Significance
G protein–coupled receptors (GPCRs) are important regulators of cellular and biological functions and are primary targets of therapeutic drugs. About 100 mammalian GPCRs are still considered orphan receptors because they lack a known endogenous ligand. We report the deorphanization of GPR182, which is expressed in endothelial cells of the microvasculature. We show that GPR182 is an atypical chemokine receptor, which binds CXCL10, 12, and 13. However, binding does not induce downstream signaling. Consistent with a scavenging function of GPR182, mice lacking GPR182 have increased plasma levels of chemokines. In line with the crucial role of CXCL12 in hematopoietic stem cell homeostasis, we found that loss of GPR182 results in increased egress of hematopoietic stem cells from the bone marrow.
Keywords: GPCR, orphan, chemokine
Abstract
G protein–coupled receptor 182 (GPR182) has been shown to be expressed in endothelial cells; however, its ligand and physiological role has remained elusive. We found GPR182 to be expressed in microvascular and lymphatic endothelial cells of most organs and to bind with nanomolar affinity the chemokines CXCL10, CXCL12, and CXCL13. In contrast to conventional chemokine receptors, binding of chemokines to GPR182 did not induce typical downstream signaling processes, including Gq- and Gi-mediated signaling or β-arrestin recruitment. GPR182 showed relatively high constitutive activity in regard to β-arrestin recruitment and rapidly internalized in a ligand-independent manner. In constitutive GPR182-deficient mice, as well as after induced endothelium-specific loss of GPR182, we found significant increases in the plasma levels of CXCL10, CXCL12, and CXCL13. Global and induced endothelium-specific GPR182-deficient mice showed a significant decrease in hematopoietic stem cells in the bone marrow as well as increased colony-forming units of hematopoietic progenitors in the blood and the spleen. Our data show that GPR182 is a new atypical chemokine receptor for CXCL10, CXCL12, and CXCL13, which is involved in the regulation of hematopoietic stem cell homeostasis.
G protein–coupled receptors (GPCRs) represent the largest group of transmembrane receptors encoded in the genome, and they are the largest group of proteins targeted by approved drugs (1, 2). GPCRs are very versatile and can bind ligands of different physicochemical properties, including ions, lipids, biogenic amines, peptides, or proteins, such as chemokines (3). Primarily by activation of heterotrimeric G proteins, GPCRs regulate multiple functions in basically all cells of mammalian organisms (4). Despite their large physiological and pharmacological relevance, the endogenous ligands, activating mechanisms and physiological functions of more than 100 GPCRs, are still not known and these receptors are therefore referred to as “orphan” receptors (3, 5). G protein–coupled receptor 182 (GPR182) is an orphan receptor, although it has been suggested to bind adrenomedullin (6), but this observation could not be confirmed (7). GPR182 was initially described to be widely expressed in various organs (8). More-detailed analyses in developing zebrafish and in mice revealed that Gpr182 is preferentially expressed in the vascular endothelium (9, 10). Widespread expression in endothelial cells of adult mice was shown using a mouse line expressing β-galactosidase under the control of the Gpr182-promoter (11), and expression of GPR182 in sinusoidal endothelial cells was reported based on immunohistochemical analysis (12). Whereas the role of GPR182 in endothelial cells is unknown, GPR182 expression was also reported in intestinal stem cells, where the receptor was shown to negatively regulate proliferation during regeneration and adenoma formation (11).
Chemokine receptors are a family of 22 GPCRs that respond to 52 chemokines (13). Upon activation, they induce G protein–mediated intracellular signaling processes which, in many cases, regulate the migration of leukocytes (14). However, more recent work has shown that the function of chemokines goes beyond the regulation of leukocyte migration and can also affect other cell functions and cell types (13, 15, 16). In addition, and in contrast to other groups of GPCRs, the chemokine receptor family contains several members, which bind chemokines but are unable to signal through G proteins. These so-called “atypical chemokine receptors” (ACKRs) can indirectly regulate the interactions between chemokines and conventional chemokine receptors by controlling chemokine localization, distribution, and abundance (13, 16, 17). As most conventional chemokine receptors, ACKRs typically bind subgroups of chemokines. For instance, ACKR1 binds various chemokines and transports them across endothelial cells or, when expressed on erythrocytes, buffers chemokine levels in the blood (18). ACKR2 functions as a scavenger receptor by binding several C-C motif chemokine ligand (CCL) chemokines and plays various roles in the immune system (19). ACKR3 only binds C-X-C motif chemokine ligand 11 (CXCL11) and CXCL12 and controls the CXCL12–CXCR4 signaling axis by direct interaction with CXCR4 and by scavenging CXCL12 (20, 21). ACKR4 binds the conventional chemokine receptor ligands CCL19, CCL21, CCL25, and CXCL19 (18, 19).
Here, we show that GPR182 functions as an atypical chemokine receptor for CXCL10, CXCL12, and CXCL13 and that it is involved in preventing hematopoietic stem cell egress from bone marrow.
Results
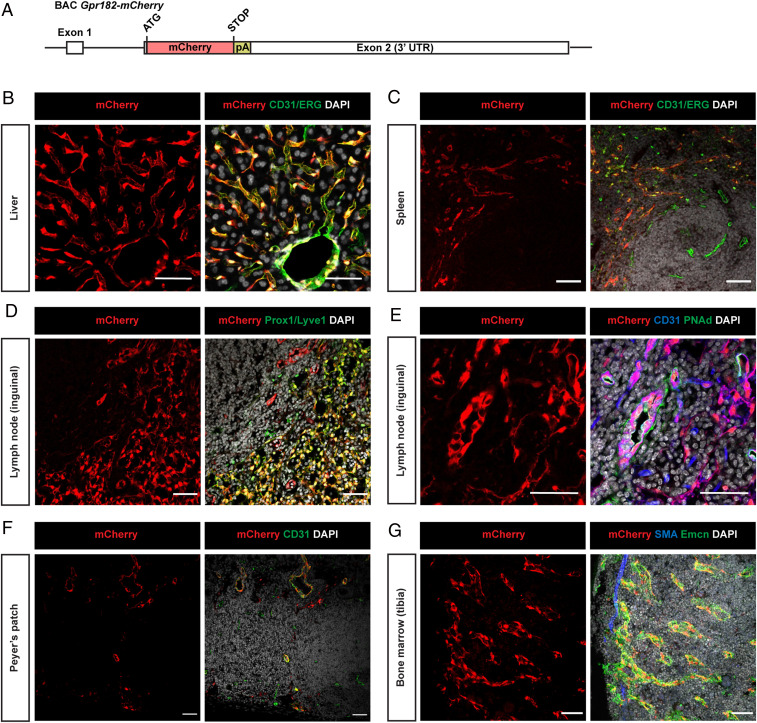
To analyze the in vivo expression of GPR182, we generated a bacterial artificial chromosome (BAC)–based transgenic mouse line expressing the monomeric red fluorescent protein mCherry under the control of the mouse Gpr182 promoter (Fig. 1A). In Gpr182mCherry mice, we observed GPR182 expression in most organs. Costaining with different markers confirmed expression of GPR182 in vascular endothelial cells of lungs, bone marrow, lymph nodes, Peyer’s patches, liver, and spleen (Fig. 1 and SI Appendix, Fig. S1A). GPR182 was not expressed in vascular endothelial cells from conductive arterial vessels (SI Appendix, Fig. S1 B and C). In the liver and spleen, GPR182 was expressed in basically all sinusoidal cells as well as in endothelial cells of central veins and the hepatic artery (Fig. 1 B and C). Also, sinusoidal endothelial cells and high endothelial cells of the lymph node expressed GPR182 (Fig. 1 D and E), as well as endothelial cells of Peyer’s patches (Fig. 1F). In the bone marrow, endomucin-positive endothelial cells of the sinusoids express GPR182, whereas endomucin-negative endothelial cells of arteries, which are identified by staining for alpha-smooth muscle actin (αSMA), do not express GPR182 (Fig. 1G). This could be confirmed by single-cell sequencing of bone marrow endothelial cells, which showed expression of Gpr182 in cells positive for the sinusoidal endothelial cell marker stabilin2 but not for arterial markers, whereas the related receptor Ackr3 was not found to be expressed in sinusoidal endothelial cells (SI Appendix, Fig. S1D). In the small and large intestine, we found GPR182 expression mainly in endothelial cells of the lamina propria, and there was no indication for expression in epithelial cells (SI Appendix, Fig. S1 E and F). We also found GPR182 to be expressed by various lymphatic endothelial cells including those of the skin, the intestine, and lymph nodes (Fig. 1D and SI Appendix, Fig. S2).
