment (Figure 3D).
Figure 3. Inhibition of miR-128-1 by miR-128-1 ASO prevents diet-induced obesity (DIO), reduces adipocyte hypertrophy and inflammation, prevents liver steatosis and inflammation, increases whole body energy expenditure and improves glucose homeostasis.
(A) Representative picture of DIO mice treated with miR-128-1 ASO or control anti-miR at 18 weeks. (B) Body weight, (C) body composition and (D) DIO mouse tissue images from miR-128-1 ASO and control treatment groups (n=10 per group). Representative histology data (H&E and IHC) of BAT (E), EPI-WAT (G) and liver (I) from the miR-128-1 ASO and control treatment groups (n=6 per group). Expression of marker genes related to (F) BAT identity (H) epididymal WAT inflammation and (J) liver steatosis was determined by qRT-PCR from the DIO mice. Expression of marker genes in muscle related to glucose homeostasis (K) and energy expenditure (L) was determined by qRT-PCR and western blotting (M). (N) whole body energy expenditure differences were measured in metabolic cages between the miR-128-1 ASO and control group (n=10 per group). Intraperitoneal (IP)-glucose tolerance (O) and insulin tolerance tests (P) were performed after 14 weeks of miR-128-1 ASO and control treatment. Error bars represent SEM. Student’s t test, *P<0.05, **P<0.01, ***P<0.001, compared to mice injected with control ASO.
Next, we evaluated the phenotypic effects of anti-miR-128-1 treatment in metabolic tissues (e.g., BAT, white adipose tissue (WAT), liver and skeletal muscle). In accordance with the gross appearance of the BAT depot, histological analysis revealed strongly decreased accumulation of fat, and markedly increased staining with an antibody recognizing the BAT hallmark, uncoupling protein 1 (Ucp1), in immunohistological analysis of anti-miR-128-1 treated mice as compared with control treated mice (Figure 3E). Evaluation of gene expression changes in BAT revealed that anti-miR-128-1 treatment resulted in an increase in mRNA levels of several predicted and verified direct targets of miR-128-1 (www.targetscan.org), including Prdm16, Ppargc1a, and Ppara, which are known to promote BAT cell fate determination and stimulate mitochondrial biogenesis and fatty acid β-oxidation (FAO) (Ohno et al., 2013; Seale et al., 2011), as well as the expression of a number of mitochondrial and energy expenditure genes such as Dio2, Cidea, and Cox family members (Cannon and Nedergaard, 2004; Seale et al., 2007) (Figure 3F and Figure S4E).
WAT hypertrophy, as well as inflammatory cell infiltration in WAT, are frequently associated with obesity and T2D (Sun et al., 2011a; Wensveen et al., 2015). We found that anti-miR-128-1 treatment of DIO mice led to smaller fat cells and decreased “crown-like” accumulation of macrophages surrounding the fat cells in epididymal WAT (Figure 3G), evidence of a positive effect of anti-miR-128-1 treatment on WAT fat storage and inflammation. In agreement, the expression of Pparg and Adipoq (adiponectin), two hallmark genes of healthy fat (Ohno et al., 2012; Rosen and Spiegelman, 2014), was increased in epididymal WAT in response to anti-miR-128-1 treatment, whereas genes encoding inflammatory cytokines and macrophage markers such as Tnfa, Il-1b, Il-6 and Mcp1 (Hotamisligil et al., 1993; Odegaard and Chawla, 2008; Weisberg et al., 2003) were downregulated (Figure 3H and Figure S4F).
Metabolic dysfunction associated with obesity and T2D typically also manifests as excess fat accumulation and inflammation in the liver, hallmarks of nonalcoholic fatty liver disease and steatohepatitis (NAFLD/NASH) (Shulman, 2000, Suzuki and Diehl, 2017). We found that anti-miR-128-1 treatment caused a dramatic decrease in hepatic lipid accumulation and inflammation in DIO mice (Figure 3I). The marked reduction in liver fat correlated with strongly decreased levels of the Srebp1 transcription factor, a master regulator of lipogenic gene expression (Horton et al., 2002). Accordingly, expression of Srebp1 target genes encoding lipid synthesis enzymes (Acc1 and Fasn) was coordinately downregulated in response to anti-miR-128-1 treatment (Figure 3J and Figure S4G). We also observed a decrease in the expression of genes linked to adipogenesis and fat storage (Pparg and Fabp4) and increased expression of Ppara, encoding a transcriptional regulator of genes involved in FAO (Pawlak et al., 2015) (Figure 3J).
Skeletal muscle represents another highly metabolically active tissue, and anti-miR-128-1 treated DIO mice also show potent stimulation of genes encoding proteins broadly involved in controlling cellular metabolic circuits, such as Pparα, Pgc-1α, Ampkα2, Ucp3 and many others in this tissue as compared to controls (Figure 3K-M). Taken together, these findings support the notion that miR-128-1 acts to coordinately restrict gene expression programs governing energy expenditure across multiple metabolically active organs and tissues. Accordingly, metabolic cage studies revealed that inhibition of miR-128-1 caused markedly enhanced energy expenditure, without significantly affecting locomotor activity levels (Figure 3N and Figure S4H-J). Consistent with miR-128-1 mediating these metabolic effects directly through regulating expression of target genes involved in metabolic control, we found that miR-128-1 negatively regulates expression of luciferase reporters fused with 3’ UTRs of human PPARGC1, PPARG, PPARA and PRDM16, with effects abolished by mutation of conserved miR-128-1 target sites (Figure S4K).
miR-128-1 is a key regulator of glucose homeostasis and insulin sensitivity
Obesity and ectopic fat deposition in liver and skeletal muscle is frequently linked to insulin resistance and poor glucose control, and is a major risk factor for the development of T2D (Shulman, 2000, Kahn et al., 2006). DIO mice present with hyperglycemia and moderate insulin resistance; these abnormal metabolic traits were significantly ameliorated by anti-miR-128-1 treatment as revealed by glucose and insulin tolerance tests (Figure 3O, P). To further ascertain the role of miR-128-1 in obesity-associated glucose abnormalities we carried out hyperinsulinemic-euglycemic clamp studies (Kim, 2009; Perry et al., 2015) in DIO mice treated with anti-miR-128-1 and control anti-miR. Accompanying the decreased body weight and lowered fat mass (Figure 4A, B), anti-miR-128-1-treated mice exhibited marked improvements in insulin-stimulated whole-body glucose metabolism, as revealed by substantially increased glucose infusion rates required to maintain euglycemia during the clamp (Figure 4C-E). This improvement in whole body insulin-stimulated glucose metabolism could be attributed to marked improvements in muscle glucose uptake as reflected by increased 2-deoxyglucose uptake in skeletal muscle (Figure 4F) as well as increased hepatic insulin responsiveness as reflected by greater suppression of hepatic glucose production during the clamp (Figure 4G). In addition, anti-miR-128-1 treatment also manifested increased WAT insulin sensitivity as reflected by increased 2-deoxyglucose uptake in WAT (Figure 4F) and increased suppression of non-esterified fatty acid (NEFA) release from WAT (Figure 4H) during the clamp, both of which are hallmarks of T2D and contributors to increased rates of hepatic gluconeogenesis (Karpe et al., 2011; Thabit et al., 2014, Perry et al., 2015). These results together demonstrate a key role for miR-128-1 in modulating metabolic circuits that govern glucose homeostasis, consistent with the genetic link of the miR-128-1 locus to T2D.
Figure 4. Anti-miR-128-1 treatment of DIO mice improves hepatic, skeletal muscle, and adipocyte insulin sensitivity.
(A) Body weight and (B) body fat percentage in anti-miR-128-1 treated and control HFD-fed mice. (C) Plasma glucose concentrations during the hyperinsulinemic-euglycemic clamp. (D) Glucose infusion rate required to maintain euglycemia during the clamp studies. (E) Steady state (100–140 min) glucose infusion rates required to maintain euglycemia during the clamp. (F) Insulin-stimulated glucose uptake in skeletal muscle and white adipose tissue. (G) Basal and insulin mediated suppression of endogenous glucose production during the clamp. (H) Insulin-mediated suppression of plasma NEFA during the clamp. In all panels, data are expressed as the mean±SEM of n=7 mice per group, with comparisons by the 2-tailed paired (basal vs. clamp) or unpaired (control vs. miR-128-1 ASO) Student’s t-test.
Role of miR-128-1 in metabolic dysregulation in leptin-deficient ob/ob mice
Next, we sought to evaluate whether miR-128-1 might also contribute to metabolic abnormalities in the classic leptin-deficient ob/ob genetic obesity mouse model (Wang et al., 2014). These mice exhibit profound hyperphagia due to the lack of the satiety hormone leptin, which is normally secreted from WAT in response to feeding and acts in the hypothalamus to suppress feeding behavior (Wang et al., 2014). Treatment of ob/ob mice fed a standard chow diet with anti-miR-128-1 resulted in a modest but significant decrease in body weight and overall fat mass, and decreased size of liver and visceral (epididymal) WAT depots (Figure S5A, B). Metabolic cage analysis demonstrated markedly elevated energy expenditure in response to anti-miR-128-1 treatment as compared with control anti-miR treatment (Figure S5C). Accordingly, histological and molecular analyses of BAT revealed decreased fat accumulation and strongly increased expression of BAT markers and energy expenditure genes such as Prdm16, Ppargc1a, Ppara (all direct targets of miR-128-1; Figure S4K) as well as Ucp1, Cidea, Dio2 and Elovl3 (Figure S5D, E), indicative of improved BAT function. We also observed decreased adipocyte hypertrophy in the subcutaneous (inguinal) WAT depot, accompanied by elevated expression of browning markers and energy expenditure genes (e.g., Prdm16, Ppargc1a, Ppara, Ucp1) in animals treated with anti-miR-128-1, consistent with browning or beigeing of subcutaneous WAT depots (Kajimura et al., 2015). Additionally, expression of Srebf1 and lipogenic genes (Acc1 and Fasn) was diminished, and expression of inflammatory cytokines (Tnfα and Il1b) was lowered (Figure S5F, G). Anti-miR-128-1 treatment also resulted in improved hepatic steatosis and led to decreased liver expression of markers of inflammation and lipogenesis in ob/ob mice (Figure S5H, I). These beneficial metabolic effects were accompanied by markedly improved glucose tolerance and insulin sensitivity in anti-miR-128-1-treated ob/ob mice (Figure S5J, K), confirming a potent metabolic regulatory function of miR-128-1 in both diet-induced and genetic obesity and T2D models.
