Significance
Two deadly human retroviruses, human T cell leukemia virus type 1 (HTLV-1) and HIV type 1 (HIV-1), enter latency in vivo, rendering viral countermeasures ineffective. Recently, novel retroviral genes have been discovered to be expressed from the antisense strand of retroviruses even during latency; they are called antisense genes, including the HBZ gene for HTLV-1 and ASP gene for HIV-1. We employed RNA-fluorescence in situ hybridization technology and discovered that human retroviral antisense messenger RNAs (mRNAs) are predominantly localized in the nucleus of infected cells, despite their coding function. Moreover, human retroviral antisense mRNAs are constantly expressed in latent retroviruses and retained in nucleus to support retroviral persistence; this may allow them to become novel feasible targets for retrovirus elimination.
Keywords: HTLV-1, HIV-1, HBZ, ASP, RNA-FISH
Abstract
Human retroviruses, including human T cell leukemia virus type 1 (HTLV-1) and HIV type 1 (HIV-1), encode an antisense gene in the negative strand of the provirus. Besides coding for proteins, the messenger RNAs (mRNAs) of retroviral antisense genes have also been found to regulate transcription directly. Thus, it has been proposed that retroviruses likely localize their antisense mRNAs to the nucleus in order to regulate nuclear events; however, this opposes the coding function of retroviral antisense mRNAs that requires a cytoplasmic localization for protein translation. Here, we provide direct evidence that retroviral antisense mRNAs are localized predominantly in the nuclei of infected cells. The retroviral 3′ LTR induces inefficient polyadenylation and nuclear retention of antisense mRNA. We further reveal that retroviral antisense RNAs retained in the nucleus associate with chromatin and have transcriptional regulatory function. While HTLV-1 antisense mRNA is recruited to the promoter of C-C chemokine receptor type 4 (CCR4) and enhances transcription from it to support the proliferation of HTLV-1–infected cells, HIV-1 antisense mRNA is recruited to the viral LTR and inhibits sense mRNA expression to maintain the latency of HIV-1 infection. In summary, retroviral antisense mRNAs are retained in nucleus, act like long noncoding RNAs instead of mRNAs, and contribute to viral persistence.
Retroviruses have an RNA genome that can be reverse transcribed into DNA and subsequently integrated into the host genome. HIV-1 and HTLV-1 are two retroviruses pathogenic to humans. Whereas HIV-1 is globally pandemic and infects over 30 million people, HTLV-1 is epidemic in areas such as southwestern Japan, sub-Saharan Africa, and South America and infects 10 million people worldwide (1). CD4 T cells appear to be the major target of both HIV-1 and HTLV-1 in vivo. However, while HIV-1 eventually kills CD4 T cells and leads to the development of AIDS, HTLV-1 drives the clonal proliferation of CD4 T cells, causing an adult T cell leukemia (ATL) in 5% of infected individuals (2). In vivo, both HIV-1 and HTLV-1 infections can enter latency where viral transcription or replication is barely detected, rendering viral countermeasures ineffective.
Retroviruses share a similar genome structure. The integrated retroviral genome, called the provirus, has two identical long terminal repeats (LTR) located at its 5′ and 3′ ends, respectively. The 5′ LTR acts as the promoter of almost all retroviral genes and thus is indispensable for viral transcription and replication. However, selective methylation of the 5′ LTR and the subsequent viral latency have been observed in HIV-1 and HTLV-1 (3–6). In contrast, the 3′ LTR of HIV-1 and HTLV-1 remains nonmethylated (3–6), and recent findings have shown that novel retroviral genes are transcribed from the 3′ LTR in an antisense direction (7, 8). HTLV-1 encodes an antisense gene named HTLV-1 bZIP factor (HBZ) (9), which is expressed in all HTLV-1–infected individuals (2). The HBZ protein has versatile functions and plays important roles in HTLV-1 pathogenesis (9). HIV-1 also encodes an antisense protein (ASP) gene. Although its function remains less characterized so far, several studies have demonstrated that ASP protein is likely a structural protein of HIV-1 virion that associates with gp120 (10) and is also functionally involved in autophagy regulation (11, 12). In addition, the ASP protein can be recognized by the host immune system and induce a specific CD8 T cell response (13, 14). More importantly, the presence of the ASP gene has been suggested to be associated with the HIV-1 pandemic (7, 15).
An intriguing finding about retroviral antisense genes is that their messenger RNAs (mRNAs) harbor regulatory functions. For example, HBZ RNA has been found to promote transcription of host genes such as E2F transcription factor 1 (E2F1), survivin, and CCR4 (16–18), whereas ASP RNA is able to suppress HIV-1 sense RNA transcription (19, 20). It has thus been suspected that retroviral antisense mRNAs are likely localized in nucleus in order to regulate transcription; however, this idea is challenged by their protein-coding roles that require them to be cytoplasmic. To better understand the nature of retroviral antisense mRNAs, we systemically examined their expression and subcellular localization by using a highly sensitive single-molecule RNA fluorescence in situ hybridization (FISH) assay (SI Appendix, Fig. S1 A and B) (21). Here, we provide direct evidence that endogenous retroviral antisense mRNAs are predominantly localized in the nucleus and associate with chromatin, which strongly suggests that they act more like nuclear long noncoding RNAs (lncRNAs) to regulate transcription.
Results
The HTLV-1 Antisense mRNA, HBZ, Is Predominantly Localized in the Nucleus in Infected Cells.
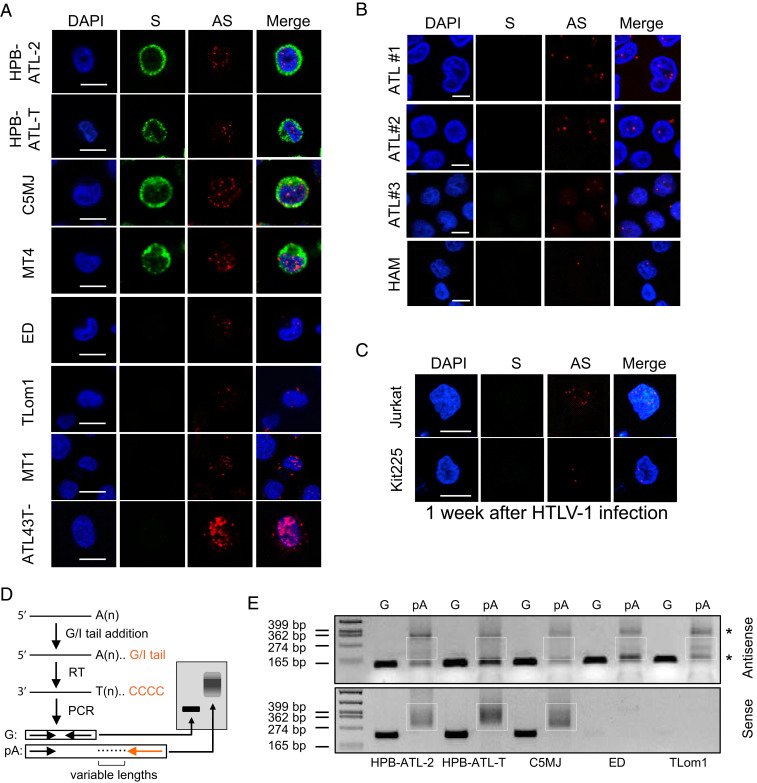
By using RNA fluorescence in situ hybridization (RNA-FISH), we first examined eight HTLV-1–infected cell lines, including those with or without expression of HTLV-1 Tax, the activator of HTLV-1 sense transcription. As shown in Fig. 1A, HBZ mRNA, despite coding for a pathogenic HBZ protein, is localized predominantly in the nucleus in HTLV-1–infected cell lines, regardless of the presence of Tax. The average percentage of HBZ RNA that was nuclear localized in each of the above cell lines was calculated and ranged from 59 (MT1) to 95% (ATL-43T-) (SI Appendix, Fig. S1C). In contrast, HTLV-1 sense mRNAs were detected mostly in the cytoplasm in Tax+ cell lines (Fig. 1A). In accordance with a recent study (22), we observed nuclear localization of HBZ RNA as well in peripheral blood mononuclear cells (PBMC) isolated from HTLV-1–infected patients with HTLV-1–associated myelopathy (HAM) (23) or ATL (24) (Fig. 1B). In these four cases, the percentages of HBZ RNA spots that were nuclear localized were all over 80% (SI Appendix, Fig. S1D). Moreover, we performed de novo HTLV-1 infection of two HTLV-1–negative T cell lines, Jurkat and Kit225, and detected HBZ RNA mainly in the nuclei of these infected cells as well (Fig. 1C and SI Appendix, Fig. S1E); thus, nuclear retention of HBZ RNA is independent of late events in HTLV-1 infection such as cellular transformation (25). Collectively, HBZ RNA was consistently detected in the nucleus in HTLV-1–infected primary cells or cell lines, independent of viral sense transcription.
