Significance
Plants produce approximately 300 O-prenylated aromatics, with their O-prenyl moieties often being crucial to their bioactivities. This study identified a gene from grapefruit encoding an aromatic O-prenyltransferase (O-PT) belonging to the UbiA superfamily. The O-PT was shown responsible for the biosynthesis of pharmaceutically active O-prenylated coumarins that cause grapefruit–drug interactions, an adverse effect disturbing the pharmacokinetics of more than 85 medications. Another UbiA O-PT for coumarins was isolated from Angelica keiskei, an apiaceous medicinal plant. Phylogenetic analysis of the rutaceous and apiaceous O-PTs suggested that aromatic O-prenylation activity emerged in parallel in these distant plant taxa. The molecular evolution of aromatic O-PTs from plant UbiA proteins may aid citrus breeding and a synthetic biology approach to bioactive O-prenylated coumarins.
Keywords: plant-specialized metabolism, aromatic O-prenyltransferase, parallel evolution, coumarin, grapefruit–drug interactions
Abstract
Plants produce ∼300 aromatic compounds enzymatically linked to prenyl side chains via C–O bonds. These O-prenylated aromatic compounds have been found in taxonomically distant plant taxa, with some of them being beneficial or detrimental to human health. Although their O-prenyl moieties often play crucial roles in the biological activities of these compounds, no plant gene encoding an aromatic O-prenyltransferase (O-PT) has been isolated to date. This study describes the isolation of an aromatic O-PT gene, CpPT1, belonging to the UbiA superfamily, from grapefruit (Citrus × paradisi, Rutaceae). This gene was shown responsible for the biosynthesis of O-prenylated coumarin derivatives that alter drug pharmacokinetics in the human body. Another coumarin O-PT gene encoding a protein of the same family was identified in Angelica keiskei, an apiaceous medicinal plant containing pharmaceutically active O-prenylated coumarins. Phylogenetic analysis of these O-PTs suggested that aromatic O-prenylation activity evolved independently from the same ancestral gene in these distant plant taxa. These findings shed light on understanding the evolution of plant secondary (specialized) metabolites via the UbiA superfamily.
Plants produce many O-prenylated aromatic molecules possessing prenyl side chains attached to the aromatic cores via C–O bonds. These aromatic core structures include flavonoids, coumarins, xanthones, and aromatic alkaloids, with roughly half of them (ca. 150 structures) being classified as coumarins (1, 2). Some O-prenylated aromatic compounds have pharmaceutically beneficial activities, whereas others are deleterious to human health (1, 2). These beneficial/detrimental activities are often due to or enhanced by O-prenyl moieties (3–6).
Native coumarin O-prenyltransferase (O-PT) activities of Rutaceae and Apiaceae, plants that accumulate large amounts of O-prenylated coumarins, have been characterized biochemically, with membrane-bound proteins found to be involved in coumarin O-prenylation (7, 8). To date, ∼50 UbiA superfamily genes encoding membrane-bound proteins have been found to encode aromatic C-PTs (9), which transfer prenyl moieties to aromatic cores via C–C bonds. Although these genes were shown to encode enzymes involved in plant primary and specialized metabolism (10), no gene encoding an aromatic O-PT has yet been identified in plants.
O-prenylated aromatic compounds have been detected in several distant plant families, including Asteraceae, Boraginaceae, Fabaceae, Hypericaceae, Rutaceae, and Apiaceae, but are not ubiquitous throughout the plant kingdom (1). The lack of knowledge of genes encoding aromatic O-PTs has prevented a determination of the appearance of aromatic O-prenylation activity during plant speciation.
Among Rutaceae, Citrus species accumulate large amounts of O-prenylated coumarins, especially in their flavedo (outer pericarp) (11–14). Citrus O-prenylated coumarins have shown various pharmaceutical properties (2), including anticancer (4, 15), antimicrobial (3), and anti-inflammatory (16) activities, although some of these derivatives have shown undesirable effects in humans. Citrus fruits and juices enhance the bioavailability of orally administrated medications, which can lead to overdoses and increased side effects (5, 17). These “grapefruit–drug interactions” have been found to alter the pharmacokinetics of more than 85 medications, including statins and calcium channel blockers (17). The US Food and Drug Administration has cautioned consumers not to consume grapefruits or grapefruit juice at times close to taking such drugs (18). Citrus species are thought to alter drug pharmacokinetics by inactivating CYP3A4, the major xenobiotic-metabolizing enzyme in the intestines and liver (5, 17).
Furanocoumarins (FCs) are tricyclic coumarins containing a furan ring. O-geranylated forms of FCs, including bergamottin and its oxidative derivatives (Fig. 1), are promising candidates responsible for grapefruit–drug interactions due to their potent inhibition of CYP3A4 (5, 17, 19). CYP3A4-catalyzed metabolism of their furan rings produces reactive chemicals that inactivate this enzyme itself (20), whereas their O-geranyl side chains contribute to the interaction with CYP3A4 (6), enhancing the metabolism of their furan rings. Bergamottin and 6′,7′-dihydroxybergamottin showed seven- and 160-fold higher in vitro inhibitory activity, respectively, than the nongeranylated form bergaptol (5). Furthermore, O-geranyl moieties act as linkers to form FC dimers, called paradisins, which are more potent CYP3A4 inactivators than monomeric O-geranylated FCs (5). These findings suggest that paradisins, along with O-geranylated FC monomers, may be involved in grapefruit–drug interactions.
Fig. 1.
The biosynthetic pathway of the major O-prenylated aromatic compounds in grapefruit. Biosynthetic steps catalyzed by O-PTs and a C-PT are shown in red and blue, respectively. Metabolites derived from umbelliferone 7-O-GT and bergaptol 5-O-GT are highlighted in yellow and orange, respectively. Bergamottin and its downstream metabolites are considered promising candidates responsible for grapefruit–drug interactions.
Starting with transcriptome analysis of flavedo tissues, this study describes the isolation of a gene encoding a coumarin O-PT involved in bergamottin biosynthesis in grapefruit. The gene product was characterized functionally, and the contribution of O-PT orthologs to coumarin biosynthesis was assessed in various Citrus species. In addition, an aromatic O-PT was isolated from Angelica keiskei, an apiaceous medicinal plant producing O-prenylated coumarins (21). The evolutionary development of aromatic O-prenylation activity in plants was assessed by phylogenetic analysis of O-PTs from the taxonomically distant families Rutaceae and Apiaceae.
Results
Construction of a Transcriptome Dataset from Grapefruit Flavedo Tissues.
Because the native enzymes catalyzing coumarin O-prenylation in lemon flavedo were shown to possess characteristics common to PTs in the UbiA superfamily, that is, membrane association and divalent cation requirement (8), the genomes of Citrus sinensis (sweet orange, the male parent of grapefruit) and Citrus clementina (clementine) in the public Phytozome database were searched to identify genes in this family. The search term “UbiA” identified 26 and 27 loci in the sweet orange and clementine genomes, respectively. In contrast, a search of the Arabidopsis thaliana genome identified only six loci, which form a minimal gene set only for the six primary metabolic pathways relevant to the UbiA superfamily (10). These in silico searches identified members of the UbiA superfamily potentially involved in the specialized metabolism of Citrus genus, as exemplified by the synthesis of 8-C-geranylumbelliferone by a lemon UbiA C-PT, ClPT1 (22). To better identify aromatic O-PT candidates, we performed transcriptome analysis of grapefruit, which is rich in O-prenylated coumarins that are involved in grapefruit–drug interactions.