Whole animal genetic ablation of miR-128-1 confirms its “thrifty” role in DIO mice
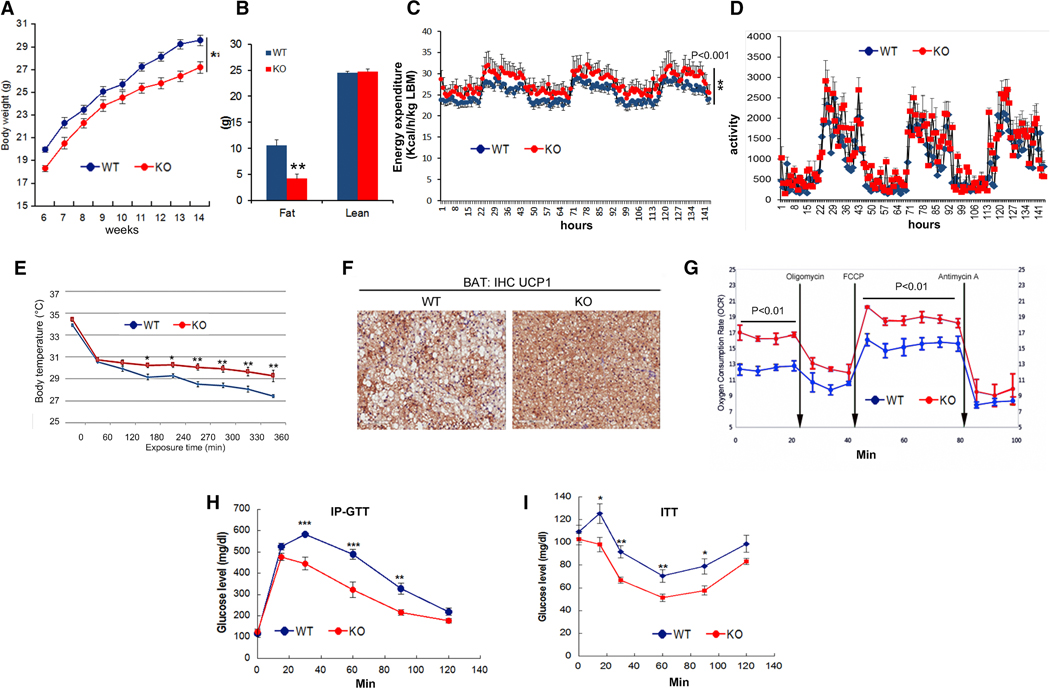
To confirm the role of miR-128-1 in controlling energy expenditure in vivo, we next analyzed the effects of miR-128-1 whole animal genetic ablation (Tan et al., 2013; Wark et al., 2017) in mice using the DIO model. Most KO animals were viable and fertile, with no obvious developmental, physiological or behavioral phenotypes. Upon challenge with a HFD, the miR-128-1 KO mice exhibited significantly decreased weight gain as compared with WT littermates (Figure 5A). After 12 weeks on the HFD, the miR-128-1 KO mice WAT depots were >60% smaller that WT controls, as revealed by EchoMRI (Figure 5B). Metabolic cage studies also showed that miR-128-1 KO mice exhibit markedly increased energy expenditure, without apparent effect on activity (Figure 5C, D). Consistently, the miR-128-1 KO mice were significantly more tolerant to cold exposure as compared with WT littermates (Figure 5E). Examination of the inter-scapular BAT depot revealed strongly decreased intracellular fat (Figure 5F), and primary BAT cells isolated from miR-128-1 KO mice exhibited greatly increased mitochondrial oxygen consumption rate and energy expenditure, as shown by Seahorse studies (Figure 5G), as well as elevated expression of BAT and energy expenditure marker genes such as Prdm16, Ppargc1a, and Ppargc1b, and Ucp1 (Figure S6). Beneficial effects on metabolic gene expression were also observed in WAT, skeletal muscle and liver of miR-128-1 KO mice (Figure S6). Finally, similar to the results in DIO mice treated with anti-miR-128-1, the HFD-fed miR-128-1 KO mice exhibited markedly improved glucose homeostasis and response to insulin (Figure 5H, I). These findings strongly support the notion that miR-128-1 represents a crucial regulator of whole animal metabolism by acting as a “thrifty” miRNA to suppress energy expenditure through apparent effects in multiple metabolic tissues, such as WAT, BAT, liver and skeletal muscle.
Figure 5. miR-128-1 deficiency prevents adiposity, increases whole body energy expenditure and improves glucose homeostasis.
(A) Body weight and (B) body composition of WT and miR-128-1 KO mice (n=10 per group). (C) Whole body energy expenditure and (D) activity differences between miR-128-1 KO and WT mice were measured in metabolic cages (n=10 per group). (E) Rectal temperatures of cold (4°C) exposed miR-128-1 KO mice and WT animals (n=6 per group). (F) Representative IHC of Ucp1 in BAT KO group were compared to WT mice (n=6 per group). (G) Energy expenditure differences of brown adipocytes isolated from miR-128-1 KO and WT mice. IP-Glucose tolerance (H) and insulin tolerance tests (I) of miR-128-1 KO and WT mice were performed at 10 weeks of HFD feeding. Error bars represent SEM. Student’s t test, *P<0.05, **P<0.01, ***P<0.001, compared to control mice.
miR-128-1 modulates white adipose differentiation and function
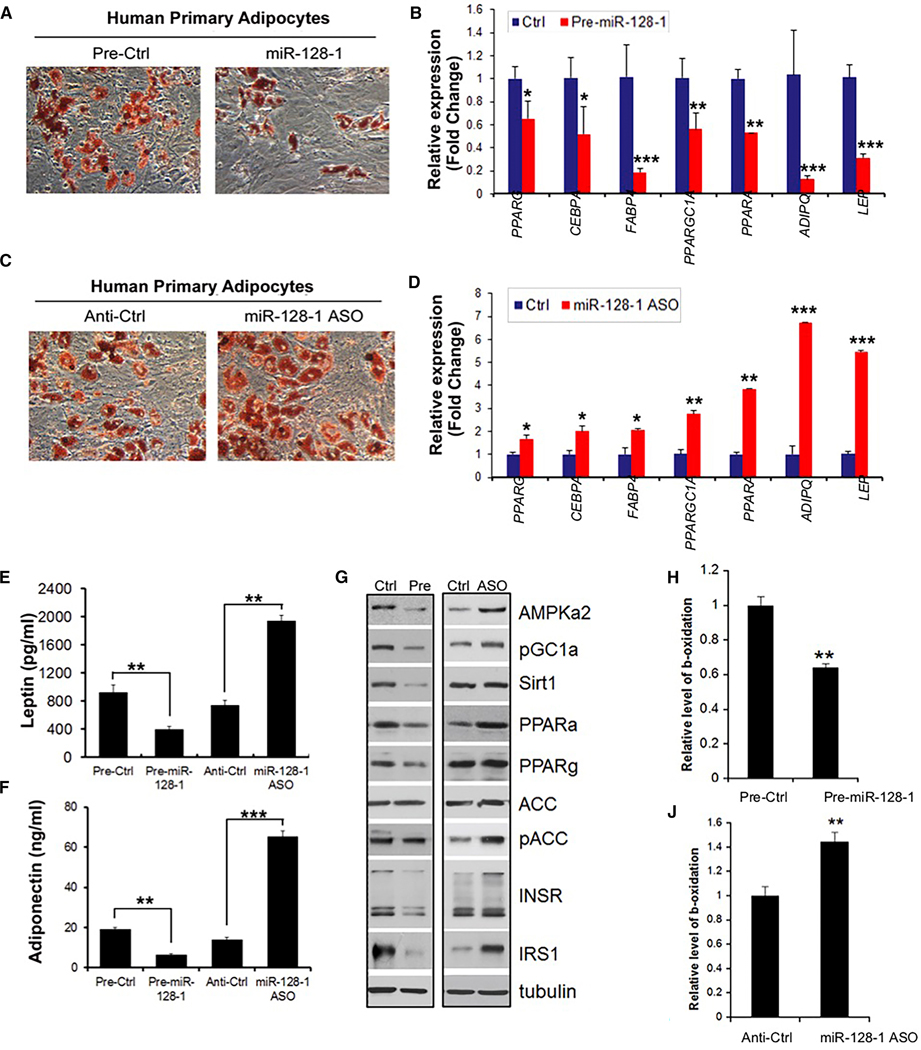
Next, we further investigated the molecular mechanism(s) of miR-128-1 regulation of cellular metabolism, and determined whether the regulatory effects observed in metabolic tissues are cell autonomous. Introduction of excess miR-128-1 precursor into human abdominal subcutaneous WAT-derived pre-adipocytes resulted in decreased adipogenesis in response to a differentiation cocktail, with lowered numbers of mature, fat-storing adipocytes, and decreased expression of white adipocyte differentiation markers such as PPARG, CEBPA, FABP4, PPARGC1A and PPARA, as well as the adipokines adiponectin (ADIPOQ) and leptin (LEP) (Figure 6A, B). Conversely, treatment of pre-adipocytes with anti-miR-128-1 caused increased adipocyte differentiation and elevated expression of genes associated with mature WAT (Figure 6C, D). In particular, the expression of adiponectin and leptin was markedly affected by changing miR-128-1 levels (both mRNAs are predicted miR-128-1 targets in humans but not in mice; www.targetscan.org). These adipokines have been shown to represent hallmarks of healthy WAT and exert beneficial metabolic effects in multiple tissues (Meier and Gressner, 2004). In accord with the gene expression data, we observed potent regulation by miR-128-1 of leptin and adiponectin secreted into the culture medium (Figure 6E, F). In addition to being targets of miR-128-1, it is likely that leptin and adiponectin are induced indirectly in WAT through the miR-128-1 target and pro-adipogenesis transcription factor PPARγ. We also verified that altering miR-128-1 levels had concordant effects on levels of target proteins, such as AMPKα2, PGC-1α, SIRT1, PPARα, PPARγ, INSR and IRS1 (Figure 6G). In keeping with the known role of AMPKα2 as a key stress kinase regulator of the lipogenic enzyme ACC1 by direct inhibitory phosphorylation, altered AMPKα2 levels in response to miR-128-1 manipulations resulted in corresponding changes in ACC1 phosphorylation status (Figure 6G). A number of miR-128-1 targets (e.g., AMPKα2, PGC-1α, SIRT1, PPARα) regulate FAO, and we found that miR-128-1 overexpression decreased, whereas miR-128-1 inhibition increased, FAO in human WAT-derived adipocytes (Figure 6H, I). These findings are consistent with in vivo data from WAT in DIO mice, where anti-miR-128-1 treatment results in decreased WAT hypertrophy, accompanied by lowered expression of Srebf1 and lipogenic target genes (Figure S7A-D), as well as significant de-repression of Pparg, Adipoq, and Ppargc1a, and elevated expression of Prdm16, Ppara and Ucp1 (Figure S7E-J), suggesting increased browning of WAT in response to inhibition of miR-128-1 in DIO mice.
Figure 6. Manipulation of miR-128-1 level in primary human adipocytes alters differentiation state, energy expenditure and leptin/adiponectin secretion.