Fig. 1.
GPR182 is expressed in microvascular endothelial cells. (A) Schematic of part of the BAC-based mouse Gpr182-mCherry reporter transgene, which had a total length of 234 kb. UTR: untranslated region. (B–G) Representative immunofluorescence confocal images of cryosections of the indicated organs from Gpr182-mCherry BAC transgenic mice. The mCherry signal corresponds to endogenous mCherry fluorescence. Cryosections were stained with antibodies against vascular or lymphatic endothelial markers (CD31 and ETS-related gene (ERG), as well as Prox1 and Lyve1, respectively). PNAd antibody was used to specifically mark high endothelial venules from lymph nodes. mCherry was often predominantly localized in the nucleus. Shown are results of representative experiments of at least three independently performed experiments. (Scale bars, 50 µm.)
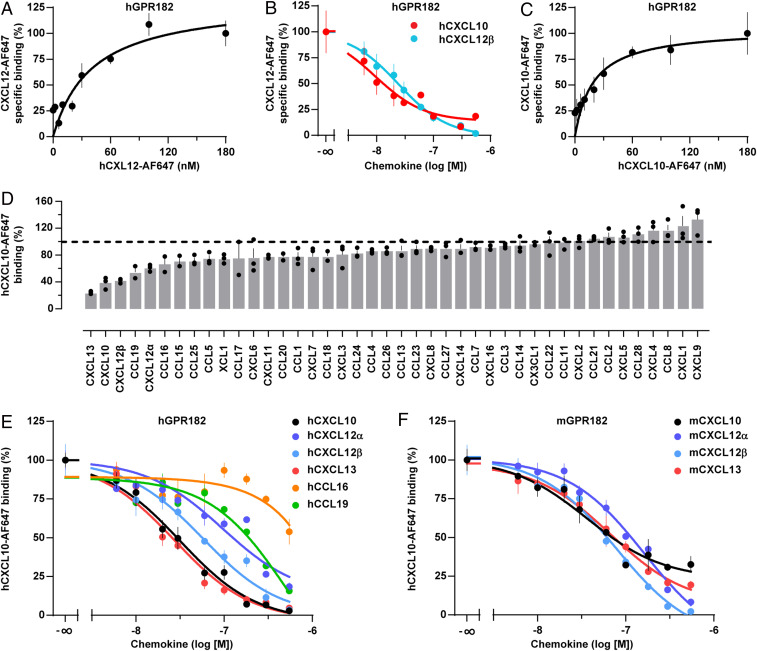
Since the closest paralogue of GPR182, the atypical chemokine receptor 3 (ACKR3, also known as CXCR7), binds CXCL11 and CXCL12, we studied binding of fluorescently labeled human CXCL12 to human GPR182. Saturation binding kinetics revealed that CXCL12 indeed bound to the receptor expressed in HEK293 cells with a dissociation constant (KD) of 41 nM (Fig. 2A). We then tested whether several related human chemokines are able to compete with fluorescently labeled CXCL12 for GPR182 and found that CXCL10 was able to do so (Fig. 2B). Incubation of HEK293 cells expressing GPR182, with increasing concentrations of fluorescently labeled CXCL10, revealed that CXCL10 bound to GPR182 with a slightly higher affinity (KD: 19 nM) than CXCL12 (Fig. 2C). We then systematically studied the ability of 42 different chemokines to bind to GPR182. When tested for their ability to displace fluorescently labeled CXCL10 from GPR182 expressed on HEK293 cells, most chemokines tested at 120 nM had no effect (Fig. 2D). However, in addition to CXCL12, CXCL13 and (to some degree) CCL16 and CCL19, were also able to compete with CXCL10 (Fig. 2D). We then performed systematic competition binding experiments (Fig. 2E), which revealed the highest binding affinity for CXCL13 and CXCL10 with an inhibition constant (Ki) of 9 and 10 nM, respectively. The two CXCL12 isoforms α and β had a slightly lower binding affinity with Ki values of 31 and 19 nM, respectively. The binding affinity of CCL19 was much lower (Ki: 260 nM), and the affinity of CCL16 was too low to be analyzed (Fig. 2E). Very similar affinities were found for murine chemokines CXCL10, CXCL12, and CXCL13 binding to mouse GPR182 (Fig. 2F and SI Appendix, Fig. S3A).
Fig. 2.
GPR182 binds the chemokines CXCL10, CXCL12, and CXCL13. (A) Binding of the fluorescently labeled human chemokine, CXCL12-AF647, to human GPR182, expressed by HEK293T cells (n = 3). (B) Competition of hCXCL12-AF647 and unlabeled hCXCL10 and hCXCL12β for binding to human GPR182 (n = 3). (C) Binding of hCXCL10-AF647 to hGPR182 (n = 3). (D) Effect of various chemokines (120 nM) on binding of hCXCL10-AF647 to hGPR182. (E) Competition of hCXCL10-AF647 and indicated unlabeled human chemokines for binding to GPR182. (F) Competition of human CXCL10-AF647 and various unlabeled murine chemokines for binding to mGPR182. Individual experiments were performed in triplicates; shown is one, representative of four (C), two (D) and three (E and F) independently performed experiments. Shown are mean values ± SEM of one experiment.
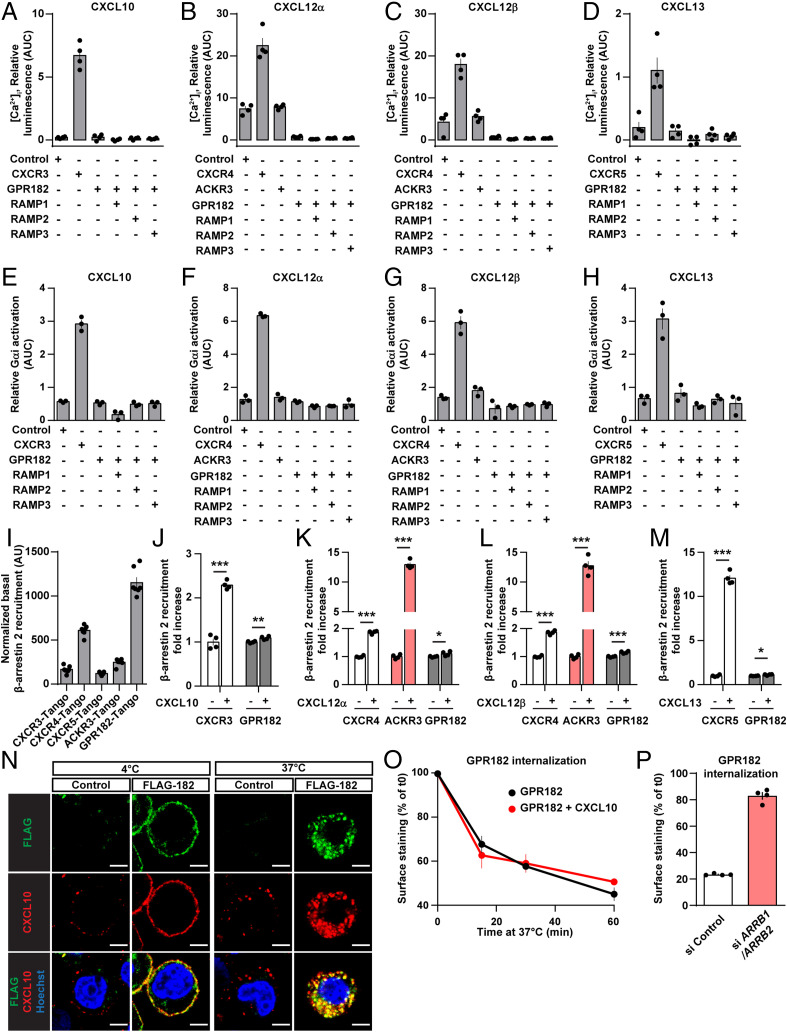
We then tested whether binding of chemokines to GPR182 resulted in G protein–mediated signaling by testing the ability of CXCL10, CXCL12, and CXCL13 to induce Ca2+ transients in cells expressing GPR182, their conventional chemokine receptors, and known ACKRs, together with a promiscuous G protein α-subunit. As shown in Fig. 3 A–D, CXCL10, CXCL12α/β, as well as CXCL13 induced Ca2+ transients in cells expressing their conventional receptors CXCR3, CXCR4, and CXCR5, respectively. In contrast, no effect was seen in cells expressing GPR182 when exposed to each of the four chemokines (Fig. 3 A–D). Cells expressing ACKR3 also showed no response to CXCL12 compared to control cells (Fig. 3 B and C). Since GPR182 has recently been described to be able to interact with receptor activity–modifying proteins (RAMPs) (22), we tested whether coexpression of RAMP-1, -2 or, -3 with GPR182 resulted in chemokine-induced G protein–mediated responses. However, none of the RAMPs promoted downstream signaling of GPR182 (Fig. 3 A–D). Very similar results were obtained when we determined receptor-mediated activation of Gi using the NanoBiT-G protein dissociation assay (23) (Fig. 3 E–H). To study potential effects of chemokines on GPR182-dependent β-arrestin recruitment, we used the Parallel Receptorome Expression and Screening via Transcriptional Output (PRESTO-Tango) system, which measures the recruitment of protease-tagged arrestin to GPCRs (24). When we expressed the GPR182-Tango construct, we noticed that GPR182 showed strong recruitment of β−arrestin in the absence of any ligand, indicating high constitutive activity (Fig. 3I). This constitutive activity was considerably higher than that of other chemokine receptors (SI Appendix, Fig. S3B). We then tested the effect of chemokines on β-arrestin recruitment. All three chemokines increased β-arrestin recruitment through their conventional receptors, CXCR3, CXCR4, and CXCR5, whereas they had hardly any effect on β-arrestin recruitment by GPR182 (Fig. 3 J–M). However, as shown before, CXCL12 strongly induced β-arrestin recruitment through its atypical receptor ACKR3 (Fig. 3 K and L). Consistent with its strong basal and almost absent ligand-dependent recruitment of β-arrestin, GPR182, together with its ligand CXCL10, strongly internalized at 37 °C (Fig. 3N). The internalization of GPR182 was, however, not affected by the ligand (Fig. 3O) but was significantly reduced in cells with suppressed expression of β-arrestin 1 and β-arrestin 2 (Fig. 3P and SI Appendix, Fig. S3C).