Grapefruits primarily accumulate two types of O-prenylated phenolics, auraptene- and bergamottin-related compounds, which are likely synthesized by distinct O-geranylation pathways and catalyzed by umbelliferone 7-O-geranyltransferase (U7OGT) and bergaptol 5-O-GT (B5OGT), respectively (Fig. 1) (11, 12). Quantification of these major O-prenylated coumarins in different grapefruit organs revealed that they are most abundant in flavedo tissues of immature and mature fruits (SI Appendix, Figs. S1 and S2), from which we constructed a transcriptome dataset (Datasets S1 and S2).
Isolation of Candidate Genes Encoding O-PTs from Grapefruit.
To comprehensively identify UbiA PTs involved in plant-specialized metabolism in the grapefruit flavedo transcriptome, in silico screening was performed using seven query sequences, that is, ClPT1 (22) and six sweet orange proteins probably orthologous to Arabidopsis thaliana UbiA PTs functionally involved in primary metabolic pathways (10). Candidates for coumarin O-PTs were selected based on three criteria (SI Appendix, Fig. S3A): 1) low to moderate amino acid identity to UbiA proteins involved in plant primary metabolism (SI Appendix, Table S1), 2) the presence in another public transcriptome dataset prepared from grapefruit leaves that accumulate O-geranylated coumarins (SI Appendix, Table S2), and 3) transcripts per million (TPM)-based expression levels of grapefruit flavedo contigs to remove those with zero TPM (Dataset S2). Nine contigs, mapped to four genes in the sweet orange genome, were selected (SI Appendix, Fig. S3B).
Using RT-PCR and primers for corresponding sweet orange sequences, we isolated the full coding sequences (CDSs) of three candidate genes from grapefruit flavedo, called C. × paradisi PT 1–3 (CpPT1–3) (SI Appendix, Fig. S3B). Although we failed to amplify a CDS for the other candidate gene, the transcript corresponding to c22985_g1_i1 seemed to be nonfunctional as shown by the lack of a coding region containing the second aspartate-rich motif that is essential for prenylation reactions in UbiA proteins (23, 24). In silico analysis predicted that the polypeptides CpPT1 and CpPT2 each contains two aspartate-rich motifs, multiple transmembrane regions, and N-terminal signal peptides to plastids (transit peptides, TPs), all of which are characteristics of plant UbiA-PT proteins (9, 10, 22, 25) (SI Appendix, Fig. S4). Although CpPT3 was not predicted to have a TP, its score was just below the threshold for the detection of a TP.
Functional Screening of CpPTs.
These individual PTs were transiently expressed in Nicotiana benthamiana leaves by agroinfiltration because this system provides an appropriate functional expression of those membrane proteins (9, 25), while heterologous expression of plant UbiA proteins embedded in plastid membranes often fails in microbial hosts. Microsomes prepared from these leaves were subjected to B5OGT and U7OGT assays in the presence of Mg2+ as a cofactor. Neither CpPT2 nor CpPT3 was able to synthesize any O-geranylated products in B5OGT assays, in which bergaptol was used as the prenyl acceptor substrate (SI Appendix, Fig. S5 A and B). CpPT2 was also unable to synthesize any product in U7OGT assays, in which umbelliferone was the aromatic substrate (SI Appendix, Fig. S5 A and B). CpPT3, like ClPT1, was found to catalyze the synthesis of two products, 8-C-geranylumbelliferone and a byproduct, but not auraptene (SI Appendix, Fig. S5C) (22). Because CpPT3 and ClPT1 show 95% amino acid identity (SI Appendix, Fig. S4A), these two enzymes may have orthologous functions in Citrus. CpPT2 and CpPT3 were also incubated in the presence of various substrate pairs, but no clear O-prenylation activity was detected (SI Appendix, Fig. S5 A and B).
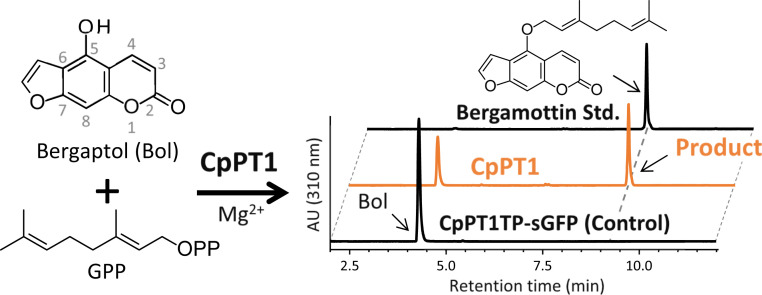
Although CpPT1-expressing microsomes did not yield any products in U7OGT assays, high-performance liquid chromatography analysis showed that these microsomes generated a product in B5OGT assays (Fig. 2). This product had the identical retention time and mass spectrometry (MS) and MS2 spectra as bergamottin, a finding confirmed by direct comparison with a standard specimen (SI Appendix, Fig. S6 A and B). MS2 analysis using the positive ion mode found that the major peak after fragmentation of the molecular ion of bergamottin (m/z = 339) was at m/z = 203, with the difference of 136 daltons corresponding to the molecular weight of a geranyl chain (SI Appendix, Fig. S6C). This total loss of a prenyl moiety is possibly unique to O-prenylated aromatics, as one carbon at the benzyl position remains after the fragmentation of C-prenyl moieties (26), resulting in a loss from C-geranyl moieties of 124 daltons (22, 26). These biochemical findings suggested that CpPT1 is a strong B5OGT candidate.
Fig. 2.
The B5OGT activity of CpPT1. Microsomes prepared from N. benthamiana leaves expressing CpPT1 were used as crude enzymes, with the negative control being microsomes prepared from N. benthamiana leaves expressing a chimeric protein consisting of N-terminal amino acids 1 to 70 of CpPT1, including the TP and synthetic green fluorescence protein (CpPT1TP-sGFP). Ultraviolet chromatograms at 310 nm of the full assay and the negative control assay are shown at a comparable scale.
Enzymatic Properties of CpPT1.
The specificity of CpPT1 for coumarin molecules as prenyl acceptors was analyzed in the presence of the prenyl donor geranyl diphosphate (GPP; Table 1 and SI Appendix, Fig. S6). CpPT1 was able to transfer prenyl moieties to coumarin molecules with hydroxy groups at the C5 (no. 3 and 7) and C8 (no. 13 and 16) positions. These enzymatic products were identified as O-geranylated forms by direct comparison with available standards or, if standards were unavailable, were predicted to be the O-geranylated forms by their MS2 fragmentation patterns (SI Appendix, Fig. S6 D–K). Coumarin and FC derivatives without hydroxy groups at C5 and C8, as well as molecules in other phenolic classes, were not recognized as substrates.
Table 1.