Over-expression of pre-miR-128-1 prevents primary human adipocyte differentiation (A) and reduces adipocyte hallmark gene expression (B) anti-miR-128-1 promotes primary human adipocyte differentiation (C) and upregulates adipocyte hallmark gene expression (D). Levels of leptin (E) and adiponectin (F) secreted from human primary adipocytes treated with miR-128-1 pre-cursor and anti-miR-128-1. (G) Cellular levels of adipocyte genes in primary human adipocytes were confirmed by western blotting. FAO changes in primary human adipocytes induced by overexpressing miR-128-1 (H) or knockdown (I) was assessed. Errors bars represent mean ± SD. Student’s t-test, *P<0.05, **P<0.01, ***P<0.001, compared to cells transfected with control ASO.
miR-128-1 inhibits brown adipose cell fate and energy expenditure
Due to the marked positive effects on BAT differentiation and energy expenditure observed in DIO and ob/ob mice treated with anti-miR-128-1 and in the HFD-fed miR-128-1 KO mice, we evaluated the molecular mechanism and functional importance of miR-128-1 in regulating brown adipose cell differentiation in vitro. Primary brown pre-adipocytes isolated from the inter-scapular BAT depot of Day 1 newborn miR-128-1 WT and KO mice were induced to differentiate and then stained with Oil-Red O as a marker of mature adipocyte differentiation. We found that loss of miR-128-1 resulted in strongly increased capacity to differentiate into mature brown adipocytes (Figure 7A). This was supported by results from Seahorse analysis showing elevated energy expenditure in brown adipocytes from miR-128-1 null mice (Figure 7B). Moreover, expression of brown adipose hallmark genes (Prdm16, Ucp1) as well as transcription factors and co-activators involved in adipose cell fate determination and regulation of gene expression programs governing lipid homeostasis and energy expenditure (Pparg, Ppara, and Ppargc1a) was elevated in differentiated brown adipocytes from miR-128-1 KO mice as compared with controls (Figure 7C-G). We also examined acute effects of introducing pre-miR-128-1 in already differentiated WT brown adipocytes, in the presence or absence of the brown adipose inducer dbcAMP (Uldry et al., 2006). Our studies revealed potent suppression of direct (e.g., Prdm16, Ppargc1a, Ppara) and indirect (e.g., Ucp1 and Cidea) miR-128-1 targets involved in brown adipocyte function and lipid homeostasis and energy expenditure programs, even in the presence of dbcAMP (Figure 7H-J). Moreover, introduction of anti-miR-128-1 into mature, differentiated mouse brown adipocytes resulted in marked de-repression of the expression of these genes (Figure 7K-M). Cell lineage tracing studies in vivo have revealed that brown adipocytes can derive from a Myf5-positive skeletal muscle precursor lineage (Sanchez-Gurmaches and Guertin, 2014; Seale et al., 2008). In vitro studies have also shown that the mouse C2C12 muscle precursor cell line can be differentiated into brown adipocytes (Sun et al., 2011b). We found that anti-miR-128-1 treatment of C2C12 cells markedly promoted brown lineage commitment and differentiation into mature brown adipocytes, as judged by elevated Oil-Red O staining and increased expression of pan-adipocyte (Pparg, aP2, Cebpa, Adipoq) and brown adipose marker genes (Prdm16, Ucp1, Cidea, Elovl3, Ppargc1a, Ppara), but decreased expression of mature myocyte genes (Figure 7N-R). In accord, induction of brown adipocyte differentiation of C2C12 cells in response to inhibition of miR-128-1 was accompanied by elevated FAO and increased energy expenditure, functional hallmarks of mature brown adipocytes (Figure 7S, T). Taken together, these findings provide strong support for a direct cell autonomous role of miR-128-1 in suppressing brown adipocyte cell fate and function (energy expenditure).
Figure 7. miR-128-1 controls primary brown adipocyte differentiation/identity and energy expenditure and restricts trans-differentiation of muscle cell to brown adipocyte lineage.
(A) miR-128-1 WT and KO primary brown adipocytes were induced to differentiate to mature brown adipocytes and stained with Oil-red O. (B) Mitochondrial respiration of miR-128-1 WT and KO brown adipocytes was measured by Seahorse analysis. (C-G) Brown adipocyte marker gene expression of miR-128-1 WT and KO brown adipocytes was measured by qRT-PCR. (H-M) Introduction of miR-128-1 pre-cursor or anti-miR-128-1 into mature brown adipocytes. (H, K) Analysis of expression of brown adipose marker genes and energy expenditure genes by qRT-PCR. (I-M) The brown adipocyte activator dbcAMP stimulates Pgc-1α and Ucp-1 expression in mature mouse brown adipocytes. (N) Anti-miR-128-1 induces C2C12 myoblast differentiation into brown adipocytes. C2C12 myoblasts were stained with Oil-red O after 8 days of treatment. (O-Q) Pan-adipocyte and brown adipose-specific marker expression was determined by qRT-PCR and Western blotting. (R) Myotube formation was assessed after 5 days of C2C12 myocyte differentiation by qRT-PCR analysis of myotube marker gene expression. FAO (S) and mitochondrial respiration (T) were measured by Seahorse analysis after induction of brown adipocyte differentiation by anti-miR-128-1 treatment. Errors bars represent mean ± SD. Student’s t-test, *P < 0.05, **P < 0.01, ***P < 0.001, compared to WT cells or cells transfected with control ASO.
Discussion
Positive selection of a long (~1 Mb) haplotype on human chromosome 2 in Europeans, harboring the lactase (LCT) gene, has been thought to be solely due to lactase persistence (LP), allowing adult milk consumption. The ability to consume fresh milk beyond infancy may indeed have provided a selective advantage in ancient populations faced with poor nutrition and famine. However, this hypothesis may not fully account for the remarkably strong positive selection signature resulting in the long haplotype in this genomic locus. For example, studies of ancient DNA and extensive archeological records have shown that herders consumed milk products for thousands of years before the emergence of the LP haplotype/phenotype around 4KY ago (Jeong et al., 2018, Mathiesen et al., 2015), challenging the importance of fresh milk consumption as a potential driver of evolutionary adaptation to famine. Importantly, the European long haplotype is also associated with metabolic disorders such as obesity and T2D. It has been difficult, however, to reconcile LP with these metabolic abnormalities, and based on our data presented here, we propose that the miR-128-1 miRNA located in the center of the positively selected locus might contribute to some of these links. Indeed, our results showing concordance of the positive selection haplotype with genetic associations of the miR-128-1 locus to obesity and T2D, as well as the phenome-wide association study using UK Biobank data revealing association of 2q21.3 variants with diverse body fat-related traits, might be consistent with metabolic regulation by miR-128-1. However, it does not rule out the possibility that some of the signal is due to LCT in humans. Self-reported fresh milk intake, depending on milk fat content, however has a complex relationship with body fat phenotypes in the UK Biobank data set, and will require further in-depth study.
In support of the links of the locus to metabolic phenotypes, we highlight data suggesting that the syntenic genomic region near R3HDM1/miR-128-1 in cattle has been under positive selection, and is associated with feed efficiency and intramuscular fat, thrifty metabolic traits selected for in certain cattle breeds (Barendse et al., 2009; Bovine HapMap et al., 2009; Mei et al., 2018). Moreover, dog breeds exhibiting a “bulky” (large size/excess backfat) phenotype have characteristic SNPs in the promoter of the miR-128-1 host gene R3HDM1 (Plassais et al., 2019). Cattle and canines are weaned and do not consume fresh milk in adulthood. While these finding may be consistent with our data showing that miR-128-1 regulates metabolic circuits and promotes fat storage in mammals, further studies will be needed to assess whether miR-128-1 contributes to the genetically selected phenotypes in cattle and dogs.
Our mechanistic analyses reveal that the European long 2q21.3 haplotype is associated with increased chromatin accessibility and elevated transcriptional activity throughout the locus, likely in part due to altered CTCF chromatin boundary factor occupancy, contributing to the changes in miR-128-1/gene expression across the locus. Although the derived allele of the rs4988235 SNP present in the MCM6 intronic enhancer is strongly correlated with increased expression of the nearby LCT gene, it is possible the activity from this enhancer, in the context of the overall chromatin architecture of the European long haplotype, is also linked to the expression of other genes in the locus, as well as miR-128-1. Future studies are needed to address this and determine which variants and targets drive the associations.
Intervention studies with anti-miR treatment targeting miR-128-1 in mouse obesity models suggest that miR-128-1 represents a crucial regulator of metabolism by suppressing the expression of a number of genes encoding critical metabolic regulators in multiple metabolic tissues. Indeed, antagonism or ablation of miR-128-1 either by antisense targeting or whole animal KO results in markedly increased energy expenditure, accompanied by reduced fat mass and improved whole body glucose metabolism, which could be attributed to increased liver, muscle and adipose tissue insulin sensitivity. Based on these observations, along with human GWAS data, we propose that elevated levels of miR-128-1 in metabolic tissues may reduce energy expenditure and promote increased fat storage in humans as well.
Taken together, our data showing a key role for miR-128-1 in programming a metabolic switch from energy expenditure to energy storage and the links of the miR-128-1 genomic locus to positive selection and abnormal metabolism complement the role of lactase persistence at the locus. Indeed, altered expression of miR-128-1 could have been beneficial in ancient times, but presently, with persistent calorie abundance, may constitute a metabolic maladaptation, predisposing to obesity and T2D. Hence, miR-128-1 may represent a therapeutic target for the treatment of metabolic diseases such as obesity and T2D.
Study Limitations
There are many limitations to this work. Comprehensive measures of selection and genome-wide association studies are not available for most species, and fewer still have well-powered GWA studies of metabolic traits. This limits our ability to evaluate the presence of, and associations with, selection at the 2q21.3 locus. In addition, the long European haplotype contains numerous variants, and determining which are causal for the observed associations is an area of future study. Direct perturbation will also enable more comprehensive characterization of the epigenetic and transcriptomic consequences of these variants, which should help determine the gene(s) and/or microRNA(s) attributable to the various observed phenotypes associated with the locus and disentangle population history, environmental correlates, and single-variant effects. Furthermore, because miR-128-1 regulates the expression of many metabolic genes coordinately in multiple metabolically active tissues, it presents a formidable challenge to fully elucidate the mechanistic links of miR-128-1 to specific targets and in specific tissues. Finally, mouse models are an important step in understanding the mechanistic basis of miR-128-1 metabolic control, however, clinical studies will be critical to applying this knowledge to human disease alleviation.
Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents may be directed to, and will be fulfilled by the lead contact Anders M. Näär (naar@berkeley.edu).
Materials Availability
Plasmids generated in this study will be freely distributed.
Data and Code Availability
Data sets analyzed: The code supporting the current study has not been deposited in a public repository because no novel algorithms or methods were employed, but is available from the corresponding author on request. Data employed include GWAS summary statistics and fine-mapped variants derived from the UK Biobank resource, Million Veteran Program, Global Lipids Genetics Consortium, and numerous other individual supporting studies; whole-genome sequencing data from gnomAD; epigenetic data from the Roadmap Epigenomics and ENCODE projects; and expression data from GDSC, CCLE, FANTOM5, ENCODE, and Civelek et al., 2013. Selection scores were derived by the Sabeti, Pritchard, and Ostrander labs, as well as the Bovine Hapmap Consortium. Underlying data from human subjects used in this study are publicly available, either without restriction or through application to dbGaP or the UK Biobank. Restrictions exist on access to underlying data from non-human studies.
Experimental Model and Subject Details
Cell culture and mouse
Cell lines:
HepG2, 3T3-L1, C2C12 and HEK293T cells were obtained from the ATCC and propagated according to ATCC’s instructions.
Brown pre-adipocytes were received from Dr. Kai Ge, NIDDK/NIH and were routinely cultured in DMEM plus 10% FBS.
Primary cells:
Primary mouse brown adipocytes were isolated from day 1 newborn pups;
Primary human white adipocytes were obtained from human subjects by surgical resection or subcutaneous adipose tissue aspiration and were cultured using regular methods described in Lee and Fried, (2014) in alpha-MEM with 10% FBS and antibiotics.