Fig. 1.
HBZ mRNA is predominantly localized in the nucleus and inefficiently polyadenylated. (A) Representative RNA-FISH results of HTLV-1–infected T cell lines showing nuclear localization of the HTLV-1 antisense (AS) mRNA HBZ (red) and cytoplasmic localization of HTLV-1 sense (S) mRNAs (green) (scale bar, 10 μm). (B) Representative RNA-FISH results of primary cells from HTLV-1–infected patients with ATL or HAM (scale bar, 5 μm). (C) Representative RNA-FISH results of Jurkat or Kit225 cells de novo infected with HTLV-1 (scale bar, 10 μm). (D) Illustration of how the poly (A) tail assay works. The assay first adds a G/I tail to mRNAs. Then, reverse transcription is performed using a universal reverse primer so that all complementary DNA will have a common 5′ end. Finally, two PCRs are performed; the first one amplifies a gene-specific product using a forward primer located at 100 to 200 bp upstream of the RNA cleavage site and a reverse primer located right before the cleavage site, and the second one amplifies a poly (A)-specific product (pA) using the same forward primer and the universal reverse primer. After electrophoresis, the G product will appear as a band while the pA product will appear as a bigger smear. (E) pA tail assay results of three Tax-positive (HPB-ATL-2, HPB-ATL-T, and C5MJ) and two Tax-negative (ED and TLom1) HTLV-1–infected cell lines indicate efficient polyadenylation of sense mRNAs, shown by smears at the Bottom. In contrast, antisense mRNA HBZ only demonstrated faint smears with some nonspecific products (*) (confirmed by sequencing). Smears indicating pA products are marked by white boxes. The assay was preoptimized as shown in SI Appendix, Fig. S3B in order to obtain equivalent amounts of gene-specific products for HTLV-1 antisense and sense mRNAs.
HBZ RNA Is Inefficiently Polyadenylated.
To understand how HBZ RNA is retained in the nucleus, we first examined its sequence to search for a possible nuclear-retention signal. We cloned the HBZ gene into a pME vector (SI Appendix, Fig. S2A), which has an enhanced SV40 promoter, SRα, that allows optimal expression in various types of cell lines (26) and an efficient SV40 polyadenylation signal (27). Then, we transfected the vectors expressing full-length or truncated HBZ RNA into HeLa cells and performed RNA-FISH to localize the HBZ RNA. However, the results indicated there seemed no nuclear-retention motif in HBZ RNA sequence (SI Appendix, Fig. S2 B and C). Next, we investigated the posttranscriptional processing of HBZ RNA including splicing and 3′-end polyadenylation because these processes are generally required for an mRNA to mature and be exported from nucleus (28). Analyses by both strand-specific RNA sequencing (RNA-seq) (SI Appendix, Fig. S2D) and RNA-FISH (SI Appendix, Fig. S2E) demonstrated that HBZ RNA was efficiently spliced; however, a semiquantitative poly (A) (pA) tail assay (Fig. 1D), which converts polyadenylated forms of RNA into DNA and presents them as a smear on a DNA gel (29, 30), indicated that HBZ RNA was inefficiently polyadenylated (Fig. 1E and SI Appendix, Fig. S3 A and B). As shown in Fig. 1E, only very faint smears along with nonspecific bands were seen with HTLV-1 antisense RNA, whereas clear smears that indicate efficient polyadenylation were observed with HTLV-1 sense mRNAs. In line with the above results, HTLV-1 sense mRNAs, which are efficiently polyadenylated, exhibited significant nuclear retention upon treatment by cordycepin (SI Appendix, Fig. S3C), an adenosine analog that inhibits polyadenylation (31). Therefore, HBZ RNA is likely retained in the nucleus due to inefficient polyadenylation, a mechanism that is commonly found in nuclear-localized lncRNAs (32).
The Native Polyadenylation Signal of the HBZ Gene Does Not Cause Inefficient Polyadenylation.
Many lncRNAs are not polyadenylated because they lack a polyadenylation signal (32), but the HBZ gene is reported to harbor a polyadenylation signal (33). To evaluate the competency of the native HBZ polyadenylation (HpA) signal, we cloned its sequence into an HBZ expression vector (SI Appendix, Fig. S2A) to replace the original simian virus 40 (SV40) polyadenylation signal (27). Then, we transfected it into HeLa cells and performed a pA tail assay (Fig. 2A and SI Appendix, Fig. S4 A and B) and RNA-FISH (Fig. 2B). We found that the complete HpA signal containing a 100-base-pair (bp) downstream element (33) (HpA-L) was competent to mediate efficient polyadenylation (Fig. 2A and SI Appendix, Fig. S4 A and B) and nuclear export (Fig. 2B) of HBZ RNA in vitro, as was the positive control SV40 polyadenylation (SpA) signal (27) (Fig. 2 A and B). Moreover, truncation of the HpA signal greatly dampened its activity (Fig. 2 A and B), further suggesting that HpA is a typical polyadenylation signal that requires downstream elements (SI Appendix, Fig. S4A) to be fully functional (34). Taking into account that its sequence is conserved in HTLV-1–infected cells (33), the native polyadenylation signal of the HBZ gene does not seem to be responsible for the inefficient polyadenylation of endogenous HBZ RNA.