Substrate specificity of CpPT1
| Class | No. | Compound | Hydroxy position on the coumarin ring (for coumarins) | DMAPP | GPP | FPP |
| Simple coumarins | 1 | Umbelliferone | 7 | N.D. | N.D. | |
| 2 | 6-Hydroxycoumarin | 6 | N.D. | |||
| 3 | 5,7-Dihydroxycoumarin | 5,7 | N.D. | + | N.D. | |
| 4 | Esculetin | 6,7 | N.D. | |||
| 5 | 5-Methoxy-7-hydroxycoumarin | 7 | N.D. | |||
| 6 | Scopoletin | 7 | N.D. | |||
| 7 | 5-Hydroxy-7-methoxycoumarin | 5 | N.D. | + | N.D. | |
| 8 | Isoscopoletin | 6 | N.D. | |||
| 9 | Daphnetin 7-methylether | 8 | N.D. | |||
| 10 | Limettin | — | N.D. | |||
| Linear FCs | 11 | Psoralen | — | N.D. | ||
| 12 | Bergaptol | 5 | N.D. | + | N.D. | |
| 13 | Xanthotoxol | 8 | N.D. | + | N.D. | |
| 14 | Bergapten | — | N.D. | |||
| 15 | Xanthotoxin | — | N.D. | |||
| 16 | 8-Hydroxybergapten | 8 | N.D. | + | N.D. | |
| Angular FCs | 17 | Isobergaptol | 5 | N.D. | ||
| 18 | Sphondinol | 6 | N.D. | |||
| Phenylpropanes | 19 | p-Coumaric acid | N.D. | N.D. | ||
| 20 | 2,4-Dihydroxycinnamic acid | N.D. | ||||
| 21 | Ferulic acid | N.D. | N.D. | |||
| Flavonoids | 22 | Isoliquiritigenin | N.D. | |||
| 23 | Genistein | N.D. | ||||
| 24 | Naringenin | N.D. | ||||
| Homogentisic acid | 25 | Homogentisic acid | N.D. | |||
Simple coumarins (no. 1 to 10), linear FCs (11 to 16), angular FCs (17 and 18), phenylpropanes (19 to 21), flavonoids (22 to 24), and homogentisic acid (25) were tested as possible prenyl acceptor substrates of CpPT1. DMAPP, GPP, and farnesyl diphosphate (FPP) were tested as possible prenyl donor substrates. Reaction mixtures (100 µL) containing microsomes, 200 µM prenyl acceptor substrate, 200 µM prenyl donor substrate, and 10 mM MgCl2, pH 7.6 were incubated at 28 °C for 20 h. Independent triplicate reactions gave the same result. The substrate pairs resulting in enzymatic products are marked with pluses. N.D., not detected. The chemical structures of the aromatic substrates are shown in SI Appendix, Fig. S6A (all molecules) and SI Appendix, Table S3 (molecules accepted by CpPT1)
The prenyl donor specificity of CpPT1 was also investigated using prenyl diphosphates of different chain lengths in the presence of coumarin derivatives accepted in GT assays, but no reaction products were observed for any combination (Table 1). Umbelliferone, p-coumaric acid, and ferulic acid were also tested in dimethylallyltransferase (DT) assays because their dimethylallylated forms have been found in Rutaceae species, with C-dimethylallylated umbelliferone molecules being precursors of FCs (27, 28). However, no reaction products were observed (Table 1). Taken together, these biochemical analyses demonstrated that CpPT1 functions as a coumarin 5/8-O-GT.
Kinetic analysis of the O-geranylation activity of CpPT1 in the presence of the five coumarin substrates demonstrated that bergaptol was the optimal prenyl acceptor (SI Appendix, Table S3). Kinetic analysis of GPP measured in the presence of bergaptol or its structural isomer, xanthtotoxol, resulted in similar apparent Km values, irrespective of the prenyl acceptor substrate (SI Appendix, Table S4). These results indicated that the recombinant CpPT1 mainly functions as B5OGT, with an optimal pH in the neutral to weak alkaline region (SI Appendix, Fig. S7A). Analysis of its divalent cation preference showed that Mg2+ was the optimal cofactor for CpPT1 (SI Appendix, Fig. S7B). The B5OGT enzymatic activities of recombinant CpPT1 and the native microsomes prepared from lemon flavedo were similar (8).
In planta Gene Expression Profile and Subcellular Localization of CpPT1.
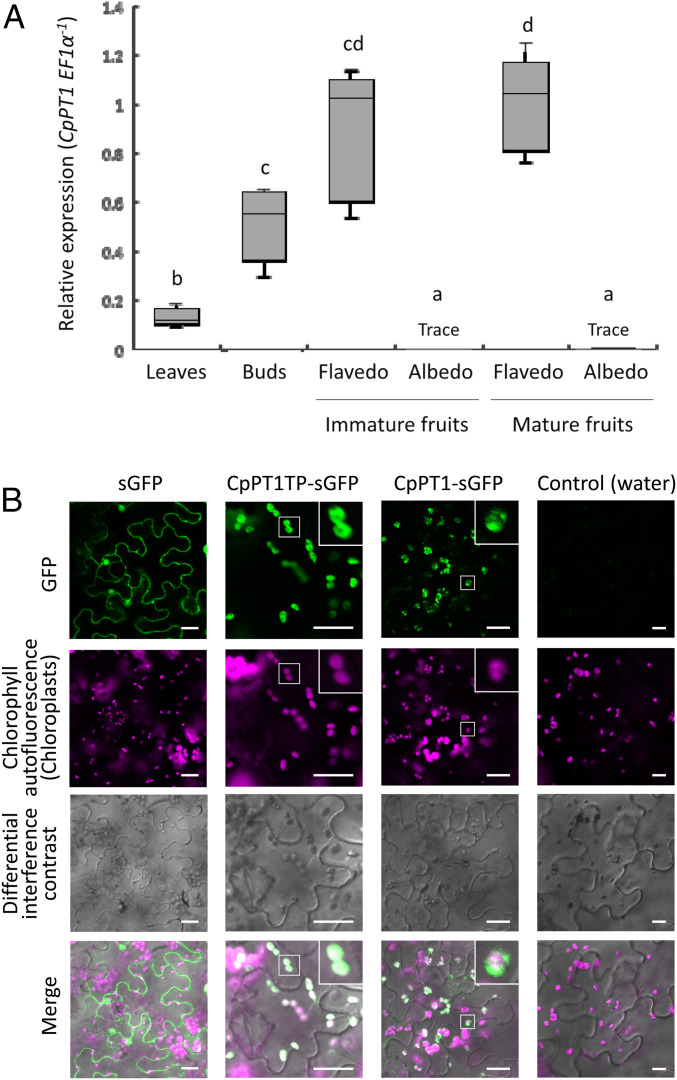
To assess the involvement of CpPT1 in the biosynthesis of O-prenylated coumarins in grapefruit, the levels of expression of CpPT1 were determined in different organs (Fig. 3A). Assessment of both immature and mature fruits showed that CpPT1 was highly expressed in flavedo but weakly expressed in albedo. This gene is also expressed in buds and leaves at similar to lower levels than in flavedo tissues. This expression profile is in agreement with the patterns of accumulation of bergamottin and its downstream derivatives (SI Appendix, Fig. S2).
Fig. 3.
Organ-specific gene expression and subcellular localization of CpPT1. (A) Organ-specific expression of CpPT1. Ratios of the relative expression of CpPT1 to CpEF1α in grapefruit leaves, buds, and the flavedo and albedo of immature and mature fruits normalized to the average CpPT1/CpEF1α ratio in mature flavedo (n = 5 biological replicates) are shown. Relative levels of expression are shown as box plots (center line, median; box limits, first and third quartiles; whiskers, minimum and maximum). Significant differences between groups are indicated by letters (P < 0.05 by Games-Howell tests). (B) Subcellular localization of CpPT1TP-sGFP and CpPT1-sGFP. Free sGFP, CpPT1TP-sGFP, and CpPT1-sGFP were transiently expressed in N. benthamiana leaves by agroinfiltration, with the negative control consisting of leaves infiltrated by water. Chloroplasts were visualized by chlorophyll autofluorescence. For merging, the brightness and contrast of the fluorescent images were adjusted in an unbiased manner, with magenta being a pseudocolor for the chlorophyll autofluorescence signal. Enlarged images are inserted for CpPT1TP-sGFP and CpPT1-sGFP. (Scale bars, 20 µm.)