Mice:
The C57BL/6J and ob/ob mice were purchased from The Jackson Laboratory, Bar Harbor, ME, all were male aged at 6–8 weeks.
miR-128-1 KO mice: experimental manipulation of miR-128-1 in vivo was achieved by crossing mice carrying miR-128-1 floxed alleles (Tan et al., 2013) to transgenic Cre-recombinase mouse lines. For germline excision, we used the ubiquitously expressed cytomegalovirus-Cre mouse line (CMV-Cre; Jackson Labs #006054). Genotyping was performed as described in Wark et al., (2017). Lines were maintained on the C57BL/6J background on a 12-hour light-dark cycle with constant access to food and water. Male mice with age from 6 weeks to 16 weeks were used in different experiments. All experimental procedures were conducted in accordance with IACUC regulations and were approved by Massachusetts General Hospital’s or Yale University School of Medicine’s IACUC.
Method Details
Mouse miR-128-1 inhibition studies
LNA–modified anti-miR oligonucleotides targeting miR-128-1 (5’-TTCACTGTG-3’a; 9-mer), (5’-GGTTCACTGTG-3’a; 11-mer) and a scrambled control LNA oligonucleotide (5’-TCATACTA-3’) were purchased from Exiqon, Denmark (now Qiagen). The 9-mer LNA ASO was used in all in vivo studies except in the hyperinsulinemic-euglycemic studies at Yale University School of Medicine, where we used the second-generation 11-mer LNA ASO. Experiments were performed using 7-week-old male C57BL/6J mice or ob/ob mice purchased from The Jackson Laboratory, Bar Harbor, ME. The diet-induced obese (DIO) mice were fed a diet containing 60% (kcal) fat (D12492, Research Diets) for 6 weeks before and during treatment. The longitudinal studies were performed in mice fed with the high-fat diet for 16 weeks, or chow-fed ob/ob mice, with once-weekly subcutaneous injection of the anti-miR-128-1 (10 mg/kg) or control scrambled LNA oligonucleotide. Body weight was measured every week. During the treatment, glucose and insulin tolerance tests are performed at around week 10 and body composition was measured at 14 weeks. Metabolic activity of the mice was analyzed in metabolic cages at around week 15 and data were collected for one week. Upon sacrifice, brown and white (inguinal and epididymal, respectively) adipose tissue and livers were collected and ~1 ml of blood was obtained from each mouse by right ventricular puncture. Blood was centrifuged at 8,000 rpm for 5 min to obtain serum, which was frozen at −80°C. Total serum cholesterol and triglyceride levels were determined with a Heska Dri-Chem 4000 Chemistry Analyzer (Heska, Loveland, CO) at Massachusetts General Hospital, Center for Comparative Medicine, Diagnostic Laboratory. FPLC analysis of pooled serum was carried out as described (Wagschal et al., 2015), the area under the curve (AUC) was calculated using the linear trapezoidal method based on a linear interpolation between data points. The AUC was calculated as follows: AUC = 1/2 (C1+C2), where C indicates the concentration of lipids in two incremental FPLC fractions. The average AUC value for the VLDL, LDL or HDL fractions was calculated and indicated with the SEM and P value. All mouse procedures were approved by the Massachusetts General Hospital or Yale University School of Medicine Institutional Animal Care and Use Committees.
Tissue sample preparation and gene expression measurement
All tissue samples were rapidly dissected, snap frozen in liquid nitrogen, and stored at −80°C until mRNA and protein extraction. Western blotting was performed using standard protocol (Wagschal et al., 2015), briefly, cells or tissues were lysed in RIPA buffer (0.5% NP-40, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl (pH 7.5)). Proteins were separated by SDS–PAGE, transferred to nitrocellulose membrane (Millipore) and probed with antibodies of the respective targets. For mRNA and miRNA quantification, TRIzol (Life Technologies/Invitrogen) was used for total RNA and miRNA extraction from either mouse tissues or cells. For miRNA extraction from human tissues mirVana™ miRNA Isolation Kit, with phenol (ThermoFisher Scientific Cat# AM1560) was used. The high Capacity cDNA Reverse Transcription Kit and the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies/Invitrogen) were used for further reverse transcription. cDNA was further evaluated by real-time PCR for gene expression using Power SYBR Green or Taqman PCR Master Mix with ABI 7500 Real-Time PCR System The amount of the indicated mRNA or miRNA was normalized to the amount of 18S and U6 RNA, respectively. Total RNA for miRNA expression levels analysis in human tissues was purchased from Ambion (FirstChoice Human Total RNA Survey Panel, AM6000). To quantify miR-128-1 levels, RNAs were reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit and quantified with the miRNA-specific TaqMan miRNA assays (Life Technologies/Invitrogen). PCR primer sequences are available from A.M.N. upon request.
Histology
Adipose and liver tissues from each study animal were sectioned for histology (Lillie and Fulmer, 1976). Paraffin-embedded sections were prepared for H&E and IHC, respectively and the samples were processed in the histology core facility of MGH. The completed slides were examined and representative pictures were taken.
Metabolic cage housing and energy expenditure measurements
After 15–16 weeks of treatment, mice were individually housed and maintained under otherwise standard housing conditions (12-hour light-dark cycle). The 240 LabMaster System (TSE Systems, Chesterfield, MO) was used to measure the calorimetry indirectly. Mice were provided ad libitum access to food and water with a 4-day acclimation period. Oxygen consumption, energy expenditure, activity (X-Y-axis movement activity and Z-axis 245 rearing activity), as well as food/water consumption were monitored in consecutive light-dark cycles over 6 successive days.
Hyperinsulinemic-euglycemic clamp
Male mice underwent surgery under general isoflurane anesthesia to implant a polyethylene catheter in the right jugular vein. After a one-week recovery period, mice were fasted overnight (14 hr) and underwent a hyperinsulinemic-euglycemic clamp as previously described (Jurczak et al., 2012). Briefly, mice received a basal infusion of [3-3H] glucose (0.05 μCi/min) for 120 min, after which blood samples were taken from the tail vein, centrifuged, and plasma stored at −80°C for further analysis. A 140 min hyperinsulinemic-euglycemic clamp was then initiated: mice were infused with insulin (3.0 mU/[kg-min]) and [3-3H] glucose (0.1 μCi/min). Blood samples were obtained from the tip of the tail at 0, 25, 45, 65, 80, 90, 110, 120, 130, and 140 min for plasma glucose measurements. To evaluate insulin-stimulated tissue-specific glucose uptake, a 10 μCi bolus of 2-deoxy-d-[1-14C] glucose (PerkinElmer) was injected after 85 minutes. After IV pentobarbital euthanasia, skeletal muscle (gastrocnemius + soleus) and epididymal white adipose tissue were isolated, and tissue [14C] 2-deoxyglucose specific activity was measured.
Biochemical analysis
Plasma glucose concentrations were measured using the YSI Glucose Analyzer, and NEFA using an enzymatic assay (Wako Diagnostics HR Series NEFA-HR(2)). Plasma insulin was measured by radioimmunoassay by the Yale Diabetes Research Center. Basal and clamp endogenous glucose turnover were calculated by measurement of steady-state plasma [3H] glucose enrichment at the end of the basal and clamp periods. Tissue 2-deoxyglucose uptake was calculated following measurement of plasma and tissue [14C] 2-deoxyglucose specific activity as previously reported (Youn and Buchanan, 1993).
Seahorse
Brown and white pre-adipocytes and C2C12 cells were plated in gelatin-coated XF24-well cell culture microplates (Seahorse Bioscience) and brown and white pre-adipocytes were then differentiated into adipocytes. Oxygen consumption and mitochondrial function were measured by XF24 Extracellular Flux Analyzer (Seahorse Biosciences) as described previously (Ahfeldt et al., 2012).
Intraperitoneal glucose tolerance test and insulin tolerance test
Mice were fasted for 12 hours before the tests. For glucose tolerance test, 1.5 mg/g D-(+)-glucose (G7528, Sigma Aldrich) was injected intra-peritoneally into the mice and the glucose levels were measured at 0 (before injection), 30, 60, and 120 min post-injection. For insulin tolerance test (ITT), 0.00075 U/g insulin (I9278-5 ML, Sigma Aldrich) was used for intra-peritoneal injection and the glucose levels were measured at 0 (before injection), 15, 45 and 60 min after injection.
Adipocyte and myocyte differentiation and Oil-red O staining
Brown and white pre-adipocytes were differentiated and Oil-red O-stained as described (Wang et al., 2010). Briefly, cells were plated at a density of 5 × 105 per 10-cm dish in differentiation medium (DMEM plus 10% FBS, 0.1 μM insulin, and 1 nM T3) 4 days before induction of adipogenesis. At day 0, cells were fully confluent and were treated with differentiation medium supplemented with 0.5 mM 3-isobutyl-1-methyl-xanthine (IBMX), 2 μg/mL dexamethasone, and 0.125 mM indomethacin. Two days later, cells were changed to the differentiation medium. The medium was replenished at 2-day intervals. At day 3–4, small lipid droplets appeared in differentiating cells. At day 6–8 postinduction, fully differentiated cells were either stained with Oil Red O or subjected to gene expression analysis by qRT-PCR or Western blot. Myocyte differentiation and staining were conducted as described (Sun et al., 2011). Briefly, to induce myoblasts for adipogenesis, C2C12 myoblasts or primary human myoblasts were infected as described below, cultured to confluence, and then exposed to brown adipocyte differentiation conditions: 10%FBS DMEM, Insulin 850 nM (Sigma), Dexamethasone 0.5 μM (Sigma), IBMX 250 μM (Sigma), Rosiglitazone 1uM (Cayman Chemical), T3 1 nM (Sigma), and Indomethacin 125 nM (Sigma). After 2 days, cells were incubated in culture medium containing insulin 160nM and Rosiglitazone 1 μM for another 2 days, and then were switched to 10% FBS DMEM. To stimulate thermogenesis, cells were incubated with dibutyryl cAMP at 0.5 mM (Sigma) for 4 h.
Fatty acid beta-oxidation analysis
Fatty acid beta-oxidation was assessed by using 14C-Palmitate with the methods described in Antinozzi et al., (1998) and Watkins et al., (1991).
Cell transfection
Cells were obtained from the ATCC and propagated according to ATCC’s instructions. Transfection of antisense and precursor oligonucleotides (35 nM final concentration) was performed in mammalian cells using Lipofectamine RNAiMAX Reagent (Life Technologies/Invitrogen. Plasmid transfection in HEK293T cells was performed with Lipofectamine 2000 (Life Technologies/Invitrogen) according to the manufacturer’s instructions.
Luciferase assays
Luciferase plasmids harboring the 3’ UTRs of PPARG, PPARA, PPARGC1A and PRDM16 were purchased from GeneCopoeia. Mutagenesis of the 3’ UTRs was performed with the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. Luciferase activity was measured according to the manufacturer’s protocol (Promega, Madison, WI) and was normalized to β-galactosidase activity.