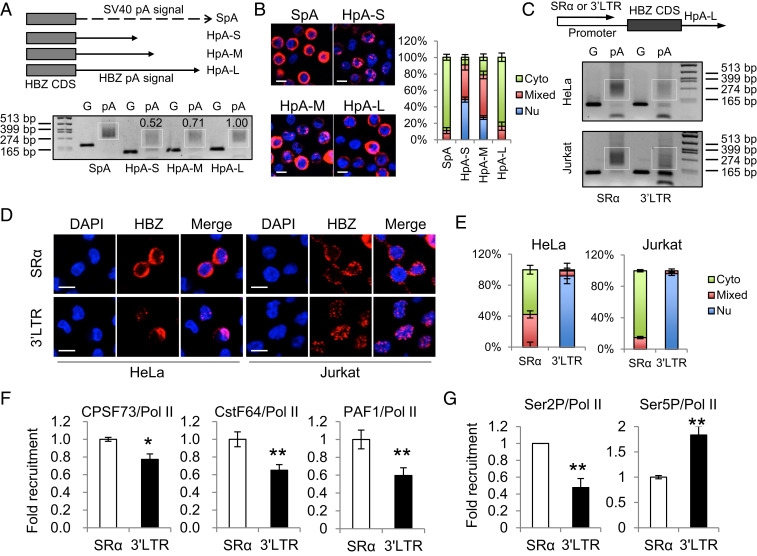
Fig. 2.
The native promoter of the HBZ gene, the HTLV-1 3′ LTR, is likely responsible for the inefficient polyadenylation and nuclear retention of HBZ RNA. (A) The Top illustrates HBZ-expression vectors with modified polyadenylation signals from the HBZ gene. SpA is the positive control SV40 pA signal. HpA is cloned from the native HBZ pA signal. HpA-S ends at the cleavage site of HpA, HpA-M ends at the G/T-rich region, and HpA-L ends 100 bp downstream the AATAAA sequence of HpA. See SI Appendix, Fig. S4A for more details. We expressed these vectors in HeLa cells and performed a pA tail assay, the result of which is shown (Lower). Smears indicating pA products are marked by white boxes and the numerals above the white boxes indicate the relative polyadenylation based on densitometry analysis by ImageJ. The optimization of the pA tail assay is shown in SI Appendix, Fig. S4B. (B) RNA-FISH was performed using HeLa cells expressing the above vectors to localize HBZ RNA (red). The percentage of cells with predominantly cytoplasmic, nuclear, or mixed localization patterns of HBZ RNA was counted using ImageJ and is shown on the Right (scale bar, 10 μm). (C–G) HBZ RNA–expressing vectors using either SR-α or the HTLV-1 3′ LTR as the promoter were transfected into HeLa or Jurkat cells. After 24 h, we performed a pA tail assay (C), RNA-FISH (D), and a chromatin immunoprecipitation (ChIP) assay (F and G). (C) The pA tail assay indicates significantly less smear products when HTLV-1 3′ LTR was the promoter. Smears indicating pA products are marked by white boxes. The optimization of the pA tail assay is shown in SI Appendix, Fig. S4C. (D) RNA-FISH suggests the use of HTLV-1 3′ LTR as the promoter caused nuclear retention of HBZ RNA (scale bar, 10 μm). (E) The percentage of cells with predominantly cytoplasmic, nuclear, or mixed localization patterns of HBZ RNA is shown. (F) A ChIP assay shows reduced recruitment of CPSF73, CstF64, and PAF1 to the HBZ coding sequence when HTLV-1 3′ LTR is used as the promoter. All ChIP results were normalized to the result of nonphosphorylated RNA Pol II. (G) A ChIP assay shows reduced recruitment of Ser2P Pol II, but not Ser5P Pol II, when HTLV-1 3′ LTR is used as the promoter. The statistical analyses were performed by Student’s t test. *P < 0.05; **P < 0.01.
The Native Promoter of the HBZ Gene, the HTLV-1 3′ LTR, Is Likely Responsible for the Inefficient Polyadenylation and Nuclear Retention of HBZ RNA.
Polyadenylation is not an independent process but is rather tightly coupled with transcription (35, 36); proteins responsible for the 3′ end cleavage and polyadenylation of RNA, such as cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulatory factor (CstF), are recruited to the C-terminal domain (CTD) of RNA polymerase (Pol) II during transcription elongation and further released at the polyadenylation signal for the processing of the premature RNA (36, 37). Thus, polyadenylation is subject to regulation by promoter (36). Indeed, recent studies have demonstrated that the promoter can affect mRNA decay and alternative polyadenylation (38, 39). Accordingly, we next investigated whether the HTLV-1 3′ LTR, the native HBZ promoter, might play a role in the inefficient polyadenylation of HBZ RNA. When we expressed HBZ under the SR-α promoter in either HeLa or Jurkat cells, we observed efficient polyadenylation and cytoplasmic localization of HBZ RNA (Fig. 2 C–E and SI Appendix, Fig. S4C). However, when we replaced the SR-α promoter with the HTLV-1 3′ LTR, we observed inefficient polyadenylation and nuclear retention of the HBZ RNA (Fig. 2 C–E and SI Appendix, Fig. S4C). This suggests the HTLV-1 3′ LTR is likely responsible for the inefficient polyadenylation of HBZ RNA. In agreement with this notion, a chromatin immunoprecipitation assay in HeLa cells showed that the recruitment of polyadenylation factors such as CPSF73 and CstF64 to the HBZ coding sequence (CDS) was impaired when the promoter was changed from SR-α to the HTLV-1 3′ LTR (Fig. 2F). Pol II–associated factor 1 (PAF1), a central component of the PAF1 complex that acts as a bridge between RNA Pol II and polyadenylation factors (40), was also recruited less (Fig. 2F).
Phosphorylation of RNA Pol II CTD is critical to its function; different phosphorylation patterns of RNA Pol II determines to which transcription factors it binds and to which promoters it is recruited (41). Two important phosphorylation patterns of RNA Pol II are Serine 5 phosphorylation (Ser5P) that initiates transcription and Serine 2 phosphorylation (Ser2P) that is responsible for transcription elongation and termination (41). Particularly, Ser2P Pol II plays a key role in RNA polyadenylation because it acts as a major component of the polyadenylation machinery and recruits various polyadenylation factors for proper transcription termination (41). In accordance with the crucial role of Ser2P Pol II in polyadenylation, we found that the HTLV-1 3′ LTR did recruit less Ser2P Pol II, but not less Ser5P Pol II, than SR-α (Fig. 2G). These results collectively demonstrate that the nuclear retention of HBZ RNA is closely associated with inefficient polyadenylation, likely caused by HTLV-1 3′ LTR being a promoter less efficient in recruiting Ser2P RNA Pol II and polyadenylation factors.
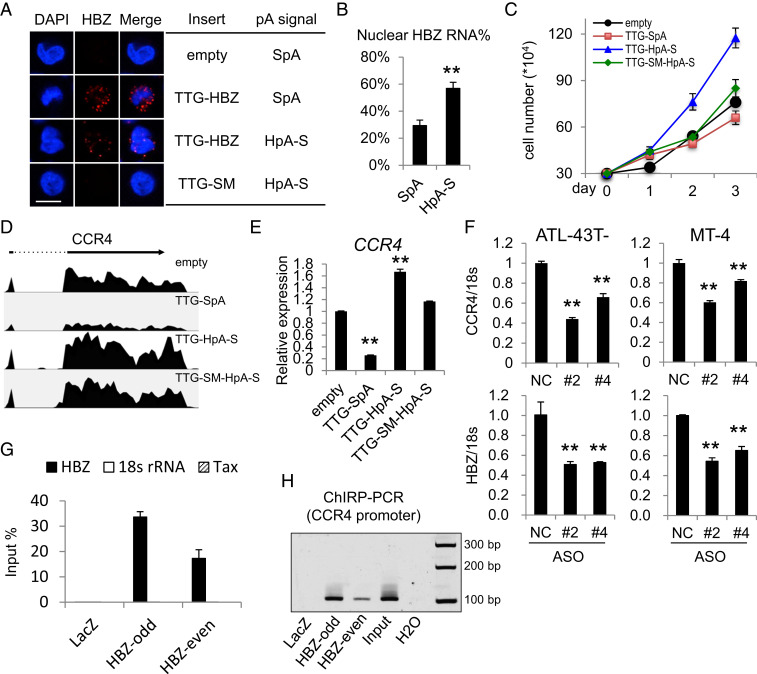
Nuclear-Localized HBZ RNA Associates with the CCR4 Promoter and Enhances CCR4 Transcription.
