Abstract
Quantitative variation in expression of the Arabidopsis floral repressor FLC influences whether plants overwinter before flowering, or have a rapid cycling habit enabling multiple generations a year. Genetic analysis has identified activators and repressors of FLC expression but how they interact to set expression level is poorly understood. Here, we show that antagonistic functions of the FLC activator FRIGIDA (FRI) and the repressor FCA, at a specific stage of embryo development, determine FLC expression and flowering. FRI antagonizes an FCA-induced proximal polyadenylation to increase FLC expression and delay flowering. Sector analysis shows that FRI activity during the early heart stage of embryo development maximally delays flowering. Opposing functions of cotranscriptional regulators during an early embryonic developmental window thus set FLC expression levels and determine flowering time.
Keywords: Arabidopsis FLC, cotranscriptional regulation, embryogenesis, transcript isoforms, sector analysis
Quantitative regulation of FLC expression influences overwintering requirement and vernalization of many Brassicaceae species. Regulators of FLC expression have been identified by genetic analysis in Arabidopsis. A series of repressors grouped in the autonomous pathway include RNA binding proteins, 3′-end processing factors, and chromatin modifiers. One of the RNA binding proteins, FCA, promotes proximal polyadenylation of many transcripts in the genome (1), including its own transcript (2). However, premature termination of sense FLC transcription has not emerged as the predominant mechanism for repression of FLC and proximally polyadenylated FLC transcripts were rare in seedlings (1). Instead, FCA was found to promote proximal polyadenylation of COOLAIR antisense transcripts at FLC (3). This resulted in H3K4 demethylation and H3K27me3 accumulation (4, 5), creating a chromatin environment supporting low transcriptional firing and slow elongation of both FLC sense and antisense transcription (6).
The major activator of FLC is FRI, with loss-of-function FRI alleles accounting for the evolution of many rapid cycling accessions, including Col-0 (7, 8). FRI is part of a COMPASS-like complex that binds near the FLC promoter and generates a chromatin environment promoting high transcription and fast elongation (9, 10). FRI acts as a scaffold for LEC2 and FUS3 to enable resetting of FLC in the embryo and induce an epigenetically stable high expression state (11).
How the initial antagonistic relationship of FCA and FRI on FLC is established has so far been unclear. The expression of endogenous FCA is unaffected by FRI and vice versa (2). However, the balance of their activities can be modulated through removal of the autofeedback regulation of FCA, which overcomes FRI-induced late flowering (2). Here, we show that they antagonistically regulate polyadenylation site usage of the FLC nascent transcript within an early developmental window during embryo development. This establishes an expression state that is then maintained by a Polycomb mechanism during the rest of development.
Results
Regulation of FLC Polyadenylation Site Usage during Embryogenesis.
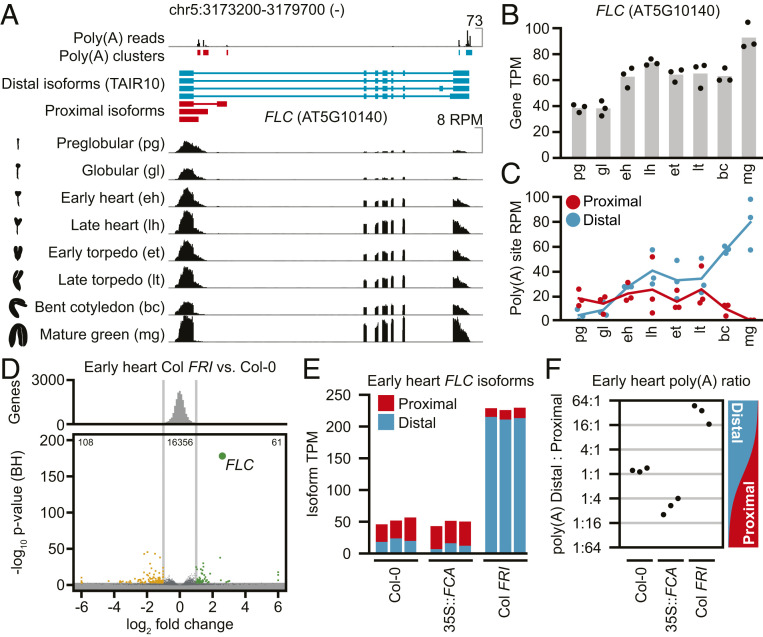
We used RNA sequencing (RNA-seq) to analyze FLC expression and map sense transcript polyadenylation sites across eight stages of Arabidopsis Col-0 embryo development (12). Three proximally polyadenylated FLC isoforms were identified that terminated within the first intron of FLC near the Polycomb nucleation region (13) (Fig. 1A and Dataset S1). Proximal FLC isoforms accounted for over 80% of poly(A) reads in preglobular Col-0 embryos (Fig. 1 B and C). From the early heart stage onwards, a majority of FLC transcripts were canonically spliced and used distal polyadenylation sites, and by the mature green stage, poly(A) reads from proximal FLC were undetectable.