Fig. 3.
GPR182 does not signal in response to ligand binding. (A–D) Effect of the indicated chemokines on [Ca2+]i in HEK293 cells expressing the indicated receptors together with Ca2+-sensitive bioluminescent fusion protein (G5A) (n = 3 replicates and 3 independent experiments). (E–H) Effect of the indicated chemokines on Gαi activity using the Gαi NanoBiT assay system, as described in Methods in HEK cells transfected with the indicated human receptors (n = 3 replicates and 1 experiment). (I) Ligand-independent recruitment of β-arrestin by GPR182 and several other conventional and ACKRs (n = 7 replicates). (J–M) Effect of the indicated chemokines on β-arrestin recruitment to GPR182 using the TANGO assay system as described in Methods (n = 4 replicates and 3 independent experiments). (N) Single-cell suspension HEK293T cells expressing FLAG-tagged human GPR182 and control cells (control) were incubated at 4 °C for 1 h in the presence of 100 nM of hCXCL10-AF647 and FITC-coupled anti-FLAG antibody, followed by 3 washes and 30 min of incubation at 37 °C. Thereafter cells were fixed and stained with Hoechst dye and fluorescence analyzed by confocal microscopy. (O) Cells expressing FLAG-tagged GPR182 were incubated at 4 °C with an anti-FLAG antibody, and subsequently incubated at 37 °C in the presence or absence of hCXCL10 (100 nM). After incubation for increasing time periods, cells were stained with a secondary antibody to reveal FLAG-GPR182 at the cell surface, which was then quantified by flow cytometry. (n = 3 replicates). Shown are mean values ± SEM of one experiment; *P ≤ 0.05; **P ≤ 0.01; and ***P ≤ 0.001; n.s., nonsignificant (unpaired two-tailed Student’s t test). (P) Cells expressing FLAG-tagged GPR182 were, in addition, transfected with control siRNA (si Control) or siRNA against β-arrestin-1 and β-arrestin-2 (si ARRB1/ARRB2). Cells were incubated at 4 °C with an anti-FLAG antibody and subsequently incubated at 37 °C for 60 min. After incubation, cells were incubated with a secondary antibody to stain FLAG-GPR182 at the cell surface. Surface staining was then quantified by flow cytometry. Data are represented as percentage of surface staining relative to basal surface staining of cells kept at 4 °C (which inhibits internalization). Shown is one representative experiment of two independently performed experiments. Shown are mean values ± SEM of one experiment.
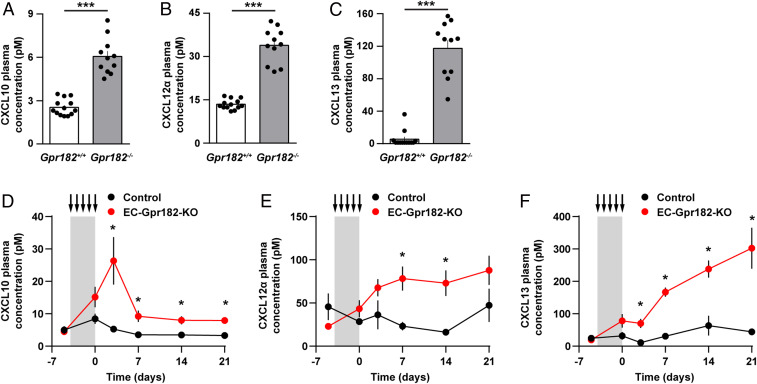
Since the in vitro data indicate that GPR182 is an atypical chemokine receptor for CXCL-10, -12, and -13, which binds chemokines but does not induce downstream signaling in response to chemokine binding, we tested whether plasma chemokine levels are affected by the loss of GPR182. In GPR182-deficient mice, plasma concentrations of all three chemokines were significantly increased. While CXCL10 and CXCL12 levels were two to threefold increased, CXCL13 levels were found to be increased more than 10-fold (Fig. 4 A–C), whereas levels of related chemokines, which do not bind GPR182, were not affected (SI Appendix, Fig. S3D). To validate these findings and to test whether the acute loss of endothelial GPR182 expression would result in elevated plasma levels of chemokines, we generated inducible endothelium-specific GPR182-deficient mice (Cdh5-CreERT2;Gpr182flox/flox [EC-Gpr182-KO]). In contrast to control mice, induction of EC-Gpr182-KO mice resulted in a rapid increase in the plasma concentration of CXCL10, CXCL12, and CXCL13 (Fig. 4 D–F). Similar to the constitutive GPR182-deficient mice, the increase in CXCL13 levels was more pronounced than increases in CXCL10 and CXCL12 plasma levels. While the increases in CXCL10 and CXCL12 plasma levels appeared to remain stable over a time period of 20 d, CXCL13 plasma levels continued to increase after induction of endothelial GPR182 deficiency (Fig. 4 D–F).
Fig. 4.
Increased free plasma concentrations of CXCL10, CXCL12α, and CXCL13 in mice lacking GPR182. (A–C) Plasma concentrations of CXCL10, CXCL12α, and CXCL13 in control Gpr182+/+ and GPR182-deficient mice (Gpr182−/−) (n = 11 to 13 mice per genotype). (D–F) Plasma concentrations of CXCL10, CXCL12α, and CXCL13 before as well as 1, 3, 7, 14, and 21 d after tamoxifen treatment on five consecutive days (arrows) in control mice and in EC-Gpr182-KO animals (n = 4 to 7 mice per genotype). Shown are mean values ± SEM; *P ≤ 0.05; **P ≤ 0.01; and ***P ≤ 0.001; n.s., nonsignificant [unpaired two-tailed Student’s t test (A–C) or multiple t test with Sidak correction (D–F)].