Quantification and statistical analysis
1. In vitro/in vivo experiments
Statistical parameters and details are reported in each figure legend. Generally, in vitro experiments were repeated at least three times and histograms represent mean ± SD unless specified. Luciferase assays were performed in quadruplicate starting from four biological replicates, and error bars represent the SD. qRT-PCR analyses were carried out in triplicate or duplicate sets and the error bars represents the SD. The number and age of animals used in each mouse study follows the pertinent literature of comparable studies in which the desired effect sizes were considered to be significant, and details could be found in each individual figure legend. There were no mice excluded in the analyses. Error bars in mouse studies indicate SEM. For both in vitro and in vivo studies, the unpaired two-sided Student’s t-test was used for statistical differences. P ≤ 0.05 was considered as statistically significant. Data analysis was performed using Excel Software Version 14.4.5.
2. Human quantification and statistical analysis
2.1. GWAS summary statistics
For Figure S1B in which summary statistics were reported, SNPs were merged by rsid with HapMap3 SNPs, filtered for strand ambiguity and alleles being different than in HapMap3, and tested for genomic position and p-value. All reported SNPs within the window were included in the locuszoom plot. Total cholesterol and LDL were downloaded from the GLGC website (http://lipidgenetics.org/); 1,5-Anhydroglucitol from the metabolomics server (http://gwas.eu/); and singleton density score from the Pritchard lab website (http://web.stanford.edu/group/pritchardlab/home.html). Plots were generated in R using ggplot2.
2.2. PheWAS plots
The UK Biobank-wide PheWAS plot was downloaded from the Oxford BIG Server (http://big.stats.ox.ac.uk/, Elliot et al. 2018 = https://pubmed.ncbi.nlm.nih.gov/30305740/). For this analysis, the PheWAS for rs1438307 was directly queried, and the resulting associations were filtered to name relevant traits in the figure.
2.3. Ancient DNA analysis
The ancient DNA analysis with estimated allele frequency trajectories and selection coefficients in ifferent ancient European populations was carried out as described (Mathieson and Mathieson, 2018).
2.4. eQTL analysis
Individuals from our cohorts were compared based on genotype at rs1446585 (the SNP which was present in all datasets). A linear model of the form mir128_Ct ~ control_RNA_Ct + age + sex + batch + Tissue + sex * Tissue + age^2 was set up in R to correct for common confounders in these regressions, to which a term for genotype dosage was added. An ANOVA F-test was used to evaluate significance (p = 0.06). Inverse-variance weighted meta-analysis was used for combining individual dosage test statistics across cohorts.
2.5. Roadmap epigenome and Hi-C analysis
Data from the 127 reference epigenomes was visualized using the WashU epigenome browser focused on the region, together with CTCF in a few cell types. Both Hi-C and Hi-ChIP data, normalized by the browser, were reported as annotated in the WashU browser.
2.6. gnomAD analysis
The chromosome 2 VCF was downloaded from the gnomAD website (http://gnomad.broadinstitute.org/) and reported allele frequency was folded (AF = min(AF, 1-AF)) and subset to either African or non-Finnish European as designated by gnomAD (Karczewski et al., 2020), then plotted using R and ggplot2.
2.7. Expression correlation analysis
Matched expression data for cell lines in GDSC and CCLE were downloaded from CellMinerCDB (https://discover.nci.nih.gov/cellminercdb/) and plotted using R and ggplot2.
2.8. FANTOM analysis
FANTOM time course miRNA data were downloaded from the FANTOM website (http://fantom.gsc.riken.jp/5) and promoters were assigned to genes based on the calls from the miRNA paper (deRie et al., 2017). Expression was plotted over time using ggplot2 in R, with error ribbons as 95% confidence intervals using the loess model.
2.9. METSIM miRNA analysis
The METSIM calls did not distinguish between miR-128-1 and miR-128–2, as only mature miRNA sequences were used. To overcome this, we downloaded the raw METSIM miRNA-seq data from SRA (linked from GEO GSE45159, Civelek et al., 2013), took miRNA hairpin sequences from the miRBase (http://www.mirbase.org/) and aligned to the human orthologs and quantified using Kallisto (Bray et al., 2016). We filtered out any samples with fewer than 500,000 total mapped reads. Clinical data were downloaded from individual GEO pages and used to compare fasting glucose levels to the measured expression with R.
2.10. Read count analyses
Raw BAMs from the CATO project (Maurano et al., 2015: https://resources.altius.org/publications/CATO/) were downloaded and filtered to those with at least 10 reads covering the rs1438307 variant. Lines with 1 or 2 reference or alternative alleles were excluded due to the potential for miscalling. Each line’s position was calculated based on the total reference + alternative read count versus their log2 ratio. Thus, the non-reference, non-alternative reads were ignored, as was the read quality (beyond that which was performed in Maurano et al., 2015). The processed genotype calls were then aggregated by genotype, scaled on a per-line basis by the total read depth in the filtered BAM file. Then each call was scaled by the number of lines with the given genotype to avoid scale differences due to the number of datasets included. Data were output as a bigWig track to the WashU browser after being converted using deeptools at 25 bp resolution.
Supplementary Material
Figure S1. Selection signals at the 2q21.3 locus. Related to Fig. 1.
(A) Composite of multiple signals (CMS) un-normalized selection signal. Normalization (Grossman et al., 2010) corrects for frequency-dependence of CMS values.
(B) 2q21.3 associations with multiple metabolic traits and positive selection. In each case, the axes show the 2 Mb section of chromosome 2 surrounding the positively selected region and the associations of all SNPs in the region. This region associates with total cholesterol, LDL cholesterol, and 1,5-AG. Furthermore, there is a strong signal of positive selection as indicated by singleton density score (Field et al., 2016).
(C) Effect of the lactase persistence allele at rs41380347 on lipid levels in European-ancestry and African-ancestry individuals from UK Biobank (Sinnott-Armstrong, Tanigawa et al., 2019) and the Million Veteran Program (Klarin et al., 2018). 95% confidence intervals shown. (D) Dramatic selection at 2q21.3 was present in European but not African individuals in gnomAD. All variants in gnomAD in the region are showed, with a heatmap of counts at different points in the folded site frequency spectrum. The total count of variants contained within a given bin of frequencies and positions corresponds to the saturation of the pixels in the heatmap.
Figure S2. Allele-specific epigenetic effects of the long European haplotype. Related to Fig. 2.
(A) Top: Positional patterns of CTCF enrichment around UBXN4 promoter, shown as the heatmap in the 3-Kbp proximity of UBXN4 transcription start site (TSS) among the ENCODE samples ranked by the rs1438307 variant frequency. CTCF enrichment in samples with the highest frequency (marked by red bar; see also red boxplot in Fig. 2D) is much higher. Bottom: This stratification of ENCODE samples by CTCF enrichment is not due to a systematic bias towards a stronger genome-wide CTCF signal. Distributions of Z-scores of CTCF enrichment at all peaks genome-wide (shown as boxplots aligned with the heatmap above) are approximately similar in all ENCODE samples, whereas the Z-score for the enrichment of CTCF at the rs1438307 locus (shown as red dots) stands out much stronger in the samples with the higher allelic frequency of the variant. The color bar shows the SNP allele frequency in individual ENCODE samples, estimated from the CTCF ChIP-seq reads at the rs1438307 locus. (B) Heatmaps of chromatin interactions in the genomic region of interest, derived from a high-resolution Hi-C experiment in the human GM12878 cell line (Rao et al., 2014). Hi-C chromatin domain contains the variant, confirming the effect shown in HiChIP. The topological domain structure does not appear to be an artifact of the cohesin pull-down used in the HiChIP assay. (C) Heatmap showing positional profiles of H3K4me3 ChIP-seq density (normalized by input) profile in the 3-Kbp proximity of UBXN4 transcription start site (TSS) among the ENCODE samples ranked by the rs1438307 variant frequency. (D) Accessibility in the promoter region upstream of miR-128-1 is altered by genotype at rs1438307. Normalized accessibility by the total read depth was averaged for each of the lines with a given genotype at rs1438307 and the window around the miR-128-1 gene was plotted. Just upstream of miR-128-1, there is an accessibility peak corresponding to the promoter. This peak is genotype dependent, with the derived T allele at rs1438307 having the highest average accessibility.
Figure S3. Role of miR-128-1 expression in regulation of metabolism in humans. Related to Fig. 2.
(A) Expression of miR-128-1 is higher in visceral than subcutaneous adipose tissue. Using all individuals from our starting cohort and correcting for Snord48, age, sex, and batch, there is substantially less expression of miR-128-1 in subcutaneous adipose tissue than visceral. (B) This effect was confirmed in our replication cohort of matched individuals. (C) Haplotype is a weak eQTL for miR-128-1 expression in adipose tissue. Samples were quantified for miR-128-1 and a trend was observed across cohorts (Methods). (D-F) Our well-phenotyped replication cohort has significant associations between miR-128-1 expression and fasting glucose (D), HbA1c (E), and Triacylglycerol/HDL (F). (G) The fasting glucose association is reproduced in the METSIM cohort, as assayed using miRNA-seq. (H) miR-128-1 expression goes up substantially over time in the FANTOM5 time course expression data. Secondary association at 2q21.3 has a high posterior probability placed on rs1446585 as being causal for Apolipoprotein A1 levels (I) and HDL-cholesterol levels (J), as inferred by FINEMAP by the Finucane lab.
Figure S4. Pharmacological inhibition of miR-128-1 by anti-miR-128-1 prevents diet-induced obesity (DIO), reduces adipocyte hypertrophy and inflammation, prevents liver steatosis and increases whole body energy expenditure and improves glucose homeostasis. Related to Fig. 3.
(A) Adipocyte miR-128-1 levels were determined by qRT-PCR in DIO mice treated with anti-miR-128-1 (n = 10 per group). (B) food intake of anti-miR-128-1 and control anti-miR treated DIO mice. Plasma triglycerides (C) and cholesterol (D) were determined by FPLC analysis. Expression of selected marker genes related to (E) BAT identity, (F) WAT inflammation, and (G) liver steatosis was determined by western blotting. Differences between the anti-miR-128-1 and control groups (n=10 per group) in whole body oxygen consumption (H) and activity (I, J) were measured using metabolic cages. (K) Luciferase activity in HEK293T cells transfected with 3′ UTR-luciferase reporters for human PPARGC1, PPARG, PPARA and PRDM16 or reporters harboring point mutations in the predicted miRNA target sites, co-transfected with miR-128-1 precursor. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to mice injected with the control ASO.
Figure S6. Analysis of gene expression in miR-128-1 KO mice and wild-type controls. Related to Fig. 5.
Quantitative RT-PCR analysis of expression of selected genes involved in metabolic control and inflammation in (A) BAT, (B) WAT, (C) skeletal muscle and (D) liver in wild-type and miR-128-1 KO mice. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to WT control mice.
Figure S5. Pharmacological inhibition of miR-128-1 in leptin-deficient (ob/ob) mice prevents adiposity, reduces adipocyte hypertrophy and inflammation, increases the brown appearance of BAT, prevents liver steatosis and inflammation, increases whole body energy expenditure and improves glucose homeostasis. Related to Fig. 3.