Fig. 1.
FLC polyadenylation site usage changes during embryo development. (A, Top) Distribution of reads with untemplated 3′-terminal poly(A) sequences across eight timepoints of embryo development and grouped into clusters. (A, Middle) Transcript models corresponding to full-length (distal) FLC and proximally polyadenylated (proximal) FLC. (A, Bottom) Mean RNA-seq read coverage at FLC from three biological replicates at eight embryo stages. (B) Mean transcripts per million (TPM) of FLC across the embryonic time series. (C) Poly(A) reads per million (RPM) contained in proximal and distal poly(A) clusters, respectively. (D) Differential gene expression between Col FRI and Col-0 embryos; genes more than twofold significantly higher (green) or lower (orange) in Col FRI embryos are marked (adjusted P value <10−3, DEseq2). (E) Estimated abundance of proximal and distal FLC isoforms in three biological replicates of Col-0, 35S::FCA, and Col FRI early heart embryos. (F) Distal:proximal poly(A) ratio in the same samples as in E, calculated as log2[proximal poly(A) RPM/distal poly(A) RPM].
To understand how the repressor FCA and activator FRIGIDA modulate FLC we analyzed polyadenylation site usage in dissected early heart stage embryos from Col-0 (mutant for FRIGIDA) (7), Col FRI, and transgenic Col-0 carrying a 35S::FCA overexpression construct (4). The latter was chosen over an fca mutant because of functional redundancy between fca and fpa (1). FLC was the most significantly up-regulated gene in Col FRI embryos, reaching higher transcript abundance at the early heart stage than any timepoint measured during Col-0 embryo development (Fig. 1 D and E and Dataset S2) (11). This higher FLC expression was associated with distal polyadenylation site usage; while total transcript abundance was 4.4-fold higher, distal isoforms were 10-fold higher and proximal isoforms were half as abundant as in Col-0 (Fig. 1E). Proximal poly(A) reads roughly equalled distal reads in Col-0, but distal poly(A) usage outweighed proximal poly(A) reads 20 to 1 in Col FRI embryos (Fig. 1F and Dataset S2). No RNA-seq reads could be unambiguously attributed to COOLAIR antisense transcripts.
There was a shift from distal to proximal polyadenylation in the FLC transcript in 35S::FCA embryos compared to Col-0 (Fig. 1 E and F), further reducing functional FLC expression. FCA thus represses FLC expression in Col-0 by promoting proximal polyadenylation via a mechanism enhanced by 35S::FCA. The shift to predominantly distal polyadenylation site usage at FLC by the end of embryogenesis in Col-0 correlates with reduction of functional FCA through increased FCA autofeedback negative regulation (Dataset S3) (1). FRI thus antagonizes this FCA-induced proximal polyadenylation by promoting use of the distal FLC polyadenylation site early in embryogenesis.
FRI Is Required in Early Embryogenesis to Fully Delay Flowering.