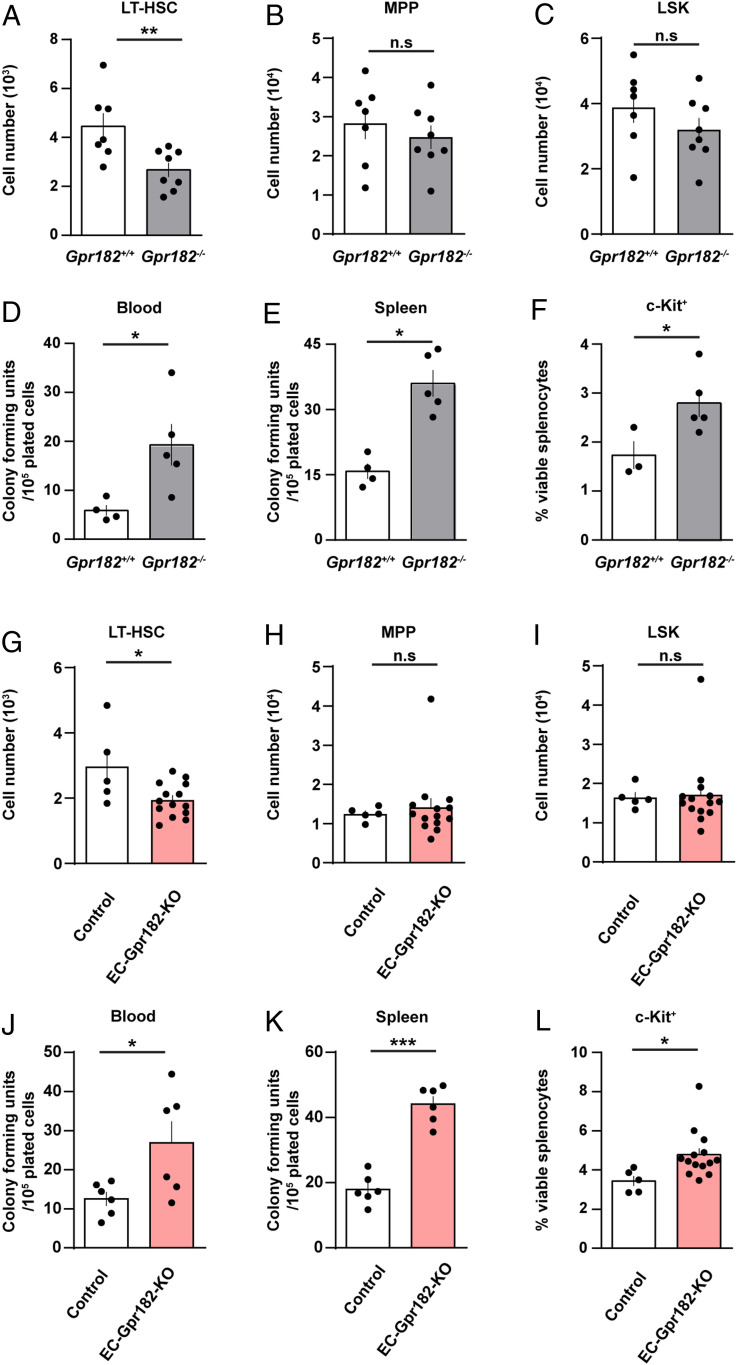
When analyzing the architecture and composition of lymph nodes, we observed no obvious differences between control and GPR182-deficient mice (SI Appendix, Fig. S4 A–C). Peripheral blood analysis showed increased levels of neutrophils and monocytes, indicating an alteration in leukocyte generation or turnover (SI Appendix, Fig. S4 D–J). Given the expression of Gpr182 in bone marrow sinusoidal cells and the known function of CXCL12 in the regulation of hematopoietic stem cell (HSC) retention to the bone marrow niche (25), we analyzed hematopoiesis in wild-type and GPR182-deficient mice by immune phenotyping using flow cytometry. The analysis revealed a significant decrease in long-term repopulating HSCs (LT-HSCs, defined as CD150+CD48−CD34low LSK) (Fig. 5A). In contrast, progenitor populations were not affected (Fig. 5 B and C and SI Appendix, Figs. S5 A–H and S6). No alteration in the distribution of CXCL12 was observed in the bone marrow of mice lacking GPR182 (SI Appendix, Figs. S7 and S8). To test whether an increased mobilization of HSCs from the bone marrow is mirrored by an increase in hematopoietic stem/progenitors outside the bone marrow, we determined colony-forming units (CFUs) of hematopoietic stem/progenitors in the blood and spleen (Fig. 5 D and E). CFUs were significantly increased both in the peripheral blood and in the spleen (Fig. 5 D and E). This was paralleled by an increase in c-Kit+ cells in the spleen (Fig. 5F). This effect was due to GPR182 expressed in endothelial cells, since tamoxifen-induced loss of endothelial GPR182 expression in inducible endothelium-specific GPR182 knockout (EC-Gpr182-KO) mice basically recapitulated the phenotype of the constitutive GPR182 knockout (Fig. 5 G–L and SI Appendix, Fig. S5 I–P).
Fig. 5.
Alteration of the HSC population in GPR182-deficient mice. (A–L) Quantitative analysis of hematopoietic stem and progenitor cells (A–C and G–I), hematopoietic progenitor cell colony–forming units (CFUs), in the spleen and peripheral blood (D, E, J, and K), and c-Kit+ splenic cells (F and L) in GPR182-deficient mice and littermate controls (A–F), as well as in EC-Gpr182-KO mice and corresponding controls 3 mo (G–I and L) or 3 wk (J–K) after induction of endothelium-specific GPR182 deficiency with tamoxifen. Shown are the long-term repopulating HSCs (LT-HSCs; A and G), multipotent progenitors (MPP; B and H) and lineage-negative SCA-1+ c-Kit+ (LSK; C and I) determined in both femura of the analyzed mice; n = 7 mice (Gpr182+/+ in A–C); n = 8 mice (Gpr182−/− in A–C); n = 4 mice (Gpr182+/+ in D and E); n = 3 mice (Gpr182+/+ in F); n = 5 mice (Gpr182−/− in D–F); n= 5 mice (control in G–I); n = 14 mice (EC-Gpr182-KO in G–I); n = 6 mice (control and EC-Gpr182-KO in [J and K]; n = 5 mice [control in L] ; and n= 14 mice [EC-Gpr182-KO in L]). Shown are mean values ± SEM; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., non significant. Two-tailed nonparametric Mann–Whitney U test (A–E, and G–K). Unpaired two-tailed Student’s t test (F and L).
Discussion
GPR182 has been shown to be expressed in endothelial cells, but its function has remained elusive, since the physiological ligand of the receptor is not known. In this study, we show that GPR182 binds CXCL10, CXCL12, and CXCL13 but does not couple binding of these ligands to G protein or β-arrestin activation or downstream signaling. This identifies GPR182 as an atypical chemokine receptor. The closest paralogue of GPR182, ACKR3, has an overlapping ligand binding spectrum interacting with CXCL11 and CXCL12. Thus, similar to CXCL11, which can bind to two atypical chemokine receptors, ACKR1 and ACKR3 (16, 19), CXCL12 also has two atypical receptors, which underlines the complexity of the chemokine system (26, 27). Similar to the ACKRs 1 through 4, GPR182 has an alteration in the canonical DRYLAIV motif in the second intracellular loop of conventional chemokine receptors, which is believed to be required for G protein activation and signaling (28). However, insertion of the corresponding region from CXCR4 into ACKR3 did not restore G protein signaling (29, 30). This indicates that additional properties distinguish conventional from ACKRs.
Expression of GPR182 resulted in a strong basal recruitment of β-arrestin. A constitutive interaction with β-arrestin has also been reported for ACKR2 and ACKR3 (31–34) but not for ACKR1 (35). In contrast to ACKR2, ACKR3, and ACKR4, which show increased interaction with β-arrestin in response to ligand binding (36–38), the constitutive β-arrestin recruitment of GPR182 was not further increased by ligand application. Consistent with a role of β-arrestin in receptor internalization, ACKR2, ACKR3, and ACKR4 show constitutive internalization (34, 37, 39, 40), which, in the case of ACKR3 and ACKR4, has been reported to be further increased by ligand binding (37, 39). This correlates with our observation that GPR182 underwent constitutive receptor internalization, which was not affected by a GPR182 ligand. Thus, we conclude that the strong β-arrestin recruitment of GPR182 leads to very efficient internalization of GPR182 and its ligands. This suggests that GPR182 may function to locally remove chemokines.