As HBZ protein is a target of host cytotoxic T cells (CTL) (42), the predominantly nuclear localization of HBZ RNA presumably maintains low abundance of the immunogenic HBZ protein and thus helps HTLV-1 evade host immune surveillance (42, 43). To explore the physiological significance of nuclear-localized HBZ RNA, we established a T cell line that stably expresses HBZ RNA and retains it in the nucleus. Using the SR-α promoter instead of the HTLV-1 3′ LTR for an optimal HBZ expression level and a truncated HpA signal (HpA-S) for inefficient polyadenylation and nuclear retention (Fig. 2 A and B), we generated an HBZ RNA–expressing Kit225 T cell line, TTG-HpA-S (18), in which both the expression and localization patterns of HBZ RNA resembled those in HTLV-1–infected cells (Fig. 3 A and B). The start codons of the HBZ CDS were altered to TTG to silence HBZ protein translation so that we could examine the function of HBZ RNA alone. We found the TTG-HpA-S line that expresses more nuclear HBZ RNA exhibited enhanced proliferation compared to all other lines including the empty control line, the TTG-SpA line expressing predominantly cytoplasmic HBZ RNA, and the TTG-SM-HpA-S line (18) in which the function of HBZ RNA is disrupted by altering its sequence with silent mutations (Fig. 3C). We further performed RNA-seq on these four lines and identified 15 genes whose expressions were regulated oppositely in the TTG-HpA-S versus TTG-SpA lines (SI Appendix, Fig. S5 and Table S1). Interestingly, we found that CCR4, which is highly expressed in HTLV-1–infected cells and promotes cell migration and proliferation (44, 45), was up-regulated only in the TTG-HpA-S line (Fig. 3 D and E). To demonstrate that CCR4 is a target gene of endogenous HBZ RNA, we performed HBZ knockdown in two HTLV-1–infected cell lines using antisense oligos (ASO) (46). Reduced CCR4 expression (Fig. 3F) and impaired cell proliferation (SI Appendix, Fig. S6A) were both observed in the knockdown cells. Importantly, HBZ protein level was not affected by the transient knockdown (SI Appendix, Fig. S6B), suggesting that HBZ RNA alone could promote CCR4 expression in HTLV-1–infected cells. To investigate if HBZ RNA might associate with the CCR4 promoter, we performed a chromatin isolation by RNA purification (ChIRP) assay that has been widely used to identify genomic binding sites of RNA (47). We designed oligo probes for HBZ RNA and separated those into two pools, each serving as a control for the other; both retrieved HBZ RNA with great efficiency and specificity in ATL-43T- cells (Fig. 3G), an HTLV-1–infected cell line with high level of nuclear HBZ RNA (Fig. 1A and SI Appendix, Fig. S1C). We then performed ChIRP-PCR and discovered that a previously described promoter region of the CCR4 gene (17) (SI Appendix, Fig. S6C) was pulled down specifically by both pools of HBZ probes (Fig. 3H). This suggests that HBZ RNA is indeed recruited to the promoter of CCR4 in HTLV-1–infected ATL-43T- cells. Therefore, by being retained in the nucleus and enhancing the expression of the growth-promoting gene CCR4, HTLV-1 antisense mRNA HBZ seems to act like a lncRNA to support the proliferation of HTLV-1–infected cells.
Fig. 3.
Nuclear-localized HBZ RNA associates with the CCR4 promoter and enhances CCR4 transcription. (A) Representative RNA-FISH results of four Kit225 lines. HBZ RNA was stained in red (scale bar, 10 μm). (B) The percentage of HBZ RNA that is nuclear in the TTG-SpA (SpA) and TTG-HpA-S (HpA-S) Kit225 lines. (C) A total of 3 × 104 cells of each line were seeded in full RPMI media with 100 U/mL IL-2, and cell number was counted every day until day three. (D) CCR4 tracks from RNA-seq results of the four cell lines. (E) qPCR verification of the RNA-seq results in D. (F) CCR4 and HBZ RNA levels in ASO-mediated HBZ-knockdown cells (#2 and #4) and negative control cells (NC). (G) qPCR result showing the retrieval efficiency of two pools of HBZ RNA probes (HBZ-odd and HBZ-even) and the negative control LacZ probes used in the ChIRP assay. Nonspecific retrieval of Tax RNA or 18s ribosomal RNA was not observed. (H) ChIRP-PCR was performed to amplify the previously reported CCR4 promoter region (−326 ∼ −220 bp) (17). Note that the negative control LacZ probes yielded no band whereas both pools of HBZ RNA probes yielded bands with the same size as input. The statistical analyses were performed by Student’s t test. **P < 0.01.
HIV-1 Antisense mRNA, ASP, Is Inefficiently Polyadenylated and Predominantly Localized in the Nucleus in Infected Cells.
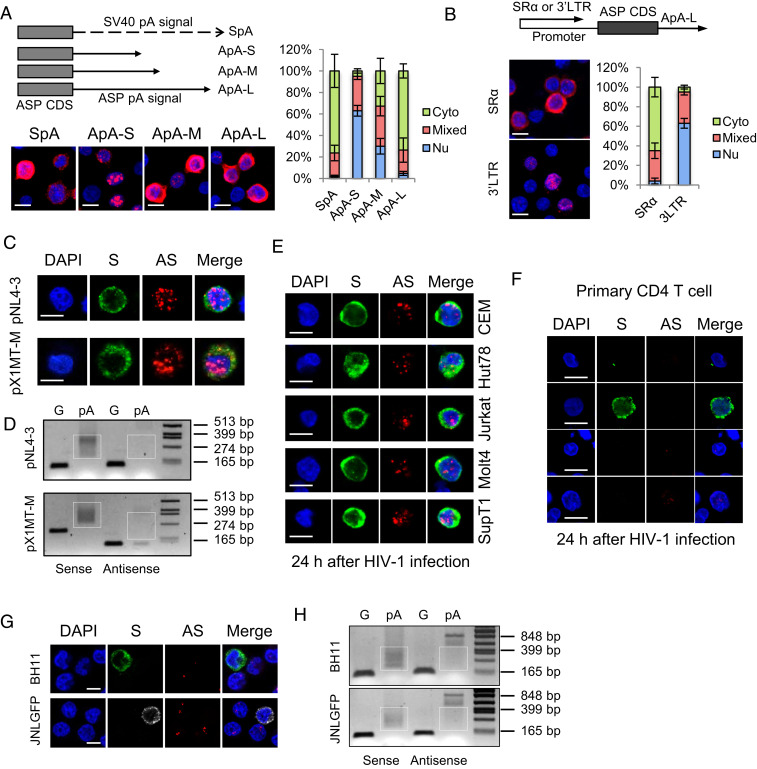
Next, we turned to examine the localization and function of the HIV-1 antisense mRNA, ASP (15), based on the resemblance between the genome structure of HTLV-1 and HIV-1. We first cloned the native polyadenylation signal of the ASP gene (SI Appendix, Fig. S7A), either truncated or complete, into the pME vector, transfected the constructs into HeLa cells, and performed RNA-FISH. As shown in Fig. 4A, the native polyadenylation signal of the ASP gene seemed efficient to mediate nuclear export as well (Fig. 4A). Moreover, when we replaced the SR-α promoter by HIV-1 3′ LTR, nuclear retention of the ASP RNA was observed (Fig. 4B). These results indicate ASP RNA may be regulated in a similar way to HBZ RNA. We next performed an RNA-FISH and a pA tail assay in HeLa cells transfected with an HIV-1–infectious clone, pNL4-3 (48), and discovered that ASP RNA was predominantly localized in nucleus and inefficiently polyadenylated as well (Fig. 4 C and D and SI Appendix, Fig. S7B). In contrast, HIV-1 sense mRNAs were efficiently polyadenylated and localized mostly in cytoplasm (Fig. 4 C and D and SI Appendix, Fig. S7B). These results resembled those observed in an HTLV-1–infectious clone pX1MT-M (49) (Fig. 4 C and D and SI Appendix, Fig. S7B). In addition, we de novo infected various T cell lines and primary CD4 T cells with wild-type HIV-1 virions, performed RNA-FISH, and found that ASP RNA was also mostly localized in the nucleus (Fig. 4 E and F).