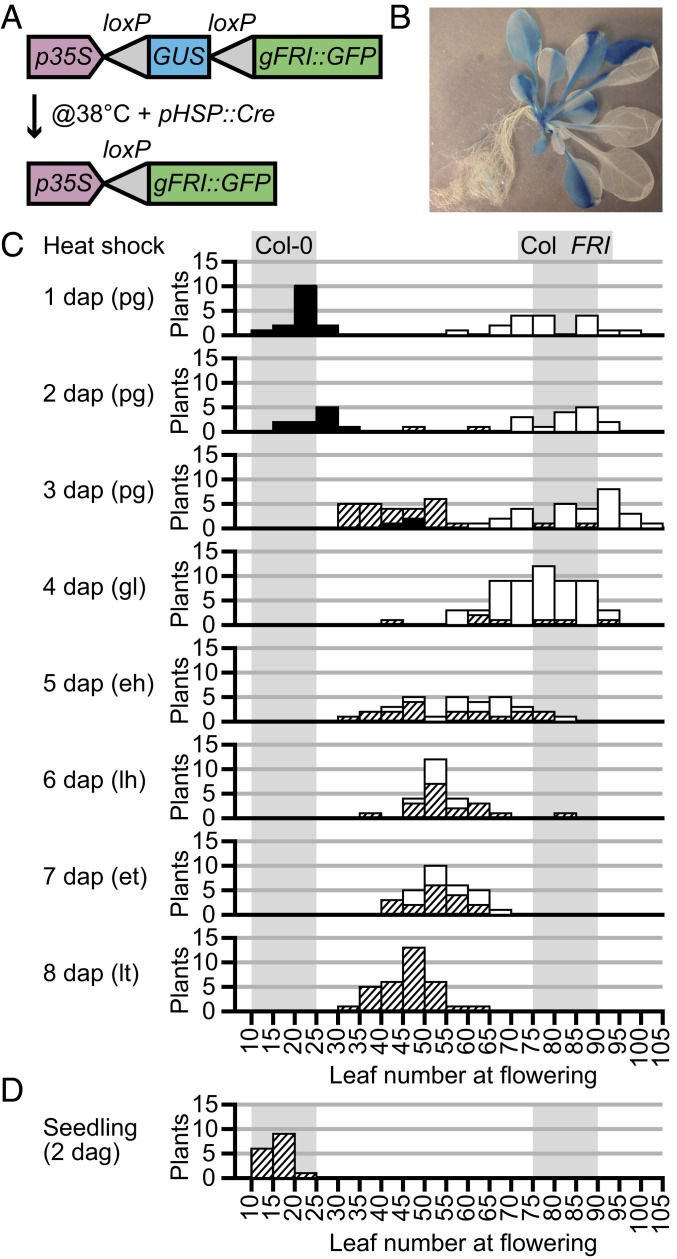
To further address the timing of the antagonism between FCA and FRI, we established a sector analysis system to address when FRI function is required to fully delay flowering. To generate FRI positive sectors we introduced a 35S:lox-GUS-lox-FRI-GFP construct containing a GUS reporter fusion flanked by two loxP sites between the 35S promoter and FRI-GFP into a heatshock-inducible Cre-loxP recombinase line (Fig. 2A). The Cre recombinase catalyses recombination between the two loxP sites leading to the excision of the GUS reporter enabling FRI to be expressed under the control of the 35S CaMV promoter. FRIGIDA active sectors are thus negative for GUS (Fig. 2B).
Fig. 2.
Induction of FRIGIDA sectors during embryogenesis and influence on flowering time. (A) Schematic of the 35S:lox-GUS-lox-FRI-GFP construct before and after heatshock. (B) GUS staining in a mosaic plant heat shocked as a 2-d-old seedling. (C) Flowering time assessed by final leaf number in plants that were heat shocked at different stages of embryo development. White bars, plants with FRI-expressing sectors covering all leaves; hatched bars, plants with FRI-expressing sectors and FRI-lacking sectors; black bars, plants with no FRI-expressing sectors. Shaded regions show the range of control plants: Columbia-0 (fri) or plants transformed with an active FRI allele. (D) Leaf number at flowering of 35S:lox-GUS-lox-FRI-GFP plants heat shocked as 2-d-old seedlings for 30 min. Control ranges are shaded as in C.
Heatshock treatment was applied to a developmental series of embryos ranging from 1 to 8 d after pollination (dap). Seeds from heatshocked embryos were sown and the flowering time was analyzed based on total leaf number (Fig. 2C). When plants began to flower, they were harvested and stained to detect the presence of the GUS reporter, thereby allowing the size and location of FRI-expressing/GUS-lacking sectors to be determined. FRI-expressing sectors generated in preglobular and globular embryos resulted in individuals that flowered as late as the Col FRI control (Fig. 2). By contrast, when FRI was induced in older embryos the resulting plants did not flower as late. Comparison of plants that had similar sized FRI-expressing sectors but generated at different times supported the conclusion that FRI induction before the heart stages of embryo development causes maximum delay in flowering. Use of the very strong estrogen-inducible promoter may account for delayed flowering seen in a previous study when FRI is induced in 5-d-old seedlings (14). In our experiments, heatshock treatments of seedlings 2 d after germination generated FRI-expressing sectors, but again, this had no effect on flowering time.