The affinity of GPR182 for CXCL12 and CXCL10 is about 10-fold and 100-fold lower than the affinity of their conventional chemokine receptors, CXCR4 and CXCR3, respectively (41, 42), whereas GPR182 binds CXCL13 with slightly higher affinity than the conventional chemokine receptor CXCR5 (43). This might explain why loss of GPR182 had a more pronounced effect on plasma levels of CXCL13 than on CXCL10 and CXCL12 plasma levels. The fact that the plasma levels of all three chemokines binding GPR182 significantly increased in mice lacking GPR182 clearly shows that GPR182 functions as a scavenger of CXCL10, CXCL12, and CXCL13. Their elevated plasma levels were still one to two orders of magnitude lower than the KD values for their conventional chemokine receptors, CXCR2, CXCR3, and CXCR5, and it remains unclear whether the increase in systemic chemokine levels per se has any effects. It appears more likely that loss of GPR182 disturbs local chemokine activities.
Gpr182 has been reported to be expressed in the epithelium of the gastrointestinal tract and to be particularly enriched in intestinal stem cells (11). This study also reported that constitutive GPR182-deficient mice show increased proliferation of intestinal epithelial cells under stress conditions, such as regeneration after injury, in the ApcMin tumor model as well as under in vivo culture conditions, which was accompanied by ERK1/2 activation. Our Gpr182 expression reporter mouse model did not show Gpr182 expression in gastrointestinal epithelial cells, a difference to the study which we currently cannot explain. However, the reported phenotype of GPR182-deficient mice would be consistent with a role of GPR182 as an atypical chemokine receptor, which has a scavenging function for CXCL12. CXCL12 has been shown to induce activation of ERK1/2 in colon carcinoma cells as well as in crypt cells of irradiated intestinal epithelium organoids and to induce proliferation of organoid crypt cells (44–46). In addition, CXCL12 promoted intestinal epithelial recovery from radiation stress (44). In the absence of GPR182, these effects are expected to be enhanced as described by Kechele et al. (11).
Our data indicate that GPR182 is involved in HSC maintenance. This may explain the recent report that mice lacking GPR182 have slightly altered blood levels of lymphocytes and neutrophils (47), an observation we could partially confirm. Overall, the consequences of the reduction of HSCs are small. Given that CXCL12 is a key factor promoting HSC maintenance and retention in the bone marrow by activating CXCR4, which is expressed on HSCs (25, 48), the ability of GPR182 to bind and inactivate CXCL12 is the most likely mechanism by which GPR182 promotes HSC maintenance. ACKR3, which also binds CXCL12, is not involved in fetal hematopoiesis (49, 50). Its role in adult hematopoiesis is unknown, but our data show no expression of ACKR3 in sinusoidal endothelial cells, suggesting that it is not involved in GPR182-related functions. The majority of HSCs is localized close to bone marrow sinusoids, where they are in close contact with endothelial cells and so-called CXCL12-abundant reticular cells, which are also known as leptin-receptor-expressing mesenchymal stromal cells, which have a perisinusoidal localization and both express CXCL12 (25, 48, 51). This is consistent with the high expression of GPR182 in sinusoidal bone marrow endothelial cells and its absence in endothelial cells of bone marrow arterioles. The importance of CXCL12 expressed by these cell types for HSC homeostasis is indicated by changed HSC maintenance and retention after conditional loss of CXCL12, specifically in endothelial and perivascular stromal cells (52, 53). How CXCL12 attracts or retains HSCs to their bone marrow niche is still poorly understood. The prevailing concept suggested that CXCL12 gradients in the bone marrow attract HSCs to their niches (54). This concept has recently been challenged, and it has been proposed that, rather, local CXCL12 hotspots are involved in HSC retention (55). Our experiment, however, revealed no gross alteration in the CXCL12 distribution pattern around bone marrow sinusoids in mice lacking GPR182. Regardless of the exact mechanism by which CXCL12 promotes HSC maintenance and retention, our data show that this is promoted by the high expression of GPR182 in sinusoidal endothelial cells. Since GPR182 scavenges CXCL12, it is likely to reduce the free CXCL12 concentration around sinusoidal endothelial cells. This may stabilize a CXCL12 gradient toward the HSC niche and promote HSC maintenance. Loss of GPR182 may have the opposite effect, which would explain reduced retention of HSCs in the bone marrow.
Methods
Reagents.
All reagents are listed in tables added to SI Appendix, Supplemental Material and Tables S1 and S2).
Cell Lines.
HEK293T cells were obtained from American Type Culture Collection. HTLA cells (a HEK293 cell line stably expressing a tTA-dependent luciferase reporter and a β-arrestin2-TEV fusion gene) were a gift from Dr. Gilad Barnea (Brown University, Providence, RI). Both cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM l-glutamine, 5 units/mL penicillin/streptomycin.
Ligand Binding Assay.
To determine equilibrium binding of CXCL10-AF647 or CXCL12-AF647 to GPR182, 5 × 105 HEK293T cells seeded in 6-well plates were transfected with a eukaryotic expression plasmid carrying the GPR182 complementary DNA (cDNA) and green fluorescent protein (GFP). 48 h later, single-cell suspension was prepared by trypsin/ethylenediaminetetraacetic acid (EDTA) treatment and incubated 1 h at room temperature in complete medium. Cells were rinsed with binding buffer (Hank's balanced salt solution (HBSS) pH 7.4, SI Appendix, Supplemental Material) and dispensed in V-bottom 96-well plate. Cells were resuspended in 100 µl binding buffer incubated at room temperature for 1 h with increasing concentration of fluorescently labeled chemokine diluted in binding buffer. Thereafter, cells were washed twice with binding buffer, fixed for 10 min with paraformaldehyde (PFA) 1% and washed twice with phosphate buffered saline (PBS). Cells were resuspended into 100 µl and transferred to a 96-well plate with black wall and transparent glass bottom. Cells were then allowed to settle down for 1 h and were analyzed with an epifluorescence Zeiss Axio Observer microscope using AxioVision software. Pictures were analyzed with Fiji software and CXCL10-AF647 or CXCL12-AF647 fluorescent signal was quantified in GFP positive cells. Competition binding assay was performed by incubating increasing concentration of nonlabeled chemokine shortly before adding the fluorescently labeled chemokine at the indicated concentration.
For quantification of binding affinity, curve fitting and Kd were determined using one site specific binding equation in GraphPad Prism 6.07 after subtracting total binding fluorescence value by the nonspecific fluorescence binding in cells transfected with control vector. In competitive experiments, curve fitting and Ki was determined using one site fit Ki equation in GraphPad Prism 6.07 by providing the concentration (40 nM) and Kd values of fluorescently labeled Alexa647 chemokine (“hot” ligand).
Cell Transfection and Determination of [Ca2+]i.
2 × 104 cells were seeded in 96-well plates with white walls and transparent bottom and transfected with plasmids containing cDNAs encoding a calcium-sensitive bioluminescent fusion protein consisting of aequorin and GFP (56), the indicated receptors and a promiscuous G protein α-subunit using Lipofectamine 2000 (Life Technologies), as described before (57). 48 h later, cells were loaded with 5 µM coelenterazine h (Promega) in HBSS containing 1.8 mM calcium and 10 mM glucose for 2 h at 37 °C. Measurements were performed using a luminometric plate reader (Flexstation 3, Molecular Devices). The area under each calcium transient was calculated by using SoftMaxPro software and expressed as area under the curve.
Gαi Activation Assay.
For determination of ligand-induced Gαi signaling, we used the recently developed NanoBiT assay (23). HEK293 cells were plated onto a 96-well plate and transfected on the same day with Galphai-LgBit, Gbeta, GgammaSmbit and the receptor (30, 6, 6, and 12 ng per well, respectively) using Lipofectamin 2000 (0.2 µl/well). After 24 h, the supernatant was aspirated and 50 µl/well of 10 µM Coelenterazine h was added and then incubated at 37 °C for 1 h. The plate was then transferred to the plate reader (Flexstation 3), and luciferase activity was determined in real-time before and after addition of the indicated chemokine for 5 min. Data were expressed relative to baseline (before addition of chemokine). The processed data were plotted in GraphPad Prism 6.07 and expressed as area under the curve.
β-Arrestin Assay.