Fig. 4.
HIV-1 antisense mRNA, ASP, is inefficiently polyadenylated and predominantly localized in the nucleus in infected cells. HeLa cells were transfected with ASP-expression vectors, HIV-1–infectious clone pNL4-3 or HTLV-1–infectious clone pX1MT-M, and RNA-FISH (A–C) and a pA tail assay (D) were performed. (A) The Left Top illustrates ASP-expression vectors with modified polyadenylation signals from the ASP gene. ApA is cloned from the native ASP pA signal. ApA-S ends at the cleavage site of ApA, ApA-M ends at the G/T-rich region, and ApA-L ends 100 bp downstream the AATAAA sequence of ApA. See SI Appendix, Fig. S7A for more details. The Left Bottom shows representative RNA-FISH results from the transfected HeLa cells detecting ASP RNA (red). The percentage of cells with predominantly cytoplasmic, nuclear, or mixed localization patterns of ASP RNA was counted using ImageJ and is shown on the Right (scale bar, 10 μm). (B) RNA-FISH suggests the use of HIV-1 3′ LTR as the promoter caused nuclear retention of ASP RNA. Bar = 10 μm. (C) Representative RNA-FISH results from the transfected HeLa cells detecting viral sense (green) and antisense (red) mRNAs (scale bar, 10 μm). (D) pA tail assay results of transfected HeLa indicating efficient polyadenylation of viral sense mRNAs but inefficient polyadenylation of antisense mRNAs. Smears indicating pA products are marked by white boxes. The assay was preoptimized in order to obtain equivalent amounts of gene-specific products for sense and antisense mRNAs (SI Appendix, Fig. S7B). (E) Representative RNA-FISH results of five T cell lines de novo infected with HIV-1. HIV-1 sense mRNAs are shown in green, and ASP RNA is shown in red (scale bar, 10 μm). (F) Representative RNA-FISH results of primary human CD4 T cells de novo infected with HIV-1. HIV-1 sense mRNAs are shown in green, and ASP RNA is shown in red (scale bar, 10 μm). (G) Representative RNA-FISH results of BH11 or JNLGFP cells. HIV-1 sense mRNAs are shown in green, except in JNLGFP they are stained in white to avoid interference from GFP. HIV-1 antisense mRNA is shown in red (scale bar, 10 μm). (H) pA tail assay results of BH11 and JNLGFP cells indicating efficient polyadenylation of HIV-1 sense mRNAs but inefficient polyadenylation of antisense mRNAs. Smears indicating pA products are marked by white boxes. The assay was preoptimized in order to obtain equivalent amounts of gene-specific products for sense and antisense mRNAs (SI Appendix, Fig. S7D).
As HIV-1 infection becomes latent in vivo, we further examined two HIV-1 latently infected cell lines, BH11 (50) and JNLGFP (51), which express a blue fluorescent protein (BFP) and a green fluorescent protein (GFP), respectively, as a marker for HIV-1 expression (SI Appendix, Fig. S7C). As expected, ASP RNA was detected mostly in the nucleus and inefficiently polyadenylated, whereas HIV-1 sense mRNAs were efficiently polyadenylated and localized mostly in cytoplasm (Fig. 4 G and H and SI Appendix, Fig. S7D). Therefore, like that of HTLV-1, HIV-1 antisense mRNA is mostly localized in nucleus. It is likely that both HTLV-1 and HIV-1 share a common mechanism for the nuclear retention of antisense mRNAs.
ASP RNA Is Recruited to HIV-1 LTR to Inhibit Viral Transcription.