To assess the subcellular localization of CpPT1 in planta, synthetic green fluorescent protein (sGFP) was fused to the C terminus of the first 70 amino acids representing the predicted TP of CpPT1 (CpPT1TP-sGFP) or to the C terminus of the full-length polypeptide (CpPT1-sGFP) (SI Appendix, Fig. S4B). Confocal microscopy of epidermal cells of N. benthamiana leaves expressing these GFP fusion proteins showed that both chimeric proteins localized to chloroplasts (Fig. 3B). These results strongly suggest that CpPT1 functions in plastids in grapefruit, consistent with the plastid localization of the MEP pathway that provides GPP in plant cells.
FC Chemotypes Related to the Gene Structures of CpPT1 Orthologs in Citrus.
Citrus domestication involved crossing of the four ancestral species, citron (C. medica), pure (or ancestral) mandarin (C. reticulata), papeda (C. micrantha), and pummelo (C. grandis), among themselves and/or with their descendants, generating most of the currently cultivated varieties, such as sweet orange, lemon, and grapefruit (29–31). The concentrations and compositions of coumarins in the flavedo of these species vary, with papeda, pummelo, and citron varieties producing high quantities, and mandarin varieties producing low quantities, of coumarins (Fig. 4A) (12). Interestingly, the major O-geranylated FCs in citrus, such as bergamottin and its downstream metabolites, are undetectable in citron varieties, despite their high contents of FCs (12).
Fig. 4.
Conservation of CpPT1 orthologs in Citrus genus. (A) Total FC and O-geranylated FC contents in the flavedo of papeda (C. micrantha, the blue circle); in different varieties of pummelo (C. grandis, red circles), citron (C. medica, yellow circles), and mandarin (C. reticulata, orange circles); and in marsh grapefruit (the gray circle). Quantitative data, expressed as means ± SEs of four samples each of Reinking and Tahiti pummelo and five samples each of all other varieties, have been reported previously (12). Trace amounts of metabolites were set at zero for figure construction. The varieties of pummelo tested included Chandler, Deep Red, Kao Pan, Pink, Reinking, Seedless, and Tahiti pummelo; the varieties of citron tested included Buddha’s hand, Corsican, and Etrog citron; and the varieties of mandarin tested included Beauty, Cleopatra, Dancy, Fuzhu, Nan Feng Mi Chu, Owari Satsuma, San Hu Hong Chu, Shekwasha, Sunki, Wase Satsuma, and Willowleaf mandarins. All mandarin varieties tested were domesticated, possessing pummelo-derived genomic segments. The FC molecules assayed included 6′,7′-dihydroxybergamottin, 8-geranyloxypsoralen, bergamottin, bergapten, bergaptol, byakangelicin, byakangelicol, cnidicin, cnidilin, epoxybergamottin, heraclenin, heraclenol, imperatorin, isoimperatorin, isopimpinellin, oxypeucedanin, oxypeucedanin hydrate, phellopterin, psoralen, xanthotoxin, and xanthotoxol, with 6′,7′-dihydroxybergamottin, 8-geranyloxypsoralen, bergamottin, and epoxybergamottin being O-geranylated FC derivatives. (B) Isolation of functional CpPT1 orthologs from papeda. Ultraviolet chromatograms at 310, 300, and 330 nm of GT assay mixtures of C. micrantha PT1a/b (CmiPT1a/b) with the aromatic substrates bergaptol (Bol, no. 12), xanthotoxol (Xol, no. 13), and 5-hydroxy-7-methoxycoumarin (5H7M, no. 7), respectively, with CpPT1TP-sGFP used as a negative control. All chromatograms are shown at a comparable scale except for that of the standard. The chemical structures of the aromatic substrates are shown in SI Appendix, Fig. S6A and Table S3.
The relationship between CpPT1 orthologs and the coumarin profile was investigated in members of the genus Citrus. A blastn search using the CpPT1 CDS detected a single close homolog each in the pummelo, citron, and pure mandarin genomes (SI Appendix, Fig. S8). Because genomic information on papeda was unavailable, we isolated two full-length CDSs 98% identical to CpPT1 from papeda by RT-PCR, named CmiPT1a/b (SI Appendix, Fig. S9A).
The pummelo genome contains a putative CpPT1 orthologous gene, in accordance with the pummelo being the female parent of grapefruit (SI Appendix, Fig. S8A) (29). Biochemical characterization demonstrated that, like CpPT1, papeda CmiPT1a/b encode functional O-GTs for both bergaptol and xanthotoxol (Fig. 4B and SI Appendix, Fig. S9 B–D). In contrast, the citron CpPT1 ortholog contains an insertion consisting of an 8 base pair (bp) repeat containing an in-frame stop codon at the 3′ end of its third exon (SI Appendix, Fig. S8 A and B). This insertion was confirmed by PCR sequencing of the genomes of three citron varieties previously shown to be devoid of O-geranylated FCs (SI Appendix, Fig. S8B) (12). These citron varieties were also found to contain two other O-geranylated coumarins, 5G7M and auraptene (12), suggesting that these varieties possess GPP pools available for coumarin prenylation. Together with previous findings (12), these results suggest that the 8 bp insertion causes loss of function of the citron CpPT1 ortholog, resulting in the absence of O-geranylated FCs from this species. The CpPT1 ortholog in pure mandarin was found to contain a deletion and an insertion (SI Appendix, Fig. S8 A and C), consistent with findings showing the undetectable or very low accumulation of O-geranylated FCs in domesticated mandarin varieties (12).
Isolation of a Coumarin O-PT from Apiaceae.
O-prenylated coumarins have also been detected in vegetables and medicinal plants in the family Apiaceae (2, 21). Because this family is taxonomically distant from Rutaceae in angiosperms (32) and to obtain evolutionary insight into the emergence of aromatic O-prenylation activity in plants, we sought to determine whether a UbiA-PT gene was involved in the synthesis of O-prenylated coumarins in Angelica keiskei, a medicinal plant endemic to Japan that is locally consumed as a vegetable (21). We selected a variety of A. keiskei accumulating O-dimethylallylated bergaptol (isoimperatorin) and its oxidative derivatives as well as C-prenylated chalcones (21). Biochemical characterization of the O-DT activities for FCs using crude enzymes prepared from the leaves of A. keiskei showed that the native bergaptol 5-O-DT (B5ODT) activity leading to the synthesis of isoimperatorin required divalent cations as a cofactor and was associated with the cell membrane (SI Appendix, Fig. S10 A–D). Native A. keiskei microsomes also possessed xanthotoxol 8-O-DT activity (X8ODT), resulting in the production of imperatorin, although this product was not detected in the A. keiskei plants used in this study (SI Appendix, Fig. S10 E and F). These findings suggest that members of the UbiA protein superfamily are involved in the O-prenylation of FCs in Apiaceae as well as in Rutaceae.