To determine ligand-induced interaction of receptors and β-arrestin, we used the TANGO assay (24). The Tango plasmid library, a gift from Bryan Roth, was obtained from Addgene Kit #1000000068. HTLA cells were transfected with receptor Tango in 96-well plates with white walls and transparent bottom. Thereafter, cells were incubated for 4 h in serum-free medium and were then stimulated for 16h with the indicated ligands diluted in sterile HBSS buffer containing 1.8 mM Ca2+ and 10 mM glucose. The next day, the supernatant was removed and replaced by 100 µL assay buffer containing 10% Bright-Glo reagent (Promega). After a 20 min incubation period at room temperature, luminescence was counted in a plate reader and expressed as relative luminescence units.
For comparison of basal activity of TANGO-CXCR3, TANGO-CXCR4, TANGO-CXCR5, TANGO-ACKR3, and TANGO-GPR182, transfected cells (in quadruplicates) were fixed with 4% PFA in PBS for 10 min at room temperature. The cells were then washed three times with PBS and blocked and permeabilized with PBS containing 0.1% triton x100 and 5% horse serum for 5 min at room temperature. To detect total receptor expression, the cells were then incubated with a mouse anti-FLAG M2 antibody (1:1,000) diluted in PBS containing 1% bovine serum albumin (BSA) for 1 h at room temperature. After three washes with PBS, the cells were incubated with Alexa488 coupled anti-mouse secondary antibodies (1:500) and DAPI (1:1,000) in PBS containing 1% BSA for 30 min at room temperature. Cells were then imaged using an epifluorecence microscope Zeiss Axio Observer and Axio Vision software. One picture per replicate was recorded by selecting a zone containing confluent cells as visualized by DAPI staining. The mean fluorescence intensity of the FLAG staining per field of view was quantified using ImageJ and corrected by subtracting the background fluorescence from untransfected cells. The results of quadruplicates were averaged, and the obtained value was used to normalize the basal TANGO bioluminescence measured in seven independent wells from the same transfection.
Analysis of CXCL10 and GPR182 Internalization.
Adherent HEK293T cells were transfected with the pcDNA3 eukaryotic expression vector, encoding the N-terminally FLAG-tagged hGPR182 or with control pcDNA3 vector using Lipofectamine 2000. After 1 d, single-cell suspensions were prepared by trypsin/EDTA treatment. Cells were then incubated for 1 h at room temperature in complete culture medium. After two washes with ice-cold binding medium (serum-free DMEM, 0.5% BSA, and 10 mM Hepes), cells were incubated at a concentration of 106 per mL with 100 nM of fluorescently labeled chemokine, CXCL10-AF647, and 1/250 fluorescein isothiocyanate (FITC)-coupled anti-FLAG-M2 monoclonal antibody in ice-cold binding medium for 1 h at 4 °C with gentle rocking to allow for chemokine and antibody binding to the cell surface. After three washes with ice-cold binding medium, cells were resuspended in ice-cold binding medium and incubated at 4 °C or 37 °C for 30 min with gentle rocking followed by two washes with ice-cold binding medium. Cells were then fixed in PFA (1% [vol/vol]) for 10 min at room temperature, washed, and stained with Hoechst (1 µg/mL final concentration) in PBS for 15 min. Cells were then allowed to settle down in Ibidi 8-well chamber slides and were analyzed using an inverted Leica SP8 confocal microscope.
For analysis of GPR182 internalization, cells were transfected and seeded as described above. Thereafter, cells were incubated at a concentration of 106 per ml with anti-FLAG-M2 antibody (Sigma [1:500]) in ice-cold binding medium for 1 h at 4 °C with gentle rocking. After three washes with ice-cold binding medium, cells were resuspended with or without 100 nM recombinant hCXCL10 in binding medium, as indicated, and incubated at 37 °C for 15, 30, or 60 min with gentle rocking or maintained at 4 °C for determination of basal surface expression, followed by two washes with binding medium. Cells were then incubated with AlexaFluor 488 conjugated donkey anti-mouse IgG (ThermoFisher Scientific [1:1,500]) in ice-cold binding buffer containing the cell viability dye 7AAD (1:10) for 30 min at 4 °C with gentle rocking. Finally, cells were washed three times with ice-cold PBS before capture and analysis of 2 × 105 total events by flow cytometry using a FacsCanto (BD Biosciences). The total amount of receptor on the cell surface was determined by median fluorescence intensity and expressed as the percent of basal cell surface GPR182 expression in cells maintained at 4 °C without chemokine treatment.
For analysis of the effect of β-arrestin knockdown on GPR182 internalization, adherent HEK293T cells were transfected with 20 nM control small interfering RNA (siRNA) or a mix of 10 nM siRNA targeting ARRB1 and 10 nM of siRNA targeting ARRB2 using lipofectamine RNAiMAX (Thermofisher) following manufacturer’s instructions. Two days after siRNA transfection, cells were transfected with the pcDNA3 eukaryotic expression vector encoding the N-terminally FLAG-tagged human GPR182 or with control pcDNA3 vector using Lipofectamine 2000. At 72 h after siRNA transfection, 106 cells per mL were incubated with anti-FLAG-M2 antibody (Sigma [1:500]) in ice-cold binding medium for 1 h at 4 °C with gentle rocking. After three washes with ice-cold binding medium, cells were incubated for 60 min at 37 °C with gentle rocking or were maintained at 4 °C for determination of basal surface expression, followed by two washes with binding medium. Cells were then incubated with AlexaFluor 488 conjugated donkey anti-mouse IgG (ThermoFisher Scientific [1:1,500]) in ice-cold binding buffer containing the cell viability dye 7AAD (1:10) for 30 min at 4 °C with gentle rocking. Finally, cells were washed three times with ice-cold PBS before capture and analysis of 2 × 105 total events by flow cytometry using a FacsCanto flow cytometer (BD Biosciences). The total amount of receptor on the cell surface was determined by median fluorescence intensity and expressed as the percent of basal cell surface GPR182 expression in cells maintained at 4 °C. Knockdown efficiency of β-arrestin was determined by western blotting using standard methods.
Determination of Chemokine Levels.
After puncture of the facial vein, mouse blood was collected into EDTA-coated tubes (Microvette, NC0973120), placed on ice, and spun down at 2,000 × g for 10 min at 4 °C. Plasma was then collected and frozen at −80 °C until analysis. Plasma cytokine and chemokine levels were determined by MILLIPLEX MAP magnetic bead–based multianalyte panel mouse cytokine/chemokine 25 Plex (MCYTOMAG-70K-PMX) or a custom designed Mouse Magnetic Luminex Assay 3 plex panel for CXCL10, CXCL12α, and CXCL13 (R&D). Samples were analyzed using the MAGPIX system and MILLIPLEX Analyst 5.1 software (Merck Millipore).
Lineage and Progenitor Staining of Bone Marrow Cells.
Both femurs were collected from each mouse for single mouse analysis and cleaned to remove excess muscle tissue and tendons. Bone marrow cells were extracted by flushing out from bones in 3 mL cold 1 × PBS by using a 5 mL syringe applied with needle.
To enrich for bone marrow mononuclear cells (BMMNCs) the resuspended cells (∼3 mL) were carefully placed on top of 3 mL Histopaque 1083 (Sigma) and isolated by Ficoll density gradient centrifugation at 400 × g for 30 min at room temperature, without break. The interphase was collected and washed twice with 1x PBS before the cells were counted. For stem and progenitor cell analyses, the BMMNCs were first stained with biotinylated antibodies against mature blood cells (CD3, CD11b, CD19, CD41, B220, and Gr-1 and Ter119) for 15 min prior to 30 min progenitor staining (Sca1-PerCPCy5.5 CD48-APCCy7, Streptavidin-BV711, cKit-BV421, CD150-PECy7, CD34-eF660, and Fixable Viability Dye eF506) on ice in the dark. After washing them once with 1x PBS, the cells were resuspended in fluorescence-activated cell sorting (FACS) buffer and analyzed on a FACS Fortessa (Becton Dickinson) with FACS Diva 7 software (BD Biosciences).