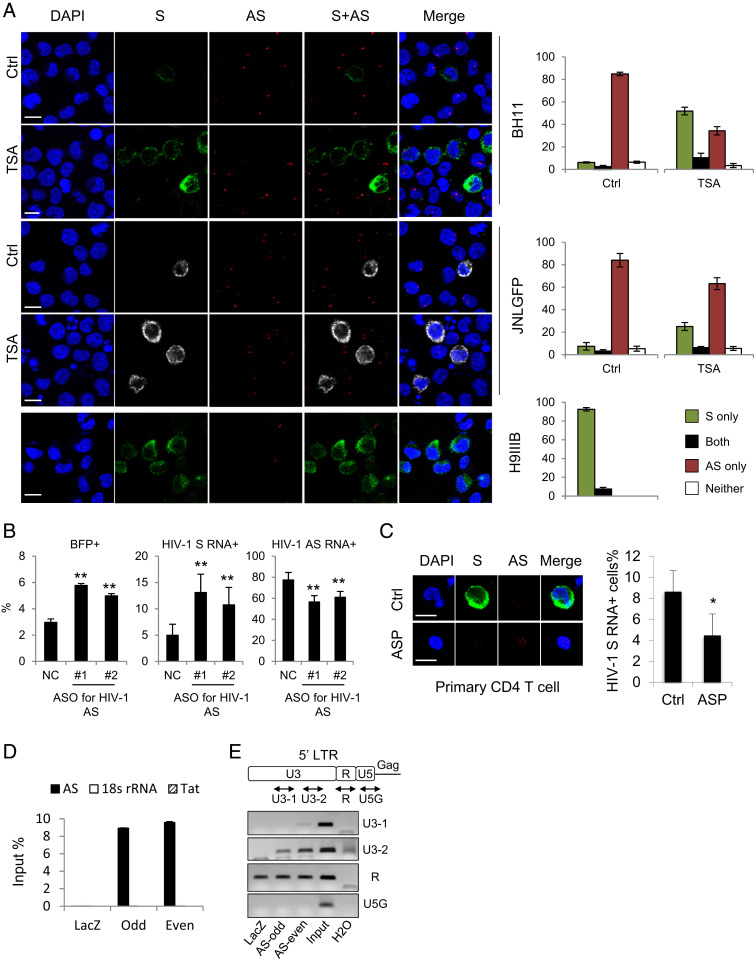
BH11 and JNLGFP are HIV-1 latently infected cell lines with less than 10% of the cells expressing HIV-1 sense mRNAs (Fig. 5A). By contrast, ASP RNA was detected in over 80% of the cells (Fig. 5A). Interestingly, an HIV-1 productively infected cell line H9IIIB (52) exhibited an opposite pattern: predominant HIV-1 sense but rare antisense mRNA expression (Fig. 5A). It appears that HIV-1 sense and antisense transcription are inversely correlated in either latently or productively infected cells. In particular, HIV-1 sense and antisense mRNAs rarely coexist in the same cells (Fig. 5A). It is likely that the expression of HIV-1 sense and antisense mRNAs is regulated in a mutually antagonistic way. Indeed, while reactivation of HIV-1 by trichostatin A (53) in latently infected BH11 or JNLGFP cells led to an expected increase of HIV-1 sense mRNAs, a concomitant decrease in ASP RNA emerged (Fig. 5A). More importantly, when we used ASOs to knock down ASP RNA in BH11 cells, HIV-1 reactivation was observed, as demonstrated by both flow cytometry (Fig. 5B and SI Appendix, Fig. S8A) and RNA-FISH results (Fig. 5B and SI Appendix, Fig. S8B). Moreover, overexpression of ASP RNA suppressed HIV-1 sense transcription in HIV-1–de novo infected CD4 T cells (Fig. 5C). These observations coincide with the reported negative regulatory role of ASP RNA in HIV-1 transcription (19, 20). As HIV-1 ASP RNA has been suggested to regulate HIV-1 5′ LTR activity, we then performed a ChIRP assay in BH11 cells to investigate if ASP RNA might associate with the HIV-1 LTR. We first pulled down ASP RNA using specific probes (Fig. 5D), then performed ChIRP-PCR and found that a region in the U3 of the HIV-1 LTR was associated with ASP RNA (Fig. 5E). Collectively, these results suggest that HIV-1 ASP RNA is recruited to the HIV-1 LTR to suppress viral transcription, thus contributing to the maintenance of HIV-1 latency.
Fig. 5.
ASP RNA is recruited to the HIV-1 LTR to inhibit viral transcription. (A) Representative RNA-FISH results of BH11, JNLGFP, or H9IIIB cells cultured in the absence or presence of trichostatin A. HIV-1 sense (S) mRNAs are shown in green, except in JNLGFP they are stained in white to avoid interference from GFP. HIV-1 antisense (AS) mRNA is shown in red. The percentage of cells expressing S mRNA only, AS mRNA only, both, or neither is shown on the Right (scale bar, 10 μm). (B) ASO-mediated knockdown of ASP RNA increased the percentage of BFP+ BH11 cells that are active for HIV-1 sense transcription (Left) and the percentage of HIV-1 sense RNA+ cells (Middle) but decreased the percentage of HIV-1 ASP RNA+ cells (Right). (C) Representative RNA-FISH results of HIV-1–infected CD4 T cells with (Lower) or without (Upper) ASP RNA overexpression (Scale bar, 10 μm). The number of HIV-1 sense RNA positive cells was counted, and the percentage is shown on the Right. (D) qPCR result showing the retrieval efficiency of two pools of ASP RNA probes (odd and even) and the negative control LacZ probes used in the ChIRP assay. Nonspecific retrieval of Tat RNA or 18s ribosomal RNA was not detected. (E) ChIRP-PCR was performed to amplify various regions in the HIV-1 LTR. Product U3-2 is considered specific because both ASP RNA probes yielded specific bands whereas LacZ probes yielded no band. The statistical analyses were all performed by Student’s t test. *P < 0.05; **P < 0.01.
Discussion
Using the highly sensitive RNA-FISH assay, we systemically analyzed the subcellular localization of mRNAs of HTLV-1 and HIV-1, two major retroviruses that are pathogenic to humans, and discover that antisense mRNAs of human retroviruses are mostly retained in the nucleus. Importantly, we prove that retroviral antisense RNAs associate with chromatin to regulate transcription in favor of viral persistence. These findings suggest a role of retroviral antisense RNAs acting more prominently as regulatory lncRNAs than as protein-coding transcripts. Although protein products of retroviral antisense genes exist and may have multifaceted functions, their recognition by the host immune system is detrimental to retroviruses, as this can lead to the elimination of infected cells. For example, the HTLV-1 HBZ protein is able to promote the proliferation of HTLV-1–infected cells and is beneficial to viral persistence, yet it can also trigger a specific CTL response that kills infected cells very effectively (42, 43). The HIV-1 ASP protein can induce a specific CD8 T cell response as well (13). Therefore, retroviruses may have evolved to retain most of their antisense RNAs in the nucleus to restrict protein production, thereby evading host immune surveillance.
A new term, bifunctional RNA, has been recently coined to describe RNA with both coding capacity and regulatory function (54, 55). Bifunctional RNAs contribute to the complexity and robustness of gene expression regulation and have been reported in many species, including humans, indicating that the bifunctional mode of RNAs has been conserved during evolution (56). For instance, the mRNA of the well-known tumor suppressor gene p53 has been found to interact with the E3 ubiquitin ligase Mdm2 and to promote p53 translation (57). Retroviral antisense mRNAs including HBZ and ASP appear to be a class of bifunctional RNAs and an example in viruses. However, it remains to be seen if the coding and regulatory functions of retroviral antisense mRNAs are independently executed or tightly coupled. In the future, it will be interesting to see if retrovirus-infected cells may change the subcellular localization of their antisense mRNAs under specific conditions or in response to certain stimuli.
Polyadenylation is a critical step of mRNA quality control and is required for mRNA export from nucleus to cytoplasm. Therefore, the inefficient polyadenylation that we observed in endogenous retroviral antisense RNAs will lead to their nuclear retention. However, whether there are additional mechanisms that might also contribute to the nuclear retention of retroviral antisense RNAs requires further investigation. It is presumed that endogenous retroviral antisense mRNAs will be efficiently exported to cytoplasm once they get efficiently polyadenylated, but it is currently technically difficult to directly increase the polyadenylation efficiency at the endogenous level. Nevertheless, we here find that inefficient polyadenylation is a conserved feature of retroviral antisense mRNAs that allows them to be retained in the nucleus and to act like lncRNAs.
It is widely appreciated that polyadenylation can increase RNA stability and prevent its degradation. Counterintuitively, polyadenylation has also been reported to induce RNA decay, although the mechanism is not yet well understood (58, 59). There was no decrease in HBZ RNA level when polyadenylation was inhibited in HTLV-1–infected cells by cordycepin (SI Appendix, Fig. S3C), suggesting that polyadenylation did not promote the stability of HBZ RNA. Indeed, the stability of HBZ RNA did not differ (SI Appendix, Fig. S9A) when it was expressed under the HTLV-1 3′ LTR or SR-α promoter and differentially polyadenylated (Fig. 2C). Therefore, inefficient polyadenylation could induce nuclear retention of endogenous HBZ RNA, but it does not necessarily impair its stability.