(A) Body weight of ob/ob mice treated with anti-miR-128-1 and control anti-miR. (B) Body composition and liver and epididymal WAT from ob/ob mice treated with anti-miR-128-1 and control anti-miR (n=10 per group). (C) Whole body energy expenditure differences were measured in metabolic cages between the anti-miR-128-1 and control group (n=10 per group). Representative histology data (H&E) of BAT (D), EPI-WAT (F) and liver (H) from the anti-miR-128-1 treated group were compared to the control group (n=6 per group). Expression of marker genes related to (E) BAT identity and energy expenditure (G) WAT hypertrophy and inflammation, and (I) liver steatosis and energy expenditure were determined by quantitative RT-PCR (qRT-PCR) from the ob/ob mice. Intraperitoneal (IP) glucose tolerance (J) and insulin tolerance tests (K) were performed after 10 weeks of anti-miR-128-1 and control anti-miR treatment. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to mice injected with the control ASO.
Figure S7. Decreased hypertrophy and increased browning of subcutaneous (inguinal/iWAT) WAT in DIO mice treated with anti-miR-128-1. Related to Figs. 3 and 7.
(A) Representative H&E-stained images of inguinal WAT from DIO mice treated with anti-miR-128-1 for 16 weeks. (B-D) Lowered expression of lipogenic genes such as Srebf1 (B), Fasn (C), and Acc1 (D) in iWAT from mice treated with anti-miR-128-1. (E-J) Increased expression of beneficial WAT genes (E) Pparg, (F) Adipoq, and energy expenditure/browning genes (G) Ppargc1a, (H) Prdm16, (I) Ppara, and (J) Ucp1 in anti-miR-128-1 treated mice. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to mice injected with the control ASO.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| UCP1 | Abcam | ab23841 |
| F4/80 | Novus biologicals | NB600-404 |
| TNF alpha | Abcam | ab6671 |
| CD36 | Abcam | ab133625 |
| PGC1 alpha | Abcam | ab54481 |
| CPT1B | Abcam | ab134988 |
| PPAR alpha | Abcam | ab24509 |
| UCP3 | Abcam | ab3477 |
| Insulin Receptor | Abcam | ab137747 |
| IRS1 | Abcam | ab52167 |
| β-Tubulin | Sigma | T8328 |
| AMPK alpha 2 | Abcam | ab3760 |
| AMPK alpha 2 (phospho S491) | Abcam | ab109402 |
| PPAR gamma | Abcam | ab45036 |
| Acetyl-CoA Carboxylase | Cell Signaling | #3662 |
| Phospho-Acetyl-CoA Carboxylase | Cell Signaling | #3661 |
| PRDM16 | Abcam | ab202344 |
| SIRT1 | Abcam | ab12193 |
| IL-1β (3A6) | Cell Signaling | #12242 |
| SREBP1 | Novus biologicals | NB100-2215 |
| SREBP1 (Phospho-S439) | Biorbyt | orb315711 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Taqman PCR Master Mix, no AmpErase UNG | Applied Biosystems | Cat# 4324018 |
| PALMITIC ACID, [1-14C]-, 50 μCi |
Perkin Elmer | NEC075H050UC |
| Dexamethasone | Sigma | D4902 |
| Indomethacin | Sigma | I7378 |
| 3-Isobutyl-1-methylxanthine | Sigma | I7018 |
| Insulin | Sigma | I2643 |
| Oil Red O | Sigma | O0625 |
| Dibutyryl cyclic-AMP | Sigma | D0260 |
| AICAR | Sigma | A9978 |
| Critical Commercial Assays | ||
| Dual-Luciferase® Reporter | Promega | E1910 |
| Assay System | ||
| Transcriptor reverse transcriptase | Roche | Cat# 03531287001 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| Pierce BCA Protein Assay Kit | Thermo Scientific | Cat# 23225 |
| Seahorse Bioassay | Agilent Technologies | Cat# 102416 |
| QuikChange™ Site-Directed Mutagenesis Kit | Agilent Technologies | Cat# 200518 |
| Experimental Models: Cell Lines and Primary cells | ||
| HepG2 | ATCC | ATCC® HB-8065™ |
| 3T3L1 | Dr. Kai Ge | ATCC® CL-173™ |
| C2C12 | ATCC | ATCC® CRL-1772™ |
| Brown preadipocytes | Dr. Kai Ge | Wang et al., 2010 |
| Primary human white preadipocyte | Dr. Susan K. Fried | |
| Primary preadipocytes | This study | |
| Recombinant DNA | ||
| PPARg | GeneCopoeia | MmiT054223 |
| PPARa | GeneCopoeia | MmiT029952 |
| PGC1a | GeneCopoeia | MmiT055331 |
| PRDM16 | GeneCopoeia | MmiT035098 |
| Oligonucleotides | ||
| PPARa-3’ UTR muta | cagagtggtttgtttgtttgtttttgttttaaaaatcgtatcattaagatatagctatgaagagcattctttttcattc gaatgaaaaagaatgctcttcatagctatatcttaatgatacgatttttaaaacaaaaacaaacaaacaaaccactctg | |
| PGC1a-3’UTR muta | Atattttgatgggctacccatcgcttctgcataagatacacaagcttcactaattttagatgca tgcatctaaaattagtgaagcttgtgtatcttatgcagaagcgatgggtagcccatcaaaatat | |
| PPARg-3’UTR muta | Ctaaaactttttgtttttaaatgctttttcatattaaatttcttaggtgtcagatttttttccctcaaaataat attattttgagggaaaaaaatctgacacctaagaaatttaatatgaaaaagcatttaaaaacaaaaagttttag | |
| Prdm16-3’UTR muta | gactgtccctagccctgagcgatgataggacaagacattcttgaattctc gagaattcaagaatgtcttgtcctatcatcgctcagggctagggacagtc | |
Highlights.
The long haplotype at 2q21.3 is linked to positive selection and metabolic disorders
The miR-128-1 microRNA is located at the center of the 2q21.3 locus
miR-128-1 controls energy expenditure and contributes to metabolic disease
Ancient adaptation to famine and metabolic diseases may be linked through miR-128-1
Acknowledgements
This work was supported by grants from the NIH (R01DK094184 and R01DK114277; both to A.M.N.). It was also supported by a grant to A.M.N. from the Boston Area Diabetes Endocrinology Research Center (P30 DK057521). A.M.N. was supported by an MGH Research Scholarship and by institutional funds at the University of California, Berkeley. L.W. and A.W. were supported by MGH ECOR Fund for Medical Discovery. A.R.W. was supported by NIH R01HD03443 (Tabin). S.K. and A.P. were supported by the Lundbeck Foundation and the Novo Nordisk Foundation. N.S.-A. was funded by the U.S. DoD through a NDSE Grant and by a Stanford Graduate Fellowship. M.C., A.S., were supported by the Broad Institute Next Generation award. S.N.D. and D.S.P.T. were supported by the Norwegian Research Council (263124/F20). Studies performed at the Yale Diabetes Research Center were supported by NIH grants P30 DK045735, R01 DK116774, R01 DK119968, R01DK114793 (G.I.S.). We thank Iain Mathieson for access to the ancient DNA data sets used for the analysis in Fig. 1D, Clifford Tabin for support of the mouse miR-128-1 KO generation and helpful guidance, the Western Norway Research Biobank for Overweight (VFO) for adipose tissue samples, and Manuel Rivas for assisting with the UK Biobank (Application #24983). The authors also thank Dylan Bennett, Xiaoxian Ma, Ali Nasiri, and Gina Butrico for technical assistance, and Sharon Grossman, Aaron Stern, Priya Moorjani, Joseph Vitti, Layla Siraj, and Iain Mathieson for helpful comments.
Footnotes
Declaration of Interests
A.M.N. has issued patents on miR-128-1 (U.S. Pat. Nos. 9,045,749; 9,476,046; 9,789,132).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahfeldt T, Schinzel RT, Lee YK, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, et al. (2012). Programming human pluripotent stem cells into white and brown adipocytes. Nature Cell Biol 14, 209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque D, Nobrega C, and Manco L. (2013). The lactase persistence −13910C>T polymorphism shows indication of association with abdominal obesity among Portuguese children. Acta paediatrica 102, e153–157. [DOI] [PubMed] [Google Scholar]
- Allison DB, Kaprio J, Korkeila M, Koskenvuo M, Neale MC, and Hayakawa K. (1996). The heritability of body mass index among an international sample of monozygotic twins reared apart. Int J Obes Relat Metab Disord 20, 501–6. [PubMed] [Google Scholar]
- Almon R, Alvarez-Leon EE, and Serra-Majem L. (2012). Association of the European lactase persistence variant (LCT-13910 C>T polymorphism) with obesity in the Canary Islands. PLoS One 7, e43978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. (2004). The functions of animal microRNAs. Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Antinozzi PA, Segall L, Prentki M, McGarry JD, and Newgard CB (1998). Molecular or pharmacologic perturbation of the link between glucose and lipid metabolism is without effect on glucose-stimulated insulin secretion. A re-evaluation of the long-chain acyl-CoA hypothesis. J Biol Chem 273, 16146–16154. [DOI] [PubMed] [Google Scholar]
- Barendse W, Harrison BE, Bunch RJ, Thomas MB, and Turner LB (2009). Genome wide signatures of positive selection: the comparison of independent samples and the identification of regions associated to traits. BMC genomics 10, 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer RC, Stylianou IM, and Rader DJ (2011). Functional validation of new pathways in lipoprotein metabolism identified by human genetics. Curr Opin Lipidol 22, 123–128. [DOI] [PubMed] [Google Scholar]
- Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, and Hirschhorn JN (2004). Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 74, 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovine HapMap C, Gibbs RA, Taylor JF, Van Tassell CP, Barendse W, Eversole KA, Gill CA, Green RD, Hamernik DL, Kappes SM, et al. (2009). Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324, 528–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–7. [DOI] [PubMed] [Google Scholar]
- Camporez JP, Wang Y, Faarkrog K, Chukijrungroat N, Petersen KF, and Shulman GI. (2017). Mechanism by which arylamine N-acetyltransferase 1 ablation causes insulin resistance in mice. Proc Natl Acad Sci U S A 114, E11285–E11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, and Nedergaard J. (2004). Brown adipose tissue: function and physiological significance. Physiol Rev 84, 277–359. [DOI] [PubMed] [Google Scholar]
- Civelek M, Hagopian R, Pan C, Che N, Yang WP, Kayne PS, Saleem NK, Cederberg H, Kuusisto J, Gargalovic PS, et al. (2013). Genetic regulation of human adipose microRNA expression and its consequences for metabolic traits. Hum Mol Genet 22, 3023–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Klocke B, Grallert H, Glunk V, Berulava T, Lee H, Oskolkov N, Fadista J, Ehlers K, et al. (2014). Leveraging cross-species transcription factor binding site patterns: from diabetes risk loci to disease mechanisms. Cell 156, 343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DL (1979). Obesity genes: beneficial effects in heterozygous mice. Science 203, 663–5. [DOI] [PubMed] [Google Scholar]
- Corella D, Arregui M, Coltell O, Portoles O, Guillem-Saiz P, Carrasco P, Sorli JV, Ortega-Azorin C, Gonzalez JI, and Ordovas JM (2011). Association of the LCT-13910C>T polymorphism with obesity and its modulation by dairy products in a Mediterranean population. Obesity 19, 1707–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornier MA, Dabelea D, Hernandez TL, Lindstrom RC, Steig AJ, Stob NR, Van Pelt RE, Wang H, and Eckel RH (2008). The metabolic syndrome. Endocr Rev 29, 777–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry A. (2013). Archaeology: The milk revolution. Nature 500, 20–22. [DOI] [PubMed] [Google Scholar]
- de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, Ashoor H, Åström G, Babina M, Bertin N, Burroughs AM, et al. (2017). An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol 35, 872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Boyle EA, Telis N, Gao Z, Gaulton KJ, Golan D, Yengo L, Rocheleau G, Froguel P, McCarthy MI, et al. (2016). Detection of human adaptation during the past 2000 years. Science 354, 760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatz G, and Rotthauwe HW (1973). Lactose nutrition and natural selection. Lancet 2, 76–7. [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, and Bernstein BE (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GBD 2015 Obesity Collaborators, Afshin A, Forouzanfar MH, Reitsma MB, Sur P, Estep K, Lee A, Marczak L, Mokdad AH, Moradi-Lakeh M. et al. (2017). Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N Engl J Med 377, 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbault P, Liebert A, Itan Y, Powell A, Currat M, Burger J, Swallow DM, and Thomas MG (2011). Evolution of lactase persistence: an example of human niche construction. Philos Trans R Soc Lond B Biol Sci 366, 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedeke L, Wagschal A, Fernandez-Hernando C, and Näär AM (2016). miRNA regulation of LDL-cholesterol metabolism. Biochim Biophys Acta 1861, 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. (2013). Identifying recent adaptations in large-scale genomic data. Cell 152, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Shlyakhter I, Karlsson EK, Byrne EH, Morales S, Frieden G, Hostetter E, Angelino E, Garber M, Zuk O, et al. (2010). A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327, 883–886. [DOI] [PubMed] [Google Scholar]
- Gupta V, Vinay DG, Sovio U, Rafiq S, Kranthi Kumar MV, Janipalli CS, Evans D, Mani KR, Sandeep MN, Taylor A, et al. (2013). Association study of 25 type 2 diabetes related Loci with measures of obesity in Indian sib pairs. PLoS One 8, e53944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard-Costa NL, Zillikens MC, Monda KL, Johansson A, Harris TB, Fu M, Haritunians T, Feitosa MF, Aspelund T, Eiriksdottir G, et al. (2009). NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet 5, e1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E, et al. (2013). Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res 41, 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, and Brown MS (2002). SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, Shargill NS, and Spiegelman BM (1993). Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 259, 87–91. [DOI] [PubMed] [Google Scholar]