Lineage-negative cells were gated out from the viable mononuclear cells. From those cells, the SCA-1, CD117 double positive (LSK: lineage−, SCA-1+, and cKIT [CD117]+) hematopoietic stem progenitor cells were subgated and contained long-term repopulating hematopoietic stem cells (LT-HSCs: CD150+, CD48−, CD34low, LSK), multipotent progenitors (MPPs: CD34+, CD150−, LSK). The SCA-1− CD117+ lineage-myeloid committed progenitors (LK) were further subgated on megakaryocyte-erythroid-progenitors (MEP: CD150+ LK) and pregranulocyte-macrophage-progenitors (pre-GMP; CD150− CD34+ LK). A graphical example of the gating strategy is shown in SI Appendix, Fig. 3A.
c-Kit Staining of Spleen Cells.
Spleens were collected from each mouse separately for single mouse analysis and meshed through 40 μM cell straining to collect spleen cells with 5 mL PBS. Approximately 1 × 107 of total spleen cells were used to stain with cKit-BV421 and Fixable Viability Dye eF506 for 30 min. After washing them once with 1x PBS to remove excessive antibodies, the cells were resuspended in FACS buffer and analyzed on a FACS Fortessa (Becton Dickinson) with FACS Diva 7 software (BD Biosciences). A graphical example of the gating strategy is shown in SI Appendix, Fig. 3B.
Histology and Microscopy.
For analysis of mCherry reporter fluorescence, mice were sacrificed by CO2 and perfused with 20 mL PBS through the left ventricle, followed by 4% PFA in PBS. After perfusion, tissues were postfixed in PFA 4% at 4 °C for 1 h (lymph nodes, Peyer’s Patches, aorta, ear skin, and part of jejunum) or for 16 h. After fixation, organs were washed at least three times with PBS and transferred to PBS containing 30% (wt/vol) sucrose at 4 °C for 24 h. Tissues were then embedded in optimal cutting temperature (OCT) compound and stored at −80 °C until further processing. OCT blocks were sectioned using cryostat, and 12 µm or 20 µm sections were mounted on SuperFrost PLUS microscope slides. For aorta, ear skin, or jejunum whole-mount immunostaining, tissues were fixed as described above and washed three times with PBS and further processed for immunofluorescence staining.
For immunofluorescence staining, cryosections were allowed to dry for 30 min at room temperature, washed three times with PBS, and blocked/permeabilized with antibody diluent (PBS containing 0.1% Triton X-100 and 5% horse serum) for 1 h at room temperature. Then cryosections were incubated with primary antibodies diluted in antibody diluent overnight at 4 °C (SI Appendix, Supplementary Material). After three washes with PBS, sections were incubated for 1 h at room temperature with AlexaFluor-488, -594, or -647 conjugated secondary antibodies (1:500) in antibody diluent containing 4′,6-diamidino-2-phenylindole (DAPI, 1:1,000). After three washes with PBS, sections were mounted in Fluoromount and covered with glass coverslips and analyzed by confocal microscopy using Leica True Confocal Scanning (TCS) SP8 or Zeiss laser scanning microscope (LSM) 880 confocal microscopes. Whole-mount aorta immunostaining protocol was similar to cryosections. For analysis of mCherry endogenous fluorescence and respective mCherry negative samples, all procedures were performed by minimizing exposure to ambient light.
Hematopoietic Progenitor Cell CFU Assay.
CFU assay was performed using buffers, medium, and dishes from StemCell Technologies (MethoCult GFM3434). Mouse blood was collected using a heparin-coated syringe and was placed in tube containing heparin (100 mg/mL, Merck). Spleens were subsequently collected and placed in Iscove’s modified Dulbecco’s medium (IMDM) medium supplemented with 2% FBS (StemCell Technologies). Spleens were minced with surgical scissors and smashed on a 40 µm cell-strainer in order to obtain single-cell suspension. Splenocytes were washed twice with IMDM medium containing 2% FBS, resuspended in IMDM medium, and counted using a Neubauer hemocytometer. Whole blood was treated with nine volumes of ammonium chloride solution (StemCell Technologies). Thereafter, blood cells were washed twice with IMDM medium and counted again. 2 to 3 × 105 splenocytes and erythrocyte-depleted blood cells were resuspended into Methocult medium and plated in Smart 6-well plates (StemCell Technologies). The number of colonies formed was assessed after 12 d of incubation at 37 °C, 5% CO2, and ≥ 95% humidity.
Blood Analysis.
For complete blood count analysis, 10 to 12 wk old mice were sacrificed by CO2,and blood was immediately collected by cardiac puncture and transferred to EDTA-coated microtubes. The collected blood as well as blood smears were analyzed by IDEXX laboratories.
Animal Models.
All mice were backcrossed onto a C57BL/6J background at least 8 to 10 times, and experiments were performed with littermates as controls. Male and female animals (8 to 12 wk old) were used unless stated otherwise. Mice were housed under a 12-h light–dark cycle, with free access to food and water under specific pathogen-free conditions unless stated otherwise. Transgenic mice expressing mCherry under the control of the Gpr182 promoter (Gpr182-mCherry) were generated using the BAC clone RP23-119B1 from mouse chromosome 10 containing the Gpr182 gene. The coding sequence of the Gpr182gene on the BAC was replaced by a cassette carrying the mCherry cDNA, followed by a polyadenylation signal and a FLP recognition target (FRT)-flanked ampicillin resistant gene (β-lactamase) using Red/ET recombination kit (Gene Bridges). Correct targeting was verified by restriction digests and DNA sequencing. After Flp-mediated excision of the ampicillin resistant gene and linearization, the recombined BAC, which had a total length of 234 kb, was injected into pronuclei of Friend Virus B NIH Jackson (FVB/N) oocytes. Transgenic offspring was genotyped for BAC insertion by genomic PCRs. In total, two independent BAC transgenic lines were produced, which showed an identical mCherry expression pattern. Mice lacking GPR182 were obtained from the Knockout Mouse Project (KOMP) Repository (knockout first allele, Gpr182tm2a(KOMP)Wtsi). To generate a conditional allele of Gpr182, in which the coding sequence of exon 2 of Gpr182 is flanked by loxP-sites, a cassette flanked by FRT sites was removed by crossing mice with the Flp-deleter mouse line (58). After Flp-mediated recombination, mice were crossed with Cdh5-CreERT2 mice (59) to obtain animals with inducible, endothelium-specific deficiency. Maintenance of the animals was in agreement with German animal welfare legislations.
Statistics.
Statistical analysis was performed using the GraphPad Prism software v.6.07 from GraphPad Software Inc. (La Jolla). Values are presented as mean ± SEM; n represents the number of independent experiments or animals. Statistical analysis between two groups were performed with an unpaired two-tailed Student’s t test or nonparametric Mann–Whitney U test when appropriate, while multiple group comparisons were analyzed with one-way ANOVA followed by Tukey’s post hoc test, unless stated otherwise, and comparisons between multiple groups at different time points were performed using two-way ANOVA followed by Bonferroni’s post hoc test. A P value of less than 0.05 was considered to be statistically significant.
Study Approval.
All procedures involving animal care and use in this study were approved by the local animal ethics committee (Regierungspräsidium Darmstadt).