In this study, we initially aimed to perform ChIRP-seq in order to screen for potential target genes of retroviral antisense mRNAs. Unfortunately, we were not able to obtain the amount of DNA optimal for high-throughput sequencing. Instead, we performed ChIRP-PCR, which has been successfully used in recent publications (60–62) and allowed us to reveal the association of retroviral antisense mRNAs with specific genomic areas. Nevertheless, in the future, novel RNA purification assays with improved sensitivity are expected to help identify more genes that are targeted by retroviral antisense mRNAs. So far, HBZ RNA has been found to promote the transcription of host genes including E2F1, survivin, and CCR4 (9); however, it is unclear through which protein partners HBZ RNA acts to do so. ASP RNA likely associates with the polycomb repressor complex 2 (20), yet it has only been reported to regulate HIV-1 LTR activity. Future studies are needed to unravel more details about how retroviral antisense mRNAs exert their regulatory functions.
HTLV-1 and HIV-1 employ distinct mechanisms for viral persistence. As an oncovirus, HTLV-1 persists in vivo by immortalizing infected CD4 T cells. Our observations demonstrate HBZ RNA is able to support HTLV-1 persistence through up-regulating the expression of CCR4 (Fig. 3F) and promoting the proliferation of HTLV-1–infected cells (SI Appendix, Fig. S6A). On the other hand, HIV-1 persists in vivo as a latent reservoir where active HIV-1 production is not detectable. Our results suggest that ASP RNA likely facilitates HIV-1 persistence by suppressing HIV-1 transcription and maintaining the latent infection (Fig. 5 B and C). Collectively, both HTLV-1 and HIV-1 antisense transcripts in their RNA form can support viral persistence. It is worth noting that nuclear localization is a prerequisite for HBZ or ASP RNA to regulate transcription to favor viral persistence.
Here, we discover a common mechanism present in human retroviruses including HTLV-1 and HIV-1: they retain their antisense mRNAs in the nucleus by causing inefficient polyadenylation. The same mechanism might exist in other retroviruses such as bovine leukemia virus, which has been reported to encode an antisense RNA that seems to be nuclear as well (63). Encoding an antisense mRNA might be a conserved feature of retroviruses due to the presence of the 3′ LTR antisense promoter and a potential antisense polyadenylation signal observed in many retroviruses (SI Appendix, Fig. S9B). In contrast to the silenced sense transcription during retroviral latency, retroviral antisense transcription seems widely active. Unraveling the functions of retroviral antisense mRNAs may help us better understand retroviral persistence and allow us to develop new therapeutics for retroviral latency.
Materials and Methods
Primary Cells.
PBMCs of HTLV-1–infected individuals including ATL and HAM patients or a healthy donor were isolated by Ficoll-Paque PLUS (GE). The experiments using primary samples in this study were conducted according to the principles expressed in the Declaration of Helsinki and approved by the Institutional Review Board of Kyoto University (permit numbers G310 and E2005). All ATL and HAM patients and healthy individuals provided written informed consent for the collection of samples and subsequent analyses.
Cell Lines.
HTLV-1–infected cell lines, including Tax-negative ED, TLom1, MT1, and ATL-43T-, and Tax-positive ATL-2, ATL-T, C5MJ, and MT4, were cultured in Roswell Park Memorial Institute medium (RPMI) supplemented with 10% fetal bovine serum (FBS). The HTLV-1–uninfected T cell lines Jurkat, CEM, Hut78, Molt4, and SupT1 were cultured in RPMI supplemented with 10% FBS. HeLa cells were cultured in Dulbecco's Modified Eagle Medium supplemented with 10% FBS. HIV-1–infected cell lines, including H9IIIB, BH11, and JNLGFP were cultured in RPMI supplemented with 10% FBS. H9IIIB is an H9 (derivative of Hut78) line infected with HIV-1 and actively produces HIV-1 virions. BH11 and JNLGFP are both Jurkat clones chronically infected with HIV-1 and only have a small proportion of cells with active HIV-1 expression (SI Appendix, Fig. S7C). All cells were maintained in a humidified incubator at 37 °C with 5% CO2.
Plasmids.
The HBZ CDS was cloned into pME18S, which uses an SR-α promoter and a SpA. To prevent production of the HBZ protein, the start codon of HBZ CDS was altered to TTG beforehand. To examine the function of the HBZ native polyadenylation signal (HpA), the complete (long) HpA (HpA-L) signal that starts from 67 bp upstream of the AATAAA signal and reaches 100 bp downstream was cloned from pX1MT-M into pME18S to replace the SpA in the vector. Two truncated (short and medium) HpA signals (HpA-S and HpA-M) with shortened downstream sequences were subcloned by PCR from HpA-L. To examine the effect of promoters on RNA polyadenylation and localization, the HTLV-1 3′ LTR was cloned by PCR from pX1MT-M and inserted into pME18S-HBZ to replace the original SR-α promoter. The complete HBZ gene was subcloned from pX1MT-M (SI Appendix, Fig. S2A). The ASP CDS (11), the native polyadenylation signal of ASP (SI Appendix, Fig. S7A), and HIV-1 3′ LTR were all cloned from pNL4-3. To prevent ASP protein translation, the start codon of cloned ASP CDS was mutated to TTG.
The rest methods including RNA-FISH, pA tail assay, ChIRP-PCR, and de novo HTLV-1 or HIV-1 infection are described in detail in the SI Appendix.
Supplementary Material
Acknowledgments
We thank D. Derse for providing pX1MT-M; C. Ohnishi and Y. Mitobe for help in the experiments; K. Toyoda and T. Shichijo for helpful discussions; and L Kingsbury for proofreading. This study was supported by the Project for Cancer Research And Therapeutic Evolution (19cm0106611h0003 to J.-i.Y. and 19cm0106306h0004 to M.M.), the Research Program on Emerging and Re-emerging Infectious Diseases (18fk0108027h0003 and 19fk0108088h0001 to J.-i.Y, and M.M.) from the Japan Agency for Medical Research and Development, and Japan Society for the Promotion of Science (JSPS) KAKENHI (19H03689 to M.M. and JP17K07166 to J.-i.Y) and in part by the JSPS Core-to-Core Program A, Advanced Research Networks. This study was also supported by the National Natural Science Foundation of China (32070155 to G.M.) and a start-up fund from China Pharmaceutical University (3154070031 to G.M.) G.M. was supported by the JSPS Postdoctoral Fellowship for Overseas Researchers and the Tokyo Biochemical Research Foundation Postdoctoral Fellowships for Asian Researchers. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014783118/-/DCSupplemental.
Data Availability
The RNA-Seq data reported in this paper have been deposited in the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA692527) (64).