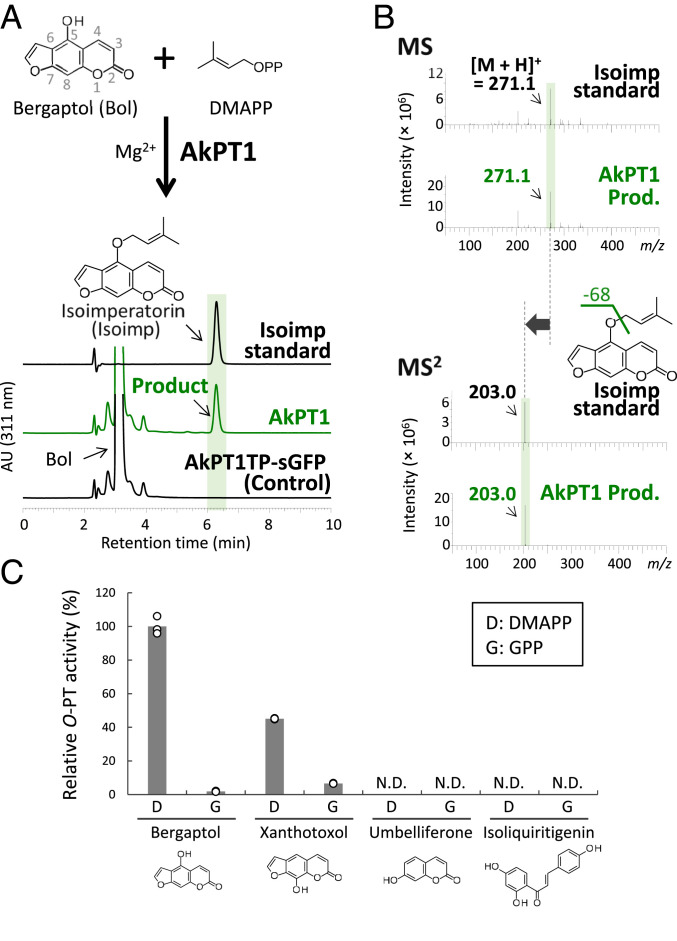
Using degenerate primers that were designed based on conserved amino acid regions among UbiA PTs, a full-length CDS was isolated from A. keiskei leaves by RT-PCR and subsequent rapid amplification of complementary DNA ends. This gene, named AkPT1, was found to encode a protein with the three conserved polypeptide features of UbiA PTs, similar to CpPT1 (SI Appendix, Figs. S11 and S12). In vitro enzymatic characterization using the N. benthamiana transient expression system showed that AkPT1 has B5ODT and X8ODT activities (Fig. 5 and SI Appendix, Fig. S13 A and B). AkPT1, however, did not prenylate umbelliferone or isoliquiritigenin, the prenyl acceptor involved in the biosynthesis of C-prenylated chalcones in A. keiskei (Fig. 5C). For bergaptol and xanthotoxol, this enzyme accepted GPP less efficiently than dimethylallyl diphosphate (DMAPP) as a prenyl donor (Fig. 5C and SI Appendix, Fig. S13 C–F), suggesting that, in A. keiskei, AkPT1 acts primarily as a coumarin 5/8-O-DT. The enzymatic properties associated with the B5ODT activity of AkPT1 were also determined (SI Appendix, Fig. S14). The apparent Km values of AkPT1 for bergaptol and DMAPP were found to be 3.1 ± 0.3 and 1.8 ± 0.1 µM, respectively. In addition, the optimal pH for AkPT1 was in the alkaline region (pH 7.5 to 9.0), and its optimal divalent cation cofactor was Mg2+.
Fig. 5.
Biochemical characterization of AkPT1. (A) Ultraviolet chromatograms at 311 nm of B5ODT reaction mixtures with N. benthamiana leaf microsomes containing recombinant AkPT1 and AkPT1TP-sGFP (negative control). (B) MS2 spectrum of the reaction product in the positive ion mode. The loss of 68 daltons probably corresponds to fragmentation caused by the loss of the O-dimethylallyl moiety attached to the FC structure. (C) Substrate specificity of AkPT1. Bars represent AkPT1 relative to the average B5ODT activity in triplicate samples. N.D., not detected.
Phylogenetic Relationship of Aromatic O-PTs from Distant Angiosperm Families.
To determine whether enzymatic properties reflect a phylogenetic relationship and to obtain evolutionary insight on the emergence of aromatic O-prenylation ability in plants, the O-PTs identified in Rutaceae and Apiaceaee were subjected to phylogenetic analysis. A neighbor-joining phylogenetic tree showed that, of the six primary metabolism clades, O-PTs from both plant families were located closest to the VTE2-1 clade responsible for tocopherol biosynthesis (Fig. 6 and SI Appendix, Table S5). These findings suggest that the molecular evolution of VTE2-1 is responsible for the emergence of O-PT genes in both Rutaceae and Apiaceae. However, the O-PTs from Rutaceae and Apiacee did not form a common clade but were grouped in independent clades together with C-PTs from the same plant families, that is, all rutaceous O-PTs were included in one clade together with citrus C-PTs (ClPT1 and CpPT3) and CpPT2 (22), whereas the apiaceous O-PT was included in another clade, together with the other apiaceous C-PTs (PcPT1, PsPT1, and PsPT2) (33, 34). A search for orthologs of the rutaceous O-PTs in public transcriptomes of apiaceous species that produce O-prenylated FCs detected no candidates (SI Appendix, Tables S6 and S7) (35). Similarly, no candidate orthologs of the apiaceous O-PT were detected in the grapefruit flavedo transcriptome (SI Appendix, Table S8). These in silico analyses suggest that the O-PT genes of Rutaceae and Apiaceae each evolved independently from VTE2-1 in a parallel manner.
Fig. 6.
Phylogenetic relationship of aromatic O-PTs in the UbiA superfamily from Rutaceae and Apiaceae. A neighbor-joining phylogenetic tree of UbiA proteins. The tree was constructed with 1,000 bootstrap tests based on a ClustalW multiple alignment of UbiA proteins, including CpPT1, CmiPT1a/b, and AkPT1. Bootstrap values are shown for nodes separating clades and for nodes between C-PTs and O-PTs in Rutaceae and Apiaceae. The bar represents an amino acid substitution rate per site of 0.20. The clades of primary metabolism-related proteins are marked with gray circles. The clades of specialized metabolism-related PTs are highlighted with differently colored circles depending on their possible ancestors, with VTE2-1-, VTE2-2-, and PPT-related clades indicated in orange, green, and blue, respectively, together with their aromatic substrates at the family scale. The PTs of Rutaceae and Apiaceae isolated in this study are highlighted in red and bold. The plant species and the accession numbers of the input sequences are provided in SI Appendix, Table S5.
Two aromatic O-PT genes belonging to the UbiA superfamily have been identified in bacteria (36, 37). Although the enzymes encoded by these bacterial genes showed less than 16% amino acid identities with the plant O-PTs, the amino acid identities among plant O-PTs were greater than 40% even when comparing O-PTs from different plant families (SI Appendix, Table S9). This homology difference between kingdoms also suggests that O-PTs in Rutaceae and Apiaceae emerged in a plant taxon-specific manner.
Discussion
Aromatic prenylation diversifies the chemical structures of plant metabolites, as these enzymes vary widely in substrate specificity for both prenyl donors and acceptors and in regio specificity (38). In addition, the structures of these metabolites are further altered by the chemical modifications of transferred prenyl moieties through, for example, hydroxylation, cyclization, and dimerization (38). These diversities in prenylation and subsequent chemical modifications resulted in the diversification of their biological activities (39).