Discussion
Mechanistic dissection of FLC regulation has previously focused on postembryonic development. FLC expression is low in Col-0 seedlings through an FCA-induced antisense-mediated chromatin silencing mechanism. Here, we now find that FCA promotes proximal polyadenylation of the sense nascent FLC transcript in Col-0 early embryos. This is developmentally specific as by the end of embryogenesis and during the rest of postembryonic development, the sense transcript is polyadenylated predominantly at the distal site.
We found that FRIGIDA up-regulates FLC expression by changing FLC polyadenylation site usage in heart stage embryos. Overexpression of FCA promotes proximal polyadenylation of the sense transcript, whereas FRI shifts polyadenylation to the distal site. Based on our sector analysis data, FRI antagonizes FCA-induced proximal polyadenylation during early embryogenesis. How FRI blocks the premature polyadenylation is an interesting question. FRI promotes a high transcriptional state and a chromatin environment involving H3K36me3 and H3K4me3, which promotes transcriptional initiation and faster elongation (10). The faster elongation may influence polyadenylation site choice through kinetic coupling of transcription and cotranscriptional processing, with stronger 3′-polyadenylation sites being chosen if already transcribed (15). Alternatively, FRI could act as an antiterminator more directly by regulating what associates with the RNA Pol II carboxy terminal domain via a transcriptional checkpoint.
By integrating these data with our previous mechanistic understanding, we propose that a low transcriptional state is induced by FCA-mediated proximal polyadenylation of FLC to establish a specific FLC chromatin state during early embryo development. This would be propagated in seedlings by the antisense-mediated Polycomb inheritance mechanism (5). A separation of an establishment phase and a maintenance phase fits with the original paradigm established for Polycomb-silenced genes. However, if an active FRIGIDA is present, FLC proximal polyadenylation is prevented in the embryo, the switch to the Polycomb-repressed state does not occur, and the locus is maintained in a high transcriptional state that persists through vegetative development (10). Opposing functions of cotranscriptional regulators at a very specific developmental stage thus set the quantitative expression state of FLC.
Materials and Methods
All methods are given in SI Appendix. Materials and associated protocols are available from corresponding authors.
Supplementary Material
Acknowledgments
We thank Silvia Gazzani and Clare Lister for construction of the lines for sector analysis and the Vienna Biocenter Core Facilities GmbH for sequencing. We acknowledge funding from a Royal Society Professorship, UK Biotechnology and Biological Sciences Research Council Institute Strategic Programme GEN (BB/P013511/1), a Wellcome Trust Investigator Award to C.D., and support by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program grant 637888 to M.D.N.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102753118/-/DCSupplemental.
Data Availability
RNA-seq data have been deposited in Gene Expression Omnibus (GEO) (GSE166728).
References
- 1.Sonmez C., et al., RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc. Natl. Acad. Sci. U.S.A. 108, 8508–8513 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quesada V., Macknight R., Dean C., Simpson G. G., Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22, 3142–3152 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu F., Marquardt S., Lister C., Swiezewski S., Dean C., Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327, 94–97 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Liu F., et al., The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Fang X., et al., The 3′ processing of antisense RNAs physically links to chromatin-based transcriptional control. Proc. Natl. Acad. Sci. U.S.A. 117, 15316–15321 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., et al., Quantitative regulation of FLC via coordinated transcriptional initiation and elongation. Proc. Natl. Acad. Sci. U.S.A. 113, 218–223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johanson U., et al., Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Gazzani S., Gendall A. R., Lister C., Dean C., Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132, 1107–1114 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geraldo N., Bäurle I., Kidou S., Hu X., Dean C., FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol. 150, 1611–1618 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z., Jiang D., He Y., FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat. Plants 4, 836–846 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Tao Z., et al., Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat. Plants 5, 424–435 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Hofmann F., Schon M. A., Nodine M. D., The embryonic transcriptome of Arabidopsis thaliana. Plant Reprod. 32, 77–91 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Qüesta J. I., Song J., Geraldo N., An H., Dean C., Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353, 485–488 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Kong X., et al., Expression of FRIGIDA in root inhibits flowering in Arabidopsis thaliana. J. Exp. Bot. 70, 5101–5114 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Giono L. E., Kornblihtt A. R., Linking transcription, RNA polymerase II elongation and alternative splicing. Biochem. J. 477, 3091–3104 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in Gene Expression Omnibus (GEO) (GSE166728).