- Itan Y, Powell A, Beaumont MA, Burger J, and Thomas MG (2009). The origins of lactase persistence in Europe. PLoS Comput Biol 5, e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong C, Wilkin S, Amgalantugs T, Bouwman AS, Taylor WTT, Hagan RW, Bromage S, Tsolmon S, Trachsel C, Grossmann J, et al. (2018). Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe. Proc Natl Acad Sci U S A 115, E11248–E11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczak MJ, Lee AH, Jornayvaz FR, Lee HY, Birkenfeld AL, Guigni BA, Kahn M, Samuel VT, Glimcher LH, and Shulman GI (2012). Dissociation of inositol-requiring enzyme (IRE1α)-mediated c-Jun N-terminal kinase activation from hepatic insulin resistance in conditional X-box-binding protein-1 (XBP1) knockout mice. J Biol Chem 287, 2558–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn SE, Hull RL, and Utzschneider KM (2006). Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444, 840–846. [DOI] [PubMed] [Google Scholar]
- Kajimura S, Spiegelman BM, and Seale P. (2015). Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab 22, 546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe F, Dickmann JR, and Frayn KN (2011). Fatty acids, obesity, and insulin resistance: time for a reevaluation. Diabetes 60, 2441–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Snoek J, and Rinn JL (2016). Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res 26, 990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettunen J, Silander K, Saarela O, Amin N, Muller M, Timpson N, Surakka I, Ripatti S, Laitinen J, Hartikainen AL, et al. (2010). European lactase persistence genotype shows evidence of association with increase in body mass index. Hum Mol Genet 19, 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK (2009). Hyperinsulinemic-euglycemic clamp to assess insulin sensitivity in vivo. Methods Mol Biol 560, 221–238. [DOI] [PubMed] [Google Scholar]
- Kim WJ, and Park CY (2013). 1,5-Anhydroglucitol in diabetes mellitus. Endocrine 43, 33–40. [DOI] [PubMed] [Google Scholar]
- Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM et al. (2018). Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet 50, 1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles JW, Xie W, Zhang Z, Chennamsetty I, Assimes TL, Paananen J, Hansson O, Pankow J, Goodarzi MO, and Carcamo-Orive I. (2015). Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. J Clin Invest 125, 1739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Jeong KH, Choi CS, and International Mouse Phenotyping C. (2014). In-depth metabolic phenotyping of genetically engineered mouse models in obesity and diabetes. Mamm Genome 25, 508–521. [DOI] [PubMed] [Google Scholar]
- Lee MJ and Fried SK (2014). Optimal protocol for the differentiation and metabolic analysis of human adipose stromal cells. Methods Enzymol 538, 49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsky RH, Jensen TG, Moller J, Stensballe A, Olsen J, and Troelsen JT (2005). T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet 14, 3945–3953. [DOI] [PubMed] [Google Scholar]
- Lillie RD and Fulmer HM (1976). Histopathologic Technique and Practical Histochemistry. 4th ed. New York: McGraw-Hill. pp. 559–610. [Google Scholar]
- Ma L, Yang J, Runesha HB, Tanaka T, Ferrucci L, Bandinelli S, and Da Y. (2010). Genome-wide association analysis of total cholesterol and high-density lipoprotein cholesterol levels using the Framingham heart study data. BMC Med Genet 11, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes HH, Neale MC, and Eaves LJ (1997). Genetic and environmental factors in relative body weight and human adiposity. Behav Genet 27, 325–51. [DOI] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M. et al. (2015). Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson S, and Mathieson I. (2018). FADS1 and the Timing of Human Adaptation to Agriculture. Mol Biol Evol 35, 2957–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C, Wang H, Liao Q, Wang L, Cheng G, Wang H, Zhao C, Zhao S, Song J, Guang X. et al. (2018). Genetic Architecture and Selection of Chinese Cattle Revealed by Whole Genome Resequencing. Mol Biol Evol 35, 688–699. [DOI] [PubMed] [Google Scholar]
- Meier U, and Gressner AM (2004). Endocrine regulation of energy metabolism: review of pathobiochemical and clinical chemical aspects of leptin, ghrelin, adiponectin, and resistin. Clin Chem 50, 1511–1525. [DOI] [PubMed] [Google Scholar]
- Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, and Chang HY (2016). HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nature Methods 13, 919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, and Näär AM (2010). MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel JV (1962). Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet 14, 353–362. [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, and Chawla A. (2008). Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat Clin Pract Endocrinol Metab 4, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Ohyama K, Sharp LZ, and Kajimura S. (2013). EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 504, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Shinoda K, Spiegelman BM, and Kajimura S. (2012). PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab 15, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak M, Lefebvre P, and Staels B. (2015). Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol 62, 720–733. [DOI] [PubMed] [Google Scholar]
- Perry RJ, Camporez JG, Kursawe R, Titchenell PM, Zhang D, Perry CJ, Jurczak MJ, Abudukadier A, Han MS, Zhang XM, et al. (2015). Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell 160, 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, Peng L, Cline GW, Wang Y, Rabin-Court A, Song JD, Zhang D, Zhang XM, Nozaki Y, Dufour S, et al. (2018). Mechanisms by which a Very-Low-Calorie Diet Reverses Hyperglycemia in a Rat Model of Type 2 Diabetes. Cell Metab 27, 210–217 e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassais J, Kim J, Davis BW, Karyadi DM, Hogan AN, Harris AC, Decker B, Parker HG, and Ostrander EA (2019). Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat Commun 10, 1489. doi: 10.1038/s41467-019-09373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum M, and Leibel RL (1998). Leptin: a molecule integrating somatic energy stores, energy expenditure and fertility. Trends Endocrinol Metab 9, 117–24. [DOI] [PubMed] [Google Scholar]
- Rottiers V, and Näär AM (2012). MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rung J, Cauchi S, Albrechtsen A, Shen L, Rocheleau G, Cavalcanti-Proenca C, Bacot F, Balkau B, Belisle A, Borch-Johnsen K, et al. (2009). Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat Genet 41, 1110–1115. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. (2002). Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. (2007). Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salque M, Bogucki PI, Pyzel J, Sobkowiak-Tabaka I, Grygiel R, Szmyt M, and Evershed RP (2013). Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature 493, 522–525. [DOI] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J, and Guertin DA (2014). Adipocyte lineages: tracing back the origins of fat. Biochim Biophys Acta 1842, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, Muller TD, Grallert H, Wichmann HE, Balkau B, et al. (2010). Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genetics 6, e1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. (2008). PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, and Spiegelman BM (2011). Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 121, 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, and Spiegelman BM (2007). Transcriptional control of brown fat determination by PRDM16. Cell Metab 6, 38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurel L, and Bon C. (2017). On the Evolution of Lactase Persistence in Humans. Annu Rev Genomics Hum Genet 18, 297–319. [DOI] [PubMed] [Google Scholar]
- Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, et al. (2014). An atlas of genetic influences on human blood metabolites. Nat Genet 46, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GI (2014). Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. New Engl J of Med 371, 1131–1141. [DOI] [PubMed] [Google Scholar]
- Shulman GI (2000). Cellular mechanisms of insulin resistance. J Clin Invest. 106, 171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott Armstrong N, Tanigawa Y, Amar D, Mars NJ, Aguirre M, Venkataraman GR, Wainberg M, Ollila HM, Pirruccello JP, Qian J. et al. (2019). Genetics of 38 blood and urine biomarkers in the UK Biobank. bioRxiv preprint doi: 10.1101/660506. [DOI] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. (2007). A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445, 881–885. [DOI] [PubMed] [Google Scholar]
- Soukas A, Cohen P, Socci ND, and Friedman JM (2000). Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev 14, 963–980. [PMC free article] [PubMed] [Google Scholar]
- Speakman JR (2013). Evolutionary perspectives on the obesity epidemic: adaptive, maladaptive, and neutral viewpoints. Annu Rev Nutr 33, 289–317. [DOI] [PubMed] [Google Scholar]
- Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, Altmaier E, CardioGram, Deloukas P, Erdmann J, et al. (2011). Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Kusminski CM, and Scherer PE (2011a). Adipose tissue remodeling and obesity. J Clin Invest 121, 2094–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, and Lodish HF (2011b). Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol 13, 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, and Diehl AM (2017). Nonalcoholic Steatohepatitis. Annual Review of Medicine 68, 85–98. [DOI] [PubMed] [Google Scholar]
- Sverrisdottir OO, Timpson A, Toombs J, Lecoeur C, Froguel P, Carretero JM, Arsuaga Ferreras JL, Gotherstrom A, and Thomas MG (2014). Direct estimates of natural selection in Iberia indicate calcium absorption was not the only driver of lactase persistence in Europe. Mol Biol Evol 31, 975–983. [DOI] [PubMed] [Google Scholar]
- Swallow DM (2003). Genetics of lactase persistence and lactose intolerance. Annu Rev Genet 37, 197–219. [DOI] [PubMed] [Google Scholar]
- Tan CL, Plotkin JL, Veno MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O’Carroll D, et al. (2013). MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342, 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thabit H, Kumareswaran K, Haidar A, Leelarathna L, Caldwell K, Elleri D, Allen JM, Nodale M, Wilinska ME, Jackson NC, et al. (2014). Glucose turnover after replacement of usual therapy by insulin in insulin-naive type 2 diabetes subjects. J Clin Endocrinol Metab 99, 2225–2232. [DOI] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, and Spiegelman BM (2006). Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab 3, 333–341. [DOI] [PubMed] [Google Scholar]
- Wagschal A, Najafi-Shoushtari SH, Wang L, Goedeke L, Sinha S, deLemos AS, Black JC, Ramirez CM, Li Y, Tewhey R, et al. (2015). Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat Med 21, 1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chandrasekera PC, and Pippin JJ (2014). Leptin- and leptin receptor-deficient rodent models: relevance for human type 2 diabetes. Curr Diabetes Rev 10, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L Jin Q, Lee J, Su I, and Ge K. (2010). Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis PNAS 2010 107 (16) 7317–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, and Speakman JR (2016). Analysis of Positive Selection at Single Nucleotide Polymorphisms Associated with Body Mass Index Does Not Support the “Thrifty Gene” Hypothesis. Cell Metab 24, 531–541. [DOI] [PubMed] [Google Scholar]
- Wark AR, Terman EJ, and Tabin CJ (2017). miR-128-1 is not required for hair pigmentation in mice. Exp Dermatol 26, 940–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA, Ferrell EV Jr, Pedersen JI, and Hoefler G (1991). Peroxisomal fatty acid beta-oxidation in HepG2 cells. Arch Biochem Biophys 289, 329–36. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, and Ferrante AW Jr. (2003). Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112, 1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensveen FM, Valentic S, Sestan M, Turk Wensveen T, and Polic B. (2015). The “Big Bang” in obese fat: Events initiating obesity-induced adipose tissue inflammation. Eur J Immunol 45, 2446–2456. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. (2013). Discovery and refinement of loci associated with lipid levels. Nat Genet 45, 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JH, and Buchanan TA (1993). Fasting does not impair insulin-stimulated glucose uptake but alters intracellular glucose metabolism in conscious rats. Diabetes 42, 757–63. [DOI] [PubMed] [Google Scholar]
- Zhou J, and Troyanskaya OG (2015). Predicting effects of noncoding variants with deep learning-based sequence model. Nature Methods 12, 931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X and Wang T. (2012). Using the Wash U Epigenome Browser to examine genome-wide sequencing data. Curr Protoc Bioinformatics Chapter 10, Unit10.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Selection signals at the 2q21.3 locus. Related to Fig. 1.