To date, several UbiA PTs catalyzing aromatic C-prenylations have been reported to be involved in primary and specialized metabolism in various plant families (10). Phylogenetic analyses of plant UbiA proteins have demonstrated that all C-PTs reportedly involved in specialized metabolism evolved from one of three primary metabolic C-PT groups, that is, VTE2-1 for tocopherol biosynthesis, VTE2-2 for plastoquinone biosynthesis, and polyprenyl diphosphate transferase (PPT) for ubiquinone biosynthesis, with the ancestor chosen in a taxon-specific manner (9). The present study showed the evolution of aromatic O-PTs was most likely due to divergence from VTE2-1. Similar to aromatic C-prenylation by other UbiA PTs, CpPT1 was found to catalyze both substrate- and regio-specific O-prenylations. UbiA C-PTs transfer prenyl moieties to carbons at the ortho-positions of phenolic hydroxy moieties, with VTE2-1 showing this regio specificity (40). CpPT1 was found to generate only O-geranylated products from two simple coumarin derivatives possessing the C5-hydroxy moiety (no. 3 and no. 7), despite the availability of the C6 position on these molecules for C-prenylation. Thus, CpPT1 being able to specifically catalyze O-prenylation may have derived from a neofunctionalized form of C-PT. The C- and O-PTs involved in the specialized metabolism of coumarins in Rutaceae likely originated from the common ancestor VTE2-1, suggesting that gene duplication and neofunctionalization events of VTE2-1 have produced a set of PTs catalyzing a variety of aromatic prenylation reactions. This molecular evolution could also apply to Apiaceae. Further identification of genes encoding UbiA PTs is necessary to determine the details of evolutionary pathways from VTE2-1 in these plant families.
The enzymatic properties of recombinant CpPT1 and the native microsomes of citrus flavedo toward B5OGT were similar (8). The pattern of CpPT1 expression in grapefruit organs matched the pattern of accumulation of its reaction products, including bergamottin and its derivatives. The predicted or biochemically verified functions of CpPT1 orthologs were associated with the accumulation of major citrus O-geranylated FCs in the flavedo of various ancestral Citrus species, strongly suggesting that CpPT1 and its orthologs play pivotal roles in the O-geranylation of FCs in Citrus species. FCs also accumulate in the pulp or flesh of citrus fruits, although these concentrations are generally lower than those in flavedo (12). However, O-geranylated FCs are undetectable in the pulp of citron varieties (SI Appendix, Fig. S15), which possess CpPT1 orthologs containing an 8 bp insertion, resulting in an in-frame stop codon (12). Thus, targeting this gene during the breeding of citrus fruits may weaken grapefruit–drug interactions. Accumulation of other prenylated coumarins, such as auraptene and O-dimethylallylated FCs, in citron varieties containing an insertion in the CpPT1 gene suggests the presence in citrus genomes of coumarin PT(s) distinct from CpPT1 and ClPT1 orthologs.
Although CpPT1 is well conserved among members of the genus Citrus, phylogenetic analysis of rutaceous and apiaceous O-PTs suggested that these plant taxa independently acquired aromatic O-prenylation activity in a parallel evolutionary manner, providing an example of repeated molecular evolution of plant UbiA proteins. Because O-prenylation enhances the relevant biological activities of FCs, both Rutaceae and Apiaceae likely acquired coumarin O-prenylation ability for chemical defenses (5, 41). Interestingly, the coumarin accumulation patterns of these plant families were found to be similar. Rutaceae and Apiaceae store large quantities of O-prenylated coumarins in oil cavities and oil ducts, respectively, with both being extracellular compartments filled with hydrophobic metabolites, such as essential oil terpenes (13, 42, 43). O-Prenylation largely increases the hydrophobicity of aromatic molecules because of the masking of hydroxyl residues by hydrophobic prenyl chains. This reaction may enhance the accumulation of coumarins in hydrophobic extracellular compartments, although the mechanisms by which hydrophobic metabolites are exported to such compartments remain undetermined. In addition, FCs are generally toxic, being responsible for photo-induced genotoxicity and P450 inactivation (20, 41, 44), suggesting that sequestering these molecules from vital organelles by storing them in extracellular compartments reduces the risk of self-toxicity. This strategy would be complementary to a glycosylation-associated self-resistance mechanism that increases the hydrophilicity of specialized metabolites, allowing their sequestration in intracellular vacuoles (45).
The results of the present study suggest that aromatic O-prenylation has developed independently among several plant families other than Rutaceae and Apiaceae (1). For example, Hypericaceae and Boraginaceae were shown to possess VTE2-2- and PPT-derived C-PTs, respectively (46, 47). Based on the molecular evolution of C- and O-PTs in Rutaceae and Apiaceae, the discovery in future of new aromatic O-PTs derived from VTE2-2 and PPT1 can be expected. The identification and characterization of genes encoding novel aromatic O-PTs in as yet unexplored plant families, including Hypericaceae and Boraginaceae, may provide deeper insights into the genetic diversity of aromatic O-prenylation in plants and the common/divergent physiological roles of these reactions in plant taxa.
In addition to their O-prenylation activity, UbiA proteins were found to evolve independently to become specialized PTs accepting similar or the same aromatic substrates, such as flavonoids and coumarins, in a convergent evolutionary manner (9, 48). Therefore, the repeated molecular evolution of members of the UbiA superfamily in pathways enabling different types of aromatic prenylation ability may underlie the biosynthesis of no less than 1,000 prenylated aromatics in plants (38). Repeated molecular evolution of different enzymatic properties has been reported in other transferase families involved in specialized metabolism in plants. For example, methyltransferases in the salicylic acid benzoic acid theobromine synthase family catalyze the N-methylation of xanthine alkaloids during caffeine biosynthesis. Both the ability to recognize purine alkaloids as methyl acceptors and N-methylation activity were acquired evolutionarily by multiple caffeine-producing plant taxa in an independent manner (49). Similarly, the repeated evolution of proteins of the plant uridine diphosphate-glycosyltransferase superfamily has included alterations in sugar acceptor (50) and sugar donor (51) specificity and in C-glycosylation (52). These examples suggest that the repeated molecular evolution of enzymes largely contributed to the current distribution of specialized metabolites in the plant kingdom.
In summary, the present study provides experimental evidence for the functional diversification of plant UbiA proteins to aromatic O-PTs, an evolutionary process that likely occurred independently in Rutaceae and Apiaceae. Identification of CpPT1 may enable the efficient creation of citrus varieties showing reduced grapefruit–drug interactions. Correlation of the expression patterns and/or genotypes of the FC O-GT gene with coumarin profiles may help determine coumarin metabolism in this agronomically important genus in which few relevant genes have been identified to date (53). Knockout of genes responsible for the formation of FC backbones may reduce not only grapefruit–drug interactions but citrus phototoxicity caused by FC photosensitization (54), which limits the application of citrus essential oils as cosmetic ingredients (55, 56). The diverse pharmaceutical activities of O-prenylated coumarins, often because of their O-prenyl moieties (2–4, 15, 28), suggest that the coumarin O-PT genes identified in this study could encode proteins producing valuable coumarin varieties.
Materials and Methods
Plant Materials, Reagents, and Experimental Procedures.