Supplementary Material
Acknowledgments
We thank Svea Hümmer for secretarial help, and Dagmar Magalei, Ulrike Krüger, and Daniel Heil for expert technical assistance. We are also grateful to Carola Meyer and Nadine Rink for help with animal ethics protocols. H.C. was a recipient of a Humboldt Foundation fellowship. This work was supported by a Challenge Grant from the Novo Nordisk Foundation and by the Collaborative Research Center 1039 of the German Research Foundation. M.A.R. was supported by the German Research Foundation (Sonderforschungsbereich [SFB] 834 [Z1] and project RI2462/1–1), by the Jose Carreras Leukemia Foundation (Grant 10R/2017), the Hessisches Ministerium für Wissenschaft und Kunst (III L 5 - 519/03/03.001 – [0015]), and the Wilhelm Sander-Stiftung (Grant 2018.116.1).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021596118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Sriram K., Insel P. A., G protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol. Pharmacol. 93, 251–258 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauser A. S., Attwood M. M., Rask-Andersen M., Schiöth H. B., Gloriam D. E., Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 16, 829–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander S. P. H.et al.; CGTP Collaborators , The concise guide to pharmacology 2019/20: G protein-coupled receptors. Br. J. Pharmacol. 176 (suppl. 1), S21–S141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wettschureck N., Offermanns S., Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–1204 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Laschet C., Dupuis N., Hanson J., The G protein-coupled receptors deorphanization landscape. Biochem. Pharmacol. 153, 62–74 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kapas S., Catt K. J., Clark A. J., Cloning and expression of cDNA encoding a rat adrenomedullin receptor. J. Biol. Chem. 270, 25344–25347 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Kennedy S. P., et al., Expression of the rat adrenomedullin receptor or a putative human adrenomedullin receptor does not correlate with adrenomedullin binding or functional response. Biochem. Biophys. Res. Commun. 244, 832–837 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Regard J. B., Sato I. T., Coughlin S. R., Anatomical profiling of G protein-coupled receptor expression. Cell 135, 561–571 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao L., Harrell J. C., Perou C. M., Dudley A. C., Identification of a stable molecular signature in mammary tumor endothelial cells that persists in vitro. Angiogenesis 17, 511–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumanas S., Jorniak T., Lin S., Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood 106, 534–541 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kechele D. O., et al., Orphan Gpr182 suppresses ERK-mediated intestinal proliferation during regeneration and adenoma formation. J. Clin. Invest. 127, 593–607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid W., Rosland J. H., von Hofacker S., Hunskår I., Bruvik F., Patient’s and health care provider’s perspectives on music therapy in palliative care–An integrative review. BMC Palliat. Care 17, 32 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachelerie F., et al., New nomenclature for atypical chemokine receptors. Nat. Immunol. 15, 207–208 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Griffith J. W., Sokol C. L., Luster A. D., Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Chen K., et al., Chemokines in homeostasis and diseases. Cell. Mol. Immunol. 15, 324–334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes C. E., Nibbs R. J. B., A guide to chemokines and their receptors. FEBS J. 285, 2944–2971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonecchi R., Garlanda C., Mantovani A., Riva F., Cytokine decoy and scavenger receptors as key regulators of immunity and inflammation. Cytokine 87, 37–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonecchi R., Graham G. J., Atypical chemokine receptors and their roles in the resolution of the inflammatory response. Front. Immunol. 7, 224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nibbs R. J., Graham G. J., Immune regulation by atypical chemokine receptors. Nat. Rev. Immunol. 13, 815–829 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Koenen J., Bachelerie F., Balabanian K., Schlecht-Louf G., Gallego C., Atypical chemokine receptor 3 (ACKR3): A comprehensive overview of its expression and potential roles in the immune system. Mol. Pharmacol. 96, 809–818 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Quinn K. E., Mackie D. I., Caron K. M., Emerging roles of atypical chemokine receptor 3 (ACKR3) in normal development and physiology. Cytokine 109, 17–23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzen E., et al., Multiplexed analysis of the secretin-like GPCR-RAMP interactome. Sci. Adv. 5, eaaw2778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue A., et al., Illuminating G-protein-coupling selectivity of GPCRs. Cell 177, 1933–1947.e25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kroeze W. K., et al., PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol. 22, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinho S., Frenette P. S., Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol. 20, 303–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balabanian K., et al., The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280, 35760–35766 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Burns J. M., et al., A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J. Exp. Med. 203, 2201–2213 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachelerie F., et al., An atypical addition to the chemokine receptor nomenclature: IUPHAR review 15. Br. J. Pharmacol. 172, 3945–3949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naumann U., et al., CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One 5, e9175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann F., et al., Rapid uptake and degradation of CXCL12 depend on CXCR7 carboxyl-terminal serine/threonine residues. J. Biol. Chem. 287, 28362–28377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leick M., et al., CCL19 is a specific ligand of the constitutively recycling atypical human chemokine receptor CRAM-B. Immunology 129, 536–546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coggins N. L., et al., CXCR7 controls competition for recruitment of β-arrestin 2 in cells expressing both CXCR4 and CXCR7. PLoS One 9, e98328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCulloch C. V., et al., Multiple roles for the C-terminal tail of the chemokine scavenger D6. J. Biol. Chem. 283, 7972–7982 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Galliera E., et al., beta-Arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J. Biol. Chem. 279, 25590–25597 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Chakera A., Seeber R. M., John A. E., Eidne K. A., Greaves D. R., The duffy antigen/receptor for chemokines exists in an oligomeric form in living cells and functionally antagonizes CCR5 signaling through hetero-oligomerization. Mol. Pharmacol. 73, 1362–1370 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Rajagopal S., et al., Beta-arrestin- but not G protein-mediated signaling by the “decoy” receptor CXCR7. Proc. Natl. Acad. Sci. U.S.A. 107, 628–632 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watts A. O., et al., β-Arrestin recruitment and G protein signaling by the atypical human chemokine decoy receptor CCX-CKR. J. Biol. Chem. 288, 7169–7181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borroni E. M., et al., β-arrestin-dependent activation of the cofilin pathway is required for the scavenging activity of the atypical chemokine receptor D6. Sci. Signal 6, ra30.1–ra30.11, S1–S3 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Alampour-Rajabi S., et al., MIF interacts with CXCR7 to promote receptor internalization, ERK1/2 and ZAP-70 signaling, and lymphocyte chemotaxis. FASEB J. 29, 4497–4511 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Luker K. E., Steele J. M., Mihalko L. A., Ray P., Luker G. D., Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene 29, 4599–4610 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crump M. P., et al., Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 16, 6996–7007 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loetscher M., Loetscher P., Brass N., Meese E., Moser B., Lymphocyte-specific chemokine receptor CXCR3: Regulation, chemokine binding and gene localization. Eur. J. Immunol. 28, 3696–3705 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Barroso R., et al., EBI2 regulates CXCL13-mediated responses by heterodimerization with CXCR5. FASEB J. 26, 4841–4854 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Chang P., et al., Mesenchymal stem cells over-expressing cxcl12 enhance the radioresistance of the small intestine. Cell Death Dis. 9, 154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brand S., et al., CXCR4 and CXCL12 are inversely expressed in colorectal cancer cells and modulate cancer cell migration, invasion and MMP-9 activation. Exp. Cell Res. 310, 117–130 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Smith J. M., Johanesen P. A., Wendt M. K., Binion D. G., Dwinell M. B., CXCL12 activation of CXCR4 regulates mucosal host defense through stimulation of epithelial cell migration and promotion of intestinal barrier integrity. Am. J. Physiol. Gastrointest. Liver Physiol. 288, G316–G326 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Kwon H. B., et al., The orphan G-protein coupled receptor 182 is a negative regulator of definitive hematopoiesis through leukotriene B4 signaling. ACS Pharmacol. Transl. Sci. 3, 676–689 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crane G. M., Jeffery E., Morrison S. J., Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 17, 573–590 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Sierro F., et al., Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc. Natl. Acad. Sci. U.S.A. 104, 14759–14764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerrits H., et al., Early postnatal lethality and cardiovascular defects in CXCR7-deficient mice. Genesis 46, 235–245 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Kiel M. J., Radice G. L., Morrison S. J., Lack of evidence that hematopoietic stem cells depend on N-cadherin-mediated adhesion to osteoblasts for their maintenance. Cell Stem Cell 1, 204–217 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Ding L., Morrison S. J., Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature 495, 231–235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greenbaum A., et al., CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature 495, 227–230 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aiuti A., Webb I. J., Bleul C., Springer T., Gutierrez-Ramos J. C., The chemokine SDF-1 is a chemoattractant for human CD34+ hematopoietic progenitor cells and provides a new mechanism to explain the mobilization of CD34+ progenitors to peripheral blood. J. Exp. Med. 185, 111–120 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kunz L., Schroeder T., A 3D tissue-wide digital imaging pipeline for quantitation of secreted molecules shows absence of CXCL12 gradients in bone marrow. Cell Stem Cell 25, 846–854.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Baubet V., et al., Chimeric green fluorescent protein-aequorin as bioluminescent Ca2+ reporters at the single-cell level. Proc. Natl. Acad. Sci. U.S.A. 97, 7260–7265 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tunaru S., et al., 20-HETE promotes glucose-stimulated insulin secretion in an autocrine manner through FFAR1. Nat. Commun. 9, 177 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farley F. W., Soriano P., Steffen L. S., Dymecki S. M., Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28, 106–110 (2000). [PubMed] [Google Scholar]
- 59.Sörensen I., Adams R. H., Gossler A., DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113, 5680–5688 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.