(A) Composite of multiple signals (CMS) un-normalized selection signal. Normalization (Grossman et al., 2010) corrects for frequency-dependence of CMS values.
(B) 2q21.3 associations with multiple metabolic traits and positive selection. In each case, the axes show the 2 Mb section of chromosome 2 surrounding the positively selected region and the associations of all SNPs in the region. This region associates with total cholesterol, LDL cholesterol, and 1,5-AG. Furthermore, there is a strong signal of positive selection as indicated by singleton density score (Field et al., 2016).
(C) Effect of the lactase persistence allele at rs41380347 on lipid levels in European-ancestry and African-ancestry individuals from UK Biobank (Sinnott-Armstrong, Tanigawa et al., 2019) and the Million Veteran Program (Klarin et al., 2018). 95% confidence intervals shown. (D) Dramatic selection at 2q21.3 was present in European but not African individuals in gnomAD. All variants in gnomAD in the region are showed, with a heatmap of counts at different points in the folded site frequency spectrum. The total count of variants contained within a given bin of frequencies and positions corresponds to the saturation of the pixels in the heatmap.
Figure S2. Allele-specific epigenetic effects of the long European haplotype. Related to Fig. 2.
(A) Top: Positional patterns of CTCF enrichment around UBXN4 promoter, shown as the heatmap in the 3-Kbp proximity of UBXN4 transcription start site (TSS) among the ENCODE samples ranked by the rs1438307 variant frequency. CTCF enrichment in samples with the highest frequency (marked by red bar; see also red boxplot in Fig. 2D) is much higher. Bottom: This stratification of ENCODE samples by CTCF enrichment is not due to a systematic bias towards a stronger genome-wide CTCF signal. Distributions of Z-scores of CTCF enrichment at all peaks genome-wide (shown as boxplots aligned with the heatmap above) are approximately similar in all ENCODE samples, whereas the Z-score for the enrichment of CTCF at the rs1438307 locus (shown as red dots) stands out much stronger in the samples with the higher allelic frequency of the variant. The color bar shows the SNP allele frequency in individual ENCODE samples, estimated from the CTCF ChIP-seq reads at the rs1438307 locus. (B) Heatmaps of chromatin interactions in the genomic region of interest, derived from a high-resolution Hi-C experiment in the human GM12878 cell line (Rao et al., 2014). Hi-C chromatin domain contains the variant, confirming the effect shown in HiChIP. The topological domain structure does not appear to be an artifact of the cohesin pull-down used in the HiChIP assay. (C) Heatmap showing positional profiles of H3K4me3 ChIP-seq density (normalized by input) profile in the 3-Kbp proximity of UBXN4 transcription start site (TSS) among the ENCODE samples ranked by the rs1438307 variant frequency. (D) Accessibility in the promoter region upstream of miR-128-1 is altered by genotype at rs1438307. Normalized accessibility by the total read depth was averaged for each of the lines with a given genotype at rs1438307 and the window around the miR-128-1 gene was plotted. Just upstream of miR-128-1, there is an accessibility peak corresponding to the promoter. This peak is genotype dependent, with the derived T allele at rs1438307 having the highest average accessibility.
Figure S3. Role of miR-128-1 expression in regulation of metabolism in humans. Related to Fig. 2.
(A) Expression of miR-128-1 is higher in visceral than subcutaneous adipose tissue. Using all individuals from our starting cohort and correcting for Snord48, age, sex, and batch, there is substantially less expression of miR-128-1 in subcutaneous adipose tissue than visceral. (B) This effect was confirmed in our replication cohort of matched individuals. (C) Haplotype is a weak eQTL for miR-128-1 expression in adipose tissue. Samples were quantified for miR-128-1 and a trend was observed across cohorts (Methods). (D-F) Our well-phenotyped replication cohort has significant associations between miR-128-1 expression and fasting glucose (D), HbA1c (E), and Triacylglycerol/HDL (F). (G) The fasting glucose association is reproduced in the METSIM cohort, as assayed using miRNA-seq. (H) miR-128-1 expression goes up substantially over time in the FANTOM5 time course expression data. Secondary association at 2q21.3 has a high posterior probability placed on rs1446585 as being causal for Apolipoprotein A1 levels (I) and HDL-cholesterol levels (J), as inferred by FINEMAP by the Finucane lab.
Figure S4. Pharmacological inhibition of miR-128-1 by anti-miR-128-1 prevents diet-induced obesity (DIO), reduces adipocyte hypertrophy and inflammation, prevents liver steatosis and increases whole body energy expenditure and improves glucose homeostasis. Related to Fig. 3.
(A) Adipocyte miR-128-1 levels were determined by qRT-PCR in DIO mice treated with anti-miR-128-1 (n = 10 per group). (B) food intake of anti-miR-128-1 and control anti-miR treated DIO mice. Plasma triglycerides (C) and cholesterol (D) were determined by FPLC analysis. Expression of selected marker genes related to (E) BAT identity, (F) WAT inflammation, and (G) liver steatosis was determined by western blotting. Differences between the anti-miR-128-1 and control groups (n=10 per group) in whole body oxygen consumption (H) and activity (I, J) were measured using metabolic cages. (K) Luciferase activity in HEK293T cells transfected with 3′ UTR-luciferase reporters for human PPARGC1, PPARG, PPARA and PRDM16 or reporters harboring point mutations in the predicted miRNA target sites, co-transfected with miR-128-1 precursor. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to mice injected with the control ASO.
Figure S6. Analysis of gene expression in miR-128-1 KO mice and wild-type controls. Related to Fig. 5.
Quantitative RT-PCR analysis of expression of selected genes involved in metabolic control and inflammation in (A) BAT, (B) WAT, (C) skeletal muscle and (D) liver in wild-type and miR-128-1 KO mice. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to WT control mice.
Figure S5. Pharmacological inhibition of miR-128-1 in leptin-deficient (ob/ob) mice prevents adiposity, reduces adipocyte hypertrophy and inflammation, increases the brown appearance of BAT, prevents liver steatosis and inflammation, increases whole body energy expenditure and improves glucose homeostasis. Related to Fig. 3.
(A) Body weight of ob/ob mice treated with anti-miR-128-1 and control anti-miR. (B) Body composition and liver and epididymal WAT from ob/ob mice treated with anti-miR-128-1 and control anti-miR (n=10 per group). (C) Whole body energy expenditure differences were measured in metabolic cages between the anti-miR-128-1 and control group (n=10 per group). Representative histology data (H&E) of BAT (D), EPI-WAT (F) and liver (H) from the anti-miR-128-1 treated group were compared to the control group (n=6 per group). Expression of marker genes related to (E) BAT identity and energy expenditure (G) WAT hypertrophy and inflammation, and (I) liver steatosis and energy expenditure were determined by quantitative RT-PCR (qRT-PCR) from the ob/ob mice. Intraperitoneal (IP) glucose tolerance (J) and insulin tolerance tests (K) were performed after 10 weeks of anti-miR-128-1 and control anti-miR treatment. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to mice injected with the control ASO.
Figure S7. Decreased hypertrophy and increased browning of subcutaneous (inguinal/iWAT) WAT in DIO mice treated with anti-miR-128-1. Related to Figs. 3 and 7.
(A) Representative H&E-stained images of inguinal WAT from DIO mice treated with anti-miR-128-1 for 16 weeks. (B-D) Lowered expression of lipogenic genes such as Srebf1 (B), Fasn (C), and Acc1 (D) in iWAT from mice treated with anti-miR-128-1. (E-J) Increased expression of beneficial WAT genes (E) Pparg, (F) Adipoq, and energy expenditure/browning genes (G) Ppargc1a, (H) Prdm16, (I) Ppara, and (J) Ucp1 in anti-miR-128-1 treated mice. Error bars represent SEM. Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to mice injected with the control ASO.
Data Availability Statement
Data sets analyzed: The code supporting the current study has not been deposited in a public repository because no novel algorithms or methods were employed, but is available from the corresponding author on request. Data employed include GWAS summary statistics and fine-mapped variants derived from the UK Biobank resource, Million Veteran Program, Global Lipids Genetics Consortium, and numerous other individual supporting studies; whole-genome sequencing data from gnomAD; epigenetic data from the Roadmap Epigenomics and ENCODE projects; and expression data from GDSC, CCLE, FANTOM5, ENCODE, and Civelek et al., 2013. Selection scores were derived by the Sabeti, Pritchard, and Ostrander labs, as well as the Bovine Hapmap Consortium. Underlying data from human subjects used in this study are publicly available, either without restriction or through application to dbGaP or the UK Biobank. Restrictions exist on access to underlying data from non-human studies.