References
- 1.Gessain A., Cassar O., Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 3, 388 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giam C. Z., Semmes O. J., HTLV-1 infection and adult T-cell leukemia/lymphoma-A tale of two proteins: Tax and HBZ. Viruses 8, 161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kauder S. E., Bosque A., Lindqvist A., Planelles V., Verdin E., Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5, e1000495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishida T., Hamano A., Koiwa T., Watanabe T., 5′ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology 3, 69 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koiwa T., et al., 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 76, 9389–9397 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blazkova J., et al., CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5, e1000554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landry S., et al., Detection, characterization and regulation of antisense transcripts in HIV-1. Retrovirology 4, 71 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudray G., et al., The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J. Virol. 76, 12813–12822 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma G., Yasunaga J., Matsuoka M., Multifaceted functions and roles of HBZ in HTLV-1 pathogenesis. Retrovirology 13, 16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Affram Y., et al., The HIV-1 antisense protein ASP is a transmembrane protein of the cell surface and an integral protein of the viral envelope. J. Virol. 93, 93 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torresilla C., et al., Detection of the HIV-1 minus-strand-encoded antisense protein and its association with autophagy. J. Virol. 87, 5089–5105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z., et al., HIV-1 antisense protein of different clades induces autophagy and associates with the autophagy factor p62. J. Virol. 93, e01757-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bet A., et al., The HIV-1 antisense protein (ASP) induces CD8 T cell responses during chronic infection. Retrovirology 12, 15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger C. T., et al., Immune screening identifies novel T cell targets encoded by antisense reading frames of HIV-1. J. Virol. 89, 4015–4019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassan E., Arigon-Chifolleau A. M., Mesnard J. M., Gross A., Gascuel O., Concomitant emergence of the antisense protein gene of HIV-1 and of the pandemic. Proc. Natl. Acad. Sci. U.S.A. 113, 11537–11542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitobe Y., Yasunaga J., Furuta R., Matsuoka M., HTLV-1 bZIP factor RNA and protein impart distinct functions on T-cell proliferation and survival. Cancer Res. 75, 4143–4152 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Sugata K., et al., HTLV-1 viral factor HBZ induces CCR4 to promote T-cell migration and proliferation. Cancer Res. 76, 5068–5079 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Satou Y., Yasunaga J., Yoshida M., Matsuoka M., HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc. Natl. Acad. Sci. U.S.A. 103, 720–725 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saayman S., et al., An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol. Ther. 22, 1164–1175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zapata J. C., et al., The human immunodeficiency virus 1 ASP RNA promotes viral latency by recruiting the Polycomb repressor complex 2 and promoting nucleosome assembly. Virology 506, 34–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F., et al., RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Billman M. R., Rueda D., Bangham C. R. M., Single-cell heterogeneity and cell-cycle-related viral gene bursts in the human leukaemia virus HTLV-1. Wellcome Open Res. 2, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Araujo A. Q., Silva M. T., The HTLV-1 neurological complex. Lancet Neurol. 5, 1068–1076 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Watanabe T., Adult T-cell leukemia: Molecular basis for clonal expansion and transformation of HTLV-1-infected T cells. Blood 129, 1071–1081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boxus M., Willems L., Mechanisms of HTLV-1 persistence and transformation. Br. J. Cancer 101, 1497–1501 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takebe Y., et al., SR alpha promoter: An efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 8, 466–472 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carswell S., Alwine J. C., Efficiency of utilization of the simian virus 40 late polyadenylation site: Effects of upstream sequences. Mol. Cell. Biol. 9, 4248–4258 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hocine S., Singer R. H., Grünwald D., RNA processing and export. Cold Spring Harb. Perspect. Biol. 2, a000752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin G., Keller W., Tailing and 3′-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA 4, 226–230 (1998). [PMC free article] [PubMed] [Google Scholar]
- 30.Bazzini A. A., Lee M. T., Giraldez A. J., Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 336, 233–237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller W. E., et al., Effect of cordycepin on nucleic acid metabolism in L5178Y cells and on nucleic acid-synthesizing enzyme systems. Cancer Res. 37, 3824–3833 (1977). [PubMed] [Google Scholar]
- 32.Sun Q., Hao Q., Prasanth K. V., Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 34, 142–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavanagh M. H., et al., HTLV-I antisense transcripts initiating in the 3'LTR are alternatively spliced and polyadenylated. Retrovirology 3, 15 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proudfoot N. J., Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCracken S., et al., The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385, 357–361 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Proudfoot N., O’Sullivan J., Polyadenylation: A tail of two complexes. Curr. Biol. 12, R855–R857 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Hirose Y., Manley J. L., RNA polymerase II and the integration of nuclear events. Genes Dev. 14, 1415–1429 (2000). [PubMed] [Google Scholar]
- 38.Bregman A., et al., Promoter elements regulate cytoplasmic mRNA decay. Cell 147, 1473–1483 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Oktaba K., et al., ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol. Cell 57, 341–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Oss S. B., Cucinotta C. E., Arndt K. M., Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem. Sci. 42, 788–798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsin J. P., Manley J. L., The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macnamara A., et al., HLA class I binding of HBZ determines outcome in HTLV-1 infection. PLoS Pathog. 6, e1001117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bangham C. R. M., Human T cell leukemia virus type 1: Persistence and pathogenesis. Annu. Rev. Immunol. 36, 43–71 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Yoshie O., et al., Frequent expression of CCR4 in adult T-cell leukemia and human T-cell leukemia virus type 1-transformed T cells. Blood 99, 1505–1511 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Sato T., et al., Mogamulizumab (Anti-CCR4) in HTLV-1-associated myelopathy. N. Engl. J. Med. 378, 529–538 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Wheeler T. M., et al., Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 488, 111–115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu C., Qu K., Zhong F. L., Artandi S. E., Chang H. Y., Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adachi A., et al., Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59, 284–291 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derse D., Mikovits J., Ruscetti F., X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology 237, 123–128 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Shimura K., Miyazato P., Oishi S., Fujii N., Matsuoka M., Impact of HIV-1 infection pathways on susceptibility to antiviral drugs and on virus spread. Virology 484, 364–376 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Kutsch O., Benveniste E. N., Shaw G. M., Levy D. N., Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76, 8776–8786 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maldarelli F., Sato H., Berthold E., Orenstein J., Martin M. A., Rapid induction of apoptosis by cell-to-cell transmission of human immunodeficiency virus type 1. J. Virol. 69, 6457–6465 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Lint C., Emiliani S., Ott M., Verdin E., Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15, 1112–1120 (1996). [PMC free article] [PubMed] [Google Scholar]
- 54.Hubé F., Francastel C., Coding and non-coding RNAs, the frontier has never been so blurred. Front. Genet. 9, 140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumari P., Sampath K., cncRNAs: Bi-functional RNAs with protein coding and non-coding functions. Semin. Cell Dev. Biol. 47-48, 40–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nam J. W., Choi S. W., You B. H., Incredible RNA: Dual functions of coding and noncoding. Mol. Cells 39, 367–374 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Candeias M. M., et al., P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol. 10, 1098–1105 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Bresson S. M., Conrad N. K., The human nuclear poly(a)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 9, e1003893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tudek A., Lloret-Llinares M., Jensen T. H., The multitasking polyA tail: Nuclear RNA maturation, degradation and export. Philos. Trans. R. Soc. Lond. B Biol. Sci. 373, 373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trimarchi T., et al., Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montes M., et al., The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat. Commun. 6, 6967 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Li Z., et al., The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U.S.A. 111, 1002–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durkin K., et al., Characterization of novel Bovine Leukemia Virus (BLV) antisense transcripts by deep sequencing reveals constitutive expression in tumors and transcriptional interaction with viral microRNAs. Retrovirology 13, 33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma G., et al., Stranded RNA-seq data of HTLV-1 infected cell lines and RNA-seq data of HBZ-RNA-expressing Kit225 cells. Sequence Read Archive database. https://www.ncbi.nlm.nih.gov/sra/PRJNA692527. Deposited 15 January 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data reported in this paper have been deposited in the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA692527) (64).