Grapefruits (Citrus × paradisi cv. Marsh) grown at the Yuasa farm of Kindai University were collected, and their organs (e.g., young leaves, mature leaves, buds, and the albedo and flavedo of immature and mature fruits) were prepared as described (SI Appendix, Fig. S1). Other citrus plants were grown and their fruits collected at the Agronomic Research Station National Institute for Agronomic and Environmental Research/International Center for Agronomic Research and Development of San Giuliano in Corsica. Angelica keiskei plants (the Oshima variety) for pilot experiments were maintained in the soil field of the Yamashina Botanical Research Institute, and the same variety of A. keiskei for main experiments were commercially purchased in Japan. Plant tissues were immediately frozen in liquid nitrogen and stored at –80 °C, if necessary. Phenolic compounds and prenyl diphosphates for the characterization of CpPT1 were purchased from Sigma-Aldrich, Herboreal Ltd, Extrasynthese, Tokyo Chemical Industry Co., Ltd, and Indofine Chemical Company, Inc. For the other experiments, DMAPP was synthesized as described (57), and GPP was kindly provided by T. Kuzuyama of the University of Tokyo and T. Kawasaki of Kyoto University. Auraptene standards were kindly provided by A. Murakami of the University of Hyogo and Y. Uto of Tokushima University, and 8-geranylumbelliferone was also generously provided by Y. Uto. The details of the experimental procedures used for transcriptomic analysis, phytochemical analysis, gene isolation, plasmid construction, microsome preparation, in vitro enzyme assays, liquid chromatography/MS analysis, qRT-PCR, subcellular localization analysis, in silico analysis, and statistical analysis are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Yann Froelicher (International Center for Agronomic Research and Development, San Giuliano), Dr. Patrick Ollitrault (Centro de Protección Vegetal y Biotecnología, Valencia), and Dr. Nobumasa Nito (Kindai University) for providing citrus samples. We also thank Dr. David Baulcombe (Cambridge University) for the pBIN61-P19 plasmid, Dr. Tsuyoshi Nakagawa (Shimane University) for the pGWB vectors, Dr. Toshiaki Mitsui (Niigata University) for the pWxTP-DsRed plasmid, and Dr. Hiroshi Kouchi (International Christian University) for the pHKN29 plasmid. We are grateful to Dr. Akira Murakami (University of Hyogo) and Dr. Yoshihiro Uto (Tokushima University) for the prenylated coumarin standards and Dr. Tomohisa Kuzuyama (The University of Tokyo) and Dr. Takashi Kawasaki (Kyoto University) for GPP. We also thank Ms. Keiko Kanai (Kyoto University), Mr. Patrick Riveron, and Mr. Clément Charles (Université de Lorraine) for technical assistance. Liquid chromatography-ion trap-time of flight/MS analyses of the enzymatic characteristics of AkPT1 were performed in collaboration with the Development and Assessment of Sustainable Humanosphere system of the Research Institute for Sustainable Humanosphere (RISH), Kyoto University. Plants were grown on the Plant Experimental Platform in Lorraine (Université de Lorraine). Transcriptome analysis of grapefruit flavedo tissues was performed with the technical support of Dr. Tomoaki Sakamoto (Kyoto Sangyo University) in the Plant Global Education Project of Nara Institute of Science and Technology. The Plant Global Education Project was supported by a Grant-in-Aid for Scientific Research for Plant Graduate Students from the Nara Institute of Science and Technology, supported by the Ministry of Education, Culture, Sports, Science and Technology. This work was also financially supported by the SAKURA program of the Japan Society for the Promotion of Science (JSPS) Research Fellowship for Young Scientists (to R.M.), by JSPS Overseas Research Fellowships (to R.M.), by Grants-in-Aid for Scientific Research (26712013 to A.S. and 16H03282 to K.Y.), by the New Energy and Industrial Technology Development Organization Project (16100890 to K.Y.), by the Precursory Research for Embryonic Science and Technology program from the Japan Science and Technology Agency (no. JPMJPR20D7 to R.M), by the French Grand Est Region and the French Research Ministry (to C.V.), by the “Bioprolor2” project (Grand Est Region, to A.H.), and by the “Impact Biomolecules” project of the Lorraine Université d'Excellence (Investissements d’avenir, the Agence Nationale de la Recherche , to A.H.). Additional support was provided by RISH, Kyoto University (Mission 5, to K.Y.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022294118/-/DCSupplemental.
Data and Materials Availability
All data needed to evaluate the conclusions in this paper are present in the paper and/or in SI Appendix. “Nucleotide” and RNA sequencing data have been deposited in the DNA Data Bank of Japan Sequence Read Archive. The accession numbers of CpPT1–3, CmiPT1a/b, and AkPT1 are LC557129–LC557134. The raw RNA sequencing reads are available as DRA010472.
References
- 1.Epifano F., Genovese S., Menghini L., Curini M., Chemistry and pharmacology of oxyprenylated secondary plant metabolites. Phytochemistry 68, 939–953 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Curini M., Cravotto G., Epifano F., Giannone G., Chemistry and biological activity of natural and synthetic prenyloxycoumarins. Curr. Med. Chem. 13, 199–222 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Adams M., et al., Antimycobacterial activity of geranylated furocoumarins from Tetradium daniellii. Planta Med. 72, 1132–1135 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Murakami A., et al., Auraptene, a citrus coumarin, inhibits 12-O-tetradecanoylphorbol-13-acetate-induced tumor promotion in ICR mouse skin, possibly through suppression of superoxide generation in leukocytes. Jpn. J. Cancer Res. 88, 443–452 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanley M. J., Cancalon P., Widmer W. W., Greenblatt D. J., The effect of grapefruit juice on drug disposition. Expert Opin. Drug Metab. Toxicol. 7, 267–286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sevrioukova I. F., Structural insights into the interaction of cytochrome P450 3A4 with suicide substrates: Mibefradil, azamulin and 6′,7′-dihydroxybergamottin. Int. J. Mol. Sci. 20, 4245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamerski D., Schmitt D., Matern U., Induction of two prenyltransferases for the accumulation of coumarin phytoalexins in elicitor-treated Ammi majus cell suspension cultures. Phytochemistry 29, 1131–1135 (1990). [DOI] [PubMed] [Google Scholar]
- 8.Munakata R., et al., Characterization of coumarin-specific prenyltransferase activities in Citrus limon peel. Biosci. Biotechnol. Biochem. 76, 1389–1393 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Munakata R., et al., Convergent evolution of the UbiA prenyltransferase family underlies the independent acquisition of furanocoumarins in plants. New Phytol. 225, 2166–2182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkelblech J., Fan A., Li S. M., Prenyltransferases as key enzymes in primary and secondary metabolism. Appl. Microbiol. Biotechnol. 99, 7379–7397 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Dugrand A., et al., Coumarin and furanocoumarin quantitation in citrus peel via ultraperformance liquid chromatography coupled with mass spectrometry (UPLC-MS). J. Agric. Food Chem. 61, 10677–10684 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Dugrand-Judek A., et al., The distribution of coumarins and furanocoumarins in Citrus species closely matches Citrus phylogeny and reflects the organization of biosynthetic pathways. PLoS One 10, e0142757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voo S. S., Grimes H. D., Lange B. M., Assessing the biosynthetic capabilities of secretory glands in Citrus peel. Plant Physiol. 159, 81–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durand-Hulak M., et al., Mapping the genetic and tissular diversity of 64 phenolic compounds in Citrus species using a UPLC–MS approach. Ann. Bot. 115, 861–877 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun S., Phrutivorapongkul A., Dibwe D. F., Balachandran C., Awale S., Chemical constituents of Thai Citrus hystrix and their antiausterity activity against the PANC-1 human pancreatic cancer cell line. J. Nat. Prod. 81, 1877–1883 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Okuyama S., et al., Anti-inflammatory and neuroprotective effects of auraptene, a citrus coumarin, following cerebral global ischemia in mice. Eur. J. Pharmacol. 699, 118–123 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Bailey D. G., Dresser G., Arnold J. M., Grapefruit-medication interactions: Forbidden fruit or avoidable consequences? CMAJ 185, 309–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The United States Food and Drug Administration , Grapefruit Juice and Some Drugs Don’t Mix. https://www.fda.gov/consumers/consumer-updates/grapefruit-juice-and-some-drugs-dont-mix. Accessed 16 April 2021.
- 19.Wangensteen H., Molden E., Christensen H., Malterud K. E., Identification of epoxybergamottin as a CYP3A4 inhibitor in grapefruit peel. Eur. J. Clin. Pharmacol. 58, 663–668 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Lin H. L., Kenaan C., Hollenberg P. F., Identification of the residue in human CYP3A4 that is covalently modified by bergamottin and the reactive intermediate that contributes to the grapefruit juice effect. Drug Metab. Dispos. 40, 998–1006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baba K., Studies on the chemical components and biological activities of Angelica keiskei Koidzumi. Bull. Osaka Univ. Pharmaceut. Sci. 7, 55–87 (2013). [Google Scholar]
- 22.Munakata R., et al., Molecular cloning and characterization of a geranyl diphosphate-specific aromatic prenyltransferase from lemon. Plant Physiol. 166, 80–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., et al., A heteromeric membrane-bound prenyltransferase complex from hop catalyzes three sequential aromatic prenylations in the bitter acid pathway. Plant Physiol. 167, 650–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng W., Li W., Structural insights into ubiquinone biosynthesis in membranes. Science 343, 878–881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munakata R., et al., Isolation of Artemisia capillaris membrane-bound di-prenyltransferase for phenylpropanoids and redesign of artepillin C in yeast. Commun. Biol. 2, 384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simons R., Vincken J. P., Bakx E. J., Verbruggen M. A., Gruppen H., A rapid screening method for prenylated flavonoids with ultra-high-performance liquid chromatography/electrospray ionisation mass spectrometry in licorice root extracts. Rapid Commun. Mass Spectrom. 23, 3083–3093 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Brown S. A., Steck W., 7-Demethylsuberosin and osthenol as intermediates in furanocoumarin biosynthesis. Phytochemistry 12, 1315–1324 (1973). [Google Scholar]
- 28.Epifano F., et al., Neuroprotective effect of prenyloxycoumarins from edible vegetables. Neurosci. Lett. 443, 57–60 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Wu G. A., et al., Genomics of the origin and evolution of Citrus. Nature 554, 311–316 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Wang L., et al., Genome of wild mandarin and domestication history of mandarin. Mol. Plant 11, 1024–1037 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Curk F., et al., Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 117, 565–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chase M. W., et al., An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linnean Soc. 181, 1–20 (2016). [Google Scholar]
- 33.Karamat F., et al., A coumarin-specific prenyltransferase catalyzes the crucial biosynthetic reaction for furanocoumarin formation in parsley. Plant J. 77, 627–638 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Munakata R., et al., Molecular evolution of parsnip (Pastinaca sativa) membrane-bound prenyltransferases for linear and/or angular furanocoumarin biosynthesis. New Phytol. 211, 332–344 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Murray R. D. H., Méndez J., Brown S. A., The Natural Coumarins (Wiley & Sons, New York, USA, 1982). [Google Scholar]
- 36.Awakawa T., Fujita N., Hayakawa M., Ohnishi Y., Horinouchi S., Characterization of the biosynthesis gene cluster for alkyl-O-dihydrogeranyl-methoxyhydroquinones in Actinoplanes missouriensis. ChemBioChem 12, 439–448 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Zeyhle P., et al., A membrane-bound prenyltransferase catalyzes the O-prenylation of 1,6-dihydroxyphenazine in the marine bacterium Streptomyces sp. CNQ-509. Chembiochem 15, 2385–2392 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Yazaki K., Sasaki K., Tsurumaru Y., Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70, 1739–1745 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Alhassan A. M., Abdullahi M. I., Uba A., Umar A., Prenylation of aromatic secondary metabolites: A new frontier for development of novel drugs. Trop. J. Pharm. Res. 13, 307–314 (2014). [Google Scholar]
- 40.Collakova E., DellaPenna D., Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 127, 1113–1124 (2001). [PMC free article] [PubMed] [Google Scholar]
- 41.Neal J. J., Wu D., Inhibition of insect cytochromes P450 by furanocoumarins. Pestic. Biochem. Physiol. 50, 43–50 (1994). [Google Scholar]
- 42.Maggi F., et al., Essential oil chemotypification and secretory structures of the neglected vegetable Smyrnium olusatrum L. (Apiaceae) growing in central Italy. Flavour Fragr. J. 30, 139–159 (2015). [Google Scholar]
- 43.Reinold S., Hahlbrock K., In situ localization of phenylpropanoid biosynthetic mRNAs and proteins in parsley (Petroselinum crispum). Bot. Acta 110, 431–443 (1997). [Google Scholar]
- 44.Bourgaud F., et al., Biosynthesis of coumarins in plants: a major pathway still to be unravelled for cytochrome P450 enzymes. Phytochem. Rev. 5, 293–308 (2006). [Google Scholar]
- 45.Sirikantaramas S., Yamazaki M., Saito K., Mechanisms of resistance to self-produced toxic secondary metabolites in plants. Phytochem. Rev. 7, 467–477 (2008). [Google Scholar]
- 46.Nagia M., et al., Sequential regiospecific gem-diprenylation of tetrahydroxyxanthone by prenyltransferases from Hypericum sp. New Phytol. 222, 318–334 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Yazaki K., Kunihisa M., Fujisaki T., Sato F., Geranyl diphosphate: 4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and characterization of a key enzyme in shikonin biosynthesis. J. Biol. Chem. 277, 6240–6246 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Wang R., et al., Molecular characterization and phylogenetic analysis of two novel regio-specific flavonoid prenyltransferases from Morus alba and Cudrania tricuspidata. J. Biol. Chem. 289, 35815–35825 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang R., O’Donnell A. J., Barboline J. J., Barkman T. J., Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc. Natl. Acad. Sci. U.S.A. 113, 10613–10618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson A. E., Wu S., Tian L., PgUGT95B2 preferentially metabolizes flavones/flavonols and has evolved independently from flavone/flavonol UGTs identified in Arabidopsis thaliana. Phytochemistry 157, 184–193 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Noguchi A., et al., Local differentiation of sugar donor specificity of flavonoid glycosyltransferase in Lamiales. Plant Cell 21, 1556–1572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ito T., Fujimoto S., Suito F., Shimosaka M., Taguchi G., C-Glycosyltransferases catalyzing the formation of di-C-glucosyl flavonoids in citrus plants. Plant J. 91, 187–198 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Limones-Mendez M., et al., Convergent evolution leading to the appearance of furanocoumarins in citrus plants. Plant Sci. 292, 110392 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Naganuma M., Hirose S., Nakayama Y., Nakajima K., Someya T., A study of the phototoxicity of lemon oil. Arch. Dermatol. Res. 278, 31–36 (1985). [DOI] [PubMed] [Google Scholar]
- 55.Buzek J., Ask B., Regulation (EC) No 1223/2009 of the European parliament and of the council of 30 November 2009 on cosmetic products. Off. J. Eur. Union L 342, 59–209 (2009). [Google Scholar]
- 56.IFRA standard, “Citrus oils and other furocoumarins containing essential oils,” 48th Amendment (2015). https://ifrafragrance.org/standards/IFRA_STD48_0174.pdf. Accessed 16 April 2021.
- 57.Cornforth R. H., Popjak G., Chemical syntheses of substrates of sterol biosynthesis. Methods Enzymol. 15, 359–390 (1969). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in this paper are present in the paper and/or in SI Appendix. “Nucleotide” and RNA sequencing data have been deposited in the DNA Data Bank of Japan Sequence Read Archive. The accession numbers of CpPT1–3, CmiPT1a/b, and AkPT1 are LC557129–LC557134. The raw RNA sequencing reads are available as DRA010472.